
International Journal of
Experimental Botany

 | Phyton- International Journal of Experimental Botany |  |
DOI: 10.32604/phyton.2021.013805
ARTICLE
Differential Responses of NHX1 and SOS1 Gene Expressions to Salinity in two Miscanthus sinensis Anderss. Accessions with Different Salt Tolerance
1Asian Natural Environmental Science Center (ANESC), The University of Tokyo, Tokyo, 188-0002, Japan
2College of Health Science and Environmental Engineering, Shenzhen Technology University, Shenzhen, 518118, China
3Field Science Center for Northern Biosphere, Hokkaido University, Sapporo, 060-0811, Japan
*Corresponding Authors: Yulai Han. Email: hanyulai@sztu.edu.cn; Tetsuo Takano. Email: takano@anesc.u-tokyo.ac.jp
Received: 20 August 2020; Accepted: 01 December 2020
Abstract: The lignocellulosic crop Miscanthus spp. has been identified as a good candidate for biomass production. The responses of Miscanthus sinensis Anderss. to salinity were studied to satisfy the needs for high yields in marginal areas and to avoid competition with food production. The results indicated that the relative advantages of the tolerant accession over the sensitive one under saline conditions were associated with restricted Na+ accumulation in shoots. Seedlings of two accessions (salt-tolerant ‘JM0119’ and salt-sensitive ‘JM0099’) were subjected to 0 (control), 100, 200, and 300 mM NaCl stress to better understand the salt-induced biochemical responses of genes involved in Na+ accumulation in M. sinensis. The adaptation responses of genes encoding for Na+/H+ antiporters, NHX1 and SOS1 to NaCl stress were examined in JM0119 and JM0099.The cDNA sequences of genes examined were highly conserved among the relatives of M. sinensis based on the sequencing on approximate 600 bp-long cDNA fragments obtained from degenerate PCR. These salt-induced variations of gene expression investigated by quantitative real-time PCR provided evidences for insights of the molecular mechanisms of salt tolerance in M. sinensis. The expression of NHX1 was up-regulated by salt stress in JM0119 shoot and root tissues. However, it was hardly affected in JM0099 shoot tissue except for a significant increase at the 100 mM salt treatment, and it was salt-suppressed in the JM0099 root tissue. In the root tissue, the expression of SOS1 was induced by the high salt treatment in JM0119 but repressed by all salt treatments in JM0099. Thus, the remarkably higher expression of NHX1 and SOS1 were associated with the resistance to Na+ toxicity by regulation of the Na+ influx, efflux, and sequestration under different salt conditions.
Keywords: Salinity; Miscanthus sinensis; NHX1; SOS1
Perennial plants with high net energy output and little CO2 emission are needed to meet the challenge of developing second-generation energy crops capable of growing on marginal lands. Miscanthus sinesis Anderss., the C4 perennial lignocellulosic grass, shows a source of genetic diversity for the development of new hybrid bioenergy crops. Furthermore, the capability of M. sinesis to easily propagate and produce high-yields in various abiotic stress environments implies less management for its establishment or production and well-adaptation to marginal lands as a bioenergy crop.
Salinity affected plants through two stress components: an osmotic stress and an ionic toxicity, especially sodium toxicity at elevated NaCl concentrations. Influx of Na+ could be achieved through plasma-membrane nonselective cation channels (NSCCs) or anatomical ‘leaks’ in the root endodermis [1–4]. A striking increase in cytoplasmic Na+ disrupts enzymatic functions and is toxic to both cells and the whole plant. Therefore, decades of study have been dedicated to the characterization of Na+ transport and distribution in plants [2,4–7]. Sodium transport involves a group of genes that play critical roles in ion homeostasis, and herein in the Na+/H+ antiporters. These genes include the SOS1 (located at the plasma membrane) and NHX1 (located in the tonoplast) which facilitates the maintenance of appropriate Na+ concentrations in the cytosol to minimize cytotoxicity. At the cellular level, SOS3, SOS2, and SOS1 are essential components of the Salt Overly Sensitive (SOS) signaling pathway that mediates cellular signaling under salt stress [8]. The primary calcium sensor SOS3 is a calcineurin-like, myristoylated Ca2+-binding protein. It perceives the increase in cytosolic Ca2+ induced by the excessive cytosolic Na+, and activates the serine/threonine protein kinase SOS2 [9–11]. Thereafter, the SOS1 is activated which causes the extrusion of excessive Na+ from the cytosol [12–15]. At the whole-plant level, SOS1 has been proposed previously to promote Na+ efflux from roots [16–18] and facilitate Na+ retrieval from or delivery to the xylem [17,19,20], maintaining a low-sodium zone at the root [18,21]. Compartmentalization of Na+ into vacuoles is assumed to lower (1) toxic Na+ concentrations in the cytoplasm, and (2)vacuolar osmotic potentials. This osmotic adjustment will allow to maintain turgor pressure and cell expansion under salt stress [4]. The sequestration of Na+ in the vacuole was proved to be important to salt tolerance by overexpression of NHX in various species, such as tomato (Solanum lycopersicum L.), soybean [Glycine max (Linn.) Merr.], rapeseed (Brassica napus L.), rice (Oryza sativa L.), maize (Zea mays L.), wheat (Triticum aestivum L.), cotton (Gossypium hirsutum L.), and tobacco (Nicotiana tabacum L.) [22–29]. In addition to the function of vacuolar sequestration of Na+, NHX1 transporter also plays a role in (1) pH homeostasis [30]; (2) plant development [30,31]; (3) the transport of other monovalent cations like K+, Li+, Rb+, and Cs+ [32–34]; (4)vesicle trafficking and protein targeting [35]; and (5) regulation of stomatal function [36].
In our previous greenhouse study, two M. sinensis accessions, JM0119 and JM0099, showed variable salt tolerance on plant dry weight, leaf chlorophyll content, total leaf area, tiller number, photosynthetic parameters, and shoot ion content [37]. The objective of the present study was to investigate the differential responses of genes encoding Na+/H+ antiporters NHX1 and SOS1 to NaCl stress in JM0119 (salt-tolerant) and JM0099 (salt-sensitive). The salt-induced variation of gene expression provided evidences for insights of the molecular mechanisms of salt tolerance in M. sinensis.
2.1 Plant Materials and Growth Conditions
Seeds of two M. sinensis accessions with distinct salt-tolerance, JM0119 (salt-tolerant) and JM0099 (salt-sensitive), were surface-sterilized with 70% ethanol for 30 s and with 1% (v/v) sodium hypochlorite (NaClO) for 10 min, and then rinsed three times with distilled water. Seeds of each accession were germinated on filter paper in a closed, 90-mm Petri dish filled with 20 ml distilled water in a growth chamber at 12 h photoperiod with 500 μmol m-2s-1 PPF, 70% humidity, and 28/22°C day/night regime. Seven-day old uniform seedlings were transplanted into a 500-ml pot and hydroponically acclimated in distilled water for seven days. Seedlings were thinned to ten per pot and grown in salt-free 1/2 Hoagland solution (concentration was gradually increased during five days) for 14 days. Thereafter, seedlings were exposed to salt treatments of 1/2 Hoagland solution containing 0 (control), 100, 200, and 300 mM NaCl. Salt addition gradually increased at a progressive rate of 50 mM per day until the final treatment levels were reached to avoid osmotic shock. Leaf and root samples were harvested 24 h after salt application for each treatment, and three days after salt application for the 300 mM NaCl treatment. These samples were then immediately immersed in liquid nitrogen and stored at –80°C.
2.2 RNA Extraction, Degenerate PCR, Cloning and Sequencing
Frozen samples of leaf tissues of M. sinensis harvested atdifferent dates were ground in liquid nitrogen, and the total RNA from each sample was extracted using a TRIzol reagent according to the protocol (Life technologies Corp. Carlsbad, USA). First-strand cDNA was synthesized with 2 μg of total RNA using a high capacity RNA-to-cDNA kit (Applied Biosystems, Inc. Foster, USA). The first-strand cDNA served as a template for a 50-μl PCR (PrimeSTAR HS DNA Polymerase; TaKaRa Bio Inc. Otsu, Shiga, Japan) primed with gene-specific degenerate primers (Tab. S1). Primers of DP 1 and DP 2 targeting NHX1 were designed based on an alignment of the NHX1 peptide sequences of Aeluropus lagopoides (GU199336), Arabidopsis thaliana (NM_122597), Diplachne fusca (JF933902), Eutrema halophilum (DQ995339), O. sativa (AB021878), Phragmites australis (AB211145), Puccinellia tenuiflora (AB628206), and Z. mays (NM_001111751). Primers of DP 3 and DP 4 targeting SOS1 were designed based on an alignment of the SOS1 peptide sequences of Aegilops speltoides (FN356230), Aeluropus littoralis (JN936862), A. thaliana (AF256224), Brachypodium sylvaticum (FJ234838), O. sativa (AY785147), Ph. australis (AB244217), P. tenuiflora (AB628205), and T. aestivum (AY326952). The PCR was conducted under a regime of 94°C for 4 min, followed by 35 cycles of 98°C for 10 s, 54°C for 15 s (5 s for SOS1 primers), and 72°C for 60 s. Purified amplicon (Agarose Gel DNA Extraction Kit; TaKaRa Bio Inc. Otsu, Japan) was ligated into pBluescript II SK (-) vector (Agilent Technologies, Inc. Santa Clara, USA), and transformed into competent DH5α Escherichia coli cells (Life technologies Corp. Carlsbad, USA). Plasmid DNA was isolated from E. coli culture using a GenElute plasmid miniprep kit (Sigma-Aldrich St. Louis, USA).
2.3 Quantitative Real-Time PCR Analysis
The extracted total RNA was purified and treated with RNase-free DNase (Recombinant DNase I; TaKaRa Bio Inc., Otsu, Japan). Quantitative real-time PCR (qRT-PCR) assay was performed on an Applied Biosystems 7500 real-time PCR system (Applied Biosystems, Inc., Foster, USA), with SYBR Premix Ex Taq II (TaKaRa Bio Inc., Otsu, Japan) under the procedure of the manufacturer’s instructions. Gene-specific primers for real-time PCR analysis were designed using the obtained approximately 600 bp-long sequence of each gene (Tab. S2). The Actin gene of M. sinensis (JN983213) was used as the reference gene. The PCR program consisted of a denaturation at 95°C for 30 s, then 45 cycles of 95°C for 5 s, a combined annealing/extension phase of 60°C for 30 s followed by a melt curve analysis. Ct (cycle threshold) values were (1) Determined in auto Ct mode using the Applied Biosystems 7500 real time PCR system software and (2) Used to calculate the gene expression relative to an internal control gene.
All data was analyzed by Statistical Product and Service Solutions (SPSS Statistics, Version 20; IBM Corp. New York, USA). The significance of the differences between salt treatments means was determined by a Student’s t-test (α < 0.05) run on quantitative real-time PCR analysis.
3.1 Sequences of NHX1 and SOS1
The cDNA sequences of genes NHX1 and SOS1 were highly conserved. Compared with the other monocotyledonous species, the translated putative protein sequence of NHX1 showed 96%, 94%, 92%, 92%, and 79% identity to that of Cenchrus americanus (L.) Morrone, Diplachne fusca (L.) Beauv., common reed (Phragmites australis (Cav.) Trin. ex Steud.), rice, and maize (Fig. 1). The translated putative protein sequence of MsSOS1 showed 80%, 82%, 82%, 82%, and 79% identity to that of Distichlis spicata (L.) Greene, rice, common reed, Brachypodium sylvaticum (Huds.) Beauv., and common wheat (T. aestivum L.) (Fig. 2).

Figure 1: Alignment of partial putative translated polypeptide of NHX1 from M. sinensis (MISI), Dip. fusca (DIFU), C. americanus (CEAM), reed (PHAU), rice (ORSA), and maize (ZEMA). Amino acid sequences were deduced by translation of cDNA from the partial sequence of MsNHX1 and from GenBank accession numbers JF933902 (DIFU), DQ071264 (CEAM), AB211145 (PHAU), AB021878 (ORSA), and NM_001111751 (ZEMA)

Figure 2: Alignment of partial putative translated polypeptide of SOS1 from M. sinensis (MISI), rice (ORSA), reed (PHAU), B. sylvaticum (BRSY), wheat (TRAE), and Dis. spicata (DISP). Amino acid sequences were deduced by translation of cDNA from the partial sequence of MsSOS1 and from GenBank accession numbers AY785147 (ORSA), AB244217 (PHAU), FJ234838 (BRSY), AY326952 (TRAE), and GU480079 (DISP)
3.2 The Relative Expression Levels of NHX1 and SOS1
Plant growth of both M. sinensis accessions was severely affected by salt stress. However, the salt-induced phenotypic symptoms such as delay in leaf emergence, burning of leaf tips, yellowing of leaves, and reduction of tiller numbers were observed more prominently in JM0099 than in JM0119 after 200 mM and 300 mM NaCl treatments (Fig. 3).
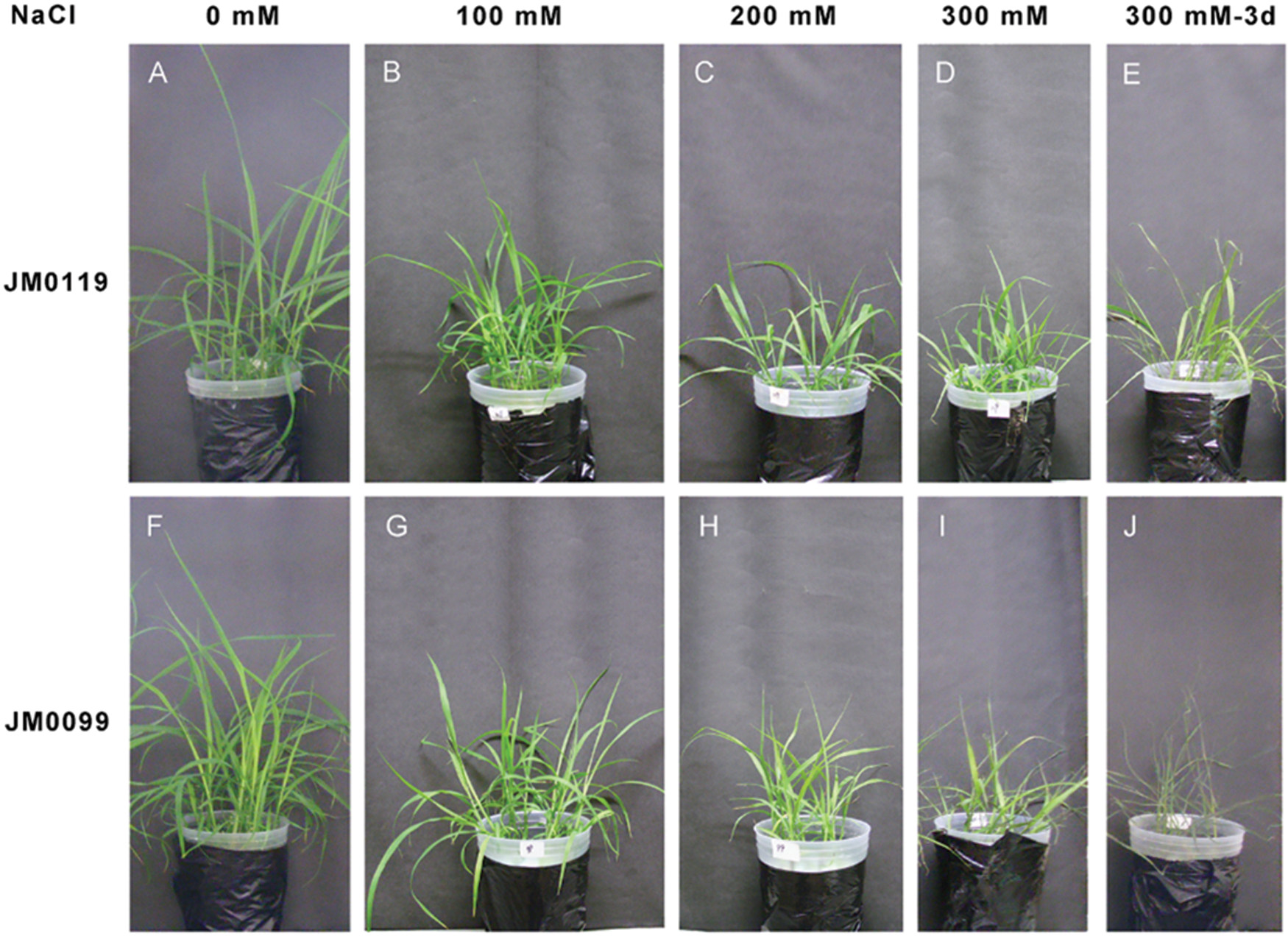
Figure 3: Phenotypic responses of two M. sinensis accessions to NaCl stress. A and F, seedlings grown in the absence of stress; B–D and G–I, seedlings exposed to increasing NaCl concentrations; E and J, seedlings after three-day-exposure to 300 mM NaCl
The relative expression of the genes encoding the Na+/H+ antiporters NHX1 and SOS1 varied with salt stress treatments and treatment duration, and showed remarkable differences between accessions. In plant shoots, the expression of NHX1 was dramatically up-regulated by salt stress in JM0119, while this increase was reduced after 3-day-exposure to 300 mM NaCl concentration (Fig. 4A). In JM0119, the expression of shoot NHX1 was 13-fold higher than that in the control after 24 h exposure to 300 mM NaCl, and fell to about 4-fold higher than that in the control after 3-day-exposure to 300 mM NaCl. A significant increase in the expression of NHX1 was also observed in the root tissue of JM0119 after exposure to the 300 mM salt treatment, after the repression at low and moderate salt levels (Fig. 4B). However, NHX1 expression was hardly changed in the shoot tissue of JM0099 except for a significant increase at 100 mM salt treatment (Fig. 4A), and it was suppressed by salt stress in the root tissue of JM0099 (Fig. 4B). The expression of SOS1 was around the control levels in the shoot tissues of both accessions (Fig. 5A). However, in the root tissue, it was markedly induced by the high salt treatment in JM0119, and repressed by all salt treatments in JM0099 (Fig. 5B).
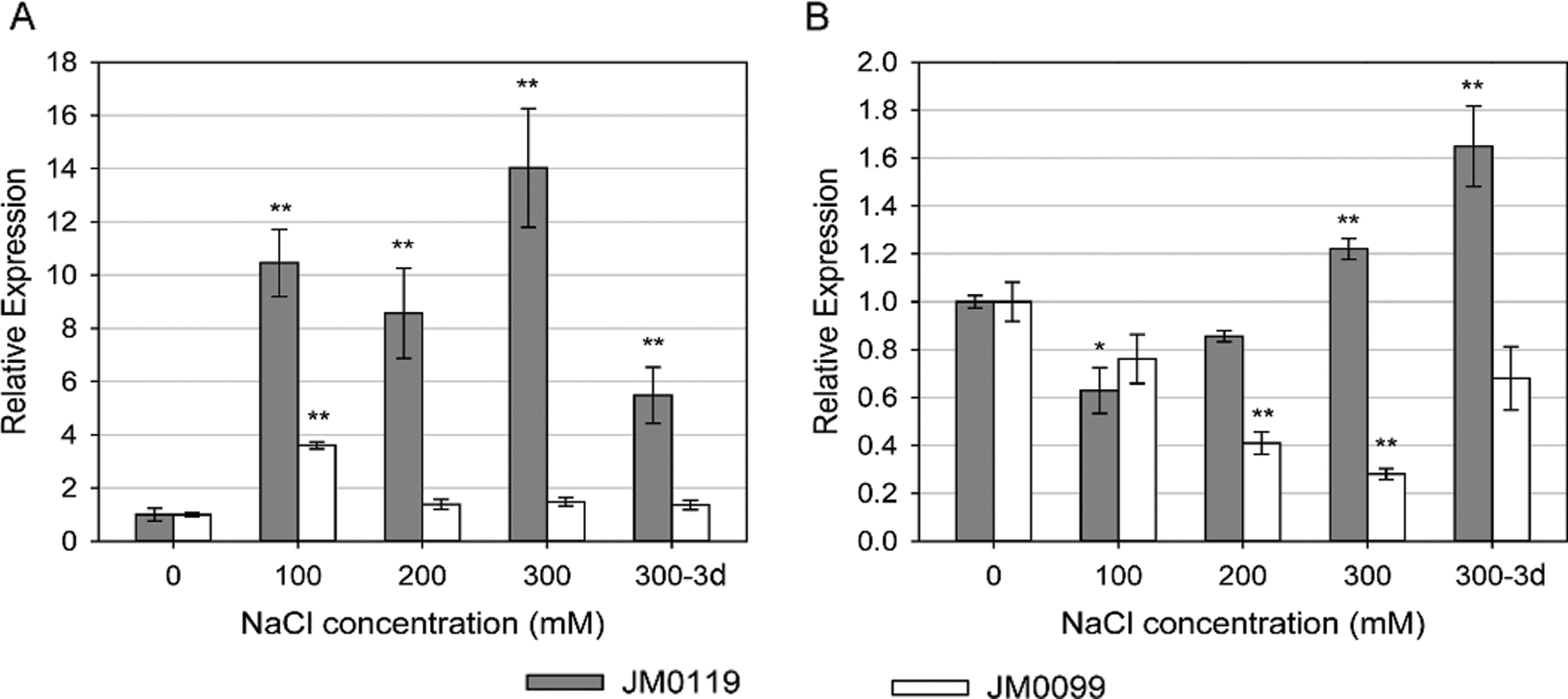
Figure 4: Effects of salinity on the expression of MsNHX1 in plant shoot (A) and root (B) to increasing NaCl concentrations in two M. sinensis accessions. Values are means ± SE (n = 3). Asterisks indicate significant differences relative to the control in each accession, * P ≤ 0.05; ** P ≤ 0.01. Absence of an asterisk denotes a non-significant effect (Student’s t-test0.05)
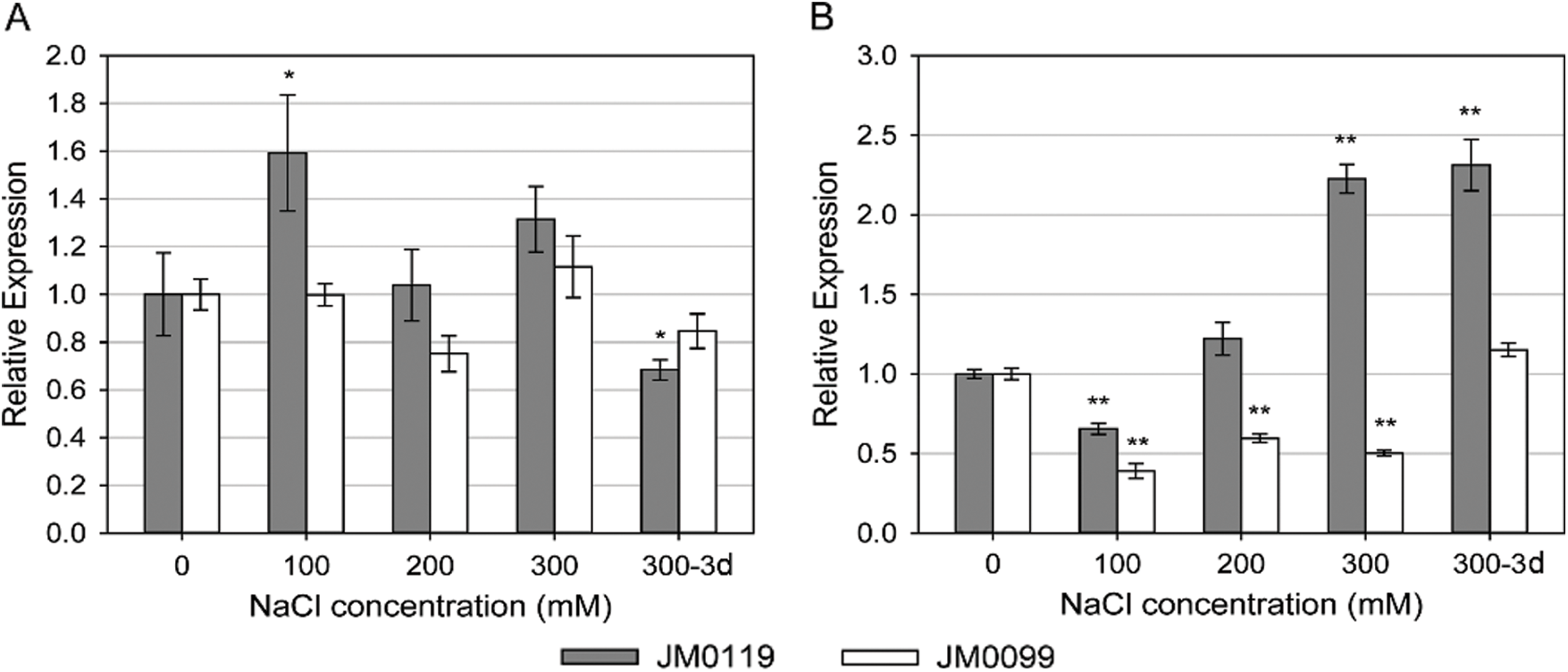
Figure 5: Effects of salinity on the expression of MsSOS1 in plant shoot (A) and root (B) to increasing NaCl concentrations in two M. sinensis accessions. Values are means ± SE (n = 3). Asterisks indicate significant differences relative to the control in each accession, * P ≤ 0.05; ** P ≤ 0.01. Absence of an asterisk denotes a non-significant effect (Student’s t-test0.05)
The important mechanisms coping with high salinity in plants are the regulation of Na+ influx, efflux and vacuolar compartmentation [4]. The distinct expression patterns of SOS1 and NHX1 during the increase of salt concentration in the two studied accessions indicate different salt-resistant mechanisms in M. sinensis. Under 100 mM NaCl stress, SOS1 and NHX1 expressions were repressed in the salt-treated root tissue of JM0119 and remarkably promoted in the shoot tissue. In this case, Na+ could not be efficiently sequestered into the root cell vacuole and would not act as a cellular osmoticum. Thus, to avoid the increase of environmental osmotic stress by root sodium extrusion, SOS1 expression was also restricted as observed in the NHX1 expression [38,39]. The increased expression of SOS1 and NHX1 in plant shoots may be a prompt response to maintain a low cytosolic Na+ concentration in the leaf to protect photosynthesis [17,18]. When salt stress became more severe (200 mM NaCl), the SOS1 and NHX1 expression recoverment in the root and reduction in the shoot may be the consequence of a controlled delivery of Na+ from the root to the shoot [19]. Under high salt stress (300 mM NaCl), both SOS1 and NHX1 expressions were up-regulated in the roots of JM0119, together with a dramatic increase of NHX1 expression in the shoots. This suggested that there were greatly activated Na+ extrusion out of the root cells and a strong sequestration of Na+ into the cell vacuoles in the salt-tolerant accession as it has been observed in other plants [7,18,21,23,29]. After 3-d under high salinity, shoot NHX1 and SOS1 expressions were both lower than those measured 24 h after treatment with 300 mM NaCl. This may be due to the severe damage to the shoot by Na+ accumulation. Compared with JM0119, the salt-sensitive accession, JM0099, showed suppressed expression of both genes in the root and changeless expression in the shoot (apart from a significant promotion at 100 mM salt concentration). This was consistent with the dramatic increase in Na+ content in the JM0099 shoots as described in a previous study [37]. It indicated the lack of Na+ efflux and compartmentalization which resulted in a high Na+ accumulation in the shoots of the salt-sensitive M. sinensis even under moderate salt stress.
In conclusion, the remarkably high expressions of NHX1 and SOS1 were associated with the regulation of Na+ accumulation in M. sinensis by mechanisms involved in Na+ influx, efflux, and sequestration.
Funding Statement: This study was supported by grants from Shenzhen Fundamental Research Program (Grant Nos. JCYJ20170818140058675 and JCYJ20170818140127741) and Natural Science Foundation of Top Talent of SZTU (Grant Nos. 2019010801010 and 2019010801009).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report in the present study.
1. Davenport, R. J., Tester, M. (2000). A weakly voltage-dependent, nonselective cation channel mediates toxic sodium influx in wheat. Plant Physiology, 122(3), 823–834. DOI 10.1104/pp.122.3.823. [Google Scholar] [CrossRef]
2. Hasegawa, P. M., Bressan, R. A., Zhu, J. K., Bohnert, H. J. (2000). Plant cellular and molecular responses to high salinity. Annual Review of Plant Physiology and Plant Molecular Biology, 51(1), 463–499. DOI 10.1146/annurev.arplant.51.1.463. [Google Scholar] [CrossRef]
3. Demidchik, V., Tester, M. (2002). Sodium fluxes through nonselective cation channels in the plasma membrane of protoplasts from Arabidopsis roots. Plant Physiology, 128(2), 379–387. DOI 10.1104/pp.010524. [Google Scholar] [CrossRef]
4. Munns, R., Tester, M. (2008). Mechanisms of salinity tolerance. Annual Review of Plant Biology, 59(1), 651–681. DOI 10.1146/annurev.arplant.59.032607.092911. [Google Scholar] [CrossRef]
5. Kronzucker, H. J., Britto, D. T. (2011). Sodium transport in plants: A critical review. New Phytologist, 189(1), 54–81. DOI 10.1111/j.1469-8137.2010.03540.x. [Google Scholar] [CrossRef]
6. Agarwal, P. K., Yadav, N. S., Jha, B. (2013). Role of Na+/H+ antiporters in Na+ homeostasis in halophytic plants. In: Tuteja, N., Gill, S. S.(eds.Weinheim, Germany: Climate change and plant abiotic stress tolerance. pp. 685–704, Wiley-VCH Verlag GmbH & Co., KGaA. [Google Scholar]
7. Fahmideh, L., Fooladvand, Z. (2018). Isolation and semi quantitative PCR of Na+/H+ antiporter (SOS1 and NHX) genes under salinity stress in Kochia scoparia. Biological Procedures Online, 20(1), 429. DOI 10.1186/s12575-018-0076-7. [Google Scholar] [CrossRef]
8. Ji, H. T., Pardo, J. M., Batelli, G., Van Oosten, M. J., Bressan, R. A. et al. (2013). The salt overly sensitive (SOS) pathway: Established and emerging roles. Molecular Plant, 6(2), 275–286. DOI 10.1093/mp/sst017. [Google Scholar] [CrossRef]
9. Liu, J. P., Zhu, J. K. (1998). A calcium sensor homolog required for plant salt tolerance. Science, 280(5371), 1943–1945. DOI 10.1126/science.280.5371.1943. [Google Scholar] [CrossRef]
10. Halfter, U., Ishitani, M., Zhu, J. K. (2000). The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proceedings of the National Academy of Sciences of the United States of America, 97(7), 3735–3740. DOI 10.1073/pnas.97.7.3735. [Google Scholar] [CrossRef]
11. Liu, J. P., Ishitani, M., Halfter, U., Kim, C. S., Zhu, J. K. (2000). The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proceedings of the National Academy of Sciences of the United States of America, 97(7), 3730–3734. DOI 10.1073/pnas.97.7.3730. [Google Scholar] [CrossRef]
12. Qiu, Q. S., Guo, Y., Dietrich, M. A., Schumaker, K. S., Zhu, J. K. (2002). Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proceedings of the National Academy of Sciences of the United States of America, 99(12), 8436–8441. DOI 10.1073/pnas.122224699. [Google Scholar] [CrossRef]
13. Quintero, F. J., Ohta, M., Shi, H. Z., Zhu, J. K., Pardo, J. M. (2002). Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proceedings of the National Academy of Sciences of the United States of America, 99(13), 9061–9066. DOI 10.1073/pnas.132092099. [Google Scholar] [CrossRef]
14. Martínez-Atienza, J., Jiang, X. Y., Garciadeblas, B., Mendoza, I., Zhu, J. K. et al. (2007). Conservation of the salt overly sensitive pathway in rice. Plant Physiology, 143(2), 1001–1012. DOI 10.1104/pp.106.092635. [Google Scholar] [CrossRef]
15. Quintero, F. J., Martinez-Atienza, J., Villalta, I., Jiang, X. Y., Kim, W. Y. et al. (2011). Activation of the plasma membrane Na/H antiporter Salt-Overly-Sensitive 1 (SOS1) by phosphorylation of an auto-inhibitory C-terminal domain. Proceedings of the National Academy of Sciences of the United States of America, 108(6), 2611–2616. DOI 10.1073/pnas.1018921108. [Google Scholar] [CrossRef]
16. Elphick, C. H., Sanders, D., Maathuis, F. J. M. (2001). Critical role of divalent cations and Na+ efflux in Arabidopsis thaliana salt tolerance. Plant Cell and Environment, 24(7), 733–740. DOI 10.1046/j.0016-8025.2001.00713.x. [Google Scholar] [CrossRef]
17. Olías, R., Eljakaoui, Z., Li, J., Alvarez De Morales, P., Marín-Manzano, M. C. et al. (2009). The plasma membrane Na+/H+ antiporter SOS1 is essential for salt tolerance in tomato and affects the partitioning of Na+ between plant organs. Plant Cell and Environment, 32(7), 904–916. DOI 10.1111/j.1365-3040.2009.01971.x. [Google Scholar] [CrossRef]
18. Gao, J. J., Sun, J., Cao, P. P., Ren, L. P., Liu, C. et al. (2016). Variation in tissue Na+ content and the activity of SOS1 genes among two species and two related genera of Chrysanthemum. BMC Plant Biology, 16(1), 651. DOI 10.1186/s12870-016-0781-9. [Google Scholar] [CrossRef]
19. Shi, H. Z., Quintero, F. J., Pardo, J. M., Zhu, J. K. (2002). The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell, 14(2), 465–477. DOI 10.1105/tpc.010371. [Google Scholar] [CrossRef]
20. Yadav, N. S., Shukla, P. S., Jha, A., Agarwal, P. K., Jha, B. (2012). The SbSOS1 gene from the extreme halophyte Salicornia brachiata enhances Na+ loading in xylem and confers salt tolerance in transgenic tobacco. BMC Plant Biology, 12(1), 188. DOI 10.1186/1471-2229-12-188. [Google Scholar] [CrossRef]
21. Oh, D. H., Leidi, E., Zhang, Q., Hwang, S. M., Li, Y. Z. et al. (2009). Loss of Halophytism by interference with SOS1 expression. Plant Physiology, 151(1), 210–222. DOI 10.1104/pp.109.137802. [Google Scholar] [CrossRef]
22. Zhang, H. X., Blumwald, E. (2001). Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit. Nature Biotechnology, 19(8), 765–768. DOI 10.1038/90824. [Google Scholar] [CrossRef]
23. He, Y., Fu, J. L., Yu, C. L., Wang, X. M., Jiang, Q. S. et al. (2015). Increasing cyclic electron flow is related to Na+ sequestration into vacuoles for salt tolerance in soybean. Journal of Experimental Botany, 66(21), 6877–6889. DOI 10.1093/jxb/erv392. [Google Scholar] [CrossRef]
24. Zhang, H. X., Hodson, J. N., Williams, J. P., Blumwald, E. (2001). Engineering salt-tolerant Brassica plants: Characterization of yield and seed oil quality in transgenic plants with increased vacuolar sodium accumulation. Proceedings of the National Academy of Sciences of the United States of America, 98(22), 12832–12836. DOI 10.1073/pnas.231476498. [Google Scholar] [CrossRef]
25. Ohta, M., Hayashi, Y., Nakashima, A., Hamada, A., Tanaka, A. et al. (2002). Introduction of a Na+/H+ antiporter gene from Atriplex gmelini confers salt tolerance to rice. FEBS Letters, 532(3), 279–282. DOI 10.1016/S0014-5793(02)03679-7. [Google Scholar] [CrossRef]
26. Yin, X. Y., Yang, A. F., Zhang, K. W., Zhang, J. R. (2004). Production and analysis of transgenic maize with improved salt tolerance by the introduction of AtNHX1 gene. Acta Botanica Sinica, 46(7), 854–861. [Google Scholar]
27. Xue, Z. Y., Zhi, D. Y., Xue, G. P., Zhang, H., Zhao, Y. X. et al. (2004). Enhanced salt tolerance of transgenic wheat (Tritivum aestivum L.) expressing a vacuolar Na+/H+ antiporter gene with improved grain yields in saline soils in the field and a reduced level of leaf Na+. Plant Science, 167(4), 849–859. DOI 10.1016/j.plantsci.2004.05.034. [Google Scholar] [CrossRef]
28. He, C. X., Yan, J. Q., Shen, G. X., Fu, L. H., Holaday, A. S. et al. (2005). Expression of an Arabidopsis vacuolar sodium/proton antiporter gene in cotton improves photosynthetic performance under salt conditions and increases fiber yield in the field. Plant and Cell Physiology, 46(11), 1848–1854. DOI 10.1093/pcp/pci201. [Google Scholar] [CrossRef]
29. Lu, S. Y., Jing, Y. X., Shen, S. H., Zhao, H. Y., Ma, L. Q. et al. (2005). Antiporter gene from Hordum brevisubulatum (Trin.) Link and its overexpression in transgenic tobaccos. Journal of Integrative Plant Biology, 47(3), 343–349. DOI 10.1111/j.1744-7909.2005.00027.x. [Google Scholar] [CrossRef]
30. Bassil, E., Tajima, H., Liang, Y. C., Ohto, M., Ushijima, K. et al. (2011). The Arabidopsis Na+/H+ antiporters NHX1 and NHX2 control vacuolar pH and K+ homeostasis to regulate growth, flower development, and reproduction. Plant Cell, 23(9), 3482–3497. DOI 10.1105/tpc.111.089581. [Google Scholar] [CrossRef]
31. Hanana, M., Cagnac, O., Yamaguchi, T., Hamdi, S., Ghorbel, A. et al. (2007). A grape berry (Vitis vinifera L.) cation/proton antiporter is associated with berry ripening. Plant and Cell Physiology, 48(6), 804–811. DOI 10.1093/pcp/pcm048. [Google Scholar] [CrossRef]
32. Venema, K., Quintero, F. J., Pardo, J. M., Donaire, J. P. (2002). The Arabidopsis Na+/H+ exchanger AtNHX1 catalyzes low affinity Na+ and K+ transport in reconstituted liposomes. Journal of Biological Chemistry, 277(4), 2413–2418. DOI 10.1074/jbc.M105043200. [Google Scholar] [CrossRef]
33. Wu, Y. Y., Chen, Q. J., Chen, M., Chen, J., Wang, X. C. (2005). Salt-tolerant transgenic perennial ryegrass (Lolium perenne L.) obtained by Agrobacterium tumefaciens-mediated transformation of the vacuolar Na+/H+ antiporter gene. Plant Science, 169(1), 65–73. DOI 10.1016/j.plantsci.2005.02.030. [Google Scholar] [CrossRef]
34. Leidi, E. O., Barragán, V., Rubio, L., El-Hamdaoui, A., Ruiz, M. T. et al. (2010). The AtNHX1 exchanger mediates potassium compartmentation in vacuoles of transgenic tomato. Plant Journal, 61(3), 495–506. DOI 10.1111/j.1365-313X.2009.04073.x. [Google Scholar] [CrossRef]
35. Sottosanto, J. B., Saranga, Y., Blumwald, E. (2007). Impact of AtNHX1, a vacuolar Na+/H+ antiporter, upon gene expression during short-term and long-term salt stress in Arabidopsis thaliana. BMC Plant Biology, 7(1), 18. [Google Scholar]
36. Barragán, V., Leidi, E. O., Andres, Z., Rubio, L., De Luca, A. et al. (2012). Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis. Plant Cell, 24(3), 1127–1142. DOI 10.1105/tpc.111.095273. [Google Scholar] [CrossRef]
37. Sun, Q., Yamada, T., Takano, T. (2014). Salinity effects on germination, growth, photosynthesis, and ion accumulation in wild Miscanthus sinensis Anderss. populations. Crop Science, 54(6), 2760–2771. DOI 10.2135/cropsci2013.09.0636. [Google Scholar] [CrossRef]
38. Cuin, T. A., Bose, J., Stefano, G., Jha, D., Tester, M. et al. (2011). Assessing the role of root plasma membrane and tonoplast Na+/H+ exchangers in salinity tolerance in wheat: in planta quantification methods. Plant Cell Environment, 34(6), 947–961. [Google Scholar]
39. Zivcak, M., Brestic, M., Sytar, O. (2016). Osmotic adjustment and plant adaptation to drought stress. In: Hossain, M. A., Wani, S. H., Bhattacharjee, S., Burritt, D. J., Tran, L. S. P. (eds.Switzerland: Drought Stress Tolerance in Plants, pp. 105-143. Springer International Publishing. [Google Scholar]
Table S1: Sequences of primer pairs used for degenerate PCR in Miscanthus sinensis
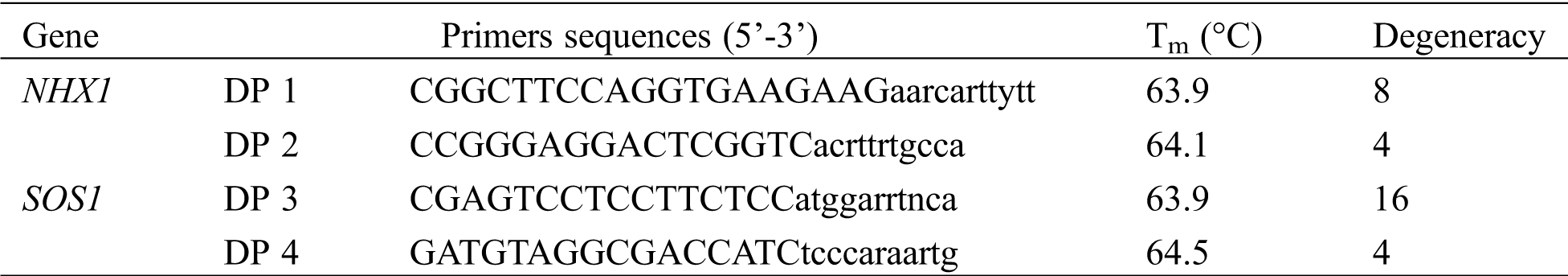
Table S2: Sequences of gene specific primer pairs used for real-time PCR in Miscanthus sinensis

 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |