DOI:10.32604/phyton.2020.011545

| Phyton-International Journal of Experimental Botany DOI:10.32604/phyton.2020.011545 |  |
| Article |
Reference Gene Selection for qRT-PCR Normalization in Iris germanica L.
Institute of Botany, Jiangsu Province and Chinese Academy of Sciences, Nanjing, 210014, China
*Corresponding Author: Haiyan Yuan. Email: yuanhaiyan416@163.com
Received: 15 May 2020; Accepted: 07 July 2020
Abstract: Quantitative real-time PCR (qPCR) is an effective and widely used method to analyze expression patterns of target genes. Selection of stable reference genes is a prerequisite for accurate normalization of target gene expression by qRT-PCR. In Iris germanica L., no studies have yet been published regarding the evaluation of potential reference genes. In this study, nine candidate reference genes were assessed at different flower developmental stages and in different tissues by four different algorithms (GeNorm, NormFinder, BestKeeper, and Ref-Finder). The results revealed that ACT11 (Actin 11) and EF1α (Elongation factor 1 alpha) were the most stable reference genes in different tissues, whereas TUA (Tubulin alpha) and UBC9 (Ubiquitin-protein ligase 9) were the most stable ones in different flower developmental stages. UBC9 and ACT11 were the most stable reference genes in all of the tested samples, while the SAMDC (S-Adenosylmethionine decarboxylase) showed the least stability. Finally, to validate the suitability of the selected reference genes, the relative expression level of IgTPS (beta-caryophyllene synthase) was assessed and highlighted the importance of suitable reference gene selection. This work constitutes the first systematic evaluation of potential reference genes in I. germanica and provides guidelines for future research on gene function and molecular mechanisms on I. germanica and related species.
Keywords: Iris germanica L; reference genes; floral scent; qRT-PCR
Quantitative real time PCR (qRT-PCR) is a reliable technique for quantifying target gene expression. It is the most commonly used technique for studying the expression of genes because of its increased sensitivity, specificity, and accuracy and high throughput compared with reverse transcription PCR (RT-PCR) and Northern blot [1]. However, the accuracy of qRT-PCR is dependent on the stability of the reference genes employed, the RNA integrity and purity, the enzymatic efficiency in cDNA synthesis, and the efficacy of PCR amplification [2]. Therefore, it is necessary to select reliable reference genes to normalize the relative expression of target genes.
A number of common reference genes, such as actin (ACT), ubiquitin (UBQ), beta-tubulin-4 (TUB4), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and elongation factor 1 alpha (EF1α), involved in primary metabolism or other basic cellular processes have been widely adopted for gene expression analyses in different tissues or organs [3–7]. However, recent studies have found that some of these genes are not consistently expressed in different plant species, tissues and developmental stages and various experimental conditions [8,9]. Therefore, an increasing number of studies have focused on the identification and evaluation of potential reference genes under various experimental conditions in different plant species, including cineraria (Senecio cruentus Masson ex L’ Herit) [10], rose (Rosa hybrida) [11], Chrysanthemum lavandulifolium [12], tree peony (Paeonia suffruticosa Andr.) [13], Rhododendron molle [14], lettuce (Lactuca sativa) [15], seashore paspalum (Paspalum vaginatum) [2] and poplar (Populus alba × Populus glandulosa) [9].
Iris, a kind of perennial herbaceous plants, consists of approximately 300 species distributed in North America, North Africa and Eurasia [16,17]. Many of them are popular ornamental plants due to their diversity of flower color, and range of volatile organic compounds (VOCs) [18,19]. Iris germanica L., belonging to the family Iridaceae, is the best known and most horticulturally important type of bearded iris, and its distinctive floral fragrance makes this species a good model for floral scent research in the Iris genus. Recently, chemical components of the floral scent in I. germanica have been reported [20], although the molecular mechanisms of floral scent have not yet been described. Revealing the molecular mechanisms of floral scent production requires clarifying the expression pattern of crucial genes involved in floral development. To obtain accurate expression data, a set of reference genes are essential as normalization factors. However, no systematic analysis of suitable reference genes in I. germanica has been conducted. Therefore, evaluation of the stability of candidate reference genes in I. germanica flower developmental stages and different tissues is necessary.
In this study, based on homolog comparisons between Iris lactea var. chinensis transcriptome data and Arabidopsis microarray data, 9 candidate reference genes, including EF1α, GAPDH, ACT11, UBQ, Ubiquitin-protein ligase 9 (UBC9), S-Adenosylmethionine decarboxylase (SAMDC), Phosphoglycerate kinase (PGK), Tubulin alpha (TUA), Protein phosphatase 2A (PP2A) were identified. Four different algorithms, GeNorm [21], BestKeeper [22], NormFinder [23], and Ref-Finder (http://www.leonxie.com/referencegene.php) were used to evaluate the most suitable reference genes for normalization. To verify the suitability of the selected reference genes in I. germanica, we assayed the relative expression levels of IgTPS in different floral development stages and tissues; this gene encodes the beta-caryophyllene synthase, an important enzyme in the terpenoid biosynthesis pathway [24]. This work provides a valuable resource for reference gene selection for qRT-PCR normalization for future gene expression studies in I. germanica.
Iris germanica L. plants were obtained from the Iris Resource Collection Garden of Institute of Botany, Nanjing Sun Yat-Sen Memorial Botanical Garden. Self-selected cultivars ‘Huangjinjia’, derived from the crossbreeding of the female parent ‘93E41076-8’ and the male parent ‘Jinwuwa’, were used as the plant materials. Three different tissues, including roots, stems, and leaves were collected, frozen immediately in liquid nitrogen and stored at −80°C freezer prior to use. Samples of flowers at four developmental stages were collected: Stage 1 (S1), flower bud and bracts are not open. Stage 2 (S2), flower bud has a cleft, and bracts are opening. Stage 3 (S3), flower bud is open, and the tips of florets are visible. Stage 4 (S4), flower is fully open (Fig. 1).

Figure 1: Four stages of I. germanica flower development. Scale bars = 1 cm
2.2 RNA Extraction and cDNA Synthesis
Total RNA from all samples was extracted using the RNA simple Total kit (TaKaRa Dalian, China) according to the manufacturer’s instructions, and then treated with RNase-free DNase I (TaKaRa, Dalian, China) at 37°C for 30 min to eliminate potential DNA contamination. The concentration and integrity of the total RNA were evaluated by the procedures described by Gu et al. [25]. The cDNA first strand was synthesized using 1 μg total RNA in a volume of 20 μl with the PrimeScript RT reagent kit (TaKaRa, Dalian, China) according to the manufacturer’s instructions.
Nine candidate reference genes, including EF1α, GAPDH, ACT11, UBQ, UBC9, SAMDC, PGK, TUA and PP2A were selected from the transcriptome data sequences of Iris lactea var. chinensis [26]. All candidate reference genes were cloned and confirmed through sequencing. The gene sequences are stored in GenBank (Tab. 1). Primers were designed using Primer 5.0 software (Premier Biosoft International) to have melting temperatures between 55 and 65°C, primer lengths between 19 and 23 bp, and amplicon lengths between 81 and 280 bp (Tab. 1).
Table 1: Primer sequences and characteristics of PCR amplifications in I. germanica
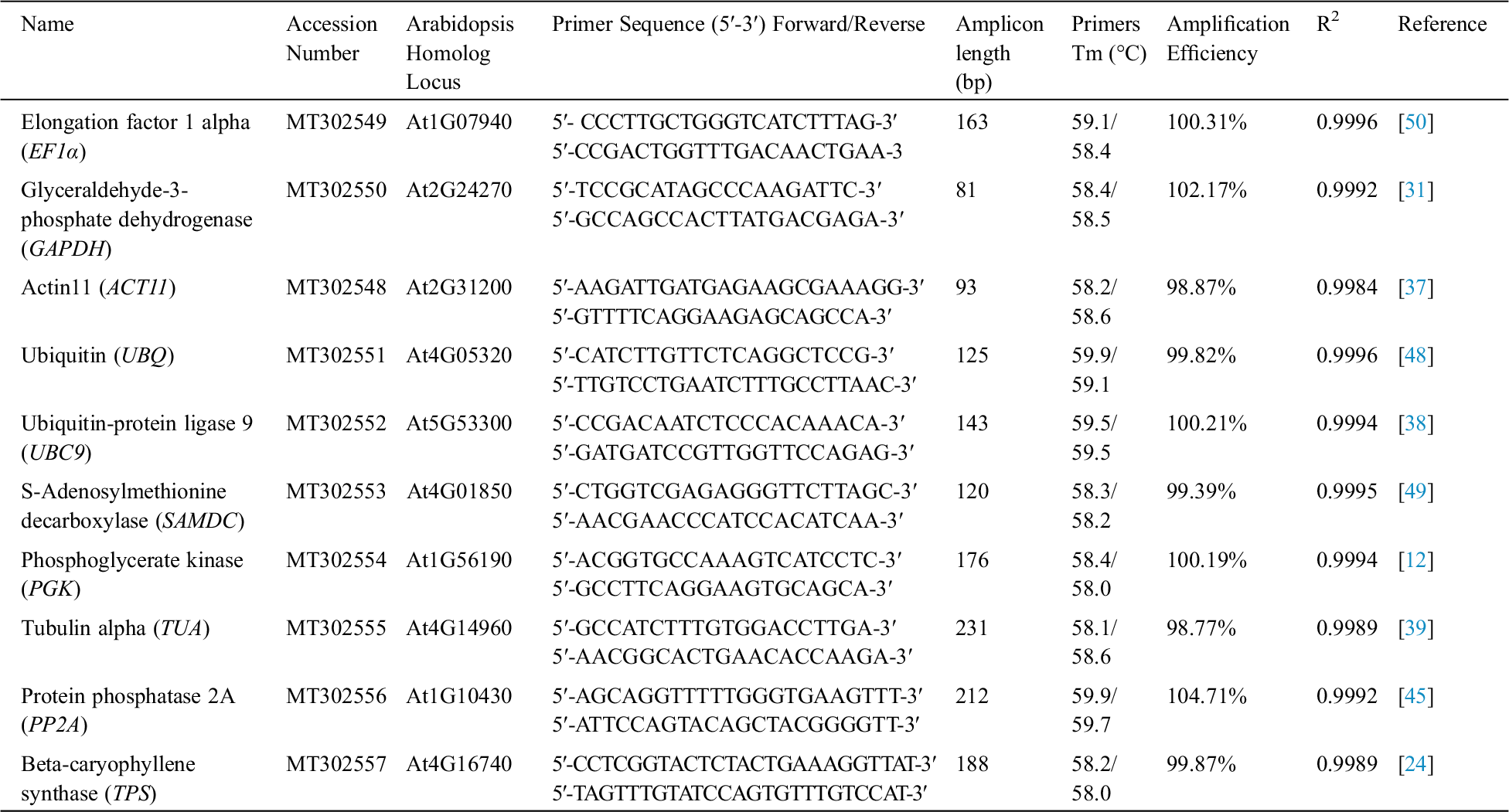
qPCR reactions were performed using a Mastercycler ep realplex 2 S device (Eppendorf, Germany) with S YBR® Premix Ex TaqTM II (TaKaRa, Dalian, China). Reactions with 20 μL mixture containing 5 μL of diluted cDNA, 0.6 μL of each amplification primer (10 μM), 10 μL of 2 × SYBR Premix and 3.8 μL of ddH2O. The amplification program comprised an initial denaturation step (95°C for 2 min), followed by 40 cycles of 95°C for 5 s and 60°C for 30 s, and 72°C for 30 s, and a melting curve protocol (60–95°C with a temperature increment of 0.5°C s−1). Each qRT-PCR reaction was performed in triplicate.
PCR efficiency based on the raw fluorescence data was evaluated for each target with the software LinRegPCR program [27]. The stability of the nine potential reference genes were assessed with four programs, including GeNorm [21], NormFinder [23], BestKeeper [22], and Ref-finder (http://www.leonxie.com/referencegene.php). GeNorm, NormFinder and BestKeeper were used to analyze the stability of nine potential reference genes. Finally, Ref-Finder was used to sequence the reference genes and select the highest stability gene in different tissues and during four flower developmental stages. The top two ranked genes and the lowest ranked gene were used to calculate IgTPS gene expression to assess the effectiveness of the reference genes. Primers used to amplify IgTPS are presented in Tab. 1. The fold change of gene expression was calculated using the 2−∆∆Ct method [28].
3.1 Performance of Primers for Nine Reference Genes
A total of nine genes, including EF1α, GAPDH, ACT11, UBQ, UBC9, SAMDC, PGK, TUA and PP2A, were selected as candidate reference genes. The specificities of primers were confirmed by analyzing melting curve assays. The presence of a single peak indicated that the expected amplicons were amplified. A single DNA band in each gel electrophoresis demonstrated that all nine primer pairs amplified a specific product with the expected size and no primer dimers were found. The correlation coefficients (R2) values were ranged between 0.9984 and 0.9996, and PCR amplification efficiencies ranged between 0.9877 and 1.0471 (Tab. 1).
The quantification cycle (Cq) values of nine reference genes were assayed by qRT-PCR analysis (Fig. 2). Mean Cq values of all reference genes were between 19.84 and 24.87, which represented the different expression levels in Iris germanica L. UBC9 showed the highest expression level and the lowest Cq value (19.84). GAPDH presented the lowest expression level and the highest Cq value.
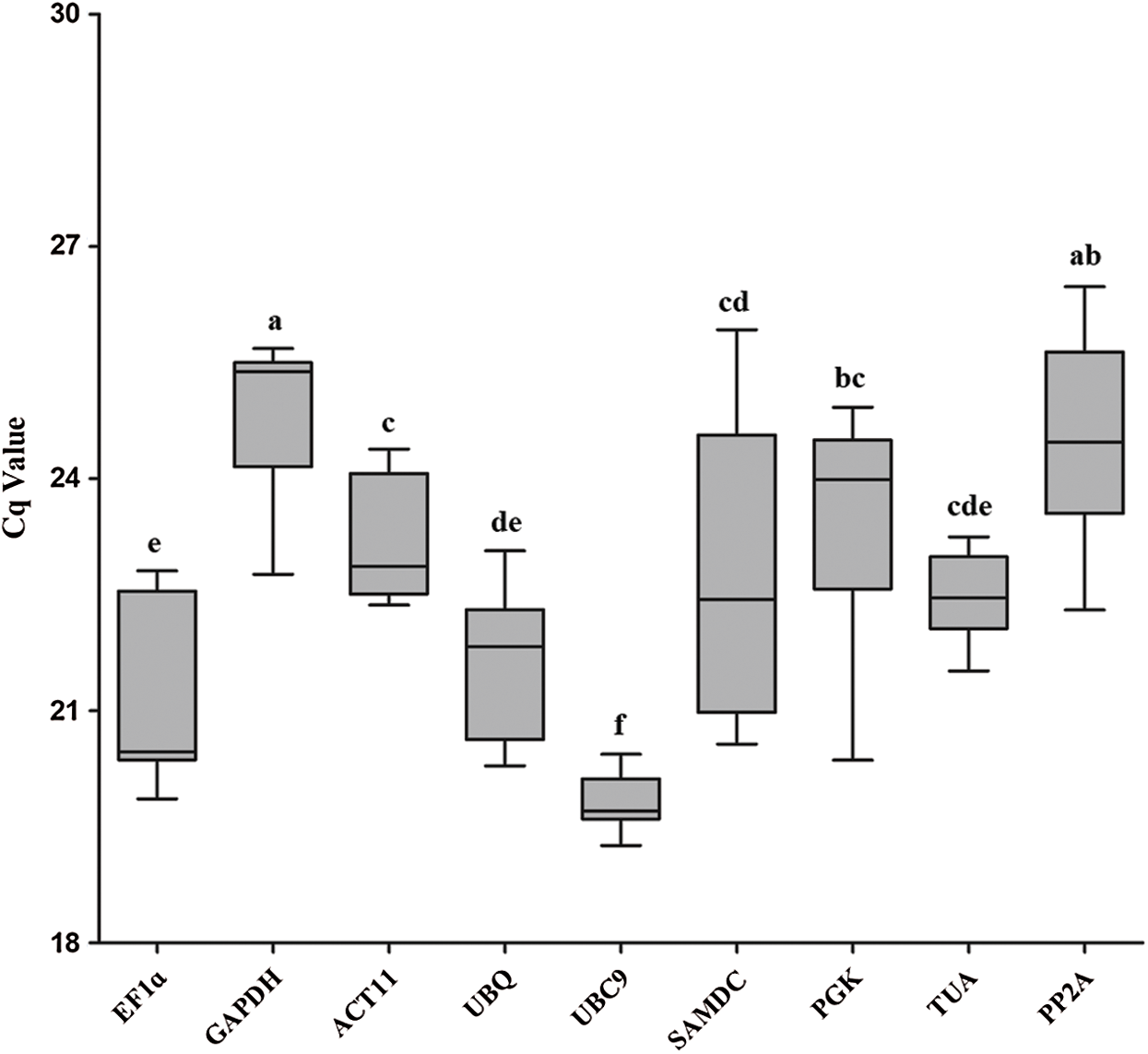
Figure 2: Cq values for 9 candidate reference genes in all samples of I. germanica. A line across the box-plot of Cq value indicates the median values. The outside boxes indicate the 25th and 75th percentiles. Whiskers represent the maximum and minimum values. Values are means ± s.d. (n = 7), p < 0.05 (student’s t-test)
3.2 Stability of Reference Genes
In our study, four software programs included GeNorm, NormFinder, BestKeeper and Ref-Finder were used to assess and rank the expression stability of each reference genes.
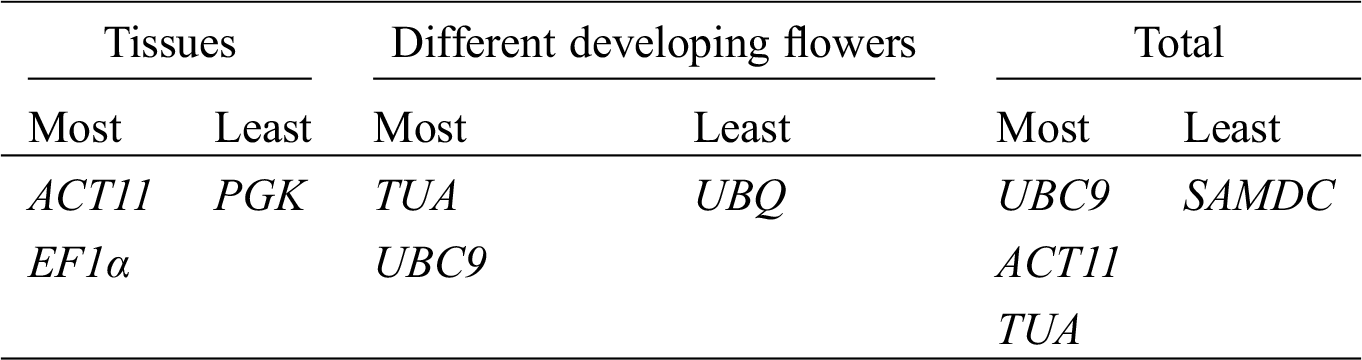
GeNorm software was used to rank the expression stability of the 9 candidate reference genes by calculating the average expression stability (M) values. Lower value of M indicates higher stability of the reference gene. Stably expressed genes have values below 1.5 [21]. Fig. 2 shows that the M values of all tested reference genes were less than 1.5, indicating that they all conformed to the basic requirements for the reference gene. ACT11 and UBQ were the most stable reference genes in different tissues, while the least stable gene was PGK. In the different developmental stages of flowers, UBC9 and TUA were the most stable reference genes, while UBQ was the least stable. In all of the samples, ACT11 and UBC9 were found to be the most stable genes, while SAMDC was found to be the least stable (Fig. 3).
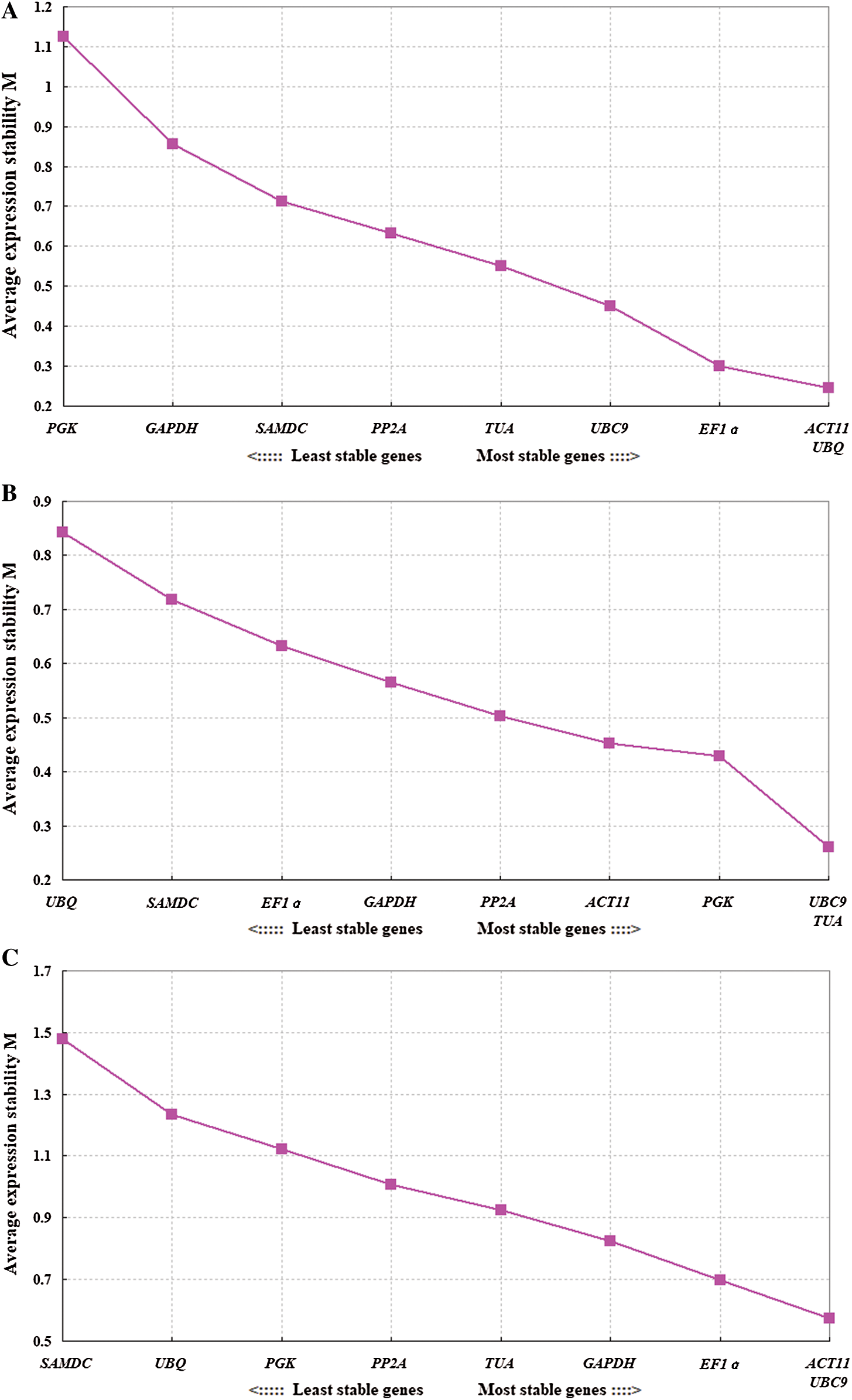
Figure 3: Average expression stability values (M) of 9 candidate reference genes as calculated by GeNorm software. The most stable genes are listed on the right, while the least stable genes are listed on the left. A: Tissues, B: Different developing flowers, C: Total
The pairwise variation (V) between sequential ranked genes (Vn/Vn + 1) based on the GeNorm algorithm was calculated to determine the optimal number of reference genes for normalization. A low level of variation between Vn/n+1 and Vn+1/Vn+2 indicates no significant effects of the addition of another gene on the normalization, and a V value of 0.15 was used as the threshold to determine whether additional reference genes are necessary [21,29]. In our study, the V2/3 value in different tissues was below 0.15 (Fig. 4), which indicated that two reference genes are sufficient for accurate normalization. The V3/4 value of 0.103 for the four developmental stages of flowers indicated that three genes are necessary for normalization (Fig. 4). However, the value of 0.15 should not hold as an inflexible threshold, and higher cutoff values of Vn/n+1 have been used in several reports [30–32]. The low variation between V2/3 and V3/4 in all of the samples (Fig. 4) indicated that two reference genes are sufficient for normalization.
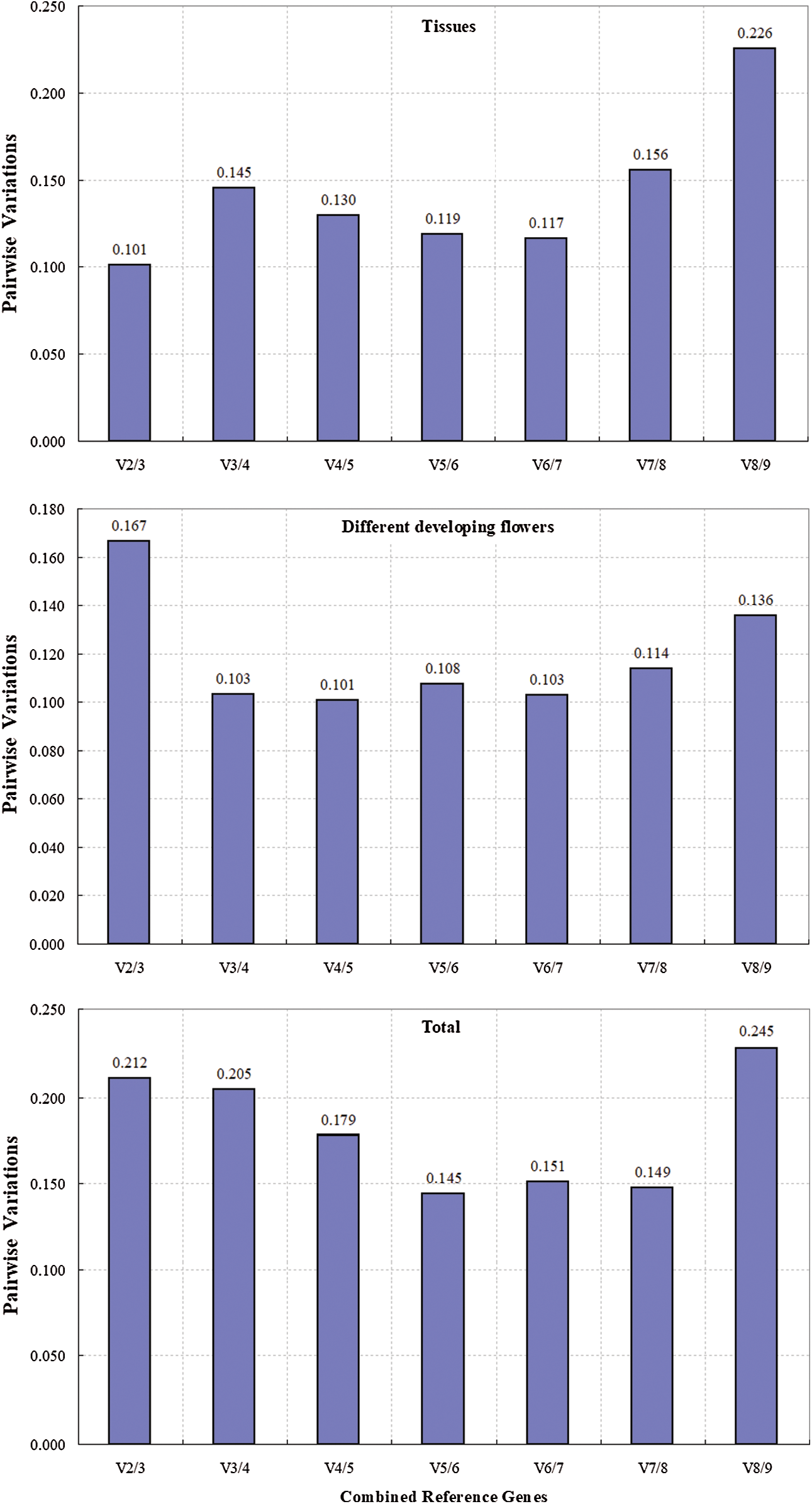
Figure 4: Pairwise variation (V) of 9 candidate reference genes were calculated using GeNorm. Vn/Vn+1 value were used to determine the optimal number of reference genes
Evaluation values detected by the NormFinder algorithm are shown in Tab. 2, with lower values indicating higher stability [34]. ACT11 and UBQ were ranked as the most stable genes in different tissues. TUA and UBC9 were identified as the most stable reference genes in the four developmental stages of flower. According to NormFinder analysis, UBC9 and ACT11 performed as the most stable genes in all of the samples. The ranking order generated by this method was somewhat similar to that determined by GeNorm.
Table 2: Expression stability analysis of 9 candidate reference genes calculated using NormFinder software
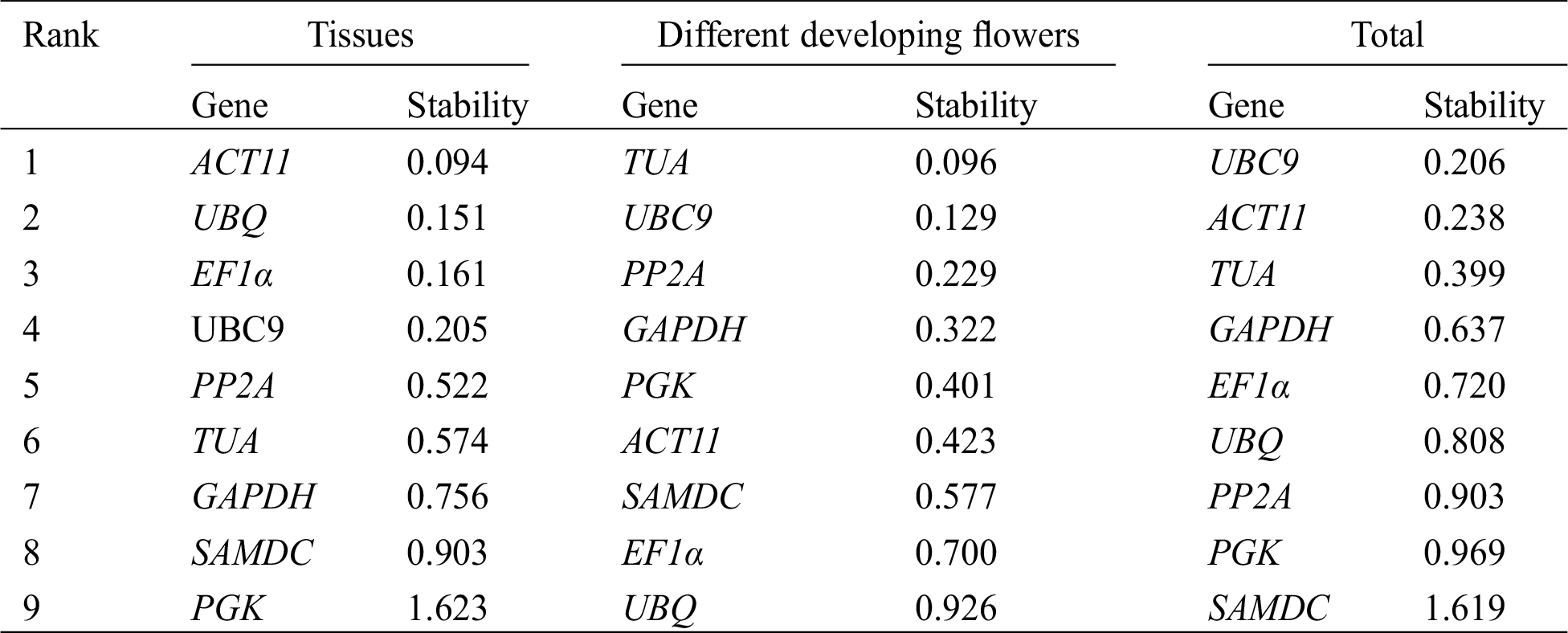
The stability of 9 reference genes was also evaluated by BestKeeper algorithm based on SD and CV of the Cq values, with lower SD and CV representing higher stability [22]. Tab. 3 shows the results of the BestKeeper analysis. In different tissues, TUA and ACT11 were identified as the most stable genes by BestKeeper analysis, whereas TUA was ranked fifth by GeNorm and sixth by NormFinder. In the four developmental stages of flowers, TUA was identified as the most stable reference gene according to BestKeeper analysis, which was similar to the result by GeNorm and NormFinder analysis. In all of the samples, TUA and UBC9 were identified as the most stable reference genes by BestKeeper, whereas TUA was ranked fifth by GeNorm and third by NormFinder.
Table 3: Expression stability analysis of 9 candidate reference genes calculated using BestKeeper software
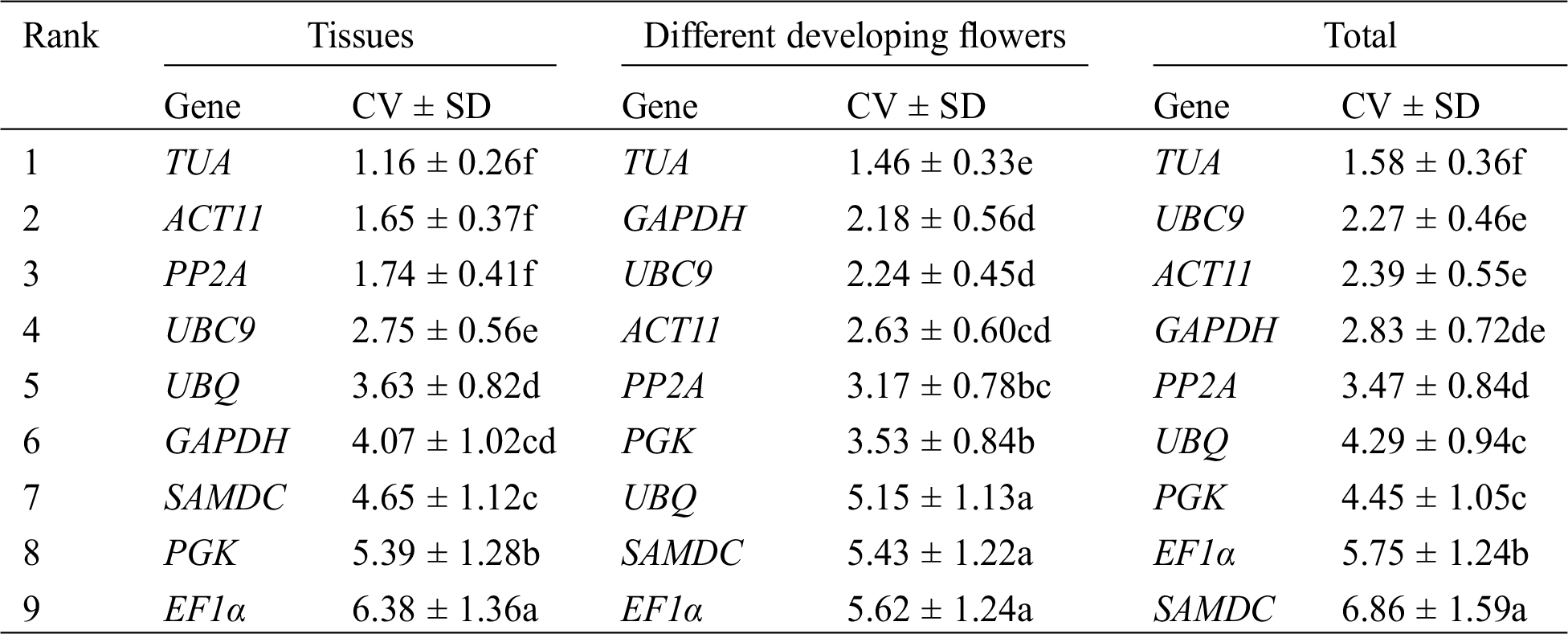
Finally, Ref-Finder was used to calculate the integrated ranking of the most stable reference genes by GeNorm, NormFinder, and BestKeeper (Tab. 4). Through Ref-Finder analysis, ACT11 and EF1α were the most stable reference genes in different tissues. TUA and UBC9 showed the two highest rankings for the four developmental stages of flowers. UBC9, ACT11, and TUA were found to be three most stable reference genes in all of the samples, whereas SAMDC was the least stable gene in all of the samples. PGK and UBQ was the least stable gene in three different tissues and the four developmental stages of flowers, respectively.
Table 4: Expression stability ranking of 9 candidate reference genes
3.3 Validation of Reference Genes
To confirm the reliability of reference genes, the expression patterns of IgTPS were evaluated using different reference genes in different tissues and four developmental stages of flowers. The two most stable genes (ACT11 and EF1a for different tissues, TUA and UBC9 for the different l stages) and the least stable reference gene (PGK for different tissues, UBQ for the different flower stages) were used in the validation test. When ACT11 or EF1a was used as the reference gene, IgTPS expression was highest in leaves among the different tissues, whereas the highest expression level was detected in stems when PGK was used as a reference (Fig. 5A). For the four developmental stages of flowers, IgTPS expression normalized by TUA or UBC9 was highest in S3 and second highest in S4, but that normalized by UBQ was highest in S4 (Fig. 5B). These results showed that the unstable genes PGK and UBQ failed to effectively standardize the expression data. These results indicate that the accuracy of qRT-PCR analysis could be altered by using different reference genes.
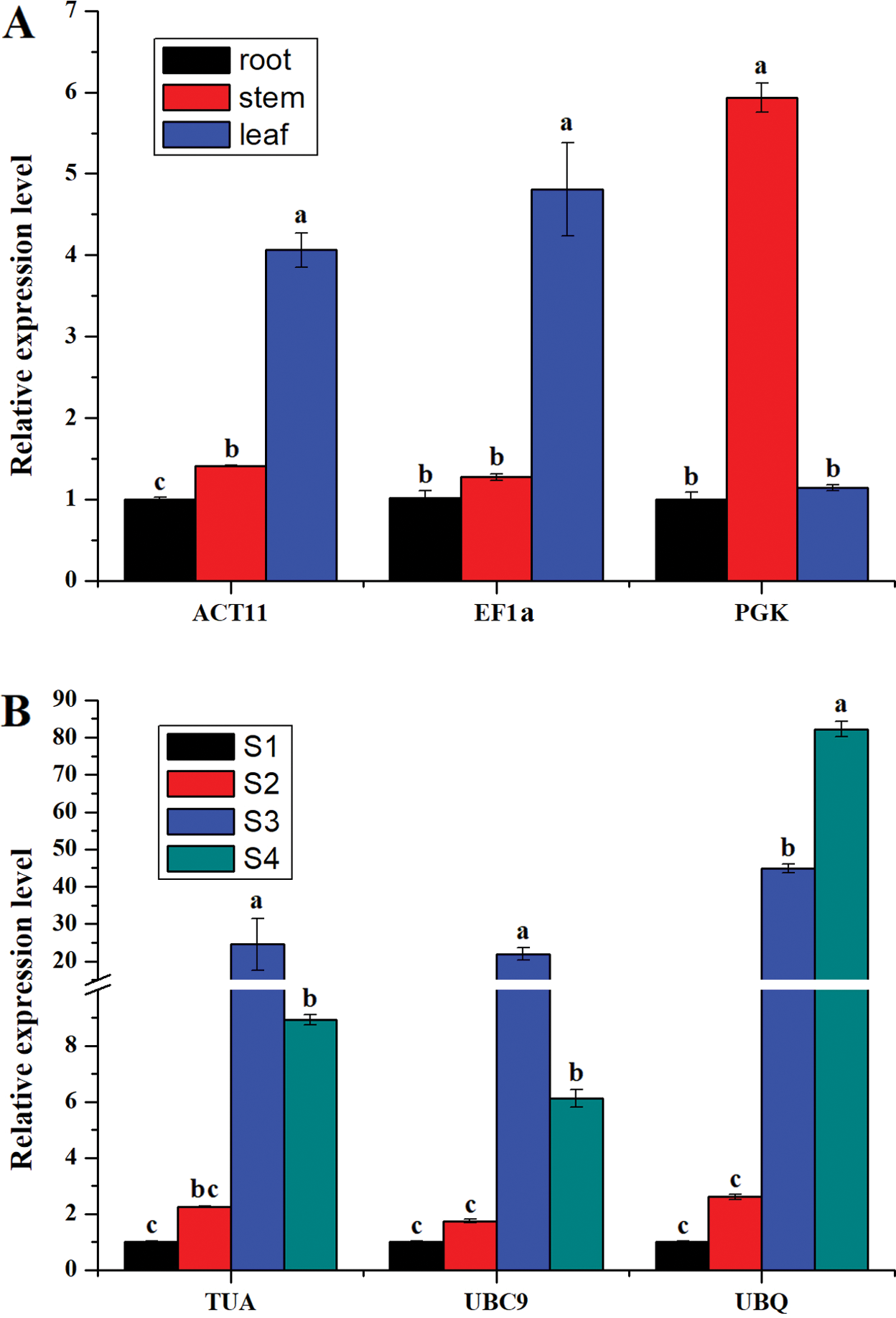
Figure 5: Relative quantification of IgTPS gene expression for flower development samples (A) and different organs samples (B) using validated reference genes, including the most and the least stable reference genes for normalization
Although powerful modern technologies, including microarrays and high-throughput sequencing have been used to test gene expression levels, qRT-PCR is still widely used for this purpose owing to its sensitivity, accuracy, and efficiency [34]. Selecting the appropriate reference genes is a fundamental prerequisite for reliable qRT-PCR analysis. The ideal reference genes should have stable expression in different tissues, organs, developmental stages, and treatments [35]. The selection of reference genes that are stably expressed in different samples can improve the accuracy and reliability of qRT-PCR results [14].
GeNorm, NormFinder, BestKeeper, Ref-Finder and delta Ct have been developed to estimate the expression stability of reference genes, but the rankings of most reference genes differ among different programs [36]. In this study, we used four different statistical algorithms, GeNorm, NormFinder, BestKeeper and Ref-Finder, to analyze the stability of candidate reference genes. The gene stability rankings created by GeNorm were similar to those of NormFinder, but they showed quite differences from the ranking obtained with BestKeeper. ACT11 and UBC9 were identified as the most stable reference genes in all of the samples by GeNorm and NormFinder. TUA ranked in the top two stable genes by BestKeeper, but was ranked fifth by GeNorm and third by NormFinder. The results are similar to many previous studies of different species such as were identified as Cynodon dactylon [31], Chrysanthemum morifolium [12], Lilium regale [37], and Tolypocladium guangdongense [38]. Finally, Ref-Finder was used to combine and validate the results. There are some differences among algorithms, but they are not obvious.
In this study, the stability of 9 reference genes in different tissues and different developmental stages of flowers were analyzed. Our study demonstrated that UBC9, ACT11 and TUA were good genes for normalization when considering all of the samples. Similar results have been obtained in Eucommia ulmoides Oliver [39], Malus domestica [40], Fragaria vesca [41], and Annona muricata L. [42]. GAPDH and PP2A were identified as the most stable reference genes for normalization in seedlings of Robinia pseudoacacia L. [43], in tissues of different developmental stages in Gentiana macrophylla [44], and in flower development and different organs of Primula forbesii [45]. However, these two genes did not perform well in our study. PGK plays an important role in glycolytic pathway [46], and it has been widely used as a reliable reference gene by qRT-PCR in Iris. lacteal var. chinensis [25] and chrysanthemum [12], while this gene was the least stable reference gene in different tissues in our study. Furthermore, UBQ was reported to be one of the most stable reference genes in Vitis vinifera L. [47]; however, UBQ exhibited the least stable expression in different developing flowers in this study, and similar studies were found in Apium graveolens [48]. In our study, SAMDC was ranked least in all of the samples. Consistent with the result in Hordeum [49], however, SAMDC was appropriate for gene normalization in Neolamarckia cadamba [50]. In our study, EF1α only performed well in different tissues, which was similar to finding in Betula platyphylla [51]. Therefore, these results suggested that different reference genes are required depending on the plant species, tissue types and developmental stages.
Stability of reference genes was further validated by checking the expression patterns analysis of a target gene, IgTPS, in different tissues and different flower samples. The expression patterns of IgTPS gene showed similar trends when the two most stable reference genes ACT11/EF1a or TUA/UBC9 were used as internal controls. However, severe disparities occurred when the least stable gene PGK or UBQ was used for normalization. These results suggest that selecting suitable internal control genes is critically important for the reliable normalization of target gene expression data by qRT-PCR.
To our knowledge, this is the first systematic research to validate candidate reference genes for the normalization of gene expression data using qRT-PCR in different tissues or developmental stages of flowers in I. germanica. Nine candidate reference genes were tested and their stability was assessed by GeNorm, NormFinder, BestKeeper, and Ref-Finder. Results revealed that UBC9, ACT11, and TUA were identified as the most suitable reference genes in all of the samples, while SAMDC showed the least stability. This work will provide useful information for reliable qRT-PCR data normalization in I. germanica gene expression studies.
Funding Statement: The work was supported by the National Natural Science Foundation of China (31801901), the Natural Science Foundation of Jiangsu (BK20180314), the Foundation of Key Laboratory of Landscaping (KF201901), Ministry of Agriculture and Rural Affairs, China, and the Jiangsu Key Laboratory for the Research and Utilization of Plant Resources (JSPKLB201814).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Artico, S., Nardeli, S. M., Brilhante, O., Grossi-de-Sa, M. F., Alves-Ferreira, M. (2010). Identification and evaluation of new reference genes in Gossypium hirsutum for accurate normalization of real-time quantitative RT-PCR data. BMC Plant Biology, 10(1), 49–60. DOI 10.1186/1471-2229-10-49.
2. Liu, Y., Liu, J., Xu, L., Lai, H., Chen, Y. et al. (2017). Identification and validation of reference genes for Seashore paspalum response to abiotic stresses. International Journal of Molecular Sciences, 18(6), 1322. DOI 10.3390/ijms18061322. [Google Scholar] [CrossRef]
3. Kozera, B., Rapacz, M. (2013). Reference genes in real-time PCR. Journal of Applied Genetics, 54(4), 391–406. DOI 10.1007/s13353-013-0173-x. [Google Scholar] [CrossRef]
4. Xiao, X., Ma, J., Wang, J., Wu, X., Li, P. et al. (2014). Validation of suitable reference genes for gene expression analysis in the halophyte Salicornia europaea by real-time quantitative PCR. Frontiers in Plant Science, 5, 788. DOI 10.3389/fpls.2014.00788.
5. Delporte, M., Legrand, G., Hilbert, J. L., Gagneul, D. (2015). Selection and validation of reference genes for quantitative real-time PCR analysis of gene expression in Cichorium intybus. Frontiers in Plant Science, 6(e91474), 651. DOI 10.3389/fpls.2015.00651.
6. Wang, C., Cui, H. M., Huang, T. H., Liu, T. K., Hou, X. L. et al. (2016). Identification and validation of reference genes for RT-qPCR analysis in non-heading Chinese cabbage flowers. Frontiers in Plant Science, 7(651), 811. DOI 10.3389/fpls.2016.00811.
7. Bao, W. L., Qu, Y. L., Shan, X. Y., Wan, Y. L. (2016). Screening and validation of housekeeping genes of the root and cotyledon of Cunninghamia lanceolata under abiotic stresses by using quantitative real-time PCR. International Journal of Molecular Sciences, 17(8), 1198. DOI 10.3390/ijms17081198. [Google Scholar] [CrossRef]
8. Zhang, Y. X., Han, X. J., Chen, S. S., Zheng, L., He, X. L. et al. (2017). Selection of suitable reference genes for quantitative real-time PCR gene expression analysis in Salix matsudana under different abiotic stresses. Scientific Reports, 7(1), 42290. DOI 10.1038/srep40290. [Google Scholar] [CrossRef]
9. Tang, F., Chu, L. W., Shu, W. B., He, X. J., Wang, L. J. et al. (2019). Selection and validation of reference genes for quantitative expression analysis of miRNAs and mRNAs in Poplar. Plant Methods, 15(1), 35. DOI 10.1186/s13007-019-0420-1. [Google Scholar] [CrossRef]
10. Jin, X. H., Fu, J. X., Dai, S. L., Sun, Y., Hong, Y. (2013). Reference gene selection for qPCR analysis in cineraria developing flowers. Scientia Horticulturae, 153, 64–70. DOI 10.1016/j.scienta.2013.01.023. [Google Scholar] [CrossRef]
11. Meng, Y., Li, N., Tian, J., Gao, J., Zhang, C. (2013). Identification and validation of reference genes for gene expression studies in postharvest rose flower (Rosa hybrida). Scientia Horticulturae, 158, 16–21. DOI 10.1016/j.scienta.2013.04.019. [Google Scholar] [CrossRef]
12. Qi, S., Yang, L. W., Wen, X. H., Yan, H., Song, X. B. et al. (2016). Reference gene selection for RT-qPCR analysis of flower development in Chrysanthemum morifolium and Chrysanthemum lavandulifolium. Frontiers in Plant Science, 7(651), 287. DOI 10.3389/fpls.2016.00287. [Google Scholar] [CrossRef]
13. Li, J., Han, J. G., Hu, Y. H., Yang, J. (2016). Selection of reference genes for quantitative real-time PCR during flower development in tree peony (Paeonia suffruticosa Andr.). Frontiers in Plant Science, 7(1259), 516. DOI 10.3389/fpls.2016.00516. [Google Scholar] [CrossRef]
14. Xiao, Z., Sun, X. B., Liu, X. Q., Li, C., He, L. S. et al. (2016). Selection of reliable reference genes for gene expression studies on Rhododendron molle G. Don. Frontiers in Plant Science, 7, e0141853. [Google Scholar]
15. Sgamma, T., Pape, J., Massiah, A., Jackson, S. (2016). Selection of reference genes for diurnal and developmental time-course real-time PCR expression analyses in lettuce. Plant Methods, 12(1), 21. DOI 10.1186/s13007-016-0121-y. [Google Scholar] [CrossRef]
16. Mavrodiev, E. V., Martínez-Azorín, M., Dranishnikov, P., Crespo, M. B. (2014). At least 23 genera instead of one: the case of Iris L. sl (Iridaceae). PLoS One, 9(8), e106459. DOI 10.1371/journal.pone.0106459. [Google Scholar] [CrossRef]
17. Tehrani, M. M., Nasr-Esfahani, M., Mousavi, A., Mortezaiinezhad, F., Azim, H. (2020). Regulation of related genes promoting resistant in Iris against root rot disease, Fusarium oxysporum f. sp. gladioli. Genomics, 112(5), 3013–3020. DOI 10.1016/j.ygeno.2020.05.013. [Google Scholar] [CrossRef]
18. Wang, H., Talavera, M., Min, Y., Flaven, E., Imbert, E. (2016). Neutral processes contribute to patterns of spatial variation for flower colour in the Mediterranean Iris lutescens (Iridaceae). Annals of Botany, 117(6), 995–1007. DOI 10.1093/aob/mcw036. [Google Scholar] [CrossRef]
19. Pellegrino, G., Bellusci, F., Palermo, A. M. (2016). Who helps whom? Pollination strategy of Iris tuberosa and its relationship with a sexually deceptive orchid. Journal of Plant Research, 129(6), 1051–1059. DOI 10.1007/s10265-016-0853-9. [Google Scholar] [CrossRef]
20. Yuan, Y., Sun, Y., Zhao, Y. C., Liu, C. G., Chen, X. L. et al. (2019). Identification of floral scent profiles in bearded irises. Molecules, 24(9), 1773. DOI 10.3390/molecules24091773. [Google Scholar] [CrossRef]
21. Vandesompele, J., DePreter, K., Pattyn, F., Poppe, B., VanRoy, N. et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology, 3(7). DOI 10.1186/gb-2002-3-7-research0034. [Google Scholar] [CrossRef]
22. Pfaffl, M. W., Tichopad, A., Prgomet, C., Neuvians, T. P. (2004). Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: bestkeeper-Excel-based tool using pair-wise correlations. Biotechnology Letters, 26(6), 509–515. DOI 10.1023/B:BILE.0000019559.84305.47. [Google Scholar] [CrossRef]
23. Brunner, A. M., Yakovlev, I. A., Strauss, S. H. (2004). Validating internal controls for quantitative plant gene expression studies. BMC Plant Biology, 4(1), 14. DOI 10.1186/1471-2229-4-14. [Google Scholar] [CrossRef]
24. Köllner, T. G., Held, M., Lenk, C., Hiltpold, I., Turlings, T. C. J. et al. (2008). A maize (E)-β-Caryophyllene synthase implicated in indirect defense responses against herbivores is not expressed in most American maize varieties. Plant Cell, 20(2), 482–494. DOI 10.1105/tpc.107.051672. [Google Scholar] [CrossRef]
25. Gu, C. S., Liu, L. Q., Deng, Y. M., Zhu, X. D., Lu, X. Q. et al. (2014). Validation of reference genes for RT-qPCR normalization in Iris. lacteavar. chinensis leaves under different experimental conditions. Scientia Horticulturae, 175, 144–149. DOI 10.1016/j.scienta.2014.06.011. [Google Scholar] [CrossRef]
26. Gu, G. S., Liu, L. Q., Deng, Y. M., Zhang, Y. X., Wang, Z. Q. et al. (2017). De novo characterization of the Iris lactea var. chinensis transcriptome and an analysis of genes under cadmium or lead exposure. Ecotoxicology and Environmental Safety, 144, 507–513. DOI 10.1016/j.ecoenv.2017.06.071. [Google Scholar] [CrossRef]
27. Ramakers, C., Ruijter, J., Deprez, R., Moorman, A. (2003). Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neuroscience Letters, 339(1), 62–66. DOI 10.1016/S0304-3940(02)01423-4. [Google Scholar] [CrossRef]
28. Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-∆∆Ct method. Methods, 25(4), 402–408. DOI 10.1006/meth.2001.1262. [Google Scholar] [CrossRef]
29. Gimeno, J., Eattock, N., Deynze, A. V., Blumwald, E. (2014). Selection and validation of reference genes for gene expression analysis in Switchgrass (Panicum virgatum) using quantitative real-time RT-PCR. PLoS One, 9(3), e91474. DOI 10.1371/journal.pone.0091474. [Google Scholar] [CrossRef]
30. Silveira, É. D., Alves-Ferreira, M., Guimarães, L. A., da Silva, F. R., Carneiro, V. T. (2009). Selection of reference genes for quantitative real-time PCR expression studies in the apomictic and sexual grass Brachiaria brizantha. BMC Plant Biology, 9(1), 84. DOI 10.1186/1471-2229-9-84. [Google Scholar] [CrossRef]
31. Marum, L., Miguel, A., Ricardo, C. P., Miguel, C., Wu, S. B. (2012). Reference gene selection for quantitative real-time PCR normalization in Quercus suber. PLoS One, 7(4), e35113. DOI 10.1371/journal.pone.0035113. [Google Scholar] [CrossRef]
32. Chen, Y., Tan, Z. Q., Hu, B. Y., Yang, Z. M., Xu, B. et al. (2015). Selection and validation of reference genes for target gene analysis with quantitative RT-PCR in leaves and roots of bermudagrass under four different abiotic stresses. Physiologia Plantarum, 155(2), 138–148. DOI 10.1111/ppl.12302. [Google Scholar] [CrossRef]
33. Andersen, C. L., Jensen, J. L., Ørntoft, T. F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Research, 64(15), 5245–5250. DOI 10.1158/0008-5472.CAN-04-0496.
34. Nicot, N., Hausman, J. F., Hoffmann, L., Evers, D. (2005). Reference gene selection for RT-qPCR normalization in potato during biotic and abiotic stress. Journal of Experimental Botany, 56(421), 2907–2914. DOI 10.1093/jxb/eri285. [Google Scholar] [CrossRef]
35. Jacobsen, K. S., Nielsen, K. O., Winther, T. N., Glebe, D., Pociot, F. et al. (2016). Identification of valid reference genes for microRNA expression studies in a hepatitis B virus replicating liver cell line. BMC Research Notes, 9(1), 38. DOI 10.1186/s13104-016-1848-2. [Google Scholar] [CrossRef]
36. Huggett, J., Dheda, K., Bustin, S., Zumla, A. (2005). Real-time RT-PCR normalisation; strategies and considerations. Genes & Immunity, 6(4), 279–284. DOI 10.1038/sj.gene.6364190. [Google Scholar] [CrossRef]
37. Arya, S. K., Jain, G., Upadhyay, S. K., Sarita, Y., Singh, H. et al. (2017). Reference genes validation in Phenacoccus solenopsis under various biotic and abiotic stress conditions. Scientific Reports, 7, 13520. DOI 10.1038/s41598-017-13925-9. [Google Scholar] [CrossRef]
38. Du, W. K., Hu, F. R., Yuan, S. X., Liu, C. (2019). Selection of reference genes for quantitative real-time PCR analysis of photosynthesis-related genes expression in Lilium regale. Physiology and Molecular Biology of Plants, 25(6), 1497–1506. DOI 10.1007/s12298-019-00707-y. [Google Scholar] [CrossRef]
39. Wang, G. Z., Cheng, H. J., Li, M., Zhang, C. H., Deng, W. Q. et al. (2020). Selection and validation of reliable reference genes for Tolypocladium guangdongense gene expression analysis under differentially developmental stages and temperature stresses. Gene, 734, 144380. DOI 10.1016/j.gene.2020.144380. [Google Scholar] [CrossRef]
40. Ye, J., Jin, C. F., Li, N., Liu, M. H., Fei, Z. X. et al. (2018). Selection of suitable reference genes for qRT-PCR normalisation under different experimental conditions in Eucommia ulmoides Oliv. Scientific Reports, 8(1), 15043. DOI 10.1038/s41598-018-33342-w. [Google Scholar] [CrossRef]
41. Zhu, L. F., Yang, C. Q., You, Y. H., Liang, W., Wang, N. N. et al. (2019). Validation of reference genes for qRT-PCR analysis in peel and flesh of six apple cultivars (Malus domestica) at diverse stages of fruit development. Scientia Horticulturae, 244, 165–171. DOI 10.1016/j.scienta.2018.09.033. [Google Scholar] [CrossRef]
42. Liu, D. C., Huang, X. R., Lin, Y., Wang, X. J., Yan, Z. M. et al. (2020). Identification of reference genes for transcript normalization in various tissue types and seedlings subjected to different abiotic stresses of woodland strawberry Fragaria vesca. Scientia Horticulturae, 261, 108840. DOI 10.1016/j.scienta.2019.108840. [Google Scholar] [CrossRef]
43. Berumen-Varela, G., Palomino-Hermosillo, Y. A., Bautista-Rosales, P. U., Peña-Sandoval, G. R., López-Gúzman, G. G. et al. (2020). Identification of reference genes for quantitative real-time PCR in different developmental stages and under refrigeration conditions in soursop fruits (Annona muricata L.). Scientia Horticulturae, 260, 108893. DOI 10.1016/j.scienta.2019.108893. [Google Scholar] [CrossRef]
44. Wang, J. X., Abbas, M., Wen, Y. Z., Niu, D. S., Wang, L. et al. (2018). Selection and validation of reference genes for quantitative gene expression analyses in black locust (Robinia pseudoacacia L.) using real-time quantitative PCR. PLoS One, 13(3), e0193076. DOI 10.1371/journal.pone.0193076. [Google Scholar] [CrossRef]
45. He, Y. H., Yan, H. L., Hua, W. P., Huang, Y. Y., Wang, Z. Z. (2016). Selection and validation of reference genes for quantitative real-time PCR in Gentiana macrophylla. Frontiers in Plant Science, 07(619), 945. DOI 10.3389/fpls.2016.00945. [Google Scholar] [CrossRef]
46. Jia, Y., Liu, S. C., Zhao, J., Chen, X. X., Sun, X. L. et al. (2020). Reference gene selection and validation by qRT-PCR during flower development and in different organs of Primula forbesii. Journal of Horticultural Science and Biotechnology, 95(3), 383–394. DOI 10.1080/14620316.2019.1681909. [Google Scholar] [CrossRef]
47. Anderson, L. E., Carol, A. A. (2005). Enzyme co-localization in the pea leaf cytosol: 3-P-glycerate kinase, glyceraldehyde-3-P dehydrogenase, triose-P isomerase and aldolase. Plant Science, 169(3), 620–628. DOI 10.1016/j.plantsci.2005.05.012. [Google Scholar] [CrossRef]
48. Luo, M., Gao, Z., Li, H., Li, Q., Zhang, C. X. et al. (2018). Selection of reference genes for miRNA qRT-PCR under abiotic stress in grapevine. Scientific Reports, 8(1), 4444. DOI 10.1038/s41598-018-22743-6. [Google Scholar] [CrossRef]
49. Li, M. Y., Wang, F., Jiang, Q., Wang, G., L., Tian, C. et al. (2016). Validation and comparison of reference genes for qPCR normalization of celery (Apium graveolens) at different development stages. Frontiers in Plant Science, 7, 313. DOI 10.3389/fpls.2016.00313. [Google Scholar] [CrossRef]
50. Cai, J., Li, P. F., Luo, X., Chang, T. L., Li, J. X. et al. (2018). Selection of appropriate reference genes for the detection of rhythmic gene expression via quantitative real-time PCR in Tibetan hulless barley. PLoS One, 13(1), e0190559. DOI 10.1371/journal.pone.0190559. [Google Scholar] [CrossRef]
51. Huang, T., Long, J. M., Liu, W. S., Yang, Z. W., Zhu, Q. J. et al. (2018). Selection and validation of reference genes for mRNA expression by quantitative Real-Time PCR analysis in Neolamarckia cadamba. Scientific Reports, 8(1), 9311. DOI 10.1038/s41598-018-27633-5. [Google Scholar] [CrossRef]
52. Li, Z. Y., Lu, H. J., He, Z. H., Wang, C., Wang, Y. C. et al. (2019). Selection of appropriate reference genes for quantitative real-time reverse transcription PCR in Betula platyphylla under salt and osmotic stress conditions. PLoS One, 14(12), e0225926.
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |