DOI:10.32604/phyton.2021.013262

| Phyton-International Journal of Experimental Botany DOI:10.32604/phyton.2021.013262 |  |
| Article |
Thylakoid Transit Peptide Is Related to the Expression and Localization of NdhB Subunits in Soybean
1State Key Laboratory of Subtropical Silviculture, Zhejiang A&F University, Hangzhou, 311300, China
2China Science and Technology Exchange Center, Beijing, 100045, China
3State Key Laboratory of Plant Physiology and Biochemistry, College of Life Sciences, Zhejiang University, Hangzhou, 310058, China
*Corresponding Author: Yi He. Email: heyi@zju.edu.cn
#These authors have contributed equally to this work
Received: 31 July 2020; Accepted: 07 September 2020
Abstract: The chloroplast NAD(P)H dehydrogenase (NDH) complex, as one of the most important photosynthesis protein complexes in thylakoid membrane, is involved in photosystem I (PSI) cyclic electron transport (CEF). Under abiotic environmental stress, the photosynthetic apparatus is susceptible to the damage caused by the strong light illumination. However, the enhancement of NDH-dependent CEF could facilitate the alleviation of the damage to the photosynthetic apparatus. The NdhB subunit encoded by chloroplast genome is one of most important subunits of NDH complex and consists of 510 amino acids. Here, according to cloning ndhB from Melrose (cultivated soybean), ACC547 (wild salt-tolerant soybean), S113-6 and S111-9 (hybrid descendant), based on the comparison and analysis of the sequences of NdhB subunits, we found that there is a novel thylakoid transit peptide of NdhB subunit in S111-9. In addition, crosslink immunoprecipitation, immunogold labeling and co-expression of GFP fusion protein indicated that the novel thylakoid transit peptide is favorable to the expression and localization of NdhB subunit in chloroplast. Therefore, we suggest that this novel thylakoid transit peptide plays the same role as chaperonin and contributes to facilitating the expression and localization of NdhB subunit.
Keywords: NDH complex; NdhB subunit; thylakoid transit peptide
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |
In land plants, the chloroplast NAD(P)H dehydrogenase (NDH) complex is involved in chlororespiration and drives cyclic electron flow (CEF) around PSI [1,2]. It is structurally and functionally divergent from the bacterial and mitochondrial complexes [1]. By analogy to Escherichia coli complex I, at least eleven subunits of chloroplast NDH have been found in Arabidopsis, which are homologous to the subunits of mitochondrial complex I (NADH dehydrogenase) and eubacterial NDH-1 [3], whereas NdhB and NdhH are considered to be the most important subunits of NDH [4,5]. Nevertheless, at present, there were some debates about the function and localization mechanism of NDH.
In the history of plant evolution, the photosynthetic apparatus and thylakoid membrane systems has developed mechanisms to adapt to some environmental stress [6–8]. One of the available pathways for alleviating the damage to the photosynthetic apparatus and thylakoid membrane systems is to accelerate cyclic electron flow (CEF) around PSI [7,8]. Plants have two known pathways for CEF: the protein gradient regulation 5 (PGR5)/PGR-like 1 (PGRL1)-ferredoxin (Fd)-dependent pathway and the NAD(P)H dehydrogenase (NDH) complex-dependent pathway [6,9–11]. Actually, many results about biological functions of NDH and subunits have been reported. Munekage et al. [6] reported that cyclic electron flow around photosystem I is essential for photosynthesis. In addition, the adaptation mechanisms of photosynthetic thylakoid membrane systems to abiotic stress have also been reported in some studies [7–9]. The enhancement of NDH activity in tobacco facilitates the alleviation of oxidative damage caused by temperature stress [9]. Increasing NDH-dependent CEF in soybean is related to Na+ sequestration into vacuoles for the improvement of salt tolerance [11]. These studies demonstrate that cyclic electron transport mediated by NDH complex could play important roles in protecting photosynthetic apparatus from oxidative damage caused by temperature and salt stresses [9–11]. Moreover, functional assembly and localization of NDH subunits in chloroplast is a coordinated mechanism as it has been illustrated that mutant plants deficient for a single subunit could impair the function and activity of NDH complexes [12–14].
Currently, in Arabidopsis thaliana, a minor subunit Cpn60β4, which forms a heterooligomeric Cpn60 complex with Cpn60α1 and Cpn60β1-β3, is specifically required for the folding of NdhH [15]. It is the first report that a chaperonin plays a critical role in the folding of subunits of NDH complex. Meanwhile, some studies suggest that chloroplast stromal proteins, CRR6 and CRR7, are required for assembly of the NDH complex [16]. CRR41 and CRR42 are involved in the assembly of a stroma-protruding arm of NDH complex [17]. Further interactive proteomic analyses indicate that CRR7 is likely to be involved in the final step of the stroma-protruding arm of NDH complex [17]. Although much has been learned about the assembly of NDH complex, less has been known about how single subunit of NDH utilizes their signal peptide or chaperonin to sort each subunit to the accurate location in thylakoid membrane. Here, a novel thylakoid transit peptide was found in the NdhB subunit differing from our cultivar soybean Melrose, which not only contributes to the localization of NdhB, but also increases the mRNA and protein relative abundance of NdhB. Moreover, our results further indicate that although the novel NdhB subunit is shorter in size than that of Melrose, it could be directed to thylakoid membrane with thylakoid transit peptide. Therefore, the novel thylakoid transit peptide further lead to a strong association with the expression and localization of NdhB subunit in the somatic hybrid descendant S111-9. It provides new insights for deepening the understanding of how NdhB plays a significant role during the stresses.
The soybean Glycine max Melrose (cv. Melrose), wild salt-tolerant Glycine cyrtoloba (series number ACC547), S111-9 and S113-6, the stable somatic hybrid descendants of the wild salt-tolerant Glycine cyrtoloba (ACC547) [18] were used in this study. When the first pair of leaves was fully expanded, the well-grown seedlings were selected and transferred into nutrient solution [19] and cultured at 25/22°C (12 h light/12 h dark) with ~70% relative humidity in the growth chamber. The well-grown and 25-day-old plants were used in our experiments.
2.2 RNA Extraction, Cloning of ndhB and qRT-PCR
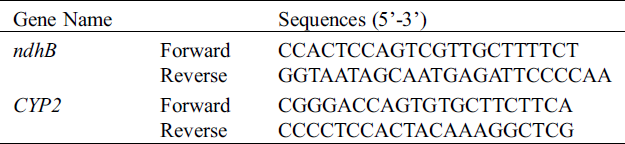
RNA was either extracted from the soybean’s leaves (Melrose, ACC547, S111-9 and S113-6) using RNAiso Plus reagent (TaKaRa, Cat#D9108B, Dalian, China) according to maufacturer’s protocol. RNA samples (2 μg) were incubated with DNase (TaKaRa) to remove any contaminating genomic DNA and converted to cDNA using M-MLV RTase cDNA Synthesis Kit following manufacturer’s instructions (TaKaRa, Japan). Then, ndhB was amplified from the diluted samples of reverse transcription products by PCR with primers 5’-TGCAGTTACTAATTCATGATCT-3’ (forward) and 5’-AAGGTAAGAGTTGAACTAAGAA-3’ (reverse). Products from the PCR reaction were ligated into pMD19-T vector, and then the pMD19-T with ndhB was used for sequencing. The above reverse transcription products were also used for qRT-PCR with forward primer 5’-CCACTCCAGTCGTTGCTTTTCT-3’ and reverse primer 5’-GGTAATAGCAATGAGATT CCCCAA-3’. The qRT-PCR protocol was described in details elsewhere [11] and primer information were in Table S2.
2.3 Vector Construction for 35S::B1::sGFP and 35S::B3::sGFP Fusion Protein
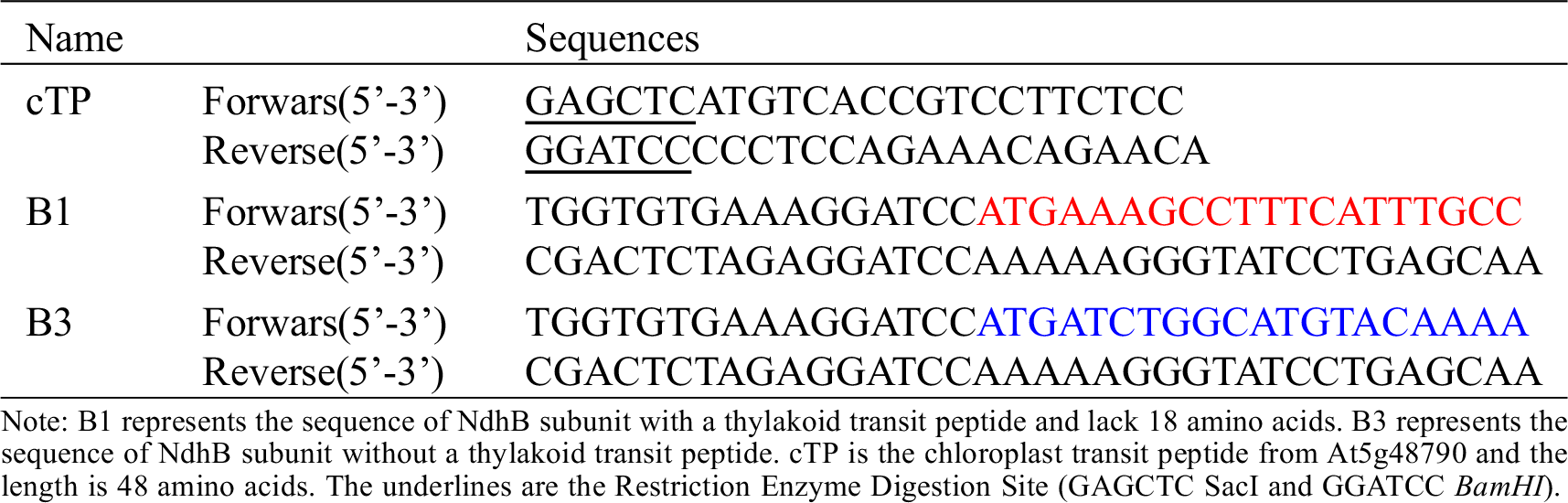
The sequences of B1 and B3 were amplified from the full-length cDNA of NdhB subunit using the B1/B3 primers (B1 and B3 represent the NdhB subunit with and without transit peptide respectively) (Table S1), and then cloned into the binary vector pCAMBIA 1300 containing a CaMV 35S promoter::sGFP cassette to create B-sGFP fusion proteins vector (Clontech: In-Fusion Advantage PCR Cloning Kit, Cat#639619, Beijing, China).
2.4 Agrobacterium-Mediated Transient Expression in N. benthamiana Leaves
After constructing the B-sGFP fusion protein vector, pt-rk CD3-999 vector (Chloroplast Localization Marker), 35S::sGFP, 35S::B1::sGFP and 35S::B3::sGFP were introduced into A. tumefaciens strain EHA105 by electroporation. And then A. tumefaciens strains EHA105 respectively with pt-rk CD3-999 and 35S::sGFP, 35S::B1::sGFP or 35S::B3::sGFP co-infected the leaves of N. benthamiana plants. After three days, the fluorescence was visualized under a Nikon microscope equipped with NIS-Elements Basic Research 3.0 software (http://www.nis-elements.com). The detailed method was described by Qi et al. [20].
Amino acid sequences of NdhB subunits from four varieties were aligned by using ClustalW (http://www.ebi.ac.uk/Tools/msa/clustalw2/), phylogenetic analysis of NdhBs in different species was performed using the MEGA4 software package, and further analyzed by TargetP, SignalP, ChloroP and TMHMM (http://www.cbs.dtu.dk/services), all of which are the amino acid sequence-based predictors [21].
2.6 Crosslink Immunoprecipitation and Western Blotting
The preparation of NdhB antibody was described by He et al. [11]. Before the antibody of NdhB was used for immunolocalization, western blotting was done to test the specificity of antibody.
In order to investigate whether there is a difference in NdhB sizes of two genes, NdhB proteins were isolated and purified from the leaves of Melrose and S111-9 extracted with Crosslink IP Kit (Product No. 26147, Thermo Scientific, USA) according to the manufacturer instructions. Firstly, NdhB antibody was immobilized on a protein A/G plus agarose, which formed antibody-crosslinked resin by crosslink reaction. Secondly, this antibody-crosslinked resin would be used to pull down the NdhB protein in the leaves of Melrose and S111-9. Finally, the eluted products were analyzed by SDS-PAGE. NdhB protein was analyzed by immunoblotting modified from the method described previously [10,22]. Total protein samples (30 μg of each sample) from the leaves of Melrose and S111-9 were separated on 12% SDS gel electrophoresis and transferred to polyvinylidene fluoride (PVDF) membrane. The PVDF membrane was incubated with a primary antibody against NdhB (1:2000 dilution). The final detection was performed using secondary fluorescent anti-rabbit IgG, followed by the SuperSignal West Pico Chemiluminescent Western blotting (Thermo Scientific, USA) [11].
2.7 Fixation and Immunolocalization
Subcellular distribution of NdhB was analyzed by electron microscopy using a previously described method [11,22], and BL containing anti-NdhB (1:300 dilution) was used for incubating the sections. The photos of immunolocalization were determined on electron micrographs at ×40,000 magnification. It was analyzed 7-8 individual cells of palisade and spongy layers from different immunolabelled sections of each variety. No significant labeling for NdhB was present in the vacuole, cell wall or cytosol [22].
All assays described above were repeated at least three times on three biological replicates. Tukey’s multiple comparison tests were used to determine the significant difference (P < 0.05) of means with the SAS 8.0 statistical software package (SAS Institute, Inc., Cary, NC, 2000) [23,24].
3.1 Molecular Cloning of ndhB, Alignment and Phylogeny of NdhB Subunits
Cloning and sequencing allowed the verification of discrepancy for ndhB among the varieties. Hence, the primers were designed from 5’UTR and 3’UTR according to the whole genomic sequences of ndhB and used to clone ndhB. Then, the ndhB from the varieties was sequenced and aligned respectively (Figs. 1A and 2A). The results of sequencing indicated that there was a point mutation in the ninth base from ‘G’ to ‘A’ (Fig. 2A) between Melrose and the other three varieties (lines). The encoded nucleotides sequences TGG (tryptophan) was replaced by TGA (a stop codon) in wild soybean ACC547 and two somatic hybrid descendants S111-9 and S113-6. According to ORF search, a new start codon (pink) was found between the site of 55th and 57th base pairs (Fig. 2A), which indicates the length of ndhB in the above three lines is 54 base pairs shorter than that in Melrose. An alignment and phylogeny of NdhB (Fig. 1) indicated that although the amino acid identity of NdhB among Melrose, S111-9, S113-6 and wild soybean (ACC547) is high, and that the genetic relationship of shortened 18 amino acid NdhB in two stable somatic hybrid descendants is exactly similar to ACC547 and lotus japonics. These results indicated that NdhB subunits in S113-6 and S111-9 was inherited from the wild soybean ACC547. It can explain, at least partly, why the above three lines bear a similarity in several photosynthetic responses to salinity resistance [11,19,22].
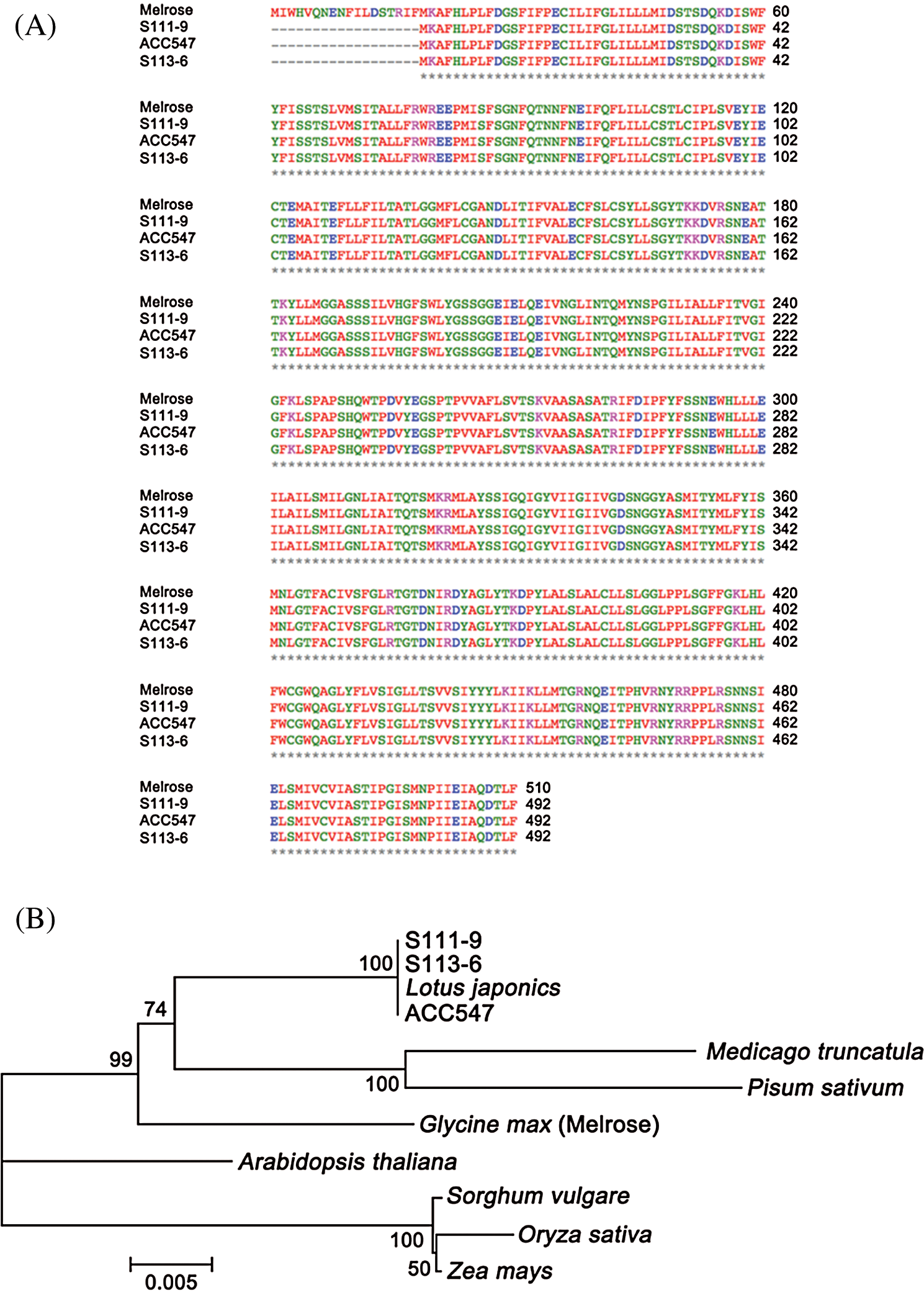
Figure 1: Multi-alignment and phylogenetic analyses of NdhB subunits. ACC547 represents an accession number of wild soybean grew in Australian seashore. Melrose is a cultivated soybean variety in Australia. S111-9 and S113-6 are the stable somatic hybrid descendants of the wild salt-tolerant Glycine cyrtoloba (ACC547) and Melrose. (A) Multi-alignment of NdhB subunits from Melrose, S111-9, ACC547 and S113-6. The amino acids length of NdhB subunit in Melrose, S111-9, ACC547 and S113-6 is 510, 492, 492 and 492, respectively. Asterisk indicates strictly conserved amino acids. (B) Phylogenetic analysis of NdhBs in difference species. The tree was constructed by using the neighbor-joining (N-J) method, and the phylogeny was tested with 1,000 bootstrap samples. Bar represents the p distance of the N-J method
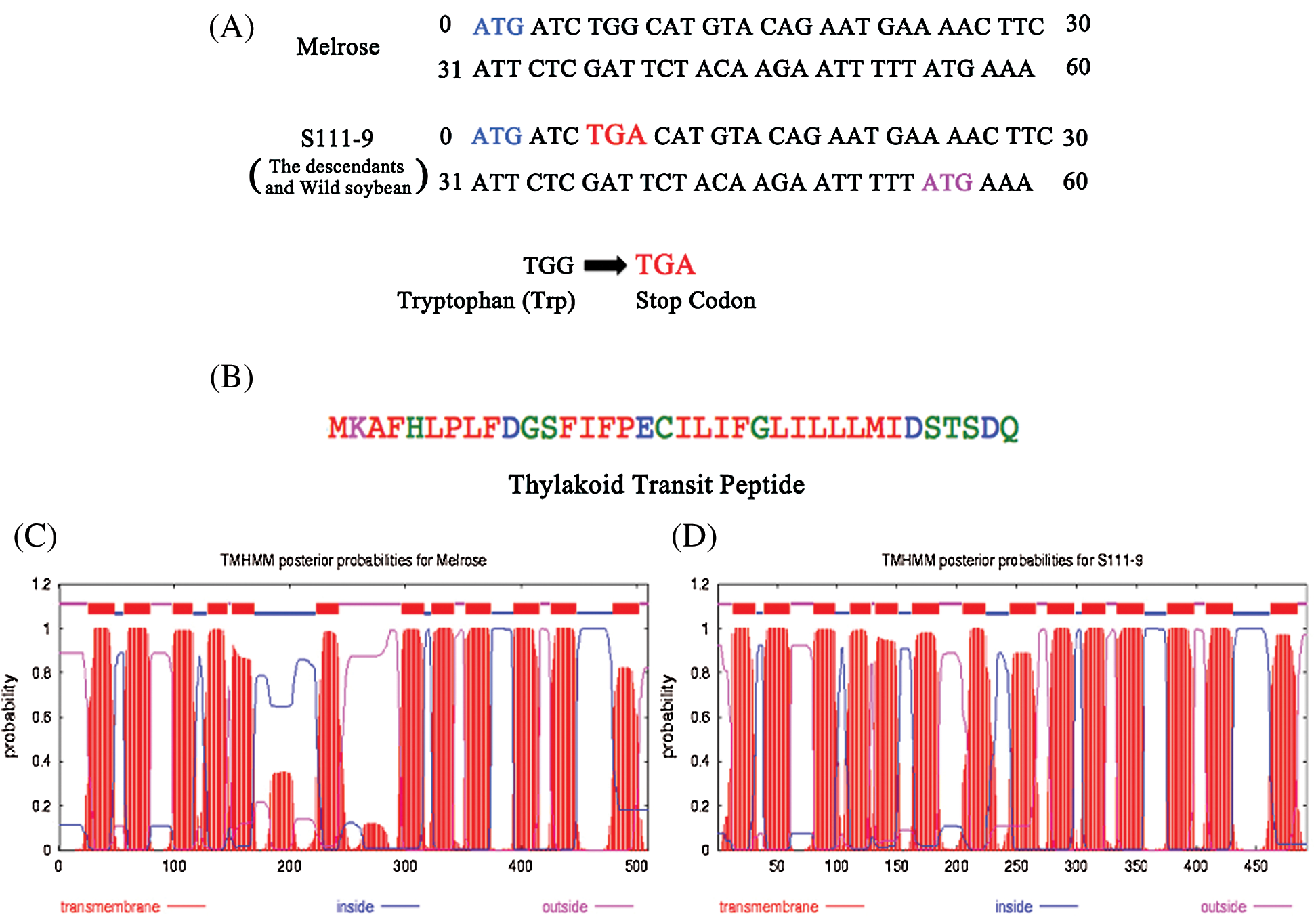
Figure 2: Structure and sequence analysis of the nucleotide and amino acids from Melrose and S111-9. (A) Analysis of the nucleotide of ndhB was performed from 0 to 60 bp in Melrose and S111-9. In S111-9, there is a point mutation in the ninth base from ‘G’ to ‘A’. So, the sequences of encoded nucleotide convert from tryptophan (TGG) to stop codon (TGA). The blue ATG is the start codon in Melrose, and the pink ATG is the new start codon in S111-9. The point mutation in the descendants (S113-6) and wild type soybean (ACC547) is in accordance with S111-9. (B) The sequence of thylakoid transit peptide in S111-9 and the length is 36 amino acids. The thylakoid transit peptide was predicted by SignalP software (http://www.cbs.dtu.dk/services/SignalP/). (C) and (D) are the results of transmembrane domain analyzed by TMHMM software (http://www.cbs.dtu.dk/services/TMHMM/). The number of transmembrane domains in Melrose (C) and S111-9 (D) is 12 and 14 respectively
3.2 Analysis of the Differences in NdhB Subunits Among Melrose, Descendants and Wild Soybean
To further understand the discrepancy of NdhB subunits between Melrose and the other three varieties, we analyzed NdhB subunits in Melrose, S11-9, S113-6 and ACC547 by TargetP, SignalP, ChloroP and TMHMM [21]. The results indicated that NdhB in S111-9, S113-6 and ACC547 has a thylakoid transit peptide with 36 amino acid (Fig. 2B). In addition, the number of transmembrane domains is different, it is 12 and 14 transmembrane domains in Melrose and other three lines, respectively (Figs. 2C and 2D). Crosslink immunoprecipitation and SDS-PAGE were used for comparing their sizes between Melrose and S111-9. The results showed that the size of NdhB subunit in S111-9 was about 54 KD and 2 KD smaller than that of Melrose (Fig. 3A) and also confirmed the difference at the level of proteins. Taken together, all of these results suggested that a novel thylakoid transit peptide exists in wild soybean and it could be passed to the somatic hybrid descendants.
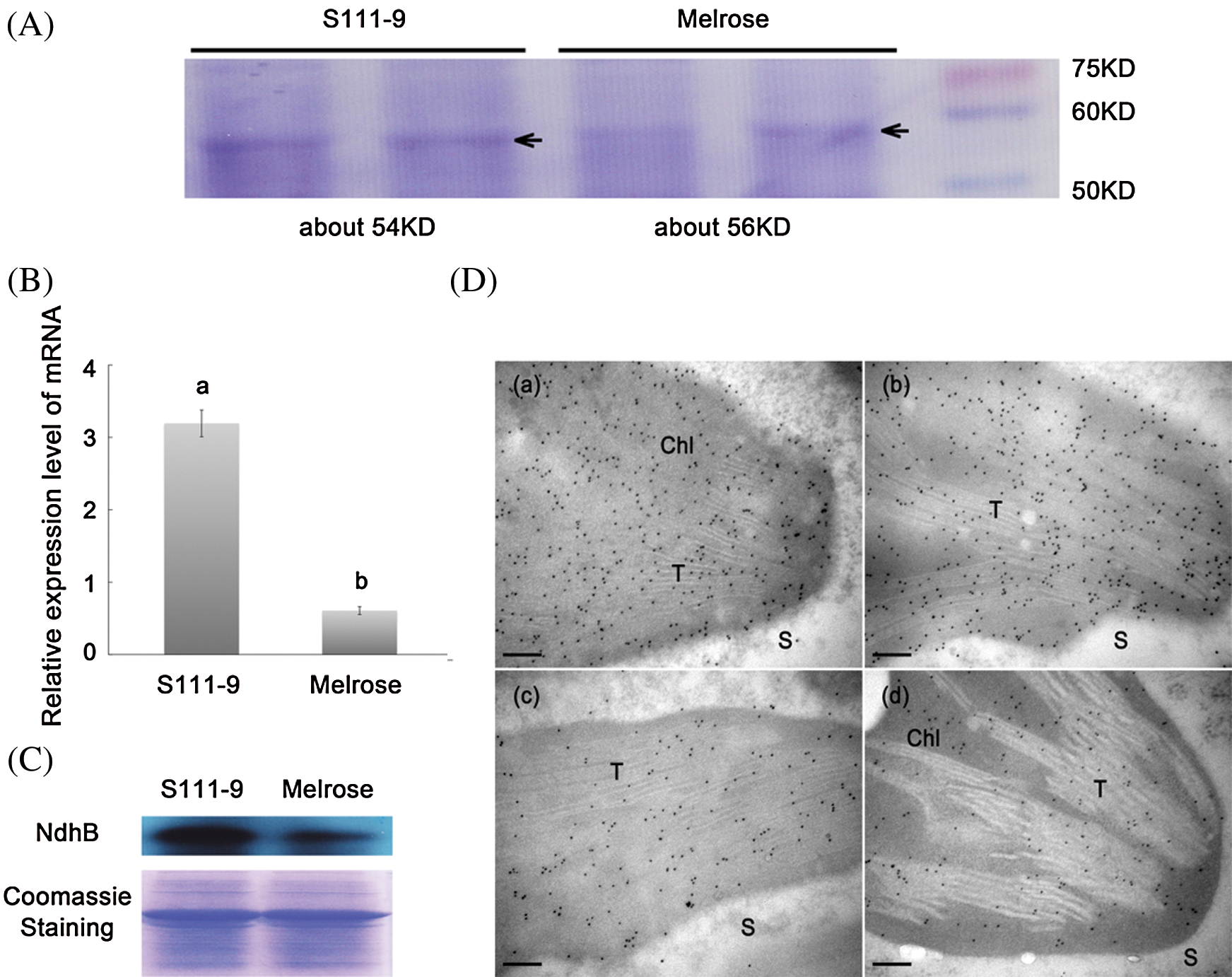
Figure 3: Comparisons of sizes, relative abundance of mRNA and protein, and immunolocalization for NdhB subunits in Melrose and S111-9. (A) Sizes of NdhB subunits were analyzed by SDS-PAGE. NdhB proteins were isolated and purified from the leaves of Melrose and S111-9 extracted with Crosslink IP Kit. (B) Relative mRNA expression level of ndhB in Melrose and S111-9. Bars represent the mean ± SD of three biological replicates. Student t-test was applied to assess difference of means between two varieties. Different letters indicate a significant difference between varieties (P < 0.05). (C) The content of NdhB were analyzed using western blotting in the leaves of Melrose and S111-9. The equal loading of lower pictures is shown by coomassie staining. (D) Immunolocalization of NdhB subunits in S111-9 (a,b) and Melrose (c,d). All immunogold particles were mainly concentrated in chloroplast, not in the cytosol. T, Thylakoid; S, Cytosol Stroma. Scale bars = 0.2 μm
3.3 Comparison of Relative Abundance and Localization for NdhB Subunits
To understand whether or not the structural differences lead to the changes in the relative abundance of NdhB subunits in chloroplasts, qRT-PCR (Fig. 3B), western blotting (Fig. 3C), and immunogold labeling (Fig. 3D) were used for determining the relative expression abundance of NdhB in Melrose and S111-9. The relative mRNA abundance of ndhB increased significantly (P < 0.05) in S111-9 than that in Melrose (Fig. 3B), its relative protein expression level also obviously increased much more in S111-9 than that in Melrose (Fig. 3C). The immunogold labeling showed more NdhB subunit particles in S111-9 (Fig. 3D(a,b)) than that in Melrose (Fig. 3D(c,d)), suggesting that the thylakoid transit peptide favor the NdhB subunit to locate in thylakoid membrane of chloroplast. It might enhance NDH complex assembly and activity.
In addition, we constructed the vectors of GFP fusion protein with B1 and B3 target fragments (B1 represents NdhB with thylakoid transit peptide from S111-9; B3 represents NdhB from Melrose) (Fig. 4A). Then, pt-rk CD3-999 vector (chloroplast localization marker) and B1::sGFP or B3::sGFP were co-expressed in the leaves of tobacco. The results indicated that the control vector with GFP is not expressed in chloroplast (Fig. 4B(a–c)), but B1::sGFP and B3::sGFP could stably co-localize in chloroplast (yellow colors) with pt-rk CD3-999 (Fig. 4B(d–i)). Therefore, these data demonstrated that the relative mRNA and protein abundance of NdhB in S111-9 is significantly increased due to the existence of the thylakoid transit peptide. Furthermore, it also could contribute to the localization of NdhB.
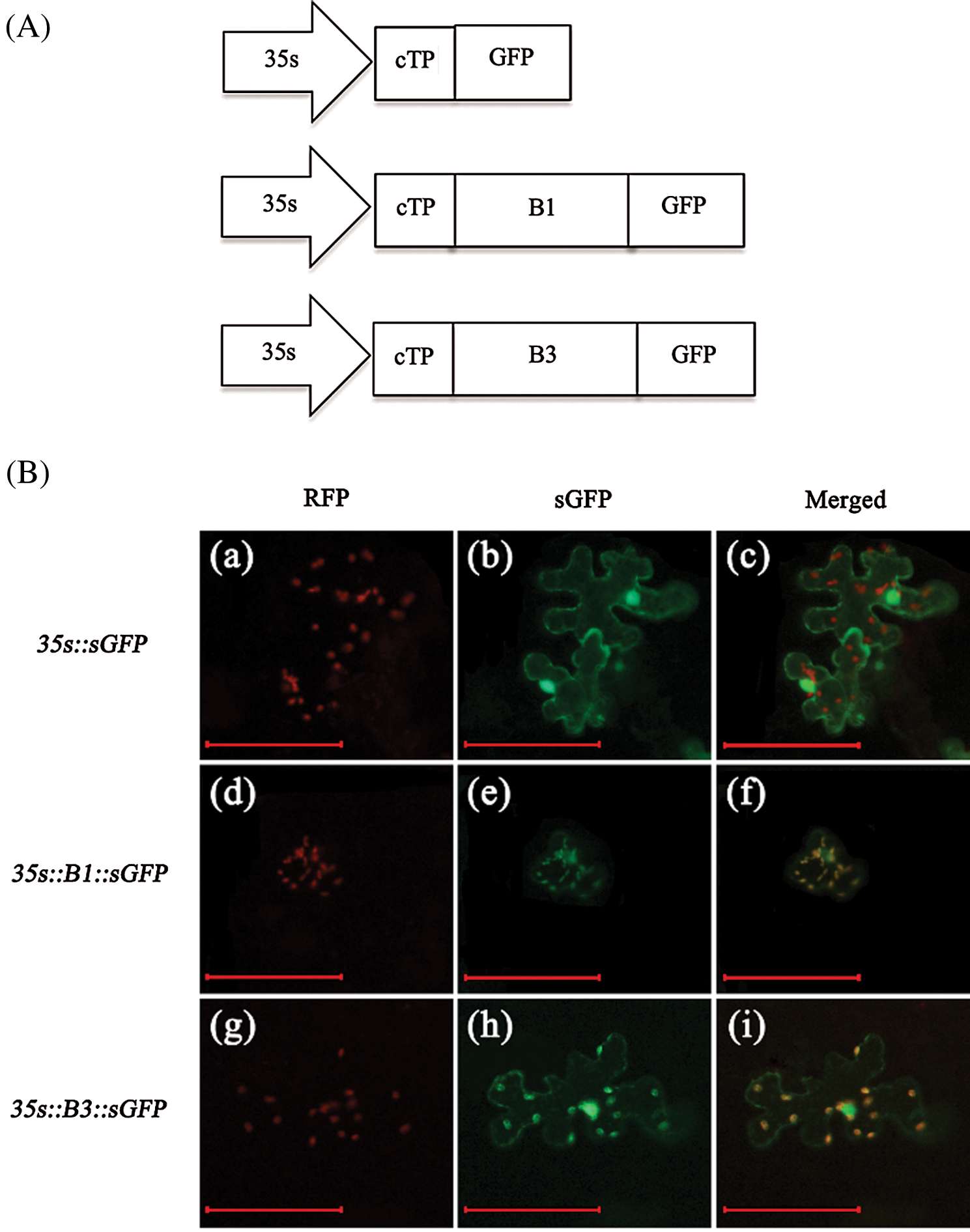
Figure 4: Coexpression of pt-rk CD3-999 vector (chloroplast localization marker) and B1::sGFP or B3::sGFP in Nicotiana benthamiana. (A) Scheme of GFP fusion protein vector construction. cTP is the chloroplast transit peptide from At5g48790 and the length is 48 amino acids. B1 and B3 represent the sequence of NdhB subunit with and without a thylakoid transit peptide respectively. And B1 is shorter 18 amino acids than B3. (B) a–c is control, coexpression of pt-rk CD3-999 and 35s::sGFP; d–f is coexpression of pt-rk CD3-999 and 35s::B1::sGFP; g–i is coexpression of pt-rk CD3-999 and 35s::B3::sGFP. Scale bars = 100 μm
Complete chloroplast genomes sequencing of Glycine max revealed that at least 11 encoded genes are involved in the assembly of NDH complex in soybean [25]. In addition, it has been reported previously that NDH functions in protecting photosynthetic apparatus from oxidative damage caused by temperature and salt stresses [9,11,26]. However, NdhB, as one of the most important subunits of NDH, its subcellular localization mechanism still remains unclear. At present, protein translocation and localization pathways in bacteria [27–29] and plants [30–33] have been the focus of many studies and continued to be of great interest in future research. In bacteria, most secreted proteins carry an N-terminal signal sequence that directs them to the inner membrane protein translocation machinery to be localized into the membrane, or internalized into the membrane, translocated to the periplasm or secreted [34,35]. In plants, however, the sorting of proteins in the cell is somewhat more complex due to the presence of different organelles, and different types of membranes [29]. The most proteins have hydrophilic surfaces and therefore do not readily pass through the hydrophobic core of the lipid bilayer. Some chaperonins or signal peptides are required to facilitate the translocation and localization of a protein through the lipid bilayer [32]. And only when finely orchestrated gene regulation and tissue gene expression along with proper subcellular compartmentalization of plant proteins allow cells to become specialized and differentiated to fulfill their role in tissue specificity, organ identity, and organism performance [29].
Because of the salt tolerant somatic hybrid descendants and their wild soybean (Glycine cyrtoloba ACC547), a cytoplasm donor parent with high NDH-dependent CEF activity demonstrated in our previous studies [11,19,22], we determined the sequence of NdhB subunits in them. Intriguingly, the sequencing results indicated that NdhB of the hybrid descendants and their wild soybean are 18 amino acid shorter than that in Melrose (Figs. 1A and 2A), whereas bioinformatics analysis further implied that there is a novel thylakoid transit peptide in their NdhB. In addition, their transmembrane domains also differ from Melrose (Figs. 2B–2D). To confirm the effects of thylakoid transit peptide on the expression and localization of NdhB, the expression and localization analysis of NdhB are performed. Interestingly, the crosslink IP and SDS-PAGE results indicated that the size of NdhB in S111-9 is actually 2 KD smaller than that in Melrose (Fig. 3A), and the relative mRNA and protein abundance of NdhB in S111-9 was significantly increased. These results suggested that the existence of thylakoid transit peptide led to the difference in the expression level of NdhB between S111-9 and Melrose. Our previous study further supported that the existence of thylakoid transit peptide in S111-9 may enhance the ability of NdhB localization, and further contributing to its higher relative mRNA and protein abundance [11]. In most plants, NdhB might mainly depend on chaperonins to facilitate protein translocation, folding and localization. Co-localization of NdhB with or without the thylakoid transit peptide showed a fact that both could locate in chloroplasts, indicating that the novel thylakoid transit peptide might play the same role as the chaperonins at least.
Taken together, the novel thylakoid transit peptide from wild soybean ACC547 contributes to enhancing the ability of localization of NdhB, and to increasing the relative mRNA and protein abundance of NdhB. It may play an important physiological role in the resistance of the soybean to salt stress. Future studies on the physiological function of NdhB with thylakoid transit peptide may unravel the important role of NDH complex under stress, and will provide a new insight for further understanding the expression and localization mechanism of thylakoid membrane proteins.
Funding Statement: This work was supported by the National Natural Science Foundation of China (31801273), the Zhejiang A&F University Scientific Research and Development Fund Project (2018FR049), the Department of Science and Technology of Ningbo (DSTNB, Project No. 2019C10008), and the China Postdoctoral Foundation (2016M591984).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Shikanai, T. (2007). Cyclic electron transport around photosystem I: genetic approaches. Annual Review of Plant Biology, 58(1), 199–217. DOI 10.1146/annurev.arplant.58.091406.110525.
2. Shikanai, T. (2007). The NAD(P)H dehydrogenase complex in photosynthetic organisms: subunit composition and physiological function. Functional Plant Science and Biotechnology, 1, 129–137. [Google Scholar]
3. Friedrich, T., Scheide, D. (2000). The respiratory complex I of bacteria, archaea and eukarya and its module common with membrane-bound multisubunit hydrogenases. FEBS Letters, 479(1–2), 1–5. DOI 10.1016/S0014-5793(00)01867-6. [Google Scholar] [CrossRef]
4. Peng, L. W., Yamamoto, H., Shikanai, T. (2011). Structure and biogenesis of the chloroplast NAD(P)H dehydrogenase complex. Biochimica et Biophysica Acta-Bioenergetics, 1807(8), 945–953. DOI 10.1016/j.bbabio.2010.10.015. [Google Scholar] [CrossRef]
5. Suorsa, M., Sirpio, S., Aro, E. M. (2009). Towards characterization of the chloroplast NAD(P)H dehydrogenase complex. Molecular Plant, 2(6), 1127–1140. DOI 10.1093/mp/ssp052. [Google Scholar] [CrossRef]
6. Munekage, Y., Hashimoto, M., Miyake, C., Tomizawa, K., Endo, T. et al. (2004). Cyclic electron flow around photosystem I is essential for photosynthesis. Nature, 429(6991), 579–582. DOI 10.1038/nature02598. [Google Scholar] [CrossRef]
7. Horvath, E. M., Peter, S. O., Joet, T., Rumeau, D., Cournac, L. et al. (2000). Targeted inactivation of the plastid ndhB gene in tobacco results in an enhanced sensitivity of photosynthesis to moderate stomatal closure. Plant Physiology, 123(4), 1337–1350. DOI 10.1104/pp.123.4.1337. [Google Scholar] [CrossRef]
8. Munne-Bosch, S., Shikanai, T., Asada, K. (2005). Enhanced ferredoxin-dependent cyclic electron flow around photosystem I and alpha-tocopherol quinone accumulation in water-stressed ndhB-inactivated tobacco mutants. Planta, 222(3), 502–511. DOI 10.1007/s00425-005-1548-y. [Google Scholar] [CrossRef]
9. Wang, P., Duan, W., Takabayashi, A., Endo, T., Shikanai, T. et al. (2006). Chloroplastic NAD(P)H dehydrogenase in tobacco leaves functions in alleviation of oxidative damage caused by temperature stress. Plant Physiology, 141(2), 465–474. DOI 10.1104/pp.105.070490. [Google Scholar] [CrossRef]
10. Wang, D., Li, X. F., Zhou, Z. J., Feng, X. P., Yang, W. J. et al. (2010). Two Rubisco activase isoforms may play different roles in photosynthetic heat acclimation in the rice plant. Physiologia Plantarum, 139(1), 55–67. DOI 10.1111/j.1399-3054.2009.01344.x. [Google Scholar] [CrossRef]
11. He, Y., Fu, J. L., Yu, C. L., Wang, X. M., Jiang, Q. S. et al. (2015). Increasing cyclic electron flow is related to Na+ sequestration into vacuoles for salt tolerance in soybean. Journal of Experimental Botany, 66(21), 6877–6889. DOI 10.1093/jxb/erv392. [Google Scholar] [CrossRef]
12. Burrows, P. A., Sazanov, L. A., Svab, Z., Maliga, P., Nixon, P. J. (1998). Identification of a functional respiratory complex in chloroplasts through analysis of tobacco mutants containing disrupted plastid ndh genes. EMBO Journal, 17(4), 868–876. DOI 10.1093/emboj/17.4.868. [Google Scholar] [CrossRef]
13. Rumeau, D., Becuwe-Linka, N., Beyly, A., Louwagie, M., Garin, J. et al. (2005). New subunits NDH-M, -N, and -O, encoded by nuclear genes, are essential for plastid Ndh complex functioning in higher plants. Plant Cell, 17(1), 219–232. DOI 10.1105/tpc.104.028282.
14. Rumeau, D., Peltier, G., Cournac, L. (2007). Chlororespiration and cyclic electron flow around PSI during photosynthesis and plant stress response. Plant, Cell & Environment, 30(9), 1041–1051. DOI 10.1111/j.1365-3040.2007.01675.x. [Google Scholar] [CrossRef]
15. Peng, L. W., Fukao, Y., Myouga, F., Motohashi, R., Shinozaki, K. et al. (2011). A chaperonin subunit with unique structures is essential for folding of a specific substrate. PLoS Biology, 9(4), e1001040. DOI 10.1371/journal.pbio.1001040. [Google Scholar] [CrossRef]
16. Peng, L. W., Cai, W., Shikanai, T. (2010). Chloroplast stromal proteins, CRR6 and CRR7, are required for assembly of the NAD(P)H dehydrogenase subcomplex A in Arabidopsis. Plant Journal, 63(2), 203–211. DOI 10.1111/j.1365-313X.2010.04240.x. [Google Scholar] [CrossRef]
17. Peng, L. W., Fukao, Y., Fujiwara, M., Shikanai, T. (2010). Multistep assembly of chloroplast NADH dehydrogenase-like subcomplex A requires several nucleus-encoded proteins, including CRR41 and CRR42, in Arabidopsis. Plant Cell, 24(1), 202–214. DOI 10.1105/tpc.111.090597. [Google Scholar] [CrossRef]
18. Neumann, G., Massonneau, A., Martinoia, E., Römheld, V. (1999). Physiological adaptations to phosphorus deficiency during proteoid root development in white lupin. Planta, 208(3), 373–383. DOI 10.1007/s004250050572. [Google Scholar] [CrossRef]
19. Yang, Y., Yan, C. Q., Cao, B. H., Xu, H. X., Chen, J. P. et al. (2007). Some photosynthetic responses to salinity resistance are transferred into the somatic hybrid descendants from the wild soybean Glycine cyrtoloba ACC547. Physiologia Plantarum, 129(3), 658–669. DOI 10.1111/j.1399-3054.2006.00853.x. [Google Scholar] [CrossRef]
20. Qi, Y. H., Wang, S. K., Shen, C. J., Zhang, S. N., Chen, Y. et al. (2012). OsARF12, a transcription activator on auxin response gene, regulates root elongation and affects iron accumulation in rice (Oryza sativa). New Phytologist, 193(1), 109–120. DOI 10.1111/j.1469-8137.2011.03910.x. [Google Scholar] [CrossRef]
21. Emanuelsson, O., Brunak, S., Von, H. G., Nielsen, H. (2007). Locating proteins in the cell using TargetP, SignalP and related tools. Nature Protocol, 2(4), 953–971. DOI 10.1038/nprot.2007.131. [Google Scholar] [CrossRef]
22. He, Y., Yu, C. L., Zhou, L., Chen, Y., Liu, A. et al. (2014). Rubisco decrease is involved in chloroplast protrusion and Rubisco-containing body formation in soybean (Glycine max) under salt stress. Plant Physiology and Biochemistry, 74, 118–124. DOI 10.1016/j.plaphy.2013.11.008. [Google Scholar] [CrossRef]
23. He, Y., Zhou, K. Y., Wu, Z. M., Li, B. X., Fu, J. L. et al. (2019). Highly efficient nanoscale analysis of plant stomata and cell surface using polyaddition silicone Rubber. Frontiers in Plant Science, 10, 1569. DOI 10.3389/fpls.2019.01569. [Google Scholar] [CrossRef]
24. Yang, Y., Xie, X. T., Tao, S. C., Zhou, K. Y., Yu, Y. X. et al. (2020). Plasmodesmata play a critical role in promoting the germination of floral buds in Ilex verticillata. Plant Growth Regulation, 91(3), 349–357. DOI 10.1007/s10725-020-00609-0. [Google Scholar] [CrossRef]
25. Saski, C., Lee, S. B., Daniell, H., Wood, T. C., Tomkins, J. et al. (2005). Complete chloroplast genome sequence of Glycine max and comparative analyses with other legume genomes. Plant Molecular Biology, 59(2), 309–322. DOI 10.1007/s11103-005-8882-0. [Google Scholar] [CrossRef]
26. Yamori, W., Sakata, N., Suzuki, Y., Shikanai, T., Makino, A. (2011). Cyclic electron flow around photosystem I via chloroplast NAD(P)H dehydrogenase (NDH) complex performs a significant physiological role during photosynthesis and plant growth at low temperature in rice. Plant Journal, 68(6), 966–976. DOI 10.1111/j.1365-313X.2011.04747.x. [Google Scholar] [CrossRef]
27. Driessen, A. J. M., Nouwen, N. (2008). Protein translocation across the bacterial cytoplasmic membrane. Annual Review of Biochemistry, 77(1), 643–667. DOI 10.1146/annurev.biochem.77.061606.160747. [Google Scholar] [CrossRef]
28. Papanikou, E., Karamanou, S., Economou, A. (2007). Bacterial protein secretion through the translocase nanomachine. Nature Reviews Microbiology, 5(11), 839–851. DOI 10.1038/nrmicro1771.
29. Moeller, L., Gan, Q. L., Wang, K. (2009). A bacterial signal peptide is functional in plants and directs proteins to the secretory pathway. Journal of Experimental Botany, 60(12), 3337–3352. DOI 10.1093/jxb/erp167. [Google Scholar] [CrossRef]
30. Brown, L. A., Baker, A. (2008). Shuttles and cycles: transport of proteins into the peroxisome matrix. Molecular Membrane Biology, 25(5), 363–375. DOI 10.1080/09687680802130583. [Google Scholar] [CrossRef]
31. Hanton, S. L., Matheson, L. A., Chatre, L., Rossi, M., Brandizzi, F. (2007). Post-Golgi protein traffic in the plant secretory pathway. Plant Cell Report, 26(9), 1431–1438. DOI 10.1007/s00299-007-0390-z.
32. Raikhel, N., Chrispeels, M. J. (2000). Protein sorting and vesicle traffic. In: Buchanan, B. B., Gruissem, W., Jones, R. L. (eds.Biochemistry and Molecular Biology of Plants. Rockville, MD: American Society of Plant Physiologist, 160–201. [Google Scholar]
33. Rojo E., Denecke J. (2008). What is moving in the secretory pathway of plants?. Plant Physiology, 147(4), 1493–1503. DOI 10.1104/pp.108.124552. [Google Scholar] [CrossRef]
34. Saier, M. H. (2006). Protein secretion and membrane insertion systems in gram-negative bacteria. Journal of Membrane Biology, 214(1–2), 75–90. DOI 10.1007/s00232-006-0049-7. [Google Scholar] [CrossRef]
35. Saier, M. H., Ma, C., Rodgers, L., Tamang, D., Yen, M. et al. (2008). Chapter 6 protein secretion and membrane insertion systems in bacteria and eukaryotic organelles. Advances in Applied Microbiology, 65, 141–197. [Google Scholar]
Supplementary Files
Table S1: Primers Sequences for GFP Fusion Protein Vector Construction