DOI:10.32604/phyton.2020.013933

| Phyton-International Journal of Experimental Botany DOI:10.32604/phyton.2020.013933 |  |
| Article |
1Key Laboratory of Plant Resources Conservation and Germplasm Innovation in Mountainous Region, Ministry of Education, Institute of Agro-Bioengineering and College of Life Sciences, Guizhou University, Guiyang, 550025, China
2Guiyang Station for DUS Testing Center of New Plant Varieties of China in Guizhou Academy of Agricultural Sciences, Guiyang, 550006, China
*Corresponding Authors: Xiaofang Zeng. Email: zengxiaofang@foxmail.com; Degang Zhao. Email: degangzhao@yahoo.com
Received: 26 August 2020; Accepted: 28 September 2020
Abstract: Plant architecture is a vital agronomic trait to control yield in rice (Oryza sativa L.). A dwarf and small seed 1 (dss1) mutant were obtained from the ethyl methanesulfonate (EMS) mutagenized progeny of a Guizhou glutinous landrace cultivar, Lipingzabianhe. The dss1 mutant displayed phenotypes similar to those of brassinosteroid (BR) deficient mutants, such as dwarfing, dark green and rugose erect leaves, small seeds, and loner neck internode panicles with primary branching. In our previous study, the underlying DSS1 gene was isolated, a novel allele of OsDWARF (OsBR6ox) that encodes a cytochrome P450 protein involved in the BR biosynthetic pathway by MutMap technology. In this work, we confirmed that a Thr335Ile amino acid substitution residing in DSS1/OsDWARF was responsible for the dwarf, panicle architecture, and small seed phenotypes in the dss1 mutants by genetic transformation experiments. The overexpression of OsDWARF in the dss1 mutant background could not only recover dss1 to the normal plant height and panicle architecture but also rescued normal leaf angles, seed size, and leaf color. Thus, the specific mutation in DSS1/OsDWARF influenced plant architecture, seed size, and chlorophyll biosynthesis.
Keywords: Brassinosteroid; chlorophyll biosynthesis; dss1 mutant; OsDWARF gene; panicle architecture
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |
Rice (Oryza sativa L.), is one of a vital food crops, and feed more than 21% of the world’s population [1]. Rice plant architecture contained plant height, tilling number, tilling angle, leaf angle, and panicle morphology [2]. Among them, branching pattern, seed size and plant height are key agronomic traits that affect grain yield and, therefore, are the major purpose of rice yield improvement programs [3]. Several rice important plant architecture genes, including the Ghd7, Ghd8, OsSPL14, OsDEP1, OsCPB1, LA1, LPA1, and OsNAC2, are involved in controlling plant height, tiller angle, panicle morphology, seed size and tilling [3–8].
Brassinosteroids (BRs), a class of steroid-type phytohormones, play vital roles in plant growth and development, such as cell elongation and division, leaf angle formation, panicle morphology modulation, senescence, photomorphogenesis and grain size [9–13]. BRs play key roles in regulating the yield determination agronomic trait such as plant height, leaf angle and gain size etc. in rice [14]. In rice, loss-of-function mutants with deficiencies in BR biosynthesis or perception, including d61, d2, d11 and brd1, exhibit dwarf phenotypes, dark-green and epinastic leaves, reduced apical dominance, altered vascular patterning, male sterility and small seeds [10,15–17]. Evidence indicates that the optimized expression of BR-related genes by the DEP1 promoter could improve grain size and yield in rice [7,18].
BR synthesis from campesterol can be divided into three steps: (1) Formation of campestanol from campesterol; (2) Two parallel pathways from campestanol to castasterone (CS); and (3) The final conversion of CS into 24-epibrassinolide (BL) by the lactonization of the B-ring. OsDWARF (OsBR6ox), is a key gene involved in the BR biosynthetic pathway. The product of OsDWARF catalyzes a late step in bioactive BR synthesis, as a combination between the early and late C-6 oxidation pathways, and encodes BR-6-oxidase that may catalyze 6-deoxo typhasterol (TY) and 6-deoxoCS into TY and CS, respectively [10,11]. The loss of OsDWARF function leads to the reduction of TY and CS, which causes the BR-deficient phenotype.
The creation and identification of plant height mutants and then isolating their regulatory genes are important for understanding the mechanism of rice growth and development, and for revealing the molecular regulatory mechanisms of rice plant height. These processes can also provide new germplasm to generate ideal rice types. In our previous study, Lipingzabianhe (LPZBH, Oryza sativa L. ssp. japonica) a glutinous rice landrace from Guizhou, China was treated with EMS to construct mutant libraries. A rice stably inherited BR-deficient mutant, dwarf and small seed1 (dss1), which was characterized as having erect, dark green leaves and small round grains, was isolated. In our previous work, we found the OsDWARF, encoding a cytochrome P450 protein involved in the BR biosynthetic pathway on chromosome 3 was one of the candidate gene for the dss1 mutant, by employing the Mutmap method [19]. Here we further confirmed and revealed that a specific modification in the DSS1/OsDWARF protein sequence simultaneously altered the development of panicle architecture, plant height, and seed size.
2.1 Plant Materials and Growth Conditions
The dss1 mutant was isolated from the M2 generation of LPZBH by EMS treatment. The wild type (WT) and dss1 mutant seeds were surface sterilized with 0.3% (w/v) NaClO for 30 min, soaked in water for 3 d at 37°C, and then placed in a seedbed for 20 d and grown in the paddy field of Guizhou University, Guiyang, Guizhou Province, China, in the summer and Sanya, Hainan Province, China, in the winter. The mutants were differentiated from normal segregants by the dwarf phenotype. The following morphological characteristics were measured.
To analyze the anatomical features of the uppermost internode, the middle parts of the internodes were fixed in FAA (formalin: Glacial acetic acid: 70% ethanol, 1:1:18), and dehydrated through a graded ethanol series using an automatic dehydrator TP1020 (Leica, Germany). After substitution with xylene, the samples were embedded in paraffin wax in a Paraffin-embedding station EG1150 (Leica, Germany) and sectioned at 10 μm using a rotary microtome. Sections were de-paraffinized in xylene, dehydrated through a graded ethanol series, and then stained with 0.05% toluidine blue O and observed with a light microscope (Leica Microsystems, Germany).
For the root and coleoptile investigation, the seeds were sterilized with 0.3% (w/v) NaCl and germinated in 1/2 Murashige and Skoog (MS) medium (0.6% agar w/v) plates containing a range of 24-epibrassinolide (BL) concentrations. For the lamina joint and root test, the second leaf laminas of 7-day-old seedlings were excised and submerged in water containing BL for 7 d. For the coleoptile and root test, the rice seeds were germinated in a dark chamber at 30°C for 3 d.
2.4 Pigment and Chlorophyll (Chl) Precursor Determination
Pigments were extracted from fresh leaf tissues with 80% acetone. The extract was measured with a spectrophotometer at 470, 645, and 663 nm (Varioskan Flash, Thermo Fisher, USA). Total Chl, Chl a, and Chl b contents were determined as described by Arnon [20]. The determination of the ALA content was based on the methods of Morton [21]. The precursors of the Chl biosynthetic pathway, including Proto IX, Mg-Proto IX, Pchlide, and Chlide, were assayed as described by Santiago-Ong et al. [22] and Masuda et al. [23]. Leaves (approximately 100 mg fresh weight) of the WT and dss1 mutant were cut and homogenized in 5 mL 9:1 acetone: 0.1 M NH4OH, and centrifuged at 3,000 g for 10 min. The supernatants were combined and washed successively with an equal volume of hexane three times before spectrophotometric analysis. Chl precursors in the acetone phase were quantified with a Varioskan Flash (Thermo Fisher, USA) using Ex400:Em632 for Proto IX, Ex440:Em633 for Pchlide, Ex440:Em672 for Chlide and Ex420:Em595 for Mg-Proto.
2.5 Plasmid Construction and Rice Transformation
For the construction of the overexpression vector, fragments containing the cauliflower mosaic virus 35S (CaMV 35S) promoter, the whole-coding region of OsDWARF, and the terminator of the 10-kDa prolamine gene were amplified by chemical synthesis and inserted into the pCambia1301 binary vector. The construct was introduced into dss1 mutants using the Agrobacterium tumefaciens-mediated genetic transformation method as described by Zeng et al. [24].
RNA samples were collected from the 4 weeks old dss1 and LPZBH. Total RNA was extracted from leaves using an RNA extraction kit (Omega Bio-Tek, Doraville, GA, USA) according to the Plant RNA Protocol II (for difficult samples). cDNA libraries were constructed and sequenced on the BGISEQ-500 platform at the Beijing Genomics Institute (www.genomics.org.cn, BGI, Shenzhen, China). Three biological replicates were prepared for each sequencing library. The DEGseq method was used to screen DEGs between groups [25]. DEGs were selected according to the following default criteria: Fold Change ≥ 2 and Adjusted P-value ≤ 0.001. Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) metabolic pathways annotation and enrichment analyses were conducted as described by GO and KEGG databases respectively.
Total RNA was extracted from leaves using an RNA extraction kit (Omega Bio-Tek, Doraville, GA, USA) according to the Plant RNA Protocol II (for difficult samples). The Reverse transcription and qRT-PCR were perform as described by Livik et al. [26]. Threshold cycles (Ct) for gene expression were standardized to OsActin1 Ct (−ΔCt). The relative expression levels of genes were determined using the 2−ΔΔCt calculation method [26]. The real-time PCR primer were listed in Tab. S1.
The statistical significance of differences in the mean values of the examined parameters between WT and dss1 were determined using the Student t-test function in software of Microsoft Excel software and SPSS PASW statistics 18.
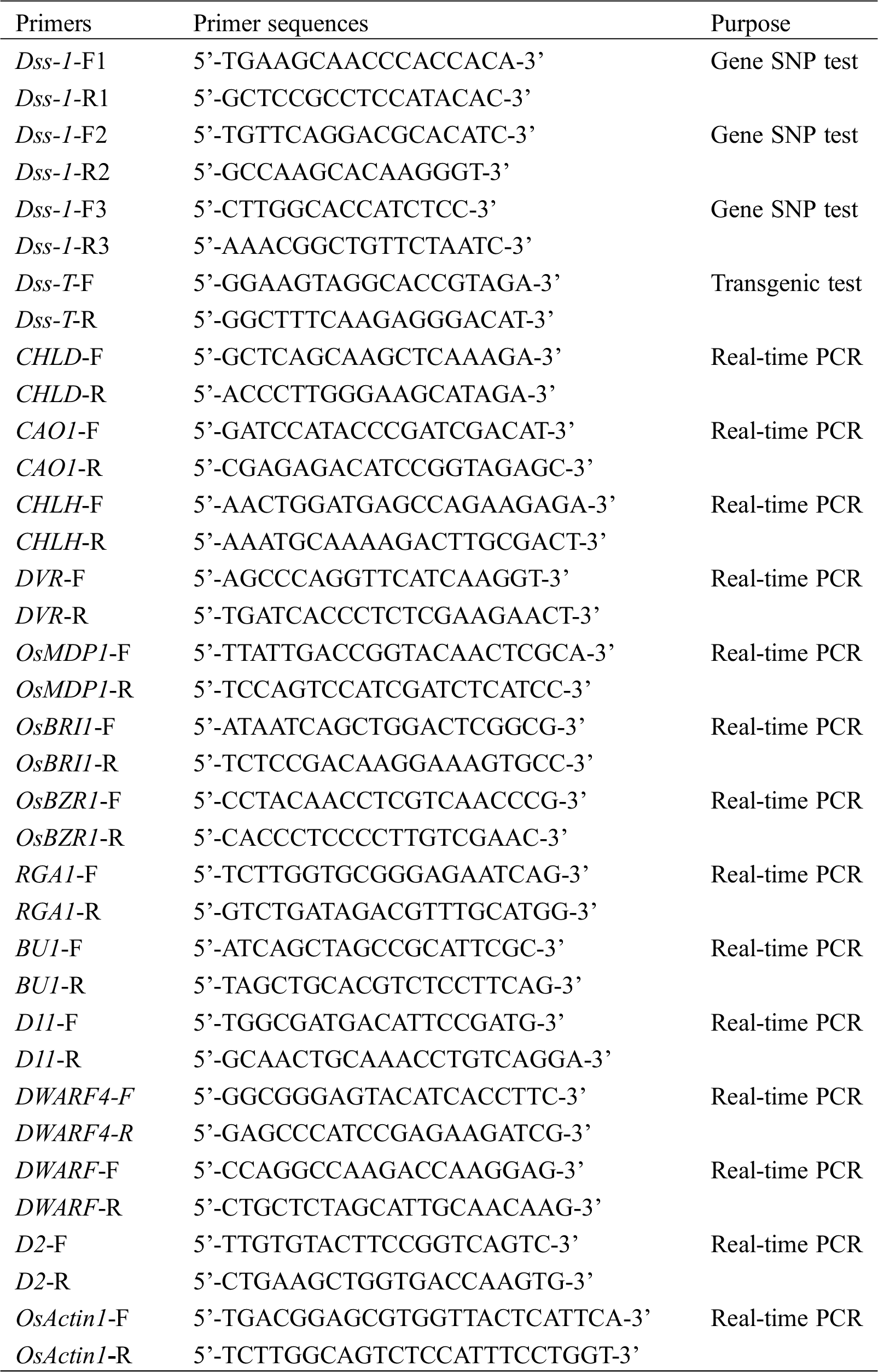
The dss1 mutant was isolated from the M2 generation of the Guizhou landrace cultivar LPZBH after EMS treatment. The dss1 plants retained the typical dwarf phenotype of BR-deficient plants, with erect and dark green leaves (Figs. 1A and 1B). After heading, the dss1 plants were 56.36% the height of the WT plants (Fig. 1A and Tab. 1). In the grain-filling stage, the flag leaf angle of dss1 (10.88°) was much less than that of the WT (94.57°) (Figs. 1A and 1D, Tab. 1). The WT plant showed the dn elongation pattern, but the second internode did not elongate in dss1 and a typical dm pattern existed (Figs. 1B and 1F). Contrary to the second internode’s growth inhibition, the neck internode, and the basal rachis internodes of the mutants were longer than those of WT plants (Figs. 1E and 1G, Tab. 1). The mutants’ phenotype was also abnormal in that the lengths and widths of dss1 seeds were significantly less than those of LPZBH seeds (Fig. 1C, Tab. 1). Besides, the 1,000-grain weight of dss1 was dramatically reduced (Tab. 1). To investigate whether the change in plant height is caused by changes in cell division or cell length, we examined sections of the fourth internode from adult plants under the microscope. The stem parenchymal cells of the dss1 were significantly shorter than those of the WT (Fig. 1H). The results showed that dwarf phenotype of dss1 was caused by the shortening of cell length.
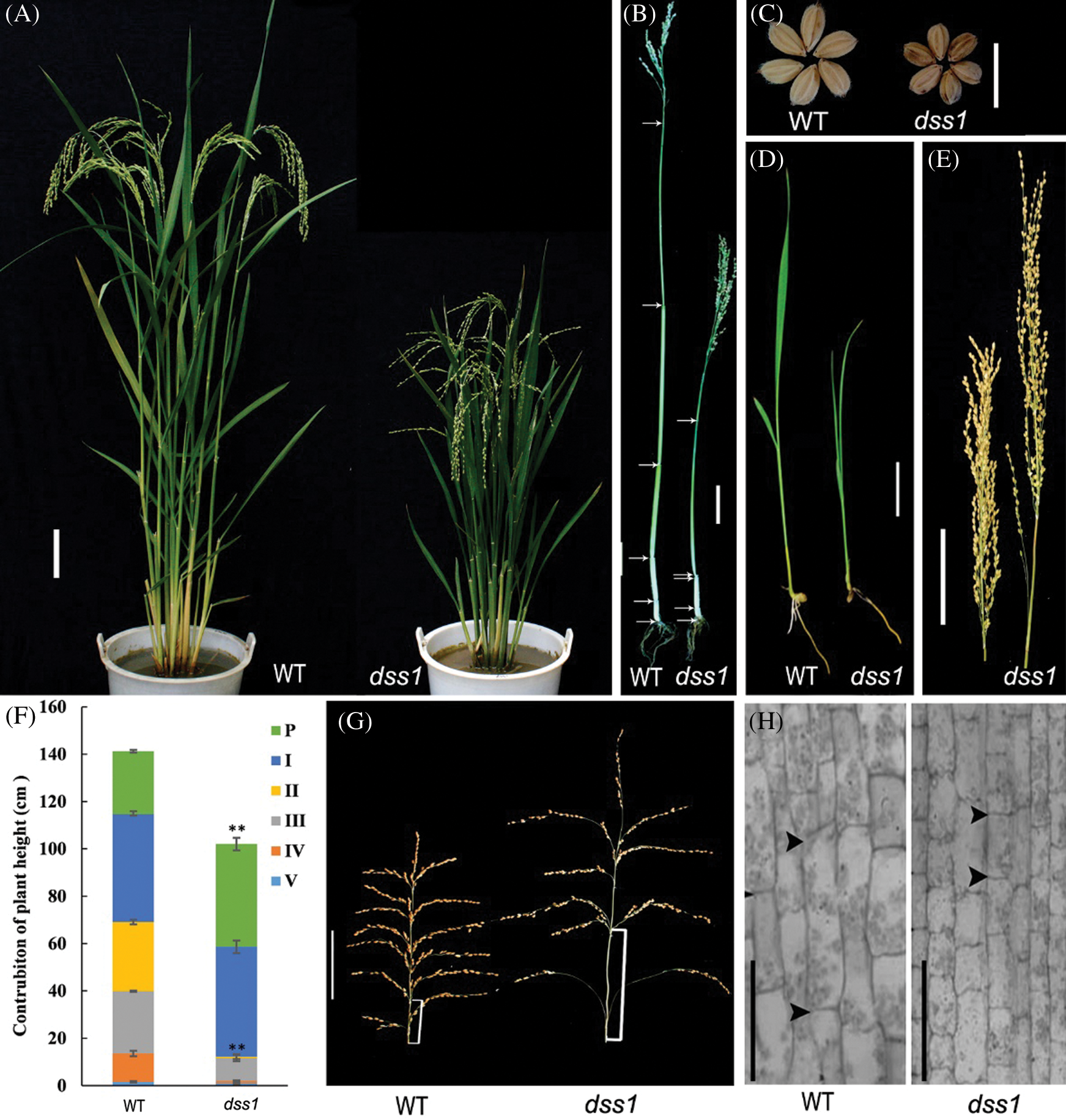
Figure 1: Phenotype of the dss1 mutant. (A) Gross morphology of a wild type (WT) plant and dss1 mutant at the grain-filling stage; Bar = 10 cm. (B) Culm elongation of the WT and dss1 mutant plants; arrows indicate the nodes; Bar = 10 cm. (C) Seed morphology of the WT and dss1 mutant. (D) Seedlings of the WT and dss1 mutant grown for 14 d; Bar = 5 cm. (E) Panicle structures of the WT and dss1 mutant; Bar = 10 cm. (F) Internode lengths of the WT and dss1 mutant. n = 30, ** indicates significant differences at P < 0.01. (G) Panicle comparison of the WT and dss1 mutant at the grain-filling stage. Brackets indicate the basal rachis internodes; Bar = 10 cm. (H) Parenchymal cells in the fourth internode in the WT and dss1 mutant; Bars = 100 μm
Table 1: Agronomic characteristics of the WT and dss1 rice mutant
3.2 The dss1 Mutant was Sensitive to BL Treatment
To verify whether dss1 is a BR deficient or insensitive mutant, we treat dss1 with BL. When exposed to increasing BL concentrations, dss1 mutant and WT were both sensitive to exogenous BR treatment, resulting in increasing coleoptile lengths and lamina joint angles, while the root growth was inhibited at higher BL concentrations (Fig. 2). This indicated that the dwarf phenotype of dss1 mutant was not caused by BR-signaling pathway, but by the deficiency in active BRs.
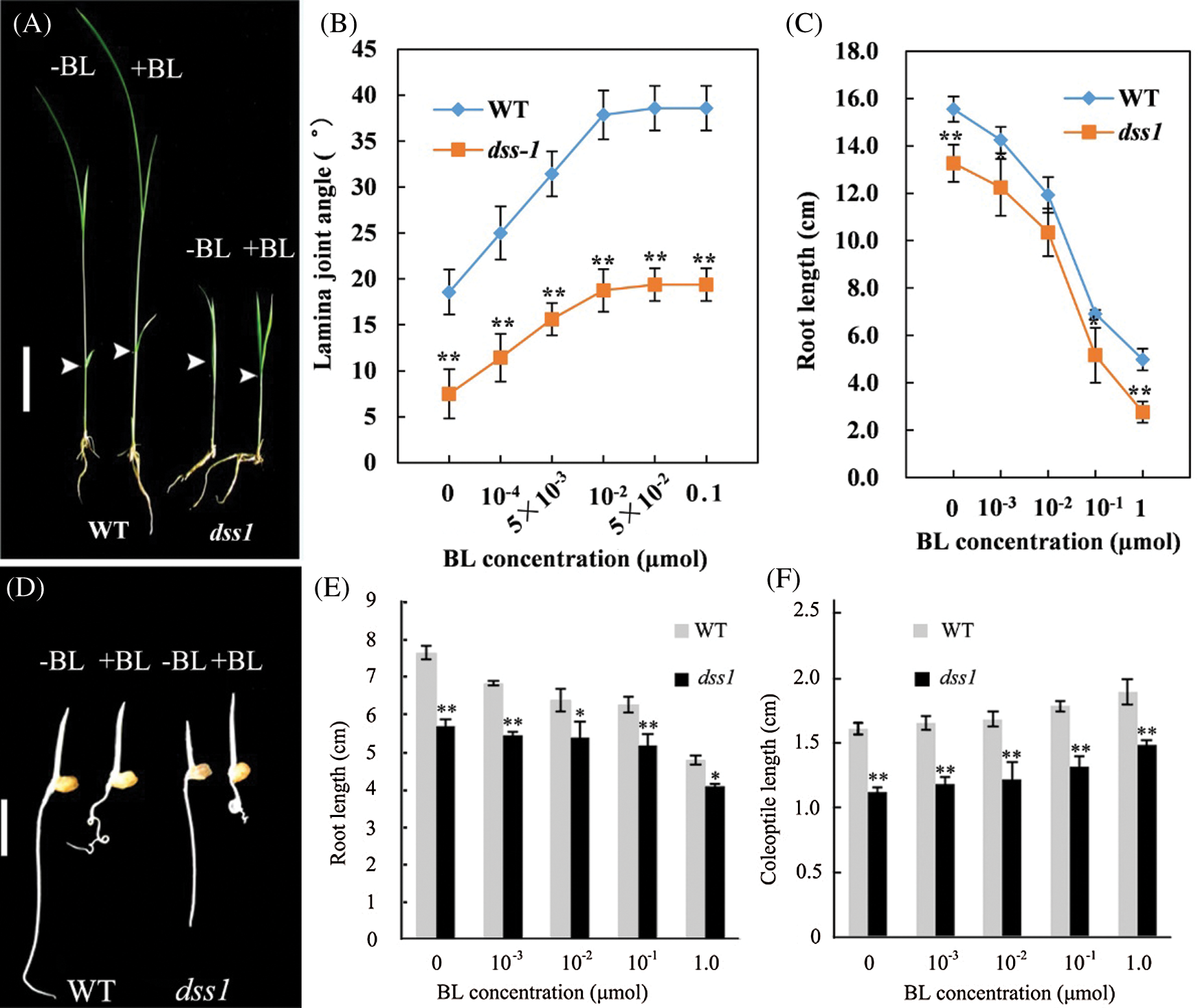
Figure 2: Responses of dss1 mutant and wild type (WT) seedlings to BL. The arrow shows the position measuring the angle between the leaves and stem. (A) Responses of the second leaf lamina joints from WT and dss1 plants. (B) The BL dose responses of the bending angles in the WT and dss1 mutants. (C) Effects of BL on the roots of WT and dss1 seedlings. (D) Seeds of the WT and the dss1 mutants were germinated on agar plates with or without BL. Seedlings were examined 3 d after germination; Bar = 1 cm. (E) Effects of BL on the extent of root elongation in WT and dss1 seedlings. (F) Effects of BL on the extent of coleoptile elongation in WT and dss1 seedlings. The plants were germinated in the same conditions as in (D), with the indicated BL concentration. Bars indicate SDs. (n = 10), * and ** indicate the significant differences at P < 0.05 and P < 0.01, respectively
3.3 DSS1 Regulates Chl Biosynthesis
The content of Chl a and Chl b levels in 40-d seeding stage of dss1 were significantly upper than in WT, while the Chl a/Chl b ratios and the content of carotene(Car) were not significantly different (Figs. 3A and 3B). Thus, DSS1 might affect Chl biosynthesis. To determine which steps were affected, we measured the contents of Chl intermediates. Compared with WT plants, the levels of the Chl intermediates 5-aminolevulinic acid (ALA), Porphobilinogen (PBG), UrogenIII, CoprogenIII and Protoporphyrin IX (ProtoIX) in dss1 mutants were reduced (Figs. 3C and 3D). While the contents of Mg-Proto IX and Pchlide were markedly higher in dss1 than in WT seedling leaves (Fig. 3E). Therefore, in dss1, the synthesis of higher levels of Proto IX and Pchlide caused a decrease in the precursor accumulation, including ALA, PGB, UrogenIII, CoprogenIII, and ProtoIX in the Mg-Proto IX biosynthetic pathway. This suggested that dss1 may have an effect on Chl biosynthesis in the Mg-Proto IX synthesis steps. To confirm this, we examined the expression levels of genes encoding the enzymes in the Chl biosynthetic pathway in rice. Compared with WT, most of the genes encoding the CHLH, DVR, and CAO enzymes were upregulated in the dss1 mutant. CHLH, which encodes the magnesium chelatase H subunit, was upregulated to more than four-fold that of the WT. The expression of CHLD was downregulated in the dss1 mutant (Fig. 3F). These changes might cause the higher Chl contents of the dss1 leaves.
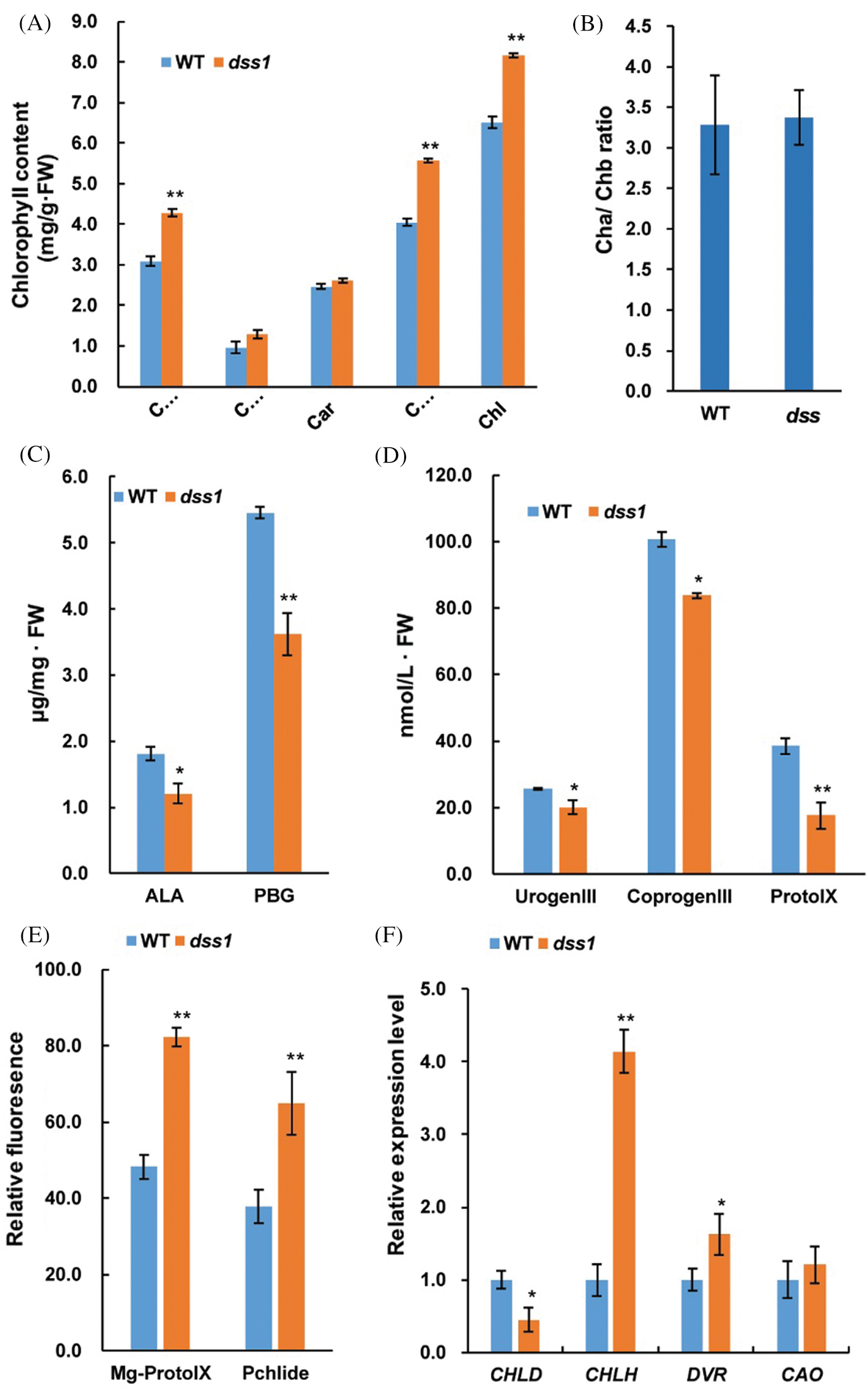
Figure 3: Analysis of chlorophyll (Chl) contents, Chl intermediates contents, and expression levels of Chl biosynthetic genes in the wild type (WT) and dss1 mutant. (A) Chl contents of the 40-day seedlings. (B) Chl a/Chl b ratios. (C) Levels of the Chl intermediates ALA and PBG measured in second leaves from 14-d-old WT and dss1 mutant seedlings. (D) Levels of UrogenIII, CoprogenIII, and ProtoIX. (E) Relative fluorescence of Mg-Proto IX and Pchlide. (F) Expression levels of Chl biosynthetic genes. Data are mean SDs (n = 3). * and ** indicate the significant differences between WT and dss1 at P < 0.05 and P < 0.01, respectively
3.4 Gene Expression Differentiation between DSS1 and WT
To further explore the biological function of DSS1, RNA-Seq analysis was carried out to compare the transcriptional profiles of differentially expressed genes (DEGs) using leaf samples obtained from 4-weeks-old seeding of WT and dss1. A total of 2,236 DEGs were identified from the 3,6324 genes in the dss1 group and the WT group, including 1217 up-regulated and 1019 down-regulated ones (Fig. 4A). All the identified differentially expressed genes were then clustered according to their biological roles. The GO enrichment and functional classification analysis were performed (Figs. 4B and 4C). Go enrichment level of genes in the molecular function were mainly found to be significantly enriched for the oxidoreductase activity and hydrolase activity, acting on ester bonds. 21 DEG genes were enrichment in hormone metabolic process in the biological process. The GO functional annotation was further divided into three components: molecular function, cellular component, and biological process (Fig. 5A). The maximum number of genes was involved in the molecular function of catalytic activity while it was also an integral part of the cell and cellular components. The result of KEGG pathway enrichment analysis showed 3 significant KEGG pathways, containing mRNA surveillance pathway, phenylpropanoid biosynthesis, starch, and sucrose metabolism (Fig. 5B). The changes in transcriptional levels of genes involved in BR biosynthesis, the BR-signaling pathway, and chlorophyll synthesis pathway, were further analyzed between dss1 and WT. In general, dss1 showed relatively high expressions in the BR signaling pathway (Fig. 6A). Among them, three genes (OsBRL1, BU1, OsBZR1) displayed larger relative changes in their expression levels between dss1 and WT. The expression of genes related to chlorophyll synthesis was up-regulated, especially OsCHLH, which was the same as our previous qRT-PCR results (Fig. 6B). The results indicated that the increase in chlorophyll content in dss1 was caused by the up-regulation of chlorophyll synthesis gene expression.
We further checked the expression of eight BR-related genes in BR biosynthesis and the BR-signaling pathway by qRT-PCR. The relative mRNA amounts of all eight BR-related genes, including in the biosynthesis and signaling pathway, were significantly increased to varying degrees in dss1 compared with WT (Fig. 7). The biosynthetic gene DWARF4 and signaling genes OsBZR1 and OsBRI1 were significantly up-regulated in the dss1 mutant. Thus, dss1 appears to be involved in the BR pathway. The DEGs involved in various pathways suggest that DSS1 triggers a change in complex regulatory networks in rice architecture, gain size, and Chl biosynthesis.
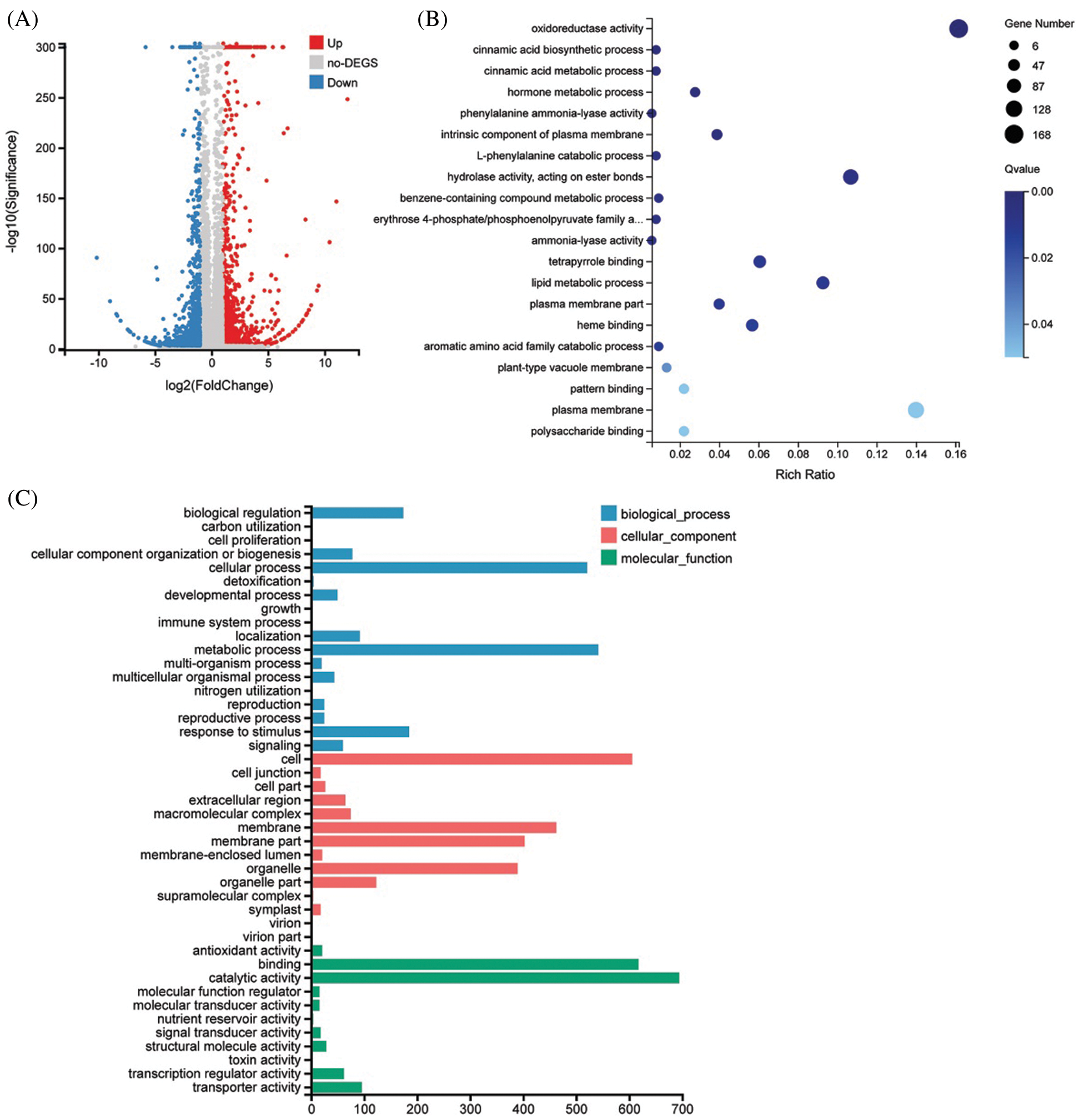
Figure 4: Transcriptional changes between dss1 and WT. (A) Volcano map of differentially expressed genes (DEGs) between dss1 and WT. (B) Go enrichment of DEGs between dss1 and WT. (C) The GO functional classification of DEGs between dss1 and WT
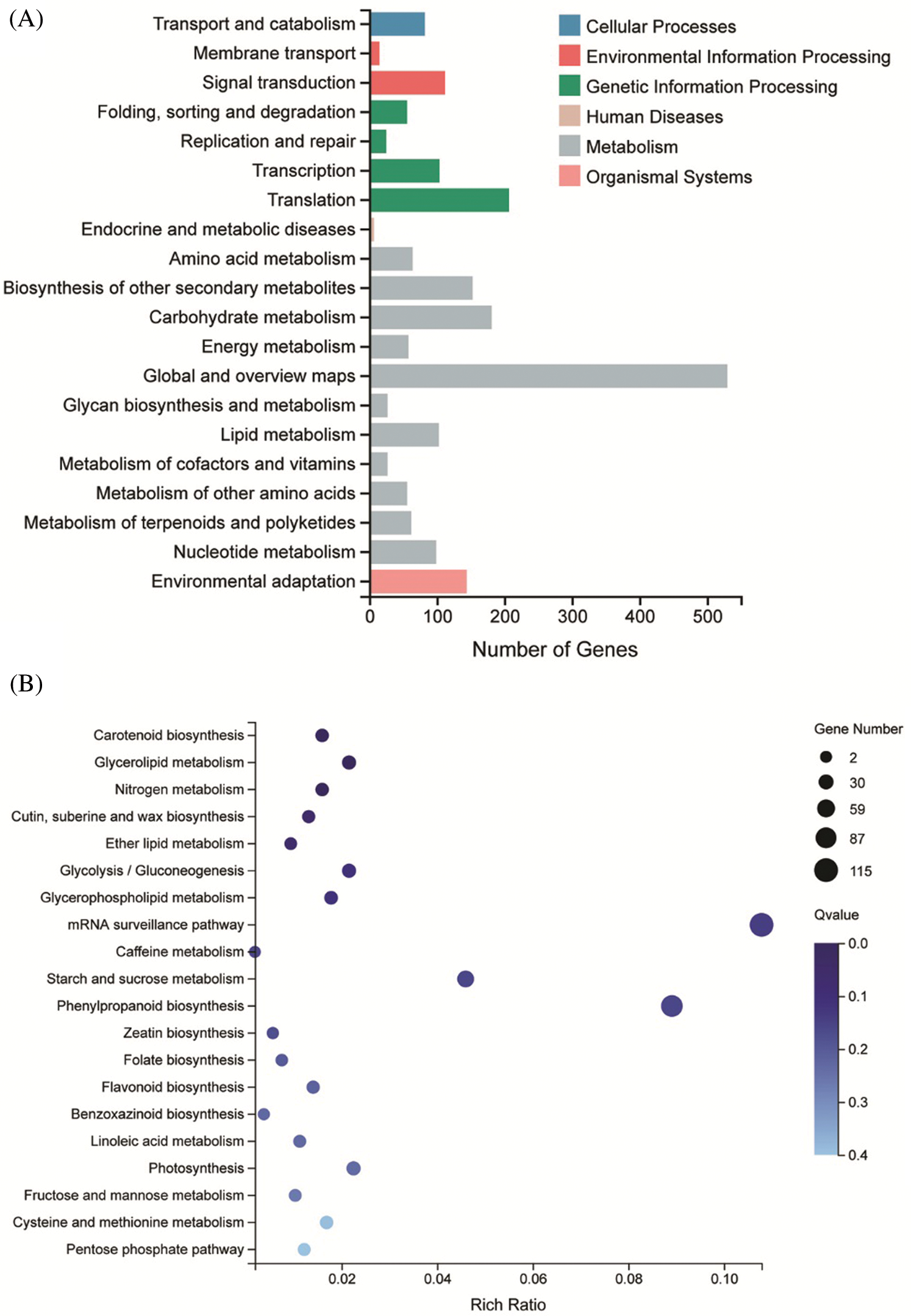
Figure 5: KEGG pathway functional classification and enrichment of DEGs between WT and dss1. (A) The KEGG functional classification of DEGs between dss1 and WT. (B) KEGG enrichment of DEGs between dss1 and WT
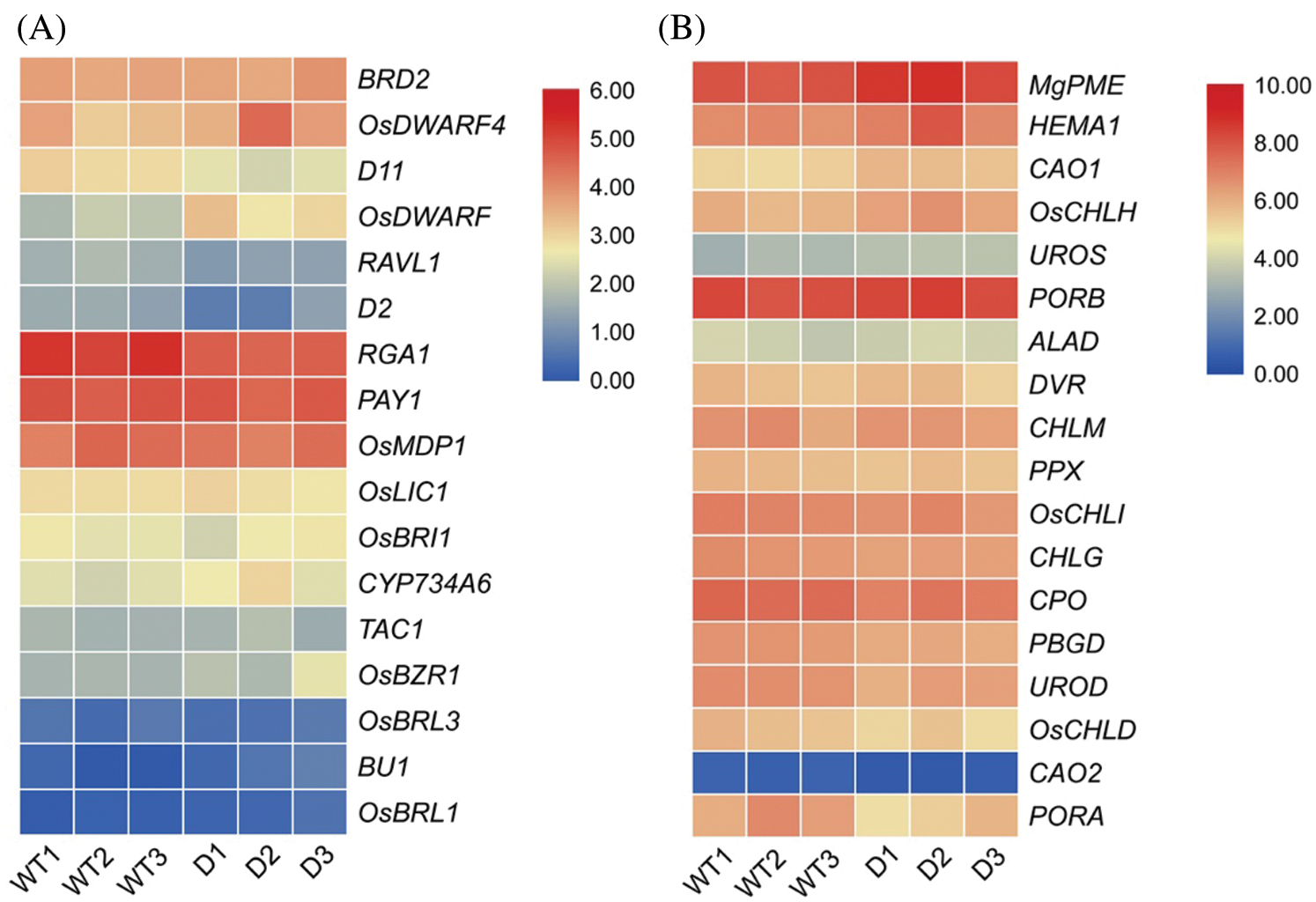
Figure 6: Expression analysis of BR-related genes (A) and chlorophyll synthesis related genes. (B) by the RNA-seq analysis. Genes were displayed by different colors. Relative levels of expression were shown by a color gradient form low (blue) to high (red). WT1-WT3, wild type; D1-D3, dss1 mutant
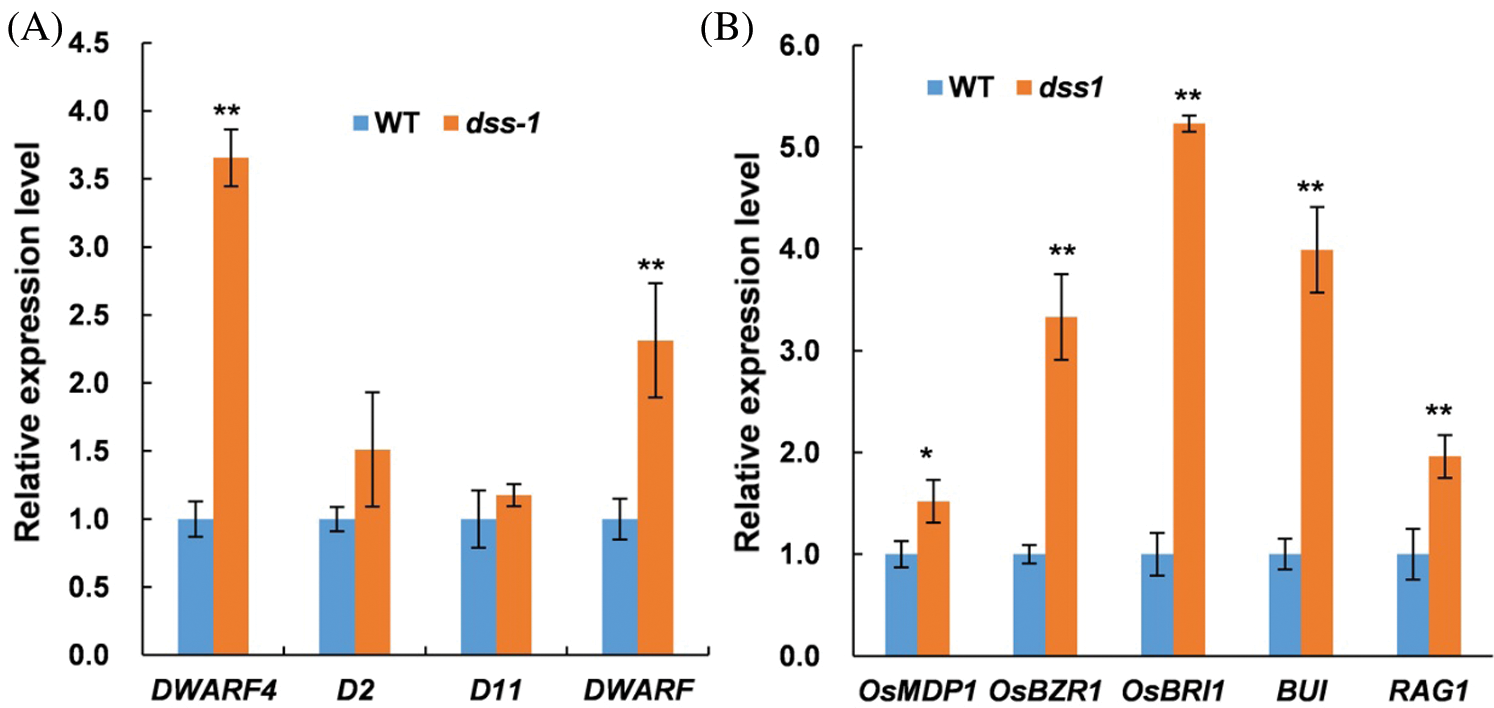
Figure 7: Expression levels of BR-related genes by qRT-PCR. Gene expression was normalized to that of the rice OsActin1 gene, and levels in wild type (WT) were set as 1.0. Transcript levels of BR-related genes assayed using RNA from leaves of seedlings 14 d after germination. (A) Transcript levels of four BR biosynthetic genes in WT and dss1. (B) Transcript levels of five BR signaling genes in WT and dss1
3.5 The Verification of the DSS1 Gene
To understand the molecular function of DSS1, in our previous work, we isolated one of the dss1 candidate gene OsDWARF, a key gene involved in the BR biosynthetic pathway by employing the MutMap method. The SNP represented a nonsense mutation of a threonine (ACT) codon to isoleucine (ATT) at residue 335 in OsDWARF1 [21]. To verify the function of OsDWARF in rescuing the dss1 phenotype, an OsDWARF overexpression vector driven by the cauliflower mosaic virus (CaMV) 35S promoter were constructed and transformed into dss1 using Agrobacterium-mediated transformation. All 22 independent T0 transgenic lines containing the OsDWARF showed overall phenotypes, including plant height, leaf angle, panicle architecture, and grain size, resembling that of the WT (Figs. 8 and 9). Also, the leaf color, Chl level, and Chl a/Chl b ratio were all restored to those of WT plants upon transformation with the OsDWARF gene (Figs. 8A and 10). Thus, it was demonstrated that the dss1 phenotypes are caused by the mutation in OsDWARF. DSS1 is a novel weak allele of OsDWARF that influences the development of panicle architecture and other traits related to BR defects.

Figure 8: Phenotypic overexpression (OE) by introducing the OsDWARF gene into dss1 rice. (A) Gross morphology at the productive phase of WT, OE-OsDWARF transgenic, and dss1 mutant plants; Bar = 10 cm. (B) Culm elongation of the WT, dss1 mutant, and OE-OsDWARF transgenic plants; arrows indicate the nodes; Bar = 10 cm. (C) Seed morphology of the WT, OE-OsDWARF transgenic and dss1 mutant; Bar = 1 cm. (D, E) Panicle structure of the WT and dss1 mutant; Bar = 5 cm
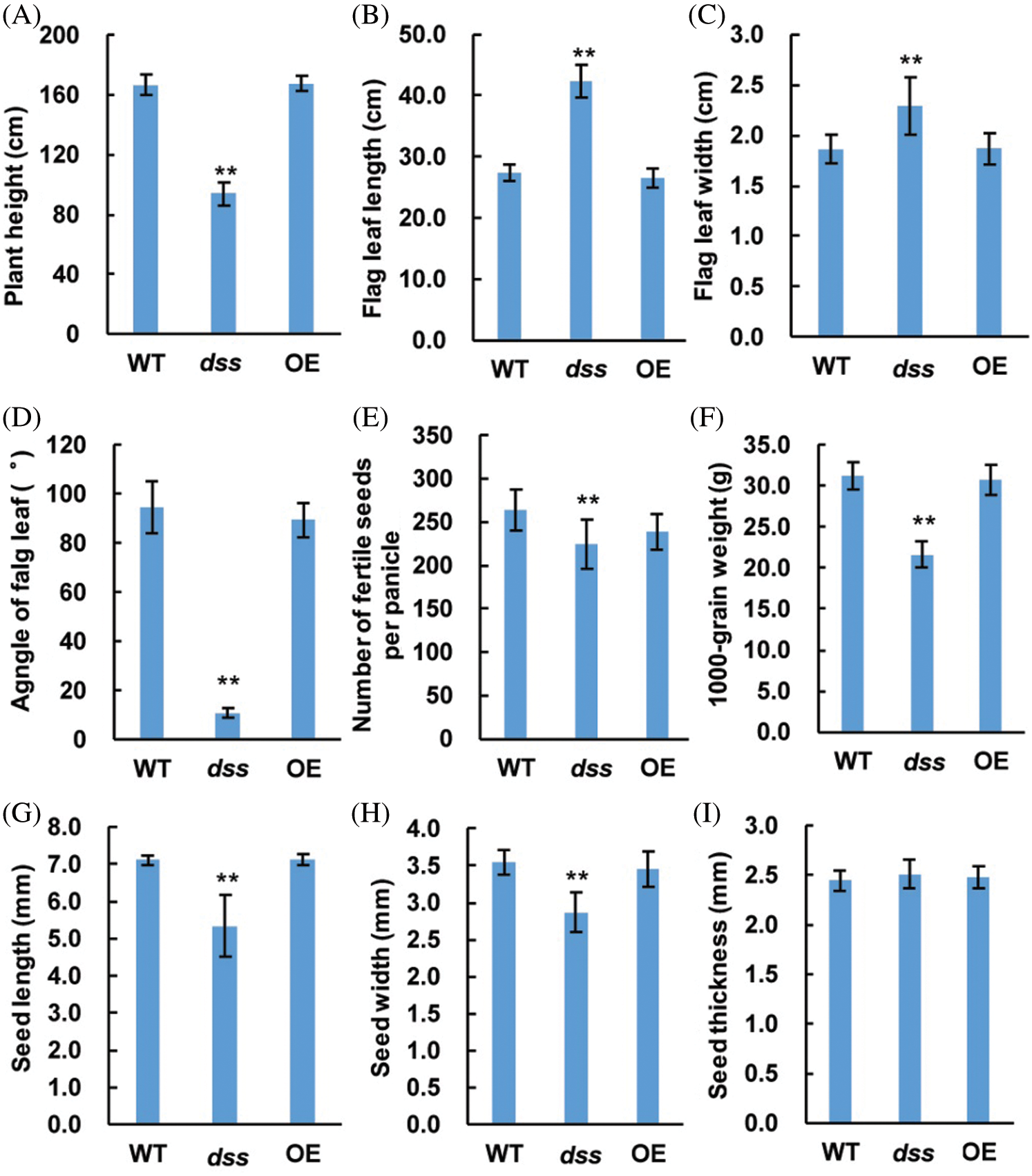
Figure 9: Phenotypes of the wild type (WT), overexpression (OE)-OsDWARF transgenic, and dss1 mutant plants. (A) Plant height comparison of the WT, OE-OsDWARF transgenic, and dss1 mutant plants. (B) Flag leaf length comparison of the WT, OE-OsDWARF transgenic, and dss1 mutant plants. (C) Flag leaf width comparison of the WT, OE-OsDWARF transgenic, and dss1 mutant plants. (D) Flag leaf angle comparison of the WT, OE-OsDWARF transgenic, and dss1 mutant plants. (E) Number of fertile seeds per panicle among the WT, OE-OsDWARF transgenic, and dss1 mutant plants. (F) The 1,000-grain weights of the WT, OE-OsDWARF transgenic, and dss1 mutant seeds. (G) Seed lengths of the WT, OE-OsDWARF transgenic, and dss1 mutant. (H) Seed widths of the WT, OE-OsDWARF transgenic, and dss1mutant. (I) Seed thicknesses of the WT, OE-OsDWARF transgenic, and dss1 mutant
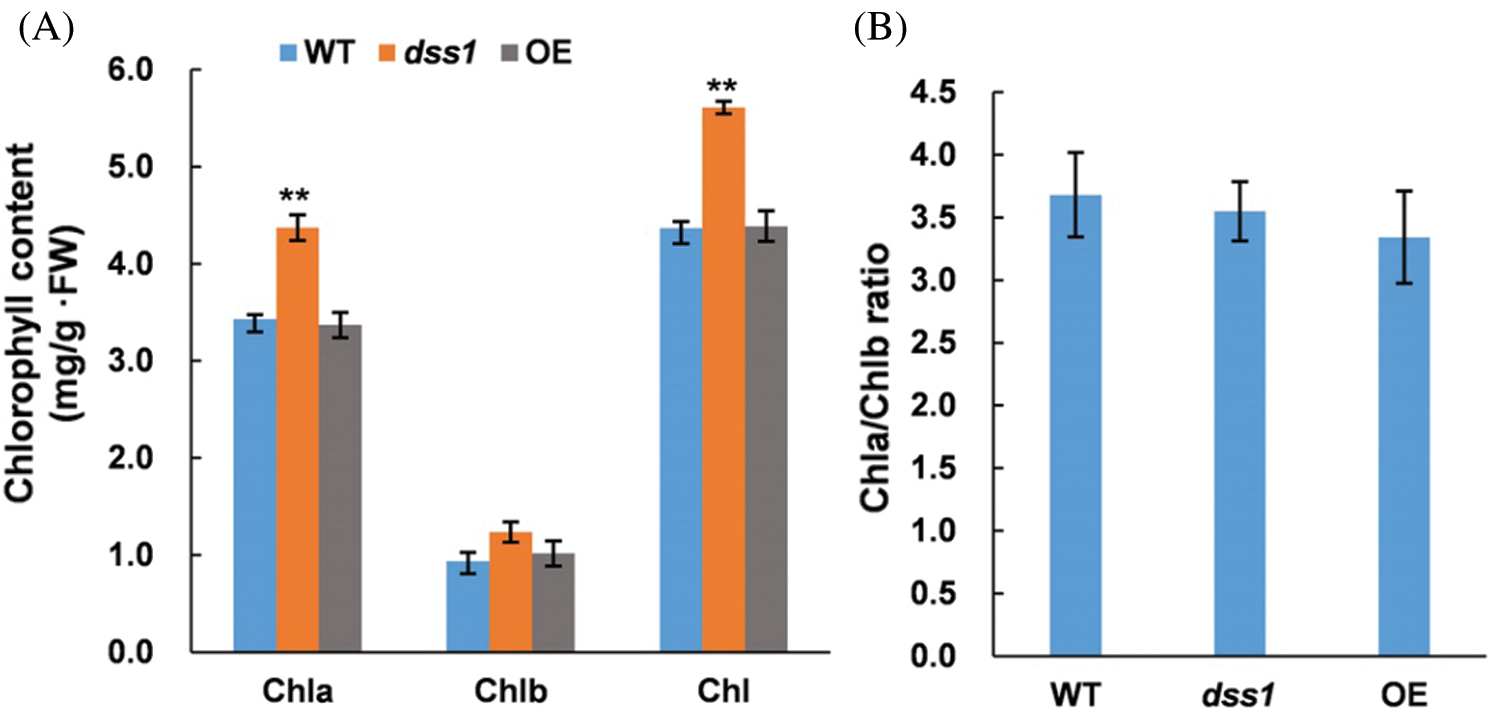
Figure 10: Complementation of the dss1 mutant with the OsDWARF gene. (A) Chlorophyll (Chl) levels in the second leaves of 60-day-old rice plants. (B) The Chl a/Chl b ratio in WT, dss1 and OE-OsDWARF transgenic seedlings
OsDWARF gene encoding a cytochrome P450 protein involved in the BR biosynthetic pathway has pleiotropic effects on plant height, grain number, fertility and root elongation [10,11]. The frameshift mutation caused by base deletion and the key conserved domain base mutations often resulted in a loss of function of the protein. Four OsDWARF mutant alleles have already been reported, brd1, brd1-1, brd1-2, and brd1-3 [10,11]. The brd1-1 mutant had a 113-bp deletion from the intron 6 to exon 7, which caused the product of brd1-1 to lack the essential heme-binding site required for P450 activity. The brd1 mutant showed a 193-bp deletion and a 5-bp insertion and then caused a frameshift. A single nucleotide substitution to exchange Gly111Val in brd1-2. This 111-residue glycine, which is conserved among all members of CYP85, CYP90, and CYP88, is important for the expression of the enzymatic activity. The brd1-3 allele’s single nucleotide substitution to exchange the 101 glycine with valine does not occur among the related P450 members. Thus, brd1, brd1-1, and brd1-2 mutants showed severe phenotypes with tortuous and stiff blades, while brd1-3 displayed a less severe phenotype than those of the other three mutants and reached an ~30-cm plant height [10,11]. In this work, a new mutation is a single nucleotide substitution to exchange Thr with Ile at residue 335 in dss1. This Thr residue was not conserved among the CYP85A1 groups when compared with Arabidopsis DWARF proteins [27]. Our results indicated that dss1 is a weak allelic mutant of OsDWARF, which is similar to, but not completely the same as, the four mutants brd1, brd1-1, brd1-2 and brd1-3 reported by Hong et al. [10] and Mori et al. [11]. While dss1 mutants showed a semi-dwarf phenotype, with long panicles, which is similar to the d2 and d61 mutants [15,16].
BR regulates its homeostasis through feedback expression of its biosynthesis and inactivation genes [28]. BR-deficient mutants in rice, such as d2, brd1, and lldd10, have increased expression levels of OsBRI1, the BR receptor kinase gene, relative to that in WT seedlings [10,16,17,29]. Sakamoto et al. [29] also reported that the expression of DWARF4 can be enhanced in BR-deficient mutants. In the present study, the OsBRI1 and DWARF4 genes were significantly upregulated in the dss1 mutant. The BL treatment caused the dss1 mutant to be hypersensitive in the root test (Figs. 2C and 2E). This may be why dss1 showed greater sensitivity to the BL treatment than did WT plants in the root test.
In rice, several BR-deficient and BR-insensitive mutants exhibit dark-green leaves [7,8,10,16,30–32]. In the d2 and lhdd10 mutants, the mutants had higher Chl contents [16,17]. Liu et al. [17] suggested that in a loss-of-function mutant for brd2, lhdd10, deficient of BR upregulates the expression of Chl biosynthetic genes. In our study, dss1 leaves also showed the darker green color. Chl contents in leaves of dss1 were significantly higher than that in WT (40 d), while the Chl a/b ratios were not significantly different. The contents of Mg-Proto and Pchlide were significantly higher than those in WT. The CHLH gene encodes the largest subunit of magnesium chelatase. Our results indicated that the high expression level of CHLH caused the increased Mg-pro IX and Chl contents in the dss1 mutant. The overexpression of the OsDWARF gene in dss1 can reduce the leaf Chl content. Thus, dss1 may affect Chl biosynthesis, and caused the darker green leaves in the dss1.
In this work, we confirmed that a Thr335Ile amino acid substitution residing in the highly conserved region of DSS1/OsDWARF was responsible for the dwarf phenotype, panicle architecture, and seed size changes in the dss1 mutants.
Author Contributions: Xiaofang Zeng and Degang Zhao designed the research and conducted the experiments. Yan Li collected and analyzed the data. All authors wrote the final text and approved the final version of manuscript.
Funding Statement: This work was supported by grants from the Genetically Modified Organisms Breeding Major Projects of China (2016ZX08010003), The Science and Technology Foundation of Guizhou Province (20181043), The Science and Technology Cooperation Project of Guizhou Province ([2016]7448) and Guizhou university talent introduced project (2015 [25]).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Fitzgerald, M. A., McCouch, S. R., Hall, R. D. (2009). Not just a grain of rice, the quest for quality. Trends Plant Science, 14(3), 133–139. DOI 10.1016/j.tplants.2008.12.004.
2. Miura, K., Ikeda, M., Matsubara, A., Song, X. J., Ito, M. et al. (2010). OsSPL14 promotes panicle branching and higher grain productivity in rice. Nature Genetics, 42(6), 545–549. DOI 10.1038/ng.592. [Google Scholar] [CrossRef]
3. Xue, W., Xing, Y., Weng, X., Zhao, Y., Tang, W. et al. (2008). Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nature Genetics, 40(6), 761–767. DOI 10.1038/ng.143. [Google Scholar] [CrossRef]
4. Huang, X., Qian, Q., Liu, Z., Sun, H., He, S. et al. (2009). Natural variation at the DEP1 locus enhances grain yield in rice. Nature Genetics, 41(4), 494–497. DOI 10.1038/ng.352.
5. Jiao, Y., Wang, Y., Xue, D., Wang, J., Yan, M. et al. (2010). Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nature Genetics, 42(6), 541–544. DOI 10.1038/ng.591.
6. Yan, W. H., Wang, P., Chen, H. X., Zhou, H. J., Li, Q. P. et al. (2011). A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Molecular Plant, 4(2), 319–330. DOI 10.1093/mp/ssq070.
7. Wu, Y., Fu, Y., Zhao, S., Gu, P., Zhu, Z. et al. (2016). CLUSTERED PRIMARY BRANCH 1, a new allele of DWARF11, controls panicle architecture and seed size in rice. Plant Biotechnology Journal, 14(1), 377–386. DOI 10.1111/pbi.12391. [Google Scholar] [CrossRef]
8. Jiang, D., Chen, W., Dong, J., Li, J., Yang, F. et al. (2018). Overexpression of OsmiR164b-resistant OsNAC2 improves plant architecture and grain yield in rice. Journal of Experimental Botany, 7, 1533–1543. [Google Scholar]
9. Nolan, T. M., Vukašinović, N., Liu, D., Russinova, E., Yin, Y. (2020). Brassinosteroids: Multidimensional regulators of plant growth, development, and stress responses. Plant Cell, 32(2), 295–318. DOI 10.1105/tpc.19.00335. [Google Scholar] [CrossRef]
10. Hong, Z., Ueguchi-Tanaka, M., Shimizu-Sato, S., Inukai, Y., Fujioka, S. et al. (2002). Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant Journal, 32(4), 495–508. DOI 10.1046/j.1365-313X.2002.01438.x. [Google Scholar] [CrossRef]
11. Mori, M., Nomura, T., Ooka, H., Ishizaka, M., Yokota, T. et al. (2002). Isolation and characterization of a rice dwarf mutant with a defect in brassinosteroid biosynthesis. Plant Physiology, 130(3), 1152–1161. DOI 10.1104/pp.007179. [Google Scholar] [CrossRef]
12. Zhao, B., Lv, M., Feng, Z., Campbell, T., Liscum, E. et al. (2016). TWISTED DWARF 1 associates with BRASSINOSTEROID INSENSITIVE 1 to regulate early events of the brassinosteroid signaling pathway. Molecular Plant, 9(4), 582–592. DOI 10.1016/j.molp.2016.01.007.
13. Tong, H., Chu, C. (2018). Functional specificities of brassinosteroid and potential utilization for crop improvement. Trends in Plant Science, 23(11), 1016–1028. DOI 10.1016/j.tplants.2018.08.007. [Google Scholar] [CrossRef]
14. Xiao, Y., Zhang, G., Liu, D., Niu, M., Tong, H. et al. (2020). GSK2 stabilizes OFP3 to suppress brassinosteroid responses in rice. Plant Journal, 102(6), 1187–1201. DOI 10.1111/tpj.14692. [Google Scholar] [CrossRef]
15. Yamamuro, C., Ihara, Y., Wu, X., Noguchi, T., Fujioka, S. et al. (2000). Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell, 12(9), 1591–1605. DOI 10.1105/tpc.12.9.1591. [Google Scholar] [CrossRef]
16. Hong, Z., Ueguchi-Tanaka, M., Umemura, K., Uozu, S., Fujioka, S. et al. (2003). A rice brassinosteroid-deficient mutant, ebisu dwarf, is caused by a loss of function of a new member of cytochrome P450. Plant Cell, 15(12), 2900–2910. DOI 10.1105/tpc.014712. [Google Scholar] [CrossRef]
17. Liu, X., Feng, Z. M., Zhou, C. L., Ren, Y. K., Mou, C. L. et al. (2016). Brassinosteroid (BR) biosynthetic gene lhdd10 controls late heading and plant height in rice (Oryza sativa L.). Plant Cell Report, 35(2), 357–368. DOI 10.1007/s00299-015-1889-3. [Google Scholar] [CrossRef]
18. Wu, C. Y., Trieu, A., Radhakrishnan, P., Kwok, S. F., Harris, S. et al. (2008). Brassinosteroids regulate grain filling in rice. Plant Cell, 20(8), 2130–2145. DOI 10.1105/tpc.107.055087. [Google Scholar] [CrossRef]
19. Zeng, X. F., Huang, Q., Chen, X., Li, Y., Li, J. R. et al. (2018). Identification and gene mapping of a dwarf and small seed mutant dss1 in ‘Lipingzabianhe’, a landrace cultivar rice in Guizhou. Plant Physiology Journal, 54, 1186–1194. [Google Scholar]
20. Arnon, D. I. (1949). Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiology, 24(1), 1–15. DOI 10.1104/pp.24.1.1. [Google Scholar] [CrossRef]
21. Morton, R. A. (1975). Biochemical spectroscopy, vol. 1. London: Adam Hilger Ltd. [Google Scholar]
22. Santiago, O. M., Green, R. M., Tingay, S., Brusslan, J. A., Tobin, E. M. (2001). shygrl1 is a mutant affected in multiple aspects of photomorphogenesis. Plant Physiology, 126(2), 587–600. DOI 10.1104/pp.126.2.587. [Google Scholar] [CrossRef]
23. Masuda, T., Fusada, N., Oosawa, N., Takamatsu, K., Yamamoto, Y. Y. et al. (2003). Functional analysis of isoforms of NADPH: Protochlorophyllide oxidoreductase (PORPORB and PORC, in Arabidopsis thaliana. Plant Cell Physiology, 44(10), 963–974. DOI 10.1093/pcp/pcg128. [Google Scholar] [CrossRef]
24. Zeng, X. F., Li, L., Li, J. R., Zhao, D. G. (2016). Constitutive expression of McCHIT1-PAT enhances resistance to rice blast and herbicide, but does not affect grain yield in transgenic glutinous rice. Applied Biochemistry and Biotechnology, 63(1), 77–85. DOI 10.1002/bab.1342. [Google Scholar] [CrossRef]
25. Wang, L., Feng, Z., Wang, X., Wang, X., Zhang, X. (2009). DEGseq, an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics, 26(1), 136–138. DOI 10.1093/bioinformatics/btp612. [Google Scholar] [CrossRef]
26. Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods, 25(4), 402–408. DOI 10.1006/meth.2001.1262. [Google Scholar] [CrossRef]
27. Shimada, Y., Fujioka, S., Miyauchi, N., Kushiro, M., Takatsuto, S. et al. (2001). Brassinosteroid-6-oxidases from Arabidopsis and tomato catalyze multiple c-6 oxidations in brassinosteroid biosynthesis. Plant Physiology, 126(2), 770–779. DOI 10.1104/pp.126.2.770. [Google Scholar] [CrossRef]
28. Siddiqui, H., Hayat, S., Bajguz, A. (2018). Regulation of photosynthesis by brassinosteroids in plants. Acta Physiology Plant, 40(3), 1–15. DOI 10.1007/s11738-018-2639-2. [Google Scholar] [CrossRef]
29. Sakamoto, T., Morinaka, Y., Ohnishi, T., Sunohara, H., Fujioka, S. et al. (2005). Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nature Biotechnology, 24(1), 105–109. DOI 10.1038/nbt1173. [Google Scholar] [CrossRef]
30. Tanabe, S., Ashikari, M., Fujioka, S., Takatsuto, S., Yoshida, S. et al. (2005). A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. Plant Cell, 17(3), 776–790. DOI 10.1105/tpc.104.024950. [Google Scholar] [CrossRef]
31. Jiang, Y., Bao, L., Jeong, S. Y., Kim, S. K., Xu, C. et al. (2012). XIAO is involved in the control of organ size by contributing to the regulation of signaling and homeostasis of brassinosteroids and cell cycling in rice. Plant Journal, 70(3), 398–408. DOI 10.1111/j.1365-313X.2011.04877.x.
32. Shi, Z., Rao, Y., Xu, J., Hu, S., Fang, Y. et al. (2015). Characterization and cloning of SMALL GRAIN 4, a novel DWARF11 allele that affects brassinosteroid biosynthesis in rice. Science Bulletin, 60(10), 905–915. DOI 10.1007/s11434-015-0798-8. [Google Scholar] [CrossRef]
Table S1: Primer used in this work