DOI:10.32604/phyton.2020.011680

| Phyton-International Journal of Experimental Botany DOI:10.32604/phyton.2020.011680 |  |
| Review |
Applications of Molecular Markers in Fruit Crops for Breeding Programs—A Review
1Department of Horticulture, Bahauddin Zakariya University, Multan, 60800, Pakistan
2Department of Horticulture, University of Sargodha, Sargodha, 40100, Pakistan
*Corresponding Author: Muhammad Akbar Anjum. Email: akbaranjum@bzu.edu.pk
Received: 23 May 2020; Accepted: 04 August 2020
Abstract: Selection and use of molecular markers for evaluation of DNA polymorphism in plants are couple of the most important approaches in the field of molecular genetics. The assessment of genetic diversity using morphological markers is not sufficient due to little differentiating traits among the species, genera or their individuals. Morphological markers are not only highly influenced by environmental factors but skilled assessment is also prerequisite to find the variations in plant genetic resources. Therefore, molecular markers are considered as efficient tools for detailed DNA based characterization of fruit crops. Molecular markers provide new directions to the efforts of plant breeders particularly in genetic variability, gene tags, gene localization, taxonomy, genetic diversity, phylogenetic analysis and also play an important role to decrease the time required for development of new and excellent cultivars. The success of molecular markers technology in genetic improvement programs depends on the close relationship among the plant breeders, biotechnologists, skilled manpower and good financial support. The present review describes application and success of molecular markers technology used for genetic improvement in different fruit crops.
Keywords: DNA fingerprinting; genetic diversity; genetic improvement programs; germplasm characterization; marker assisted selection
Molecular markers were introduced during the last two decades of the 20th century to accomplish the various aspects of breeding programs in fruit crops. The uses of phenotypic based markers are inadequate due to less differentiating traits and being influenced by environmental factors, which led to development of DNA based markers [1]. Molecular markers are genes or particular segments of DNA that are representative of differences at DNA level [2]. Molecular markers probably may or may not correlate with the expressing morphological traits [3]. All markers have their own strengths and weaknesses based on their selection and aim of study [4]. Molecular markers have many advantages as compared to phenotypic markers because they are stable, detectable in all tissues irrespective of growth, development and differentiation, and also remain unaffected by fluctuations in environmental conditions, cultural impacts and pleiotropic effects [5]. Molecular markers have been applied to detect variations in DNA sequences of varieties, parents or individual plants used in crop improvement programs. The most significant revolution in agricultural biotechnology derived from the research of genomes’ structure and the genetic mechanism behind economically important traits. Molecular markers are most efficient tools for study of genetic variability, gene tags, gene localization, taxonomy, phylogenetic analysis and development of new and excellent cultivars [2].
Molecular markers should have the following ideal properties, i.e., must be simple, inexpensive, polymorphic, with co-dominant inheritance, provide adequate resolution of genetic variations, easy to access, frequent occurrence in genome, require little amounts of DNA sample, have relationship with diverse phenotypes and easy exchange of data among laboratories [6]. Unfortunately, molecular markers technique may differ in every situation and their application depends on work purpose. Molecular markers vary from each other due to following features, i.e., genomic richness, detection level of polymorphism, specificity of locus, reproducibility and costs assay [7]. It is not easy to find a molecular technique which can fulfill all above requirements. However, techniques can be modified according to purpose of study which is undertaken. In this study, importance of molecular markers has been reviewed for genetic improvement programs in fruit crops. Present study is divided into two parts, firs part is about brief description of available molecular markers, while second part includes their application in numerous fruit crops for their improvement programs. Classification of different molecular markers used in fruit crops is illustrated in Fig. 1.
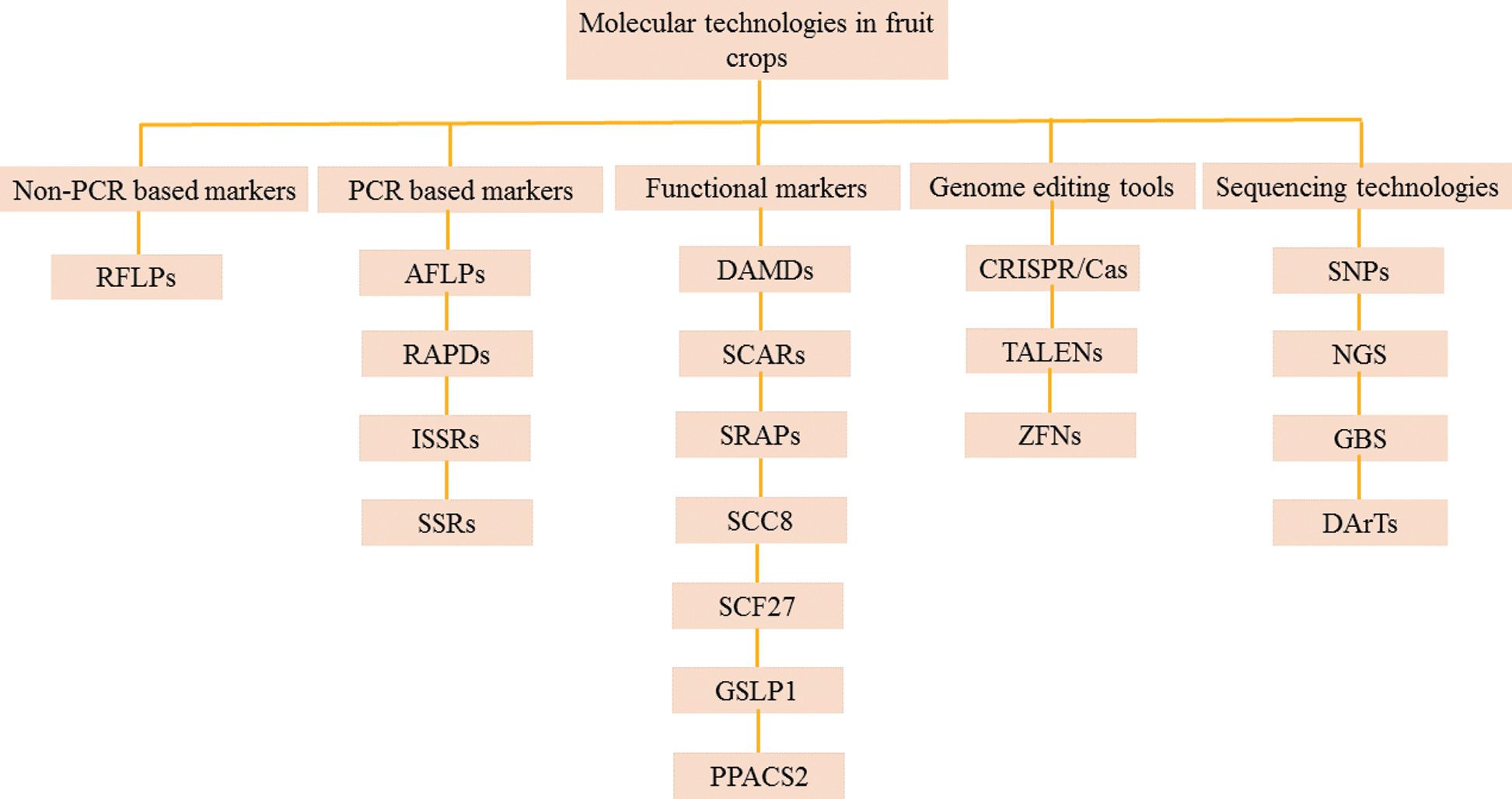
Figure 1: Classification of different molecular markers used for characterization, DNA fingerprinting, genome mapping and genome editing of fruit crops
The uniqueness in characteristics (fruit color, shape and size) of each genotype is considered as marker because it helps to obtain information regarding the genetics of other desired traits. Since ancient times, genotypic differences were measured through morphological descriptors of plants. However, there are some limitations of morphological markers in plant improvement programs such as limited to individual identification, affected by environmental factors, external impacts and developmental stage and difficulty in identifying the homozygous and heterozygous individuals.
In recent years, PCR technique and recombinant DNA technique are extensively used to track the genome regions of plants for many crop breeding programs and also highly linked with agronomic and disease resistance traits in various crops [8]. The first molecular marker used for the construction of genomic maps and detection of DNA polymorphism in humans by Botstein et al. [9] was restriction fragment length polymorphism (RFLP). Different molecular markers systems were later established for plant genome studies [10].
RFLP was the first molecular marker used for genome mapping of humans and then it was used in plant studies [10]. Generally, RFLPs are applied for hybridization studies, genetic variability, genome mapping and to investigate the association of closely linked taxa [11].
These are the main class of molecular markers because of high utility and manipulation. PCR based markers include random amplified polymorphic DNA (RAPD) [12], amplified fragment length polymorphism (AFLP) [13], microsatellites/simple sequence repeats (SSRs) [14], and inter simple sequence repeats (ISSRs) [15].
Single nucleotide polymorphisms (SNPs) are reliable sequenced based markers successfully used in fruit crops [2]. The co-dominance property of SNPs like SSRs makes these markers capable to detect the heterozygous and homozygous fragments [16]. These markers have been successfully involved in next generation sequencing and can be consistently used for various purposes, i.e., association genetics and plant breeding. The SSRs have been used along with SNPs in marker assisted-selection (MAS) to fetch desired attributes from wild germplasm. Great efforts have been made for identification of SNPs markers inside the actual diversity range of breeding materials and confirmed SNPs must be mapped accurately in large segregating populations [17]. SNPs are now dominating markers being applied all over the world due to improvements in DNA sequencing technologies facilitating the construction of high throughput analyses. The limiting factor for SNPs implementation is high initial costs for their development. SNPs have been used for evaluation of variations between two sequences [18].
7 Diversity Arrays Technology (DArT)
These markers can be utilized to detect simultaneously variations at several genomic loci as well as to develop whole genome profiling of several crops without any prior sequencing information. These markers are considered as prospective tools for detailed and powerful evaluation of structure of collected germplasm. High polymorphism information content and very high scoring reproducibility are ideal properties of DArT markers. These arrays were used for determination of genetic associations between wild as well as cultivated germplasm [19]. These markers also allow DNA fingerprinting and identification of genomic regions shared between related genotypes. These are very helpful for evaluation of genetic diversity and genomic studies and found to be more effective for further breeding purposes [19]. However, the easy approach to DArT marker sequences provides an excellent potential to incorporate the diversity knowledge with genomics as well as physical mappings.
8 Next Generation Sequencing (NGS)
NGS has many prospective uses in crop genetics as well as breeding including of fruit crops. These are very useful and efficient to construct genome mapping and to detect QTLs (quantitative trait locus) during wide crosses [20]. These QTLs linked markers can be successfully used for desired traits during progeny selection carrying promising alleles through MAS. To develop the functional markers, next generation sequencing of cDNAs of divergent genotypes for the trait of interest can be applied to detect candidate genes involved in or linked with the trait. Therefore, QTLs must serve as the perfect markers for MAS in crop breeding programs. NGS technologies have been efficiently used in association genetics or population biology and for identification of SNPs [20].
9 Functional or Diagnostic Markers (FMs)
Development of FMs needs allele sequences of functionally characterized genes from which functional motifs disturbing plant phenotype can be recognized. Moreover, application of functional markers mainly depends upon accessibility of robust marker assay techniques. Generally, FMs are developed from polymorphic sites within genes normally involved in phenotypic trait variability. Direct functional markers (DFM) and indirect functional markers (IFM) may possibly differ in development techniques as well as in the level of functional characterization of polymorphisms. It is also necessary to consider the ideal number of genes to be involved in development of functional markers for quantitative traits. Using numerous “key genes” confirms a good control of the trait and gives flexibility during pleiotropic effects of particular genes. However, using limited FMs will significantly decrease developmental cost as well as threat of linkage drag of adverse alleles [2]. Therefore, selection of appropriate “key genes” will be a critical and limiting factor in FMs development. There are following other useful characteristics of functional markers:
• Much effective fixation of alleles in populations;
• Screening for various alleles in natural and breeding populations;
• Combination of functional markers alleles which affect diverse traits in plant breeding;
• Construction of associated functional markers for haplotypes.
10 Applications of Molecular Markers in Fruit Crops
Molecular markers play an important role in breeding programs, i.e., finding diverse parents, increasing selection of elite alleles at loci governing important characters, germplasm characterization, and intellectual property defense. The major objective of breeding program is improvements in development of new commercial and high yielding cultivars; marker-locus-trait combinations may possibly be used as selection criteria for diverse parent selection and selection in segregating populations during commercialization. These combinations are not only biologically and technically important but also more helpful for excellent cultivar performance in important target markets to fetch higher prices. Marker assisted breeding is more helpful in horticultural crops by utilization of model plants in related crop species through adoption of numerous breeding methods such as backcrossing as well as genomic selection known as novel techniques. When important breeding programs attain proper attention, then their efficiency can be improved and costs can also be reduced by implementing of marker-assisted breeding.
Advent of different markers played a vital role in MAS for efficient as well as rapid studies of germplasm including trait mapping [21]. Molecular markers may increase our understanding regarding phenotypic characterization as well as their genetic relationship that can be utilized in further breeding strategies. MAS is more efficient and useful for breeders to attain early selection of a trait. When the trait is under complex genetic control, or when phenotypic traits are unreliable, MAS has significant use to resolve such issues. Phenomics, genomics and proteomics data along with efficient markers system can be utilized for further breeding purpose. Physical, cytogenetic and genetic maps have been developed for detection of population size through different molecular markers using different sequencing technique [2].
Genetic diversity is the variations within or among populations or individuals of a species or genus [22]. The assessment of genetic variations in plant species is pre-requisite for excellent crop production. Huge genetic diversity occurs in some plant species including fruit crops [23]. Diversity may occur in fruit size and quality or in any other trait [24]. Few studies revealed that biotic and abiotic stresses are main factors to lose the valuable germplasm [25]. The success of crop improvement or breeding programs mostly depends on evaluation of genetic variability in the available germplasm. Genetic diversity assessment among various genotypes is an excellent way to know the convolution of available germplasm, identification of hybrid mixtures with maximum heterozygosity and heterosis, and to find relationship of available germplasm using molecular markers for useful conservations plans [26]. Achievements made in breeding of different fruit crops through different molecular systems are presented in Tab. 1.
Table 1: Achievements made in breeding of fruit crops through molecular approaches

RFLPs, RAPDs, AFLPs, SSRs, ISSRs and DArTs can be used for the correct identification of citrus species, genera or individuals. In citrus, RAPDs have been used for assessment of genetic diversity [12], identification of citrus hybrids [27], characterization of cultivars [28], construction of genetic maps [29] and identification of markers linked with agronomic traits of the cultivars [30]. The development of first SSRs was reported in citrus by Kijas et al. [31]. Later, many studies were carried out on the development and application of SSRs for DNA finger printing [14,32]. Yaly et al. [33] used twenty four SSRs for evaluation of twelve genotypes of Citrus, Poncirus and an intergeneric hybrid. DArT markers along with next generation sequencing have successfully been used for construction of high-resolution genetic maps as well as QTLs mapping in citrus. Curtolo et al. [34] identified QTLs linked with fruit traits in F1 progeny resulting from a controlled cross of Murcott × Pera sweet orange. Association mapping can detect QTLs in citrus. Imai et al. [35] detected seven QTLs comprising four novel ones including four important QTLs for fruit weight and one QTL for each, i.e., fruit skin color and pulp firmness through genotyping-by-sequencing. The development of high resolution genomic map of citrus provides greater significance to plant breeders and to link genomic regions with desired traits. So, valuable information was attained regarding polymorphism levels, genetic maps and QTLs in citrus using DArTs [36].
Collections and maintenance of germplasm are very important sources for phenotypic and genotypic analysis in breeding, particularly in fruit crops having long period of juvenile phase [59]. Mango is an economically important fruit crop; therefore many molecular approaches have been developed to study the diversity in collected germplasm. Gomathi et al. [60] developed an easy and effective protocol for extraction of DNA from healthy as well as malformed floral tissue of mango. Molecular markers are efficient tools for characterization of relatedness or variability among the species or cultivars of mango, identification of genes for commercial importance and enhancement through gene transfer technology. Kashkush et al. [61] applied 34 AFLPs for cultivars identification and evaluation of their genetic relationship by constructing a genomic association map, which gave 13 linkage groups. Kumar et al. [62] determined genetic relationship of 18 mango cultivars cultivated in different geological regions using RAPDs and resolved that most of the cultivars were developed from a local mango gene pool and were domesticated later. RAPDs have been used for assessment of genetic relationship among 50 commercial cultivars. Chance seedling hybrids and selections showed close relationship but the genotypes exhibited variations in morphological characteristics due to different geographical regions [62]. SSRs are DNA based markers with high level of polymorphism, co-dominant inheritance, highly abundant in nature, easy to use and readily transferable. These are reported to be more efficient as compared to RFLPs and RAPDs, and widely used in plant genomic studies of mango [61]. Pandit et al. [37] used ISSRs in mango and found clear polymorphisms among various cultivars and also recommended efficient use of these molecular markers for mango improvement in cultivar identification, confirmation of parents, estimation of genetic variability in existing populations, and classification of rootstocks.
Excellent banana production mostly depends on triploid and tetraploid cultivars. However, diploid cultivars are good source of alleles responsible for resistance against biotic as well as abiotic stresses. The cross of tetraploid and improved wild diploids produce triploids hybrids, which have essential agronomic traits, i.e., reduced plant height, insects and disease resistance and good fruit quality [63]. The development of fruit without fertilization is known as parthenocarpy; the resultant fruit is seedless, which is an excellent character of banana cultivars. However, most of the current breeding population is non-parthenocarpic which cannot be recognized until near to harvest. Therefore, marker assisted selection for parthenocarpy at seedling stage would have an excellent impact on efficacy of breeding program. Many series of molecular markers have been developed on the basis of PCR methods, i.e., different markers systems (co-dominant and dominant) were widely used for assessment of genetic variations and phylogenetic relationship among banana genotypes [38]. SSRs have high potential for use in banana due to greater polymorphism, co-dominant nature, highly reproducible and easy interpretation. These are valuable tools in banana breeding programs for germplasm characterization, genome maps construction and MAS. Therefore, many series of SSRs has been developed for breeding programs. However, the number of SSRs available for genetic analysis is still limited, some authenticated and highly polymorphic SSRs are still required [64]. Mattos et al. [65] studied genetic variability for most agronomic traits as well as fruit characteristics in 26 banana cultivars through SSRs. Rout et al. [66] reported that ISSRs are also efficient markers for assessment of genetic variations among banana genotypes. Lamare et al. [67] demonstrated the comparison of SSRs with RAPDs and ISSRs and found that ISSRs and RAPD are also effective markers for genetic studies for breeding purposes. Lu et al. [68] revealed that it is possible to categorize banana genotypes or clones through ISSRs. In a previous study, Ortiz et al. [69] concluded that crosses between divergent genotypes and selecting better hybrids are important factors for the production of new banana cultivars. Jesus et al. [70] found that banana cultivars are mostly derived from crosses between wild diploid sub-species of Musa acuminata (Genoma A) and Musa balbisiana (Genoma B) and exhibit different levels of genome ploidy in their genetic make-up.
Apple germplasm can be easily distinguished with different molecular markers. Goulao et al. [71] used 13 SSRs and 7 ISSRs for assessment of DNA finger printing and determination of genetic relationship among 41 genotypes of apple. Zhang et al. [72] also evaluated genetic diversity in apple cultivars and wild apple species using SSRs. Due to higher reproducibility, SSRs and ISSRs are useful markers for varietal characterization and assessment of genotypic relationship. Diversity was also found to be quite similar within populations from different countries [73]. The presence of high genetic diversity in apple germplasm can be further utilized in selection of trees and hybridization [74]. Woolly aphid is a major economic problem for apple growers all over the globe. Bus et al. [39] constructed a genome map of woolly aphid and detected three resistance genes such as Er1, Er2 and Er3. Apple growers face major losses due to apple scab all over the world, which is caused by a fungal pathogen (Venturia inaequalis). Molecular markers have been widely used to study the pathogenic fungi [75]. RAPDs and ISSRs were used for determination of genetic variability in V. inaequalis, obtained from various orchards. The isolates were observed to be intermixed and not area specific, however cultivar specific isolates were also identified. Similarly, previous studies were also conducted in Venturia populations using RAPDs and RFLPs [76]. Therefore, resistance breeding offers a sustainable way to control this disease. Generally, conventional breeding of apple requires approximately up to twenty years from initial crossing to commercial release, while genomics-assisted breeding can be useful and helpful to accelerate such breeding programs. Apple breeding is also a challenging aspect due to slow findings of various QTLs as well as development of efficient markers. For MAS applications in apple, RAPDs, SSRs, AFLPs, ISSRs, SCARs and SNPs resulting from next generation sequencing were applied to construct high-density molecular marker map. These were applied to find out more precise and correct QTLs. Lee et al. [77] studied GWAS that has potential to detect target loci without any preparing molecular marker map in apple and also proved that it decreased time to find out efficient marker and also solved the issues of QTLs analysis. Moriya et al. [78] identified QTLs controlling flesh mealiness and evaluated their uses in apple breeding. McClure et al. [79] evaluated 172 genotypes under genome-wide association with SNPs. Some important genes were identified for resistance of different fungal pathogens. A signal for firmness retention has also been detected on chromosome 10 after storage.
Production of seedless grape cultivars is a challenging goal of table grape breeding. Low seeded cultivars can be produced through various traditional methods. Recently, modern technologies have been used to improve the production potential of seedless cultivars. Li et al. [42] used different markers, i.e., SCC8, SCF27 and GSLP1 for identification of seedless parents. Only, GSLP1 identified the seeded parents among all the studied markers. Similarly, Liu et al. [80] also used both GLSP1 and SCF27 markers to detect the seedless progeny of five cross combinations. Recently, GSLP1 marker was used to detect the seedless progeny and 14 plantlets were identified as seedless [81]. Markers development for sex expression brought a big revolution by allowing the breeding programs to select desired hermaphrodite progenies before planting in field for excellent performance [82]. Conner et al. [83] found that the primer VR00 9 has greater ability to detect the presence of female allele in seedling progenies of muscadine grapes. Many previous studies have successfully distinguished sex expression in grape vines [84]. SSRs are mainly applied for identification of diverse parents and DNA finger printing of grape cultivars [43]. Rao [43] also studied 42 genotypes of grape vines through seven SSRs for identification of hybrids. Li et al. [85] studied phylogenetic relationship among 16 Chinese genotypes and 13 foreign genotypes by using 9 SSRs. Dendrogram showed close relationship between Chinese genotypes and foreign genotypes. Guo et al. [86] constructed a high density genetic map for Vitis vinifera L. × Vitis amurensis Rupr. This genetic map contains 7,199 SNPs and In-Del markers that may be used for QTLs mapping of desired traits, i.e., resistance against cold and diseases in grapes. Such information can be further utilized for maker-assisted breeding as well as for gene cloning. Recently, Divilov et al. [44] also identified QTLs association with downy mildew resistance phenotypes and used RNA sequencing to find out candidate genes for two QTLs on chromosome 14. Meanwhile, there is more significance to develop new markers for seedless germplasm identification as well as for muscat flavor.
Correct identification and assessment of genetic relationships among genotypes is important for management of genetic variation. The biotic and abiotic stresses and urbanization are the major causes for genetic erosion of valuable germplasm [87]. In recent years, several studies have been carried out using accurate molecular techniques for determination of genetic variability, phylogenetic relationship and genome mapping of pomegranate genetic resources [45]. Caliskan et al. [88] used six SSRs for characterization of 78 pomegranate genotypes. These markers detected high level of polymorphism in pomegranate germplasm. The genotype Sayfi was found to be a clone of genotype Hicaznar, which is considered as excellent genotype on the basis of fruit and juice consumption. Many studies indicated that co-dominant markers have successfully been used for DNA fingerprinting of pomegranate germplasm [89]. Conclusively, it necessitates that this highly diverse genetic collection might be used for further breeding purposes.
Release of early ripening cultivar is one of the most essential breeding purposes for Japanese pear (Pyrus pyrifolia Nakai). Ahmed et al. [90] evaluated sixty genotypes of pear using SDS-PAGE analysis. To enhance MAS for fruit ripening day, different markers, i.e., PPACS 2 and BGA 35 were used in six populations through variance components. The 263-bp allele of PPACS 2 and the 136-bp allele of BGA 35 had early-ripening effects in six populations [91]. The 263-bp allele was previously shown to increase ethylene production and have negative effects on fruit storage [92]. In previous studies, Molecular markers, i.e., PPACS 2 and BGA 35 have been well employed for Japanese pear breeding purposes [91]. Kalkisim et al. [46] collected 31 pear genotypes for assessment of their genetic relationships by using 45 RAPDs and 25 ISSRs. Dendrogram was constructed on the basis of combination data of RAPD and ISSRs. The similarity matrix was ranged from 0.105 to 0.968. Wolko et al. [93] determined genetic diversity among six pear cultivars (P. communis) and two wild pears (P. pyraster) through SSRs. While, Song et al. [94] used a genome-wide set of 134 SSR markers and found that these were enough for evaluation of genetic relationships in P. pyrifolia. Many other studies have used RAPDs and SSRs markers to evaluate genetic variability among various pear genotypes [95]. A high density linkage map is an important tool for satisfactory QTLs mapping as well as map-based gene cloning. Genetic map was constructed through SNPs for fastening scaffolds [96]. SNPs and SSRs have been used to construct a high density genetic map to analyze QTLs of fruit quality related traits [97]. Recently, Wang et al. [98] developed a high-density genetic linkage map in pear (P. communis × P. pyrifolia) through SNPs and SSRs generated from SLAF-seq. The data obtained by using different markers have been utilized for assessment of genetic analyses such as evaluating the pear genetic resources, breeding as well as growing.
Molecular approaches are used to examine genetic diversity within or among the species or genotypes and to identify the desired genes and their improvement through genetic transformation technology [99]. However, genetic transformation information for guava is still not well developed [100]. Molecular markers have been used to determine the out crossing rates, outside pollen contamination in seeded orchards, genetic variations, identification of genotypes and their relationship in breeding populations and are also helpful to reduce the limitations of all other traditional methods [2]. The assessment of genetic variability is important during micropropagation and in vitro germplasm conservation to reduce unwanted somaclonal variants. Rai et al. [100] evaluated genetic similarity of guava plants which were produced from somatic embryogenesis. Recently, SSRs have been extensively used for estimation of genetic diversity and construction of genomic map to exploit and improve the breeding program in guava [99]. It is imperative to increase guava yield and its fruit quality through widening its available genetic resources. The diverse parents can be used for further breeding programs of guava.
Discrimination among closely related cultivars and clones is very difficult [101]. Therefore, molecular markers are the most efficient tools for accurate identification of date palm cultivars, evaluation of genetic diversity and phylogenetic relationships. Zehdi et al. [102] carried out a study on 18 date palm (Phoenix dactylifera L.) genotypes to determine their genetic variability and phylogenetic relationship. A large number of polymorphic DNA bands were generated by using appropriate set of 12 ISSRs. Adawy et al. [103] also applied 7 primers of ISSRs on 4 date palm cultivars, i.e., Samany, Hayany, Siwi and Zaghloul. Highly reproducible and scorable patterns were generated and amplified bands were calculated for presence of polymorphism. Khierallah et al. [104] studied genetic diversity in 30 date palm cultivars by using 22 SSRs. The cultivar “Jamal Al-Dean” was very closely related to “Qitaz,” while Ghanami Akhder was highly divergent from the cultivar “Ghanami Ahmer.” Elmeer et al. [105] tested 30 SSRs for DNA fingerprinting of 11 date palm cultivars. Genetic diversity ranged from 60% to 90% among the genotypes. They also suggested that new co-dominant markers could be used by researchers for further genetic mapping and diversity analysis of date palm germplasm. Several studies have been carried out for precise assessment of genetic diversity and DNA finger printing in date palm by using different molecular markers [48]. Khierallah et al. [106] also examined genetic diversity among 17 date palm cultivars by using 30 RAPDs and 12 ISSRs. ISSRs revealed more number of polymorphic bands as compared to RAPDs. So, ISSR marker system is considered as highly reproducible and more efficient as compared to RAPD marker system. Similar results were achieved by Mirbahar et al. [107], who found high level of polymorphism (84%) in studied germplasm through ISSRs. Ahmad et al. [108] used 30 SSRs and 30 ISSRs for characterization of date palm germplasm.
Molecular markers provide an extensive range of analytical methods to examine the prevailing olive germplasm. Different molecular markers, i.e., AFLPs, RAPDs, ISSRs and SNPs were successfully used for evaluation of genetic variability in olive genotypes. Besnard et al. [109] used 45 RAPDs to distinguish 102 olive genotypes which clustered them on the basis of their geographic origin and specific use. Belaj et al. [49] collected 51 olive genotypes from diverse countries and examined with 46 RAPDs on the basis of their polymorphism and discrimination capacity. Similarly, Cortes et al. [110] applied 34 RAPDs to analyze 40 olive genotypes and also obtained a good correlation between similarity data and their geographic origin. Casas et al. [111] used 8 SSRs for DNA finger printing of 65 olive genotypes from different geological regions. Neighbor-joining dendrogram revealed wide genetic variations in the collected germplasm. High level of genetic variations was also observed by previous researchers who examined olive germplasm through SSRs [112]. The co-dominant nature of SSRs gave maximum fragments per locus and a high level of heterozygosity. Thus, microsatellites are more useful tools for identification of cultivars and also important to assess inter and intra-cultivar variability [113]. RAPDs, AFLP, RFLP and SSRs were used to construct first genomic map between two highly heterozygous olive cultivars, i.e., Leccino et al. [114]. Similarly, Wu et al. [115] also developed a genetic linkage map of olive through RAPDs, SSRs and SCARs markers. Muleo et al. [116] used many SNPs in the Phytochrome A gene through high-resolution melting analysis of DNA. This technique played an important role in detecting the presence of mutations and concluded that SNPs with high-resolution melting analysis are more informative, easy, time saving and low cost method. Hakim et al. [117] also detected nine new SNPs using direct sequencing of the lupeol synthase and cycloartenol synthase genes in sixteen olive cultivars. Aabidine et al. [118] used AFLPs and SSRs to construct a genetic linkage map of olive genotypes.
Pineapple cultivars are often produced from somatic mutations. Genetic diversity within commercial cultivars provides important information necessary for crop improvement programs as well as cultivar protection purposes. Kato et al. [119] conducted phylogenetic studies on 148 genotypes of Ananas comosus and 14 genotypes of related species by using AFLPs. A high level of genetic variation was observed within existing pineapple germplasm. Self-incompatibility, intraspecific hybridization and high levels of somatic mutation are the major causes for this high degree of genetic variability [120]. Duval et al. [120] used RFLPs for analysis of genetic diversity in 301 pineapple accessions and estimated variation at intraspecific level, mostly in wild species A. ananassoides and A. parguazensis. Similarly, Ruas et al. [121] estimated moderate intraspecific genetic variation between the genera Ananas and Pseudananas with RAPDs. Wang et al. [50] also conducted a study on assessment of genetic diversity in 36 genotypes of pineapple from ten different countries using 20 SSRs and 13 ISSRs and the results indicated existence of high level of genetic variability in the studied germplasm. Thus, SSRs are extensively used in plant genomic studies [52].
The evolutionary history of many genotypes of Chinese jujube is still unclear due to frequent exchange of germplasm among various cultivated regions and insufficient information regarding cultivar historical documentation [51]. So, genetic studies using molecular markers could provide new information for evolutionary history of various genotypes. SSRs are extensively used for genetic diversity appraisal, phylogenetic relationship, evolutionary studies and breeding of various populations [122]. Liu et al. [123] used 66 SSRs for evaluation of polymorphism in six genotypes of Chinese jujube. A set of 551 SSRs was developed and applied on six Chinese jujube cultivars for their characterization [124]. Now, the availability of jujube genome sequence permits quick identification of highly polymorphic markers for population genetics and necessitates the development of SSRs primers. Molecular markers provide huge data regarding the polymorphism which is important for significant improvement in breeding purposes and development of new cultivars from available genetic resources.
Molecular markers are very useful due to long juvenile phase (up to seven years) of Indian jujube trees for the identification of cultivars during propagation and growth. Identification of cultivars plays an important role in their authenticity and management [125]. The similar genotypes are known by different names in different geographical regions possibly due to effects of environmental factors [126]. Therefore, morphological and biochemical markers are not efficient source of genetic diversity among jujube genotypes as confirmed by Anjum et al. [127]. There is little information regarding identification or relationship among Indian jujube cultivars through molecular markers [128]. Singh et al. [125] assessed genetic diversity among 47 Indian jujube cultivars with one wild accession using forty ISSRs. Obeed et al. [6] applied 15 ISSRs to identify the association among five Indian jujube cultivars and found that the cultivar Um-salem was genetically different from all other four cultivars, while the two cultivars Pakistany and Komethry were genetically similar. Anjum et al. [127] used different biochemical markers for evaluation of biodiversity among thirteen Indian jujube genotypes. Ahmad et al. [129] also used 20 ISSRs for assessment of genetic relationship among seventeen genotypes of Indian jujube and one accession of wild type.
Recent improvements have decreased the cost of different sequencing techniques while increasing their throughput analyses. The goals set in this area have not been fully met yet and the discovery of highly appropriate and more efficient markers system is still needed. The current study encompasses the applications of numerous markers’ systems used to assess genetic diversity on DNA basis in fruit crops. Molecular markers are able to enhance the effectiveness of breeding new and adapted cultivars in terms of time and cost. Generally, all molecular markers reveal useful information on DNA polymorphism and are used to describe in depth the plants’ genetic make-up. Fruit breeding requires more time compared to other crops due to long juvenile phase, high level of heterozygosity and self-incompatibility between cultivars. Conclusively, MAS plays a significant role in the construction of high-density molecular marker maps of fruit crops using RFLPs, RAPDs, SSRs, ISSRs, SNPs, SCARs, In-Del, RAMPs, SSCPs and DArT resulting from sequencing technologies.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Ahmad, R., Anjum, M. A. (2018). Applications of molecular markers to assess genetic diversity in vegetable and ornamental crops—a review. Journal of Horticultural Science and Technology, 1, 1–7.
2. Ahmad, R., Anjum, M. A., Balal, R. M. (2020). From markers to genome based breeding in horticultural crops: an overview. Phyton-International Journal of Experimental Botany, 89(2), 183–204. [Google Scholar]
3. Saqib, M., Khalid, M. F., Ahmad, R., Anjum, M. A., Hussain, S. (2019). Evaluation of genetic diversity through morphological characteristics of olive (Olea europea L.) germplasm in Pakistan. Science, Technology and Development, 38, 7–12. [Google Scholar]
4. Pena, D. N., Moura, E. F., Rodrigues, S. M., Oliveira, M. S. P., Sanches, J. P. et al. (2020). Molecular characterization of a germplasm bank of Platonia insignis Mart.: a fruit tree. Genetic Resources and Crop Evolution, 67(2), 411–420. DOI 10.1007/s10722-019-00855-w. [Google Scholar] [CrossRef]
5. Gosal, S. S., Wani, S. H., Kang, M. S. (2010). Biotechnology and crop improvement. Journal of Crop Improvement, 24(2), 153–217. DOI 10.1080/15427520903584555. [Google Scholar] [CrossRef]
6. Obeed, R. S., Harhash, M. M., Mawgood, A. A. L. (2008). Fruit properties and genetic diversity of five ber (Ziziphus mauritiana Lamk.) cultivars. Pakistan Journal of Biological Sciences, 11(6), 888–893. DOI 10.3923/pjbs.2008.888.893. [Google Scholar] [CrossRef]
7. Liu, Y., Ge, Y., Zhan, R., Lin, X., Zang, X. et al. (2020). Molecular markers and a quality trait evaluation for assessing the genetic diversity of Avocado landraces from China. Agriculture, 10(4), 102. DOI 10.3390/agriculture10040102. [Google Scholar] [CrossRef]
8. Gupta, P. K., Rustgi, S. (2004). Molecular markers from the transcribed/expressed region of the genome in higher plants. Functional & Integrative Genomics, 4(3), 139–162. DOI 10.1007/s10142-004-0107-0. [Google Scholar] [CrossRef]
9. Botstein, D., White, R. L., Skolnick, M., Davis, R. W. (1980). Construction of a genetic linkage map in man using restriction fragment length polymorphisms. American Journal of Human Genetics, 32, 314–333. [Google Scholar]
10. Zietkiewicz, E., Rafalski, A., Labuda, D. (1994). Genome fingerprinting by simple sequence repeat (SSR) anchored polymerase chain reaction amplification. Genomics, 20(2), 176–183. DOI 10.1006/geno.1994.1151. [Google Scholar] [CrossRef]
11. Jena, S. N., Kumar, S., Nair, N. K. (2009). Molecular phylogeny in Indian Citrus L.(Rutaceae) inferred through PCR-RFLP and trnL-trnF sequence data of chloroplast DNA. Scientia Horticulturae, 119(4), 403–416. DOI 10.1016/j.scienta.2008.08.030. [Google Scholar] [CrossRef]
12. El-Khayat, H. M. (2020). Genetic diversity variation based on RAPD and specific markers of some Citrus species. Middle East Journal, 9(1), 171–185. [Google Scholar]
13. Campos, T. E., Espinosa, G. M. A., Warburton, M. L., Varela, S. A., Monter, V. Á. (2005). Characterization of mandarin (Citrus spp.) using morphological and AFLP markers. Interciencia, 30, 687–693. [Google Scholar]
14. Oliveira, D. A. C., Garcia, A. N., Cristofani, M., Machado, M. A. (2002). Identification of citrus hybrids through the combination of leaf apex morphology and SSR markers. Euphytica, 128(3), 397–403. DOI 10.1023/A:1021223309212. [Google Scholar] [CrossRef]
15. Reddy, M. P., Sarla, N., Siddiq, E. A. (2002). Inter simple sequence repeat (ISSR) polymorphism and its application in plant breeding. Euphytica, 128(1), 9–17. DOI 10.1023/A:1020691618797. [Google Scholar] [CrossRef]
16. Sachidanandam, R., Weissman, D., Schmidt, S. C., Kakol, J. M., Stein, L. D. et al. (2001). A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature, 409(6822), 928–933. DOI 10.1038/35057149. [Google Scholar] [CrossRef]
17. Ganal, M. W., Altmann, T., Röder, M. S. (2009). SNP identification in crop plants. Current Opinion in Plant Biology, 12(2), 211–217. DOI 10.1016/j.pbi.2008.12.009. [Google Scholar] [CrossRef]
18. Duran, C., Appleby, N., Edwards, D., Batley, J. (2009). Molecular genetic markers: discovery, applications, data storage and visualisation. Current Bioinformatics, 4(1), 16–27. DOI 10.2174/157489309787158198. [Google Scholar] [CrossRef]
19. Sardos, J., Perrier, X., Doležel, J., Hřibová, E., Christelova, P. et al. (2016). DArT whole genome profiling provides insights on the evolution and taxonomy of edible Banana (Musa spp.). Annals of Botany, 118(7), 1269–1278. DOI 10.1093/aob/mcw170. [Google Scholar] [CrossRef]
20. Varshney, R. K., Nayak, S. N., May, G. D., Jackson, S. A. (2009). Next-generation sequencing technologies and their implications for crop genetics and breeding. Trends in Biotechnology, 27(9), 522–530. DOI 10.1016/j.tibtech.2009.05.006. [Google Scholar] [CrossRef]
21. Abouzari, A., Solouki, M., Golein, B., Fakheri, B. A., Sabouri, A. et al. (2020). Screening of molecular markers associated to cold tolerance-related traits in Citrus. Scientia Horticulturae, 263, 109145. DOI 10.1016/j.scienta.2019.109145. [Google Scholar] [CrossRef]
22. Rao, V. R., Hodgkin, T. (2002). Genetic diversity and conservation of plant genetic resources. Plant Cell, Tissue and Organ Culture, 68(1), 1–19. DOI 10.1023/A:1013359015812. [Google Scholar] [CrossRef]
23. Bhargava, R., Shukla, A. K., Chauhan, N., Vashishtha, B. B., Dhandar, D. G. (2005). Impact of hybridity on flavonoid spectrum of ber (Ziziphus mauritiana Lamk.). Environmental and Experimental Botany, 53(2), 135–138. DOI 10.1016/j.envexpbot.2004.03.008. [Google Scholar] [CrossRef]
24. Mondini, L., Noorani, A., Pagnotta, M. A. (2009). Assessing plant genetic diversity by molecular tools. Diversity, 1(1), 19–35. DOI 10.3390/d1010019. [Google Scholar] [CrossRef]
25. Hajjar, R., Jarvis, D. I., Herren, G. B. (2008). The utility of crop genetic diversity in maintaining ecosystem services. Agriculture, Ecosystems & Environment, 123(4), 261–270. DOI 10.1016/j.agee.2007.08.003. [Google Scholar] [CrossRef]
26. Venkatachalam, K., Long, A. A., Elsaesser, R., Nikolaeva, D., Broadie, K. et al. (2008). Motor deficit in a Drosophila model of mucolipidosis type IV due to defective clearance of apoptotic cells. Cell, 135(5), 838–851. DOI 10.1016/j.cell.2008.09.041. [Google Scholar] [CrossRef]
27. Bastianel, M., Schwarz, S. F., Coleta Filho, H. D., Lin, L. L., Machado, M. et al. (1998). Identification of zygotic and nucellar tangerine seedlings (Citrus spp.) using RAPD. Genetics and Molecular Biology, 21(1), 123–127. DOI 10.1590/S1415-47571998000100020. [Google Scholar] [CrossRef]
28. Deng, Z. N., Gentile, A., Nicolosi, E., Domina, F., Vardi, A. et al. (2015). Identification of in vivo and in vitro lemon mutants by RAPD markers. Journal of Horticultural Science, 70(1), 117–125. DOI 10.1080/14620316.1995.11515281. [Google Scholar] [CrossRef]
29. Cai, Q. G. C. L., Guy, C. L., Moore, G. A. (1994). Extension of the linkage map in citrus using random amplified polymorphic DNA (RAPD) markers and RFLP mapping of cold-acclimation-responsive loci. Theoretical and Applied Genetics, 89(5), 606–614. DOI 10.1007/BF00222455. [Google Scholar] [CrossRef]
30. Nicolosi, E., Deng, Z. N., Gentile, A., La Malfa, S., Continella, G. et al. (2000). Citrus phylogeny and genetic origin of important species as investigated by molecular markers. Theoretical and Applied Genetics, 100(8), 1155–1166. DOI 10.1007/s001220051419. [Google Scholar] [CrossRef]
31. Kijas, J. M., Fowler, J. C., Garbett, C. A., Thomas, M. R. (1994). Enrichment of microsatellites from the citrus genome using biotinylated oligonucleotide sequences bound to streptavidin-coated magnetic particles. Biotechniques, 16, 656–660. [Google Scholar]
32. Ahmed, S., Rattanpal, H. S., Kumari, P., Singh, J. (2017). Study of genetic variability in citrus fruit crop by molecular markers-a review. International Journal of Pure and Applied Bioscience, 5(1), 111–128. DOI 10.18782/2320-7051.2480. [Google Scholar] [CrossRef]
33. Yaly, C. M., Novelli, V. M., Bastianel, M., Machado, M. A. (2011). Transferability and level of heterozygosity of microsatellite markers in Citrus species. Plant Molecular Biology Reporter, 29(2), 418–423. DOI 10.1007/s11105-010-0241-x. [Google Scholar] [CrossRef]
34. Curtolo, M., Yaly, C. M., Gazaffi, R., Takita, M. A., Figueira, A. et al. (2017). QTL mapping for fruit quality in Citrus using DArTseq markers. BMC Genomics, 18(1), 289. DOI 10.1186/s12864-017-3629-2. [Google Scholar] [CrossRef]
35. Imai, A., Nonaka, K., Kuniga, T., Yoshioka, T., Hayashi, T. (2018). Genome-wide association mapping of fruit-quality traits using genotyping-by-sequencing approach in citrus landraces, modern cultivars, and breeding lines in Japan. Tree Genetics and Genomes, 14(2), 24. DOI 10.1007/s11295-018-1238-0. [Google Scholar] [CrossRef]
36. Curtolo, M., Soratto, T. A. T., Gazaffi, R., Takita, M. A., Machado, M. A. et al. (2018). High-density linkage maps for Citrus sunki and Poncirus trifoliata using DArTseq markers. Tree Genetics and Genomes, 14(1), 5. DOI 10.1007/s11295-017-1218-9. [Google Scholar] [CrossRef]
37. Pandit, S. S., Mitra, S., Giri, A. P., Pujari, K. H., Patil, B. P. et al. (2007). Genetic diversity analysis of mango cultivars using inter simple sequence repeat markers. Current Science, 93, 1135–1141. [Google Scholar]
38. Amorim, E. P., Vilarinhos, A. D., Cohen, K. O., Amorim, V. B., Santos-Serejo, J. A. D. et al. (2009). Genetic diversity of carotenoid-rich bananas evaluated by Diversity Arrays Technology (DArT). Genetics and Molecular Biology, 32(1), 96–103. DOI 10.1590/S1415-47572009005000024. [Google Scholar] [CrossRef]
39. Bus, V. G. M., Chagné, D., Bassett, H. C. M., Bowatte, D., Calenge, F. et al. (2008). Genome mapping of three major resistance genes to woolly apple aphid (Eriosoma lanigerum Hausm.). Tree Genetics and Genomes, 4(2), 223–236. DOI 10.1007/s11295-007-0103-3. [Google Scholar] [CrossRef]
40. Chagné, D., Carlisle, C. M., Blond, C., Volz, R. K., Whitworth, C. J. et al. (2007). Mapping a candidate gene (MdMYB10) for red flesh and foliage colour in apple. BMC Genomics, 8(1), 212. DOI 10.1186/1471-2164-8-212. [Google Scholar] [CrossRef]
41. Costa, F., Stella, S., Weg, V. W. E., Guerra, W., Cecchinel, M. et al. (2005). Role of the genes Md-ACO1 and Md-ACS1 in ethylene production and shelf life of apple (Malus domestica Borkh). Euphytica, 141(1–2), 181–190. DOI 10.1007/s10681-005-6805-4. [Google Scholar] [CrossRef]
42. Li, Z. Q., Li, T. M., Wang, Y. J., Xu, Y. (2015). Breeding new seedless grapes using in ovulo culture and marker-assisted selection. Vitro Cellular and Developmental Biology—Plant, 51, 241–248. [Google Scholar]
43. Rao, V. (2017). Role of SSR markers in characterization of grape (Vitis vinifera L.) genotypes and hybrids. International Journal of Agriculture Innovations and Research, 5, 854–858. [Google Scholar]
44. Divilov, K., Barba, P., Davidson, C. L., Reisch, B. I. (2018). Single and multiple phenotype QTL analyses of downy mildew resistance in interspecific grapevines. Theoretical and Applied Genetics, 131(5), 1133–1143. DOI 10.1007/s00122-018-3065-y. [Google Scholar] [CrossRef]
45. Ophir, R., Sherman, A., Rubinstein, M., Eshed, R., Schwager, M. S. et al. (2014). Single-nucleotide polymorphism markers from de-novo assembly of the pomegranate transcriptome reveal germplasm genetic diversity. PLoS One, 9(2), e88998. DOI 10.1371/journal.pone.0088998. [Google Scholar] [CrossRef]
46. Kalkisim, O., Okcu, M., Okcu, Z., Karabulut, B., Yildirim, N. et al. (2016). Relationships among some Pears Genotypes (Pyrus Communis L.) based on ISSR and RAPD analysis. Erwerbs-Obstbau, 58(4), 259–264. DOI 10.1007/s10341-016-0287-5. [Google Scholar] [CrossRef]
47. Pessanha, P. G. D. O., Viana, A. P., Amaral Júnior, A. T. D., Souza, R. M. D., Teixeira, M. C. et al. (2011). Assessment of genetic diversity in access to Psidium spp. via RAPD markers. Revista Brasileira de Fruticultura, 33(1), 129–136. DOI 10.1590/S0100-29452011000100018. [Google Scholar] [CrossRef]
48. Yusuf, A. O., Culham, A., Aljuhani, W., Ataga, C. D., Hamza, A. M. et al. (2015). Genetic diversity of Nigerian date palm (Phoenix dactylifera) germplasm based on microsatellite markers. Journal of Bioscience and Biotechnology, 7, 121–132. [Google Scholar]
49. Belaj, A., Trujillo, I., Rosa, D. L. R., Rallo, L., Gimenez, M. J. (2001). Polymorphism and discrimination capacity of randomly amplified polymorphic markers in an olive germplasm bank. Journal of the American Society for Horticultural Science, 126(1), 64–71. DOI 10.21273/JASHS.126.1.64. [Google Scholar] [CrossRef]
50. Wang, J. S., He, J. H., Chen, H. R., Chen, Y. Y., Qiao, F. (2017). Genetic diversity in various accessions of pineapple [Ananas comosus (L.) Merr.] using ISSR and SSR markers. Biochemical Genetics, 55(5–6), 347–366. DOI 10.1007/s10528-017-9803-z. [Google Scholar] [CrossRef]
51. Wang, S., Liu, Y., Ma, L., Liu, H., Tang, Y. et al. (2014). Isolation and characterization of microsatellite markers and analysis of genetic diversity in Chinese jujube (Ziziphus jujuba Mill.). PLoS One, 9(6), e99842. DOI 10.1371/journal.pone.0099842. [Google Scholar] [CrossRef]
52. Baranski, R., Kaul, M. A., Nothnagel, T., Cavagnaro, P. F., Simon, P. W. et al. (2012). Genetic diversity of carrot (Daucus carota L.) cultivars revealed by analysis of SSR loci. Genetic Resources and Crop Evolution, 59(2), 163–170. DOI 10.1007/s10722-011-9777-3. [Google Scholar] [CrossRef]
53. Pérez, V., Larrañaga, N., Abdallah, D., Wünsch, A., Hormaza, J. I. (2020). Genetic diversity of local peach (Prunus persica) accessions from La Palma Island (Canary Islands, Spain). Agronomy, 10(4), 454–457. DOI 10.3390/agronomy10040454. [Google Scholar] [CrossRef]
54. Ozkilinc, H., Yildiz, G., Silan, E., Arslan, K., Guven, H. et al. (2020). Species diversity, mating type assays and aggressiveness patterns of Monilinia pathogens causing brown rot of peach fruit in Turkey. European Journal of Plant Pathology, 157(4), 1–16. DOI 10.1007/s10658-020-02040-7. [Google Scholar] [CrossRef]
55. Rajapakse, S., Belthoff, L. E., He, G., Estager, A. E., Scorza, R. et al. (1995). Genetic linkage mapping in peach using morphological, RFLP and RAPD markers. Theoretical and Applied Genetics, 90(3–4), 503–510. DOI 10.1007/BF00221996. [Google Scholar] [CrossRef]
56. Jiao, Y., Ma, R. J., Shen, Z. J., Yu, M. L. (2014). Development of Ty1-copia retrotransposon-based SSAP molecular markers for the study of genetic diversity in peach. Biochemical Systematics and Ecology, 57, 270–277. DOI 10.1016/j.bse.2014.08.010. [Google Scholar] [CrossRef]
57. Lee, Y. R., Kim, J., Lee, S. Y., Lee, J. (2020). Diallelic SNP marker development and genetic linkage map construction in octoploid strawberry (Fragaria × ananassa) through next-generation resequencing and high-resolution melting analysis. Horticulture, Environment, and Biotechnology, 61(2), 371–383. DOI 10.1007/s13580-019-00223-8. [Google Scholar] [CrossRef]
58. Miller-Butler, M. A., Smith, B. J., Kreiser, B. R., Blythe, E. K. (2019). Comparison of anthracnose resistance with the presence of two SCAR markers associated with the Rca2 gene in strawberry. HortScience, 54(5), 793–798. DOI 10.21273/HORTSCI13805-18. [Google Scholar] [CrossRef]
59. García-García, A. L., Grajal-Martín, M. J. González-Rodríguez, Á. M. (2020). Polyploidization enhances photoprotection in the first stages of Mangifera indica. Scientia Horticulturae, 264, 189–198. [Google Scholar]
60. Gomathi, K. A., Nailwal, T. K., Bains, G., Shukla, A., Pant, R. C. (2005). A rapid and efficient procedure for extracting high quality genomic DNA from mango (Mangifera indica L.). Physiology and Molecular Biology of Plants, 11, 169–171. [Google Scholar]
61. Kashkush, K., Gui, J. F., Tomer, E., Hillel, J., Lavi, U. (2001). Cultivar identification and genetic map of mango (Mangifera indica). Euphytica, 122(1), 129–136. DOI 10.1023/A:1012646331258. [Google Scholar] [CrossRef]
62. Kumar, H., Narayanaswamy, P., Prasad, T., Mukunda, G. K., Sondur, S. (2001). Estimation of genetic diversity of commercial mango (Mangifera indica L.) cultivars using RAPD markers. Journal of Horticultural Science and Biotechnology, 76(1), 529–533. DOI 10.1080/14620316.2001.11511322. [Google Scholar] [CrossRef]
63. Silva, S. D. O., Flores, J. C. D. O., Neto, L. F. P. (2002). Evaluation of banana cultivars and hybrids in four production cycles. Pesquisa Agropecuária Brasileira, 37(11), 1567–1574. DOI 10.1590/S0100-204X2002001100007. [Google Scholar] [CrossRef]
64. Amorim, E. P., Silva, P. H., Ferreira, C. F., Amorim, V. B. O., Santos, V. J. et al. (2012). Research note new microsatellite markers for bananas (Musa spp). Genetics and Molecular Research, 11(2), 1093–1098. DOI 10.4238/2012.April.27.8. [Google Scholar] [CrossRef]
65. Mattos, L. A., Amorim, E. P., Amorim, V. B. D. O., Cohen, K. D. O., Ledo, C. A. D. S. (2010). Agronomical and molecular characterization of banana germplasm. Pesquisa Agropecuária Brasileira, 45(2), 146–154. DOI 10.1590/S0100-204X2010000200005. [Google Scholar] [CrossRef]
66. Rout, G. R., Senapati, S. K., Aparajita, S., Palai, S. K. (2009). Studies on genetic identification and genetic fidelity of cultivated banana using ISSR markers. Plant Omics, 2, 250–258. [Google Scholar]
67. Lamare, A., Rao, S. R. (2015). Efficacy of RAPD, ISSR and DAMD markers in assessment of genetic variability and population structure of wild Musa acuminata colla. Physiology and Molecular Biology of Plants, 21(3), 349–358. DOI 10.1007/s12298-015-0295-1. [Google Scholar] [CrossRef]
68. Lu, Y., Zhang, X., Pu, J., Qi, Y., Xie, Y. (2011). Molecular assessment of genetic identity and genetic stability in banana cultivars (Musa spp.) from China using ISSR markers. Australian Journal of Crop Science, 5, 25–31. [Google Scholar]
69. Ortiz, R., Vuylsteke, D. (1996). Recent advances in Musa genetics, breeding and biotechnology. Plant Breeding Abstracts, 66, 1355–1363. [Google Scholar]
70. De Jesus, O. N., De Oliveira E. Silva, S., Amorim, E. P., Ferreira, C. F., de Campos, Jé M. S. et al. (2013). Genetic diversity and population structure of Musa accessions in ex situ conservation. BMC Plant Biology, 13(1), 41. DOI 10.1186/1471-2229-13-41. [Google Scholar] [CrossRef]
71. Goulao, L., Oliveira, C. M. (2001). Molecular characterisation of cultivars of apple (Malus× Domestica Borkh.) using microsatellite (SSR and ISSR) markers. Euphytica, 122(1), 81–89. DOI 10.1023/A:1012691814643. [Google Scholar] [CrossRef]
72. Zhang, Q., Li, J., Zhao, Y., Korban, S. S., Han, Y. (2012). Evaluation of genetic diversity in Chinese wild apple species along with apple cultivars using SSR markers. Plant Molecular Biology Reporter, 30(3), 539–546. DOI 10.1007/s11105-011-0366-6. [Google Scholar] [CrossRef]
73. Liebhard, R., Koller, B., Gianfranceschi, L., Gessler, C. (2003). Creating a saturated reference map for the apple (Malus × Domestica Borkh.) genome. Theoretical and Applied Genetics, 106(8), 1497–1508. DOI 10.1007/s00122-003-1209-0. [Google Scholar] [CrossRef]
74. Fazeli, S., Sheidai, M., Farahani, F., Noormohammadi, Z. (2016). Looking for genetic diversity in iranian apple cultivars (Malus × domestica Borkh.). Journal of Sciences, Islamic Republic of Iran, 27(3), 205–215. [Google Scholar]
75. Gladieux, P., Caffier, V., Devaux, M., Cam, L. B. (2010). Host-specific differentiation among populations of Venturia inaequalis causing scab on apple, pyracantha and loquat. Fungal Genetics and Biology, 47(6), 511–521. DOI 10.1016/j.fgb.2009.12.007. [Google Scholar] [CrossRef]
76. Kumar, A., Bennetzen, J. L. (1999). Plant retrotransposons. Annual Review of Genetics, 33(1), 479–532. DOI 10.1146/annurev.genet.33.1.479. [Google Scholar] [CrossRef]
77. Lee, S. J., Ban, S. H., Kim, G. H., Kwon, S. I., Kim, J. H. et al. (2017). Identification of potential gene-associated major traits using GBS-GWAS for Korean apple germplasm collections. Plant Breeding, 136(6), 977–986. DOI 10.1111/pbr.12544. [Google Scholar] [CrossRef]
78. Moriya, S., Kunihisa, M., Okada, K., Iwanami, H., Iwata, H. et al. (2017). Identification of QTLs for flesh mealiness in apple (Malus× domestica Borkh.). Horticulture Journal, 86(2), 159–170. DOI 10.2503/hortj.MI-156. [Google Scholar] [CrossRef]
79. McClure, K. A., Gardner, K. M., Douglas, G. M., Song, J., Forney, C. F. et al. (2018). A genome-wide association study of apple quality and scab resistance. Plant Genome, 11(1), 1–14. DOI 10.3835/plantgenome2017.08.0075. [Google Scholar] [CrossRef]
80. Liu, Q., Zhang, J. X., Wang, Y. J., Yu, D. D., Xia, H. Y. (2016). Breeding for cold-resistant, seedless grapes from Chinese wild Vitis amurensis using embryo rescue. New Zealand Journal of Crop and Horticultural Science, 44(2), 136–151. DOI 10.1080/01140671.2016.1153489. [Google Scholar] [CrossRef]
81. Li, T., Li, Z., Yin, X., Guo, Y., Wang, Y. et al. (2018). Improved in vitro Vitis vinifera L. embryo development of F 1 progeny of ‘Delight’בRuby seedless’ using putrescine and marker-assisted selection. In Vitro Cellular and Developmental Biology—Plant, 54, 291–301. [Google Scholar]
82. Picq, S., Santoni, S., Lacombe, T., Latreille, M., Weber, A. et al. (2014). A small XY chromosomal region explains sex determination in wild dioecious V. vinifera and the reversal to hermaphroditism in domesticated grapevines. BMC Plant Biology, 14(1), 59–246. DOI 10.1186/s12870-014-0229-z. [Google Scholar] [CrossRef]
83. Conner, P., Conner, J., Catotti, P., Lewter, J., Clark, J. R. et al. (2017). Development and characterization of molecular markers associated with female plants in muscadine grape. Journal of the American Society for Horticultural Science, 142(2), 143–150. DOI 10.21273/JASHS04012-16. [Google Scholar] [CrossRef]
84. Battilana, J., Lorenzi, S., Moreira, F. M., Sanz, M. P., Failla, O. et al. (2013). Linkage mapping and molecular diversity at the flower sex locus in wild and cultivated grapevine reveal a prominent SSR haplotype in hermaphrodite plants. Molecular Biotechnology, 54(3), 1031–1037. DOI 10.1007/s12033-013-9657-5. [Google Scholar] [CrossRef]
85. Li, B., Jiang, J., Fan, X., Zhang, Y., Sun, H. et al. (2017). Molecular characterization of chinese grape landraces (Vitis L.) using microsatellite DNA markers. HortScience, 52(4), 533–540. DOI 10.21273/HORTSCI11802-17. [Google Scholar] [CrossRef]
86. Guo, Y., Shi, G., Liu, Z., Zhao, Y., Yang, X. et al. (2015). Using specific length amplified fragment sequencing to construct the high-density genetic map for Vitis (Vitis vinifera × Vitis amurensis). Frontiers in Plant Science, 6, 393. [Google Scholar]
87. Caliskan, O., Bayazit, S. (2013). Morpho-pomological and chemical diversity of pomegranate accessions grown in Eastern mediterranean region of Turkey. Journal of Agricultural Science and Technology, 15, 1449–1460. [Google Scholar]
88. Caliskan, O., Bayazit, S., Oktem, M., Ergul, A. (2017). Evaluation of the genetic diversity of pomegranate accessions from Turkey using new microsatellite markers. Turkish Journal of Agriculture and Forestry, 41, 142–153. DOI 10.3906/tar-1606-124. [Google Scholar] [CrossRef]
89. Pirseyedi, S. M., Valizadehghan, S., Mardi, M., Ghaffari, M. R., Mahmoodi, P. et al. (2010). Isolation and characterization of novel microsatellite markers in pomegranate (Punica granatum L.). International Journal of Molecular Sciences, 11(5), 2010–2016. DOI 10.3390/ijms11052010. [Google Scholar] [CrossRef]
90. Ahmed, M., Anjum, M. A., Rabbani, M. A., Hassan, L. (2009). Characterization of indigenous Pyrus germplasm of Azad Jammu and Kashmir revealed by SDS-PAGE analysis. African Journal of Biotechnology, 8(22), 6442–6452. [Google Scholar]
91. Nishio, S., Hayashi, T., Yamamoto, T., Yamada, M., Takada, N. et al. (2016). Validation of molecular markers associated with fruit ripening day of Japanese pear (Pyrus pyrifolia Nakai) using variance components. Scientia Horticulturae, 199, 9–14. DOI 10.1016/j.scienta.2015.12.032. [Google Scholar] [CrossRef]
92. Yamamoto, T., Terakami, S., Takada, N., Nishio, S., Onoue, N. et al. (2014). Identification of QTLs controlling harvest time and fruit skin color in Japanese pear (Pyrus pyrifolia Nakai). Breeding Science, 64(4), 351–361. DOI 10.1270/jsbbs.64.351. [Google Scholar] [CrossRef]
93. Wolko, Ł., Antkowiak, W., Lenartowicz, E., Bocianowski, J. (2010). Genetic diversity of European pear cultivars (Pyrus communis L.) and wild pear (Pyrus pyraster (L.) Burgsd.) inferred from microsatellite markers analysis. Genetic Resources and Crop Evolution, 57(6), 801–806. DOI 10.1007/s10722-010-9587-z. [Google Scholar] [CrossRef]
94. Song, Y., Fan, L., Chen, H., Zhang, M., Ma, Q. et al. (2014). Identifying genetic diversity and a preliminary core collection of Pyrus pyrifolia cultivars by a genome-wide set of SSR markers. Scientia Horticulturae, 167, 5–16. DOI 10.1016/j.scienta.2013.12.005. [Google Scholar] [CrossRef]
95. Puskas, M., Höfer, M., Sestraş, R. E., Peil, A., Sestraş, A. F. et al. (2016). Molecular and flow cytometric evaluation of pear (Pyrus L.) genetic resources of the German and Romanian national fruit collections. Genetic Resources and Crop Evolution, 63(6), 1023–1033. DOI 10.1007/s10722-015-0298-3. [Google Scholar] [CrossRef]
96. Wu, J., Wang, Z., Shi, Z., Zhang, S., Ming, R. et al. (2013). The genome of the pear (Pyrus bretschneideri Rehd.). Genome Research, 23(2), 396–408. DOI 10.1101/gr.144311.112. [Google Scholar] [CrossRef]
97. Wu, J., Li, L. T., Li, M., Khan, M. A., Li, X. G. et al. (2014). High-density genetic linkage map construction and identification of fruit-related QTLs in pear using SNP and SSR markers. Journal of Experimental Botany, 65(20), 5771–5781. DOI 10.1093/jxb/eru311. [Google Scholar] [CrossRef]
98. Wang, L., Li, X., Wang, L., Xue, H., Wu, J. et al. (2017). Construction of a high-density genetic linkage map in pear (Pyrus communis × Pyrus pyrifolia nakai) using SSRs and SNPs developed by SLAF-seq. Scientia Horticulturae, 218, 198–204. DOI 10.1016/j.scienta.2017.02.015. [Google Scholar] [CrossRef]
99. Nimisha, S., Kherwar, D., Ajay, K. M., Singh, B., Usha, K. (2013). Molecular breeding to improve guava (Psidium guajava L.current status and future prospective. Scientia Horticulturae, 164, 578–588. DOI 10.1016/j.scienta.2013.10.017. [Google Scholar] [CrossRef]
100. Rai, M. K., Asthana, P., Jaiswal, V. S., Jaiswal, U. (2010). Biotechnological advances in guava (Psidium guajava L.recent developments and prospects for further research. Trees, 24(1), 1–12. DOI 10.1007/s00468-009-0384-2. [Google Scholar] [CrossRef]
101. Akkak, A., Scariot, V., Marinoni, D. T., Boccacci, P., Beltramo, C. et al. (2009). Development and evaluation of microsatellite markers in Phoenix dactylifera L. and their transferability to other Phoenix species. Biologia Plantarum, 53(1), 164–166. DOI 10.1007/s10535-009-0026-y. [Google Scholar] [CrossRef]
102. Zehdi, S., Trifi, M., Salem, O. M. A., Marrakchi, M., Rhouma, A. (2002). Survey of inter simple sequence repeat polymorphisms in Tunisian date palms (Phoenix dactylifera L.). Journal of Genetics and Breeding, 56, 77–83. [Google Scholar]
103. Adawy, S. S., Hussein, E. H., El-Khishin, D., Moharam, H., El-Itriby, H. A. (2002). Genetic variability studies and molecular fingerprinting of some Egyptian date palm (Phoenix dactylifera L.) cultivars: II. RAPD and ISSR profiling. Arab Journal of Biotechnology, 5, 225–236. [Google Scholar]
104. Khierallah, H. S. M., Bader, S. M., Baum, M., Hamwieh, A. (2011). Genetic diversity of Iraqi date palms revealed by microsatellite polymorphism. Journal of the American Society for Horticultural Science, 136(4), 282–287. DOI 10.21273/JASHS.136.4.282. [Google Scholar] [CrossRef]
105. Elmeer, K., Sarwath, H., Malek, J., Baum, M., Hamwieh, A. (2011). New microsatellite markers for assessment of genetic diversity in date palm (Phoenix dactylifera L.). 3 Biotech 1, 91–97. [Google Scholar]
106. Khierallah, H. S., Al-Sammarraie, S. K., Mohammed, H. I. (2014). Molecular characterization of some Iraqi date palm cultivars using RAPD and ISSR markers. Journal of Asian Scientific Research, 4, 490–503. [Google Scholar]
107. Mirbahar, A. A., Khan, S., Markhand, G. S., Kauser, N., Saeed, R. (2016). DNA fingerprinting of some Pakistani date palm (Phoenix dactylifera L.) Cultivars using ISSR markers. Pakistan Journal of Botany, 48, 2005–2010. [Google Scholar]
108. Ahmad, R., Malik, W., Anjum, M. A. (2020). Molecular markers systems revealed high genetic similarity among fifty date palm (Phoenix dactylifera) genotypes. International Journal of Agriculture and Biology, 4, 563–574. [Google Scholar]
109. Besnard, G., Baradat, P. H., Chevalier, D., Tagmount, A., Bervillé, A. (2001). Genetic differentiation in the olive complex (Olea europaea) revealed by RAPDs and RFLPs in the rRNA genes. Genetic Resources and Crop Evolution, 48(2), 165–182. DOI 10.1023/A:1011239308132. [Google Scholar] [CrossRef]
110. Cortes, S. F., Badenes, M. L., Paz, S., Iniguez, A., Llacer, G. (2001). Molecular characterization of olive cultivars using RAPD markers. Journal of the American Society for Horticultural Science, 126(1), 7–12. DOI 10.21273/JASHS.126.1.07. [Google Scholar] [CrossRef]
111. Casas, L. G., Scollo, F., Distefano, G., Continella, A., Gentile, A. et al. (2014). Molecular characterization of olive (Olea europaea L.) Sicilian cultivars using SSR markers. Biochemical Systematics and Ecology, 57, 15–19. DOI 10.1016/j.bse.2014.07.010. [Google Scholar] [CrossRef]
112. Debbabi, S. O., Miazzi, M. M., Elloumi, O., Fendri, M. F., Ben Amar, F. et al. (2020). Recovery, assessment, and molecular characterization of minor olive genotypes in Tunisia. Plants, 9(3), 382. DOI 10.3390/plants9030382. [Google Scholar] [CrossRef]
113. Ercisli, S., Ipek, A., Barut, E. (2011). SSR marker-based DNA fingerprinting and cultivar identification of olives (Olea europaea). Biochemical Genetics, 49(9–10), 555–561. DOI 10.1007/s10528-011-9430-z. [Google Scholar] [CrossRef]
114. de la Rosa, R., Angiolillo, A., Guerrero, C., Pellegrini, M., Rallo, L. et al. (2003). A first linkage map of olive (Olea europaea L.) cultivars using RAPD, AFLP, RFLP and SSR markers. Theoretical and Applied Genetics, 106(7), 1273–1282. DOI 10.1007/s00122-002-1189-5. [Google Scholar] [CrossRef]
115. Wu, S. B., Collins, G., Sedgley, M. (2004). A molecular linkage map of olive (Olea europaea L.) based on RAPD, microsatellite, and SCAR markers. Genome, 47(1), 26–35. DOI 10.1139/g03-091. [Google Scholar] [CrossRef]
116. Muleo, R., Colao, M. C., Miano, D., Cirilli, M., Intrieri, M. C. et al. (2009). Mutation scanning and genotyping by high resolution DNA melting analysis in olive germplasm. Genome, 52(3), 252–260. DOI 10.1139/G09-002. [Google Scholar] [CrossRef]
117. Hakim, I. R., Kammoun, N. G., Makhloufi, E., Rebaï, A. (2010). Discovery and potential of SNP markers in characterization of Tunisian olive germplasm. Diversity, 2(1), 17–27. DOI 10.3390/d2010017. [Google Scholar] [CrossRef]
118. Aabidine, A. Z. E., Charafi, J., Grout, C., Doligez, A., Santoni, S. et al. (2010). Construction of a genetic linkage map for the olive based on AFLP and SSR markers. Crop Science, 50(6), 2291–2302. DOI 10.2135/cropsci2009.10.0632. [Google Scholar] [CrossRef]
119. Kato, C. Y., Nagai, C., Moore, P. H., Zee, F., Kim, M. S. et al. (2005). Intra-specific DNA polymorphism in pineapple (Ananas comosus (L.) Merr.) assessed by AFLP markers. Genetic Resources and Crop Evolution, 51(8), 815–825. DOI 10.1007/s10722-005-0005-x. [Google Scholar] [CrossRef]
120. Duval, M. F., Noyer, J. L., Perrier, X., Eeckenbrugge, C., Hamon, P. (2001). Molecular diversity in pineapple assessed by RFLP markers. Theoretical and Applied Genetics, 102(1), 83–90. DOI 10.1007/s001220051621. [Google Scholar] [CrossRef]
121. Ruas, C. F., Ruas, P. M., Cabral, J. R. S. (2001). Assessment of genetic relatedness of the genera Ananas and Pseudananas confirmed by RAPD markers. Euphytica, 119(3), 245–252. DOI 10.1023/A:1017597911205. [Google Scholar] [CrossRef]
122. Sunnucks, P. (2000). Efficient genetic markers for population biology. Trends in Ecology & Evolution Trends, 15(5), 199–203. DOI 10.1016/S0169-5347(00)01825-5. [Google Scholar] [CrossRef]
123. Liu, J., Liu, H., Ma, L., Wang, S., Gao, J. et al. (2014). A Chinese jujube (Ziziphus jujuba Mill.) fruit-expressed sequence tag (EST) library: annotation and EST-SSR characterization. Scientia Horticulturae, 165, 99–105. DOI 10.1016/j.scienta.2013.10.033. [Google Scholar] [CrossRef]
124. Xiao, J., Zhao, J., Liu, M., Liu, P., Dai, L. et al. (2015). Genome-wide characterization of simple sequence repeat (SSR) loci in Chinese jujube and jujube SSR primer transferability. PLoS One, 10(5), e0127812. DOI 10.1371/journal.pone.0127812. [Google Scholar] [CrossRef]
125. Singh, A. K., Sharma, P., Singh, R., Singh, B., Koundal, K. R. et al. (2007). Assessment of genetic diversity in Ziziphus mauritiana using inter-simple sequence repeat markers. Journal of Plant Biochemistry and Biotechnology, 16(1), 35–40. DOI 10.1007/BF03321926. [Google Scholar] [CrossRef]
126. Singh, A. K., Singh, R., Singh, N. K. (2009). Comparative evaluation of genetic relationships among ber (Ziziphus sp.) genotypes using RAPD and ISSR markers. Indian Journal of Genetics and Plant Breeding, 69, 50–57. [Google Scholar]
127. Anjum, M. A., Rauf, A., Bashir, M. A., Ahmad, R. (2018). The evaluation of biodiversity in some indigenous Indian jujube (Zizyphus mauritiana) germplasm through physico-chemical analysis. Acta Scientiarum Polonorum Hortorum Cultus, 17(4), 39–52. DOI 10.24326/asphc.2018.4.4. [Google Scholar] [CrossRef]
128. Singh, A. K., Sharma, R. K., Singh, N. K., Bansal, K. C., Koundal, K. R. et al. (2015). Genetic diversity in ber (Ziziphus spp.) revealed by AFLP markers. Journal of Horticultural Science and Biotechnology, 81(2), 205–210. DOI 10.1080/14620316.2006.11512051. [Google Scholar] [CrossRef]
129. Ahmad, R., Malik, W., Anjum, M. A. (2019). Genetic diversity and selection of suitable molecular markers for characterization of indigenous zizyphus germplasm. Erwerbs-Obstbau, 61(4), 345–353. DOI 10.1007/s10341-019-00438-0. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |