 Open Access
Open Access
VIEWPOINT
Non-canonical BRAF variants and rearrangements in hairy cell leukemia
Cancer Molecular Diagnostics, St. James’s Hospital, Dublin, D08 W9RT, Ireland
* Corresponding Author: STEPHEN E. LANGABEER. Email:
Oncology Research 2024, 32(9), 1423-1427. https://doi.org/10.32604/or.2024.051218
Received 29 February 2024; Accepted 15 May 2024; Issue published 23 August 2024
Abstract
Hairy cell leukemia (HCL) is an uncommon mature B-cell malignancy characterized by a typical morphology, immunophenotype, and clinical profile. The vast majority of HCL patients harbor the canonical BRAF V600E mutation which has become a rationalized target of the subsequently deregulated RAS-RAF-MEK-MAPK signaling pathway in HCL patients who have relapsed or who are refractory to front-line therapy. However, several HCL patients with a classical phenotype display non-canonical BRAF mutations or rearrangements. These include sequence variants within alternative exons and an oncogenic fusion with the IGH gene. Care must be taken in the molecular diagnostic work-up of patients with typical HCL but without the BRAF V600E to include investigation of these uncommon mechanisms. Identification, functional characterization, and reporting of further such patients is likely to provide insights into the pathogenesis of HCL and enable rational selection of targeted inhibitors in such patients if required.Keywords
Classical Hairy Cell leukemia (HCL) is an uncommon B-cell malignancy morphologically characterized by the typical presence of medium-sized, mature B-lymphocytes with cytoplasmic, villous projections in the bone marrow and spleen. HCL cells usually express CD11c, CD20, CD25, and CD103 allowing a relatively rapid diagnosis of suspected cases by immunophenotyping. HCL is more prevalent in men than women with common clinical signs including recurrent infection, splenomegaly, and pancytopenia with moncytopenia prevalent [1]. The standard first-line therapy for HCL is a purine nucleoside analog (cladribine or pentostatin) with or without the chimeric monoclonal antibody rituximab that targets CD20. However, a significant number of patients, almost half, will relapse or will become refractory and require further, alternative lines of treatment [2].
A milestone in the pathobiology of HCL was the discovery of the BRAF V600E mutation (c.1799T>A; NM_004333.4) in nearly all cases of the classical form of HCL over a decade ago which was achieved by whole exome sequencing [3]. The RAS-RAF-MEK-MAPK intracellular signaling pathway is one of the most commonly mutated oncogenic pathways in cancer with the BRAF V600E mutation previously described at a high frequency in malignant melanoma, papillary thyroid cancer, and colorectal cancer [4]. Transplantation of BRAF V600E hematopoietic stem cells (HSC) into mice results in stable engraftment, revealing the functional self-renewable capacity of HCL HSC. Forced expression of the oncogene in murine HSC results in a lethal hematopoietic disorder but restricting expression to mature B cells does not result in disease [5]. Given that HCL cells display a gene expression signature similar to that of post-germinal center B cells, there is an implication that additional genetic alterations are co-operatively required to induce hairy cell development from B cells [6]. Recent murine studies have shown that concurrent mutations in tumor suppressors such as TP53 and PTEN are required for HCL ontogeny [7]. Despite the high frequency of the BRAF V600E in HCL [8–10] corroborated by several groups [11–14], there remained some patients with classical HCL in whom the BRAF V600E mutation could not be detected [15,16].
Non-Canonical BRAF Mutations and Rearrangements
In those HCL patients in whom no BRAF V600E mutation could be detected, alternative molecular mechanisms that activate the RAS-BRAF-MEK-MAPK pathway in capitulating the HCL phenotype are likely involved. Cases were identified by a National Library of Medicine search (https://pubmed.ncbi.nlm.nih.gov/). Sequencing all of BRAF exon 15 and alternative exons has demonstrated the presence of other mutations that result in HCL (Table 1) [17–19]. It is noteworthy that the V600E mutation is co-existent in three cases.
In addition to these mutations, in two BRAF V600E-negative HCL patients, an IGH-BRAF translocation was detected by fluorescent in situ hybridization (FISH) [20,21] (Table 1). The translocation occurs in the IGM switch region of the IGH locus, which is also interrupted in IGH-MYC and IGH-BCL6 fusions, that fuses to BRAF exon 10 [20,21] (Fig. 1). Both patients with this translocation showed molecular evidence of RAS-BRAF-MEK-MAPK pathway activation.
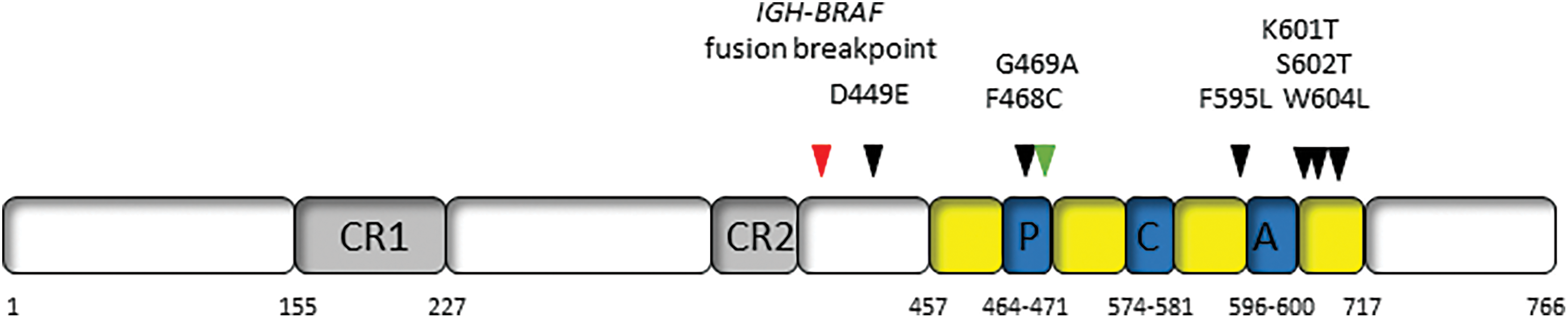
Figure 1: Location of non-canonical BRAF mutations and rearrangements in hairy cell leukemia. CR1 and CR2: highly conserved regulatory regions; P: P loop; C: catalytic loop; A: activation loop.
Characterization of the BRAF V600E mutation has now become an integral part of the diagnostic workup for suspected HCL [22,23] but recognition of these non-canonical mutations and rearrangements has implications for the molecular diagnostic approach employed. While techniques such as allele-specific quantitative PCR or droplet digital PCR can sensitively identify the V600E and consequently monitor disease burden [24,25] they will not expose BRAF variants in other exons. Consideration must therefore be given to include sequencing of at least BRAF exon 11 (in addition to exon 15), if not all coding exons of BRAF in V600E-negative cases. While this would only be an occasional practice, such sequencing approaches would need to be validated with the use of appropriate internal and external quality control. FISH with an IGH probe is also indicated in those cases of HCL that have a classical morphology, clinical features, and immunophenotype. The time and expense of such further investigations would be easily offset by the potential discovery of a non-canonical BRAF mutation that would allow appropriately selected treatment with an inhibitor or not.
Given the high rate of relapse with standard front-line therapy, the molecular defect resulting from the BRAF V600E is a highly attractive target of therapy in HCL patients with relapsed or refractory disease. As the BRAF inhibitor Vemurafenib was already available for BRAF V600E-mutated melanoma [26], proof of principle for clinical application in relapsed/refractory HCL was rapidly proven followed by that of Dabrafenib [27–29]. In HCL cells, BRAF inhibitors cause marked MEK/ERK dephosphorylation, silencing of the RAS-RAF-MEK-MAPK pathway, loss of the HCL-specific gene expression signature, and eventually apoptosis [30]. Clinical trials ensued demonstrating the efficacy of a short oral course of Vemurafenib. However, the persistence of phosphorylated ERK leukemia cells at the end of treatment suggested bypass reactivation of MEK and ERK as a resistance mechanism [31,32]. Specific inhibition of MEK activity is also a therapeutic option [33] with combinations of inhibitors of this same pathway currently being explored [34] (Fig. 2).
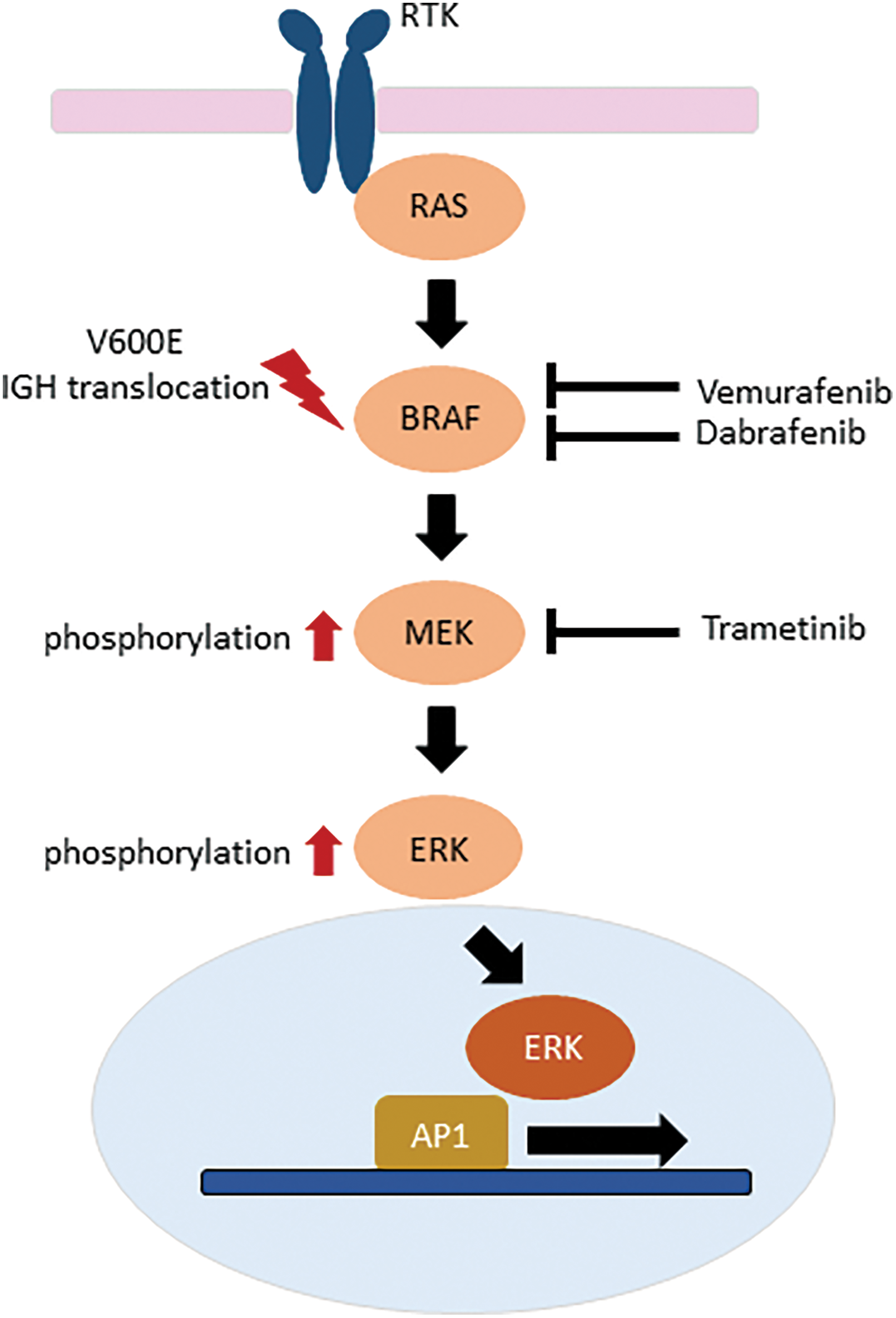
Figure 2: Inhibitors of the RAS-RAF-MEK-ERK intracellular signaling pathway in relapsed or refractory hairy cell leukemia.
How therefore do the above-described non-canonical BRAF mutations and rearrangement potentially impact targeted therapy? Amongst all cancers, a wide range of BRAF mutations have been described and can be divided into three classes based on biochemical and signaling aspects though this classification remains controversial. Class I mutations are within the V600 codon and result in strong kinase activity; Class II mutations are non-V600 variants that have weaker downstream kinase activity; and Class III mutations result in very low kinase activity and cannot directly phosphorylate MEK [35]. Given their location in exon 15, outside the activation loop (Fig. 1) and proximity to codon V600, the K601T, S602T and W604L would be likely categorized as Class II mutations displaying some response to BRAF and MEK inhibitors, whereas the exon 11 mutations of D449E and F468C might be considered Class III with limited response to inhibitors of this signaling pathway. Caution must be taken as the functional characteristics of each of these mutations have not been demonstrated ex vivo. The patients with IGH-BRAF fusions were treated with cladribine and rituximab and cladribine only, achieving long-term molecular and clinical responses [20,21]. However, in one of the latter patients, HCL cells harboring the IGH-BRAF fusion were resistant to Vemurafenib with the authors suggesting it may be advisable not to administer BRAF inhibitors to such patients [21].
While this review has focused on non-canonical abnormalities in patients with classical HCL, such a mutation has been recently described in a patient with the variant form of HCL (CD25-negative). The BRAF G469A is within the protein kinase domain which results in increased BRAF dimerization, kinase activity, and ERK activation [36].
Testing for the presence of the BRAF V600E mutation is necessary for patients with relapsed or refractory HCL to assign appropriate inhibitor therapy. The above-described non-canonical means of BRAF de-regulation must in some form resemble that of the V600E as they all result in an HCL phenotype. Identification, further functional characterization, and reporting of more HCL patients with non-canonical mutations and rearrangements are required to better understand how they disrupt the individual components of the RAS-RAF-MEK-MAPK signaling pathway and provide an opportunity for rationalized selection of inhibitors.
Acknowledgement: None.
Funding Statement: The author received no specific funding for this study.
Author Contributions: The author confirms sole responsibility for the following: study conception and design, data collection, analysis and interpretation of results, and manuscript preparation.
Availability of Data and Materials: Not applicable.
Ethics Approval: Not applicable.
Conflicts of Interest: The author declares that he has no conflicts of interest to report regarding the present study.
References
1. Mendez-Hernandez, A., Moturi, K., Hanson, V., Andritsos, L. A. (2023). Hairy cell leukemia: Where are we in 2023? Current Oncology Reports, 25(8), 833–840. https://doi.org/10.1007/s11912-023-01419-z. [Google Scholar] [PubMed] [CrossRef]
2. Robak, T., Robak, P. (2023). Refractory and relapsed hairy-cell leukemia (HCLCasting light on promising experimental drugs in clinical trials. Expert Opinion on Investigational Drugs, 32(4), 311–324. https://doi.org/10.1080/13543784.2023.2193323. [Google Scholar] [PubMed] [CrossRef]
3. Tiacci, E., Trifonov, V., Schiavoni, G., Holmes, A., Kern, W. et al. (2011). BRAF mutations in hairy cell leukemia. New England Journal of Medicine, 364(24), 2305–2315. https://doi.org/10.1056/NEJMoa1014209. [Google Scholar] [PubMed] [CrossRef]
4. Cantwell-Dorris, E. R., O’Leary, J. J., Sheils, O. M. (2011). BRAFV600E: Implications for carcinogenesis and molecular therapy. Molecular Cancer Therapeutics, 10(3), 385–394. https://doi.org/10.1158/1535-7163.MCT-10-0799. [Google Scholar] [PubMed] [CrossRef]
5. Chung, S. S., Kim, E., Park, J. H., Chung, Y. R., Lito, P. et al. (2014). Hematopoietic stem cell origin of BRAFV600E mutations in hairy cell leukemia. Science Translational Medicine, 6(238), 238ra71. https://doi.org/10.1126/scitranslmed.3008004. [Google Scholar] [PubMed] [CrossRef]
6. Maitre, E., Cornet, E., Debliquis, A., Drenou, B., Gravey, F. et al. (2022). Hairy cell leukemia: A specific 17-gene expression signature points to new targets for therapy. Journal of Cancer Research and Clinical Oncology, 148(8), 2013–2022. https://doi.org/10.1007/s00432-022-04010-4. [Google Scholar] [PubMed] [CrossRef]
7. Yap, J., Yuan, J., Ng, W. H., Chen, G. B., Sim, Y. R. M. et al. (2023). BRAF (V600E) mutation together with loss of Trp53 or pTEN drives the origination of hairy cell leukemia from B-lymphocytes. Molecular Cancer, 22(1), 125. https://doi.org/10.1186/s12943-023-01817-8. [Google Scholar] [PubMed] [CrossRef]
8. Boyd, E. M., Bench, A. J., van’t Veer, M., Wright, P., Bloxham, D. M. et al. (2011). High resolution melting analysis for the detection of BRAF exon 15 mutations in hairy cell leukaemia and other lymphoid malignancies. British Journal of Haematology, 155(5), 609–612. https://doi.org/10.1111/j.1365-2141.2011.08868.x. [Google Scholar] [PubMed] [CrossRef]
9. Arcaini, L., Zibellini, S., Boveri, E., Riboni, R., Rattotti, S. et al. (2012). The BRAF V600E mutation in hairy cell leukemia and other mature B-cell malignancies. Blood, 119(1), 188–191. https://doi.org/10.1182/blood-2011-08-368209. [Google Scholar] [PubMed] [CrossRef]
10. Blombery, P. A., Wong, S. Q., Hewitt, C. A., Dobrovic, A., Maxwell, E. L. et al. (2012). Detection of BRAF mutations in patients with hairy cell leukemia and related lymphoproliferative disorders. Haematologica, 97(5), 780–783. https://doi.org/10.3324/haematol.2011.054874. [Google Scholar] [PubMed] [CrossRef]
11. Lennerz, J. K., Klaus, B. M., Marienfeld, R. B., Möller, P. (2012). Pyrosequencing of BRAF V600E in routine samples of hairy cell leukaemia identifies CD5+ variant hairy cell leukaemia that lacks V600E. British Journal of Haematology, 157(2), 267–269. https://doi.org/10.1111/j.1365-2141.2011.08963.x. [Google Scholar] [PubMed] [CrossRef]
12. Ewalt, M., Nandula, S., Phillips, A., Alobeid, B., Murty, V. V. et al. (2012). Real-time PCR-based analysis of BRAF V600E mutation in low and intermediate grade lymphomas confirms frequent occurrence in hairy cell leukemia. Hematological Oncology, 30(4), 190–193. https://doi.org/10.1002/hon.1023. [Google Scholar] [PubMed] [CrossRef]
13. Langabeer, S. E., O’Brein, D., Liptrot, S., Flynn, C. M., Hayden, P. J. et al. (2012). Correlation of the BRAF V600E mutation in hairy cell leukaemia with morphology, cytochemistry and immunophenotype. International Journal of Laboratory Hematology, 34(4), 417–421. https://doi.org/10.1111/j.1751-553X.2012.01402.x. [Google Scholar] [PubMed] [CrossRef]
14. Schnittger, S., Bacher, U., Haferlach, T., Wendland, N., Ulke, M. et al. (2012). Development and validation of a real-time quantification assay to detect and monitor BRAF V600E mutations in hairy cell leukemia. Blood, 119(13), 151–154. https://doi.org/10.1182/blood-2011-10-383323. [Google Scholar] [PubMed] [CrossRef]
15. Langabeer, S. E., O.’Brien, D., McElligott, A. M., Lavin, M., Browne, P. V. (2013). BRAF V600E-negative hairy cell leukaemia. Case Reports in Hematology, 2013, 513049. https://doi.org/10.1155/2013/513049. [Google Scholar] [PubMed] [CrossRef]
16. Gozzetti, A., Sammartano, V., Bacchiarri, F., Raspadori, D., Bocchia, M. (2020). A BRAF-negative classic hairy cell leukemia patient with long-lasting complete remisssion after rituximab and pentostatin. Turkish Journal of Haematology, 37(4), 286–287. https://doi.org/10.4274/tjh.galenos.2020.2020.0204. [Google Scholar] [PubMed] [CrossRef]
17. Tschernitz, S., Flossbach, L., Bonengel, M., Roth, S., Rosenwald, R. et al. (2014). Alternative BRAF mutations in BRAF V600E-negative hairy cell leukaemias. British Journal of Haematology, 165(4), 529–533. https://doi.org/10.1111/bjh.12735. [Google Scholar] [PubMed] [CrossRef]
18. Thomas, C., Amanuel, B., Finlayson, J., Grieu-Iacopetta, F., Spagnolo, D. V. et al. (2015). BRAF mutation detection in hairy cell leukaemia from archival haematolymphoid specimens. Pathology, 47(4), 349–354. https://doi.org/10.1097/PAT.0000000000000245. [Google Scholar] [PubMed] [CrossRef]
19. Maitre, E., Macro, M., Troussard, X. (2023). Hairy cell leukaemia with unusual BRAF mutations. Journal of Cellular and Molecular Medicine, 27(17), 2626–2630. https://doi.org/10.1111/jcmm.17890. [Google Scholar] [PubMed] [CrossRef]
20. Thompson, E. R., Lim, K. J. C., Kuzich, J. A., McBean, M., Westerman, D. et al. (2020). Detection of an IGH-BRAF fusion in a patient with BRAF Val600Glu negative hairy cell leukemia. Leukemia & Lymphoma, 61(8), 2024–2026. https://doi.org/10.1080/10428194.2020.1753045. [Google Scholar] [PubMed] [CrossRef]
21. Matsumoto, K., Kakazu, N., Imataki, O., Kondo, A., Kanaji, N. et al. (2023). Hairy cell leukaemia with an IGH-BRAF fusion gene. British Journal of Haematology, 202(6), e67–e70. https://doi.org/10.1111/bjh.18980. [Google Scholar] [PubMed] [CrossRef]
22. Wierda, W. G., Byrd, J. C., Abramson, J. S., Bhat, S., Bociek, G. et al. (2017). Hairy cell leukemia, version2.2018, NCCN clinical practice guidelines in oncology. Journal of the National Comprehensive Cancer Network, 15(11), 1414–1427. https://doi.org/10.6004/jnccn.2017.0165. [Google Scholar] [PubMed] [CrossRef]
23. Parry-Jones, N., Joshi, A., Forconi, F., Dearden, C., BSH guidelines committee (2020). Guidelines for diagnosis and management of hairy cell leukaemia (HCL) and hairy cell variant (HCL-V). British Journal of Haematology, 191(5), 730–737. https://doi.org/10.1111/bjh.17055. [Google Scholar] [PubMed] [CrossRef]
24. Broccoli, A., Terragna, C., Nanni, L., Martello, M., Armuzzi, S. et al. (2022). Droplet digital polymerase chain reaction for the assessment of disease burden in hairy cell leukemia. Hematological Oncology, 40(1), 57–62. https://doi.org/10.1002/hon.2932. [Google Scholar] [PubMed] [CrossRef]
25. Robak, T., Robak, P. (2022). Measurable residual disease in hairy cell leukemia: Technical considerations and clinical significance. Frontiers in Oncology, 12, 976374. https://doi.org/10.3389/fonc.2022.976374. [Google Scholar] [PubMed] [CrossRef]
26. Flaherty, K. T., Puzanov, I., Kim, K. B., Ribas, A., McArthur, G. A. et al. (2010). Inhibition of mutated, activated BRAF in metastatic melanoma. New England Journal of Medicine, 363(9), 809–819. https://doi.org/10.1056/NEJMoa1002011. [Google Scholar] [PubMed] [CrossRef]
27. Dietrich, S., Glimm, H., Andrulis, M., von Kalle, C., Ho, A. D. et al. (2012). BRAF inhibition in refractory hairy-cell leukemia. New England Journal of Medicine, 366(21), 2038–2040. https://doi.org/10.1056/NEJMc1202124. [Google Scholar] [PubMed] [CrossRef]
28. Follows, G. A., Sims, H., Bloxham, D. M., Zenz, T., Hopper, M. A. et al. (2013). Rapid response of bialleleic BRAFV600E mutated hairy cell leukaemia to low dose vemurafenib. British Journal of Haematology, 161(1), 150–153. https://doi.org/10.1111/bjh.12201. [Google Scholar] [PubMed] [CrossRef]
29. Vergote, V., Dierickx, D., Janssens, A., Verhoef, G., Tousseyn, T. et al. (2014). Rapid and complete hematological response of refractory hairy cell leukemia to the BRAF inhibitor dabrafenib. Annals of Hematology, 93(12), 2087–2089. https://doi.org/10.1007/s00277-014-2104-2. [Google Scholar] [PubMed] [CrossRef]
30. Pettirossi, V., Santi, A., Imperi, E., Russo, G., Pucciarini, A. et al. (2015). BRAF inhibitors reverse the unique molecular signature and phenotype of hairy cell leukemia and exert potent antileukemic activity. Blood, 125(8), 1207–1216. https://doi.org/10.1182/blood-2014-10-603100. [Google Scholar] [PubMed] [CrossRef]
31. Tiacci, E., Park, J. H., de Carolis, L., Chung, S. S., Broccoli, A. et al. (2015). Targeting mutant BRAF in relapsed or refractory hairy-cell leukemia. New England Journal of Medicine, 373(18), 1733–1747. https://doi.org/10.1056/NEJMoa1506583. [Google Scholar] [PubMed] [CrossRef]
32. Dietrich, S., Pircher, A., Endris, V., Peyrade, F., Wendtner, C. M. et al. (2016). BRAF inhibition in hairy cell leukemia with low-dose vemurafenib. Blood, 127(23), 2847–2855. https://doi.org/10.1182/blood-2015-11-680074. [Google Scholar] [PubMed] [CrossRef]
33. Caesar, R., Collord, G., Yao, W. Q., Chen, Z., Vassiliou, G. S. et al. (2019). Targeting MEK in vemurafenib-resistant hairy cell leukemia. Leukemia, 33(2), 541–545. https://doi.org/10.1038/s41375-018-0270-2. [Google Scholar] [PubMed] [CrossRef]
34. Kreitman, R. J., Moreau, P., Ravandi, F., Hutchings, M., Gazzah, A. et al. (2023). Dabrafenib plus trametinib in patients with relapsed/refractory BRAF V600E mutation-positive hairy cell leukemia. Blood, 141(9), 996–1006. https://doi.org/10.1182/blood.2021013658. [Google Scholar] [PubMed] [CrossRef]
35. Owsley, J., Stein, M. K., Porter, J., In, G. K., Salem, M. et al. (2021). Prevalence of class I–III BRAF mutations among 114,662 cancer patients in a large genomic database. Experimental Biology and Medicine, 246(1), 31–39. https://doi.org/10.1177/1535370220959657. [Google Scholar] [PubMed] [CrossRef]
36. Vijayanarayanan, A., Wang, S. A., Garces, S., Saluja, K., Medeiros, L. J. et al. (2023). Non-V600E BRAF mutation in hairy cell leukemia variant. EJHaem, 5(1), 266–268. https://doi.org/10.1002/jha2.836. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF
 Downloads
Downloads
 Citation Tools
Citation Tools