 Open Access
Open Access
ARTICLE
LncRNA PCGEM1 facilitates cervical cancer progression via miR-642a-5p/KIF5B axis
1 Department of Obstetrics and Gynecology, Shanghai First Maternity and Infant Hospital, School of Medicine, Tongji University, Shanghai, 200092, China
2 Department of Obstetrics and Gynecology, The Second People’s Hospital of Nantong City, Nantong, 226002, China
3 Department of Obstetrics and Gynecology, Affiliated Hospital of Nantong University, Nantong, 226002, China
* Corresponding Authors: DANDAN ZHANG. Email: ; HUIQIN LIU. Email:
Oncology Research 2024, 32(7), 1221-1229. https://doi.org/10.32604/or.2024.047454
Received 06 November 2023; Accepted 10 January 2024; Issue published 20 June 2024
A correction of this article was approved in:
Correction: LncRNA PCGEM1 facilitates cervical cancer progression via miR-642a-5p/KIF5B axis
Read correction
Abstract
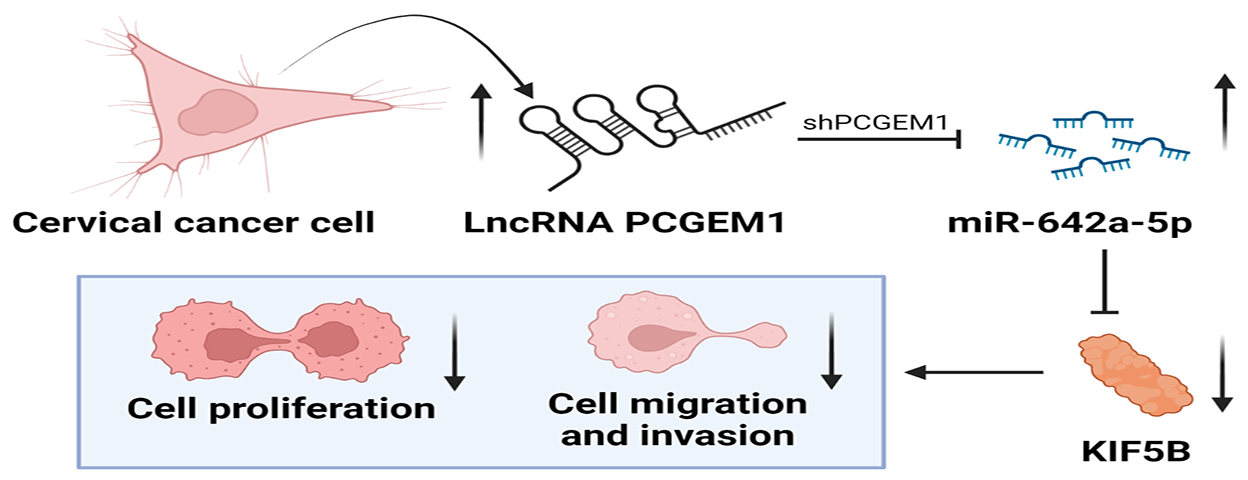
At present, the role of many long non-coding RNAs (lncRNAs) as tumor suppressors in the formation and development of cervical cancer (CC) has been studied. However, lncRNA prostate cancer gene expression marker 1 (PCGEM1), whose high expression not only aggravates ovarian cancer but also can induce tumorigenesis and endometrial cancer progression, has not been studied in CC. The objective of this study was to investigate the expression and the underlying role of PCGEM1 in CC. The relative expression of PCGEM1 in CC cells was detected by real-time PCR. After the suppression of PCGEM1 expression by shRNA, the changes in the proliferation, migration, and invasion capacities were detected via CCK-8 assay, EdU assay, and colony formation assay wound healing assay. Transwell assay and the changes in expressions of epithelial-to-mesenchymal transition (EMT) markers were determined by western blot and immunofluorescence. The interplay among PCGEM1, miR-642a-5p, and kinesin family member 5B (KIF5B) was confirmed by bioinformatics analyses and luciferase reporter assay. Results showed that PCGEM1 expressions were up-regulated within CC cells. Cell viabilities, migration, and invasion were remarkably reduced after the suppression of PCGEM1 expression by shRNA in Hela and SiHa cells. N-cadherin was silenced, but E-cadherin expression was elevated by sh-PCGEM1. Moreover, by sponging miR-642a-5p in CC, PCGEM1 was verified as a competitive endogenous RNA (ceRNA) that modulates KIF5B levels. MiR-642a-5p down-regulation partially rescued sh-PCGEM1’s inhibitory effects on cell proliferation, migration, invasion, and EMT process. In conclusion, the PCGEM1/miR-642a-5p/KIF5B signaling axis might be a novel therapeutic target in CC. This study provides a research basis and new direction for targeted therapy of CC.Graphic Abstract
Keywords
Cervical cancer (CC) ranks 2nd in the global cancer incidence and 4th in terms of worldwide mortality [1]. Traditional treatment options for CC include radical hysterectomy and chemoradiotherapy [2]. Nevertheless, in clinical settings, the majority of patients with advanced or metastatic CC have a poor prognosis [3]. Consequently, it is crucial to learn about the therapeutic targets in the progression of CC.
Long non-coding RNAs (lncRNAs), as a subclass of non-coding RNA with a length more than 200 nucleotides (nt), are located in the nucleus and cytoplasm and exert vital effects on both physiological and pathological processes by controlling the expression of genes involved in those processes [4]. At present, the role of lncRNAs as tumor suppressors in the formation and development of malignant tumors including CC has been studied. For instance, the knockdown of small nucleolar RNA host gene 14 (SNHG14) inhibited tumor cell proliferation and promoted apoptosis [5]. Higher LINC00511 expression predicted a poor prognosis of CC, and silencing LINC00511 expression suppressed CC cell proliferation, metastasis, and invasion [6]. Ectopic GAS5 could attenuate CC cell proliferation and motility [7].
A new long noncoding RNA (lncRNA), prostate cancer gene expression marker 1 (PCGEM1), is located on chromosome 2q32.3, which is overexpressed in prostate cancer [8,9]. The oncogene PCGEM1 has been reported to be activated in a variety of malignancies. For example, Chen et al. indicated that up-regulated PCGEM1 deteriorated the malignant behaviors of ovarian cancer by interacting with RhoA [10]. Previous study has shown that PCGEM1 over-expression induced tumorigenesis and endometrial carcinoma progression by regulating miR-129-5p/STAT3 signaling [11]. Zhang et al. found that silencing of PCGEM1 expression memorably modulated EMT to further descend gastric cancer metastasis [12]. Although PCGEM1 has been found to have carcinogenic effects in various malignancies, its possible role in the development of CC was unknown. This study concentrated on examining the roles played by PCGEM1 in CC as well as the underlying mechanism.
Ect1/E6E7 and CC cells (HeLa, CaSki, and SiHa) were supplied by ATCC (USA) and cultivated in DMEM media with 10% FBS (Gibico, NY, USA), which was kept in a humid incubator at 37°C and 5% CO2.
CC cell lines were cultured in six-well plates except for cell counting in 24-well plates. The cationic liposome transfection method was used according to the manufacturer’s protocol. In brief, the fresh antibiotics-free cell culture medium(Thermo Fisher Scientific, USA) was changed, and inoculated 1.0 × 106 cells into each well the day before transfection. Cells were treated with shRNA in the presence of Lipofectamine2000 (Invitrogen) and the ratio of vector/Lipofectamine2000 was 1:3, incubated at serum-free and non-antibiotics media for 6–8 h, then changed to the full media with 10% FCS till 48 h. Short hairpin RNA (shRNA)-expressing lentivirus vectors were created by Genechem to target PCGEM1, shRNA negative controls (sh-NC), NC mimic/miR-642a-5p mimic, and NC inhibitor/miR-642a-5p inhibitor (Ribobio, Shanghai, China). Each experiment was performed in triplicate and the standard deviation was obtained.
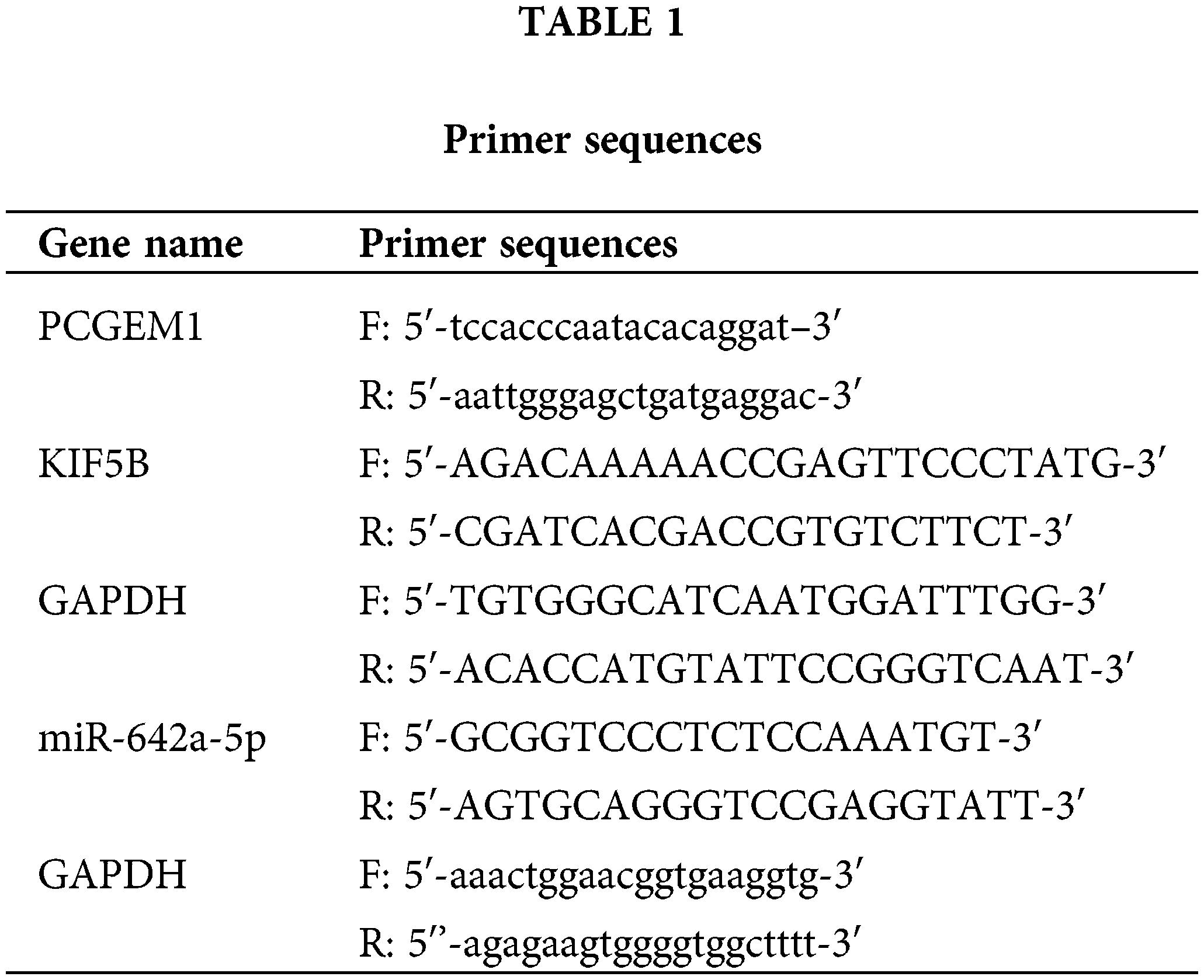
Total RNA was isolated from CC cell lines using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. cDNA synthesis for coding genes was performed with 1 μg of total RNA according to the manufacturer’s instructions (Takara, Tokyo, Japan). The mRNA levels of PCGEM1, KIF5B, and miR-642a-5p were analyzed using SYBR PCR Master Mix reagent kits (Takara, Tokyo, Japan) according to the manufacturer’s instructions. GAPDH was amplified as a control. The relative expression levels of CCKBR were calculated using the 2−ΔΔCT method. The specific primers as shown in Table 1.
Cell viability was evaluated by a CCK-8 (Beyotime Biotechnology, Shanghai, China) according to the manufacturer’s instructions. Different groups’ transfected CC cells (2 × 103) were seeded into 96-well plates. CCK-8 kit (Sigma, USA) was utilized after being incubated for 0 and 48 h. Using a spectrophotometer (Molecular Devices, San Jose, USA), absorbance (OD) (450 nm) was detected.
Transfected CC cells from various groups were given a 30-minute incubation period with the EdU reaction solution. CC cells were then trypsinized, fixed with formaldehyde, and washed in PBS containing 1% BSA. PBS was used to rinse the cells after Triton X-100 permeabilization and fluorescence microscopy was used to see the cells.
In brief, CC cells were inoculated into six-well plates. After 48 h of transfection, CC cells were cultured in a fresh medium and the medium should be updated every 3 days during culture. After 14 days, CC cells were gently washed with PBS, followed by fixing with paraformaldehyde (4%, Beyotime) for 0.5 h at 4°C. Then, these cells were again washed with PBS, followed by staining with crystal violet (0.1%, Beyotime) for 2 h. A microscope (Olympus, Tokyo, Japan) was employed for counting the colonies (>50 cells per colony).
The proper density of transfected CC cells (5 × 105/well) was added to 6-well plates. A wound was scraped once the cells had 80% confluence. A light microscope (Nikon, Japan) was used to take photos of cells pictures at 0 and 48 h (200×).
For invasion experiments, Matrigel was evenly spread on the Transwell chamber and dissolved in serum-free DMEM for one hour at 37°C. CC cells from the various groups (sh-NC, sh-PCGEM1, sh-PCGEM1+miR-642a-5p NC, sh-PCGEM1+miR-642a-5p inhibitor) were planted into the top chambers at the right density (105/well) for the migration and invasion experiments. Into the bottom chambers was put a medium containing 10% FBS. Migrated and invaded cells were stained with crystal violet (0.1%) and photographed using a light microscope (Nikon, Japan) after 48 h of incubation at 37°C at room temperature (200×).
Protein was extracted from CC cell lines using RIPA buffer (50 mM Tris (pH 7.4), 150 mM NaCl, 1% v/v Triton X-100, 1% v/v sodium deoxycholate, 0.1% v/v SDS containing protease and phosphatase inhibitors (100 mmol/L PMSF, 1 mg/mL leupeptin, 1 mg/mL aprotinin)) (Beyotime, China). Total proteins (60 µg) were resolved by SDS-PAGE and transferred to a polyvinylidene fluoride membrane (PVDF, Bio-Rad, Hercules, CA, USA). The polyvinylidene fluoride membranes were incubated with the primary and secondary antibodies. Membranes were then incubated with primary antibodies overnight at 4°C, using 5% skimmed milk. Since N-cadherin and E-cadherin have similar molecular weights, We used the 8% separation gel and selected two primary antibodies with a large distance between predicted locations, where the predicted position of N-cadherin (05-915, Sigma-Aldrich) was about 140 kDa and the predicted position of E-cadherin (MAB3199Z, Sigma-Aldrich) was about 100 kDa, in order to distinguish the two proteins as much as possible. Anti-KIF5B (1:2,000, bs-24715R, Bioss) and anti-GAPDH (1:2,000, bs-0755R, Bioss) antibodies are used, with GAPDH serving as the endogenous control. Next, membranes were treated with a secondary antibody for an additional hour at 37°C (1:2, 000, bs-0311P-HRP, Bioss). Finally, protein blots were examined using ECL (Millipore, USA). Densitometry was performed using image-J software (1.45 s, National Institutes of Health, USA).
The CC cell slices from different groups were sealed for 30 min with 3% bovine serum protein (Gibico, NY, USA) at 37°C. Then slides were incubated overnight with primary antibodies at 4°C. Then slices were incubated with second antibody and DAPI for 60 and 15 min, respectively, at room temperature with DAPI. Fields were photographed in each slice using a microscope (Leica Microsystems, Wetzlar, Germany).
In this study, PCGEM1 or KIF5B WT/MUT were sub-cloned to produce pmirGLO-PCGEM1 or pmirGLO-KIF5B WT/MUT, which were then co-transfected into CC cells along with NC mimics or miR-642a-5p mimics. The luciferase activity was assessed after co-transfection for 48 h (Promega, USA).
Data from three repeats are shown by the mean ± standard deviation (SD). The Software used is GraphPad Prism 5.0 (GraphPad Software, Inc.). The normal distribution was used S-W test. The difference with p < 0.05 was considered statistically significant, according to the t-test used to compare the two groups, the single-factor ANOVA used to compare the groups, and the Tukey post-test. Six parallels were set for each treatment group in vitro experiments, and quantification of microscopy results was performed by randomly selecting 10 representative fields from each group for quantitative analysis.
PCGEM1 knockdown suppressed CC cell proliferation
Herein, RT-PCR was employed for assessing PCGEM1 expressions within CC cells and Ect1/E6E7 cells. In CC cells, PCGEM1 expression was up-regulated, especially in Hela, SiHa and CaSki cells, as shown in Fig. 1A. As demonstrated by RT-PCR and GFP fluorescence microscopy, PCGEM1 expressions within Hela and SiHa cells was silenced by shRNA transfection (Fig. 1B). CCK-8, EdU and formation of colonies were conducted for determining sh-PCGEM1’s effects on CC cell proliferation. As shown in (Fig. 1C), the Cell viability of Hela and SiHa cells was remarkably reduced after the suppression of PCGEM1 expression by shRNA. From the results of EdU assays, the proportion of EdU-positive cells was reduced after the suppression of PCGEM1 expression by shRNA (Fig. 1D). Additionally, the knockdown of PCGEM1 significantly decreased colony formation ability in Hela and SiHa cells (Fig. 1E).
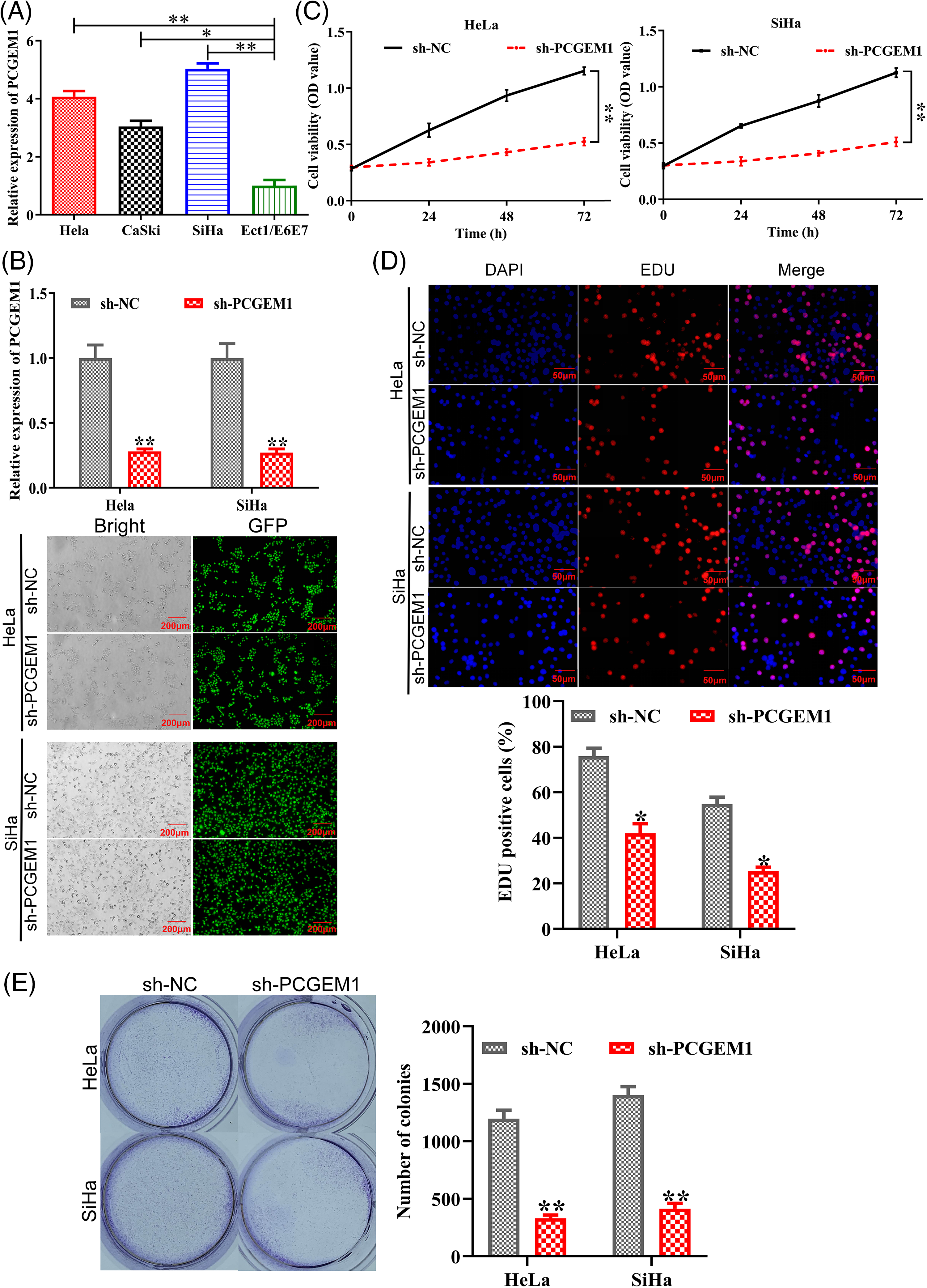
Figure 1: PCGEM1 knockdown suppressed CC cell proliferation. (A) Three human CC cell lines (HeLa, CaSki, and SiHa) and healthy human cervical epithelial Ect1/E6E7 cells were used in the real-time PCR analysis of PCGEM1 expression. *p < 0.05, **p < 0.01 vs. Ect1/E6E7 cells. (B) Sh-PCGEM1 or sh-NC were transfected into Hela and SiHa cells, respectively. Real-time PCR and the evaluation of GFP expression provided evidence of the transfection’s effectiveness (scale bar, 25 µm). (C) CCK-8, (D) EdU (scale bar, 50 µm), and (E) formation of colonies assays were adopted to ascertain the proliferation of sh-PCGEM1-transfected Hela and SiHa cells. *p < 0.05, **p < 0.01 vs. sh-NC group.
PCGEM1 depletion reduced migration and invasion of CC cells
To explore PCGEM1’s impacts on CC cell metastasis, wound healing, and Transwell, as well as western blotting analyses of associated proteins were conducted when PCGEM1 was down-regulated. The functional analysis demonstrated that PCGEM1 down-regulation reduced Hela and SiHa cell migration (Fig. 2A) and invasion (Fig. 2B) capacities by decreasing matrix metalloproteinases (MMP-2, and MMP -13) (Fig. 2C).
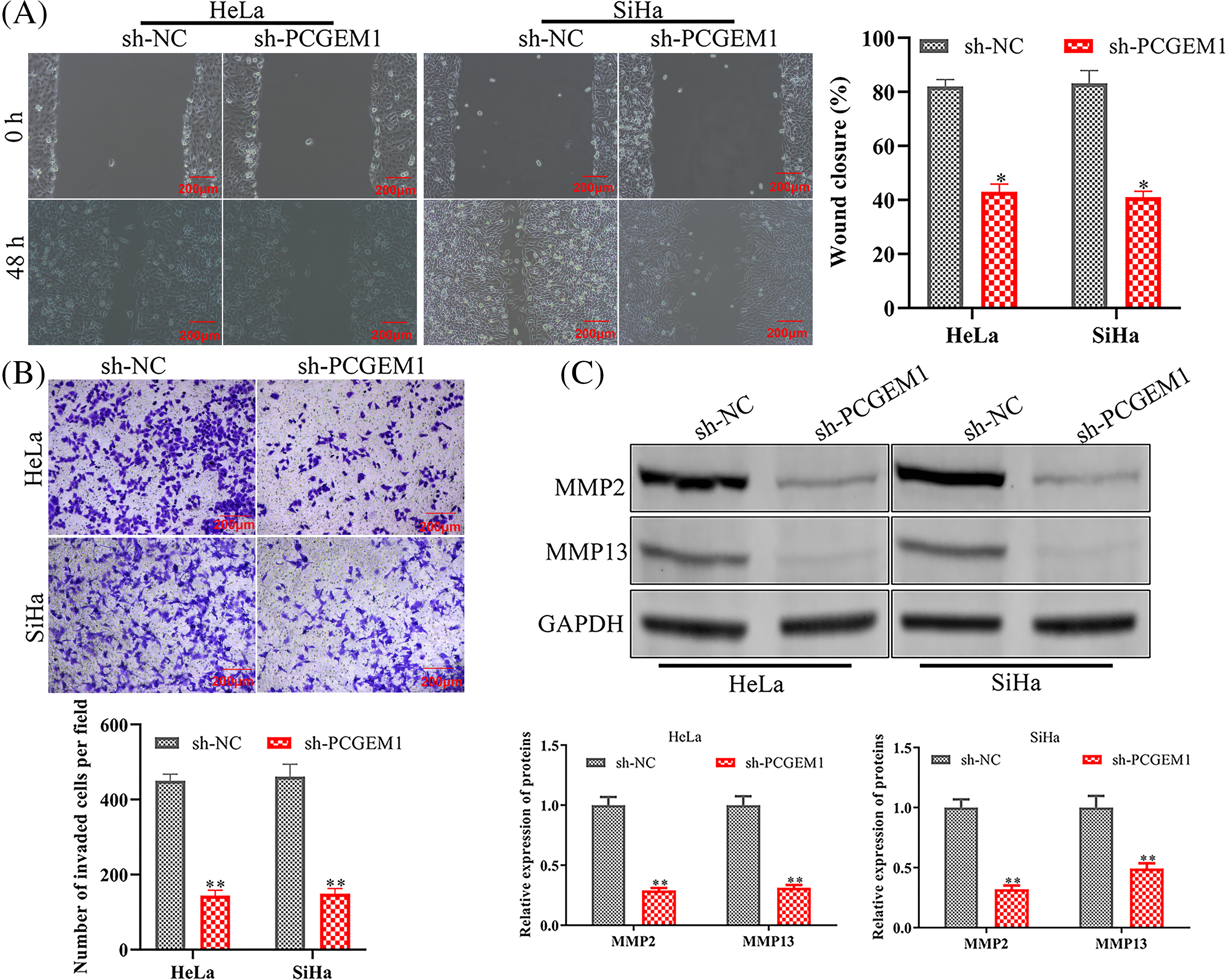
Figure 2: PCGEM1 depletion reduced migration and invasion of CC cells. (A and B) The ability of sh-PCGEM1-transfected Hela and SiHa cells to migrate and invade was evaluated using the wound-healing assay (scale bar, 100 µm) and the Transwell assay (scale bar, 50 µm). (C) The expression of MMP-2 and MMP-13 in sh-PCGEM1-transfected Hela and SiHa cells was determined using the western blot assay. *p < 0.05, **p < 0.01 vs. sh-NC group.
PCGEM1 inhibition repressed the EMT program
Whether PCGEM1 affected EMT progression in CC cells was further investigated. The western blot results indicated PCGEM1 silencing significantly down-regulated the mesenchymal marker N-cadherin, along with increasing E-cadherin expressions, an epithelial marker (Fig. 3A). Similarly, results of the immunofluorescence analysis revealed that N-cadherin was silenced, but E-cadherin expression was elevated by sh-PCGEM1 (Fig. 3B).
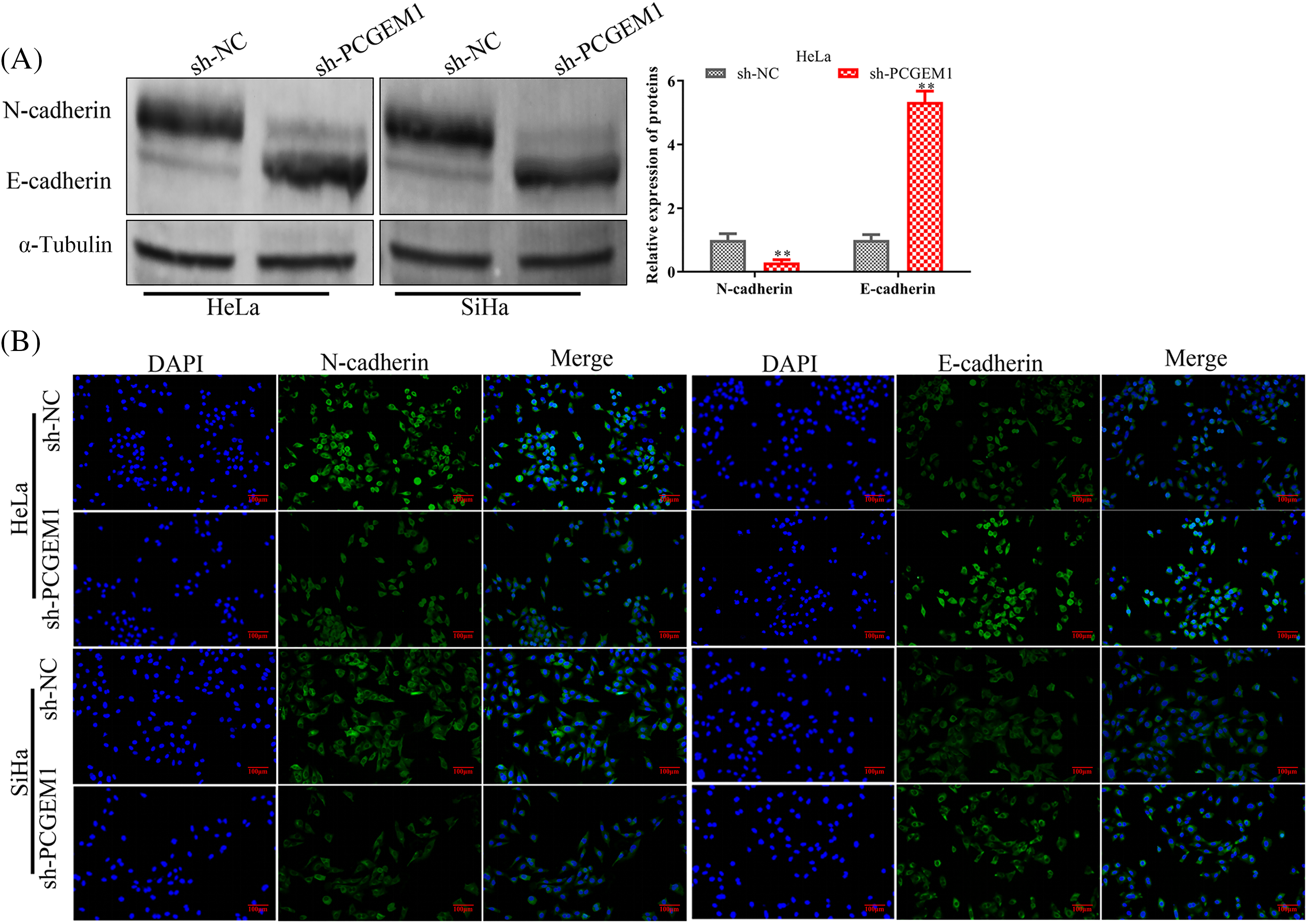
Figure 3: PCGEM1 inhibition repressed the EMT program. In Hela and SiHa cells transfected with sh-PCGEM1, the expression levels of N- and E-cadherin were measured using Western blot assay (A) and immunofluorescence (B) assays (scale bar, 50 µm). **p < 0.01 vs. sh-NC group.
PCGEM1 regulated KIF5B by competing for miR-642a-5p
LncRNAs could serve as miRNA sponges, thereby modulating mRNA expression [13]. We investigated whether PCGEM1-mediated tumorigenesis might operate through a competitive endogenous RNA (ceRNA) regulatory mechanism. PCGEM1 inhibition could significantly enhance miR-642a-5p levels within CC cells (Fig. 4A). MiR-642a-5p combining sites in PCGEM1 were displayed in Fig. 4B. The PCGEM1 mimic lowered the luciferase activity of the miR-642a-5p WT reporter vector, but not the mutant reporter, according to a dual-luciferase experiment (Fig. 4C). MiR-642a-5p over-expression could diminish KIF5B levels, an oncogene within CC cells (Figs. 4D and 4E). The 3′UTR region of KIF5B was predicted by a bioinformatics study to include numerous combining sites for miR-642a-5p (http://www.targetscan.org/) (Fig. 4F). Based on Fig. 4G, the activities of WT-KIF5B 3'-UTR luciferase were observably decreased by miR-642a-5p mimic (Fig. 4G). Collectively, these findings revealed that PCGEM1 competed with KIF5B for miR-642a-5p binding in CC cells.
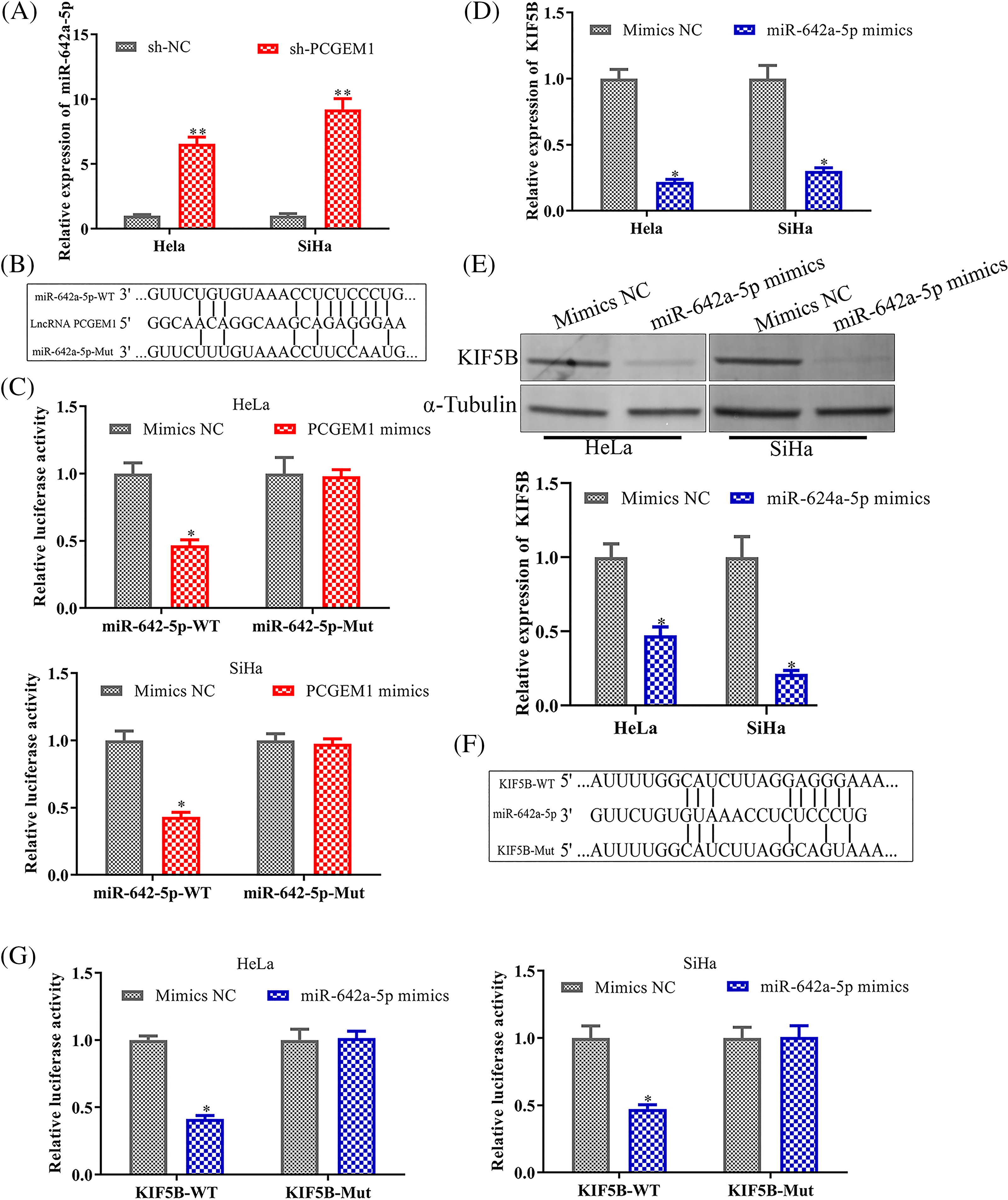
Figure 4: PCGEM1 regulated KIF5B expression by competing for miR-642a-5p. (A) The mRNA expression of miR-642a-5p was measured in Hela and SiHa cells transfected with sh-PCGEM1 or sh-NC. **p < 0.01 vs. sh-NC group. (B) The conceptual representation of the miR-642a-5p combining sites in PCGEM1. (C) In Hela and SiHa cells, the luciferase reporters harboring WT or MUT miR-642a-5p were co-transfected with PCGEM1 mimic or mimic NC. *p < 0.05 vs. Mimics NC group. (D–E) In Hela and SiHa cells transfected with miR-642a-5p mimic or mimic NC, the mRNA and protein levels of KIF5B were assessed. *p < 0.05 vs. Mimics NC group. (F) The miR-642a-5p putative targeting sites in the WT and MUT of KIF5B are shown schematically. (G) In Hela and SiHa cells, the luciferase reporters harboring WT or MUT KIF5B were co-transfected with miR-642a-5p mimic or mimic NC. *p < 0.05 vs. Mimics NC group.
PCGEM1 silencing inhibited proliferation, metastasis, invasion and EMT in CC cells by regulating the miR-642a-5p/KIF5B axis
Finally, we investigated whether miR-642a-5p inhibitor could antagonize the anti-tumor effects of sh-PCGEM1. As expected, KIF5B levels were suppressed by sh-PCGEM1, which was attenuated by transfection with a miR-642a-5p inhibitor within SiHa cells (Figs. 5A and 5B). Further investigation illustrated that miR-642a-5p down-regulation partially rescued sh-PCGEM1’s inhibitory effects on cell proliferation (Figs. 5C–5D), migration (Fig. 5E), invasion (Fig. 5F) and EMT process (Fig. 5G). Taken together, these results demonstrated that PCGEM1 exerted carcinogenic effects in CC cells via the miR-642a-5p/KIF5B axis.
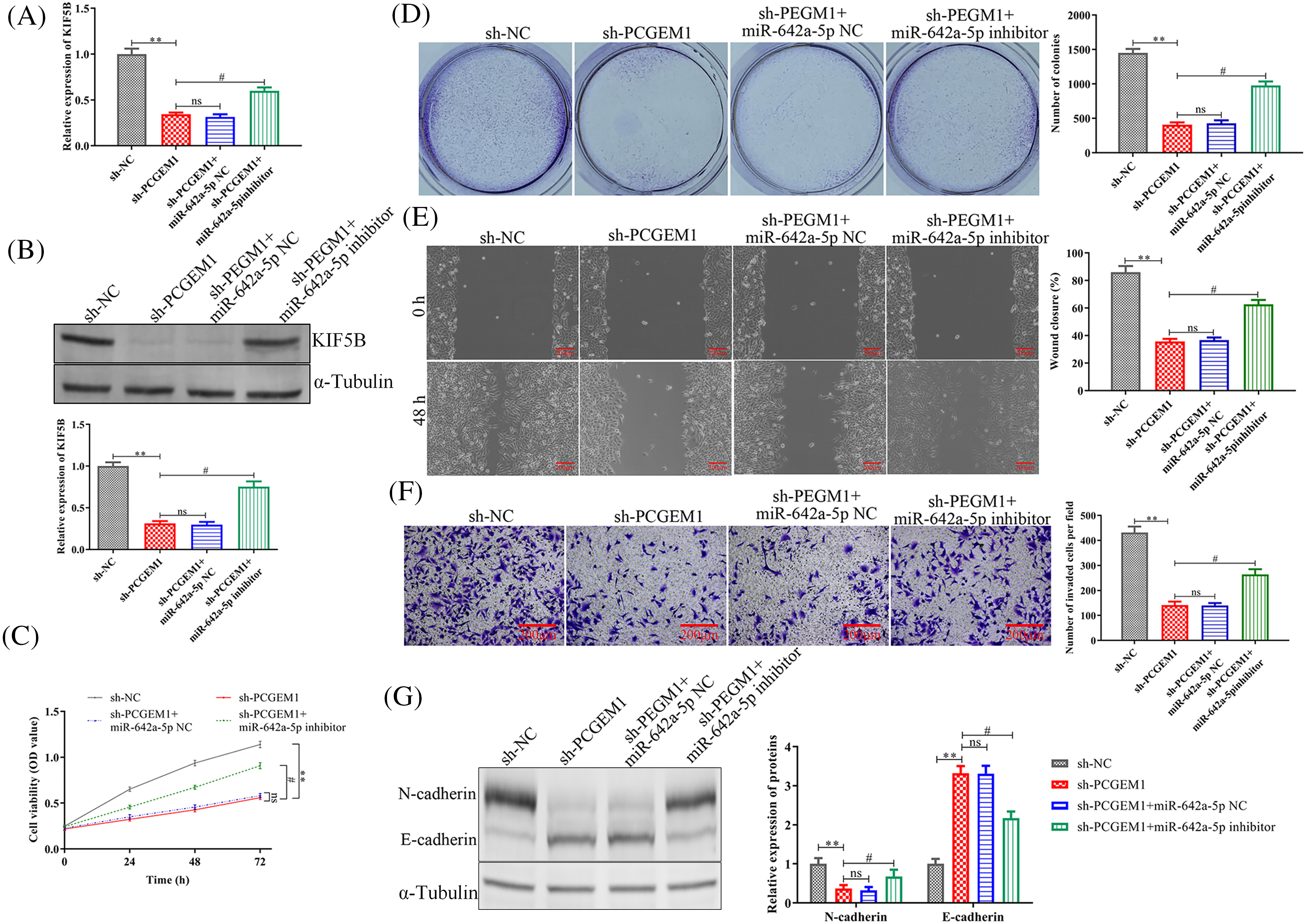
Figure 5: PCGEM1 silencing inhibited proliferation, metastasis, invasion, and EMT in CC cells by the regulation of miR-642a-5p/KIF5B axis. (A and B) The mRNA and protein levels of KIF5B, (C) CCK-8 assay and colony formation assay, (D) formation of colonies assays, (E) wound-healing assay (scale bar, 100 µm), (F) Transwell assay (scale bar, 50 µm), and (G) the protein levels of N-cadherin and E-cadherin were determined in SiHa cells transfected with sh-NC or sh-PCGEM1 alone, or together with miR-642a-5p inhibitor or inhibitor NC. **p < 0.01 vs. sh-NC group, #p < 0.01 vs. sh-PCGEM1+miR-642a-5p NC group.
As a carcinogenic lncRNA, PCGEM1 contributed to the occurrence and development of prostate cancer [14,15], ovarian cancer [10], endometrial carcinoma [11], and gastric cancer [12,16]. However, there was few report about the biological roles of PCGEM1 in CC. In this study, we for the first time found CC cells contain increased PCGEM1 levels. The majority of deaths connected to cancer are still caused by metastasis. In this research, we investigated the PCGEM1 functions on PCGEM1 in CC progression and revealed PCGEM1 down-regulation inhibited CC cell proliferation, metastasis, and invasion in vitro. EMT, an essential progress in CC, mediated by lncRNAs has been implicated in diverse tumors. For instance, SNHG7 was described as an EMT-promoter in breast cancer cells, where it sponged miR-34a, leading to the activation of the Notch-1 pathway [17]. H19 promoted esophageal cancer cell metastasis via negatively regulating let-7c, an anti-EMT miRNA [18]. Fer-1-like protein 4 (FER1L4) prevented EMT initiation, controlling osteosarcoma cell proliferation via miR-18a-5p/SOCS5/PI3K/AKT signaling pathway [19]. As indicated by western blot and immunofluorescence analysis, the knockdown of PCGEM1 expression dramatically inhibited the EMT process by down-regulating N-cadherin but up-regulating E-cadherin within CC cells.
Although increased PCGEM1 expression has been found in CC cells, the detailed regulatory mechanism by which PCGEM1 mediated the development of CC remains largely unclear. It has been demonstrated that lncRNAs act as post-transcriptional miRNA sponges. There is mounting evidence that lncRNAs act as post-transcriptional miRNA sponges in a new regulation mechanism. A previous study demonstrated that SNHG20 functioned as a ceRNA to target ZEB2 and RUNX2 by sponging miR-154 [20]. SNHG6 may contribute to breast cancer aggressiveness via the miR-26a/VASP axis [21]. Urothelial Cancer Associated 1 (UCA1) acted as ceRNA directly combining miR-498, a tumor suppressor, inhibiting its function in esophageal cancer cells [22]. Using bioinformatics databases (DIANA tools), PCGEM1 and KIF5B were predicted to share the regulatory sites for miR-642a-5p. Besides, further evidence was provided to support the regulatory connection between PCGEM1 and miR-642a-5p. (1) PCGEM1 inhibition increased miR-642a-5p levels. (2) Luciferase activity assay proved the target relationship between PCGEM1 and miR-642a-5p. Besides, miR-642a-5p could directly target KIF5B, a positive regulator of EMT [23,24]. Similarly, miR-642a-5p acted as a tumor suppressor in multiple tumor types, including Hodgkin lymphoma [25] and colon cancer [26]. These data provided evidence that PCGEM1 served as a ceRNA for miR-642a-5p to modulate KIF5B within CC cells. Furthermore, miR-642a-5p down-regulation restored the inhibition of CC cell proliferation, invasion, and EMT caused by PCGEM1 silencing.
Increasing evidence has shown that circular RNAs (circRNAs) can be used as valuable biomarkers for early detection of CC and have the potential to be a therapeutic target for intervention. The role of circRNAs in a variety of diseases such as cancer has been revealed. Not only as miRNA sponge, it affects the transport of intracellular RNA binding proteins (RBPs), regulates the expression of parental genes, and regulates protein translation, but also circRNAs have Janus-faced characteristics in cellular processes [27]. Whether PCGEM1 has a similar mechanism in CC requires further investigation.
Studies have shown that TUG1 [28], SNHG5 [29], MBNL1-AS1 [30], HOTAIR [31] and other lncRNAs are significantly increased in cervical cancer tissues and cell,associated with FIGO stage, lymph node metastasis, tumor size and differentiation in patients with cervical cancer. By regulating the expression of key genes, they regulate proliferation, apoptosis, cell cycle, migration and invasion, EMT and cancer stem cells, and participate in chemotherapy resistance, radiation resistance and immunotherapy. It is involved in the occurrence and development of cervical cancer [32]. Whether PCGEM1 has a similar mechanism in CC requires further investigation.
To sum up, PCGEM1 expressions were up-regulated within CC cells. Interference of PCGEM1 could inhibit CC cell proliferation and metastasis capacities, and EMT progression by acting as a ceRNA to modulate KIF5B levels through sponging miR-642a-5p, offering a novel perspective on the prevention and treatment of CC.
Acknowledgement: None.
Funding Statement: The authors received funding by the Nantong Municipal Health Commission Research Project (MB2021054) for this study.
Author Contributions: Study conception and design: DDZ and HQL; data collection: YLL, YL, and QW; analysis and interpretation of results: YLL; draft manuscript preparation: YLL and YW. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: All data generated or analyzed during this study are included in this published article.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Manaf, R. A., Ismail, S., Cecilia, N. C. (2017). Global burden of cervical cancer: A literature review. International Journal of Public Health & Clinical Sciences, 4(2), 10–18. [Google Scholar]
2. Kang, D. F., Gynaecology, D. O. (2014). Effect analysis of combined treatment of chemotherapy, radiotherapy and surgery to treatIIb cervical cancer. China Modern Medicine (In Chinese). [Google Scholar]
3. Barker, H. E., Paget, J. T., Khan, A. A., Harrington, K. J. (2015). The tumour microenvironment after radiotherapy: Mechanisms of resistance and recurrence. Nature Reviews Cancer, 15(7), 409–425. [Google Scholar] [PubMed]
4. Kwok, Z. H., Tay, Y. (2017). Long noncoding RNAs: Lincs between human health and disease. Biochemical Society Transactions, 45(3), 805–812. [Google Scholar] [PubMed]
5. Zhang, Y. Y., Li, M., Xu, Y. D., Shang, J. (2019). LncRNA SNHG14 promotes the development of cervical cancer and predicts poor prognosis. European Review for Medical and Pharmacological Sciences, 23(9), 3664–3671. https://doi.org/10.26355/eurrev_201905_17790. [Google Scholar] [PubMed] [CrossRef]
6. Yu, C. L., Xu, X. L., Yuan, F. (2019). LINC00511 is associated with the malignant status and promotes cell proliferation and motility in cervical cancer. Bioscience Reports, 39(9), BSR20190903. https://doi.org/10.1042/BSR20190903. [Google Scholar] [PubMed] [CrossRef]
7. Yang, W., Xu, X., Hong, L., Wang, Q., Huang, J. et al. (2019). Upregulation of lncRNA GAS5 inhibits the growth and metastasis of cervical cancer cells. Journal of Cellular Physiology, 234(12), 23571–23580. https://doi.org/10.1002/jcp.28926. [Google Scholar] [PubMed] [CrossRef]
8. Hirsch, G. E., Parisi, M. M., Martins, L. A., Andrade, C. M., Barbe-Tuana, F. M. et al. (2015). Gamma-oryzanol reduces caveolin-1 and PCGEM1 expression, markers of aggressiveness in prostate cancer cell lines. The Prostate, 75(8), 783–797. https://doi.org/10.1002/pros.22960. [Google Scholar] [PubMed] [CrossRef]
9. Zhang, A., Zhang, J., Kaipainen, A., Lucas, J. M., Yang, H. (2016). Long non-coding RNA: A newly deciphered “code” in prostate cancer. Cancer Letters, 375(2), 323–330. https://doi.org/10.1016/j.canlet.2016.03.003. [Google Scholar] [PubMed] [CrossRef]
10. Chen, S., Wang, L. L., Sun, K. X., Liu, Y., Guan, X. et al. (2018). LncRNA PCGEM1 induces ovarian carcinoma tumorigenesis and progression through RhoA pathway. Cellular Physiology and Biochemistry, 47(4), 1578–1588. https://doi.org/10.1159/000490931. [Google Scholar] [PubMed] [CrossRef]
11. Li, Q., Shen, F., Zhao, L. (2018). The relationship between lncRNA PCGEM1 and STAT3 during the occurrence and development of endometrial carcinoma. Biomedicine & Pharmacotherapy, 107, 918–928. https://doi.org/10.1016/j.biopha.2018.08.091. [Google Scholar] [PubMed] [CrossRef]
12. Zhang, J., Jin, H. Y., Wu, Y., Zheng, Z. C., Guo, S. et al. (2019). Hypoxia-induced LncRNA PCGEM1 promotes invasion and metastasis of gastric cancer through regulating SNAI1. Clinical & Translational Oncology, 21(9), 1142–1151. https://doi.org/10.1007/s12094-019-02035-9. [Google Scholar] [PubMed] [CrossRef]
13. Chen, W., Chen, X., Wang, Y., Liu, T., Liang, Y. et al. (2019). Construction and analysis of lncrna-mediated cerna network in cervical squamous cell carcinoma by weighted gene co-expression network analysis. Medical Science Monitor, 25, 2609–2622. https://doi.org/10.12659/MSM.913471. [Google Scholar] [PubMed] [CrossRef]
14. Srikantan, V., Zou, Z., Petrovics, G., Xu, L., Augustus, M. et al. (2000). PCGEM1, a prostate-specific gene, is overexpressed in prostate cancer. Proceedings of the National Academy of Sciences of the United States of America, 97(22), 12216–12221. https://doi.org/10.1073/pnas.97.22.12216. [Google Scholar] [PubMed] [CrossRef]
15. Ifere, G. O., Ananaba, G. A. (2009). Prostate cancer gene expression marker 1 (PCGEM1A patented prostate-specific non-coding gene and regulator of prostate cancer progression. Recent Patents on DNA & Gene Sequences, 3(3), 151–163. [Google Scholar]
16. Jiang, H., Guo, S., Zhao, Y., Wang, Y., Piao, H. Y. et al. (2019). Circulating long non-coding RNA PCGEM1 as a novel biomarker for gastric cancer diagnosis. Pathology, Research and Practice, 215(10), 152569. https://doi.org/10.1016/j.prp.2019.152569. [Google Scholar] [PubMed] [CrossRef]
17. Sun, X., Huang, T., Liu, Z., Sun, M., Luo, S. (2019). LncRNA SNHG7 contributes to tumorigenesis and progression in breast cancer by interacting with miR-34a through EMT initiation and the Notch-1 pathway. European Journal of Pharmacology, 856, 172407. https://doi.org/10.1016/j.ejphar.2019.172407. [Google Scholar] [PubMed] [CrossRef]
18. Chen, M. J., Deng, J., Chen, C., Hu, W., Yuan, Y. C. et al. (2019). LncRNA H19 promotes epithelial mesenchymal transition and metastasis of esophageal cancer via STAT3/EZH2 axis. The International Journal of Biochemistry & Cell Biology, 113, 27–36. https://doi.org/10.1016/j.biocel.2019.05.011. [Google Scholar] [PubMed] [CrossRef]
19. Ye, F., Tian, L., Zhou, Q., Feng, D. (2019). LncRNA FER1L4 induces apoptosis and suppresses EMT and the activation of PI3K/AKT pathway in osteosarcoma cells via inhibiting miR-18a-5p to promote SOCS5. Gene, 721, 144093. https://doi.org/10.1016/j.gene.2019.144093. [Google Scholar] [PubMed] [CrossRef]
20. Jin, L. L., Jiang, X. G., He, G. Q., Shi, J. C., Su, F. F. et al. (2019). SNHG20 knockdown suppresses proliferation, migration and invasion, and promotes apoptosis in non-small cell lung cancer through acting as a miR-154 sponge. Biomedicine & Pharmacotherapy, 112, 108648. https://doi.org/10.1016/j.biopha.2019.108648. [Google Scholar] [PubMed] [CrossRef]
21. Li, K., Ma, Y. B., Tian, Y. H., Xu, X. L., Gao, Y. et al. (2019). Silencing lncRNA SNHG6 suppresses proliferation and invasion of breast cancer cells through miR-26a/VASP axis. Pathology, Research and Practice, 152575. https://doi.org/10.1016/j.prp.2019.152575. [Google Scholar] [PubMed] [CrossRef]
22. Wang, P., Liu, X., Han, G., Dai, S., Ni, Q. et al. (2019). Downregulated lncRNA UCA1 acts as ceRNA to adsorb microRNA-498 to repress proliferation, invasion and epithelial mesenchymal transition of esophageal cancer cells by decreasing ZEB2 expression. Cell Cycle, 18(18), 2359–2376. https://doi.org/10.1080/15384101.2019.1648959. [Google Scholar] [PubMed] [CrossRef]
23. Moamer, A., Hachim, I. Y., Binothman, N., Wang, N., Lebrun, J. J. et al. (2019). A role for kinesin-1 subunits KIF5B/KLC1 in regulating epithelial mesenchymal plasticity in breast tumorigenesis. eBioMedicine, 45, 92–107. https://doi.org/10.1016/j.ebiom.2019.06.009. [Google Scholar] [PubMed] [CrossRef]
24. Jing, X., Peng, J., Dou, Y., Sun, J., Ma, C. et al. (2019). Macrophage ERalpha promoted invasion of endometrial cancer cell by mTOR/KIF5B-mediated epithelial to mesenchymal transition. Immunology and Cell Biology, 97(6), 563–576. https://doi.org/10.1111/imcb.12245. [Google Scholar] [PubMed] [CrossRef]
25. Paydas, S., Acikalin, A., Ergin, M., Celik, H., Yavuz, B. et al. (2016). Micro-RNA (miRNA) profile in Hodgkin lymphoma: Association between clinical and pathological variables. Medical Oncology, 33(4), 34. https://doi.org/10.1007/s12032-016-0749-5. [Google Scholar] [PubMed] [CrossRef]
26. Liu, B. X., Yang, J., Zeng, C., Chen, Y. (2022). MACC1 correlates with tumor progression and immune cell infiltration of colon adenocarcinoma and is regulated by the lncRNA ZFAS1/miR-642a-5p axis. Journal of Oncology, 2022, 8179208. https://doi.org/10.1155/2022/8179208 [Google Scholar] [PubMed] [CrossRef]
27. Begliarzade, S., Sufianov, A., Ilyasova, T., Shumadalova, A., Sufianov, R. et al. (2024). Circular RNA in cervical cancer: Fundamental mechanism and clinical potential. Non-Coding RNA Research, 9(1), 116–124. https://doi.org/10.1016/j.ncrna.2023.11.009. [Google Scholar] [PubMed] [CrossRef]
28. Hu, Y., Sun, X., Mao, C., Guo, G., Ye, S. et al. (2017). Upregulation of long noncoding RNA TUG1 promotes cervical cancer cell proliferation and migration. Cancer Medicine, 6(2), 471–482. https://doi.org/10.1002/cam4.994. [Google Scholar] [PubMed] [CrossRef]
29. Zhang, L., Wu, X., Li, Y., Teng, X., Zou, L. et al. (2021). LncRNA SNHG5 promotes cervical cancer progression by regulating the miR-132/SOX4 pathway. Autoimmunity, 54(2), 88–96. https://doi.org/10.1080/08916934.2020.1864731. [Google Scholar] [PubMed] [CrossRef]
30. Zhang, Q., Wu, Y., Chen, J., Tan, F., Mou, J. et al. (2022). The regulatory role of both MBNL1 and MBNL1-AS1 in several common cancers. Current Pharmaceutical Design, 28(7), 581–585. https://doi.org/10.2174/1381612827666210830110732. [Google Scholar] [PubMed] [CrossRef]
31. Kim, H. J., Lee, D. W., Yim, G. W., Nam, E. J., Kim, S. et al. (2015). Long non-coding RNA HOTAIR is associated with human cervical cancer progression. International Journal of Clinical Oncology, 46(2), 521–530. https://doi.org/10.3892/ijo.2014.2758. [Google Scholar] [PubMed] [CrossRef]
32. Wang, X., Gu, Y., Zhang, L., Ma, J., Xia, Y. et al. (2024). Long noncoding RNAs regulate intrauterine adhesion and cervical cancer development and progression. Seminars in Cell and Developmental Biology, 154, 221–226. https://doi.org/10.1016/j.semcdb.2023.02.007. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools