 Open Access
Open Access
ARTICLE
The interplay mechanism between IDH mutation, MGMT-promoter methylation, and PRMT5 activity in the progression of grade 4 astrocytoma: unraveling the complex triad theory
1 Department of Pathology, Faculty of Medicine, King Abdulaziz University, Rabigh, Saudi Arabia
2 Department of Pathology, College of Medicine, Umm Al-Qura University, Makkah, Saudi Arabia
3 Department of Surgery, Faculty of Medicine, King Abdulaziz University, Jeddah, Saudi Arabia
4 Department of Surgery, Faculty of Medicine, University of Tabuk, Tabuk, Saudi Arabia
5 Section of Neurosurgery, Department of Surgery, King Abdulaziz Medical City, Jeddah, Saudi Arabia
6 Department of Pediatrics, Faculty of Medicine, King Abdulaziz University, Jeddah, Saudi Arabia
7 Department of Clinical Biochemistry, Faculty of Medicine, King Abdulaziz University, Jeddah, Saudi Arabia
8 Department of Neurosciences, King Faisal Specialist Hospital and Research Center, Jeddah, Saudi Arabia
9 Department of Surgery, King Abdulaziz Specialist Hospital, Taif, Saudi Arabia
* Corresponding Author: MAHER KURDI. Email:
Oncology Research 2024, 32(6), 1037-1045. https://doi.org/10.32604/or.2024.051112
Received 28 February 2024; Accepted 22 March 2024; Issue published 23 May 2024
Abstract
Background: The dysregulation of Isocitrate dehydrogenase (IDH) and the subsequent production of 2-Hydroxyglutrate (2HG) may alter the expression of epigenetic proteins in Grade 4 astrocytoma. The interplay mechanism between IDH, O-6-methylguanine-DNA methyltransferase (MGMT)-promoter methylation, and protein methyltransferase proteins-5 (PRMT5) activity, with tumor progression has never been described. Methods: A retrospective cohort of 34 patients with G4 astrocytoma is classified into IDH-mutant and IDH-wildtype tumors. Both groups were tested for MGMT-promoter methylation and PRMT5 through methylation-specific and gene expression PCR analysis. Inter-cohort statistical significance was evaluated. Results: Both IDH-mutant WHO grade 4 astrocytomas (n = 22, 64.7%) and IDH-wildtype glioblastomas (n = 12, 35.3%) had upregulated PRMT5 gene expression except in one case. Out of the 22 IDH-mutant tumors, 10 (45.5%) tumors showed MGMT-promoter methylation and 12 (54.5%) tumors had unmethylated MGMT. All IDH-wildtype tumors had unmethylated MGMT. There was a statistically significant relationship between MGMT-promoter methylation and IDH in G4 astrocytoma (p-value = 0.006). Statistically significant differences in progression-free survival (PFS) were also observed among all G4 astrocytomas that expressed PRMT5 and received either temozolomide (TMZ) or TMZ plus other chemotherapies, regardless of their IDH or MGMT-methylation status (p-value=0.0014). Specifically, IDH-mutant tumors that had upregulated PRMT5 activity and MGMT-promoter methylation, who received only TMZ, have exhibited longer PFS. Conclusions: The relationship between PRMT5, MGMT-promoter, and IDH is not tri-directional. However, accumulation of D2-hydroxyglutarate (2-HG), which partially activates 2-OG-dependent deoxygenase, may not affect their activities. In IDH-wildtype glioblastomas, the 2HG-2OG pathway is typically inactive, leading to PRMT5 upregulation. TMZ alone, compared to TMZ-plus, can increase PFS in upregulated PRMT5 tumors. Thus, using a PRMT5 inhibitor in G4 astrocytomas may help in tumor regression.Graphic Abstract
Keywords
Grade 4 astrocytomas are malignant brain tumors that originate from neuroglial stem cells. It is subclassified by 2021-World Health Organization (WHO) and European Association of Neuro-Oncology (EANO) into isocitrate dehydrogenase (IDH)-mutant and IDH-wildtype tumors. IDH-wildtype tumors are isolated for glioblastomas [1,2]. There are currently no known risk factors for G4 astrocytoma aside from rare genetic mutations. The recommended initial treatment since 2005 is complete surgical removal followed by radiotherapy (RT) and temozolomide (TMZ) chemotherapy (CTx). However, in most cases, resistance to TMZ eventually develops [3]. The cause of this resistance is not yet determined. Despite this treatment, the median survival rate for G4 astrocytoma typically falls within the range of 14 to 15 months [3]. Targeting cellular pathways altered in G4 astrocytoma, such as mammalian target of rapamycin (mTOR), epidermal growth factor receptor (EGFR) gene amplification, PTEN (phosphatase and tensin homolog) mutation and TP53 mutation, have also failed to improve the outcome [4]. However, neither TMZ nor any other systemic adjuvants targeted these receptors have shown superiority over placebo in terms of efficacy [5]. Although there have been cases where certain additional therapies have provided extended periods of progression-free survival (PFS), no specific pharmacological intervention has demonstrated a significant efficacy in improving the overall course of the disease [1]. There is a growing interest in targeting not just the tumor cells but also the surrounding tumor microenvironment, including blood vessels, tumor-associated macrophages (TAMs), and tumor-infiltrating lymphocytes (TILs). PD-L1 (programmed death-ligand 1) inhibitor targeting PDL1 receptors on tumor microenvironment have also failed to prevent tumor progression [5–7].
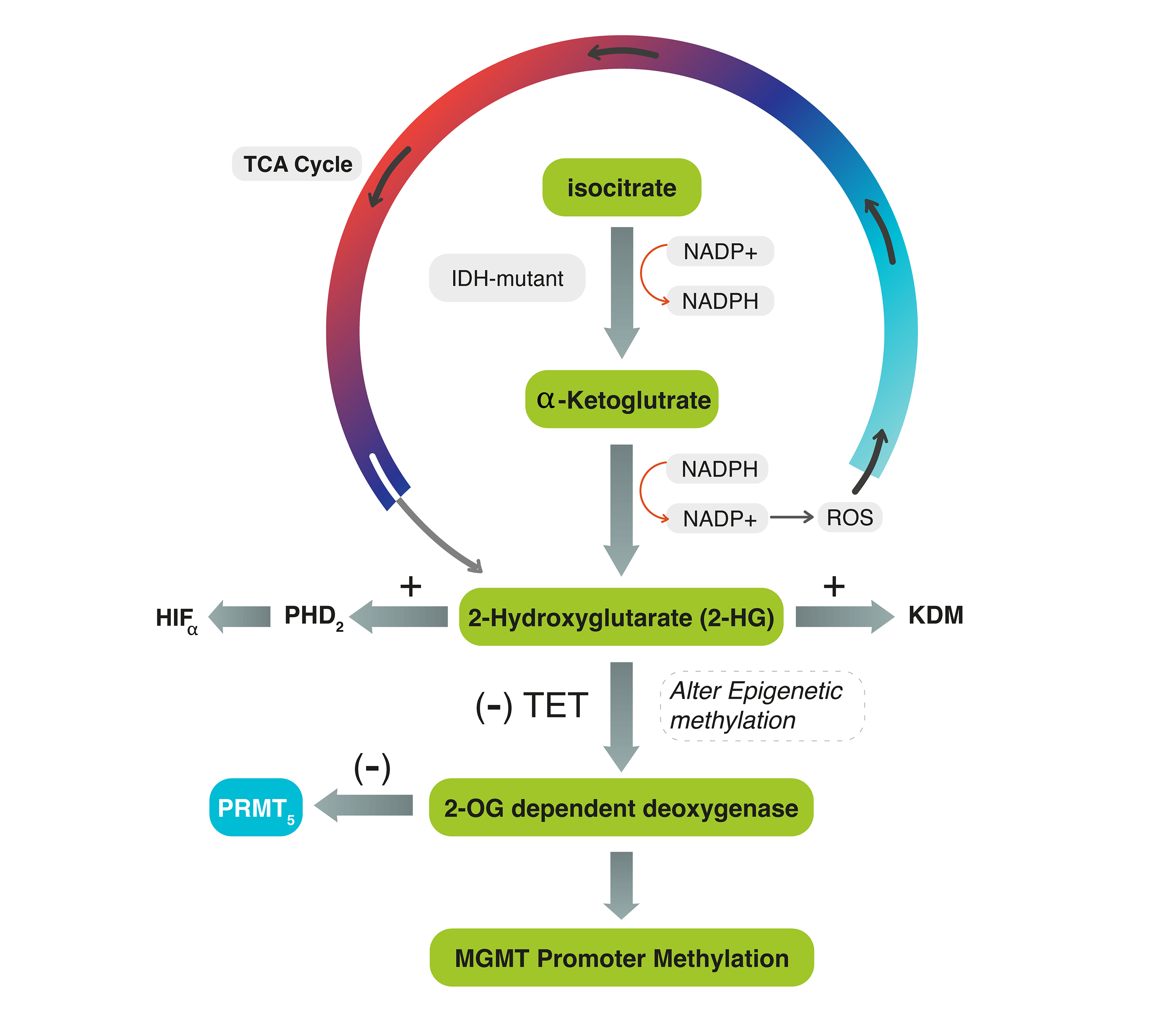
Several biomarkers are still used in clinical practice to improve the diagnostic and prognostic significance of G4 astrocytomas. One of these biomarkers are IDH mutation and O-6-methylguanine-DNA methyltransferase (MGMT) gene promotor methylation. The IDH gene was shown to correlate with outcome of patients with G4 astrocytoma [8,9]. IDH is an enzyme encoded by IDH genes that catalyzes the oxidative decarboxylation of isocitrate to α-ketoglutarate (α-KG) in the tricarboxylic acid (TCA) cycle. In this reaction, IDH utilizes Nicotinamide Adenine Dinucleotide Phosphate (NADP+) as a cofactor and generates NADPH as a product [10]. The mutant isoforms of IDH catalyze the conversion of α-KG to the oncometabolite D2-hydroxyglutarate (2-HG) instead of α-KG, which inhibits α-KG-dependent enzymes, including histone demethylases, resulting in alterations of histone and DNA methylation [10,11] (Fig. 1).
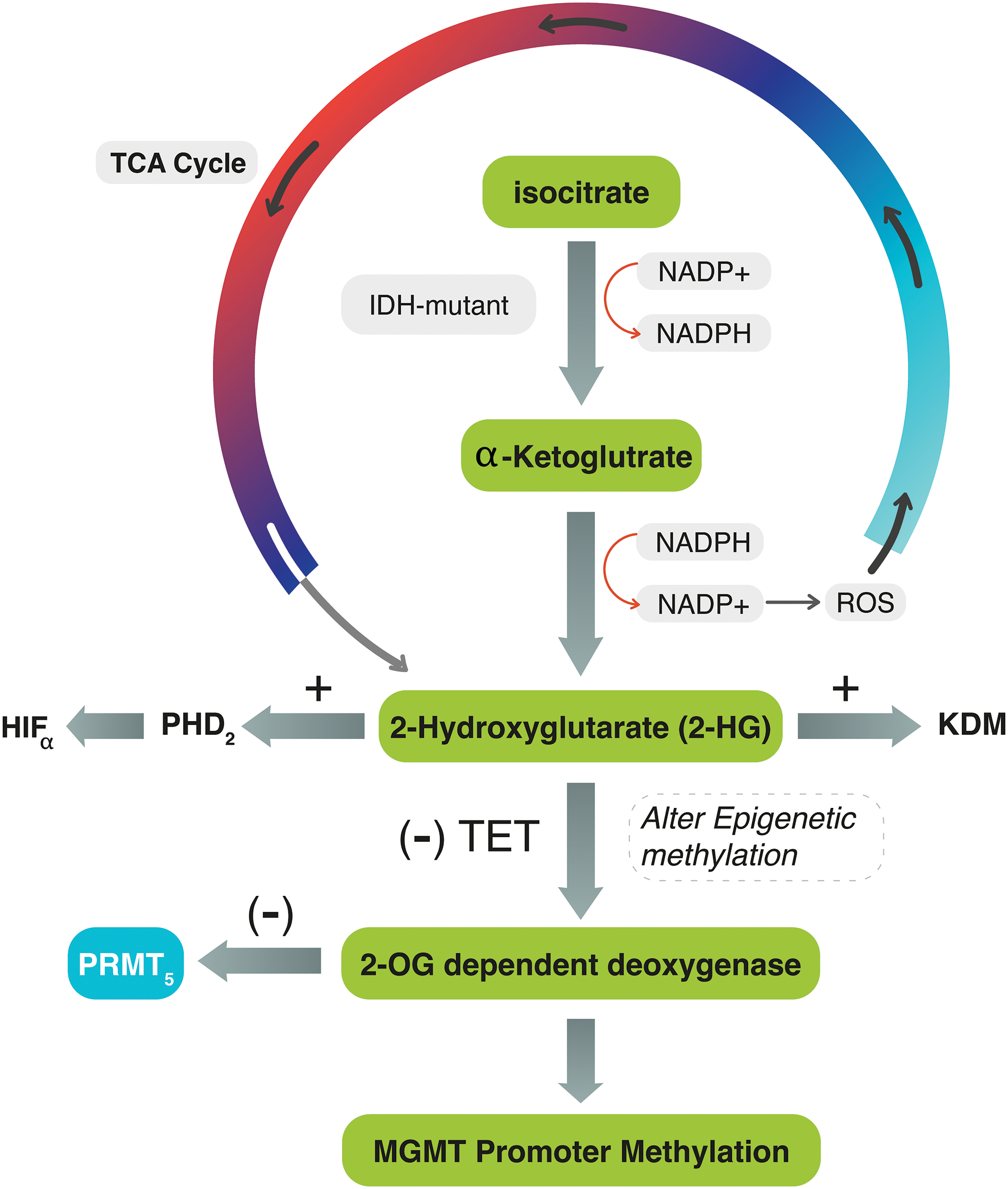
Figure 1: The diagram explains the interconnected mechanism between IDH, MGMT-promoter, and PRMT5 genes during the TCA cycle. In IDH mutant tumors, IDH converts α-KG to 2-HG, activating ROS and PHD2 and generating NADP+. The accumulation of 2-HG stimulates HIF-α and inhibits TET enzymes, preventing the conversion of 5mC to 5hmC. It also competes with 2-OG for binding to the active site of 2-OG-dependent oxygenase. These reactions can lead to DNA methylation, chromatic modification, and impair PRMT5. In IDH-wildtype tumors, 2-HG is not produced, and PRMT5 may be upregulated.
The dysregulation of IDH and the subsequent production of 2-HG may affect the balance of NADPH and NADP+ levels in the cell. Consequently, the mutant IDH enzyme consumes NAD+ and generates NADH, leading to an increased NADH/NAD+ ratio. This altered ratio possesses various effects on cellular processes, including changes in energy metabolism and redox signaling, and stimulates the production of reactive oxygen species (ROS). Simultaneously, the accumulation of 2-HG within the cells increases the activity of prolyl hydroxylase domain 2 (PHD2), leading to the dysregulation (activation) of hypoxia-inducible factor-α (HIF-α). Oncometabolite 2-HG also inhibits the function of ten-eleven translocation (TET) enzymes, which are responsible for converting 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC). It competes with 2-OG for binding to the active site of 2-OG-dependent oxygenase [11,12] (Fig. 1). All these effects may lead to a wide range of DNA methylation and chromatic modification and dysregulate epigenetic proteins such as protein methyltransferase proteins-5 (PRMT5).
The effect of IDH products on the MGM-promoter and PRMT5 was rarely investigated. MGMT, which is a DNA repair protein that removes alkyl groups from several residues, particularly the O6-position of guanine, is considered the most relevant for the action of TMZ [13,14]. The relationship between MGMT-promotor methylation and IDH mutation with treatment modalities and survival rates also showed controversial results [15]. Indeed, the potential predictive role of the MGMT status at recurrence needs to be further studied.
Protein arginine methylation (PRM) is the process of adding a methyl group to the amino acid arginine in proteins. This modification plays a crucial role in regulating various cellular processes. PRMT5 is an epigenetic modifier that expressly methylates histones, thus exerting control over gene expression [16,17]. Upregulated or highly activated PRMT5 expression has been linked to a poor prognosis in several types of cancer [18,19]. PRMT5 gene expression varies in different grades of glioma: low in low-grade gliomas and high in malignant gliomas [20]. This is because most low-grade gliomas may have mutant-IDH. IDH-mutant WHO grade-4 astrocytomas that cause excessive production of 2-HG may also alter 2-OG-dependent dioxygenase enzymes, eventually leading to PRMT5 inhibition [21] (Fig. 1). This effect would decrease the likelihood of tumor cell proliferation and progression. Suva et al. found that IDH-mutant WHO G4 astrocytomas exhibit lower levels of PRMT5 expression compared to IDH-wildtype glioblastoma, suggesting that PRMT5 may be influenced by IDH mutation [21]. In IDH-wildtype glioblastoma, the production of 2-HG is absent, allowing for PRMT5 evolution. Kurdi et al. also observed this pattern in their study conducted in 2024, where upregulated PRMT5 in brain tumor tissues was associated with earlier tumor recurrence in IDH-wildtype glioblastomas [22].
Our research study aims to explore the impact of IDH mutation, both at the mutational and non-mutational level, on MGMT-promoter methylation and PRMT5 gene expression. Additionally, it will assess the effect of IDH and MGMT-promoter on PFS in tumors with upregulated PRMT5. The findings of this study will deliberate the complex triad theory beyond these three determinants.
The study included 34 patients who were diagnosed with Grade 4 astrocytoma and underwent complete surgical resection at a single medical institution in Saudi Arabia between 2018 and 2020. Patients’ written informed consents have been previously obtained. The study was approved by the Biomedical Ethics Committee between King Faisal Specialist Hospital and King Abdulaziz University under a decree of (HA-02-J-008). The histopathological diagnosis followed the 2021-WHO classification of central nervous system (CNS) tumors [1]. Patients’ clinical data, including IDH-mutation status, treatment protocol, and PFS, were obtained from hospital records. The patients were classified into two groups: IDH-mutant and IDH-wildtype tumors. Both groups were tested for MGMT-promoter methylation and PRMT5 gene expression (Fig. 2). A 4-µm formalin-fixed and paraffin-embedded (FFPE) tissue sections were used during tissue processing for DNA and RNA extractions.
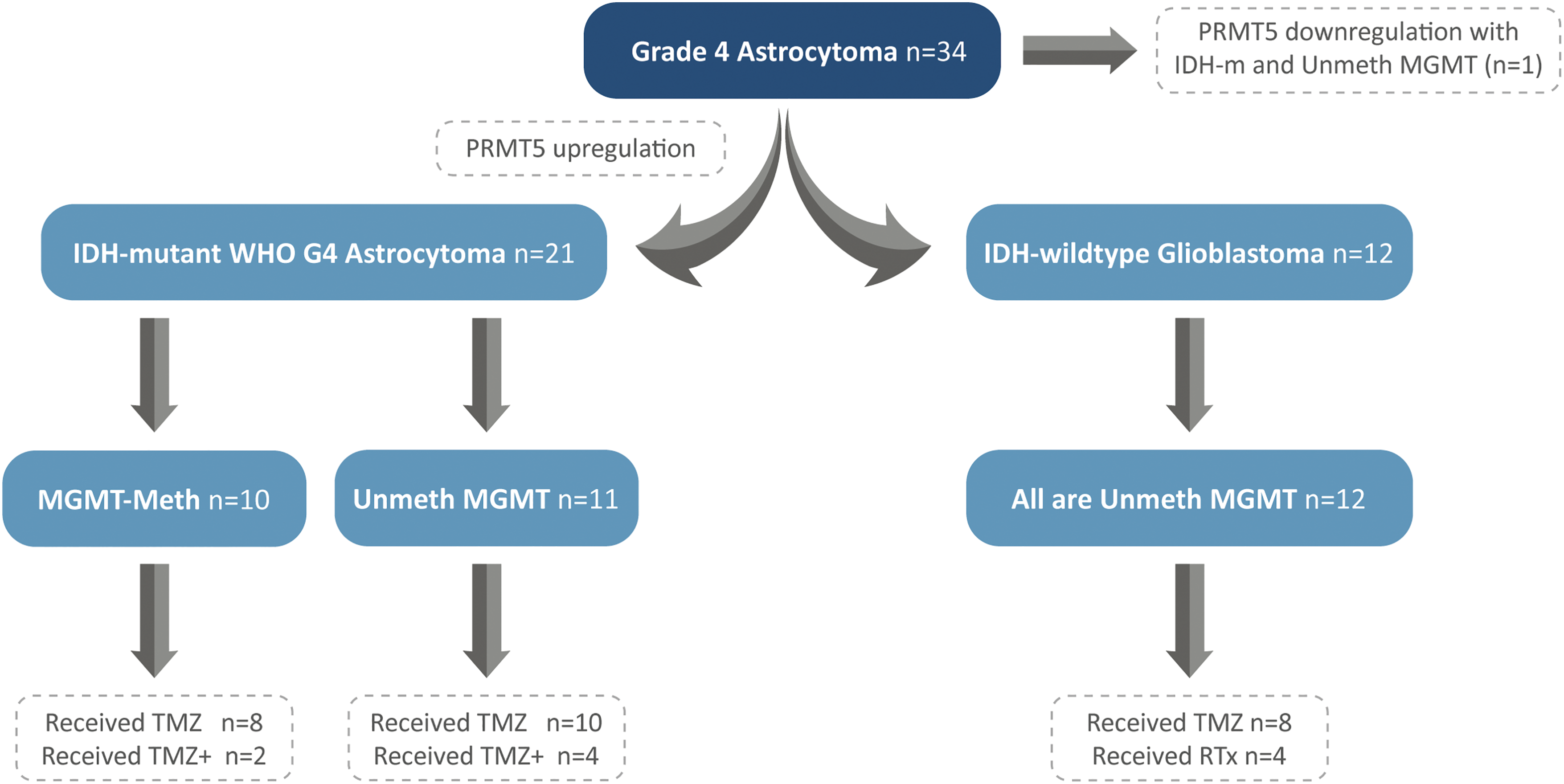
Figure 2: Patients’ stratification into IDH-mutant WHO-Grade 4 astrocytomas and IDH-wildtype glioblastomas, with segregation of MGMT-promoter and PRMT5 gene expression.
DNA extraction and MGMT-promoter methylation sequencing
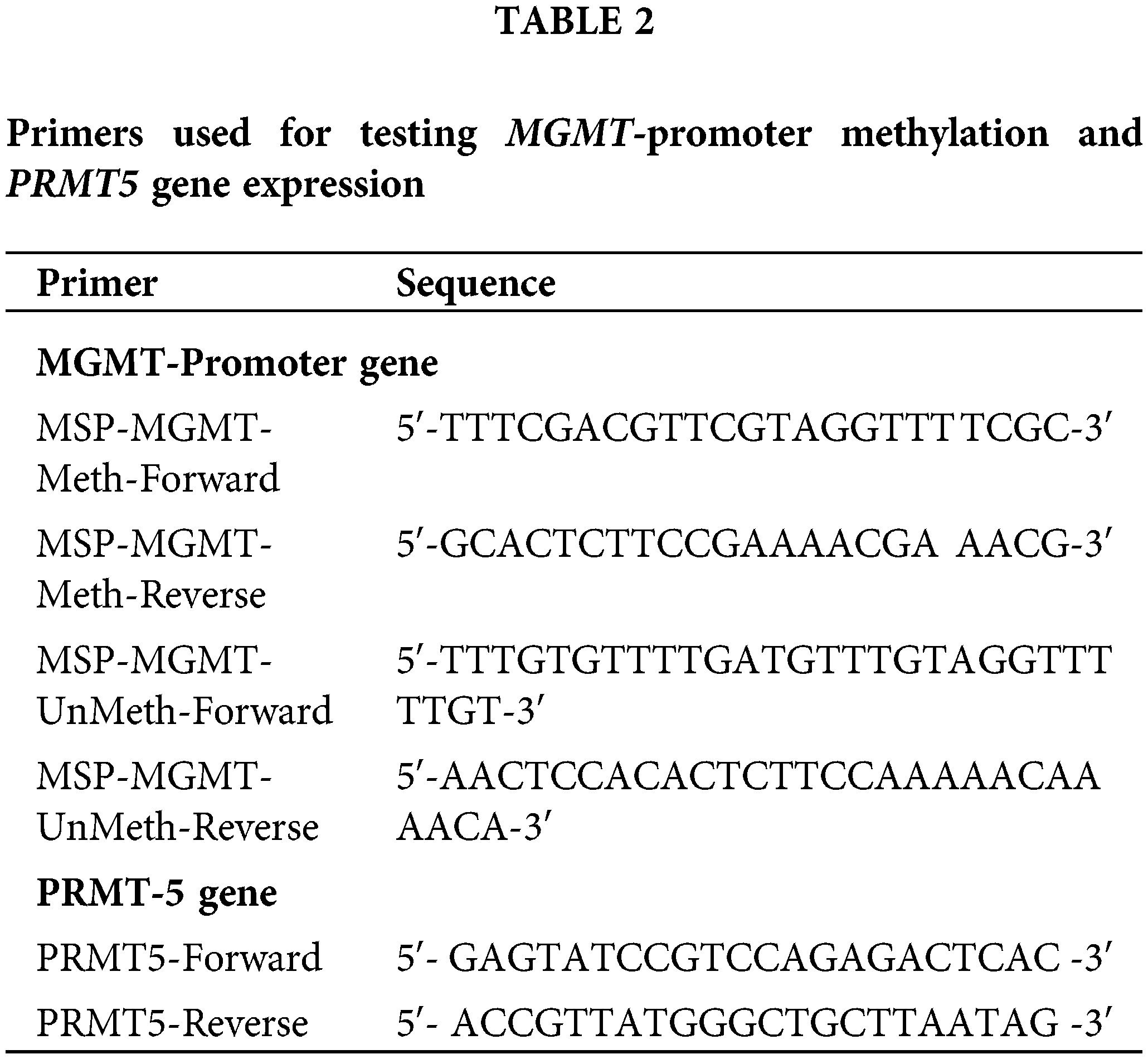
All samples in our study have been retrospectively tested for MGMT-promoter methylation using a qualitative methylation-specific polymerase chain reaction (MS PCR) to detect MGMT-promoter methylation. The MSP assay is utilized to detect CpG island methylation with high sensitivity and specificity. H&E-stained sections of 34 blocks were examined to select regions with a high content of tumor cells. DNA extraction was performed using the QIAamp DNA FFPE tissue Kit from Qiagen (Venlo, Netherlands) following the manufacturer’s instructions. The quantity and quality of the extracted DNA were assessed using a nanodrop spectrophotometer at A250/A260 and A240/A230 ratios. The concentration of each DNA sample was standardized to 45 ng and subsequently subjected to bisulfate treatment using the EpiTect bisulfited Kit from Qiagen (Maryland, USA). Qualitative detection of MGMT-promoter methylation was performed using MSP, as described by Esteller et al. [23]. The forward and reverse primers targeting methylated and unmethylated exon 1 of the human MGMT-promoter gene are described in Table 1.
The PCR Kit used in this study was the HotStarTaq plus DNA polymerase from Qiagen (Maryland, USA). The thermal cycling process was performed on a Veriti thermal cycler (Thermo-Fisher, Waltham, Massachusetts, USA). The cycling protocol consisted of an initial step at 95°C for two minutes, followed by 45 cycles of 35 s at 95°C, 30 s at 50°C, and 25 s at 70°C for 10 min. Both methylated and non-methylated control DNA samples were included in each run to ensure optimum sample accuracy. The PCR products were then visualized using 8% non-denaturing polyacrylamide gels and stained with ethidium bromide. Samples that showed amplification of a methylated sequence were considered methylation positive. The cut-off value which determined the percentage of methylated amplicons detected in an unmethylated control was used to distinguish between unmethylated and methylated tumors. The methylation results are summarized in Table 1.
RNA extraction, cDNA synthesis, and PRMT5 Gene expression assay using qPCR
RNA was extracted from 34 FFPE tissue blocks using the Qiagen Kit (Venlo, Netherlands) following the instructions provided by the manufacturer. The extraction process involved centrifugation and deparaffinization with xylene, followed by multiple washes with ethanol to ensure the cleanliness of the samples. After drying, a mixture of 150 µL Proteinase K Digestion Buffer (PKDB) and 10 µL Proteinase K (PK) was prepared and incubated at different temperatures (50°C, 75°C, and on ice). A combination of 300 µL RBC lysing buffer and ethanol was vortexed, centrifuged, and discarded. The resulting dry spin column was then transferred to a free-RNase tube. From the eluates containing RNA, a suitable sample was selected for spectrophotometric analysis. To generate complementary DNA (cDNA), the Reverse Transcription (RT) Kit (Applied Biosystems™, California, USA) was utilized. The master mix was prepared using RT buffer, dNTP mix, and random primers. The RT mixture was combined with 45ng of RNA, and the final volume was adjusted to 12 µL using RNase-free water. The primers for the experimental gene (PRMT5) and two reference genes (GAPDH and ACTB) were designed. The PRMT5 primers used for qPCR sequences (Haven Scientific, Thuwal, KSA) are described by Kurdi et al. [22] (Table 2).
RT-qPCR was performed using the EverGreen Universal qPCR Master Mix (Haven Scientific, Thuwal, KSA). The synthesized cDNA was combined with the EverGreen master mix and a small amount of each oligo to create the final PCR reaction. Triplicate reactions were performed to ensure sample accuracy. The plates were sealed with the RT-qPCR adhesive seal. For analysis, two replicates of threshold cycle (CT) values were obtained for both the target gene (PRMT5) and the reference genes (GAPDH and ACTB). The relative quantification (Rq) and fold change (FC) were estimated to measure gene expression. These values provide insights into the expression levels of the genes. The results of these data analysis can be found in the supplementary file (Suppl. Table S1) attached to this article. however, gene expression results are summarized in Table 1.
Data are described as frequencies and percentages. PFS is the time estimated after surgical resection to the first day of tumor recurrence. Pearson’s Chi-Square test was used to explore the association between MGMT-promoter and IDH in all PRMT5 expressed or non-expressed tumors. The Kaplan Meier curves (KMC) and log-rank tests were used to compare the distribution of PFS across all different groups. All statistical analyses were performed using IBM SPSS1 ver. 24 statistical software (IBM Corp., Armonk, NY, USA).
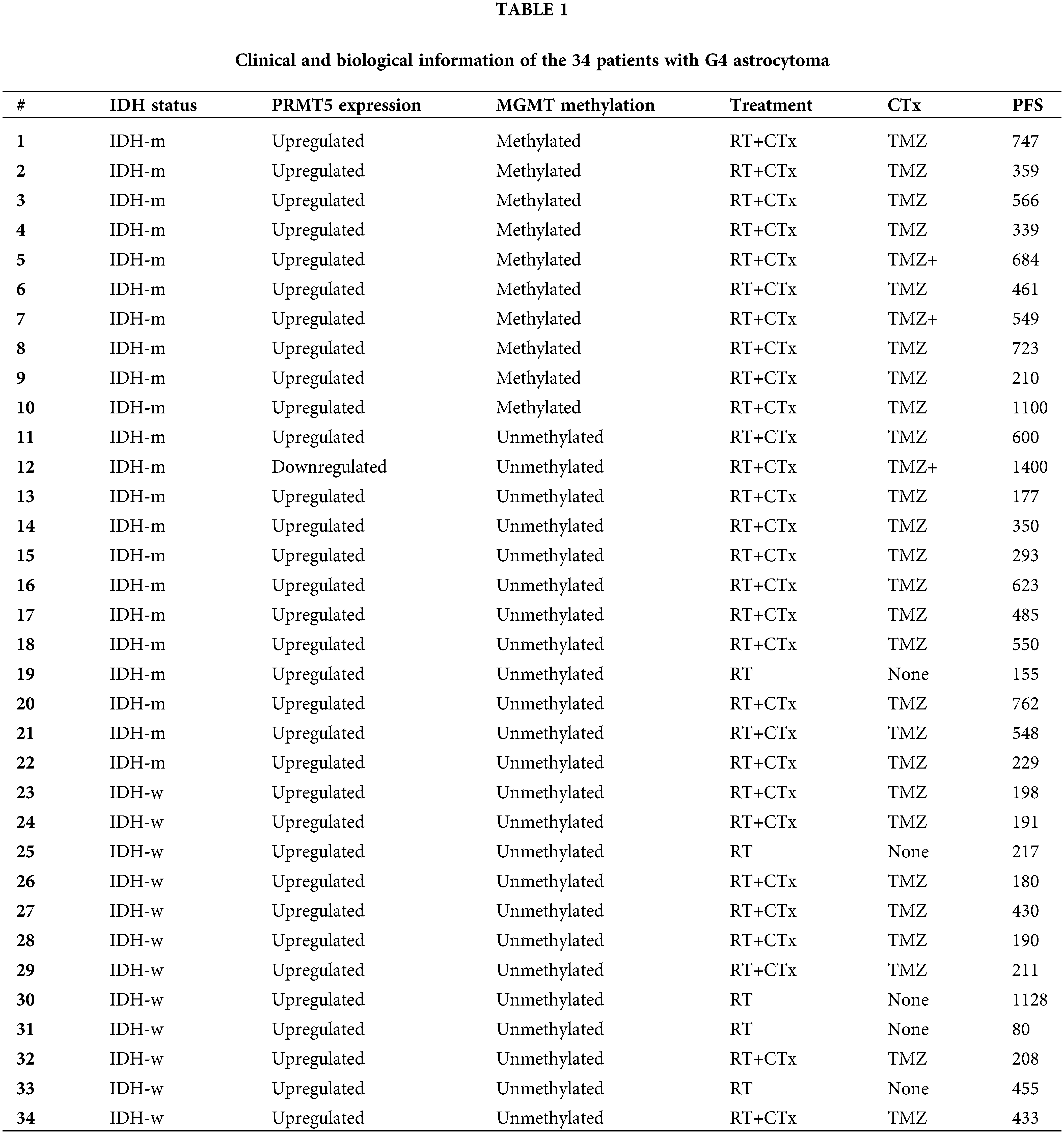
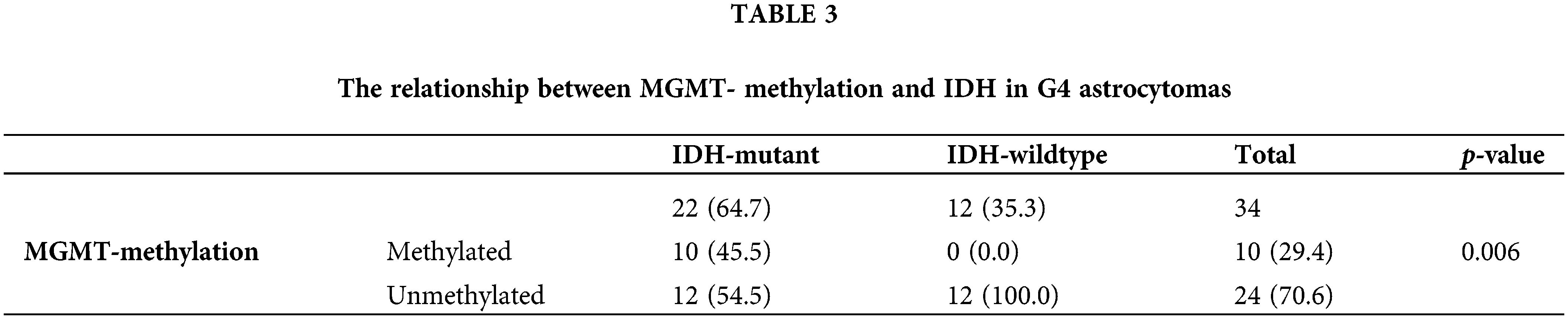
Patients are subclassified into two groups: group (a) IDH-mutant WHO grade 4 astrocytoma (n = 22, 64.7%), and group (b) IDH-wildtype glioblastoma (n = 12, 35.3%) (Fig. 2). All tumors showed PRMT5 gene upregulation (n = 33) except in one case (Fig. 2) (Table 1). Of the 22 IDH-mutant tumors, 10 (45.5%) tumors had MGMT-promoter methylation, and 12 (54.5%) tumors had unmethylated MGMT-promoter. All IDH-wildtype tumors (n = 12) had unmethylated MGMT. There was a statistically significant relationship between MGMT-promoter methylation and IDH in G4 astrocytoma (p-value = 0.006) (Table 3).
In total, 29 patients received a standard protocol of RT and CTx after surgery, while 4 patients received only RT. Patients who received CTx have been stratified based on IDH mutation and MGMT-promoter methylation (Fig. 2). They either received TMZ therapy or TMZ plus other chemotherapeutic agents such as chloroethylating agent, procarbazine, and/or bevacizumab. Standard RT was given as a total dose of 60 Gy, and the TMZ was given at 150–200 mg/m2 for 5 days every 28 days for 6–12 cycles. Statistically significant differences in PFS were observed among all G4 astrocytomas that expressed PRMT5 and received either TMZ or TMZ plus other chemotherapies, regardless of their IDH or MGMT-promoter methylation status (p-value = 0.0014) (Fig. 3).
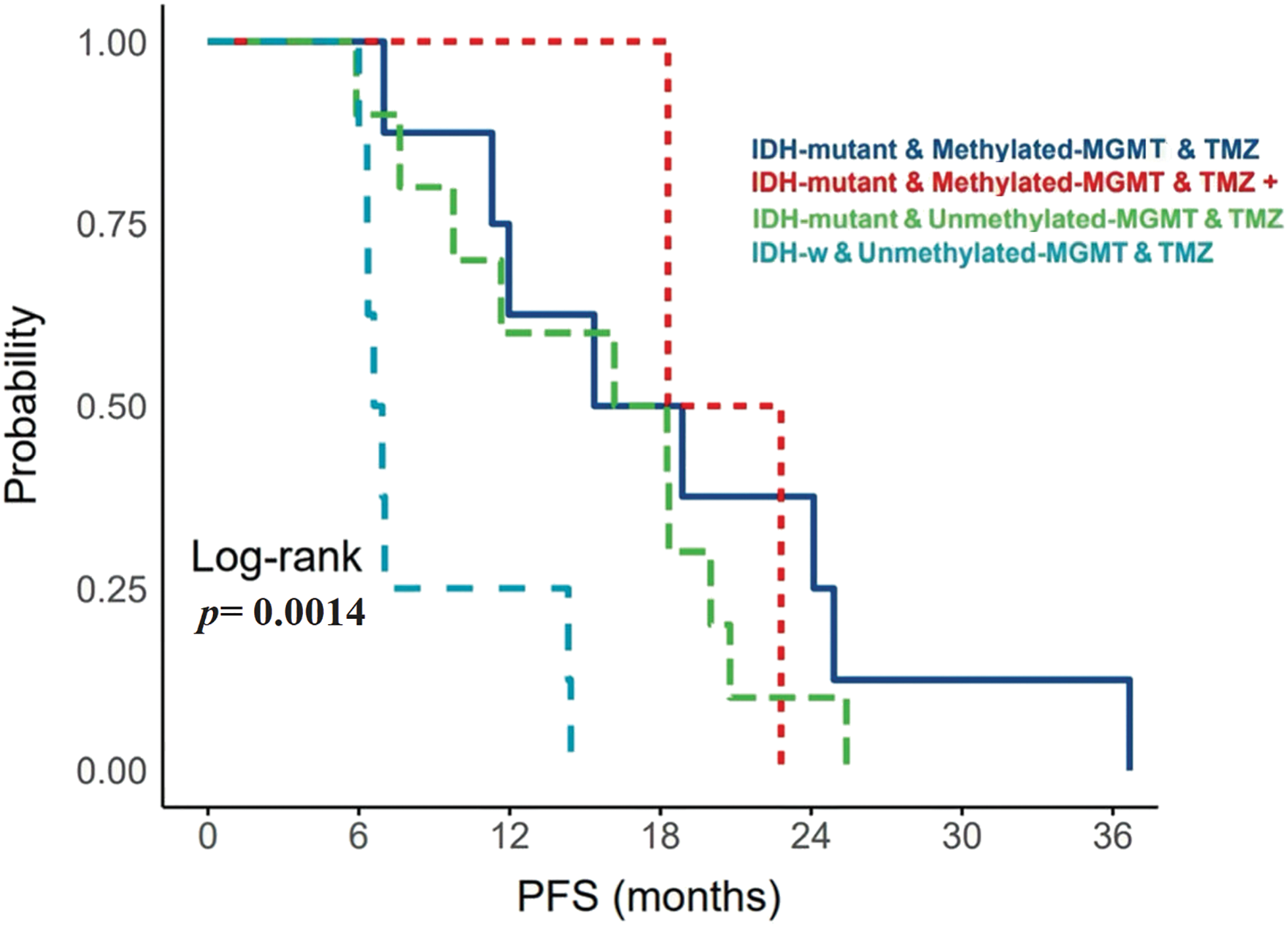
Figure 3: The relationship between MGMT-promoter and upregulated PRMT5 gene expression with PFS in patients with IDH-mutant WHO G4 astrocytoma and IDH-wildtype glioblastoma. MGMT: O-6-methylguanine-DNA methyltransferase; TMZ: Temozolomide; W: Wildtype.
Specifically, IDH-mutant tumors that had upregulated PRMT5 and MGMT-promoter methylation, who received only TMZ, have exhibited longer PFS. On the other hand, IDH-wildtype glioblastoma cases that expressed PRMT5 and had unmethylated MGMT-promoter, who received TMZ treatment, have experienced early tumor recurrence.
The end-product of isocitrate oxidative decarboxylation into α-KG by the IDH enzyme in the TCA cycle typically determines the action of protein methylation in tumor cells. When the IDH gene mutates, the metabolic product ends with 2-HG, which inhibits histone demethylases, disrupts HIF-α regulation, and alters epigenetic proteins through the 2-OG-dependent dioxygenase pathway. This alteration can potentially deactivate the DNA-repair function of MGMT and inhibit the PRMT5 gene expression [10]. On the other hand, if the IDH gene does not mutate, the oxidative decarboxylation process is interrupted, which may result in the failure to methylate MGMT-promoter. Consequently, PRMT5 may be upregulated (Fig. 1). Although this proposed theory has not been described before, the logical relationship between these determinants explains their association. The association between IDH-wildtype glioblastomas and PRMT5 upregulation has been previously reported by Kurdi et al. and Suva et al. [21,22]. The higher the grade, the more aggressive the astrocytoma with wildtype IDH, and the more significant the upregulation of PRMT5. However, this relationship does not apply to all cases of G4 astrocytoma with IDH mutation. Our current research provides a clear example, where 50% (n = 11/22) of IDH-mutant G4 astrocytoma showed upregulated PRMT5 expression, and 10 of them had unmethylated MGMT-promoter. This indicates that mutant isoforms of IDH may not directly deactivate the DNA repair function of MGMT or inhibit PRMT5. Other unidentified or non-specific gene drivers or demethylases could potentially be involved in maintaining the expression of PRMT5 or in demethylating the MGMT-promoter region.
Although the sensitivity of MGMT-methylated tumors to chemotherapeutic agents varies, MGMT-promoter is considered the most relevant for the action of TMZ [13,14]. Its relationship with G4 astrocytoma treatment modalities and clinical outcomes showed provocative results [15]. According to various studies, there is conflicting evidence regarding the sensitivity of G4 astrocytomas with methylated MGMT-promoter to TMZ treatment. Some studies suggest that G4 astrocytomas are sensitive to TMZ, while others indicate that MGMT-promoter methylation is not associated with a favorable outcome [23–26]. Despite this controversy, MGMT-promoter methylation is still considered a prognostic factor for patients with G4 astrocytoma. Kurdi et al. found that using TMZ or TMZ plus other chemotherapeutic agents did not result in significant differences in PFS for G4 astrocytomas, regardless of their IDH status [15]. However, the same study suggested that IDH-wildtype glioblastomas with unmethylated MGMT-promoter may benefit from combined chemotherapeutic modalities to improve prognosis. PRMT5 gene dysregulation can occur in IDH-mutant WHO-G4 astrocytomas and IDH-wildtype glioblastomas, regardless of MGMT-promoter status. However, inhibiting the PRMT5 activity may assist in suppressing tumor growth in both cases. Kurdi et al. briefly described how blocking the PRMT5 receptor can lead to regression of tumor cells [22]. It is worth noting that downregulation of PRMT5 in IDH-mutant WHO G4 astrocytomas or IDH-wildtype glioblastomas is associated with a more favorable prognosis.
There is evidence to suggest that the growth of glioma may be reliant on PRMT5 expression, making it a potential therapeutic target [27]. Otani et al. demonstrated that blocking PRMT5 activity led to apoptosis in stem-like glioma cells and restoration of immune response [28]. Abe et al. suggested that PRMT5 plays a role in promoting the immunosuppressive characteristics of the tumor microenvironment [29]. Consequently, inhibiting PRMT5 may contribute to the recovery of the immune cycle and enhance the immune system’s ability to combat cancer cells. Preclinical studies, in vitro and in vivo, revealed that among the PRMT5 inhibitors, treatment of glioblastoma with compound 5 (CMP5) reflects the impact of PRMT5 knockdown, which results in apoptosis of differentiated cells driven into a nonreplicated aging state [27]. PRMT5 inhibitor has also been shown to sensitize glioma cells to radiation and chemotherapies [27,30,31]. This suggests that combining a PRMT5 inhibitor with existing chemotherapies may prevent tumor progression. There is no clear evidence on whether IDH mutation or MGMT promoter methylation can affect the anti-tumor activity of PRMT5 in G4 astrocytoma when used in conjunction with other chemotherapeutic agents. However, it has been suggested that a PRMT5 inhibitor may increase the sensitivity of glioma cells to chemotherapies. It is unclear which chemotherapies interact more effectively with a PRMT5 inhibitor. Nevertheless, using multiple regimen chemotherapies can potentially enhance patients’ survival by inhibiting tumor re-growth in different ways [15].
In our current study, we have observed that there were significant differences in PFS of all G4 astrocytoma with upregulated PRMT5, receiving TMZ or TMZ plus other chemotherapeutic agents, regardless of IDH or MGMT-promoter status (Fig. 3). IDH-mutant WHO-G4 astrocytomas with upregulated PRMT5 and methylated MGMT-promoter who received only TMZ (n = 8) had the longest PFS. Additionally, IDH-mutant tumors regardless of MGMT-promoter status, who received only TMZ had better PFS than those who received TMZ plus other chemotherapies (Fig. 3). Our findings are interesting but might not be satisfactory because of the low number of samples acknowledged in our research.
Herein, two possible observations are noticed. First, our findings are consistent with previously mentioned theories emphasizing that 2-HG end-product of mutant IDH alters the epigenetic proteins in the DNA of these tumors. This alteration should frequently cause MGMT-methylation and PRMT5 inhibition. However, we found that 2-HG in most IDH-mutant cases caused partial methylation of MGMT-promoter but upregulation of the PRMT5 gene in 97% of the tumors. This probably occurred because 2-HG causes partial activation of 2-OG-dependent dioxygenases. There is no clear evidence beyond this finding. However, possible unknown factors may have participated in this mechanism of action. One of the potential theories is HIF-α. HIF-α is activated by 2-HG, a product of IDH-mutation, which indirectly affects 2-OG-dependent dioxygenase under hypoxic conditions. 2-OG is highly sensitive to oxygen, and when the oxygen level is high, HIF-α dysregulates and activates 2-OG [32]. Moreover, a lack of vitamin C levels in the body may also indirectly activate 2-OG [33].
Second, IDH-mutant tumors with upregulated PRMT5 activity responded well to TMZ than TMZ plus other chemotherapies, which subsequently improved PFS (Fig. 3). Nonetheless, using TMZ with other chemotherapies may decrease the sensitivity of the tumor cells to TMZ, regardless of MGMT-promoter status. MGMT-methylation has no substantial effect on tumor response to CTx [15]. It is unclear if one of these chemotherapies used has reduced the effect of TMZ or the sensitivity of tumor cells to TMZ had less response. This theory is mainly based on IDH-mutant WHO-G4 astrocytoma. In IDH-wildtype glioblastomas, using multiple regimes of chemotherapies regardless of MGMT-promoter status is better than using single TMZ therapy, as Kurdi et al mentioned in their study [15]. In our current study, the theory was not clearly defined as all IDH-wildtype tumors with unmethylated MGMT-promoter and increased PRMT5 activity were treated solely with TMZ, resulting in a low PFS. Additionally, we hypothesize that the efficacy of TMZ in G4 astrocytoma may improve with the use of PRMT5 inhibitors, irrespective of the methylation status of MGMT-promoter or IDH mutational status.
The relationship between PRMT5 activity and MGMT-promoter methylation with IDH mutation is not tri-directional. Partial activation of 2-OG-dependent deoxygenase due to the accumulation of 2-HG in IDH-mutant tumors may not have an impact on MGMT or PRMT5 activity. In IDH-wildtype glioblastomas, the 2HG-2OG pathway remains inactive, leading to persistent upregulation of PRMT5. TMZ alone has been observed to increase PFS in upregulated PRMT5 tumors. However, the use of TMZ with other chemotherapies may impede tumor regression for reasons that are currently unknown. PRMT5 inhibitors in both isoforms may help in tumor regression.
Acknowledgement: None.
Funding Statement: None.
Author Contributions: All authors contributed to the study conception and design. Material preparation and tissue processing were performed Maher Kurdi and Alaa Alkhotani. Data collection and analysis were performed by Maher Kurdi, Abdulrahman Sabbagh, Eyad Faizo, Ahmed Lary, Ahmed Bamaga, Majid Almansouri, Badr Hafiz Thamer Alsharif and Saleh Baeesa. The first draft of the manuscript was written by Maher Kurdi and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Availability of Data and Materials: The datasets generated during and/or analysed during the current study are available from the corresponding author (MK) on reasonable request. Dataset is represented in a supplementary file.
Ethics Approval: The study was conducted according to the guidelines of the Declaration of Helsinki and was approved by the Biomedical Ethics Committee Agreement between King Faisal Specialist Hospital and King Abdulaziz University under the Decree (HA-02-J-008) in Jeddah, Saudi Arabia. Informed consent: Patients’ written informed consents have been previously obtained.
Conflicts of Interest: The authors have no relevant conflict of interest to disclose.
References
1. Louis, D. N., Perry, A., Wesseling, P., Brat, D. J., Cree, I. A. et al. (2021). The 2021 WHO classification of tumors of the central nervous system: A summary. Neuro-Oncology, 23(8), 1231–1251. https://doi.org/10.1093/neuonc/noab106. [Google Scholar] [PubMed] [CrossRef]
2. Kurdi, M., Moshref, R. H., Katib, Y., Faizo, E., Najjar, A. A. et al. (2022). Simple approach for the histomolecular diagnosis of central nervous system gliomas based on 2021 world health organization classification. World Journal of Clinical Oncology, 13(7), 567–576. https://doi.org/10.5306/wjco.v13.i7.567. [Google Scholar] [PubMed] [CrossRef]
3. Jiapaer, S., Furuta, T., Tanaka, S., Kitabayashi, T., Nakada, M. (2018). Potential strategies overcoming the temozolomide resistance for glioblastoma. Neurologia Medico-Chirurgica, 58(10), 405–421. https://doi.org/10.2176/nmc.ra.2018-0141. [Google Scholar] [PubMed] [CrossRef]
4. Weller, M., van den Bent, M., Preusser, M., Le Rhun, E., Tonn, J. C. et al. (2021). EANO guidelines on the diagnosis and treatment of diffuse gliomas of adulthood. Nature Reviews Clinical Oncology, 18(3), 170–186. https://doi.org/10.1038/s41571-020-00447-z. [Google Scholar] [PubMed] [CrossRef]
5. Stupp, R., Wong, E. T., Kanner, A. A., Steinberg, D., Engelhard, H. et al. (2018). NovoTTF-100A versus physician’s choice chemotherapy in recurrent glioblastoma: A randomised phase III trial of a novel treatment modality. European Journal of Cancer, 48, 2192–2202. [Google Scholar]
6. Sturm, D., Witt, H., Hovestadt, V., Khuong-Quang, D. A., Jones, D. T. W. et al. (2012). Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell, 22, 425–437. https://doi.org/10.1016/j.ccr.2012.08.024. [Google Scholar] [PubMed] [CrossRef]
7. Stadler, C., Gramatzki, D., Le Rhun, E., Hottinger, A. F., Hundsberger, T. et al. (2023). Glioblastoma in the oldest old: Clinical characteristics, therapy, and outcome in patients aged 80 years and older. Neuro-Oncology Practice, 11(2), 132–141. [Google Scholar] [PubMed]
8. Hartmann, C., Meyer, J., Balss, J., Capper, D., Mueller, W. et al. (2009). Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: A study of 1,010 diffuse gliomas. Acta Neuropathologica Communications, 118, 469–474. https://doi.org/10.1007/s00401-009-0561-9. [Google Scholar] [PubMed] [CrossRef]
9. Cai, M., Zhao, J., Ding, Q., Wei, J. (2024). Oncometabolite 2-hydroxyglutarate regulates anti-tumor immunity. Heliyon, 10(2), e24454. https://doi.org/10.1016/j.heliyon.2024.e24454. [Google Scholar] [PubMed] [CrossRef]
10. Waitkus, M. S., Diplas, B. H., Yan, H. (2018). Biological role and therapeutic potential of IDH mutations in cancer. Cancer Cell, 34(2), 186–195. https://doi.org/10.1016/j.ccell.2018.04.011. [Google Scholar] [PubMed] [CrossRef]
11. Leather, T., Jenkinson, M. D., Das, K., Poptani, H. (2017). Magnetic resonance spectroscopy for detection of 2-Hydroxyglutarate as a biomarker for IDH mutation in gliomas. Metabolites, 7(2), 29. https://doi.org/10.3390/metabo7020029. [Google Scholar] [PubMed] [CrossRef]
12. Li, F. J., Li, L. M., Zhang, R. H., Xu, C., Zhou, P. et al. (2017). The role of 5-hydroxymethylcytosine in melanoma. Melanoma Research, 27(3), 175–179. https://doi.org/10.1097/CMR.0000000000000349. [Google Scholar] [PubMed] [CrossRef]
13. Fang, Q. (2024). The versatile attributes of MGMT: Its repair mechanism, crosstalk with other DNA repair pathways, and its role in cancer. Cancers, 16(2), 331. https://doi.org/10.3390/cancers16020331. [Google Scholar] [PubMed] [CrossRef]
14. Fahrer, J., Christmann, M. (2023). DNA alkylation damage by nitrosamines and relevant DNA repair pathways. International Journal of Molecular Sciences, 24(5), 4684. https://doi.org/10.3390/ijms24054684. [Google Scholar] [PubMed] [CrossRef]
15. Kurdi, M., Shafique Butt, N., Baeesa, S., Alghamdi, B., Maghrabi, Y. et al. (2021). The impact of IDH1 mutation and MGMT promoter methylation on recurrence-free interval in glioblastoma patients treated with radiotherapy and chemotherapeutic agents. Pathology & Oncology Research, 27, 1609778. https://doi.org/10.3389/pore.2021.1609778. [Google Scholar] [PubMed] [CrossRef]
16. Jarrold, J., Davies, C. C. (2019). PRMTs and arginine methylation: Cancer’s best-kept secret? Trends in Molecular Medicine, 25(11), 993–1009. https://doi.org/10.1016/j.molmed.2019.05.007. [Google Scholar] [PubMed] [CrossRef]
17. Wang, Y. J., Cao, J. B., Yang, J., Liu, T., Yu, H. L. et al. (2024). PRMT5-mediated homologous recombination repair is essential to maintain genomic integrity of neural progenitor cells. Cellular and Molecular Life Sciences, 81(1), 123. https://doi.org/10.1007/s00018-024-05154-x. [Google Scholar] [PubMed] [CrossRef]
18. Shimizu, D., Kanda, M., Sugimoto, H., Shibata, M., Tanaka, H. et al. (2017). The protein arginine methyltransferase 5 promotes malignant phenotype of hepatocellular carcinoma cells and is associated with adverse patient outcomes after curative hepatectomy. International Journal of Oncology, 50(2), 381–386. https://doi.org/10.3892/ijo.2017.3833. [Google Scholar] [PubMed] [CrossRef]
19. Chan, L. H., Wang, P., Abuhammad, S., Lim, L. R. J., Cursons, J. S. et al. (2023). PRMT5 and CDK4/6 inhibition result in distinctive patterns of alternative splicing in melanoma. PLoS One, 18(11), e0292278. https://doi.org/10.1371/journal.pone.0292278. [Google Scholar] [PubMed] [CrossRef]
20. Han, X., Li, R., Zhang, W., Yang, X., Wheeler, C. G. et al. (2014). Expression of PRMT5 correlates with malignant grade in gliomas and plays a pivotal role in tumor growth in vitro. Journal of Neuro-Oncology, 118(1), 61–72. https://doi.org/10.1007/s11060-014-1419-0. [Google Scholar] [PubMed] [CrossRef]
21. Suvà, M. L., Riggi, N., Janiszewska, M., Radovanovic, I., Provero, P. et al. (2009). EZH2 is essential for glioblastoma cancer stem cell maintenance. Cancer Research, 69(24), 9211–9218. https://doi.org/10.1158/0008-5472.CAN-09-1622. [Google Scholar] [PubMed] [CrossRef]
22. Kurdi, M., Fadul, M., Addas, B., Faizo, E., Bamaga, A. K. et al. (2024). Glioblastoma with PRMT5 gene upregulation is a key target for tumor cell regression. Oncologie. https://doi.org/10.1515/oncologie-2023-0534 [Google Scholar] [CrossRef]
23. Esteller, M., Hamilton, S. R., Burger, P. C., Baylin, S. B., Herman, J. G. (1999). Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Research, 59(4), 793–797. [Google Scholar] [PubMed]
24. Dunn, J., Baborie, A., Alam, F., Joyce, K., Moxham, M. et al. (2009). Extent of MGMT promoter methylation correlates with outcome in glioblastomas given temozolomide and radiotherapy. British Journal of Cancer, 101, 124–131. https://doi.org/10.1038/sj.bjc.6605127. [Google Scholar] [PubMed] [CrossRef]
25. Esteyrie, V., Dehais, C., Martin, E., Carpentier, C., Uro-Coste, E. et al. (2021). Radiotherapy plus procarbazine, lomustine, and vincristine vs. radiotherapy plus temozolomide for IDH-mutant anaplastic astrocytoma: A retrospective multicenter analysis of the French POLA cohort. Oncologist, 26(5), e838–e846. https://doi.org/10.1002/onco.13701. [Google Scholar] [PubMed] [CrossRef]
26. Park, C. K., Park, S. H., Lee, S. H., Kim, C. Y., Kim, D. W. et al. (2009). Methylation status of the MGMT gene promoter fails to predict the clinical outcome of glioblastoma patients treated with ACNU plus cisplatin. Neuropathology, 29, 443–449. https://doi.org/10.1111/neu.2009.29.issue-4. [Google Scholar] [CrossRef]
27. Banasavadi-Siddegowda, Y. K., Welker, A. M., An, M., Yang, X., Zhou, W. et al. (2018). PRMT5 as a druggable target for glioblastoma therapy. Neuro-Oncology, 20(6), 753–763. https://doi.org/10.1093/neuonc/nox206. [Google Scholar] [PubMed] [CrossRef]
28. Otani, Y., Sur, H. P., Rachaiah, G., Namagiri, S., Chowdhury, A. et al. (2021). Inhibiting protein phosphatase 2A increases the antitumor effect of protein arginine methyltransferase 5 inhibition in models of glioblastoma. Neuro-Oncology, 23(9), 1481–1493. https://doi.org/10.1093/neuonc/noab014. [Google Scholar] [PubMed] [CrossRef]
29. Abe, Y., Sano, T., Tanaka, N. (2013). The role of PRMT5 in immuno-oncology. Genes, 14(3), 678. [Google Scholar]
30. Yuan, Y., Nie, H. (2021). Protein arginine methyltransferase 5: A potential cancer therapeutic target. Cellular Oncology, 44(1), 33–44. https://doi.org/10.1007/s13402-020-00577-7. [Google Scholar] [PubMed] [CrossRef]
31. Yan, F., Alinari, L., Lustberg, M. E., Martin, L. K., Cordero-Nieves, H. M. et al. (2014). Genetic validation of the protein arginine methyltransferase PRMT5 as a candidate therapeutic target in glioblastoma. Cancer Resesrch, 74(6), 1752–1765. https://doi.org/10.1158/0008-5472.CAN-13-0884. [Google Scholar] [PubMed] [CrossRef]
32. Gonzalez, F. J., Xie, C., Jiang, C. (2018). The role of hypoxia-inducible factors in metabolic diseases. Nature Reviews Endocrinology, 15(1), 21–32. [Google Scholar] [PubMed]
33. Treadway, C. J., Boyer, J. A., Yang, S., Yang, H., Liu, M. et al. (2024). Using NMR to monitor TET-dependent methylcytosine dioxygenase activity and regulation. ACS Chemical Biology, 19(1), 15–21. https://doi.org/10.1021/acschembio.3c00619. [Google Scholar] [PubMed] [CrossRef]
Supplementary Materials
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools