DOI:10.32604/phyton.2020.012303

| Phyton-International Journal of Experimental Botany DOI:10.32604/phyton.2020.012303 |  |
| Review |
Do Strigolactones Regulate Bud Winter Dormancy and Charactrisitc Secondary Metabolism in Tea?
1Institute of Fruit and Tea, Hubei Academy of Agricultural Science/Hubei Tea Engineering and Technology Research Centre, Wuhan, 430064, China
2College of Life Sciences, Xinyang Normal University, Xinyang, 464000, China
*Corresponding Author: Ziming Gong. Email: ziminggong@163.com
Received: 24 June 2020; Accepted: 21 August 2020
Abstract: Tea (Camellia sinensis [L.] O. Kuntze.) is an important cash crop, which mainly uses tender shoots and young leaves for manufacturing. Due to the marketing characteristic that earlier made tea has higher price, the time of the breaking of winter dormancy buds in spring is extremely important in tea industry. Strigolactones are a group of carotenoids-derived metabolites which regulates bud outgrowth, shoot branching, tiller angle and environmental stress responses. The role of strigolactones in tea plant was briefly summarized in the current review, with an emphasis of the association of strigolactones on bud ecodormancy and shoot branching. The involvement of strigolactones on the biosynthesis of the tea characteristic metabolites flavonoids, caffeine and theanine were also discussed. Moreover, recent advances on the biosynthesis of strigolactones and its regulation by microRNAs and environmental stresses were also presented. This review provides a basis for future investigations underlying the mechanisms of strigolactones on bud winter dormancy and tea secondary metabolism.
Keywords: Camellia sinensis; strigolactones; winter dormancy; secondary metabolites
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |
Tea (Camellia sinensis [L.] O. Kuntze.) is used for producing a non-alcoholic beverage consumed worldwide with unique flavor and health benefits [1]. The quality of processed tea is basically determined by the secondary metabolisms constituents in tea leaves, include flavonoids and alkaloids (the main contributors determining tea flavour and astringency), amino acids (the main contributors towards the umami taste of tea infusions) [2–4]. As an evergreen perennial crop, tea plants initiate shoot elongation and undergo winter dormancy in late autumn in each annual growth cycle to resist against environmental stresses, especially low temperatures, for survival during the winter. When environmental conditions are suitable in the early spring, the buds break dormancy and begin a new growth cycle. Tea yield largely depends on newly sprouted shoot tips with two unfolded leaves, which are generally harvested for green tea processing. Bud outgrowth activity has long been a target of breeding selection, because it significantly affects crop yields by affecting both tiller number and inflorescence complexity [5]. Bud dormancy release time is pivotal in influencing spring tea price and yield [6]. However, winter dormancy molecular mechanisms are not well elucidated so far.
The relationship between winter dormancy and plant hormones has been intensively uncovered. The changes in the level of active gibberellins (GAs) is associated with induction of growth cessation primarily regulated by circadian clock [7]. Numerous genes related to hormone metabolism or signaling are differentially expressed in transcriptome studies of dormant bud in Euphorbia esula [8]. In addition to the well-known isoprenoid phytohormones such as cytokinins (CK), abscisic acid (ABA), brassinosteroids (BR) and gibberellic acid (GA), strigolactones, a group of carotenoids-derived metabolites, suppress shoot branching [9], bud development in addition to several additional biological functions [10,11]. Moreover, association of ABA in strigolactone pathway could facilitate to control buds paradormancy [12]. Additionally, auxin regulated strigolactone depletion is a significant aspect for branching after shoot apices removal [13,14]. These findings offer more understanding about the mechanism of tea winter dormancy. This mini-review focuses on the possible involvement of strigolactones in tea winter dormancy and the biosynthesis of some secondary metabolites.
2 Strigolactones and Winter Dormancy
Strigolactones are derived from β-carotene; their biosynthetic pathway has gradually been elucidated (Fig. 1): First, β-carotene isomerase catalyzes the 9-cis/all-trans isomerization of β-carotene, followed by the stereospecific C9′-C10′ cleavage of 9-cis-β-carotene mediated by carotenoid cleavage dioxygenase 7 [CCD7, MORE AXILLARY GROWTH 3 (MAX3) in Arabidopsis, then, the transformation of intermediate 9-cis-β-apo-10′-carotenal into carlactone is mediated by CCD8 (MAX4 in Arabidopsis) [15–18]. In addition, several other proteins such as α/β-hydrolase (AtD14 in Arabidopsis, D14/D88/HTD2 in rice) and F-box protein (MAX2 in Arabidopsis, D3 in rice) are involved in strigolactones signal transduction [13,17,18].
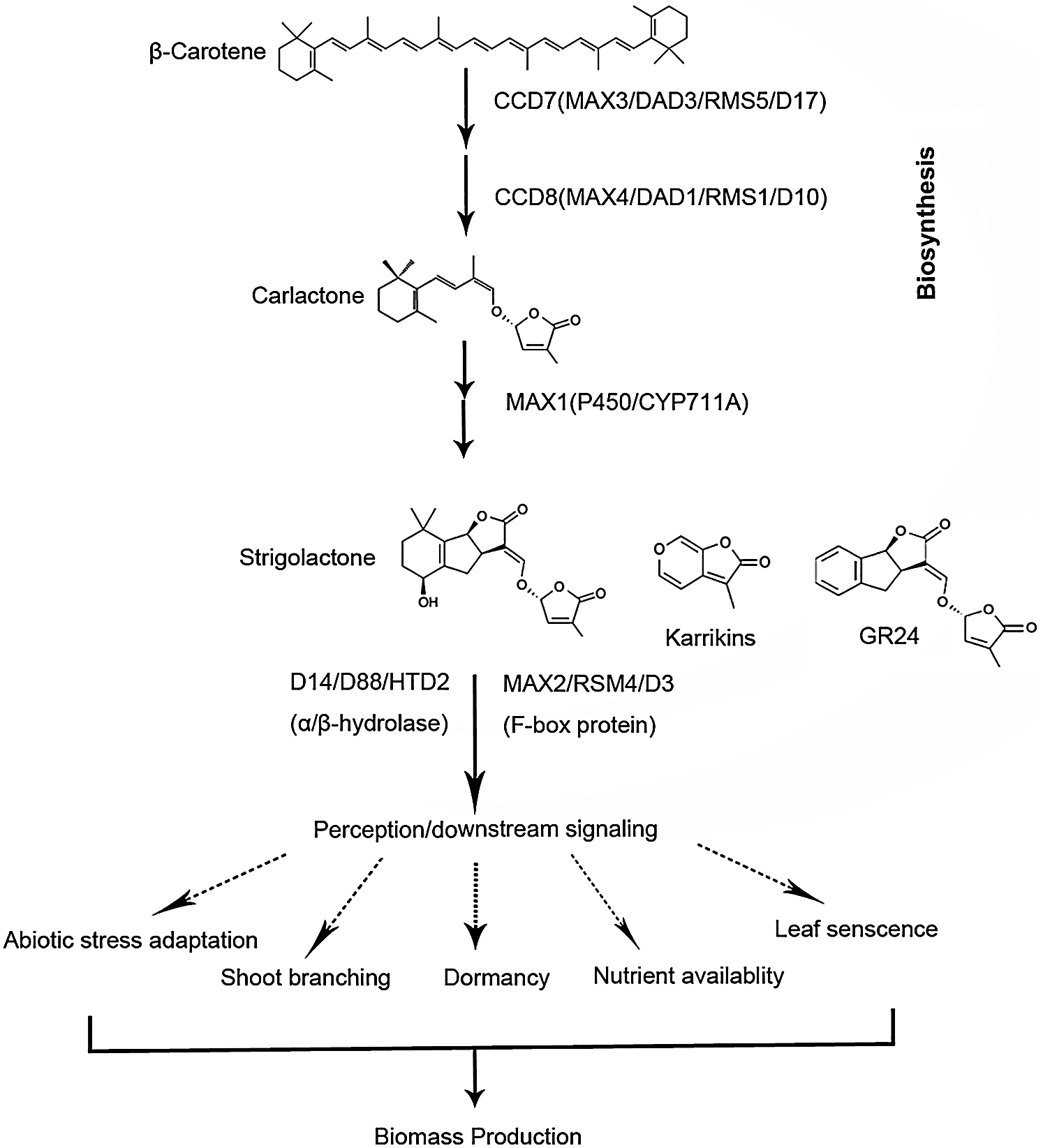
Figure 1: Major steps of strigolactones biosynthesis and signal transduction in plants.
In biosynthesis, which is included carotenoid cleavage dioxygenases7 (CCD7, MAX3, RMS5, or DAD3) [15] and CCD8 (MAX4, RMS1, D10 or DAD1) [17], and one cytochrome P450 monooxygenase (MAX1) [15]. For strigolactones signal transduction, which is involving MAX2/RMS4/D3 [13], an F-box leucinerich protein and DWARF14 (D14)/D88/HTD2, a member of the α/β-fold hydrolase superfamily [18].
Genes associated with strigolactones pathway, such as MAX genes in Arabidopsis and their orthologs in Pisum sativum is tightly correlated with bud dormancy [16–19]. MAX2 plays a significant role in conferring dormancy and boosting shoot growth [16,20]. Indeed, max2 mutant exhibit reduced expression of dormancy-associated gene 1/auxin-repressed protein (DRM1/ARP). MAX2 transcript levels were found being elevated in endodormant crown buds in leafy spurge [8], further strengthening that strigolactones function in ecodormancy induction. In the tillering dwarf Oryza sativa mutants d3, d10, d14, d17 and d27, axillary meristems develop normally but with weakened ability of the dormancy in tiller buds [21]. Strigolactones bind to α/β-hydrolase fold-containing proteins, which activates MAX2, ultimately leading to lateral bud inhibition and dormancy [22]. In rice, TB1 (Teosinte branched 1)/FC1 (Fine Culm1) and D14 interact with dormancy release marker MADS57 to regulate shoot branching [23]. In addition, MADS57 is a target of miR444a, suggesting that MADS57 plays a role in the strigolactone signaling pathway. To further investigate the regulation of tea winter dormancy, studies are required to detect interactions between tea analogs of MAX2 in the strigolactones pathway and other strigolactones signals or miRNAs.
3 Strigolactones and Secondary Metabolite Biosynthesis
3.1 Strigolactones and Flavonoid Biosynthesis
Many genes associated with the biosynthesis of flavonoid metabolites are differentially expressed during different phases of dormancy [8]. Chalcone synthase, flavone isomerase, theobromine synthase, caffeine synthase and glutamate (or theanine) synthase, were found being expressed in a dormant tea bud. These genes and their encoded enzymes can protect plants from UV damage and are upregulated during dormancy in raspberry and leafy spurge [24]. Flavonoid signaling pathway was found involved in maintaining of underground adventitious bud paradormancy and paradormancy release in leafy spurge. This signaling pathway together with the action of auxin, ABA, GA, CK, JA, ethylene and BR levels/signals, control the dormancy and the initiation of shoot growth [14,25].
Some studies strengthened a framework among auxin, flavonoids and strigolactones. For example, crosstalk modulates strigolactones and auxin levels [26], and flavonoid modulates auxin flow in stems and buds (regulated by MAX1) helps regulating bud repression in Arabidopsis [27]. Flavonoids in tea help determining its flavor and astringency [2]. In Arabidopsis, MAX1 directly or indirectly regulates the 11 flavonoid genes and transcription factor An2 [27]. One hypothesis is that in the absence of MAX1, the flavonoid level is reduced, result in some auxin transporters derepression and subsequently increase auxin transport in the bud, thereby controlling tea winter dormancy.
In biosynthesis, which is included carotenoid cleavage dioxygenases 7 (CCD7, MAX3, RMS5, or DAD3) [15] and CCD8 (MAX4, RMS1, D10 or DAD1) [17], and one cytochrome P450 monooxygenase (MAX1) [15]. For strigolactones signal transduction, which is involving MAX2/RMS4/D3 [13], an F-box leucine-rich protein and DWARF14 (D14)/D88/HTD2, a member of the α/β-fold hydrolase superfamily [18].
3.2 Strigolactones and Theanine Biosynthesis
Defective glutamine synthetase (GS1;2) and NADH-dependent glutamate synthase 1 (NADH-GOGAT1) lead to reduced bud outgrowth in rice but some studies have demonstrated that defective GS1;2 results in reduction of tiller number in the GS1;2 mutant [28–30], likely due to increased concentration of strigolactones. CsGS in tea, encoding glutamine synthetase (which is involved in theanine biosynthesis) is downregulated (42% reduced expression) during winter dormancy [31]. Further investigations are needed to determine how CsGS influences theanine biosynthesis and interacts with strigolactones, consequently affecting tea winter dormancy.
3.3 Strigolactones and Caffeine Biosynthesis
Expression of the gene encoding caffeine synthase and caffeine content decrease during tea dormancy as allantoin contents increase, indicating that caffeine degradation is activated or caffeine biosynthesis is inhibited due to tea winter dormancy [32,33]. Indeed, differential expression of theobromine synthase and caffeine synthase genes occurs during different phases of tea dormancy [34]. In addition, these genes alter bud dormancy status by controlling auxin transport, which is mediated by MAX1 [24]. It is also known that cytokinins can be transported through PUP1 (purine permease) transport system as adenine and caffeine [35]. Further studies are needed to elucidate how the transport system functions during bud dormancy and whether it interacts with strigolactones to regulate caffeine metabolism in tea.
4 Regulators of Strigolactone Functions
Overexpression of miR156 increases branching in Brassica napus [36] and Arabidopsis [36,37] and similar branching phenotype has been observed in loss-of-function max3 and max4 mutants [38], suggesting a link between strigolactones metabolism and miR156 expression. Some of strigolactones pathway genes such as MAX1, MAX3, MAX4 in Arabidopsis and D3, D10, D27 in rice holds target fragments recognized by miR156a–g [39]. Osa-miR156 overexpressing in rice results in a reduced D27 transcript level but elevated D3 and D14 transcript levels, suggesting a feedback mechanism in strigolactones pathway [40]. Altogether, these studies unveiling a new regulatory pathway of strigolactones function by osa-miR156 to control dormancy [39].
In active and dormant tea buds, numerous tea miRNAs which are differentially expressed have been detected [41]. An HXXXD-type acyl-transferase-like protein-coding gene, targeted by cs-miR414, is involved in maintaining BR homeostasis [42]. Strigolactones (possibly regulated by cs-miRNA 164), auxin (by cs-miRNA 397), CK (by cs-miRNA 1846), ABA (by cs-miRNA408) and GA (by cs-miRNA 1886) are also downstream products of regulatory gene network of cs-miRNAs [41]. Some target transcripts have been identified being differentially expressed in a dormant bud-specific suppression subtractive hybridization (SSH) library [34,43].
4.2 Regulated by Environmental Stress
Unfavorable external factors, such as nutrients, water and low temperature, would induce the growth to dormancy and will quickly resume growth in the absence of these unfavorable factors [44]. In accordance with the central roles of strigolactones in stress responses, strigolactones biosynthetic genes MAX1, MAX3 and MAX4 are influenced by drought, salt and ABA in Arabidopsis leaves, further supporting the notion that stress-mediated strigolactones homeostasis is essential for boosting plant resistance against environmental stresses [45]. Application of strigol analogue GR24 improves drought tolerance in Arabidopsis, demonstrating positive regulatory role of strigolactones for improving plant tolerance to drought in both an ABA-dependent and ABA-independent manner [46]. So, we speculated that strigolactones may play dominant roles in adaptation to abiotic stress in tea.
Nutrient starvation-induced phenotypes can be affected by the strigolactones biosynthetic pathway. Moreover, the extreme branching phenotype associated with nitrogen application levels of the strigolactone biosynthetic mutant max4 indicate that the influence of nitrogen limitation on branching is mediated (at least in part) by strigolactones. Phosphorous deficiency, which is regulated by strigolactones, affects tillering in rice [47]. GR24 application can increase wild-type pea nodule number and restore the reduced nodule number to wild-type in the strigolactone-deficient rms1 mutant. The modified strigolactones derivatives appliaction to enhance crops nodule growth would be a cost-effective way to contribute to nitrogen-fixing and plant development [48]. It is interesting to find out whether strigolactones are involved in the high ammonium utilization capacity in tea.
Strigolactones levels or signaling can be affected by light intensity [49]. strigolactones act downstream of the phyB-dependent responses to different red-to-far red light (R: FR) ratios [50]. Arabidopsis phyB mutants are defective in detecting R: FR ratios, and in high R: FR light, these mutants exhibit reduced branching, whereas max2 mutants are highly branched [51]. Similar, In sorghum, both null mutation of phyB and low R: FR ratios induced by shading and defoliation affect branching and upregulate the expression of Sbtb1 (Teosinte branched 1), SbMAX2 and SbDRM1 [52], suggesting that phyB helps determine whether buds become dormant or active via strigolactones. strigolactones might play a role in determining growth status via a series of MAX2 or BRC (branched) of its biosynthesis or signaling to preserve normal growth.
Strigolactones are positive regulators by controlling the expression of different stress or ABA-responsive genes related to plant growth and environment stress responses in stress and diverse ABA signaling pathways. Moreover, defective strigolactones signaling transmission also results in depressed CK degradation oxidase (CKX) genes expression. Altogether, these results reveal that coordinated network among strigolactones, ABA and CK regulates plants adaptation to different abotic stress [45]. strigolactones in conjunction with other phytohormones could favor nutrients and water assimilate by promoting root development [49]. Therefore, the use of strigolactones derivatives could favor the adaptation of the plant architecture to such as drought and phosphates abiotic stress [48].
Thus, we propose a model explaining the interactions between strigolactones and terpenoid hormones (auxin, cytokinin, ABA, GA, BRs), secondary metabolism and abiotic stress in tea (Fig. 2). This model will pave the way for uncovering the mechanism underlying secondary metabolism, growth and winter dormancy regulation in tea. However, further investigations should be performed to obtain direct evidence for the role of strigolactones biosynthesis and signaling in tea, especially in the regulation of tea winter dormancy.
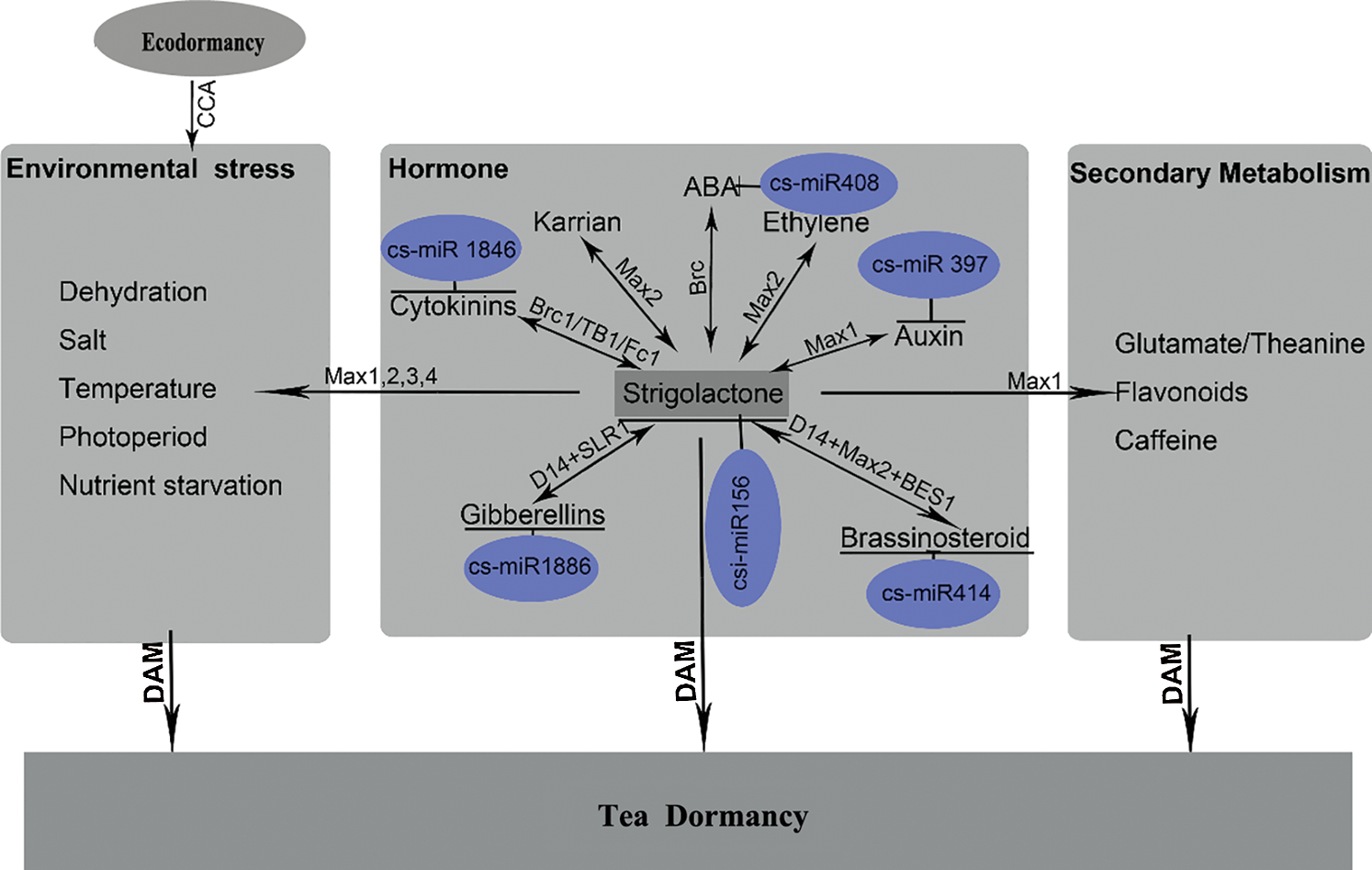
Figure 2: A predicted functional network underlying winter dormancy in tea. The phytohormones strigolactones and their integration with secondary metabolism and environmental stress regulatory pathways may control the dormancy process in tea
5 Conclusions and Future Perspective
Strigolactones are crucial for efficient nutrient allocation in plants, especially under Pi and N deficiency. Strigolactones also play essential roles in responsing to drought and salt environment. Moreover, strigolactones can also stimulate seed germination and inhibit shoot branching. The observation that strigolactone mutants exhibit weakened bud dormancy activity suggests that strigolactones play a dominate role of dormancy in plants; Strigolactones, along with ABA, GA, BR, CK, ethylene and auxin, may act directly on buds to coordinately regulate bud outgrowth and ddormancy. In tea, genes encoding caffeine synthase and flavonoid synthase deferentially express in various dormancy phases. These enzymes change bud development through regulating auxin flow, which is mediated by MAX1, a protein involved in strigolactones biosynthesis or attributed to the same transport system as strigolactones. Therefore, strigolactones may mediate secondary metabolism in tea during the winter dormancy period.
The regulation of dormancy and, more importantly, the control of growth activation at bud burst are highly important for maintaining productivity and building biomass in perennial plants [53,54]. We have merely begun to understand some aspects of the roles of strigolactones as a phytohormone, and its interactions with secondary metabolism and abiotic stress tolerance in tea are yet to be elucidated. Doing so will build a more complete picture of the role of strigolactones in the growth of perennial plants. Further elucidating how strigolactones regulates the processes of dormancy, cold acclimation, and freezing tolerance will help us develop crop plants that are better adapted to new and changing climates, thus increasing tea yields and quality.
Funding Statement: This work was financially supported by the China Postdoctoral Science Foundation (2018M632821), the Open Fund of State Key Laboratory of Tea Plant Biology and Utilization (SKLTOF 20180105), China agriculture research system (CARS-19), Hubei Natural Science Foundation Project (2019CFB178), Natural Science Foundation of Hubei Academy of Agricultural Sciences (2021NKYJJ13), the Agricultural Science and Technology Innovation Project of Hubei Province (2019-620-000-001-24).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Cabrera, C., Artacho, R., Gimênez, R. (2006). Beneficial effects of green tea—A review. Journal of the American College of Nutrition, 25(2), 79–99. DOI 10.1080/07315724.2006.10719518.
2. Chaturvedula, V. S. P., Prakash, I. (2011). The aroma, taste, color and bioactive constituents of tea. Journal of Medicinal Plants Research, 5(11), 2110–2124. [Google Scholar]
3. Alcázar, A., Ballesteros, O., Jurado, J. M., Pablos, F., Martín, M. J. et al. (2007). Differentiation of green, white, black, Oolong and Pu-erh teas according to their free amino acids content. Journal of Agricultural and Food Chemistry, 55(15), 5960–5965. DOI 10.1021/jf070601a.
4. Chen, Y. L., Duan, J., Jiang, Y. M. (2010). Production, quality and biological effects of Oolong Tea (Camellia sinensis). Food Reviews International, 27(1), 1–15. DOI 10.1080/87559129.2010.518294. [Google Scholar] [CrossRef]
5. Tian, C. H., Jiao, Y. L. (2015). A systems approach to understand shoot branching. Current Plant Biology, 3–4, 13–19. DOI 10.1016/j.cpb.2015.08.001. [Google Scholar] [CrossRef]
6. Carr, M. K. V. (1972). The climatic requirements of the tea plant: A review. Experimental Agriculture, 8(1), 1–14. DOI 10.1017/S0014479700023449. [Google Scholar] [CrossRef]
7. Arana, M. V., Marin-de la Rosa, N., Maloof, J. N., Blazquez, M. A., Alabadi, D. (2011). Circadian oscillation of gibberellin signaling in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America, 108(22), 9292–9297. DOI 10.1073/pnas.1101050108. [Google Scholar] [CrossRef]
8. Horvath, D. P., Chao, W. S., Suttle, J. C., Thimmapuram, J., Anderson, J. V. (2008). Transcriptome analysis identifies novel responses and potential regulatory genes involved in seasonal dormancy transitions of leafy spurge (Euphorbia esula L.). BMC Genomics, 9(1), 536. DOI 10.1186/1471-2164-9-536. [Google Scholar] [CrossRef]
9. Gomez-Roldan, V., Fermas, S., Brewer, P. B., Puech-Pagès, V., Dun, E. A. et al. (2008). Strigolactone inhibition of shoot branching. Nature, 455(7210), 189–194. DOI 10.1038/nature07271. [Google Scholar] [CrossRef]
10. Dun, E. A., Brewer, P. B., Beveridge, C. A. (2009). Strigolactones: Discovery of the elusive shoot branching hormone. Trends in Plant Science, 14(7), 364–372. DOI 10.1016/j.tplants.2009.04.003. [Google Scholar] [CrossRef]
11. Alder, A., Jamil, M., Marzorati, M., Bruno, M., Vermathen, M. et al. (2012). The path from β-carotene to carlactone, a strigolactone like plant hormone. Science, 335(6074), 1348–1351. DOI 10.1126/science.1218094. [Google Scholar] [CrossRef]
12. López-Ráez, J. A., Kohlen, W., Charnikhova, T., Mulder, P., Undas, A. K. et al. (2010). Does abscisic acid affect strigolactone biosynthesis? New Phytologist, 187(2), 343–354. DOI 10.1111/j.1469-8137.2010.03291.x. [Google Scholar] [CrossRef]
13. Brewer, P. B., Dun, E. A., Ferguson, B. J., Rameau, C., Beveridge, C. A. (2009). Strigolactone acts downstream of auxin to regulate bud outgrowth in pea and Arabidopsis. Plant Physiology, 150(1), 482–493. DOI 10.1104/pp.108.134783. [Google Scholar] [CrossRef]
14. Chao, W. S., Doğramaci, M., Horvath, D. P., Anderson, J. V., Foley, M. E. (2016). Phytohormone balance and stress-related cellular responses are involved in the transition from bud to shoot growth in leafy spurge. BMC Plant Biology, 16(1), 419. DOI 10.1186/s12870-016-0735-2. [Google Scholar] [CrossRef]
15. Al-Babili, S., Bouwmeester, H. J. (2015). Strigolactones, a novel carotenoid-derived plant hormone. Annual Review of Plant Biology, 66(1), 161–186. DOI 10.1146/annurev-arplant-043014-114759. [Google Scholar] [CrossRef]
16. Stirnberg, P., van De Sande, K., Leyser, H. M. (2002). MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development, 129(5), 1131–1141. [Google Scholar]
17. Sorefan, K., Booker, J., Haurogné, K., Goussot, M., Bainbridge, K., et al. (2003). MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and Pea. Genes & Development, 17(12), 1469–1474. DOI 10.1101/gad.256603. [Google Scholar] [CrossRef]
18. Booker, J., Auldridge, M., Wills, S., McCarty, D., Klee, H. et al. (2004). MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Current Biology, 14(14), 1232–1238. DOI 10.1016/j.cub.2004.06.061. [Google Scholar] [CrossRef]
19. Tatematsu, K., Ward, S., Leyser, O., Kamiya, Y., Nambara, E. (2005). Identification of cis-elements that regulate gene expression during initiation of axillary bud outgrowth in Arabidopsis. Plant Physiology, 138(2), 757–766. DOI 10.1104/pp.104.057984. [Google Scholar] [CrossRef]
20. Nelson, D. C., Scaffidi, A., Dun, E. A., Waters, M. T., Flematti, G. R. et al. (2011). F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America, 108(21), 8897–8902. DOI 10.1073/pnas.1100987108. [Google Scholar] [CrossRef]
21. Ishikawa, S., Maekawa, M., Arite, T., Onishi, K., Takamure, I. et al. (2005). Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant and Cell Physiology, 46(1), 79–86. DOI 10.1093/pcp/pci022. [Google Scholar] [CrossRef]
22. Mudgil, Y., Ghawana, S., Jones, A. M. (2013). N-MYC down-regulated-like proteins regulate meristem initiation by modulating auxin transport and MAX2 expression. PLoS One, 8(11), e77863. DOI 10.1371/journal.pone.0077863. [Google Scholar] [CrossRef]
23. Guo, S., Xu, Y., Liu, H., Mao, Z. W., Zhang, C. et al. (2013). The interaction between OsMADS57 and OsTB1 modulates rice tillering via DWARF14. Nature Communications, 4(1), 2850. DOI 10.1038/ncomms2542. [Google Scholar] [CrossRef]
24. Krishnaraj, T., Gajjeraman, P., Palanisamy, S., Subhas, C. S. R., Azad, M. A. K. (2011). Identification of differentially expressed genes in dormant (banjhi) bud of tea (Camellia sinensis (L.) O. Kuntze) using subtractive hybridization approach. Plant Physiology Biochemistry, 49(6), 569–571. [Google Scholar]
25. Anderson, J. V., Doğramacı, M., Horvath, D. P., Foley, M. E., Chao, W. S. et al. (2012). Auxin and ABA act as central regulators of developmental networks associated with paradormancy in Canada thistle (Cirsium arvense). Functional & Integrative Genomics, 12(3), 515–531. DOI 10.1007/s10142-012-0280-5. [Google Scholar] [CrossRef]
26. Hayward, A., Stirnberg, P., Beveridge, C. A., Leyser, O. (2009). Interactions between auxin and strigolactone in shoot branching control. Plant Physiology, 151(1), 400–412. DOI 10.1104/pp.109.137646. [Google Scholar] [CrossRef]
27. Lazar, G., Goodman, H. M. (2006). MAX1, a regulator of the flavonoid pathway, controls vegetative axillary bud outgrowth in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America, 103(2), 472–476. DOI 10.1073/pnas.0509463102. [Google Scholar] [CrossRef]
28. Kazuhiro, F., Soichi, K., Mayumi, T. K., Sawa, Y., Nakayama, Y. et al. (2013). Cytosolic glutamine synthetase 1;2 is responsible for the primary assimilation of ammonium in rice roots. Plant and Cell Physiology, 54(6), 934–943. DOI 10.1093/pcp/pct046. [Google Scholar] [CrossRef]
29. Yamaya, T., Kusano, M. (2014). Evidence supporting distinct functions of three cytosolic glutamine synthetases and two NADH-glutamate synthases in rice. Journal of Experimental Botany, 65(19), 5519–5525. DOI 10.1093/jxb/eru103.
30. Ohashi, M., Ishiyama, K., Kusano, M., Fukushima, A., Kojima, S. et al. (2015). Lack of cytosolic glutamine synthetase1;2 in vascular tissues of axillary buds causes severe reduction in their outgrowth and disorder of metabolic balance in rice seedlings. Plant Journal, 81(2), 347–356. DOI 10.1111/tpj.12731. [Google Scholar] [CrossRef]
31. Rana, N. K., Mohanpuria, P., Kumar, V., Yadav, S. K. (2010). A CsGS is regulated at transcriptional level during developmental stages and nitrogen utilization in Camellia sinensis (L.) O. Kuntze. Molecular Biology Reports, 37(2), 703–710. DOI 10.1007/s11033-009-9559-6. [Google Scholar] [CrossRef]
32. Mohanpuria, P., Kumar, V., Joshi, R., Gulati, A., Ahuja, P. S. et al. (2009). Caffeine biosynthesis and degradation in tea [Camellia sinensis (L.) O. Kuntze] is under developmental and seasonal regulation. Molecular Biotechnology, 43(2), 104–111. DOI 10.1007/s12033-009-9188-2. [Google Scholar] [CrossRef]
33. Mohanpuria, P., Kumar, V., Yadav, S. K. (2010). Tea caffeine: Metabolism, functions, and reduction strategies. Food Science and Biotechnology, 19(2), 275–287. DOI 10.1007/s10068-010-0041-y. [Google Scholar] [CrossRef]
34. Thirugnanasambantham, K., Prabu, G., Palanisamy, S., Chandrabose, S. R. S., Mandal, A. K. A. (2013). Analysis of dormant bud (Banjhi) specific transcriptome of tea [Camellia sinensis (L.) O. Kuntze] from cDNA library revealed dormancy-related genes. Applied Biochemistry and Biotechnology, 169(4), 1405–1417. DOI 10.1007/s12010-012-0070-5. [Google Scholar] [CrossRef]
35. Bűrkle, L., Cedzich, A., Döpke, C., Stransky, H., Okumoto, S. et al. (2003). Transport of cytokinins mediated by purine transporters of the PUP family expressed in phloem, hydathodes, and pollen of Arabidopsis. Plant Journal, 34(1), 13–26. DOI 10.1046/j.1365-313X.2003.01700.x. [Google Scholar] [CrossRef]
36. Wei, S., Yu, B., Gruber, M. Y., Khachatourians, G. G., Hegedus, D. D. et al. (2010). Enhanced seed carotenoid levels and branching in transgenic Brassica napus expressing the Arabidopsis miR156b gene. Journal of Agricultural and Food Chemistry, 58(17), 9572–9578. DOI 10.1021/jf102635f. [Google Scholar] [CrossRef]
37. Schwab, R., Palatnik, J. F., Riester, M., Schommer, C., Schmid, M. et al. (2005). Specific effects of microRNAs on the plant transcriptome. Developmental Cell, 8(4), 517–527. DOI 10.1016/j.devcel.2005.01.018. [Google Scholar] [CrossRef]
38. Wei, S., Gruber, M. Y., Yu, B. Y., Gao, M. J., Khachatourians, G. G. et al. (2012). Arabidopsis mutant sk156 reveals complex regulation of SPL15 in a miR156-controlled gene network. BMC Plant Biology, 12(1), 169. DOI 10.1186/1471-2229-12-169. [Google Scholar] [CrossRef]
39. Pandey, A., Sharma, M., Pandey, G. K. (2016). Emerging roles of strigolactones in plant responses to stress and development. Frontiers in Plant Science, 7, 434. [Google Scholar]
40. Chen, Z., Gao, X., Zhang, J. (2015). Alteration of osa-miR156e expression affects rice plant architecture and strigolactones pathway. Plant Cell Reports, 34(5), 767–781. DOI 10.1007/s00299-015-1740-x. [Google Scholar] [CrossRef]
41. Jeyaraj, A., Chandran, V., Gajjeraman, P. (2014). Differential expression of microRNAs in dormant bud of tea [Camellia sinensis (L.) O. Kuntze]. Plant Cell Reports, 33(7), 1053–1069. DOI 10.1007/s00299-014-1589-4. [Google Scholar] [CrossRef]
42. Roh, H., Jeong, C. W., Fujioka, S., Kim, Y. K., Lee, S. et al. (2012). Genetic evidence for the reduction of brassinosteroid levels by a BAHD acyltransferase-like protein in Arabidopsis. Plant Physiology, 159(2), 696–709. DOI 10.1104/pp.112.197202. [Google Scholar] [CrossRef]
43. Wang, X. C., Hao, X. Y., Ma, C. L., Cao, H., Yue, C. et al. (2014). Identification of differential gene expression profiles between winter dormant and sprouting axillary buds in tea plant (Camellia sinensis) by suppression subtractive hybridization. Tree Genetics & Genomes, 10(5), 1149–1159. DOI 10.1007/s11295-014-0749-6. [Google Scholar] [CrossRef]
44. Lang, G. A. (1987). Dormancy: A new universal terminology. HortScience, 22, 817–820. [Google Scholar]
45. Ha, C. V., Leyva-Gonzalez, M. A., Osakabe, Y., Tran, U. T., Nishiyama, R. et al. (2014). Positive regulatory role of strigolactone in plant responses to drought and salt stress. Proceedings of the National Academy of Sciences of the United States of America, 111(2), 851–856. DOI 10.1073/pnas.1322135111. [Google Scholar] [CrossRef]
46. Liu, J. W., He, H. Z., Vitali, M., Visentin, I., Charnikhova, T. et al. (2015). Osmotic stress represses strigolactone biosynthesis in Lotus japonicus roots: Exploring the interaction between strigolactones and ABA under abiotic stress. Planta, 241(6), 1435–1451. DOI 10.1007/s00425-015-2266-8. [Google Scholar] [CrossRef]
47. Umehara, M., Hanada, A., Magome, H., Takeda-Kamiya, N., Yamaguchi, S. (2010). Contribution of strigolactones to the inhibition of tiller bud outgrowth under phosphate deficiency in rice. Plant and Cell Physiology, 51(7), 1118–1126. DOI 10.1093/pcp/pcq084. [Google Scholar] [CrossRef]
48. Screpanti, C., Fonné-Pfister, R., Lumbroso, A., Rendine, S., Lachia, M. et al. (2016). Strigolactone derivatives for potential crop enhancement applications. Bioorganic & Medicinal Chemistry Letters, 26(10), 2392–2400. DOI 10.1016/j.bmcl.2016.03.072. [Google Scholar] [CrossRef]
49. Koltai, H. (2011). Strigolactones are regulators of root development. New Phytologist, 190(3), 545–549. DOI 10.1111/j.1469-8137.2011.03678.x. [Google Scholar] [CrossRef]
50. Wen, C., Zhao, Q. C., Nie, J., Liu, G. Q., Shen, L. et al. (2016). Physiological controls of chrysanthemum DgD27 gene expression in regulation of shoot branching. Plant Cell Reports, 35(5), 1053–1070. DOI 10.1007/s00299-016-1938-6. [Google Scholar] [CrossRef]
51. Shen, H., Luong, P., Huq, E. (2007). The F-Box protein MAX2 functions as a positive regulator of photomorphogenesis in Arabidopsis. Plant Physiology, 145(4), 1471–1483. DOI 10.1104/pp.107.107227. [Google Scholar] [CrossRef]
52. Kebrom, T. H., Burson, B. L., Finlayson, S. A. (2006). Phytochrome B represses teosinte branched 1 expression and induces sorghum axillary bud outgrowth in response to light signals. Plant Physiology, 140(3), 1109–1117. DOI 10.1104/pp.105.074856. [Google Scholar] [CrossRef]
53. Cooke, J. E. K., Eriksson, M. E., Junttila, O. (2012). The dynamic nature of bud dormancy in trees: Environmental control and molecular mechanisms. Plant Cell and Environment, 35(10), 1707–1728. DOI 10.1111/j.1365-3040.2012.02552.x. [Google Scholar] [CrossRef]
54. Yordanova, Y. S., Mab, C., Straussb, S. H., Busov, V. B. (2014). Early Bud-Break 1 (EBB1) is a regulator of release from seasonal dormancy in poplar trees. Proceedings of the National Academy of Sciences of the United States of America, 111(27), 10001–10006. DOI 10.1073/pnas.1405621111. [Google Scholar] [CrossRef]