

 | Phyton-International Journal of Experimental Botany |  |
DOI: 10.32604/phyton.2021.014156
ARTICLE
The Potassium Transporter AtKUP12 Enhances Tolerance to Salt Stress through the Maintenance of the K+/Na+ Ratio in Arabidopsis
Xinjiang Key Laboratory of Biological Resources and Genetic Engineering, College of Life Science and Technology, Xinjiang University, Urumqi, 830046, China
*Corresponding Author: Yan Wang. Email: wangyanxju@126.com
Received: 03 September 2020; Accepted: 25 October 2020
#These authors have contributed equally to this work
Abstract: Potassium (K+) is a necessary nutrient for plant growth and crop production. The K+ transporter plays crucial roles in the absorption and transport of K+ in plants. Most K+ transporters in Arabidopsis have been reported, but AtKUP12, which is a member of the KT/KUP/HAK family, has not yet been the subject of relevant in-depth research. In the present study, we demonstrated that AtKUP12 plays a crucial role in K+ uptake in Arabidopsis under 100 μM low-K+ and 125 mM salt stress conditions. AtKUP12 transcripts were induced by K+ deficiency and salt stress. We analyzed the K+ uptake of AtKUP12 using the K+ uptake-deficient yeast R5421 and Arabidopsis mutant atkup12. Transformation with AtKUP12 rescued the growth defect of mutant yeast and atkup12 mutant plants at the low-K+ concentration, which suggested that AtKUP12 might be involved in high-affinity K+ uptake in low-K+ environments. In comparison to the wild-type (WT) and atkup12-AtKUP12 complementation lines, atkup12 showed a dramatic reduction in potassium concentration, K+/Na+ ratio, and root and shoot growth on 12-day-old seedlings under the salt conditions; however, there was no significant difference between the complementation and WT lines. Taken together, these results demonstrate that AtKUP12 might participate in salt tolerance in Arabidopsis through K+ uptake and K+/Na+ homeostasis.
Keywords: AtKUP12; atkup12 mutant; K+ uptake; K+/Na+ homeostasis; salt tolerance
Potassium (K+) is an essential and the most abundant macroelement that plays several critical roles in plant growth and productivity [1] and accounts for approximately 2% to 10% of the dry weight in plants [2]. In contrast to the relatively high and stable K+ concentration (approximately 100–150 mM) in the cytoplasm of plant cells, the K+ concentration in soil solution is relatively low and highly variable, ranging from 0.01–1 mM [3]. The ability of plants to absorb and utilize K+ is relatively limited; thus, increasing the efficiency of K+ absorption and transport by means of molecular biology and genetics is an important method to alleviate the shortage of K+ resources for cash crops [4]. For example, overexpression of ZmHAK5 promotes K+ uptake and seedling growth in maize under low K+ levels [5]. Additionally, the expression of OsHAK5 in rice increases K+ acquisition and transport from roots to shoots facing low external K+ [6]. Plants possess a series of transport systems that play key roles in K+ absorption, redistribution and ion homeostasis [7], such as the Shaker, TPK and Kir-like K+ channel families [8] and the KT/HAK/KUP, Trk/HKT, KEA and CHX K+ transporter families [9]. Among them, KT/HAK/KUP is the largest family and is widely distributed in bacteria, fungi and plants but absent in animal cells [10]. Generally, K+ channels conduct low-affinity K+ uptake under higher K+ conditions, whereas K+ transporters perform high-affinity K+ uptake under lower K+ conditions [11]. At present, 13 and 27 members of the KT/HAK/KUP family genes have been identified in Arabidopsis and rice (Oryza sativa L.), respectively [12,13]. KT/HAK/KUP K+ transporters play versatile roles in K+ uptake and transport, K+ homeostasis, salt tolerance, osmotic regulation, and the morphogenesis of root and phenotype in plants [14]. The genes of this family can be divided into four distinct clusters (I–IV) by their amino acid sequence homology [15]. There is only one member in the Arabidopsis cluster I family, and the other 12 members are mainly distributed in cluster II and cluster III. In rice, 8 members are distributed in cluster I, 15 members are distributed in cluster II and cluster III, and 4 members are distributed in cluster IV [16]. Cluster I transporters such as AtHAK5, HvHAK1 and OsHAK1 may play key roles in high-affinity K+ uptake when external K+ is low [6,15,17,18], while cluster II transporters such as AtKT2/KUP2 and AtKT3/AtTRH1 from Arabidopsis thaliana and OsHAK7 and OsHAK10 from rice are involved in low-affinity K+ transport [15,19]. The representative members of cluster III and IV are AtKUP12 and OsHAK11, respectively. Cluster III members may function in maintaining K+/Na+ homeostasis [15], and little research has been conducted regarding their functions [9]. The results of phylogenetic analysis showed that cluster II and cluster III members are highly conserved [16], and they may be the ancestors of the KT/HAK/KUP transporter family. Cluster II and III members are crucial in completing plant life activities, while those of cluster I and cluster IV may play a positive role in adapting to the environment by regulating plant K+ concentration [20].
K+ deficiency leads to a decrease in total root length (TRL) and root length density (RLD) in the last stage of wheat growth [21]. KT/HAK/KUP transporters play significant roles not only in absorption and transport of K+ but also morphogenesis of the root and shoot in plants [17,22,23]. Mutation of AtKUP2 (shy3–1) decreases cell expansion in shoots, resulting in a dwarf phenotype in Arabidopsis, and tiny root hairs were observed in the knockout mutation of AtKUP4/TRH1 [19]. Rubén Tenorio-Berrío et al. investigated a significant reduction in embryo and cotyledon size of Arabidopsis hak/kt12, kup4, and kup9 mutants relative to wild-type Arabidopsis, and AtHAK/KT12 and AtKUP4 were regulated by auxin IAA and IAM [24]. Knockout of OsHAK5 not only decreased K+ uptake by roots and K+ upward transport from roots to shoots with low external K+ [6], but also significantly reduced tillering number irrespective of K+ status [14]. The destruction of OsHAK1 dramatically reduced the K+ concentration in both roots and shoots, and severely stunted the growth and development of rice, resulting in delayed grain filling and decreased grain yields [25]. AtKUP2, AtKUP6, and AtKUP8 are three kinds of K+ efflux transporters. Triple mutation atkup2/6/8 had a greater effect on cell expansion, which indicated that these transporters were involved in the negative regulation of turgor pressure-dependent growth [26]. Under the low-K+ level, the athak5 mutant displayed a shorter primary root length than wild-type plants. In maize, the loss of function of ZmHAK5 reduced K+ uptake and inhibited seedling growth, while opposite results were present in the overexpression lines [5].
The tolerance of plants to salt stress is shown in the adaptation to Na+ toxicity and the maintenance of their own K+ nutrition [27]. Maintaining the K+ uptake ratio at a high level of exogenous Na+ is essential for K+/Na+ homeostasis and salt tolerance [11]. There is increasing evidence of the involvement of KT/HAK/KUP members in K+ absorption, transport, and regulation of plant salt tolerance in Arabidopsis [6]. OsHAK5 and OsHAK1 may play a pivotal role in salt tolerance because high-salt treatment temporarily induces their expression in some rice organs [6,24]. Expression of OsHAK5 in tobacco BY2 cells improves salt tolerance by accumulating more K+ without affecting the Na+ concentration [28]. Knockout of OsHAK5 or OsHAK21 results in a decrease in the K+/Na+ ratio in tissues, and mutants are more sensitive than wild-type plants to salt [6,29]. AtHAK5 also plays a key role in maintaining high-affinity K+ absorption and regulating plant growth under salt conditions [30].
In previous studies, we cloned and obtained the HcKUP12 gene from a suppression subtractive library of the halophyte Halostachys caspica, and found that it responded positively to salt stress [31]. Also, sequence homology comparison revealed that its closest homology is AtKUP12. We speculated that AtKUP12 might also participate in response to salt stress. Arabidopsis thaliana is a model organism; thus, studies examining the K+ absorption, transport and salt tolerance of the KT/HAK/KUP family in Arabidopsis can provide a reference for the study of homologous gene functions in other species. Previous studies have found that atkup12 mutant plants display a sensitive phenotype under abiotic stress such as low K+, salt and oxidation [32]. However, the physiological functions of AtKUP12 in K+ uptake and salt tolerance of plants remain unclear and warrant further investigation.
In this study, we investigated the expression patterns of AtKUP12 in Arabidopsis under low-K+ and salt stress conditions. We studied the K+-uptake activity of AtKUP12 using K+ uptake-deficient yeast CY162 and atkup12 mutants. We measured the root length, fresh weight and ion concentration of wild-type (WT), atkup12 mutant and atkup12-AtKUP12 complementation lines of Arabidopsis under low-K+ or salt stress conditions. The expression of AtKUP12 completely rescued the low-K+ and salt-sensitive phenotype of atkup12 mutant. Overall, we demonstrated that AtKUP12 participated in K+ uptake, and then improved the plant salt tolerance by maintaining the K+/Na+ ratio in Arabidopsis.
2.1 Plant Materials, Growth Conditions and Stress Treatments
Arabidopsis thaliana ecotype Columbia (Col) was used as the WT plant. We obtained Arabidopsis atkup12 mutant (SALK_083613.47.35.x) seeds from the Arabidopsis Biological Resource Center (http://www.Arabidopsis.org/abrc/), Ohio State University, USA.
Seeds were stratified after surface sterilization at 4°C in the dark for 3 days. For potassium treatments, seeds were sown on low-K+ (LK, 10, 100, 200 μM KCl) or high-K+ (HK, 1000 μM KCl) Murashige and Skoog (MS) medium (pH 5.8) supplemented with 1.5% (w/v) sucrose and solidified with 0.7% (w/v) agar to grow for 12 days under conditions of 22°C, 16 h light/8 h dark and a relative humidity of 70%. LK medium was modified from MS medium [33]. For salt stress, seeds were sown on MS medium supplemented with 0, 100 and 125 mM NaCl and grown for 12 days under the above conditions.
2.2 Expression Analysis of AtKUP12
Sterile two-week-old seedlings were subjected to stress treatments by removal to filter paper supplemented with 100 mM NaCl and and 0.5 μM low-K+, and the seedlings were harvested at 0, 1, 6, 12 and 24 h, respectively. Total RNA was extracted from different samples with the RNA Plant Plus Reagent Kit (Tiangen, China) following the manufacturer’s instructions. First-strand cDNA was synthesized with M-MLV reverse transcriptase (TaKaRa, China). Quantitative real-time PCR analysis was performed with SYBR® Select Master Mix (Applied Biosystems, USA) under the following cycling conditions: 50°C for 2 min, 95°C for 2 min, 95°C for 15 s and 40 cycles at 60°C for 1 min. The β-Atactin gene of Arabidopsis was used as the internal control. The relative gene expression was calculated by using the 2–ΔΔCT method [34]. The primer pairs RqAtKUP12P1 and RqAtKUP12P2 for AtKUP12 and qAtactinP1 and qAtactinP2 for β-Atactin were listed in Tab. S1. AtCOR15A acts as a marker gene for abiotic stresses [35,36]. Each PCR was carried out for three biological replicates; each corresponded to three technological repetitions of a separate experiment.
2.3 Functional Complementation Assay of AtKUP12 in Yeast
AtKUP12 was cloned into the pYES2.0 vector, which was linearized by double digestion with Kpn I and Xba I. The vectors pYES2.0-AtKUP12 and pYES2.0 were then transformed into K+ uptake-deficient mutants of yeast strain CY162 (MATα, Δtrk1, trk2::pCK64, his3, leu2, ura3, trp1 and ade2) [37]. The transformant was selected on SD-agar plates without uracil contained additional KCl to a final K+ concentration of 100 mM. Yeast strain CY162-pYES2.0 was employed as a negative control, and yeast strain CY162-pYES2.0-AtAKT1 (gifted by Dr Yue Shen, Nanjing Agricultural University) was used as a positive control. Subsequent growth assays for yeast were performed under low-K+ (LK, 10, 20, 30 mM KCl) or high-K+ (HK, 100 mM KCl) concentrations. Three independent clones per construct were used and analyzed in all growth tests on solid medium incubated at 30°C for 2 days.
2.4 Generation of the atkup12-AtKUP12 Complementation Line
Based on the TAIR database (http://www.Arabidopsis.org), the open reading frame of AtKUP12 (GenBank: NM_104706) was amplified. The PCR product was digested with Kpn I and Pst I and ligated into the pCAMBIA1301–1 expression vector, with the CaMV35S promoter added at multiple cloning sites. The construct was introduced into Agrobacterium tumefaciens strain GV3101 and then transformed into the atkup12 mutant using the floral-dip method [38]. The transgenic lines were selected with 30 µg mL−1 hygromycin and identified by PCR. Homozygous T3 plants were selected for further analysis.
2.5 Assays for Root Length, Fresh Weight, and Mineral Determination
Under low-K+ or salt stress conditions, the seeds germinated and grew for 12 days. Primary root length was measured using ImageJ software, and the fresh weight of seedlings was recorded. The measurement of K+ and Na+ concentration followed previously described procedures [39]. Arabidopsis seedlings were rinsed in distilled water to wash away possible surface contamination of Na+ and K+ from the culture solution and soaked. The seedlings were dried in an oven at 105°C for 30 min, and then baked at 70°C for 3 days. The dried materials (ca 50 mg) were extracted in 20 mL of boiling water for 2 h. Na+ and K+ concentration was determined using ICP-AES systems (IRIS Advantage, Thermo Electron, Waltham, Massachusetts, USA). The experiment was performed four times. The phenotypic results were from one representative experiment, and the presented data were based on four biological replicates.
Statistical differences were determined by one-way ANOVA and Tukey’s multiple comparison tests using GraphPad Prism 5 software (San Diego, CA, USA). The means were considered to be significantly different at p < 0.05 or p < 0.01.
3.1 Expression Analyses of AtKUP12
Expression levels of AtKUP12 under conditions of low-K+ and NaCl stresses were detected by qRT-PCR. The Arabidopsis AtCOR15A gene is upregulated at the transcriptional level under adversie stresses, such as high salt, ABA, cold, and drought [35,36]. Therefore, we used AtCOR15A gene as a marker gene. AtCOR15A was indeed upregulated under adverse conditions, except low-K+ (Fig. 1). AtKUP12 gene was induced by low-K+ stress, reaching its peak after 12 h, and then it gradually declined until 24 h of stress (Fig. 1A). Under NaCl stress, the AtKUP12 was continuously upregulated, peaking at 24 h by 30-fold (Fig. 1B). These results indicated that the expression of AtKUP12 was stimulated by stress conditions, such as low-K+ and NaCl treatments, and may play significant roles in the response of Arabidopsis to adverse environmental conditions.
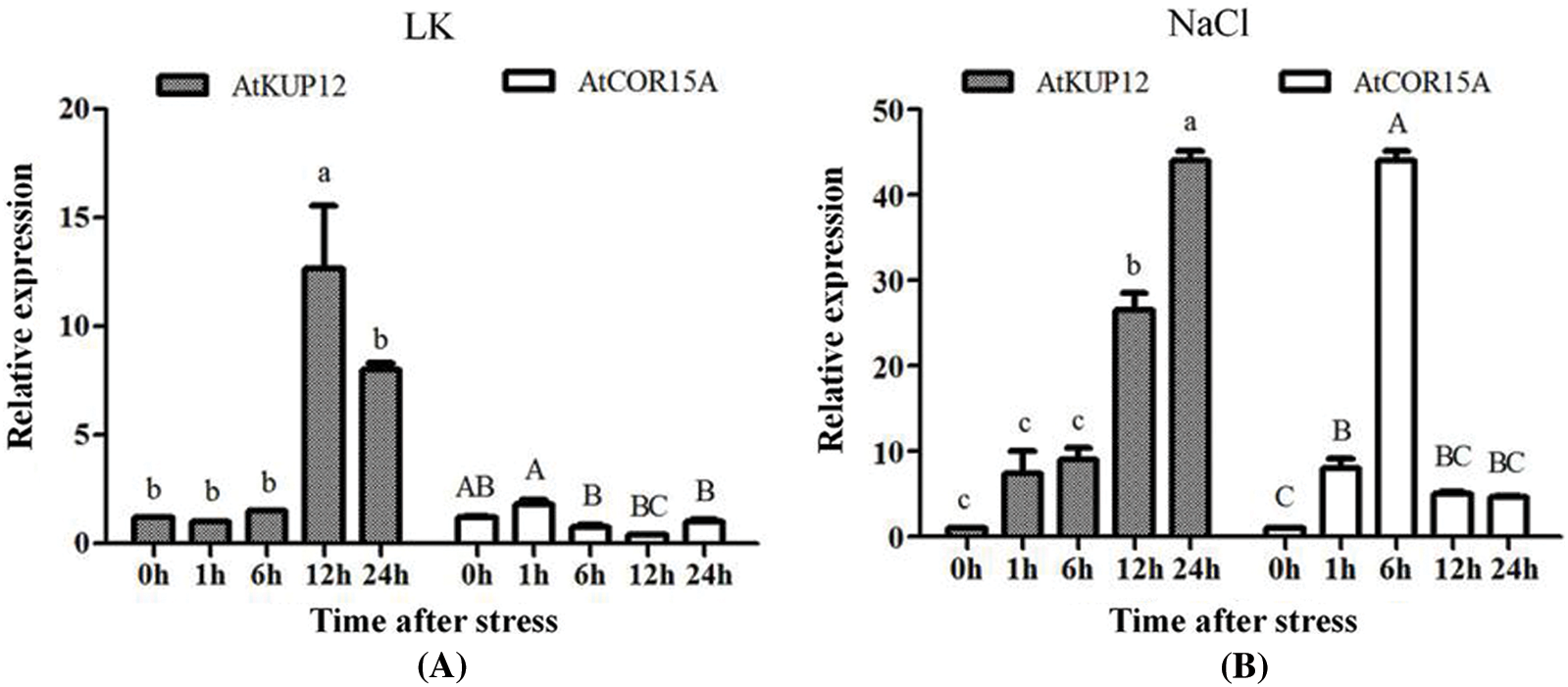
Figure 1: AtKUP12 expression levels under different conditions: Expression of AtKUP12 in Arabidopsis under 100 μM low-K+ (A) or 100 mM NaCl (B). Letters denote significantly different groups identified by Tukey’s test (p < 0.05). Data the means ± SE (n = 3)
3.2 Function of AtKUP12 in K+ Uptake in Yeast Cells
To investigate the K+-uptake activity of AtKUP12, we performed complementation tests using the mutant yeast strain CY162. We used AtAKT1 as a positive control, which mediated K+ uptake from solutions containing 10 μM K+ [40], and we used empty pYES2.0 vector as a negative control. Then, we performed yeast growth assays on SD-Ura medium containing 10, 20, 30 and 100 mM K+. We found no remarkable differences among transformants when they were grown on SD-Ura medium containing 100 mM KCl (Fig. 2A). When the concentration of K+ in SD-Ura medium was decreased to 30 mM, the growth of the yeast strain CY162 transformed with the empty pYES2.0 vector severely inhibited, but there was no visible difference between the yeasts transformed with AtAKT1 and the vector harboring AtKUP12 (Fig. 2B). When the concentration of K+ in SD-Ura medium was decreased to 20 mM and 10 mM, AtKUP12 partially rescued the growth defect of mutant yeast CY162 (Figs. 2C and 2D). These results indicated that AtKUP12 conferred significant K+ uptake and growth of yeasts at low-K+ concentrations.
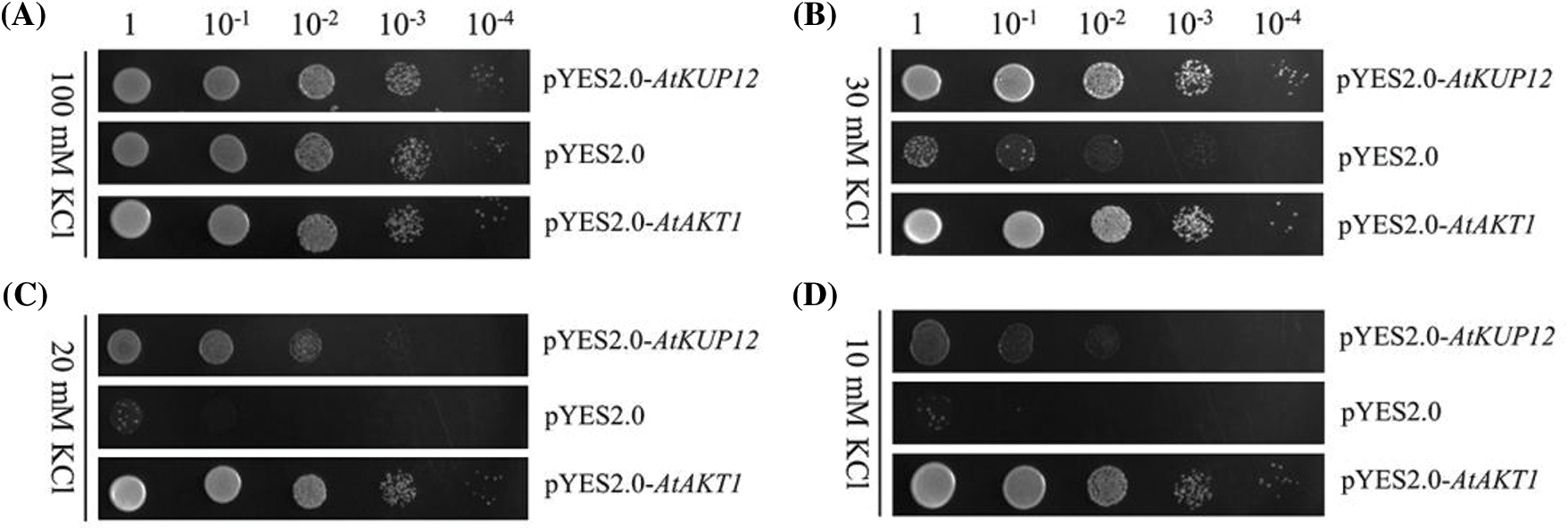
Figure 2: Functional complementation of AtKUP12 for K+ uptake. Growth status of K+ uptake-deficient yeast mutant CY162 cells expressing AtKUP12, empty pYES2.0 vector (negative control) and AtAKT1 (positive control) on SD-Ura medium containing different K+ concentrations
3.3 Functional Characterization of AtKUP12 in Arabidopsis
To test whether AtKUP12 has physiological roles in response to the low-K+ stress (10, 100, 200 μM KCl), the atkup12 mutant At1g60160 (SALK_083613) was used in this study. The T-DNA insertion site was located at the eighth exon of the AtKUP12 gene (Fig. 3A). Subsequently, we generated complementation lines for atkup12. As shown in Figs. 3B–3D, the primary root length of the atkup12 mutant was significantly shorter than that of WT plants when grown on low-K+ medium (10 and 100 μM K+), while the complementation line (atkup12-AtKUP12) completely rescued the low-K+ sensitive phenotype of the atkup12 mutant. Along with the increased K+ concentrations (200 and 1000 μM K+), the low-K+ sensitive phenotype of atkup12 disappeared (Figs. 3B–3D). These results indicated that the low-K+ sensitive phenotype of atkup12 was due to the disruption of AtKUP12.
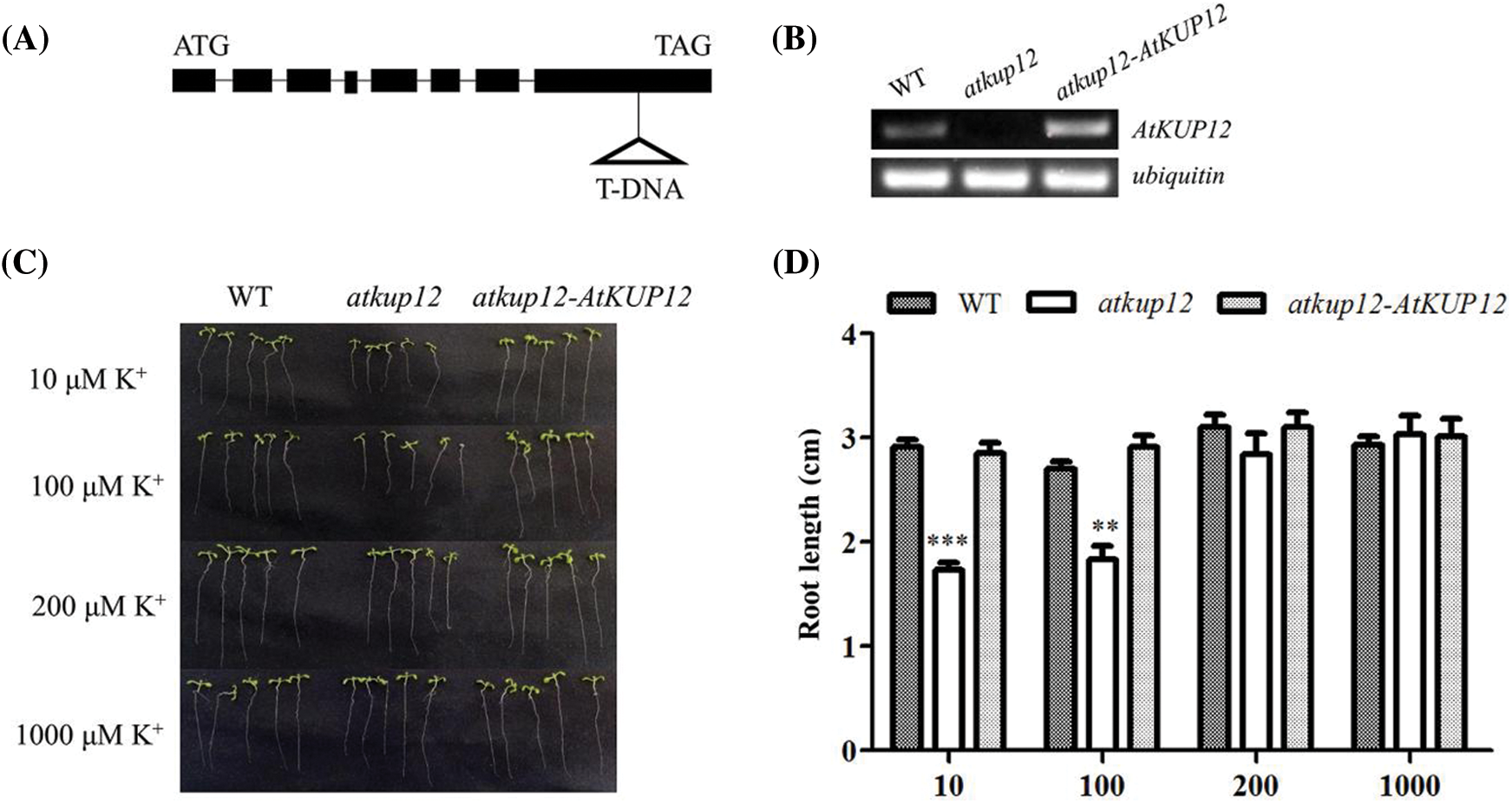
Figure 3: Expression of AtKUP12 rescued the low-K+-sensitive phenotype of the Arabidopsis atkup12 mutant. (A) Schematic diagram of the AtKUP12 gene. Black boxes indicate exons, and lines represent introns. The T-DNA insertion site in the atkup12 mutant is shown by a triangle. (B) RT-PCR verification of AtKUP12 expression in the WT, atkup12 mutant and complementation lines. Ubiquitin was used as an internal control. (C) Phenotypic comparison among WT, atkup12, and atkup12-AtKUP12 complementation lines under different K+ concentrations. Seeds were germinated and grown on low-K+ (10, 100 and 200 μM) or K+-sufficient (1000 μM) medium for 12 days. (D) The primary root length among the plant materials is shown in (C). Asterisks indicate significantly different groups identified by Tukey’s test (p < 0.05). Data means ± SE (n = 4) of 5 seedlings
3.4 Loss of Function of AtKUP12 Inhibits Seedling Growth and Reduces K+ Uptake
Under the 100 μM low-K+ condition, the primary root length of the atkup12 mutant was significantly shorter than that of the WT and atkup12-AtKUP12 complementation lines. Therefore, we selected 100 μM K+ as the low-K+ stress for the determination of fresh weight and K+ concentration. Under sufficient K+ conditions (1000 μM K+), for 12 days, we observed no substantial difference in either fresh weight or K+ concentrations among the plant materials (Figs. 4A–4C). However, under low-K+ stress, the atkup12 mutant shoots were smaller, and the fresh weight and K+ concentration were much lower than those of WT plants, while the fresh weight and K+ concentration in the complementation line recovered, which was similar to WT plants (Figs. 4A–4C). These results implied that AtKUP12 might function in K+ uptake in Arabidopsis plants.
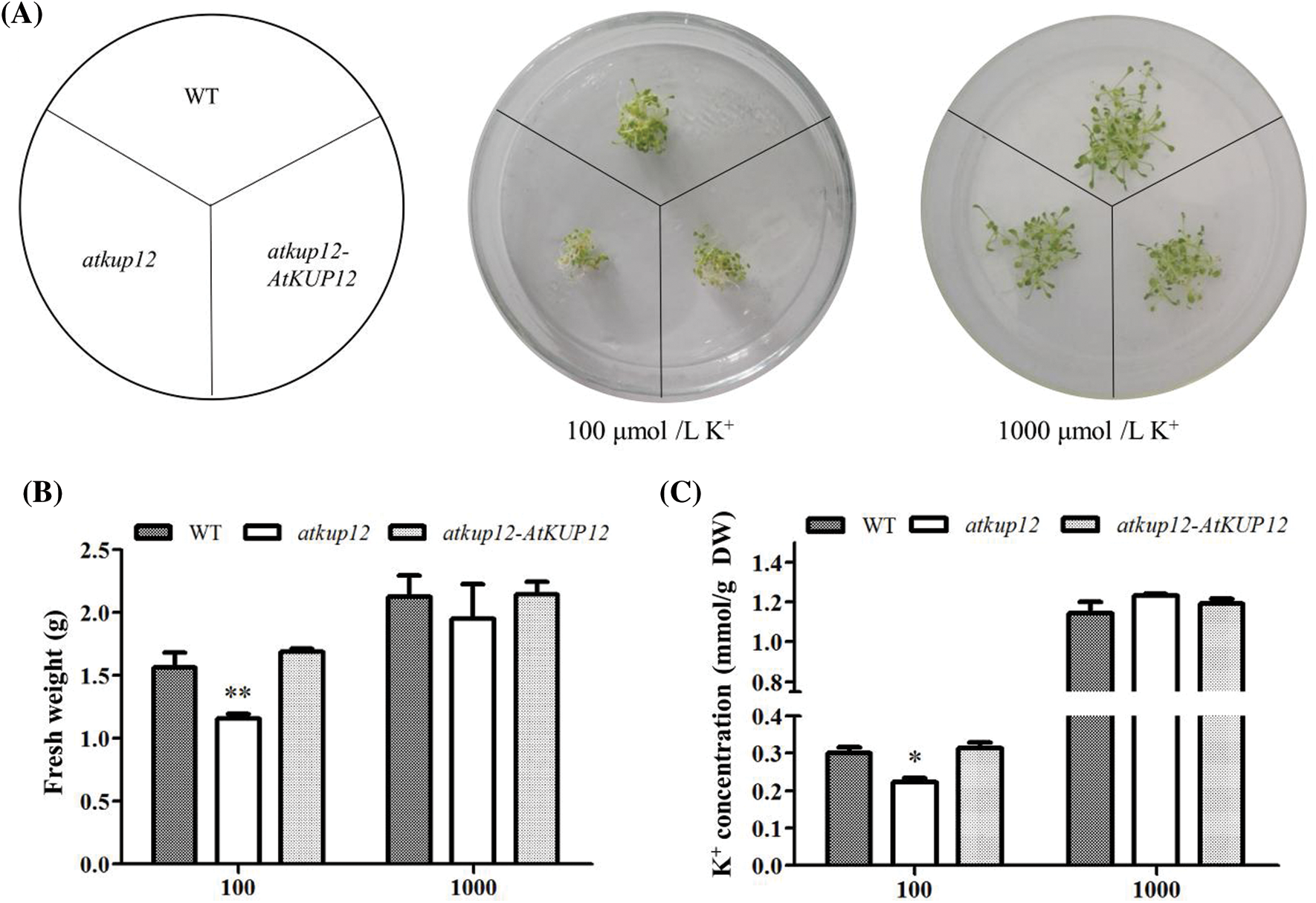
Figure 4: Effects of AtKUP12 disruption on growth and K+ concentration under low-K+ stress. Seeds germinated on medium containing 100 and 1000 μM K+ for 12 days growth. (A) Growth phenotype of shoots. (B) Fresh weight. (C) K+ concentration. Asterisks indicate significant differences from the WT at p < 0.05 by the Student’s t-test. Data are means ± SE (n = 4) of 60 seedlings
3.5 Effect of Disruption of AtKUP12 on Growth Tolerance to Salt, K+ Uptake and K+/Na+ Homeostasis
To confirm that the loss of function of AtKUP12 was responsible for the salt-sensitive phenotype, seeds of the WT, atkup12 and atkup12-AtKUP12 complementation lines were germinated and grown on medium containing 125 mM NaCl for 12 days. As shown in Fig. 5, the phenotypes among the plant materials, including the WT, atkup12 and complementation lines, did not obviously differ under control conditions. However, the primary root length of atkup12 was inhibited to a greater extent than that of the WT under 125 mM NaCl stress, and the complementation lines almost rescued the salt-sensitive phenotype of atkup12 mutants (Figs. 5A and 5B). We also observed and measured the primary root length of different plants under 100 mM NaCl stress, but there was no difference among the plant materials (Figs. S1A and S1B).
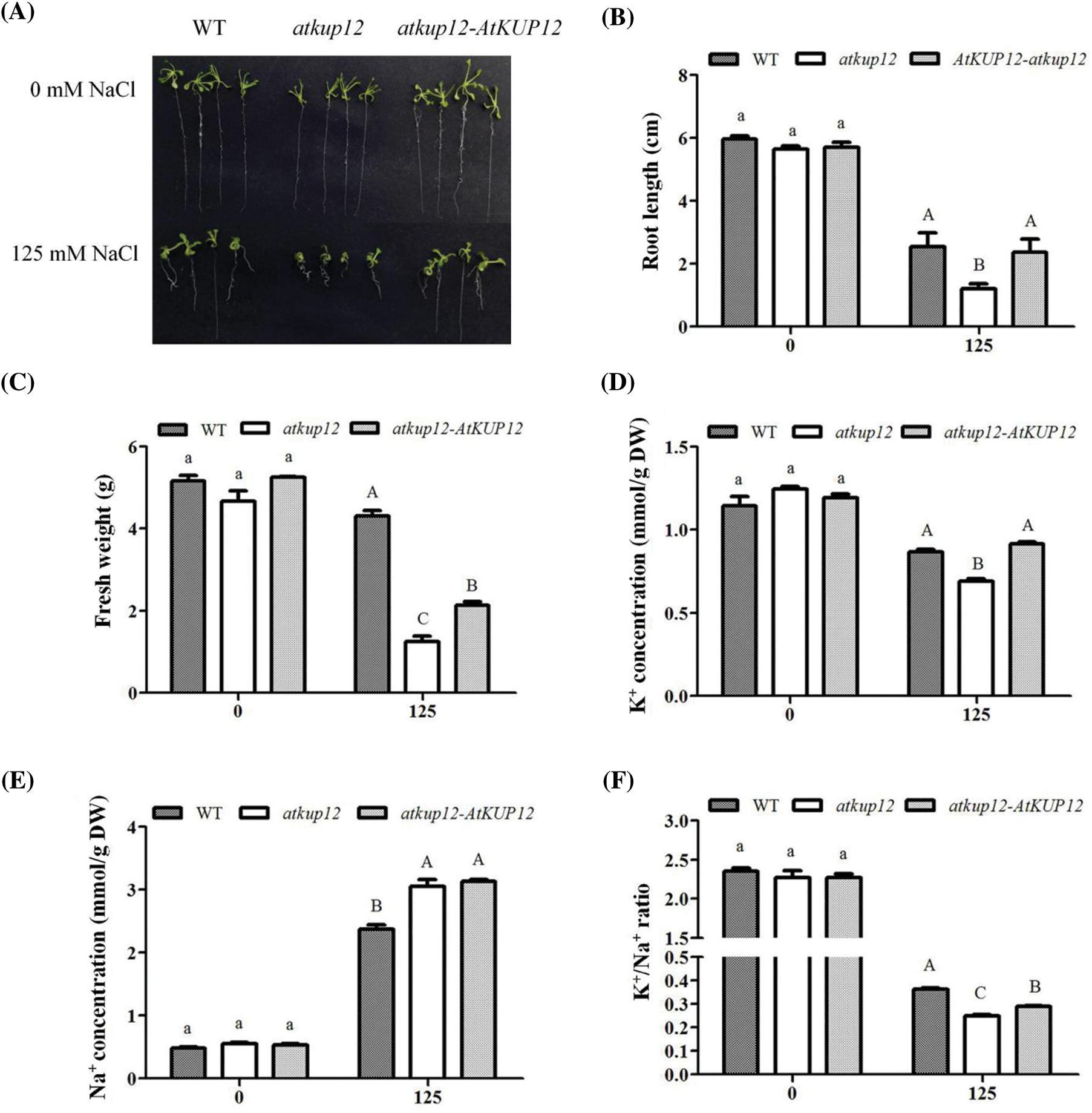
Figure 5: Complementation lines rescue the salt-sensitive phenotype of atkup12 mutants. (A) Phenotypic comparison among WT, atkup12, and atkup12-AtKUP12 complementation line under either salt stress (125 mM NaCl) or not (0 mM NaCl). Seeds germinated and grew on medium containing 0 and 125 mM NaCl for 12 days. Primary root length (B), fresh weight (C), K+ concentration (D), Na+ concentration (E) and K+/Na+ ratio (F) are shown. Letters denote significantly different groups identified by Tukey’s test (p < 0.05). Data are means ± SE (n = 4) of 60 seedlings
In addition, we measured the fresh weight and further determined the K+ and Na+ concentrations of diverse plant materials (Figs. 5C–5E). Under control conditions, there was no obvious difference in fresh weight, K+ and Na+ concentrations, and K+/Na+ ratio among various plants (Figs. 5C–5F). Under salt stress conditions, the atkup12 mutant showed a reduced fresh weight that was statistically significant, while the fresh weight of the complementation line was partly resumed (Fig. 5C). atkup12 showed lower K+ and greater Na+ concentrations than WT plants, while K+ concentration in the complementation line was restored to levels close to the corresponding WT values. However, the Na+ concentration was significantly higher in the complementation line than in the WT (Figs. 5D and 5E). Furthermore, the calculated K+/Na+ ratio in atkup12 was significantly lower than WT and partly restored in the complementation line (Fig. 5F). These data suggest that AtKUP12 may function in K+ uptake and maintenance of the K+/Na+ ratio under salt stress, which is crucial for salt tolerance in plants.
The importance of K+ is self-evident. K+ transporters play crucial roles in K+ absorption, transport and maintenance of K+ homeostasis under K+ deficient environments to ensure the normal growth of plants under abiotic stress [41]. At present, K+ transporter family genes responding to salt, drought and hormone stress have been found in a variety of plants, such as the Arabidopsis AtHAK5, AtHKT1, and AtKUP6 genes, tomato LeHAK5 gene, rice OsHAK21 gene, and pear PbHAK1, PbHAK7 and PbHAK10 genes, which are significantly enhanced by adverse stresses [26,29,42–44].
Arabidopsis AtCOR15A responds positively to adverse stresses such as high salt, ABA, cold, and drought; thus, it is often used as a marker gene of Arabidopsis under the above abiotic stresses [35,36]. In this experiment, Arabidopsis AtCOR15A was employed as a positive control under stress treatment. The expression of AtKUP12 under different stress treatments showed that AtKUP12 responded positively to abiotic stresses of salt and low-K+ treatment. The expression patterns of the AtKUP12 gene under NaCl stress was continuously upregulated, reaching a maximum value at 24 h, and the expression increased by 30 fold, compared with the control. The above experimental results show that the AtKUP12 had the strongest response to salt stress.
As a single cell eukaryote, yeast is often used for functional verification and analysis of genes because of its short culture cycle and easy genetic transformation. The CY162 yeast strain is a type of K+ uptake-deficient yeast cell caused by the deletion and insertion of Trk1 and Trk2; thus, it has become a model material for studying K+ transport and related proteins [6,29]. The inward-rectifier K+ channel of the shaker family, AtAKT1 in Arabidopsis, is involved in K+ uptake at both high- and low-affinity ranges [40]. The major regulation of the AtAKT1 activity has been described at the protein level; AtAKT1 activity is modulated by the CBL1/9–CIPK23 pathway under low-K+ supply conditions [45]. Here, we used AtAKT1 as a positive control. The heterologous expression of OsHAK21 in CY162 yeast has shown that OsHAK21 can mediate specific K+ transport under low-K+ [6]. Han et al. also indicated that AtKUP7 has K+ transport activity by using the yeast mutant but that the transformed K-deficient yeast strain can only grow at a concentration of more than 500 μM, suggesting that AtKUP7 is a low-affinity K+ transporter [46]. Qin et al. found that the heterologous expression of maize ZmHAK5 and ZmHAK1 in CY162 yeast can mediate K+ uptake under a low-K+ condition, but ZmHAK1 K+ transport activity is weaker than ZmHAK5 [5]. Similarly, our results showed that the heterologous expression of AtKUP12 in the K+ uptake-deficient yeast CY162 could mediate K+ uptake under low-K+ conditions, indicating that AtKUP12 in Arabidopsis participates in high-affinity K+ uptake and transport.
Arabidopsis AtHAK5 is a high-affinity K+ uptake transporter gene; after the destruction of its alleles, the athak5 mutant showed a certain growth inhibition under a low-K+ condition [17]. However, the heterologous expression of rice OsHAK21 and maize ZmHAK5 transporters completely restored the defect of athak5 mutants [5,23]. A complementation line of the AtKUP7 gene in Arabidopsis restored the growth inhibition of the atkup7 mutant under low-K+ conditions [46]. Our results also showed that the atkup12-AtKUP12 complementation line alleviated the growth inhibition of atkup12 under low-K+ stress (Figs. 3 and 4). Under 100 μM low-K+ treatment, the fresh weight of atkup12 mutant was significantly lower than that of WT, but there was no significant difference between the complementation and WT lines, suggesting that the AtKUP12 gene might regulate plant growth through K+ uptake. It should be noted that both atkup12 and athak5 mutants [5,17] affected the root length of plants under low-K+ stress, while atkup7 mutants exhibited chlorosis symptoms compared with WT plants [46], which may be related to the specific functions of different transporters. The K+ concentration testing showed that the K+ concentration in the complementation line resumed, which was similar to the K+ concentration in WT plants under low-K+ conditions. This finding indicated that AtKUP12 conferred significant K+ uptake capacity and improved seedling growth and root development in Arabidopsis at low-K+ concentrations.
The K+/Na+ ratio in cytoplasm is critical for the salt tolerance of plants. Knockout of OsHAK5 or OsHAK21 leads to a decreased K+/Na+ ratio in rice tissues and to salt sensitivity, while overexpression of OsHAK5 significantly improves the salt tolerance of transgenic rice [6,29]. In addition, some transporters, such as HvHAK1 and PhaHAK5, are also able to function in low-affinity Na+ uptake in the presence of NaCl [47,48]. In this study, under salt stress conditions, the atkup12 mutant also showed salt-sensitivity, and the atkup12-AtKUP12 complementation line significantly alleviated the inhibition of root length and fresh weight. Under 125 mM NaCl treatment, the K+ concentration of the atkup12 mutant was significantly lower than that of WT and complementation lines, while the Na+ concentration of atkup12 and the complementation line was significantly higher than that of WT line. This might be due to the similarity in physicochemical properties between Na+ and K+, resulting in competition between Na+ and K+ [49]. However, the K+/Na+ ratio of the atkup12 mutant was significantly lower than that of WT, while the K+/Na+ ratio in the complementation line was partially restored under salt stress. Therefore, we speculate that AtKUP12 is an important KT/HAK/KUP family member that improves the salt tolerance of plants by K+ uptake and the K+/Na+ ratio balance.
From the results presented in this study, we demonstrated that a K+ transport gene AtKUP12 was induced by low-K+ and NaCl stresses, while AtKUP12 participated in K+ acquisition and the growth of the yeast mutant strain CY162 at low-K+ concentrations. In addition, AtKUP12 also promoted plant growth, enhanced the K+ concentration under low-K+ conditions, and enhanced the salt tolerance of plants by maintenance of the K+/Na+ ratio. These results may facilitate further investigation of the function and mechanism underlying K+ uptake and utilization of AtKUP12 under low-K+ and salt stress conditions.
Acknowledgement: We gratefully thank Dr Jianhua Yang and Shen Zhang for revising the manuscript.
Funding Statement: This work was supported by the National Natural Science Foundation of China [Grant No. 31860061], Opening of Key Laboratory of Autonomous Region [Grant No. 2017D04026] and Tianshan Youth Program [Grant No. 2019Q013].
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Li, L. G., Kim, B. G., Cheong, Y. H., Pandey, G. K., Luan, S. (2006). A Ca2+ signaling pathway regulates a K+ channel for low-K response in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America, 103(33), 12625–12630. DOI 10.1073/pnas.0605129103. [Google Scholar] [CrossRef]
2. Watanabe, T., Broadley, M. R., Jansen, S., White, P. J., Takada, J. et al. (2007). Evolutionary control of leaf element composition in plants. New Phytologist, 174(3), 516–523. DOI 10.1111/j.1469-8137.2007.02078.x. [Google Scholar] [CrossRef]
3. White, P. J. (2013). Improving potassium acquisition and utilisation by crop plants. Journal of Plant Nutrition and Soil Science, 176(3), 305–316. DOI 10.1002/jpln.201200121. [Google Scholar] [CrossRef]
4. Shen, X. Y., Jiang, C. R., Wang, Z. L. (2011). Optimization of the method for screening Arabidopsis low-K+ tolerant mutants. Journal of Henan Agricultural Sciences, 40(3), 56–58. DOI 10.15933/j.cnki.1004-3268.2011.03.001. [Google Scholar] [CrossRef]
5. Qin, Y. J., Wu, W. H., Wang, Y. (2019). ZmHAK5 and ZmHAK1 function in K+ uptake and distribution in maize under low K+ conditions. Journal of Integrative Plant Biology, 61(6), 691–705. DOI 10.1111/jipb.12756. [Google Scholar] [CrossRef]
6. Yang, T. Y., Zhang, S., Hu, Y. B., Wu, F. C., Hu, Q. D. et al. (2014). The role of a potassium transporter OsHAK5 in potassium acquisition and transport from roots to shoots in rice at low potassium supply levels. Plant Physiology, 166(2), 945–959. DOI 10.1104/pp.114.246520. [Google Scholar] [CrossRef]
7. Wang, Y., Jiang, G. Q., Han, Y. N., Liu, M. M. (2013). Effects of salt, alkali and salt–alkali mixed stresses on seed germination of the halophyte Salsola ferganica (Chenopodiaceae). Acta Ecologica Sinica, 33(6), 354–360. DOI 10.1016/j.chnaes.2013.09.010. [Google Scholar] [CrossRef]
8. Lebaudy, A., Véry, A. A., Sentenac, H. (2007). K+ channel activity in plants: Genes, regulations and functions. FEBS Letters, 581(12), 2357–2366. DOI 10.1016/j.febslet.2007.03.058. [Google Scholar] [CrossRef]
9. Alexander, G. (2007). Plant KT/KUP/HAK potassium transporters: Single family multiple functions. Annals of Botany, 99(6), 1035–1041. DOI 10.1093/aob/mcm066. [Google Scholar] [CrossRef]
10. Corratgé-Faillie, C., Jabnoune, M., Zimmermann, S., Véry, A. A., Fizames, C. (2010). et al. Potassium and sodium transport in non-animal cells: The Trk/Ktr/HKT transporter family. Cellular and Molecular Life Sciences, 67(15), 2511–2532. DOI 10.1007/s00018-010-0317-7. [Google Scholar] [CrossRef]
11. Cheng, X. Y., Liu, X. D., Mao, W. W., Zhang, X. R., Chen, S. L. et al. (2018). Genome-wide identification and analysis of HAK/KUP/KT potassium transporters gene family in wheat (Triticum aestivum L.). International Journal of Molecular Sciences, 19(12), 3969–3990. DOI 10.3390/ijms19123969. [Google Scholar] [CrossRef]
12. Yang, Z. F., Gao, Q. S., Sun, C. S., Li, W. J., Gu, S. L. et al. (2009). Molecular evolution and functional divergence of HAK potassium transporter gene family in rice (Oryza sativa L.). Journal of Genetics and Genomics, 36(3), 161–172. DOI 10.1016/S1673-8527(08)60103-4. [Google Scholar] [CrossRef]
13. Mäser, P., Thomine, S., Schroeder, J. I., Ward, J. M., Hirschi, K. et al. (2001). Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiology, 126(4), 1646–1667. DOI 10.1104/pp.126.4.1646. [Google Scholar] [CrossRef]
14. Li, W. H., Xu, G. H., Alli, A., Yu, L. (2018). Plant HAK/KUP/KT K+ transporters: Function and regulation. Seminars in Cell & Developmental Biology, 74, 133–141. DOI 10.1016/j.semcdb.2017.07.009. [Google Scholar] [CrossRef]
15. Okada, T., Nakayama, H., Shinmyo, A., Yoshida, K. (2008). Expression of OsHAK genes encoding potassium ion transporters in rice. Plant Biotechnology, 25(3), 241–245. DOI 10.5511/plantbiotechnology.25.241. [Google Scholar] [CrossRef]
16. Véry, A. A., Nieves-Cordones, M., Daly, M., Khan, I., Fizames, C. et al. (2014). Molecular biology of K+ transport across the plant cell membrane: What do we learn from comparison between plant species. Journal of Plant Physiology, 171(9), 748–769. DOI 10.1016/j.jplph.2014.01.011. [Google Scholar] [CrossRef]
17. Pyo, Y. J., Gierth, M., Schroeder, J. I., Cho, M. H. (2010). High-affinity K+ transport in Arabidopsis: AtHAK5 and AKT1 are vital for seedling establishment and postgermination growth under low-potassium conditions. Plant Physiology, 153(2), 863–875. DOI 10.1104/pp.110.154369. [Google Scholar] [CrossRef]
18. Fulgenzi, F. R., Peralta, M. L., Mangano, S., Danna, C. H., Vallejo, A. J. et al. (2008). The ionic environment controls the contribution of the barley HvHAK1 transporter to potassium acquisition. Plant Physiology, 147(1), 252–262. DOI 10.1104/pp.107.114546. [Google Scholar] [CrossRef]
19. Ahn, S. J., Shin, R., Schachtman, D. P. (2004). Expression of KT/KUP genes in Arabidopsis and the role of root hairs in K+ uptake. Plant Physiology, 134(3), 1135–1145. DOI 10.1104/pp.103.034660. [Google Scholar] [CrossRef]
20. Li, X. W., You, X. L., Wang, Y. (2019). Research progress of HAK/KUP/KT potassium transporter family in plant response to salt stress. Plant Science Journal, 37(1), 101–108. DOI 10.11913/PSJ.2095-0837.2019.10101. [Google Scholar] [CrossRef]
21. Roshani, G. A., Narayanasamy, G. (2010). Effects of potassium on temporal growth of root and shoot of wheat and its uptake in different soils. International Journal of Plant Production, 4(1), 25–32. [Google Scholar]
22. Han, M. (2015). Functional analysis of potassium transporter KUP7 in Arabidopsis responses low-K+ stress(Ph.D. Thesis). China Agricultural University, Beijing. [Google Scholar]
23. Shen, Y. (2015). Functional analyses of potassium transporter OsHAK21 and channel OsKx in response to salt stress in rice (Ph.D. Thesis). Nanjing Agricultural University, Nanjing. [Google Scholar]
24. Tenorio-Berrío, R., Pérez-Alonso, M. M., Vicente-Carbajosa, J., Martín-Torres, L., Dreyer, I. et al. (2018). Identification of two auxin-regulated potassium transporters involved in seed maturation. International Journal of Molecular Sciences, 19(7), 2132–2148. DOI 10.3390/ijms19072132. [Google Scholar] [CrossRef]
25. Chen, G., Hu, Q. D., Luo, L., Yang, T. Y., Zhang, S. et al. (2015). Rice potassium transporter OsHAK1 is essential for maintaining potassium-mediated growth and functions in salt tolerance over low and high potassium concentration ranges. Plant, Cell & Environment, 38(12), 2747–2765. DOI 10.1111/pce.12585. [Google Scholar] [CrossRef]
26. Osakabe, Y., Arinaga, N., Umezawa, T., Katsura, S., Nagamachi, K. et al. (2013). Osmotic stress responses and plant growth controlled by potassium transporters in Arabidopsis. Plant Cell, 25(2), 609–624. DOI 10.1105/tpc.112.105700. [Google Scholar] [CrossRef]
27. Zhang, H. W., Xiao, W., Yu, W. W., Jiang, Y., Li, R. F. (2020). Halophytic Hordeum brevisubulatum HbHAK1 facilitates potassium retention and contributes to salt tolerance. International Journal of Molecular Sciences, 21(15), 5292–5306. DOI 10.3390/ijms21155292. [Google Scholar] [CrossRef]
28. Horie, T., Sugawara, M., Okada, T., Taira, K., Kaothien-Nakayama, P. et al. (2011). Rice sodium-insensitive potassium transporter, OsHAK5, confers increased salt tolerance in tobacco BY2 cells. Journal of Bioscience and Bioengineering, 111(3), 346–356. DOI 10.1016/j.jbiosc.2010.10.014. [Google Scholar] [CrossRef]
29. Shen, Y., Shen, L., Shen, Z. X., Jing, W., Ge, H. L. et al. (2015). The potassium transporter OsHAK21 functions in the maintenance of ion homeostasis and tolerance to salt stress in rice. Plant, Cell & Environment, 38(12), 2766–2779. DOI 10.1111/pce.12586. [Google Scholar] [CrossRef]
30. Nieves-Cordones, M., Aleman, F., Martinez, V., Rubio, F. (2010). The Arabidopsis thaliana HAK5 K+ transporter is required for plant growth and K+ acquisition from low K+ solutions under saline conditions. Molecular Plant, 3(2), 326–333. DOI 10.1093/mp/ssp102. [Google Scholar] [CrossRef]
31. Yang, Z. M., Wang, Y. (2015). Cloning of potassium transporter gene (HcKUP12) from Halostachys capsica and its expression profile under salt stress. Plant Science Journal, 33(4), 499–506. DOI 10.3969/j.issn.1004-31IX.2014.03.0074. [Google Scholar] [CrossRef]
32. You, X. L., Yang, Z. M., Wang, Y. (2017). Identification of potassium transporter mutant atkup12 from Arabidopsis and sensitivity analysis to abiotic stresses. Biotechnology, 27(5), 497–504. DOI 10.16519/j.cnki.1004-311x.2017.05.0079. [Google Scholar] [CrossRef]
33. Liu, L. L., Ren, H. M., Chen, L. Q., Wang, Y., Wu, W. H. (2013). A protein kinase, calcineurin B-like protein-interacting protein Kinase9, interacts with calcium sensor calcineurin B-like Protein3 and regulates K+ homeostasis under low-K+ stress in Arabidopsis. Plant Physiology, 161(1), 266–277. DOI 10.1104/pp.112.206896. [Google Scholar] [CrossRef]
34. Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods, 25(4), 402–408. DOI 10.1006/meth.2001.1262. [Google Scholar] [CrossRef]
35. Ishitani, M., Xiong, L. M., Lee, H., Stevenson, B., Zhu, J. K. (1998). HOS1, a genetic locus involved in cold-responsive gene expression in Arabidopsis. Plant Cell, 10(7), 1151–1161. DOI 10.1105/tpc.10.7.1151. [Google Scholar] [CrossRef]
36. Lim, G. H., Zhang, X., Chung, M. S., Lee, D. J., Woo, Y. M. et al. (2010). A putative novel transcription factor, AtSKIP, is involved in abscisic acid signalling and confers salt and osmotic tolerance in Arabidopsis. New Phytologist, 185(1), 103–113. DOI 10.1111/j.1469-8137.2009.03032.x. [Google Scholar] [CrossRef]
37. Anderson, J. A., Huprikar, S. S., Kochian, L. V., Lucas, W. J., Gaber, R. F. (1992). Functional expression of a probable Arabidopsis thaliana potassium channel in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences, 89(9), 3736–3740. DOI 10.1073/pnas.89.9.3736. [Google Scholar] [CrossRef]
38. Clough, S. J., Bent, A. F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant Journal, 16(6), 735–743. DOI 10.1046/j.1365-313x.1998.00343.x. [Google Scholar] [CrossRef]
39. Matsushita, N., Matoh, T. (1991). Characterization of Na+ exclusion mechanisms of salt-tolerant reed plants in comparison with salt-sensitive rice plants. Physiologia Plantarum, 83(1), 170–176. DOI 10.1111/j.1399-3054.1991.tb01298.x. [Google Scholar] [CrossRef]
40. Nieves-Cordones, M., Martínez, V., Benito, B., Rubio, F. (2016). Comparison between Arabidopsis and rice for main pathways of K+ and Na+ uptake by roots. Frontiers in Plant Science, 7(ra43), 992–1005. DOI 10.3389/fpls.2016.00992. [Google Scholar] [CrossRef]
41. Shabala, S., Pottosin, I. (2014). Regulation of potassium transport in plants under hostile conditions: Implications for abiotic and biotic stress tolerance. Physiologia Plantarum, 151(3), 12165–12187. DOI 10.1111/ppl.12165. [Google Scholar] [CrossRef]
42. Sun, Y. L., Kong, X. P., Li, C. L., Liu, Y. X., Ding, Z. J. (2015). Potassium retention under salt stress is associated with natural variation in salinity tolerance among Arabidopsis accessions. PLoS One, 10(5), e0124032. DOI 10.1371/journal.pone.0124032. [Google Scholar] [CrossRef]
43. Bacha, H., Ródenas, R., López-Gómez, E., García-Legaz, M. F., Nieves-Cordones, M. et al. (2015). High Ca2+ reverts the repression of high-affinity K+ uptake produced by Na+ in Solanum lycopersycum L. (var. microtom) plants. Journal of Plant Physiology, 180, 72–79. DOI 10.1016/j.jplph.2015.03.014. [Google Scholar] [CrossRef]
44. Li, Y., Peng, L. R., Xie, C. Y., Shi, X. Q., Dong, C. X. et al. (2018). Genome-wide identification, characterization, and expression analyses of the HAK/KUP/KT potassium transporter gene family reveals their involvement in K+ deficient and abiotic stress responses in pear rootstock seedlings. Plant Growth Regulation, 85(2), 187–198. DOI 10.1007/s10725-018-0382-8. [Google Scholar] [CrossRef]
45. Xu, J., Li, H. D., Chen, L. Q., Wang, Y., Liu, L. L. et al. (2006). A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell, 125(7), 1347–1360. DOI 10.1016/j.cell.2006.06.011. [Google Scholar] [CrossRef]
46. Han, M., Wu, W., Wu, W. H., Wang, Y. (2016). Potassium transporter KUP7 is involved in K+ acquisition and translocation in Arabidopsis root under K+-limited conditions. Molecular Plant, 9(3), 437–446. DOI 10.1016/j.molp.2016.01.012. [Google Scholar] [CrossRef]
47. Santa-Maria, G. E., Rubio, F., Dubcovsky, J., Rodríguez-Navarro, A. (1998). The HAK1 gene of barley is a member of a large gene family and encodes a high-affinity potassium transporter. Plant Cell, 9(12), 2281–2289. DOI 10.1105/tpc.9.12.2281. [Google Scholar] [CrossRef]
48. Takahashi, R., Nishio, T., Ichizen, N., Takano, T. (2007). High-affinity K+ transporter PhaHAK5 is expressed only in salt-sensitive reed plants and shows Na+ permeability under NaCl stress. Plant Cell Reports, 26(9), 1673–1679. DOI 10.1007/s00299-007-0364-1. [Google Scholar] [CrossRef]
49. Ashraf, M., Ali, Q. (2007). Relative membrane permeability and activities of some antioxidant enzymes as the key determinants of salt tolerance in canola (Brassica napus L.). Environmental and Experimental Botany, 63(1–3), 266–273. DOI 10.1016/j.envexpbot.2007.11.008. [Google Scholar] [CrossRef]
Appendix
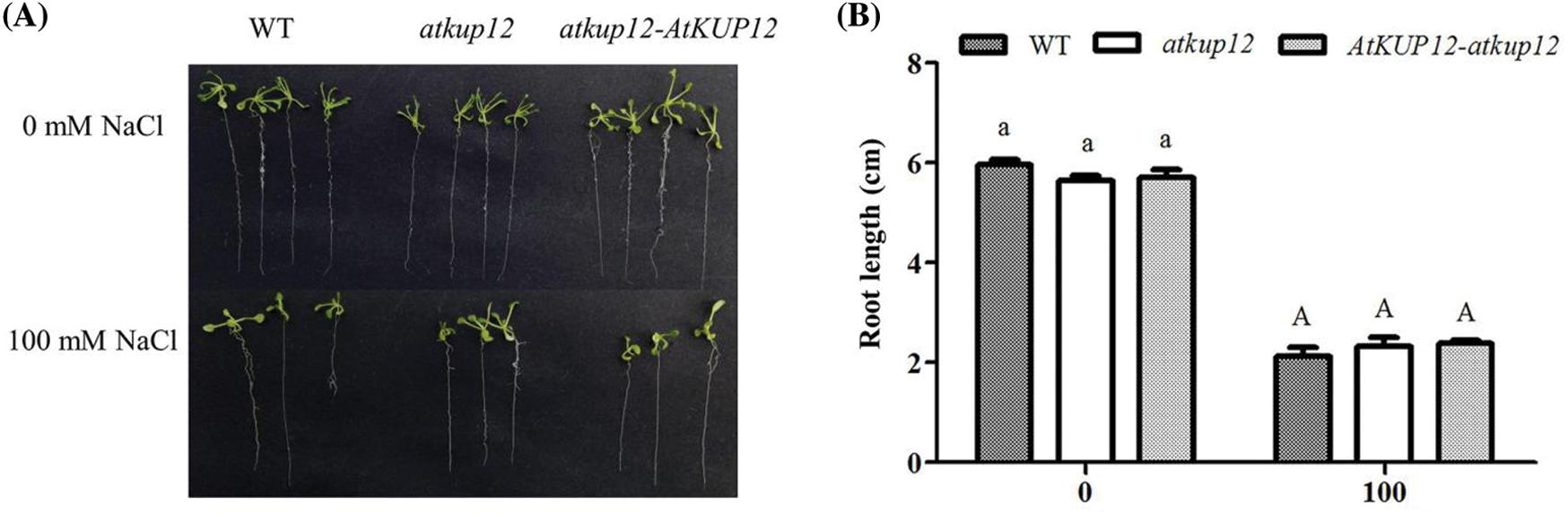
Figure S1: Phenotype test of atkup12 mutants and the atkup12-AtKUP12 complementation lines on medium containing 100 mM NaCl. (A) Phenotypic comparison among WT, atkup12, and atkup12-AtKUP12 complementation lines under 100 mM NaCl stress. Seeds germinated and grew on medium for 12 days. (B) Primary root length. Letters denote significantly different groups identified by Tukey’s test (p < 0.05). Data are means ± SE (n = 4)
Table S1: Primer sequences used in this study

 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |