
International Journal of
Experimental Botany

 | Phyton- International Journal of Experimental Botany |  |
DOI: 10.32604/phyton.2021.015532
ARTICLE
Characterization and Candidate Gene Analysis of the Yellow-Green Leaf Mutant ygl16 in Rice (Oryza sativa L.)
1Rice Research Institute, Key Laboratory of Application and Safety Control of Genetically Modified Crops, Academy of Agricultural Sciences, Southwest University, Chongqing, 400716, China
2Chongqing Three Gorges Academy of Agricultural Sciences, Chongqing, 404155, China
*Corresponding Author: Changwei Zhang. Email: zcw2013@swu.edu.cn
Received: 25 December 2020; Accepted: 26 January 2021
#These authors contribute equally to this work and should be considered co-first authors
Abstract: Leaf color mutants are ideal materials for studying many plant physiological and metabolic processes such as photosynthesis, photomorphogenesis, hormone physiology and disease resistance. In this study, the genetically stable yellow-green leaf mutant ygl16 was identified from mutated “Xinong 1B”. Compared with the wild type, the pigment concentration and photosynthetic capacity of the ygl16 decreased significantly. The ultrastructural observation showed that the distribution of thylakoid lamellae was irregular in ygl16 chloroplasts, and the grana and matrix lamellae were blurred and loose in varied degrees, and the chloroplast structure was disordered, while the osmiophilic corpuscles increased. The results of the genetic analysis and mapping showed that the phenotype of ygl16 was controlled by a pair of recessive nuclear gene. The gene located in the 56Kb interval between RM25654 and R3 on the long arm of chromosome 10. The sequencing results showed that the 121st base of the first intron of the candidate gene OsPORB/FGL changed from A to T in the interval. qRT-PCR results showed that the expression of chlorophyll synthase-related genes in the mutant decreased.
Keywords: Rice; yellow-green leaf mutant; ygl16; gene mapping; candidate gene analysis
Rice (Oryza sativa L.) is an important food crop in the world. More than half of the world’s population take rice as the staple food. Therefore, increasing rice yield is of great significance to solve the global food deficiency. Leaves are the main organs of rice photosynthesis, and leaf photosynthesis after heading provides 60% of 90% carbon in rice panicles [1]. At present, the light energy utilization of rice is only 1.5% to 2.0%, which is lower than the theoretical utilization rate of 3.0% to 5.0% [2]. Thus, it is an effective method to improve rice yield by improving photosynthetic efficiency. Leaf color mutants are ideal materials for leaf photosynthesis research [3] and an illuminating molecular mechanism of photosynthesis, chloroplast development and photosynthetic pigment metabolic pathway. So far, leaf color mutants have been found in many plant species, such as rice [4], wheat (Triticum aestivum L.) [5], barley (Hordeum vulgare L.) [6], tomato (Solanum lycopersicum) [7], rape (Blassikakapestris) [8], soybean (Glycine max (Linn.) Merr.) [9] and so on. In recent years, with the development of molecular biotechnology, in-depth studies have been taken on rice leaf color mutations, especially in the mechanism of photosynthesis, chlorophyll biosynthesis pathway, chloroplast development and genetic control mechanism [10]. According to the differences on phenotype characteristics, rice mutants can be divided into light green, albino, yellowing, stripes and zebra leaves [11]. A large number of studies has shown that mutation of genes participating in chloroplast development, chlorophyll metabolism pathways, heme feedback regulation, photosensitive pigment regulation, and plastid gene expression could result in leaf color mutations [12–14]. At present, more than 180 genes [15] related to leaf color mutation have been reported in rice, and about 68 genes have been cloned, among which the genes related to yellow-green leaves are ygl1/ygl80 [16,17], ygl7/OsChlD [18,19], OsChlH [18,20], OsChlI [18,20], OsDVR [21], OsCAO1/ygl10 [22,23], OsCAO2 [22], ygl12 [23], OsSIG1 [24], OsPORA/OsPORB [25] and so on. Among them, ygl1 is located on chromosome 5, which encodes chlorophyll synthase, that catalyzes chlorophyllin vinegar phytol, and produces chlorophyllin. YGL7 [18,19], OsChlH [18,20], and OsChlI [18,20] are located on the long arm of rice chromosome 3, encoding for the magnesium chelating enzyme D subunit gene; OsDVR [21] is located on the short arm of rice chromosome 3, encoding for the biethylene reductase; ygl10 and OsCAO2 are located on rice chromosome 10, encoding for chlorophyll an oxidase. Ygl12 [23] is located on chromosome 12 and its expressed protein ygl12 is homologous to the RNA stem-ring binding protein CSP41b in Arabidopsis thaliana. OsPORA [25] is located on chromosome 4 encoding for the prochlorophyllin oxidoreductase A. In this study, a rice yellow-green leaf mutant ygl16, with genetically stable characters induced by Xinong 1B was identified, and its whole growth process showed yellow-green leaves. The phenotypic analysis, physiological and biochemical characteristics, molecular mapping, candidate gene analysis and gene sequencing verification study of ygl16 led to a good foundation for the cloning of the target gene and the subsequent regulation of molecular mechanisms.
The indica rice maintainer line Xinong 1B selected by the Rice Research Institute of Southwest University was treated with ethyl methanesulfonate (EMS) to get the mutant library. A rice yellow-green leaf mutant ygl16, was screened from the mutant library and its phenotypic inheritance was stable after multiple generations of self-crossing. In 2019, F1 seeds were harvested after crossing with the mutant ygl16 as the female parent, and Zhonghua 11 with normal leaf color phenotype as the male parent. F1 seeds were planted in Hainan in the same year to observe the F1 phenotype and harvest F2 seeds. In 2020, the F2 population was planted for genetic analysis.
At the mature stage, 10 wild-type and ygl16 plants with good growth were randomly selected (excluding marginal effects). After harvesting, the main agronomic characters such as plant height, ear length, number of primary branches, number of secondary branches, number of filled grains, seed setting rate and 1000-grain weight were investigated, and the results were analyzed by the Student t-test (n = 10).
2.2.2 Determination of Photosynthetic Pigments and Photosynthetic Characteristics
According to the method of Lichtenthaler [26], the concentration of photosynthetic pigments in the leaves of the wild type and mutant at the seedling stage, and in the middle of the first, second and third leaf at the booting stage were determined, respectively. Five plants of the wild type and mutant with relatively consistent growth were randomly selected in the middle of the planting plot. After weighing 0.05 g of leaves, they were put in a centrifuge tube containing a 25 mL volume ratio of ethanol: acetone = 1:1, sealed in a dark treatment for 24 h, and shaked several times in the middle, until leaves completely lost their green colour and turned white. This process was repeated 3 times. Then, the absorbance values were measured at 663 nm, 645 nm and 470 nm wavelengths with an ultraviolet spectrophotometer.
The photosynthetic parameters were measured at the heading stage from 9:00 to 12:00 AM. The net photosynthetic rate (Pn), transpiration rate (Tr), stomatal conductance (Gs) and intercellular CO2 concentration (Ci) of mutant ygl16 and wild type Xinong 1B were measured using a LI-6400 portable photosynthetic instrument.
2.2.3 Chloroplast Autofluorescence Observation
The inverted second leaves of the wild type and mutant were cut at the heading stage and embedded in Tissue-Tek, SAKURA. They were then frozen at –20°C until the embedding agent was solidified, and thereafter cut into 8 μm thick slices with a frozen slice machine. The slices were washed with ddH2O for 3 times, then covered with cover slides after cleaning the embedding agent around the slices, and photographed under a fluorescence microscope.
2.2.4 Transmission Electron Microscopy
The leaves in the middle part of the second inverted leaf on the wild type and mutant ygl16 were taken at the heading stage. The veins were removed near the mid vein and cut into 1 × 2 mm cubes. These cubes were quickly placed in a 2 mL centrifuge tube filled with a 2 mL projection electron microscope plant fixed liquid. The leaves were vacuum fixed at room temperature for 48 h, and all them were then sunk into the bottom of the centrifuge tube and stored at 4°C. Tissues were stained with uranyl acetate, dehydrated in ethanol, embedded in Spur’s medium, and then cut into thin sections. The samples were then stained with uranyl acetate again and examined using an H-7500 instrument (Hitachi) for TEM.
2.2.5 Histochemistry Analysis and Measurement of Physiological Indexes
At the heading stage, the content ration on of hydroxyl radical (•OH), hydrogen peroxide (H2O2) and the activities of catalase (CAT), superoxide anion (O2-), peroxidase (POD) and total superoxide dismutase (T-SOD) were determined in the first, second and third leaves of the wild type and mutant ygl16. This procedure was repeated 3 times and the T-test was carried out. The determination method followed the instructions of the kit provided by the manufacturer (Nanjing Jiancheng Institute of Biological Engineering, Nanjing, China).
According to the phenotype differences, 10 plants of the normal and mutant phenotypes F2 plants were selected, and the same amount of leaves was cut and mixed to construct both normal and mutant gene pools. The genomic DNA of both the normal and mutant gene pools was extracted by an improved CTAB method [27]. The genomic DNA of the F2 population for linkage analysis was extracted by the alkali cooking method [28].
2.2.7 Molecular Marker Analysis
The design of the SSR primer sequence refers to the differential primer http://www.gramene.org/microsat Magi Indel derived from the differential sequence between the Zhonghua 11 of japonica rice and the Xinong 1B of indica male sterile line (maintainer line). The PCR system was 12.5 μL, including 1.25 μL 10 × PCR buffer, 0.5 μL 2.5 mmol·L−1 dNTPs, 8.65 μL ddH2O, 10 μmol·L–1 forward and reverse primers 0.5 μL, 1.2 μL template DNA and 0.1μL 5U μL−1 Taq DNA polymerase. The PCR reaction procedure was pre-denaturation at 94°C for 5 min, denaturation at 94°C for 30 s, annealing at 56°C for 30 s, extension at 72°C for 30 s, and 35 cycles and 10 min re-extension at 72°C. The PCR products were recorded by 10% non-denaturing polyacrylamide gel electrophoresis and rapid silver staining [29].
The target gene was mapped by the BSA method [30], and the F1 generation was obtained by crossing Zhonghua 11 as male parent and mutant ygl16; the separated population of F2 generation obtained by self-crossing of the F1 generation was used for genetic analysis and gene mapping. The lines with mutant phenotype in the F2 generation were selected as the population to construct the linkage map. Vector NTI Advance 11.5 software and the website (http://www.gramene.org/microsat database) were used to develop new polymorphic SSR and InDel molecular markers between the two parents to determine the interval of the target gene. The primers we used were synthesized by the Chengdu Tsingke Biotechnology Company, China.
According to the phenotype of the mutant, the mutant site and target candidate gene were determined by sequencing and sequence alignment of the genes in the location interval of the NCBI and China Rice Data Center (http://www.ricedata.cn/gene/index.htm).
3.1 The Phenotype Analysis and Agronomic Traits of ygl16
Compared with the wild type, the mutant ygl16 had a slightly lighter leaf color at the first leaf stage, and the yellow-green leaf phenotype began to appear at the second leaf stage (Fig. 1A). The upper leaves of the subsequent tillering stage (Figs. 1B and 1C), young panicle differentiation stage, heading stage, filling stage (Fig. 1D) and mature stage were yellowish green, which indicated that ygl16 is a yellow-green leaf mutant in the whole growth stage.

Figure 1: Plant morphology of the wild type (WT) and the ygl16 mutant. A: Plants of the wild type (WT) and the ygl16 mutant at the seedling stage; B: Plants of the wild type (WT) and the ygl16 mutant at the tillering stage; C: The second leaf blade of the wild type (WT) and the ygl16 mutant at the tillering stage; D: Plants of the wild type (WT) and the ygl16 mutant at the maturity stage
In terms of main agronomic characters, plant height, total grains per panicle, filled grains per panicle, a 1000-grain weight and ear length decreased significantly by 25.46%, 40.32%, 48.86%, 12.02% and 10.97% in the mature mutant ygl16, respectively, compared with the wild type. The number of effective panicles decreased by 4.00% compared with the wild type, but the difference was not significant (Tab. 1).
Table 1: Performance of agronomic traits of the wild type (WT) and the ygl16 mutant

3.2 Determination of Photosynthetic Pigment Concentration
The photosynthetic pigment concentrations of the wild type and mutant were determined at the seedling stage and booting stages. At the seedling stage, the content ration of chlorophyll a, chlorophyll b, total chlorophyll and carotenoid of the mutant ygl16 were significantly lower than those of the wild type (Fig. 2A). At the booting stage, compared with the wild type, the concentrations of chlorophyll a, chlorophyll b, total chlorophyll and carotenoid in mutant ygl16 were lower than those in the wild type. Except for the difference of chlorophyll b between the first and the third leaves, the other differences reached a very significant level (Figs. 2B–2D).

Figure 2: Photosynthetic pigments concentrations of the wild type (WT) and ygl16 mutant types at different plant developmental morphology stages. A: Photosynthetic pigment concentrations of the wild type (WT) and ygl16 mutant types at the seedling stage; B–D: Photosynthetic pigment concentrations of the middle part of the flag (B), second (C), and third (D) leaves, respectively, in the wild type (WT) and ygl16 mutant types at the booting stage. *Significantly different at P < 0.05; ** Significantly different at P < 0.01 by the Student t-test, n = 3
3.3 Photosynthetic Characterization
The net photosynthetic rate at the heading stage of ygl16 was 26.61% significantly lower than that of the wild type; differences for this parameter were not significant on the second and third leaves (Fig. 3A). The stomatal conductance of the first and second leaves decreased significantly by 22.74% and 14.81%, respectively, on the ygl16 mutant in comparison with the wild type (Fig. 3B). However, the stomatal conductance of the third leaf was 59.17% significantly higher on the ygl16 mutant than that (Fig. 3B). The intercellular CO2 concentration of the first, second and third leaves increased by 3.36%, 0.57% and 7.29%, respectively, on the ygl16 mutant in comparison with the wild type. However, the difference was only significant on the third leaf (Fig. 3C). The transpiration rates of the first and second leaves increased by 4.29% and 15.81%, respectively, in the ygl16 mutant compared with the wild type, but the differences were not significant. The transpiration rate of the third leaf significantly increased by 50.75% in the ygl16 mutant in comparison with the wildtype (Fig. 3D).

Figure 3: Photosynthetic characteristics of the ygl16 mutant (ygl16) and wild type (WT) at the heading stage. A: Net photosynthetic rate; B: Stomatal conductance; C: Intercellular CO2 concentration; D: Transpiration rate* Significantly different at P < 0.05; ** Significantly different at P < 0.01 by Student t-test, n = 3
3.4 Chloroplast Autofluorescence Observation
The middle leaves of the wild type (WT) and mutant ygl16 were selected at the heading stage to make frozen sections. Under fluorescence microscope, the chloroplast color of mutant ygl16 was weaker than that of the wild type, and the arrangement of mesophyll cells of mutant ygl16 was looser than that on the wild type (Figs. 4A and 4C). The red fluorescence produced by the chloroplast under ultraviolet light was also significantly weaker than that on the wild type (Figs. 4B and 4D).

Figure 4: Fluorescent of the wild type (WT) and the ygl16 mutant. A, B: Microstructure in cross section of the wild type under natural light and UV light; C, D: Microstructure in cross section of the ygl16 mutant under natural light and UV light; Bar = 200 μm
The ultrastructure of mesophyll cells in the middle of the second leaf on the wild type (WT) and mutant ygl16 was observed under a transmission electron microscope at the heading stage. This was to understand the cellular structure of the mutant and the morphological changes of the chloroplast. The results showed that chloroplasts of the wild type and mutant were fully developed, with regular adherent distribution of chloroplasts, and the development of membranes and cell structures were complete (Figs. 5A and 5D). However, the number of osmiophilic corpuscles increased significantly in ygl16 (Figs. 5B and 5E). The matrix inside the chloroplast of the wild-type was thicker, and the lamellar structure was arranged tightly and neatly. However, the matrix lamellar structure in the mutant chloroplast structure was loose, and its number was significantly less than that of the wild type (Figs. 5C and 5F). Thus, the chloroplast development of the mutant ygl16 was abnormal or was degraded to varying degrees.

Figure 5: Mesophyll cell and chloroplast ultrastructure of the wild type (WT) and ygl16 mutant. A: Mesophyll cell of the wild-type (WT); B: Chloroplast of the wild-type (WT); C: Stroma lamella of the wild-type (WT); D: Mesophyll cell of the ygl16 mutant; E: Chloroplast of the ygl16 mutant; F: Stroma lamella of the ygl16 mutant. n: nuclear; ch: chloroplast; og: osmiophilic granule, sl: stroma lamella. Bars: 2 μm (A, D), 1 μm (B, E), 200 nm (C, F)
3.6 Physiological Index Measurement
The concentrations of physiological and biochemical indexes on the wild type and mutant ygl16 were measured at the heading stage. The •OH concentration of the first and the second leaves on the mutant ygl16 decreased significantly by 31.02% and 37.52%, respectively, compared with the wild type; the difference of the inverted third leaves, however, was not significant (Fig. 6A). The O2– concentrations of the first, second and third leaves on the mutant were 103.14%, 32.27% and 84.76% significantly higher than those on the wild type, respectively (Fig. 6E). The H2O2 concentrations of the first, second and third leaves were higher on the ygl16 mutant than those on the wild type (Fig. 6D). However, differences were only 39.33% significantly on the second leaf of the mutant than on that of the wild type, and differences were not significant for the first and third leaves (Fig. 6D). The T-SOD activity on the mutant ygl16 was significantly lower than that on the wild type (42.21%, 41.37% and 55.60% for the first, second and third leaves, respectively) (Fig. 6F). The POD activity of the first and third leaves decreased significantly by 32.76% and 29.13%, respectively, in the ygl16 mutant compared with the wild type (Fig. 6C). The CAT activity of the first and second leaves was similar on the ygl16 mutant and the wild type; however, that activity on the third leaf was significantly higher on wild type than on the ygl16 mutant (Fig. 6B). The results showed that the YGL16 mutation may affect the normal activity of the antioxidant enzymes and decreased the scavenging ability of H2O2 and O2−, while •OH had a strong oxidation ability in the plant and could effectively combine with O2− to remove O2− in the plant. The decrease of the •OH concentration led to increased O2− concentrations in the plant.
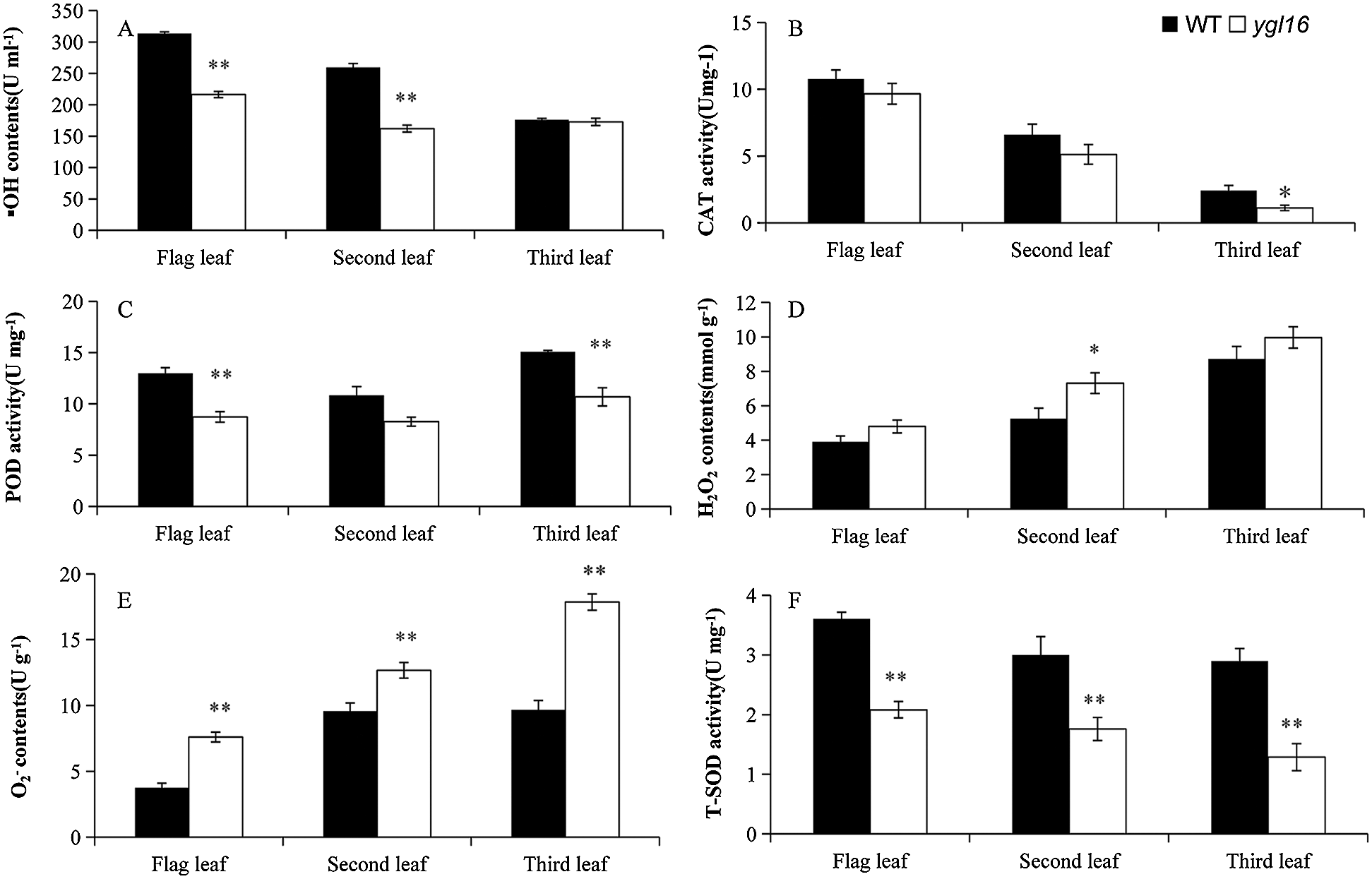
Figure 6: Physiological indexes of the wild type (WT) and the ygl16 mutant at the heading stage. * Significantly different at P < 0.05; ** Significantly different at P < 0.01 by Student t-test, n = 3
The phenotype of F1 obtained from the cross between the mutant ygl16 (female parent) and the Zhonghua 11 (male parent) was the same as that of the wild type Xinong 1B, and the yellow-green leaf phenotype was found in the F2 population obtained from the F1 inbred. The investigation and statistics were in accordance with the Mendelian segregation ratio at 3:1 (X2 = 1.20 < X20.05,1 = 3.84), which indicated that the mutation of ygl16 was controlled by a pair of recessive genes.
3.8 Molecular Location of Mutant ygl16
Ten normal leaf color plants and 10 leaf color mutants in the F2 population were selected to construct the gene pool, and polymorphic primers evenly distributed on 12 chromosomes were used for linkage analysis. The results showed that RM5471 located on rice chromosome 10 might be linked to the target gene. Five pairs of polymorphic molecular markers were developed between Xinong 1B and Zhonghua 11 to further determine the interval of the target gene (Tab. 2). Finally, the target gene was located in the region between the molecular markers RM25654 and R3; the genetic distance between the two molecular markers was 0.38 cM and 0.57 cM, respectively, and the physical distance was 56 Kb (Fig. 7).
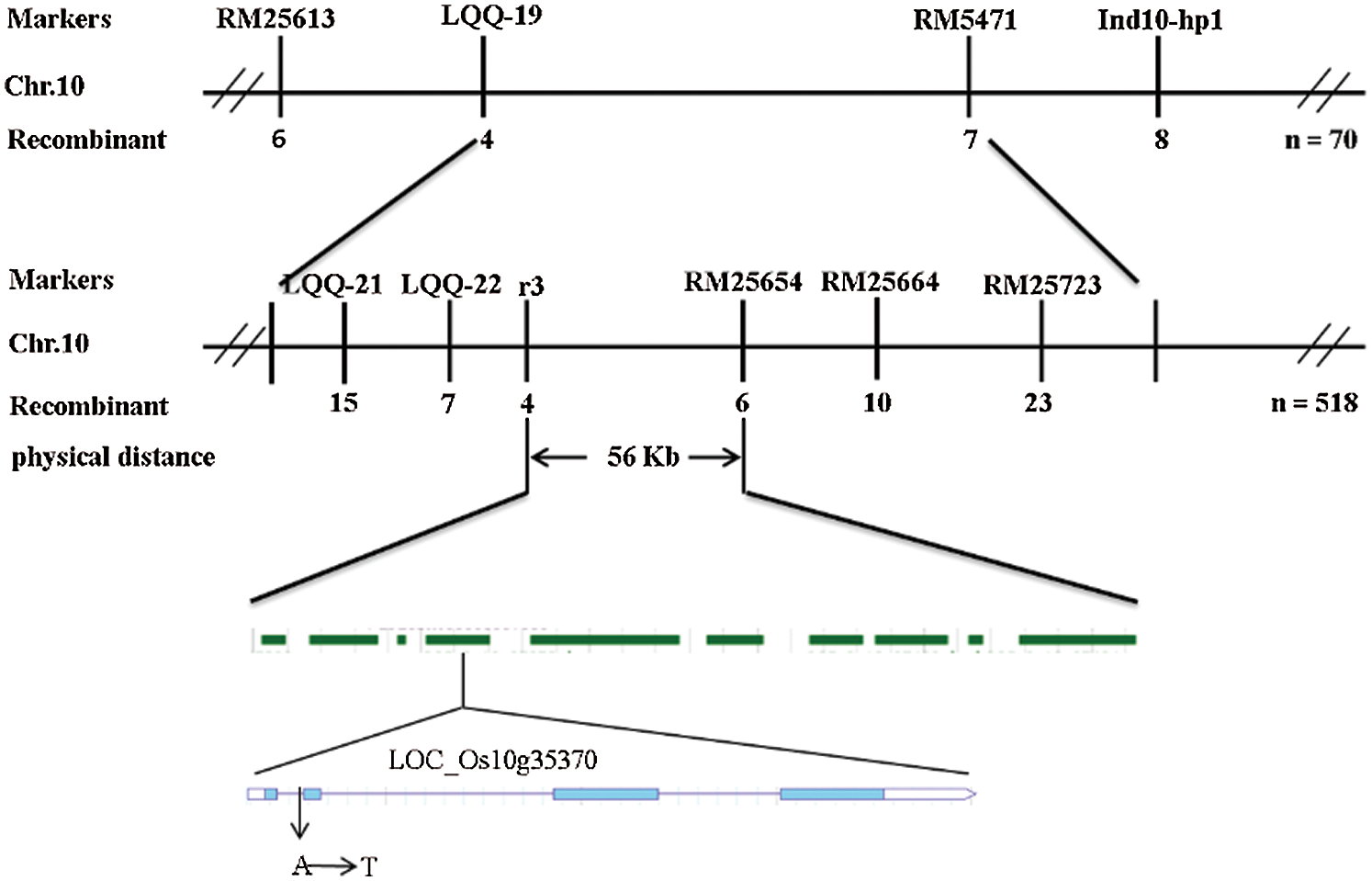
Figure 7: Linkage map of the ygl16 on rice chromosome 10
Table 2: Newly designed polymorphic markers used for gene mapping

There were 10 candidate genes in this interval. The annotations of each gene were searched in the NCBI and the National Rice data Center. It was found that there were four expressed proteins, two retrotransposon proteins, an acyltransferase protein, a transposable protein, a triangular pentapeptide repeat protein and a gene related to chlorophyll synthesis. The promoters and open reading frames of all annotated genes in this region were amplified and sequenced As a result, it was found that only the 121st base of the first intron encoded by the chlorophyll biosynthesis gene LOC_Os10g35370 mutated from A to T (Fig. 8).

Figure 8: The DNA sequencing comparison of the YGL16 gene
Real-time fluorescence quantitative PCR (quantitative Real-time PCR) was used to analyze the expression of genes in the photosynthetic pigment, carotenoid, and photosystem-related metabolic pathways at the tillering stage (Fig. 9). Compared with the wild type, the expression of the (1) photosystem-related genes (petA, petB, petD, psaA, psbA and Cab1R), (2) chlorophyll metabolism pathway genes (HEMA1, CHLD, HEME and CAO1), and (3) carotenoid metabolism-related genes (PSY1 and PSY3) was significantly decreased on the ygl16 mutant, but for HEMD, CHLM and PSY3 (Fig. 9). The expression of psbA, encoding photoreaction center protein subunit, decreased the most; this may be due to the abnormal assembly of the original parts in the photosystem reaction after mutation, while the genes PSY1 and PSY3, encoding octahydrolycopene synthase, were related to the accumulation of reactive oxygen species and the formation of chloroplast light complex proteins in the plant. The expression of psbA decreased and the reactive oxygen species accumulated, which resulted in an increased ability of reactive oxygen species to damage the photosynthetic system and further hinder the formation of light complex proteins in the chloroplast development pathway [31].
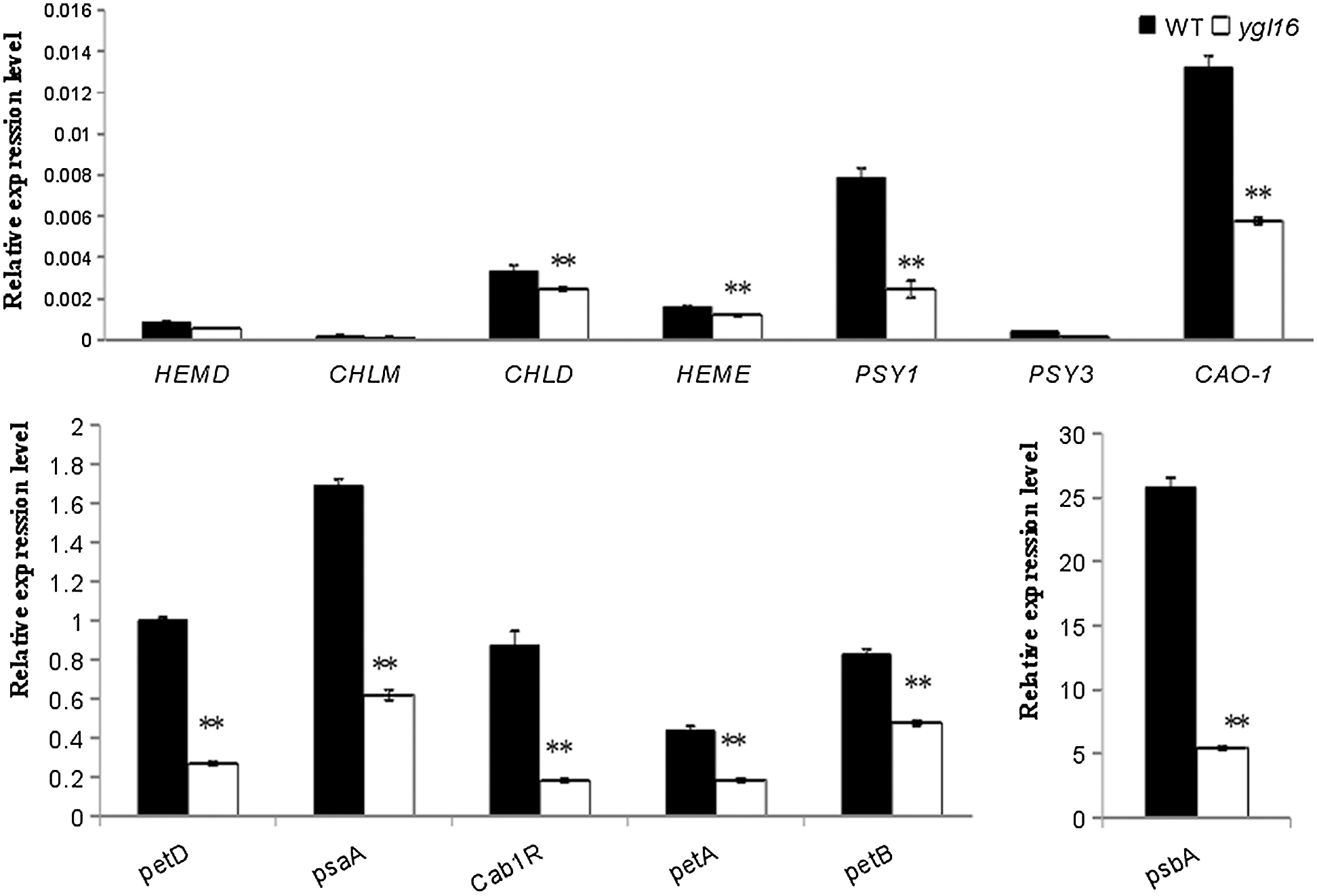
Figure 9: Relative expression levels of genes associated with pigment metabolism and photosynthesis in the wild type (WT) and ygl16 at the seedling stage. Photosystem-related genes (petA,petB,petD,psaA,psbA,Cab1R) are coded separately: cytochrome f, cytochrome b6, cytochrome b/f complex subunit IV, light reaction center protein subunits, photosystem II chlorophyll a/b binding protein; Chlorophyll metabolic pathway genes (CHLM, HEMA1, CHLD, HEME, CAO1): Mg-protoporphyrin IX methyltransferase; glutamyl-tRNA synthetase; Mg chelatase D subunit, uroporphyrinogen decarboxylase; Regulation of the coding of carotenoid metabolic pathway genes (PSY1 and PSY3): Octahydro-lycopene synthase. * Significantly different at P < 0.05; ** Significantly different at P < 0.01 by the Student t-test, n = 3
Leaf color mutants are easy to identify since the phenotypes are not easily affected by environmental factors. The genes that cause leaf color variation often have an important impact on plant growth and development. Therefore, researchers pay more and more attention in making research and utilization of leaf color mutants. The mutant gene of leaf color can directly or indirectly affect the synthesis and degradation of photosynthetic pigments, resulting in an imbalance of the proportion of photosynthetic pigments in the plant. In rice, YGL1 is a chlorophyll synthase gene, the mutation of which can block chlorophyll synthesis, decrease chlorophyll concentration and thylakoid membrane, change the content ration and proportion of various pigments, and lead to changes in leaf color [18]. OsCAO1 and OsCAO2 are genes encoding chlorophyll oxidase, which mainly play roles in the biosynthesis of chlorophyll b. The abnormal function of this enzyme can lead not only to a significant decrease in the chlorophyll concentration in plants, but also to the inability to synthesize chlorophyll [22]. Sgr encodes for a primitive protein gene containing specific chloroplast transport and regulates the activity of the pheophorbide oxygenase [32], which affects the degradation of pigment complex proteins and chloroplast cleavage [33]. Nyc encodes for the chlorophyll reductase; this is the first identified enzyme in the chlorophyll biodegradation pathway. Its mutation can inhibit the degradation of chlorophyll b and most bound carotenoids, and of the thylakoid grana [34]. OsHAP3 is a gene related to the development of chloroplast and starch; if it loses its function, it can influence the lamellar structure of chloroplast development, the accumulation of starch grains, the decreased concentration of chlorophyll, and the degradation of the chloroplast [35]. In this study, the yellow-green leaf mutant ygl16 showed yellow-green leaves throughout the growth period; the total chlorophyll concentration at the booting stage was higher than that at the seedling stage, and the etiolation phenotype was more obvious at the seedling than at the booting stage. The leaf phenotype at the seedling stage was curled, and that at the tillering stage was gradually aggravated from the leaf tip; also, other parts were light green, which was different from that reported in fgl [25]. The leaf color of mutant fgl was light green from the edge of the leaf at the seedling stage. On the other hand, the interior of the leaf in fg1 is evergreen and does not curl, and the phenotypic yellowing of the leaf increases with the growth and development of the plant, until the maturity stage. The phenotype difference between the two mutants may be due to the different mutation sites in the genes, resulting in different effects of gene expression. Chlorophyll is the main cytochrome that absorbs, transmits and converts light energy in the process of photosynthesis, which can make that plant organisms appear green [36]. Therefore, any change of the chlorophyll concentration directly affects growth and development of plants. In this study, the expression of the genes related to the chlorophyll synthesis pathway was down-regulated in the mutant ygl16, which directly affected the normal synthesis of chlorophyll. As a result, the chlorophyll concentration decreased directly, and the plant leaf phenotype was yellowish green. The genes PSY1 and PSY3, related to the carotene metabolic pathway, were also significantly down-regulated, resulting in the accumulation of reactive oxygen species in the plant, hindering the synthesis of photosystem complexes, and decreasing the ability of photoinhibition produced during light absorption. This affected the normal photosystem of the plant. In addition to the abnormal expression of the above metabolism-related genes, the expression of other photosynthesis-related genes was also down-regulated, especially psbA. Thus, encoding of the photoreaction center protein subunit in the photosystem pathway was down-regulated very seriously: This may directly leads to the abnormal functioning of the photosystem, resulting in a weakening of photosynthesis.
Artificial mutagenesis is the most common method in gene function analysis and plant breeding. Many alleles have been identified to elucidate their function in plant growth and development which will greatly promote the understanding of the mechanisms involved in leaf color regulation. In this study, the candidate gene YGL16, mutated from A to T at the 121st base of the first intron (total length 138 bp) of the coding region. According to the GT-AG recognition rule [37], the mutation may cause a mistaken splicing of the RNA, and part of the non-coding sequence may be retained, which may result in the incorrect and nonfunctional protein sequence. The mutation of YGL16 was found in LOC_Os10g35370, encoding for the prochlorophyll oxidoreductase B (OsPORB). The expression of OsPORB increases rapidly under high light conditions, and catalyzes the formation of chlorophyllin ester from prochlorophyllin in the chlorophyll synthesis. This can mainly regulate the change of the chlorophyll concentration as to affect growth and development of leaves at the seedling stage.
Leaf color mutants not only play an important role in revealing the mechanism of chlorophyll biosynthesis and photosynthesis in rice, but also can eliminate hybrids and identify hybrid purity in improved variety breeding [38]. In this study, the rice leaf color mutant ygl16 was systematically studied from the aspects of agronomic characters, physiological and biochemical characteristics, and location and expression of mutant genes. The results obtained can provide a theoretical basis for further studies on the mechanism behind leaf color mutants.
The yellow-green leaf phenotype of mutant ygl16 was controlled by a pair of recessive nuclear genes and distributed in the leaves of the whole plant from the seedling to the mature stage. Compared with the wild type, the mutant ygl16 plant became shorter, the yield decreased, and the concentrations of chlorophyll a, chlorophyll b, total chlorophyll and carotenoid decreased significantly at the seedling and heading stages; at the same time, the fluorescence of the ygl16 chlorophyll weakened, the irregular distribution of chloroplast thylakoid lamella decreased, and the grana and matrix lamellae in the chloroplast were blurred, loose, disordered and osmiophilic bodies increased significantly. The concentrations of H2O2 and O2- in the mutant ygl16 increased significantly, while the concentration of •OH, and the activities of the protective enzymes CAT, POD and T-SOD also decreased significantly (Fig. 10). YGL16 was located in the 56kb region between RM25654 and R3 at the long arm of the rice chromosome 10. The 121st base of the intron of the candidate gene LOC_Os10g35370 in this region mutated from A to T. The mutation at this site may affect the splicing of the target candidate gene mRNA, resulting in the emergence of the yellow-green leaf phenotype. Therefore, the mutant ygl16 may be an allelic mutation of the reported OsPORB/FGL gene.

Figure 10: Summary figure
Authors’ Contributions: Changwei Zhang conceived and designed the research. Linjun Cai, Junhua Liu, Han Yun, Dan Du, Xiaolong Zhong, Zhenlin Yang, and Xianchun Sang assisted in the experiments. Linjun Cai and Junhua Liu analyzed the experimental data and wrote the manuscript. All authors discussed the results and commented on the manuscript.
Data Availability: All data are fully available without restriction.
Ethics Approval: All analyses were based on previous published studies, thus no ethical approval and patient consent are required.
Funding Statement: This work was supported by grants from the Project of Creating High Quality, Disease Resistance and High Combining Ability CMS Lines (Grant No. cstc2018jscx-msybX0250) from Chongqing Technology Innovation and Application Demonstration Project and the Project of High Photosynthetic Efficiency Rice Breeding Technology System (Grant No. 2017YFD0100201) from the National Key Research and Development Program “Seven Crops Breeding”.
Conflicts of Interest: Authors have no conflict of interest to declare.
1. Zhang, B. L. (2013). Advances of research on leaf senescence in rice (Ph.D. Thesis). Tianjin Agricultural Sciences. [Google Scholar]
2. Loomis, R. S., Williams, W. A. (1963). Maximum crop productivity: An estimate. Crop Science, 3(1), 67–72. DOI 10.2135/cropsci1963.0011183X000300010021x. [Google Scholar] [CrossRef]
3. Wu, Z. M., Zhang, X. M. (2007). Map-based cloning and application of the rice mutant gene controlling yellow-green leaf. Chinese Bulletin of Life Sciences, 19(6), 614–615. [Google Scholar]
4. Zhao, S., Long, W., Wang, Y., Liu, L., Wang, Y. et al. (2016). A rice white-stripe leaf 3 (wsl3) mutant lacking an HD domain-containing protein affects chlorophyll biosynthesis and chloroplast development. Journal of Plant Biology, 59(3), 282–292. DOI 10.1007/s12374-016-0459-8. [Google Scholar] [CrossRef]
5. Wang, Y., Zheng, W., Zheng, W., Zhu, J., Liu, Z. et al. (2018). Physiological and transcriptomic analyses of a yellow-green mutant with high photosynthetic efficiency in wheat (Triticum aestivum L.). Functional & Integrative Genomics, 18(2), 175–194. DOI 10.1007/s10142-017-0583-7. [Google Scholar] [CrossRef]
6. Wang, R., Yang, F., Zhang, X. Q., Wu, D., Xu, Y. (2017). Characterization of a thermo-inducible chlorophyll-deficient mutant in barley. Frontiers in Plant Science, 8, 309. DOI 10.3389/fpls.2017.01936. [Google Scholar] [CrossRef]
7. Barry, C. S., Aldridge, G. M., Herzog, G., Ma, Q., Mcquinn, R. P. et al. (2012). Altered chloroplast development and delayed fruit ripening caused by mutations in a zinc metalloprotease at the lutescent 2 locus of tomato. Plant Physiology, 159(3), 1086–1098. DOI 10.1104/pp.112.197483. [Google Scholar] [CrossRef]
8. Stéphane, D. N., Guillaume, L. M., Frédérique, E., Olivier, C., Hervé, M. et al. (2007). Homeologous recombination plays a major role in chromosome rearrangements that occur during meiosis of brassica napus haploids. Genetics, 175(2), 487–503. DOI 10.1534/genetics.106.062968. [Google Scholar] [CrossRef]
9. Reed, S., Atkinson, T., Gorecki, C., Espinosa, K., Sandhu, D. (2014). Candidate gene identification for a lethal chlorophyll-deficient mutant in soybean. Agronomy, 4(4), 462–469. DOI 10.3390/agronomy4040462. [Google Scholar] [CrossRef]
10. Deng, X. J., Zhang, H. Q., Wang, Y., Shu, Z. F., Wang, G. H. et al. (2012). Research advances on rice leaf-color mutant genes. Hybrid Rice, 27(5), 9–14. [Google Scholar]
11. Gustafsson, Ã. K. (1942). The plastid development in various types of chlorophyll mutations. Hereditas, 28(3–4), 483–492. DOI 10.1111/j.1601-5223.1942.tb03292.x. [Google Scholar] [CrossRef]
12. Kusumi, K., Mizutani, A., Nishimura, M., Iba, K. (1997). A virescent gene V1 determines the expression timing of plastid genes for transcription/translation apparatus during early leaf development in rice. Plant Journal, 12(6), 1241–1250. DOI 10.1046/j.1365-313x.1997.12061241.x. [Google Scholar] [CrossRef]
13. Kushnir, S., Babiychuk, E., Storozhenko, S., Davey, M. W., Papenbrock, J. et al. (2001). A mutation of the mitochondrial ABC transporter Sta1 leads to dwarfism and chlorosis in the Arabidopsis mutant starik. Plant Cell, 13(1), 89–100. DOI 10.1105/tpc.13.1.89. [Google Scholar] [CrossRef]
14. Huq, E., Al-Sady, B., Hudson, M., Kim, C. H., Apel, M. et al. (2004). Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science, 305(5692), 1937–1941. DOI 10.1126/science.1099728. [Google Scholar] [CrossRef]
15. Chu, Z. Z., Guo, H. B., Liu, X. L., Chen, Y. L., Liu, Y. G. (2016). Genetic analysis and gene mapping of a yellow-green leaf mutant in rice. Acta Agronomica Sinica, 42(5), 684–689. DOI 10.3724/SP.J.1006.2016.00684. [Google Scholar] [CrossRef]
16. Li, Y. Q., Gao, J. X., Xiao, Y. H., Li, X. L., Deng, X. J. (2014). Genetic Analysis and gene fine mapping of yellow-green leaf mutant ygl80 in rice. Acta Agronomica Sinica, 40(4), 644–649. DOI 10.3724/SP.J.1006.2014.00644. [Google Scholar] [CrossRef]
17. Wu, Z. M. (2007). Map-based cloning and functional study of the rice mutant gene ygl1 controlling yellow-green leaf (Ph.D. Thesis). Nanjing Agricultural University. [Google Scholar]
18. Jung, K. H., Hur, J., Ryu, C. H., Choi, Y., Chung, Y. Y. et al. (2003). Characterization of a rice chlorophyll-deficient mutant using the T-DNA gene-trap system. Plant and Cell Physiology, 44(5), 463–472. DOI 10.1093/pcp/pcg064. [Google Scholar] [CrossRef]
19. Deng, X. J. (2014). Map-based cloning and functional study of a rice yellowy-green leave’s gene YGL7 (Ph.D. Thesis). Hunan Agricultural University. [Google Scholar]
20. Zhang, H., Li, J., Yoo, J. H., Yoo, S. C., Cho, S. H. et al. (2006). Rice chlorina-1 and chlorina-9 encode ChlD and ChlI subunits of mg-chelatase, a key enzyme for chlorophyll synthesis and chloroplast development. Plant Molecular Biology, 62(3), 325–337. DOI 10.1007/s11103-006-9024-z. [Google Scholar] [CrossRef]
21. Wang, P., Gao, J., Wan, C., Zhang, F., Xu, Z. et al. (2010). Divinyl chlorophyll(ide) a can be converted to monovinyl chlorophyll(ide) a by a divinyl reductase in rice. Plant Physiology, 153(3), 994–1003. DOI 10.1104/pp.110.158477. [Google Scholar] [CrossRef]
22. Lee, S., Kim, J. H., Yoo, E. S., Lee, C. H., Hirochika, H. et al. (2005). Differential regulation of chlorophyll a oxygenase genes in rice. Plant Molecular Biology, 57(6), 805–818. DOI 10.1007/s11103-005-2066-9. [Google Scholar] [CrossRef]
23. Yang, H. L., Liu, M., Guo, M., Li, R. D., Zhang, H. G. et al. (2014). Genetic analysis and position cloning of a yellow-green leaf 10(ygl10) gene, responsible for leaf color in rice. Chinese Journal of Rice Science, 28(1), 41–48. [Google Scholar]
24. Tozawa, Y., Teraishi, M., Sasaki, T., Sonoike, K., Hirochika, H. (2007). The plastid sigma factor SIG1 maintains photosystem I activity via regulated expression of the psaA operon in rice chloroplasts. Plant Journal for Cell & Molecular Biology, 52(1), 124–132. DOI 10.1111/j.1365-313X.2007.03216.x. [Google Scholar] [CrossRef]
25. Sakuraba, Y., Rahman, M. L., Cho, S. H., Kim, Y. S., Koh, H. J. et al. (2013). The rice faded green leaf locus encodes protochlorophyllide oxidoreductase B and is essential for chlorophyll synthesis under high light conditions. Plant Journal, 74(1), 122–133. DOI 10.1111/tpj.12110. [Google Scholar] [CrossRef]
26. Lichtenthaler, H. K. (1987). Chlorophylls and carotenoids-pigments of photosynthetic biomembranes. Methods in Enzymology, 148, 350–382. [Google Scholar]
27. Matthes, N., Westphal, K., Haldemann, C., Egert, M., Jokisch, C. et al. (2020). Validation of a modified CTAB method for DNA extraction from protein-rich maize feedstuffs. Journal of Consumer Protection and Food Safety, 15(4), 331–340. DOI 10.1007/s00003-020-01285-y. [Google Scholar] [CrossRef]
28. Li, W., Dai, Z. Y., Xia, W. W., Wang, W. W. (2017). A rapid DNA extraction method for large squash population. China Vegetable,s, 9, 36–40. [Google Scholar]
29. Zhang, T., Wang, S., Sun, S., Zhang, Y., Zhao, F. (2020). Analysis of QTL for grain size in a rice chromosome segment substitution line Z1392 with long grains and fine mapping of qGL-6. Rice, 13(1), 16. DOI 10.1186/s12284-020-00399-z. [Google Scholar] [CrossRef]
30. Chen, P., Hu, H. T., Zhang, Y., Wang, Z. W., Dong, G. J. et al. (2018). Genetic analysis and fine-mapping of a new rice mutant, white and lesion mimic leaf 1. Plant Growth Regulation, 85(3), 425–435. DOI 10.1007/s10725-018-0403-7. [Google Scholar] [CrossRef]
31. Du, W. K., Yuan, S. X., Hu, F. R. (2019). Research progress on molecular mechanisms of the leaf color mutation. Molecular Plant Breeding, 17(6), 1888–1897. [Google Scholar]
32. Panaud, O., Chen, X., Mccouch, S. R. (1996). Development of microsatellite markers and characterization of simple sequence length polymorphism (SSLP) in rice (Oryza sativa L.). Molecular and General Genetics, 252(5), 597–607. [Google Scholar]
33. Jiang, H. W., Li, M. R., Liang, N. T., Yan, H. B., Wei, Y. B. (2007). Molecular cloning and function analysis of the stay green gene in rice. Plant Journal, 52(2), 197–209. DOI 10.1111/j.1365-313X.2007.03221.x. [Google Scholar] [CrossRef]
34. Kusaba, M., Ito, H., Morita, R., Iida, S., Sato, Y. et al. (2007). Rice non-yellow coloring 1 is involved in light-harvesting complex II and grana degradation during leaf senescence. Plant Cell, 19(4), 1362–1375. DOI 10.1105/tpc.106.042911. [Google Scholar] [CrossRef]
35. Kubo, T., Serizawa, A. (2008). Identification, characterization and interaction of HAP family genes in rice. Journal of Receptor Research, 7(1–4), 279–289. [Google Scholar]
36. Fromme, P., Melkozernov, A., Jordan, P., Krauss, N. (2003). Structure and function of photosystem I: interaction with its soluble electron carriers and external antenna systems. FEBS Letters, 555(1), 40–44. DOI 10.1016/S0014-5793(03)01124-4. [Google Scholar] [CrossRef]
37. Fergany, A. A. M., Tatarskiy, V. V. (2020). RNA splicing: Basic aspects underlie antitumor targeting. Recent Patents on Anti-Cancer Drug Discovery, 15(4), 293–305. DOI 10.2174/1574892815666200908122402. [Google Scholar] [CrossRef]
38. Li, X. L., Deng, A. F., Xu, Y. R., Deng, J. L., Gu, A. Y. et al. (2011). Breeding of rice leaf margin albino leaf-color marker three sterile line Yunfeng 88 A. Seed, 30(12), 109–111. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |