 Open Access
Open Access
ARTICLE
STIL enhances the development of lung adenocarcinoma by regulating the glycolysis pathway
1 Department of Respiratory Medicine, Jen Ching Memorial Hospital, Suzhou, 215300, China
2 Department of Respiratory Medicine, Shandong Provincial Third Hospital, Jinan, 250010, China
* Corresponding Author: XIANJIN XIE. Email:
Oncology Research 2025, 33(1), 123-132. https://doi.org/10.32604/or.2024.048562
Received 12 December 2023; Accepted 19 April 2024; Issue published 20 December 2024
Abstract
Background: To investigate SCL/TAL 1 interrupting locus (STIL)’s role and prognostic significance in lung adenocarcinoma (LUAD) progression, we examined STIL and E2 promoter binding factor 1 (E2F1) expression and their impacts on LUAD prognosis using Gene Expression Profiling Interactive Analysis (GEPIA). Methods: Functional assays including CCK-8, wound-healing, 5-ethynyl-2-deoxyuridine (EdU), Transwell assays, and flow cytometry, elucidated STIL and E2F1’s effects on cell viability, proliferation, apoptosis, and migration. Gene set enrichment analysis (GSEA) identified potential pathways, while metabolic assays assessed glucose metabolism. Results: Our findings reveal that STIL and E2F1 are overexpressed in LUAD, correlating with adverse outcomes. It enhances cell proliferation, migration, and invasion, and suppresses apoptosis, activating downstream of E2F1. Silencing E2F1 reversed the promotion effect of the STIL overexpression on cell viability and invasiveness. Importantly, STIL modulates glycolysis, influencing glucose consumption, lactate production, and energy balance in LUAD cells. Conclusion: Our model, incorporating STIL, age, and disease stage, robustly predicts patient prognosis, underscored STIL’s pivotal role in LUAD pathogenesis through metabolic reprogramming. This comprehensive approach not only confirms STIL’s prognostic value but also highlights its potential as a therapeutic target in LUAD.Keywords
Supplementary Material
Supplementary Material FileLung cancer encompasses various types, with non-small cell lung cancer (NSCLC) accounting for approximately 80%–85% of cases. Within NSCLC, lung adenocarcinoma (LUAD) is the most common subtype, making up about 40%–50% of NSCLC diagnoses, highlighting its significance in the overall landscape of lung cancer [1]. According to the latest data, lung cancer remains the leading cause of cancer deaths globally, with increasing incidence in China [2,3]. The diagnosis of LUAD currently necessitates a thorough integration of clinical radiography, molecular biology, and pathological data to precisely evaluate the prognosis and identify potential treatment targets [4]. Among them, molecular biological detection is important for improving both early diagnosis and prognosis [5,6]. Some genes, such as activated CDC42-related kinase 1 (ACK1), MET, and the stimulator of interferon genes (STING) are beneficial in predicting the prognosis of lung cancer and provide effective methods for early diagnosis [7–9]. Expanding the exploration of other biomarkers will offer greater potential for discovering innovative diagnostic techniques for LUAD.
The SCL/TAL1 interrupting locus (STIL) is a gene involved in regulating the cell cycle and facilitating the duplication of centrioles in dividing cells. Its function primarily involved in centrosome formation and regulating the number of spindles for chromosome segregation [10]. Genetic instability is a common feature of tumor cells. Thus, STIL can influence the development of chromosomal instability by controlling the structure and quantity of centrosomes, which has been linked to various malignant tumors, including prostate, colon, pancreatic, and gastric cancers [10–13]. However, the expression and prognostic significance of STIL in LUAD have not been reported.
Earlier studies showed that glycolysis plays an important role in the ATP synthesis in tumor cells. The Warburg effect revealed that in the tumor, cells prefer to produce lactic acid via glycolysis even when adequate oxygen is present [14–16]. The elevation of lactate levels causes an acidic environment that promotes the growth and expansion of tumor cells. Therefore, glycolysis plays an important role in the progression of tumor cells. Studies have found that oncogenes influence the progression of cancer by participating in the glycolysis process [16]. Peng et al. discovered that USP47 promoted glycolysis and NSCLC progression through BACH1 deubiquitination [17]. Fascin enhanced the glycolysis process, thus promoting both tumor growth and metastasis [18] In this study, the prognostic impact, expression, and modulation of STIL level in LUAD were examined and validated via immunohistochemistry and bioinformatics prediction. This study revealed that STIL promoted the development of LUAD by regulating the glycolysis pathway.
A total of 10 cancer and adjacent tissue samples from LUAD patients (admitted to the hospital during March 2021 and 2022) were used in the analysis. These tissues were obtained during tumor resection surgery and obtained the samples from 7 male and 3 female patients (median age, 59 years; range, 35‒83 years). Of these, 2 had stage I + II cancers while 8 had stage III + IV. Before the study, patients were provided written consent. The study was approved by Ethics Committee of Shandong Provincial Third Hospital (DWKYLL-2021039).
The human bronchial epithelial cell line (16 HBE) and lung cancer cells (NCI-H1975, H1299, H23, A549) were purchased from Procell Life Science & Technology Co., Ltd., (Wuhan, China) and grown in RPMI-1640 media (Gibco, Carlsbad, CA, USA) with 10% fetal bovine serum (FBS, Gibco, USA). Cells were maintained at 37°C with 5% CO2. RiboBio (Guangzhou, China) synthesized siRNA molecules targeting STIL (si-STIL-1 and si-STIL-2), E2 promoter binding factor 1-1 (E2F1-1), E2F1-2, and a negative control (si-NC). The amplified forms of STIL were inserted into the pc-DNA3.1 Vector from Tsingke Biotechnology Co., Ltd., in Beijing, China. The sequences were as follow: si-STIL-1: Sense, 5′ GAAUAUUUCUCAAGUUCAAGG-3′; Anti-sense, 5′ UUGAACUUGAGAAAUAUUCAG-3′; si-STIL-2: Sense, 5′-GAACUUUGUUCAAGAGAAAUG-3′; Anti-sense, 5′-UUUCUCUUGAACAAAGUUCUU-3′; si-E2F1-1: Sense, 5′-GAUGGUUAUGGUGAUCAAAGC-3′; Anti-sense, 5′-UUUGAUCACCAUAACCAUCUG-3′; si-E2F1-2: Sense, 5′-CACUGAAUCUGACCACCAAGC-3′; Anti-sense, 5′-UUGGUGGUCAGAUUCAGUGAG-3′; si-NC: Sense, 5′-UUCUCCGAACGUGUCACGUTT-3′; Anti-sense, 5′-ACGUGACACGUUCGGAGAATT-3′.
Logarithmic growth phase H1299 and H23 cells were plated in 6-well plates (Gibco) and grown for 24 h before transfection. The si-NC, si-STIL-1, si-STIL-2, and STIL overexpression vector were tansfected using Lipofectamine 2000 transfection reagent (Invitrogen, Carlsbad, CA, USA). The transfection effectiveness was determined by Western blotting. Cells in si-STIL-1 + Z-VAD-FMK group were treated with 10 µM Z-VAD-FMK (Sigma-Aldrich, St. louis, MO, USA) for 24 h after transfection.
Cells were lysed with RIPA buffer (E-BC-R327, Elabscience, Wuhan, China) for protein extraction. Proteins (20 μg) were electrophoresed on SDS-PAGE and transferred to PVDF membranes which were treated with antibodies against STIL (ab89314, Abcam, Cambridge, UK), E-Cadherin (ab231303, Abcam), N-Cadherin (ab76011, Abcam), Vimentin (A2584, Abclonal, Wuhan, China), E2F1 (ab288369, Abcam), Hexokinase II (HK-2, ab209847, Abcam), glucose transporter 1 (Glut-1, A6982, Abclonal), Lactate dehydrogenase A (LDHA, #3582, Cell Signalling, Technology, Danvers, MA, USA), cyclin-dependent kinase 4 (CDK4, ab108357, Abcam), cyclin E1 (CCNE1, ab33911, Abcam), cyclin D1 (CCND1, ab16663, Abcam), and GAPDH (ab181603, Abcam) at 4°C overnight. It was then incubated at room temperature for 2 h with the secondary antibody (ab7090 and ab96879, Abcam). The chemiluminescent reagent Vazyme, Nanjing, China was added onto the PVDF membrane. The membrane was scanned and analyzed using the Tanon 4800 image system (Beijing Yuanpinghao Biotechnology Co., Ltd, Beijing, China), and the grayscale valure of the bands used to assess relative expression. Assessment of protein levels was performed 48 h following transfection.
All respective cell lines were allowed to grow in 96-well plates (Beyotime Biotechnology, Shanghai, China), with 3 replicate wells in each group. A growth medium (100 μL/well) was placed in each well and incubated in 5% CO2 for 24 h at 37°C. A volume of 10 μL/well of CCK-8 detection reagent (Dojindo, Kumamoto, Japan) was then added. Theabsorbance of different time (0, 24, 48, 72, and 96 h) were detected at 450 nm via a microplate reader (BIO-TEK, Biotek Winooski, Vermont, USA).
Cells were allowed to grow in 24-well plates (Thermo Fisher Scientific, Waltham, MA, USA) for 24 h. Each well was enriched with EdU medium (100 μL, 50 μmol/L, Beyotime Biotechnology) for 2 h after which cells were placed in paraformaldehyde (PFA, 4%) and 0.5% Triton X-100 solution for fixation followed by EdU staining. Nuclei were labeled with DAPI (Beyotime Biotechnology). All fluorescence microscopic images were quantified for the detection of the proportion of EdU-positive cells.
Cells were trypsinized, rinsed with PBS, and resuspended in Binding buffer to 1 × 106 cells/mL. After adding Annexin V-FITC (BD, San Jose, USA), the sample was kept for 15 min at 25°C (dark). Next, PI was added for 5 min. Results were quantified and analyzed via flow cytometry (BD) and Cell Quest software (BD).
For cell cycle detection, H1299 and H23 cells were fixed in ice-cold 70% ethanol at 4°C for 24 h, washed with PBS, incubated with a staining solution containing PI (Sigma-Aldrich, USA) and RNase A (Sigma-Aldrich) at 4°C for 30 min. Samples were loaded and assessed using a FACS Calibur flow cytometer (BD Biosciences, USA).
H1299 and H23 cells were plated in 6-well plates and grown to monolayers after which the cell surface was scratched vertically with a sterilized tip. The detached or free-floating cells were then rinsed with PBS before the serum-free medium (SFM) was added. They were kept at 37°C with 5% CO2. The images were collected at 0 and 24 h.
The dissolved Matrigel gel (BD) was combined with an serum free medium and distributed uniformly across the chamber of the transwell (upper chamber surface). To turn the gel into gelatin, it was allowed to solidify at 37°C for approximately 2 h. A specific volume of the serum-free culture medium was diluted before digesting and counting the H1299 and H23 cell lines. Cells were inoculated uniformly to the upper portion of the transwell chamber, the chamber was transferred to a plate containing 10% FBS medium. The transwell cells were removed after 24 h of cell culture. Any remaining matrigel adhesive was removed using cotton swabs, and the wells were rinsed thrice with PBS. After PFA fixation and crystal violet staining, the cells migrating the the bottom of the chamber were examined by counting analysis.
Dual-luciferase reporter assay
Initial steps involved cloning the sequence of the wild-type or mutant STIL promoter into the pGL3-luciferase reporter construct. After this, HEK-293T cells were co-transfected with pcDNA3.1-Vector (Vector, Tsingke Biotechnology Co., Ltd.) or E2F1 (Tsingke Biotechnology Co., Ltd.) via Lipofectamine3000 (Invitrogen) for 48 h before assessement of luciferase activity assay via a Promega luciferase activity kit.
Gene expression analysis via RT-qPCR
Total RNA was extracted with TRIzol (Invitrogen) and reverse-transcribed to cDNA with a PrimeScript RT Reagent Kit (TaKaRa, Tokyo, Japan). qRT-PCR was conducted using a SYBR Green PCR Kit (Qiagen, Duesseldorf, Germany), on a LightCycler 480 II (Roche, Switzerland).
Beijing Tsingke Biotech supplied primers [STIL, F primer: 5′-CTACCCTGCAAACAGACCTCA-3′, R primer: 5′-AAAGGATATATAGGCTCCATG-3′; E2F1, F primer: 5′-GGGACTTTGCAGGCAGCGGCG-3′, R primer: 5′-GACGATCTGCGAGGAGTCGAG-3′; GAPDH (internal control), F primer: 5′-CTGCCGGTGACTAACCCTGCG-3′, R primer: 5′-GCCCAATACGACCAAATCAGA-3′]. Expression was determined via the 2−ΔΔCt method.
Chromatin immunoprecipitation (ChIP) assay
This experiment was conducted using ChIP commercial kit (cat:53008; Active Motif, Carlsbad, CA, USA). Cells were cultured into a 10 cm2 plate and allowed to proliferate until reaching a state of 70%–80% confluency. They were fixed and collected for further experiments. All nuclear precipitates were re-suspended in ChIP buffer for ultrasonic treatment and incubated for 24 h with a 5 μg antibody. The DNA protein complex is cross-linked and eluted through a series of washes. The DNA was prepared in TE buffer for analysis. We used RT-qPCR to evaluate STIL’s relative enrichment after the ChIP assay. The primer sequences were: F:5′-CGCCCCGGAACTTAACCGTTG-3′; R:Primer: 5′-TCCGCGCTCGACCAATCCCAA-3′.
Glucose uptake, lactate levels, and ATP/ADP ratios were detected by Glucose Uptake Assay Kit (J1343, Promega, Beijing, China), Lactate Assay Kit (L256, Dojindo), and ADP/ATP Ratio Assay Kit (A552, Dojindo), respectively.
The oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were observed via the Seahorse XF96 Extracellular Flux Analyzer. (Agilent, USA), in a assay medium (Agilent, Santa Clara, CA, USA) within a non-CO2 incubator for 1 h under basal conditions.
The GEPIA website (http://gepia.cancer-pku.cn/index.html) was used to evaluate differences in STIL and E2F1 levels between normal lung tissue and LUAD tissue, and the effect of STIL and E2F1 on the prognosis of LUAD. Clinical data were analyzed by Cox risk regression using the survival package in R (Version 3.4.1). The relationship between STIL and E2F1 in The Cancer Genome Atlas Lung Adenocarcinoma (TCGA-LUAD) was tested by Spearman correlation. Kaplan-Meier curves with log-rank tests were used to assess survival, and then the expression of STIL and clinical indicators in TCGA-LUAD were analyzed using univariate Cox regression. LASSO regression was performed using the glmnet package method. For potential predictive factors, a Cox model was employed for estimating hazard ratios (HRs) with confidence intervals (CIs). The pROC package in R was used for drawing ROC curves. The metadata for TCGA-LUAD patients was provided in Table S1.
Gene set enrichment analysis (GSEA) was used to analyze the transcription factor for STIL based on the TCGA-LUAD database (ENCODE_TF_ChIP-seq_2015.txt). The JASPAR software (https://jaspar.genereg.net/) was used for predicting potential transcription factor interaction sites in the STIL promoter. The association between STIL and E2F1 was examined using R software. GSEA was also used to study the relationship between LUAD-related biological pathways and STIL expression based on the TCGA-LUAD database (h.all.v2023.1.Hs.entrez.gmt).
All results were analyzed via GraphPad Prism 5.0 (GraphPad Software, La Jolla, CA, USA). The data are expressed as the mean ± standard deviation (SD). Data were compared using a two independent sample t-test, and the rates were evaluated via χ2 test or Fisher’s exact probability method. Substantial differences were indicated by p < 0.05. Triplicate experiments were conducted for statistical analysis.
Overexpression of STIL in LUAD
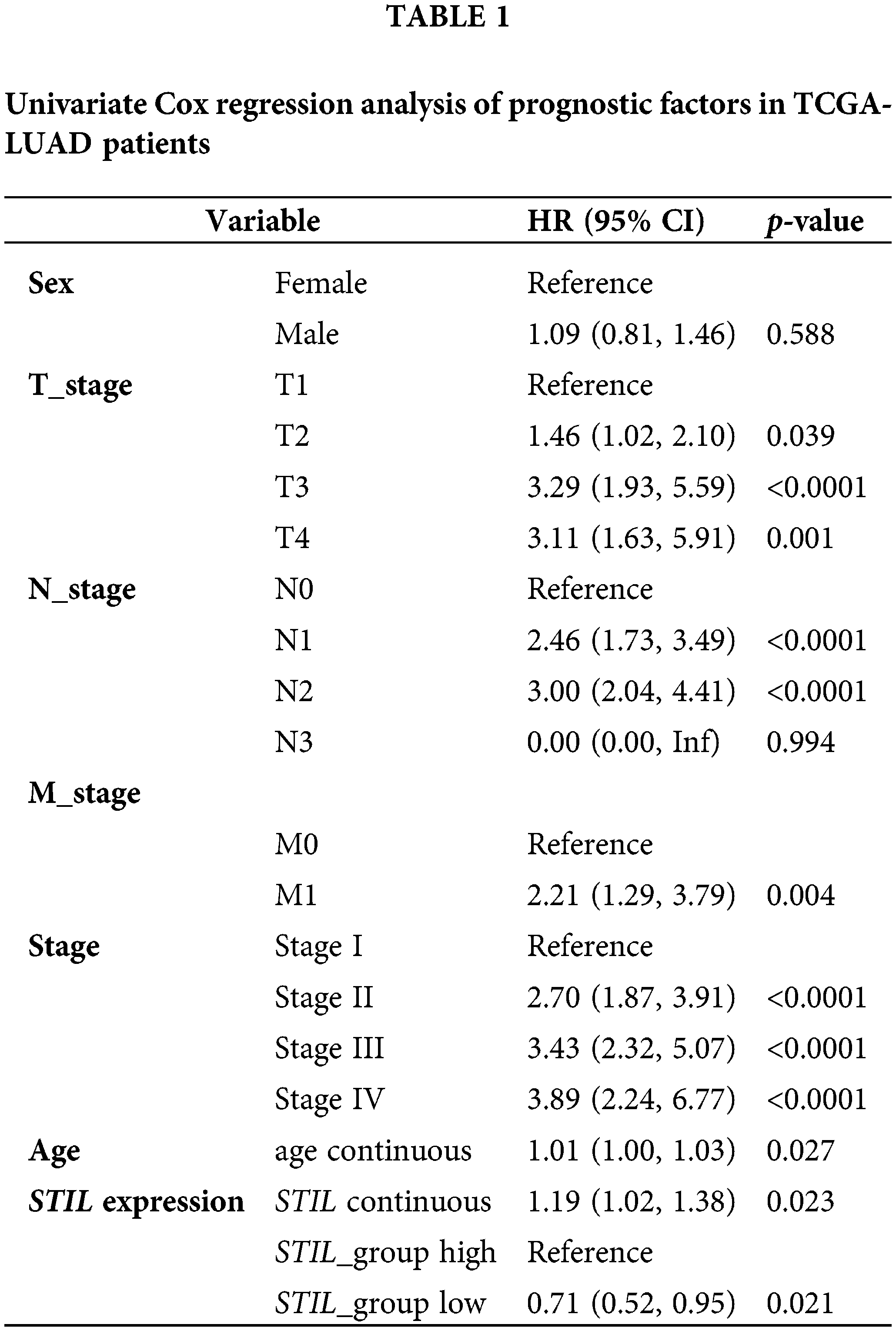
We analyzed the expression pattern of the STIL gene in LUAD tissue as compared with the normal lung tissue using the GEPIA database. The results showed that STIL expression was markedly upregulated in LUAD tissue relative to normal lung tissue (Fig. 1A). Moreover, STIL levels were significantly related to the overall survival (OS). The OS rate of LUAD patients with the STIL low expression being substantially higher compared with those with the high STIL expression (Fig. 1B). Univariate and multivariate analyses were performed using the TCGA-LUAD dataset (Table 1). Through Lasso regression analysis, we selected variables combining the expression level of STIL with clinical indicators based on data of TCGA-LUAD. Aiming to establish a predictive model, we integrated clinical data from TCGA-LUAD patients, examining a comprehensive set of seven clinical parameters including STIL expression, sex, age, stage, (T_stage, N_stage, M_stage) through Lasso regression analysis (Figs. S1A, S1B). Ultimately, three variables with positive coefficients were deemed significant: STIL expression, age, and cancer stage (Fig. 1C). Using these, a Cox proportional hazards model was constructed with the formula: Hazard Ratio = exp (0.920 × Stage II + 1.113 × Stage III + 1.361 × Stage IV + 0.172 × STIL + 0.016 × age). As shown in Fig. 1C, higher stages (II, III, IV) significantly increased risk ratios (HRs) to 2.51, 3.04, and 3.90, respectively, indicating disease progression’s impact. STIL’s elevated levels were associated with an HR of 1.19 (p = 0.034), signifying its prognostic relevance. Age influenced risk marginally yet significantly (HR of 1.02, p = 0.049). Risk scores calculated from this model stratified patients into high and low-risk groups, revealing marked differences in survival, and underscoring the model’s predictive strength (Fig. S1C). Moreover, our model’s risk score demonstrated excellent prognostic prediction for LUAD patients (AUC = 0.683, Fig. S1D). It was also found that STIL was upregulated in LUAD tissue relative to normal samples (Fig. 1D). Moreover, the level of STIL expression in cancer cells was markedly elevated compared to normal cells (16HBE), particularly in H23 and H1299 cells (Fig. 1E). Collectively, the level of STIL was significantly elevated in LUAD, and increased STIL levels were related with unfavorable prognosis.
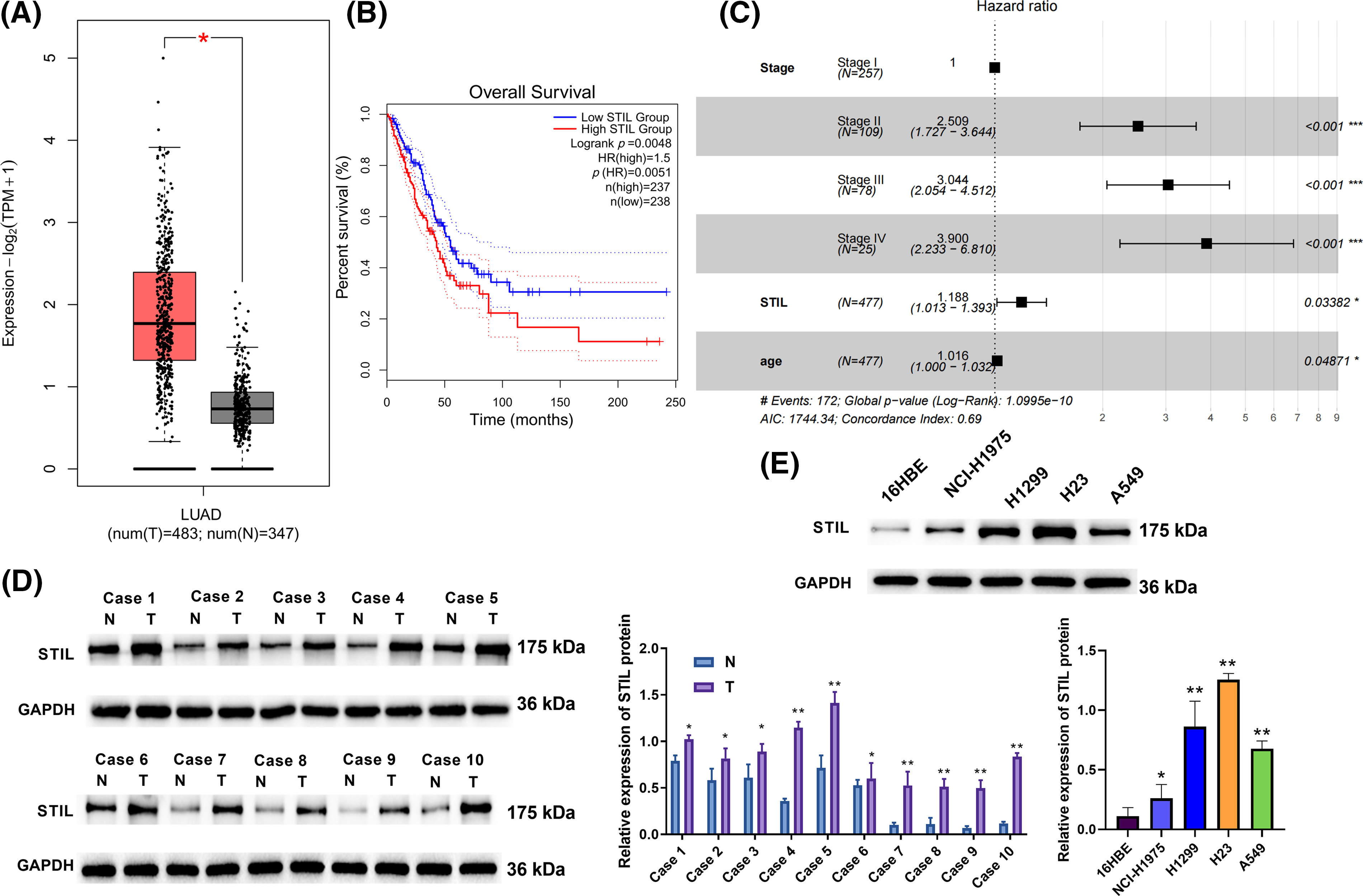
Figure 1: Overexpression of STIL in LUAD. (A) The STIL gene expression level was evaluated using the GEPIA database; (B) STIL gene association with overall LUAD patient survival; (C) Cox risk analysis of LUAD patients; (D and E) the levels of STIL in tissues and cells were quantified via Western blot (T: tumor, N: normal). Compared to normal or 16HBE group, *p < 0.05, **p < 0.01, ***p < 0.001.
Knockdown of STIL inhibited cell proliferation and promoted cell apoptosis
As seen in Fig. 2A and Fig. S2A, STIL expression was notably suppressed or elevated by transfection with siRNAs (si-STIL-1 and si-STIL-2) or STIL-overexpressing vectors. H23 and H1299 cell viability in si-STIL-1 and si-STIL-2 cells was substantially lower than si-NC group (Fig. 2B). Furthermore, the knockdown of STIL markedly reduced cell proliferation in both H23 and H1299 cell lines, as depicted in Fig. 2C. In contrast, an increase in the number of EdU upon STIL overexpression was observed in H23 and H1299 cells (Fig. S2B). Moreover, Knockdown of STIL increased cancer cell apoptosis, as shown in Fig. 2D. In contrast, STIL overexpression inhibited apoptosis (Fig. S2C). Collectively, STIL promoted LUAD cell proliferation and restrained cell apoptosis.
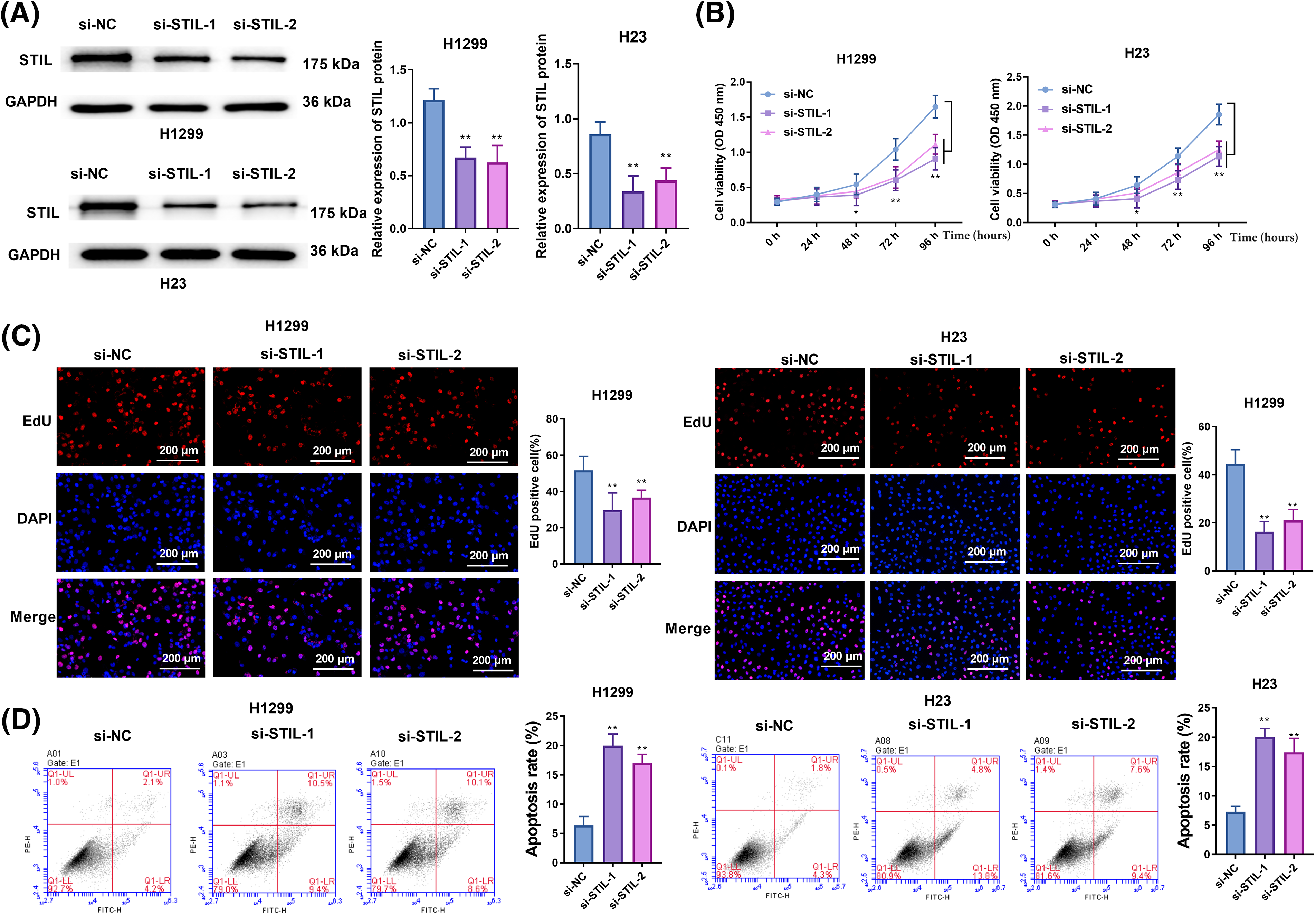
Figure 2: Knockdown of STIL suppressed cell proliferation and enhanced cell apoptosis. (A) STIL protein levels were determined by Western blot; cell viability, proliferation, and apoptosis were examined by CCK-8 (B), EdU (C), and flow cytometry (D), respectively. Compared to the si-NC group, *p < 0.05, **p < 0.01.
Knockdown of STIL restrained cell migration, invasion, and cycle progression
A significant reduction in the rate of wound healing was found in both the si-STIL-1 and si-STIL-2 groups, as assessed by wound healing assays (Fig. 3A), and invading cells, as measured by Transwell assays (Fig. 3B). In contrast, STIL overexpression increased the wound healing rate (Fig. S2D). Moreover, the results obtained from the Western blot analysis (Fig. 3C) showed the molecular mechanisms underlying these cellular changes. Markedly increased E-Cadherin levels were observed after STIL knockdown, suggesting an improvement in epithelial characteristics. Simultaneously, knockdown of STIL induced a significant reduction in the levels of N-Cadherin and Vimentin, implying a concurrent decrease in mesenchymal features. In addition, we also found that knockdown of STIL caused cell cycle arrest at the G1 phase (Fig. 3D). Further measurement of the cell cycle-related proteins, CDK4, CCNE1, and CCND1, confirmed the suppression of cell cycle progression under STIL inhibition (Fig. 3E). Collectively, these observed alterations in protein expression substantiate the conclusion that STIL effectively affected the migratory, invasive, and cycle progression potential of LUAD cells.
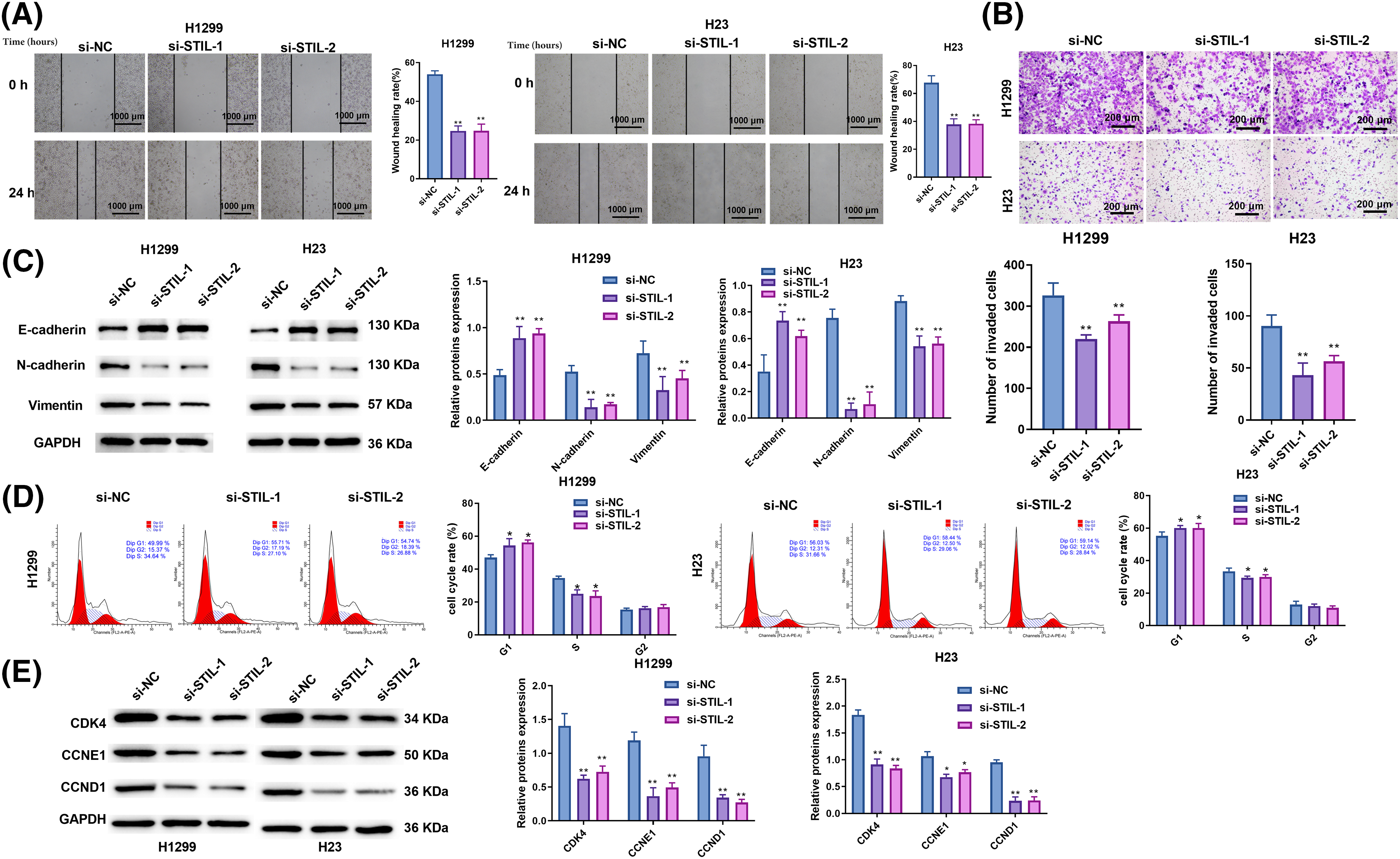
Figure 3: Knockdown of STIL restrained cell migration, invasion, and cycle progression. Invasion and migration of cells were analyzed using the wound-healing (A) and transwell (B) assays; (C) protein levels of N-Cadherin, E-Cadherin, and vimentin were measured by Western blot. (D) Cell cycle distribution was determined using flow cytometry. (E) Western blotting was used to quantify the levels of the cell cycle-associated proteins, cyclin-dependent kinase 4 (CDK4), cyclin E1 (CCNE1), and cyclin D1 (CCND1). Compared to the si-NC group, *p < 0.05, **p < 0.01.
Up-regulation of E2F1 promoted STIL expression level
Based on a single-gene GSEA analysis of TCGA-LUAD using data from “ENCODE_TF_ChIP-seq_2015.txt”, the upstream mechanism of STIL prediction was examined. The results indicate that E2F1 potentially functions as a transcription factor that positively regulates the level of STIL (Figs. 4A, 4B). It was noted that the STIL promoter contains a possible E2F1 binding site upstream of the translation initiation site using the JASPAR software to predict potential transcription factor binding sites (Fig. 4C). Further, GEPIA analysis found that E2F1 was higher expressed in LUAD patients’ samples, and higher E2F1 was correlated with poor prognosis (Figs. 4D, 4E). A positive correlation was observed between E2F1 and STIL expression in LUAD tissues (Fig. 4F). The STIL-E2F1 interaction was verified using dual-luciferase reporter experiment and ChIP assays. The results of the experiment indicated that the luciferase activity of STIL promoter-WT-transfected cells was markedly enhanced by E2F1 upregulation. In contrast, the stimulatory impacts of E2F1 upregulation on luciferase activity were considerably reduced when the binding sequence was mutated. This suggests that E2F1 may interact with the promoter of STIL to increase its expression (Fig. 4G). Relative enrichment of the STIL promoter in the E2F1 group was markedly greater than in the IgG group (Fig. 4H). Further, the down-regulation of E2F1 remarkably inhibited the level of STIL (Fig. 4I). Altogether, E2F1 promoted the STIL expression level. Moreover, the findings of Fig. 4I also confirmed the successful transfection of E2F1 siRNA into H1299 and H23 cells.
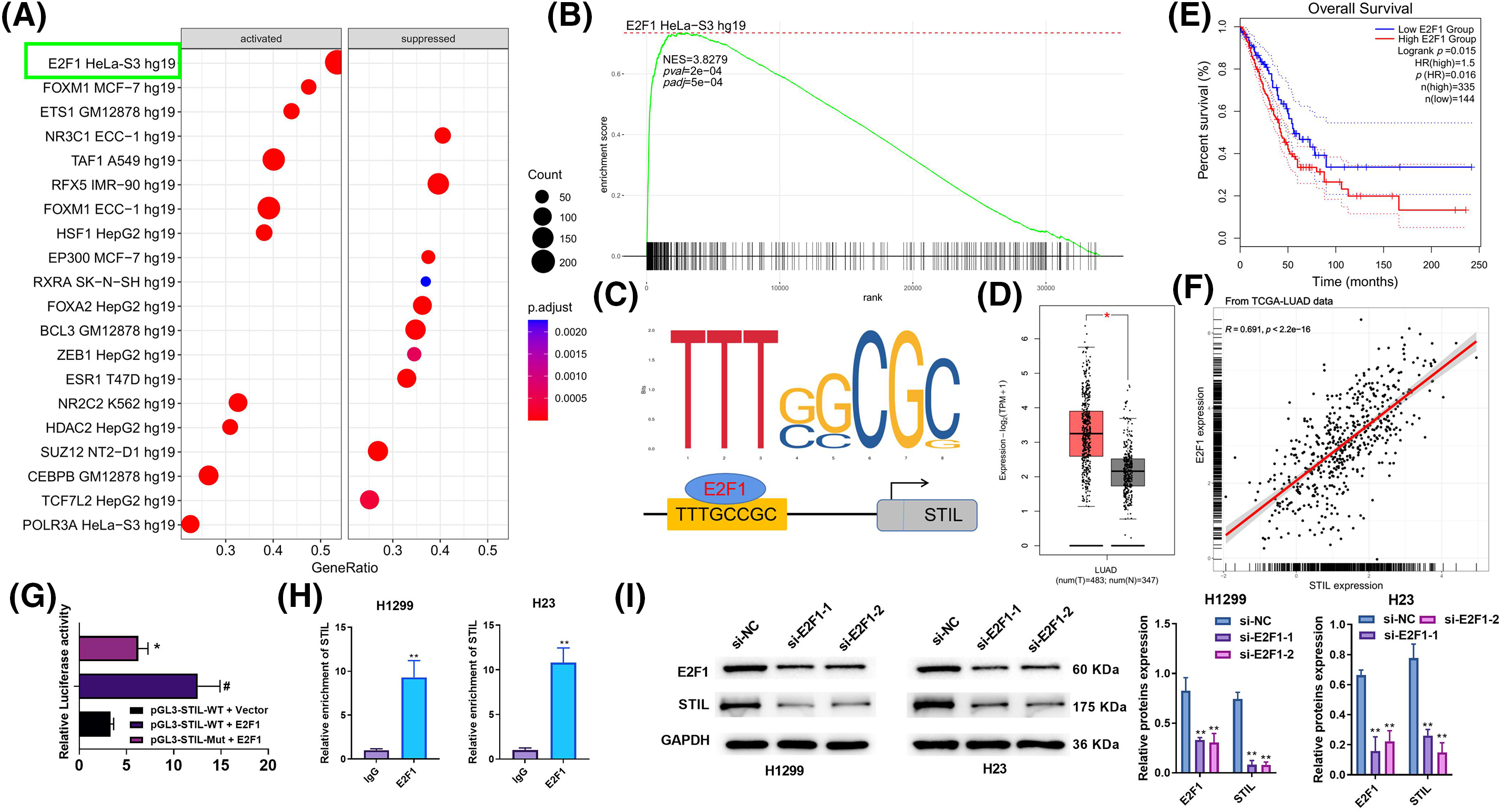
Figure 4: Upregulation of E2F1 promoted STIL expression level. (A and B) GSEA analysis of STIL in LUAD using data from “ENCODE_TF_ChIP-seq_2015.txt”; (C) possible transcription factor binding sites of STIL are predicted by the JASPAR software; (D) analysis of the E2F1 gene expression level using the GEPIA database; (E) the association between the E2F1 gene and the overall survival rates of patients with LUAD; (F) correlation analysis of E2F1 and STIL; dual-luciferase reporter assay (G) and ChIP assay (H) was used to verify the interaction of STIL and E2F1; (I) protein level of E2F1 and STIL were tested by Western blot. Compared to the pGL3-STIL-WT + Vector group, #p < 0.05. Compared to pGL3-STIL-WT + E2F1 or si-NC groups, *p < 0.05, **p < 0.01.
Effects of E2F1 on cell viability, invasion ability, and cell cycle were regulated by STIL
Associations between E2F1 and STIL in cell viability and invasion were further studied. Firstly, we verified the expression levels of E2F1 in 16HBE, H1299, and H23. This showed that E2F1 levels were markedly higher in the LUAD cells relative to the normal cells (Fig. 5A). Secondly, we transfected STIL into cells. We found that after transfection, the level of intracellular STIL was markedly up-regulated, suggesting that the transfection was successful (Fig. 5B). As shown in Fig. 5B, down-regulation of E2F1 could significantly reduce the expression level of STIL. In contrast, overexpression of STIL could reverse this reduction. Moreover, relative to the si-NC + Vector group, the knockdown of E2F1 significantly reduced cell viability, invasion, and the levels of cycle-related proteins, while overexpression of STIL could significantly increase cell viability, invasion, and the levels of cycle-related proteins. Meanwhile, compared with the si-E2F1-1 + Vector group, overexpression of STIL counteracted the effects of E2F1 knockdown on cell viability, invasion, and the levels of cycle-related proteins (Figs. 5C–5E). Collectively, the influence of E2F1 on cell viability, invasion ability, and cell cycle was mediated by STIL.
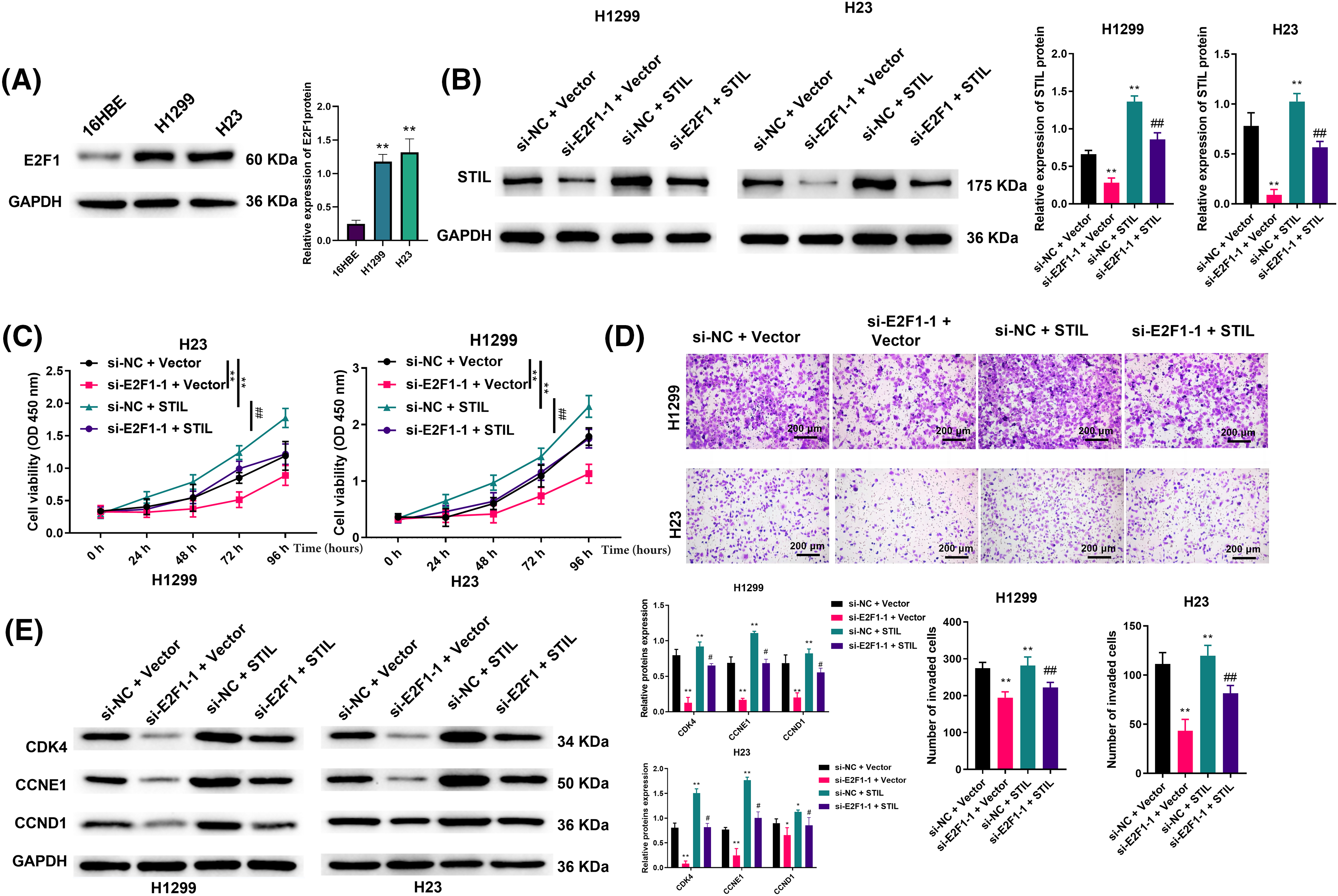
Figure 5: Effects of E2F1 on cell viability and invasion ability were regulated by STIL. (A and B) protein levels of E2F1 and STIL were determined by Western blotting; cell viability and invasion were evaluated by CCK-8 (C) and transwell (D) assays. (E) Western blotting was used to measure the expression levels of the cell cycle-related proteins, CDK4, CCNE1, and CCND1. Compared to the si-NC + Vector group, *p < 0.05, **p < 0.01. Compared to the si-E2F1-1 + Vector group; #p < 0.05, ##p < 0.01.
GSEA enrichment analysis, leveraging the TCGA-LUAD database (h.all.v2023.1.Hs.entrez.gmt), identified glycolysis as a significantly associated pathway with STIL expression (Figs. 6A, 6B). Further investigations revealed that downregulation of STIL notably inhibited intracellular glucose uptake, lactate production, and the ATP/ADP ratio (Figs. 6C–6E). Conversely, overexpression of STIL enhanced glycolytic processes, including glucose uptake, lactate levels, and theATP/ADP ratios in H1222 and H23 cells (Figs. S2E–S2G). Additionally, STIL downregulation reduced the protein levels of key glycolytic enzymes HK-2, GLUT-1, and LDHA (Fig. 6F), while their levels increased following STIL overexpression (Fig. S2H). Notably, STIL knockdown significantly diminished the ECAR (Fig. 6G) and elevated the OCR (Fig. 6H), suggesting a shift from aerobic glycolysis towards enhanced aerobic oxidation. To further explore STIL’s regulatory role in glycolysis, we employed the apoptosis inhibitor Z-VAD-FMK. The findings indicated that STIL downregulation continued to decrease the protein levels of HK-2, GLUT-1, and LDHA even after treatment with Z-VAD-FMK (Fig. 6I). The results revealed that the effect of STIL on the expression of key glycolysis enzymes was not caused by apoptosis. Collectively, the results demonstrate the involvement of STIL in regulating glycolysis in LUAD.
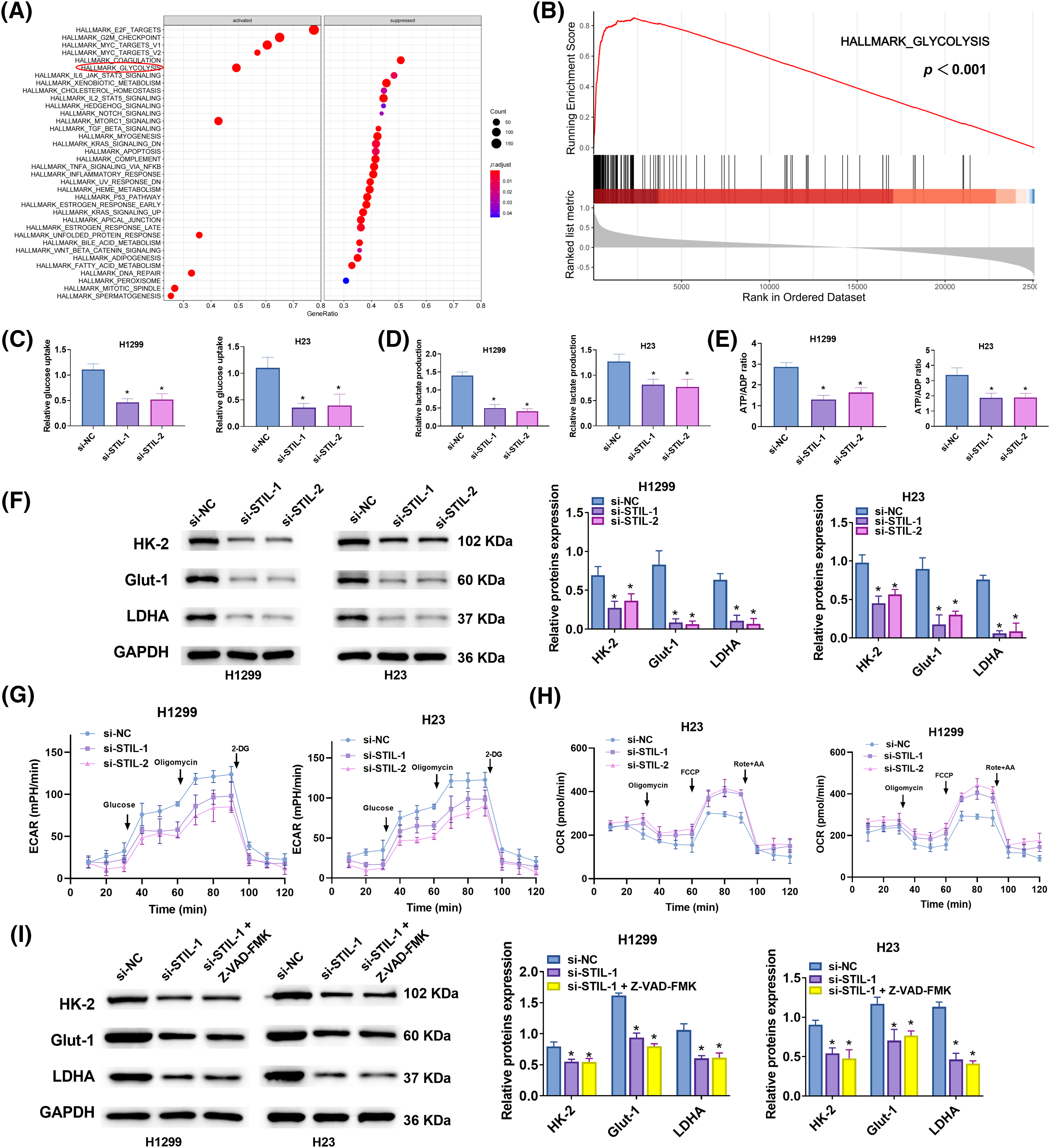
Figure 6: Glycolysis regulated by STIL. (A and B) GSEA analysis of STIL in LUAD using data from “h.all.v2023.1.Hs.symbols.gmt”; (C–E) Glucose absorption, lactate production, and ATP/ADP ratio were assessed using kits; (F) Protein levels of HK-2, GLUT-1, and LDHA were assessed by Western blot. (G) ECAR and (H) OCR in LUAD cells after STIL knockdown. (I) The protein levels of HK-2, GLUT-1, and LDHA were tested by Western blot after treatment with Z-VAD-FMK. Compared with the si-NC group; *p < 0.05.
Lung adenocarcinoma incidence is more than that of other subtypes, and the development and incidence of LUAD have demonstrated an upward trend recently [19,20]. Patients with early lung adenocarcinoma generally have no typical manifestations, which leads to the failure to detect lung cancer in time, delays the best time for treatment, and leads to poor prognosis [21].
Further, the incidence of lung cancer is substantially elevated as a result of the impact of multiple factors, including long-term smoking, working environment, climate change, etc. [22]. The association between CEP85 and STIL was identified in a study by Liu et al. This association facilitates cell migration. This discovery adds to the growing body of evidence emphasizing STIL’s multifaceted roles in cancer biology, particularly in the context of cell migration, a critical component of cancer metastasis [23]. Concurrently, Ouyang et al. reported STIL upregulation in nasopharyngeal carcinoma tissues. Further, Wang et al. discovered that in vivo and in vitro, STIL depletion blocked the growth and metastasis of lung cancer. In contrast, excess STIL promotes cancer cell invasion and migration by triggering the EMT [24]. Importantly, it was found that STIL not only promoted proliferation but also aided migration and invasion. These observations highlight the multifaceted roles of STIL in multiple types of cancer, representing that it may serve as a target for therapeutic interventions [25].
This study focused on examining the expression and prognosis of STIL in LUAD, as well as its influence on the progression of LUAD. The expression of STIL was discovered to be considerably elevated in LUAD, and this was found to be correlated with a negative prognosis. These findings suggest potential use as a biomarker and target for treatment. Meanwhile, we constructed a multifactor Cox risk regression model by combining the expression of STIL with clinical data. The risk score is a reliable predictor of the survival outcome for patients with LUAD. We also examined STIL’s role in LUAD growth and metastasis, revealing its involvement in cell propagation, dissemination, invasion, and cell death. Similar effects were observed in gastric cancer cells upon STIL inhibition [13]. Overall, STIL may function as an oncogene in LUAD and contribute to the development of tumors.
To further elucidate the mechanism underlying STIL’s action in LUAD, additional experiments were conducted. A study by Erez et al. revealed that the transcription factor E2F1 could interact with the STIL promoter. They showed that overexpression of E2F1 induced the expression of STIL, whereas knockdown of E2F1 led to downregulated STIL expression [26]. Similarly, we found that the STIL promoter was bound by transcription factor E2F1 and activated by E2F1. Moreover, E2F1 suppression inhibited cellular invasion and viability. However, STIL overexpression reversed this effect. The findings reveals a possible regulatory pathway that includes E2F1 and STIL, giving a better understanding of how STIL impacts LUAD progression.
Recently, multiple studies have emphasized the importance of active glycolysis as a key promotor in tumorigenesis [27,28]. Importantly, the function of PRDM14 in promoting chemoresistance in A549/cisplatin-resistant cells, including their progenitor A549 human lung adenocarcinoma cells, has been elucidated by He et al. [29]. Furthermore, Li et al. have revealed that heightened expression of GLUT-4 not only enhances cell propagation but also drives glycolysis, with significant effects for NSCLC patients. Collectively, these findings present convincing evidence regarding the pivotal significance of glycolysis in the advancement of lung cancer [30]. In this study, glycolysis product or key enzymes, including HK-2, GLUT-1, LDHA, and ATP/ADP ratio, was inhibited via the knockdown of STIL. Taken together, it is predicted that STIL reduction has an antitumor effect on LUAD via the inhibition of glycolysis. This finding is consistent with the findings of previous studies.
In conclusion, STIL is remarkably highly expressed in LUAD and its elevated level can be used as a marker of unfavorable prognosis in LUAD. This gene, which is anticipated to become a novel target for the detection and intervention of LUAD patients, is activated by E2F1 and influences the progression of LUAD by modulating glycolysis. However, the precise mechanisms by which STIL controls lung cancer cell migration need to be further studied. The complexities of STIL’s function in the progression of cancer biology require further investigation.
Acknowledgement: None.
Funding Statement: The authors have no funding to report.
Author Contributions: Study conception and design: LW and XJX; data collection: LW and XJX; analysis and interpretation of results: LW and XJX; draft manuscript preparation: LW and XJX. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The data that supported the findings of this study are available from the corresponding author upon reasonable request.
Ethics Approval: The protocol of this research has been approved by the Ethics Committee of Shandong Provincial Third Hospital (DWKYLL-2021039). All patients have signed written informed consent.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary material is available online at https://doi.org/10.32604/or.2024.048562.
References
1. Chen P, Liu Y, Wen Y, Zhou C. Non-small cell lung cancer in China. Cancer Commun. 2022;42(10):937–70. doi:10.1002/cac2.12359. [Google Scholar] [PubMed] [CrossRef]
2. Siegel RL, Miller KD. Cancer statistics. 2022 CA Cancer J Clin. 2022;72(1):7–33. doi:10.3322/caac.21708. [Google Scholar] [PubMed] [CrossRef]
3. Sung H, Ferlay J, Siegel RL. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49. doi:10.3322/caac.21660. [Google Scholar] [PubMed] [CrossRef]
4. Duma N, Santana-Davila R, Molina JR. Non-small cell lung cancer: epidemiology, screening, diagnosis, and treatment. Mayo Clin Proc. 2019;94(8):1623–40. doi:10.1016/j.mayocp.2019.01.013. [Google Scholar] [PubMed] [CrossRef]
5. Zhang Z, Ma Y, Guo X, Du Y, Zhu Q, Wang X, et al. FDX1 can impact the prognosis and mediate the metabolism of lung adenocarcinoma. Front Pharmacol. 2021;12:749134. doi:10.3389/fphar.2021.749134. [Google Scholar] [PubMed] [CrossRef]
6. Liao H, Lin P. The comprehensive investigation of transcription factor AP-2 alpha in lung adenocarcinoma. Transl Cancer Res. 2020;9(3):1547–57. doi:10.21037/tcr. [Google Scholar] [CrossRef]
7. Terlecka P, Krawczyk P, Grenda A, Milanowski J. MET gene dysregulation as a promising therapeutic target in lung cancer-a review. J Pers Med. 2021;11(12):1370. doi:10.3390/jpm11121370. [Google Scholar] [PubMed] [CrossRef]
8. Zhu J, Liu Y, Ao H, Liu M, Zhao M, Ma J. Comprehensive analysis of the immune implication of ACK1 gene in non-small cell lung cancer. Front Oncol. 2020;10:1132. doi:10.3389/fonc.2020.01132. [Google Scholar] [PubMed] [CrossRef]
9. Tang W, Zhou W, Ji M, Yang X. Role of STING in the treatment of non-small cell lung cancer. Cell Commun Signal. 2024;22(1):202. doi:10.1186/s12964-024-01586-x. [Google Scholar] [PubMed] [CrossRef]
10. Li J, Yang Z, Qi Y, Liu X, Liu Y, Gao X, et al. STIL Acts as an oncogenetic driver in a primary cilia-dependent manner in human cancer. Front Cell Dev Biol. 2022;10:804419. doi:10.3389/fcell.2022.804419. [Google Scholar] [PubMed] [CrossRef]
11. Pradhan T, Kumar V, Surya HE, Krishna R, John S, Jissa VT, et al. STIL Endows oncogenic and stem-like attributes to colorectal cancer plausibly by Shh and Wnt signaling. Front Oncol. 2021;11:581671. doi:10.3389/fonc.2021.581671. [Google Scholar] [PubMed] [CrossRef]
12. Ye Z, Zhu Z, Xie J, Feng Z, Li Y, Xu X, et al. Hsa_circ_0000069 knockdown inhibits tumorigenesis and exosomes with downregulated hsa_circ_0000069 suppress malignant transformation via inhibition of STIL in pancreatic cancer. Int J Nanomed. 2020;15:9859–73. doi:10.2147/IJN.S279258. [Google Scholar] [PubMed] [CrossRef]
13. Wang J, Zhang Y, Dou Z, Jiang H, Wang Y, Gao X, et al. Knockdown of STIL suppresses the progression of gastric cancer by down-regulating the IGF-1/PI3K/AKT pathway. J Cell Mol Med. 2019;23(8):5566–75. doi:10.1111/jcmm.2019.23.issue-8. [Google Scholar] [CrossRef]
14. Pascale RM, Calvisi DF, Simile MM, Feo CF, Feo F. The warburg effect 97 years after its discovery. Cancers. 2020;12(10):2819. doi:10.3390/cancers12102819. [Google Scholar] [PubMed] [CrossRef]
15. Zam W, Ahmed I, Yousef H. The warburg effect on cancer cells survival: the role of sugar starvation in cancer therapy. Curr Rev Clin Exp Pharmacol. 2021;16(1):30–8. doi:10.2174/1574884715666200413121756. [Google Scholar] [PubMed] [CrossRef]
16. Li X, Yang Y, Zhang B, Lin X, Fu X, An Y, et al. Lactate metabolism in human health and disease. Signal Transduct Target Ther. 2022;7(1):305. doi:10.1038/s41392-022-01151-3. [Google Scholar] [PubMed] [CrossRef]
17. Peng J, Li W, Tan N, Lai X, Jiang W, Chen G. USP47 stabilizes BACH1 to promote the Warburg effect and non-small cell lung cancer development via stimulating Hk2 and Gapdh transcription. Am J Cancer Res. 2022;12(1):91–107. [Google Scholar] [PubMed]
18. Lin S, Li Y, Wang D, Huang C, Marino D, Bollt O, et al. Fascin promotes lung cancer growth and metastasis by enhancing glycolysis and PFKFB3 expression. Cancer Lett. 2021;518:230–42. doi:10.1016/j.canlet.2021.07.025. [Google Scholar] [PubMed] [CrossRef]
19. Bade BC, Dela Cruz CS. Lung cancer 2020: epidemiology, etiology, and prevention. Clin Chest Med. 2020;41(1):1–24. doi:10.1016/j.ccm.2019.10.001. [Google Scholar] [CrossRef]
20. Oliver AL. Lung Cancer: epidemiology and Screening. Surg Clin North Am. 2022;102(3):335–44. doi:10.1016/j.suc.2021.12.001. [Google Scholar] [PubMed] [CrossRef]
21. Hong Y, Kim WJ. DNA methylation markers in lung cancer. Curr Genomics. 2021;22(2):79–87. doi:10.2174/18755488MTEw0NjUbx. [Google Scholar] [CrossRef]
22. Cousin F, Desir C, Ben Mustapha S, Mievis C, Coucke P, Hustinx R. Incidence, risk factors, and CT characteristics of radiation recall pneumonitis induced by immune checkpoint inhibitor in lung cancer. Radiother Oncol. 2021;157:47–55. doi:10.1016/j.radonc.2021.01.001. [Google Scholar] [PubMed] [CrossRef]
23. Liu Y, Kim J, Philip R, Sridhar V, Chandrashekhar M. Direct interaction between CEP85 and STIL mediates PLK4-driven directed cell migration. J Cell Sci. 2020;133(8):jcs238352. [Google Scholar] [PubMed]
24. Wang YW, Chen SC, Gu DL, Yeh YC, Tsai JJ, Yang KT, et al. A novel HIF1α-STIL-FOXM1 axis regulates tumor metastasis. J Biomed Sci. 2022;29(1):24. doi:10.1186/s12929-022-00807-0. [Google Scholar] [PubMed] [CrossRef]
25. Ouyang Y, Jin YB, Chen XP, Zhang GY, Mao SL, Ling F, et al. STIL is upregulated in nasopharyngeal carcinoma tissues and promotes nasopharyngeal carcinoma proliferation, migration and invasion. Neoplasma. 2020;67(1):37–45. doi:10.4149/neo_2019_190306N192. [Google Scholar] [PubMed] [CrossRef]
26. Erez A, Chaussepied M, Castiel A, Colaizzo-Anas T, Aplan PD, Ginsberg D, et al. The mitotic checkpoint gene, SIL is regulated by E2F1. Int J Cancer. 2008;123(7):1721–5. doi:10.1002/ijc.v123:7. [Google Scholar] [CrossRef]
27. Kasprzak A. Insulin-like growth factor 1 (IGF-1) signaling in glucose metabolism in colorectal cancer. Int J Mol Sci. 2021;22(12):6434. doi:10.3390/ijms22126434. [Google Scholar] [PubMed] [CrossRef]
28. El Sayed R, Haibe Y, Amhaz G, Bouferraa Y, Shamseddine A. Metabolic factors affecting tumor immunogenicity: what is happening at the cellular level? Int J Mol Sci. 2021;22(4):2142. doi:10.3390/ijms22042142. [Google Scholar] [PubMed] [CrossRef]
29. He S, Ma X, Zheng N, Wang G, Wang M, Xia W, et al. PRDM14 mediates chemosensitivity and glycolysis in drug‐resistant A549/cisplatin cells and their progenitor A549 human lung adenocarcinoma cells. Mol Med Rep. 2021;23(2):149. [Google Scholar] [PubMed]
30. Li L, Wang Y, Wang Q, Qu J, Wei X, Xu J, et al. High developmental pluripotency‐associated 4 expression promotes cell proliferation and glycolysis, and predicts poor prognosis in non‐small‐cell lung cancer. Mol Med Rep. 2019;20(1):445–54. [Google Scholar] [PubMed]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools