 Open Access
Open Access
ARTICLE
The role of polymorphic cytochrome P450 gene (CYP2B6) in B-chronic lymphocytic leukemia (B-CLL) incidence and outcome among Egyptian patients
1 Division of Biochemistry, Department of Chemistry, Faculty of Science, Mansoura University, Mansoura, 35511, Egypt
2 Hematology Unit, Department of Clinical Pathology, Faculty of Medicine, Mansoura University, Mansoura, 35511, Egypt
3 Medical Oncology Unit, Oncology Center Mansoura University, Mansoura, 35511, Egypt
4 Department of Genetics, Mansoura University Children’s Hospital, Mansoura, 35511, Egypt
* Corresponding Author: MENNA AL-ADL. Email:
Oncology Research 2024, 32(4), 785-797. https://doi.org/10.32604/or.2024.047021
Received 22 October 2023; Accepted 17 January 2024; Issue published 20 March 2024
Abstract
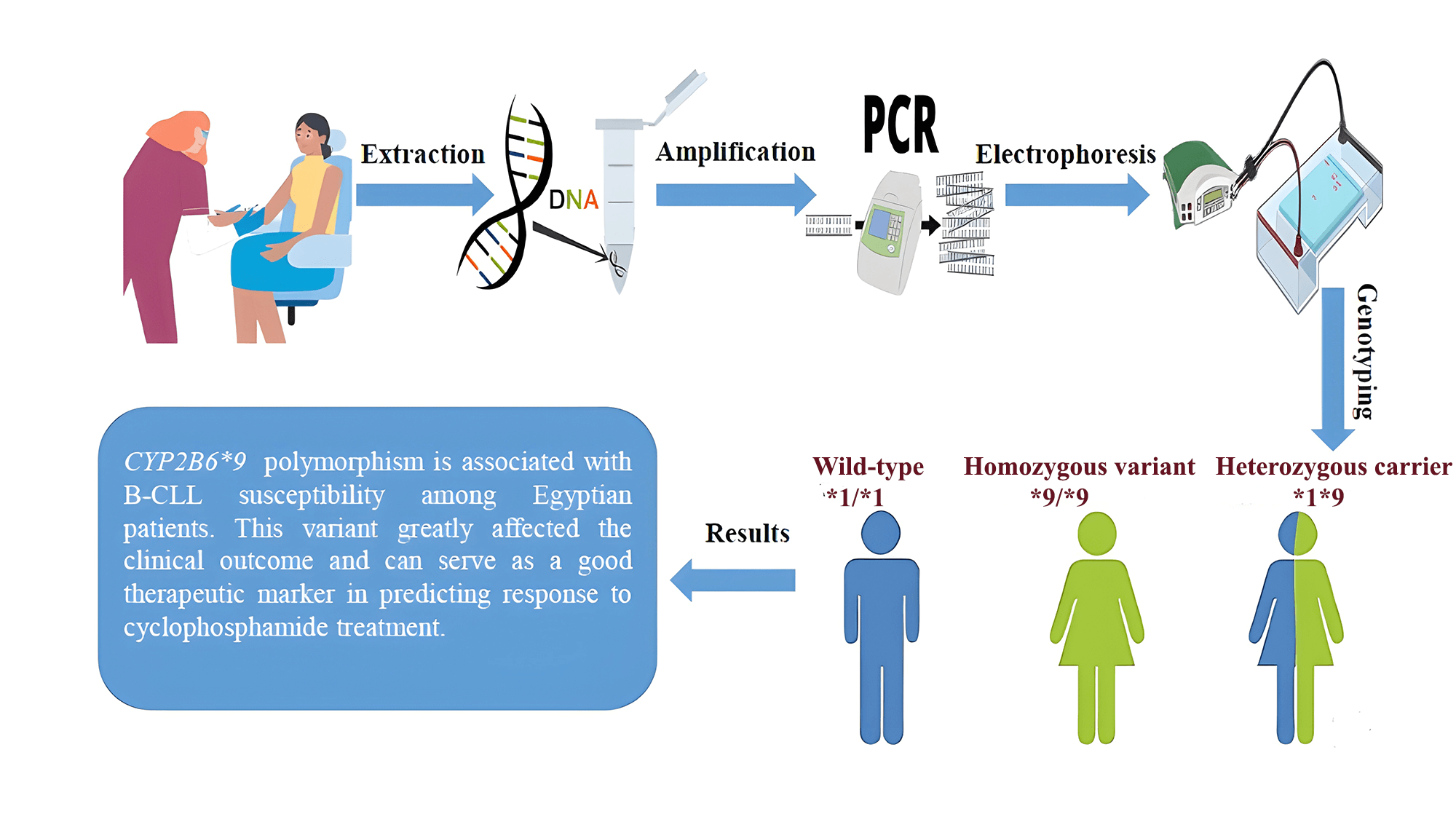
Cytochromes P450 (CYPs) play a prominent role in catalyzing phase I xenobiotic biotransformation and account for about 75% of the total metabolism of commercially available drugs, including chemotherapeutics. The gene expression and enzyme activity of CYPs are variable between individuals, which subsequently leads to different patterns of susceptibility to carcinogenesis by genotoxic xenobiotics, as well as differences in the efficacy and toxicity of clinically used drugs. This research aimed to examine the presence of the CYP2B6*9 polymorphism and its possible association with the incidence of B-CLL in Egyptian patients, as well as the clinical outcome after receiving cyclophosphamide chemotherapy. DNA was isolated from whole blood samples of 100 de novo B-CLL cases and also from 100 sex- and age-matched healthy individuals. The presence of the CYP2B6*9 (G516T) polymorphism was examined by PCR-based allele specific amplification (ASA). Patients were further indicated for receiving chemotherapy, and then they were followed up. The CYP2B6*9 variant indicated a statistically significant higher risk of B-CLL under different genetic models, comprising allelic (T-allele vs. G-allele, OR = 4.8, p < 0.001) and dominant (GT + TT vs. GG, OR = 5.4, p < 0.001) models. Following cyclophosphamide chemotherapy, we found that the patients with variant genotypes (GT + TT) were less likely to achieve remission compared to those with the wild-type genotype (GG), with a response percentage of (37.5% vs. 83%, respectively). In conclusion, our findings showed that the CYP2B6*9 (G516T) polymorphism is associated with B-CLL susceptibility among Egyptian patients. This variant greatly affected the clinical outcome and can serve as a good therapeutic marker in predicting response to cyclophosphamide treatment.Graphic Abstract
Keywords
Abbreviations
| λ-light chain | Lambda-light chain |
| κ-light chain | Kappa-light chain |
| ALL | Acute lymphoblastic leukemia |
| AML | Acute myeloblastic leukemia |
| ALT | Alanine transaminase |
| ASA | Allele specific amplification |
| AST | Aspartate transaminase |
| B-CLL | B-chronic lymphocytic leukemia |
| BCR | B-cell receptor |
| CBC | Complete blood count |
| CPA | Cyclophosphamide |
| CVP | Cyclophosphamide/Vincristine/Prednisone |
| CYP2B6 | Cytochrome P450, family 2, subfamily B, member 6 |
| CYPs | Cytochromes P450 superfamily |
| EDTA | Ethylene diamine tetra acetic acid |
| FC | Fludarabine/Cyclophosphamide |
| FISH | Fluorescence in situ hybridization |
| Hb | Hemoglobin |
| IGHV-M | Mutated immunoglobulin heavy chain variable |
| IGHV-UM | Unmutated immunoglobulin heavy chain variable |
| IPT | Immunophenotyping |
| IW-CLL | International Workshop on CLL |
| LDH | Lactate dehydrogenase |
| NCCN | National Comprehensive Cancer Network |
| NF-κB | Nuclear factor kappa B |
| OCMU | Oncology Center Mansoura University |
| PCR | Polymerase chain reaction |
| sIg | Surface immunoglobulin |
| SNP | Single nucleotide polymorphism |
| TBE | Tris-borate-EDTA |
| WBCs | White blood cells |
| β2M | Beta-2 microglobulin |
B-chronic lymphocytic leukemia (B-CLL), the most widespread lymphoproliferative disorder affecting the elderly, is characterized by an aggressive expansion of monoclonal lymphocytes in the peripheral blood, bone marrow and lymphoid organs [1]. The global incident number of B-CLL was 1,034.67 ×102 in 2019. The incidence varies by race and geographic location, which is higher among Caucasians while extremely low in Asian populations [2]. Even though the exact cause of B-CLL is unidentified, a combination of hereditary and environmental factors may contribute to the pathogenic pathways that lead to the development of the disease. For patients with B-CLL, chemotherapy remains the primary treatment option. The disease has a clinical trajectory marked by repeated recurrences and medication resistance, even if the majority of cases respond to first-line therapy [3].
Individuals are constantly exposed to high concentrations of environmental pollutants, drugs and toxins known as xenobiotics. If exposure to these xenobiotics continues, it may lead to genotoxicity due to disruptions in DNA integrity. On the other hand, one’s ability to biotransform harmful xenobiotics into harmless ones can be seen as a crucial defensive mechanism [4]. Cytochromes P450 (CYPs) are a superfamily of heam-containing enzymes catalyzing phase I biotransformation that metabolize different xenobiotics and endogenous compounds [5]. The expression of CYPs genes is affected by a unique combination of genetic and non-genetic factors that are responsible for the inter-individual variability in the enzyme’s catalytic activities; these factors include genetic polymorphisms, age, sex, hormones and disease state. Fifty-seven human cytochrome P450 enzymes have been identified and are classified into 18 families [6,7].
Cytochrome P450 2B6 (CYP2B6) is a crucial enzyme for the hepatic metabolism of fatty acids, steroids, and chemical detoxification. Consequently, endo- and xenobiotic metabolism may be affected by CYP2B6 upregulation or inhibition [8]. Numerous commercially available medications, including ß-blockers, selective serotonin reuptake inhibitors, antivirals, antidepressants, analgesics, and anticancer drugs like cyclophosphamide, are also metabolized by CYP2B6 [9].
An oxazaphosphorine prodrug called cyclophosphamide is widely used to treat CLL. A primary catalyst in the cyclophosphamide bioactivation pathways is CYP2B6. The varied pharmacokinetics of cyclophosphamide and its toxic byproducts, which can result in an inadequate response to therapy and the development of side effects that are clinically relevant, may be attributed to CYP-mediated metabolism [10].
The CYP2B6 gene is a highly polymorphic gene that exhibits ~300-fold variability in expression, with many single nucleotide polymorphisms (SNPs) that have distinct ethnic frequencies and are linked to changes in enzyme activity and gene expression [11]. The liver is the primary site of CYP2B6 expression; extrahepatic tissues such as the lungs, kidney, digestive system, and brain express it to a lower degree. CYP2B6 constitutes ~6%–10% of the whole hepatic CYPs content and contributes to the metabolism of 10%–12% of commercially available drugs [9].
A commonly studied SNP, CYP2B6*9 (G516T, rs3745274), is characterized by the presence of a G to T transversion in exon 4 and is associated with decreased gene expression and reduced enzyme activity compared to the CYP2B6*1 wild-type allele [12]. The CYP2B6*9 variant was reported to be widely occurring, from ∼25% of whites to 60% of the Asian population, and has been revealed to affect pre-mRNA splicing, resulting in a diminished level of the normal mRNA transcript and a 50%–75% reduction in CYP2B6 protein level [13]. This SNP may occur in combination with K262R in CYP2B6*6 or with other alleles (CYP2B6*7, *13, *19, *20, *26, *34, *36, *37 and *38). It can occur by itself in CYP2B6*9 as well [14].
CYP2B6*9 was previously reported to be associated with cancer risk. Justenhoven et al. concluded that the genetic variants CYP2B6*9, which are known to decrease activity of the CYP2B6 enzyme, contribute to a higher risk of breast cancer [15]. This SNP was also found to be associated with hematological malignancies as well. Yuan et al. stated that the T-allele of the CYP2B6*9 SNP was associated with the risk of ALL and AML. They suggested that the inherited impaired function of the CYP2B6 detoxification pathway may be an essential genetic determinant of leukemia risk [16]. Based on the fact that CYP2B6*9 enzyme has an important role in xenobiotic biotransformation and cyclophosphamide metabolism, we aimed to study the inter-individual variation of CYP2B6 isoform among Egyptian B-CLL patients by detecting the presence of the CYP2B6*9 SNP and its possible association with the incidence and clinical outcome of the disease following cyclophosphamide chemotherapy.
Through the clinical presentation, bone marrow aspiration and immunophenotyping, the B-CLL diagnosis was performed. The current work is a case-control study with follow-up conducted on one hundred patients diagnosed with B-cell chronic lymphocytic leukemia at the Oncology Center Mansoura University (OCMU) in accordance with the National Comprehensive Cancer Network (NCCN) guidelines for B-cell chronic lymphocytic leukemia [17]. A control group of one hundred healthy volunteers was used. This research was carried out between March 2021 and February 2023 with consent from the Mansoura University Faculty of Medicine’s Ethics Committee (Code No. MD.21.03.62). All participants provided written informed consent, which was collected. Only patients with typical B-CLL were included. The exclusion criteria involved any person in the B-CLL and control groups who had a family history of cancer, atypical B-CLL patients or other causes of lymphoproliferative disease, typical B-CLL patients with co-morbidities such as renal insufficiency, hepatitis and heart diseases, or those with autoimmune anemia or thrombocytopenia, and B-CLL patients with a Tp53 deletion or mutation.
From the 100 eligible B-CLL patients, two patients died early and four patients with the 17p13 deletion were neglected. The other 94 allocated patients were classified into two groups according to the type of treatment received: [62 were treated with the Fludarabine/Cyclophosphamide (FC) regimen and 32 were treated with the Cyclophosphamide/Vincristine/Prednisone (CVP) regimen]. Patients treated with the FC regimen received it as follows: fludarabine (30 mg/m2) and cyclophosphamide (250 mg/m2), which were given i.v. on days 1–3 every 28 days, for a maximum of six courses. Patients treated with the CVP regimen received it as follows: cyclophosphamide (750 mg/m2) and vincristine (1.4 mg/m2 on day 1) given i.v. and oral prednisone (100 mg on days 1–5). This was repeated every 3 weeks, and this was continued for eight cycles. After three months since the last chemotherapy cycle, the bone marrow examination, radiological scan and blood tests were repeated. The treatment outcome of all the patients was determined according to the response criteria defined by the International Workshop on Chronic Lymphocytic Leukemia (IW-CLL), depending on the tumor load and recovery of bone marrow functions after chemotherapy [18] (Suppl. Fig. S1).
Each participant in the B-CLL and control groups had six milliliters of blood drawn from them. Each blood sample was separated into two aliquots: 3 mL were placed in sterilized tubes containing EDTA for the hematological, genetic and flow cytometric investigations, whereas the other portion of the blood was left to clot, centrifuged at 4000 rounds per minute (rpm) for 10 min, and then serum was separated and stored at −20°C for the further biochemical measurements. Additionally, all B-CLL patients had their bone marrow aspirated, and the marrow smears were ready for the morphological analysis. For the cytogenetic study, two milliliters of the aspirate were put in heparin tubes.
From all participants, the genomic DNA was isolated from whole blood samples using a commercial extraction kit (QIAamp® DNA Extraction Kit, Cat. No. #51106, QIAGEN, Hilden, Germany), according to the manufacturer’s directives. After extraction, the DNA purity and concentration were determined by analyzing the A260/A 280 and A260/A230 ratios using (NanoDrop™ 1000 Spectrophotometer, NanoDrop Tech., Inc., Wilmington, USA). Ten μL of DNA were added to 990 μL nuclease-free water in a quartz cuvette, and a reading was taken. The absorbances of proteins and nucleic acids are 260 and 280 nm, respectively. The purity of nucleic acid extraction has been measured using the ratio of absorbance at these wavelengths. “Pure” DNA is commonly defined as having a ratio of about 1.8. A high 260/280 ratio typically suggests that there is protein contamination in the sample. The predicted value of the 260/230 ratio, which is a secondary indicator of nucleic acid purity, is typically between 2.0 and 2.2. If the ratio is noticeably lower than anticipated, this indicates the presence of contaminants that absorb at 230 nm. After that, the DNA concentration was then computed by the formula:
Amplification of CYP2B6*9 (G516T) variant
The genotyping and amplification of the CYP2B6*9 (G516T) variant were analyzed using allele specific amplification-PCR (ASA), previously described by Müller et al. The primers used were: (F1-wild-type: 5′-GACCCCACCTTCCTCTTCTAG-3′), (F2-variant: 5′-GACCCCACCTTCCTCTTCTAT-3′) and (R-common: 5′-GGTCATCCTTTTCTCGTGTG-3′). β-actin was used as a control gene, and the presence of the β-actin band was an indicator of successful PCR amplification (β-actin F: 5′-ATGTCCCCCGTCTGGCCTGG-3′) and (β-actin R: 5′-CTGTAGCCGCGCTCGGTGAG-3′) [19,20]. The PCR was applied using (SimpliAmp™ Thermal Cycler, ThermoFisher Scientific, Waltham, USA). Each PCR reaction was performed in two tubes, with a total volume of 25 μL for both. The first one contained 4 µL DNA extract (~100 ng/µL), 12.5 µL master mix (COSMO RED, Cat. No. #ND-1289-5O, Willowfort, Birmingham, UK), and 1 µL of each [F1, R, β-actin (F and R)] primer (10 pmol/µL). The second one contained 4 µL DNA extract, 12.5 µL master mix and 1 µL of each [F2, R, β-actin (F and R)] primer. The final volume in both reactions was reached with molecular-grade water. Every set of samples was run along with a negative control (containing all reaction components except for the genomic DNA).
The thermal cycler was used according to the cycling program consisting of initial denaturation (94°C for 5 min), 35 cycles of [denaturation (94°C for 30 s), annealing (53°C for 30 s), extension (72°C for 1 min)] and a final extension (72°C for 7 min). Every set of samples was run along with a negative control. Finally, the PCR products of CYP2B6*9 amplification (223 bp) and β-actin (321 bp) amplification were analyzed directly by agarose gel electrophoresis using 2.5% agarose gel and 1X Tris-borate-EDTA (TBE) buffer electrophoresis at 200 V for 30 min, stained with ethidium bromide (500 mg/L), and visualized under an ultraviolet trans-illuminator. The wild-type genotype (GG) appeared as one band at 223 bp in the 1st PCR reaction, the variant genotype (TT) appeared as one band at 223 bp in the 2nd PCR reaction and the heterozygous genotype (GT) appeared as two bands in both reactions at 223 bp.
Immunophenotyping and cytogenetics
Immunophenotyping (IPT) was carried out using a flow cytometer (BD FacsCanto II TM Clinical Flow Cytometer System, BD Bioscience, New Jersey, USA). The chromosomal anomalies were identified by fluorescence in-situ hybridization (FISH). During the FISH cytogenetics analysis, dividing cells must have their chromosomes stopped at the metaphase stage using a spindle inhibitor, such as colchicine. The cells must then be hypotonically treated and fixed with Carnoy’s fixative (1:3 ratio of acetic acid to methanol) for the optimal period. After that, the methanol/acetic acid cells were placed onto glass slides that had been cleaned and given at least seven days to mature. Further, RNAse (100 µg/mL) was added to 2X standard saline citrate and incubated for an hour at 37°C. The slides were dehydrated using a succession of ethanol concentrations of 70%, 90%, and 99%.
Fluorescent-labeled DNA probes that hybridize to particular chromosomal areas were used to detect chromosomal abnormalities. A hybridization mixture containing ten microliters was overlaid on each slide, covered with a coverslip, and sealed with a rubber solution. The hybridized probe was detected with a fluorescein isothiocyanate (FITC)-conjugated avidin (Vector), and the fluorescence intensity was reinforced with a sandwich amplification with a biotinylated anti-avidin monoclonal antibody. The cells were counterstained with propidium iodide (2 µg/mL). The slides were evaluated under a fluorescence microscope (Olympus BX60 F5, Olympus, Tokyo, Japan). At least 200 nuclei were analyzed, and cells from three healthy donors with a normal karyotype were used as controls. Direct sequencing was used after the amplification of coding exons 4–10 and flanking intronic regions to investigate the mutational status of TP53 [21].
STATA software version 17 was applied to calculate the sample size and the findings of Botros et al. when they examined the influence of CYPs polymorphisms on Egyptian patients with AML, as this is the first study to test CYPs polymorphisms in CLL patients from Egypt [22]. Analysis for a matched case-control study revealed that the sample size would be 47 participants per group (ratio 1:1), using a power of (80%) and an alpha error of (5%), and the chance of exposure in the control group would be 28%. To boost the study’s power, we used 100 cases and 100 controls in the current investigation. The Kolmogorov-Smirnov test was used to check for normality. The mean ± standard deviation (SD) was used to describe normally distributed data, the median (minimum and maximum) was used to describe non-normally distributed data, and percentages and numbers were used to represent qualitative data. The non-parametric data was processed using the Mann-Whitney and Kruskal-Wallis methods, while the parametric data was treated by the student t-test. Chi-Square was used to compare qualitative data between groups. The significance level was adjusted to (p < 0.05). The chi-squared (χ2) goodness-of-fit test was used to determine the SNP in both groups using the Hardy-Weinberg equilibrium equation. 95% confidence intervals (CI) for the odds ratio (OR) were obtained using binary logistic regression.
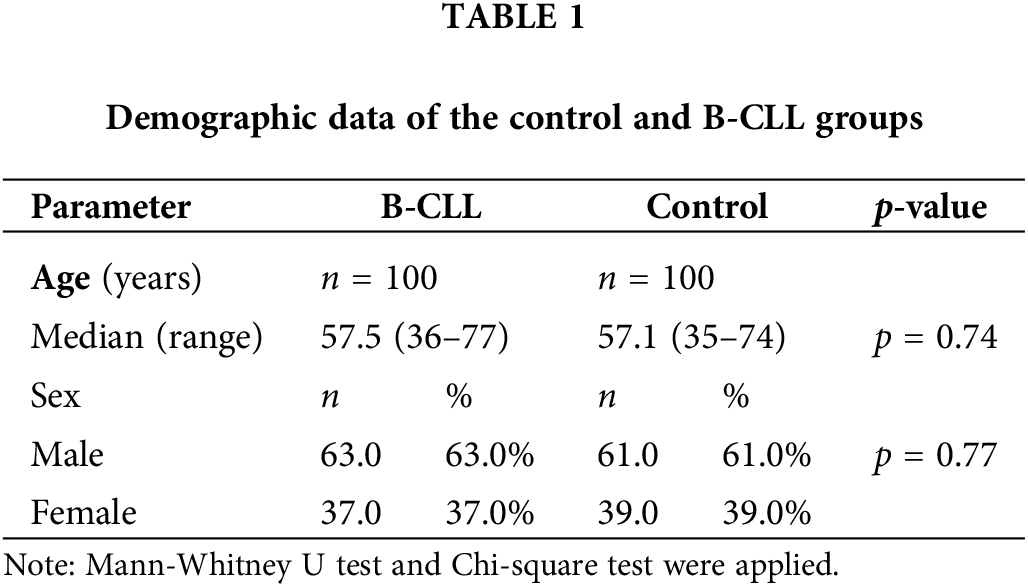
Demographic data of the control and B-CLL groups
The current work included 100 Egyptian B-CLL patients: 63% males and 37% females with a median age of 57.5 years, ranging from 36 to 77 years, and 100 age- and sex-matched healthy individuals (Suppl. Fig. S2). Regarding age and gender, we did not discover any appreciable differences between the two groups, as presented in Table 1.
Clinical features and lab measurements of B-CLL patients
The IPT determined by the flow cytometric analysis of patients’ samples showed that (97%, 100%, 100%, 95%, 7%, 10%, 6%, 35%, 42%, 6%, 43% and 57%) of the patients expressed CD5, CD19, CD20, CD23, CD10, CD22, FMC7, CD38, CD49d, sIg, ƛ-light chain and ƛ-light chain, respectively. Cytogenetic analysis by FISH was determined successfully for all the patients and showed that 21%, 18%, 4% and 1% of patients presented with 13q14, 11q23 and 17p13 deletions and trisomy 12, respectively.
The physical examination and radiological scan showed that 100% of the cases presented with enlarged lymph nodes, 97% had splenomegaly and 86% had hepatomegaly. In addition, 12%, 16% and 17% of the patients presented with night sweats, weight loss and fever, respectively. The bone marrow aspiration revealed that 95% of patients presented with hypercellular marrow, and 5% of patients presented with normocellular marrow with mature lymphocyte infiltration. In addition, results of IGHV mutational status revealed that one patient carried an unmutated IGHV gene only.
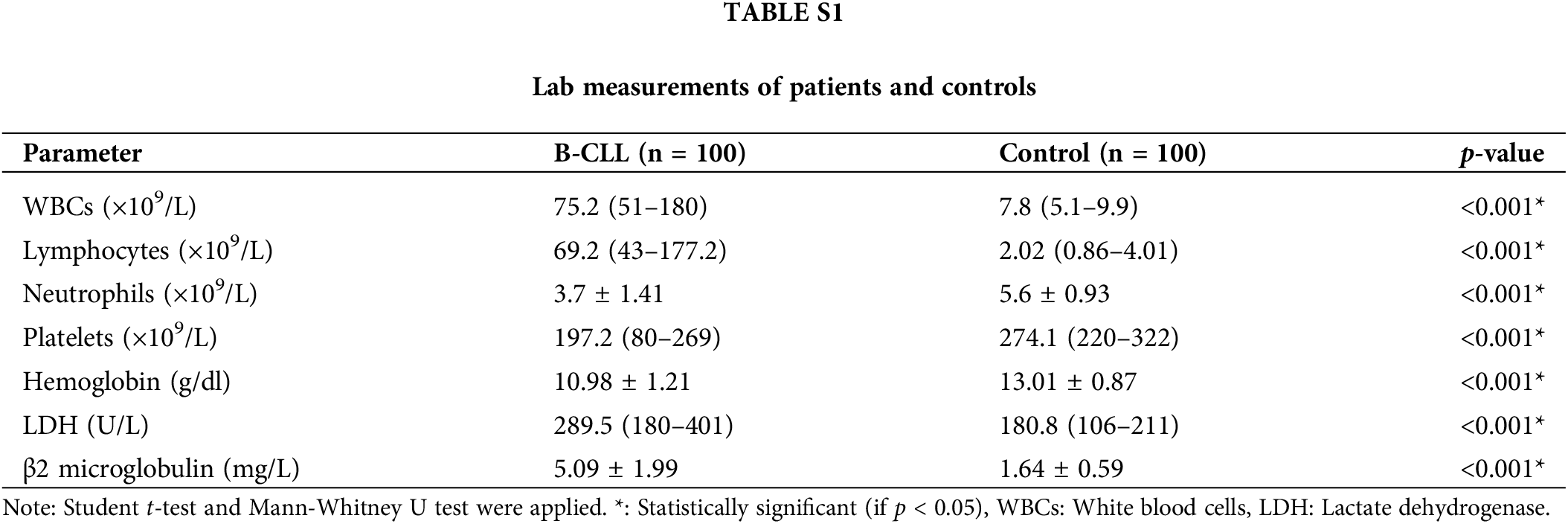
Suppl. Table S1 demonstrated that the B-CLL group had significantly higher WBCs and absolute lymphocyte counts than the control (both were p < 0.001), but significantly lower absolute neutrophil counts, hemoglobin levels, and platelet counts (p < 0.001 for each). Furthermore, there was a significant rise in both the concentration of LDH and β2 microglobulin in the B-CLL group as compared to the control (both were p < 0.001).
Depending on whether a patient has lymphocytosis, lymphadenopathy, splenomegaly, hepatomegaly, thrombocytopenia, or anemia, the disease staging was determined using the Rai and Binet staging systems (Rai et al. 1975 [23]; Binet et al. 1981 [24]). We found that 59% and 41% of patients were classified as Binet B and C, respectively, based on the Binet staging, and 51%, 22%, and 27% of cases were classified as Rai II, III, and IV, based on the Rai staging. The International Prognostic Index-Chronic Lymphocytic Leukemia (IPI-CLL) was used to assess risk stratification according to a number of prognostic markers, including age, B-CLL stage, IGHV status, 17p13 deletion, and β2 microglobulin level (Eichhorst et al. 2021). We found that (10%, 79%, and 11%) of the cases were respectively stratified as low, intermediate, and high risk, respectively. The applied Rai and Binet staging systems depend on whether a patient has or does not have lymphocytosis, lymphadenopathy, splenomegaly, hepatomegaly, thrombocytopenia and anemia [23,24]. According to the Rai staging, 51%, 22% and 27% of cases were classified as Rai II, III, and IV, respectively, and according to the Binet staging, 59% and 41% of cases were classified as Binet B and C, respectively. The International Prognostic Index-Chronic Lymphocytic Leukemia (IPI-CLL) was used to assess risk stratification according to a number of prognostic markers, including age, B-CLL stage, IGHV status, 17p13 deletion, and β2 microglobulin level [25]. We discovered that the patients were categorized as low-, intermediate-, and high-risk groups in 10%, 79%, and 11% of the cases, respectively.
Association of the CYP2B6*9 with the B-CLL risk
The CYP2B6 gene polymorphism (*9, G516T) was investigated using ASA-PCR, and three genotypes: wild-type (GG), heterozygous carrier (GT), and homozygous variation (TT) were found. After being separated on an agarose gel, the PCR result was displayed in (Fig. 1).
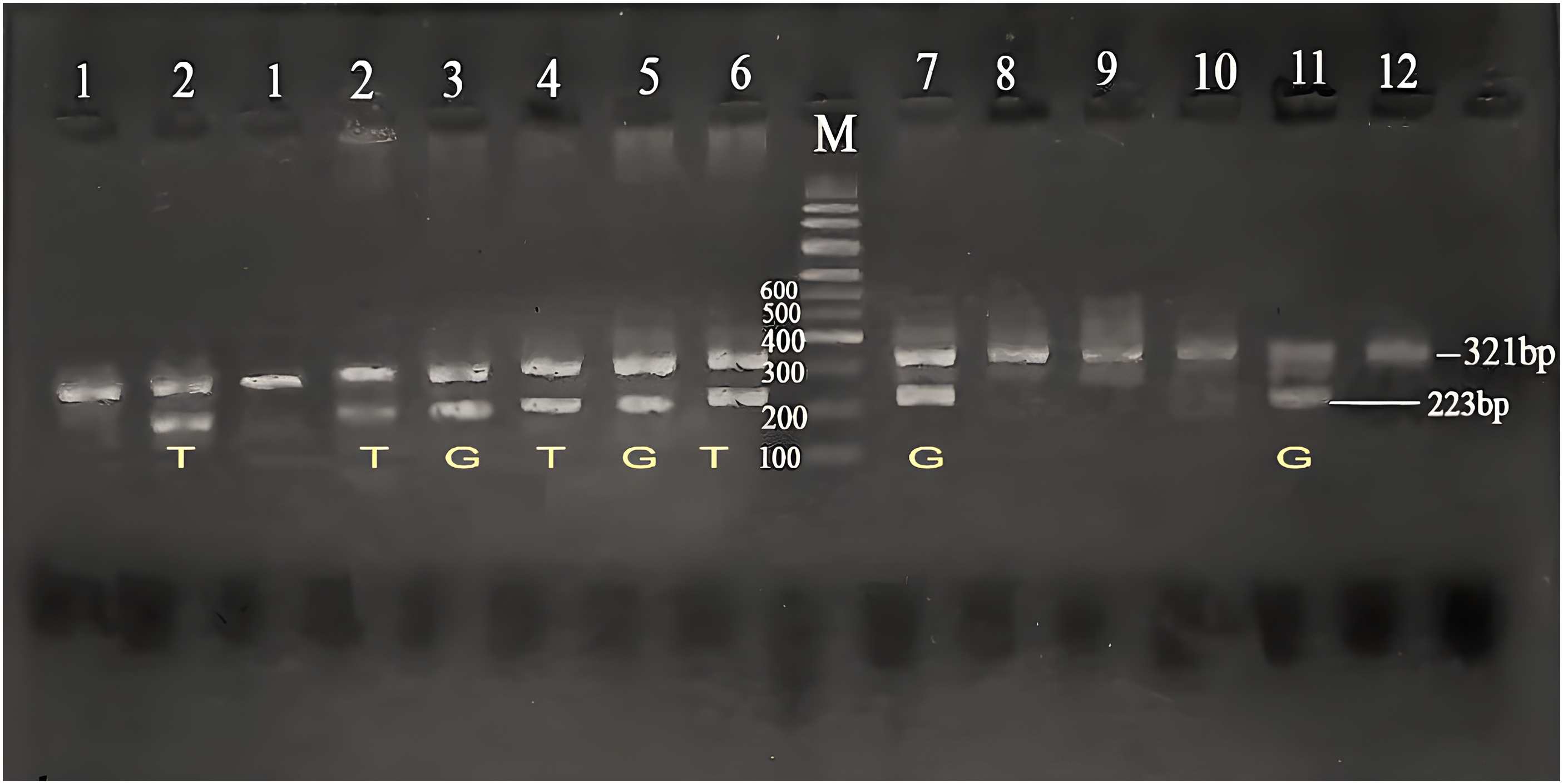
Figure 1: Agarose gel electrophoresis for CYP2B6 (G516T) polymorphism in the studied groups. Lane (M) displayed the DNA marker; lanes 7,8,11,12 displayed (GG genotype at 223 bp); lanes 3,4,5,6 showed (GT genotype at 223 bp); lanes 1,2 displayed (TT genotype at 223 bp). The β-actin band was at 321 bp.
The minor allele frequency of the T-allele was found to be 36% in patients and 6% in healthy controls. Furthermore, the common G-allele is shared by 64% of patients and 94% of healthy people. The frequency of the most common genotype (GG) was 41% in patients and 79% in healthy controls (Fig. S3). The genotypes of CYP2B6*9 in both the control and B-CLL groups were found to be in HWE by the chi-square test, meaning that there were no significant differences between the observed and predicted values (p = 0.14 and p = 0.06, respectively).
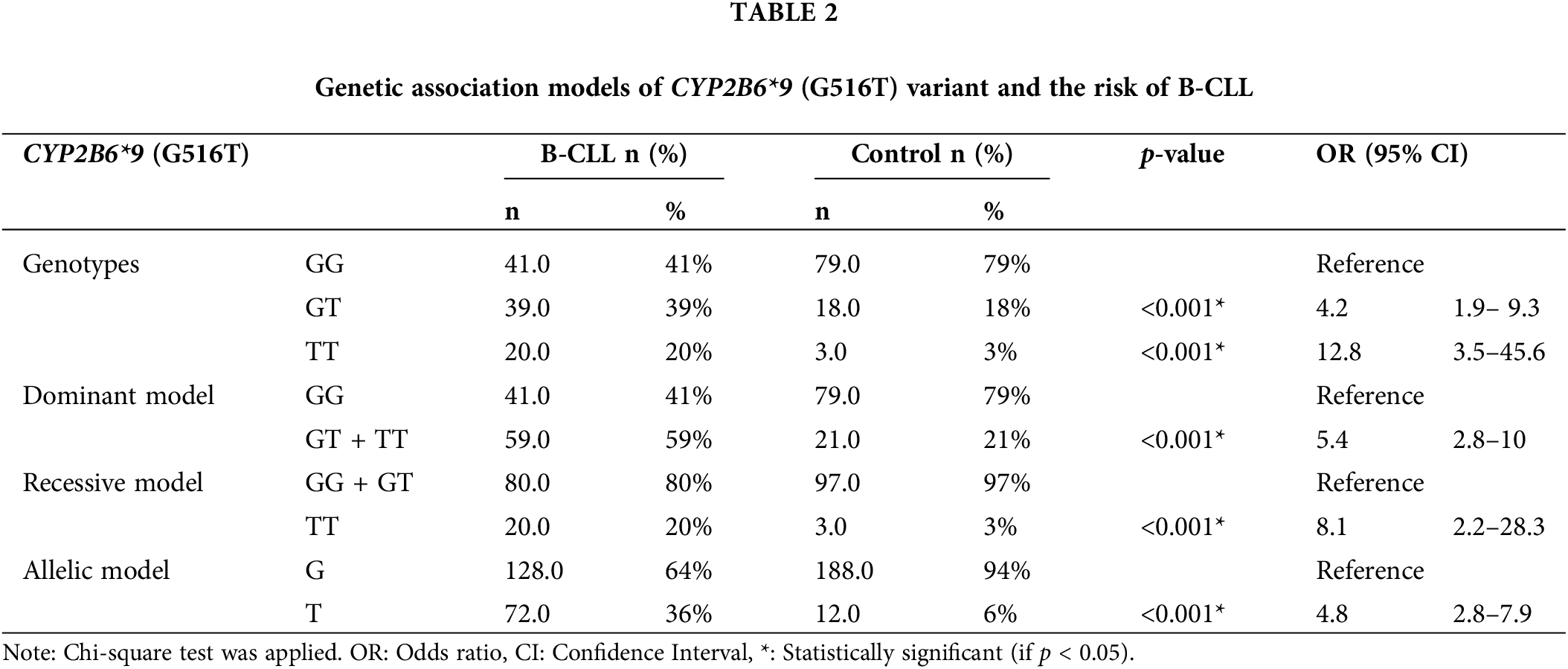
Combining various genetic association models (co-dominant, dominant, recessive, and allelic), B-CLL patients showed a strong link with the CYP2B6*9 (G516T) polymorphism. The co-dominant model (heterozygous comparison GT vs. GG, OR = 4.2, p < 0.001) and the homozygous comparison TT vs. GG, OR = 12.8, p < 0.001), the dominant model (GT + TT vs. GG, OR = 5.4, p < 0.001), the recessive model (TT vs. GG + GT, OR = 8.1, p < 0.001), and the allelic model (T-allele vs. G-allele, OR = 4.8, p < 0.001), as displayed in (Table 2).
Association of the CYP2B6*9 with the characteristics of B-CLL patients
The wild-type (GG), heterozygous (GT), and variant (TT) genotypes for the CYP2B6 (G516T) variant were evaluated to see if there were any differences in lab measurements and clinical characteristics, as indicated in (Table 3). The B-CLL patients in the (GT) and (TT) groups had a considerably lower median age than the (GG) group (p = 0.003 and p = 0.001, respectively). Gender-wise, there was a statistically significant difference in the proportion of male patients between the (GT) and (GG) groups (p = 0.009).
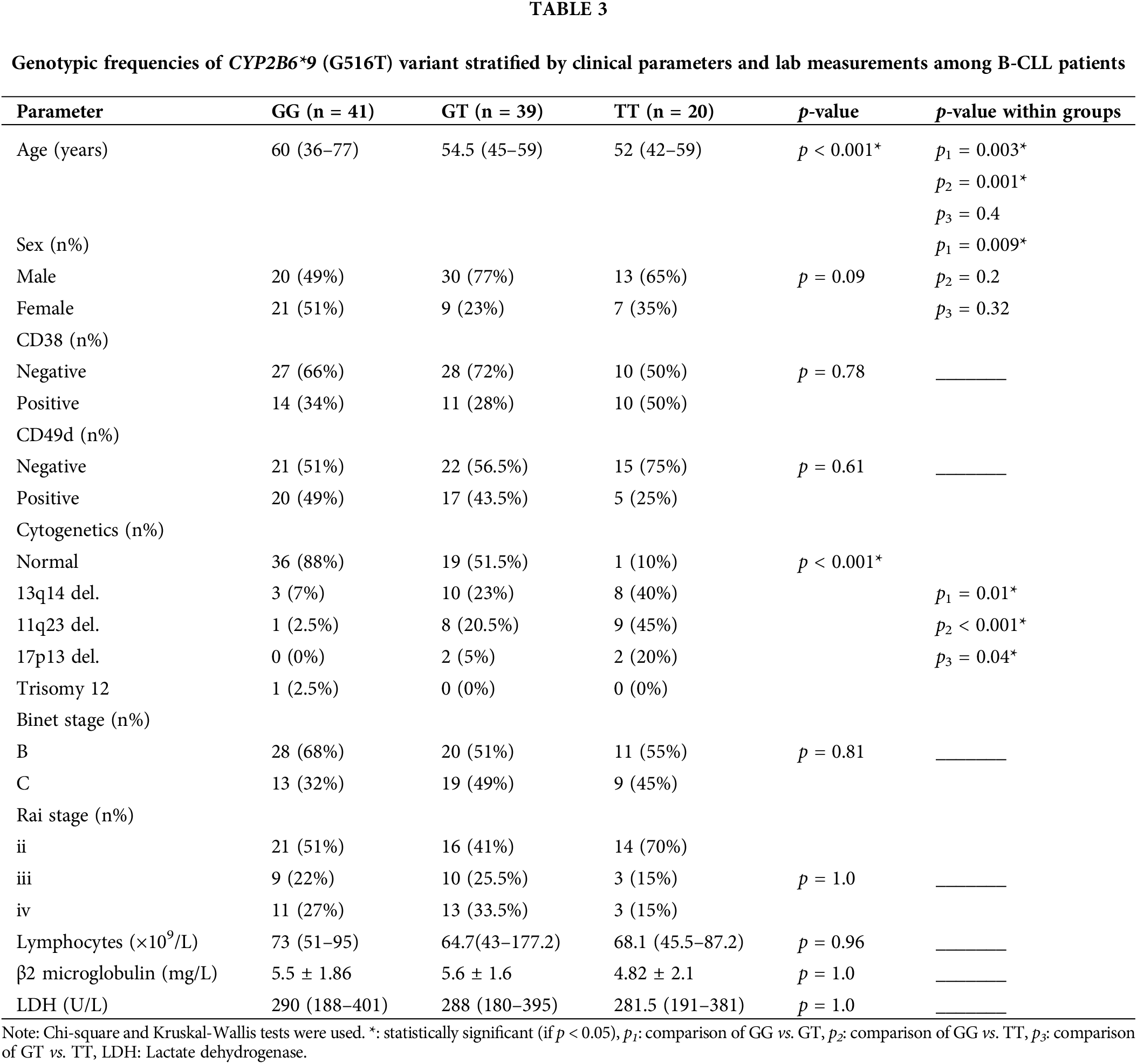
Additionally, the percentages of patients with chromosomal anomalies were greater in the (GT) and (TT) groups compared to the (GG) group (p = 0.01 and p < 0.001, respectively) and higher in the (TT) group than the (GT) group (p = 0.04). The CYP2B6*9 SNP did not significantly correlate with the illness stage, CD38 and CD49d expressions, absolute lymphocyte count, β2M, or LDH levels.
Association of the CYP2B6*9 on the clinical outcome
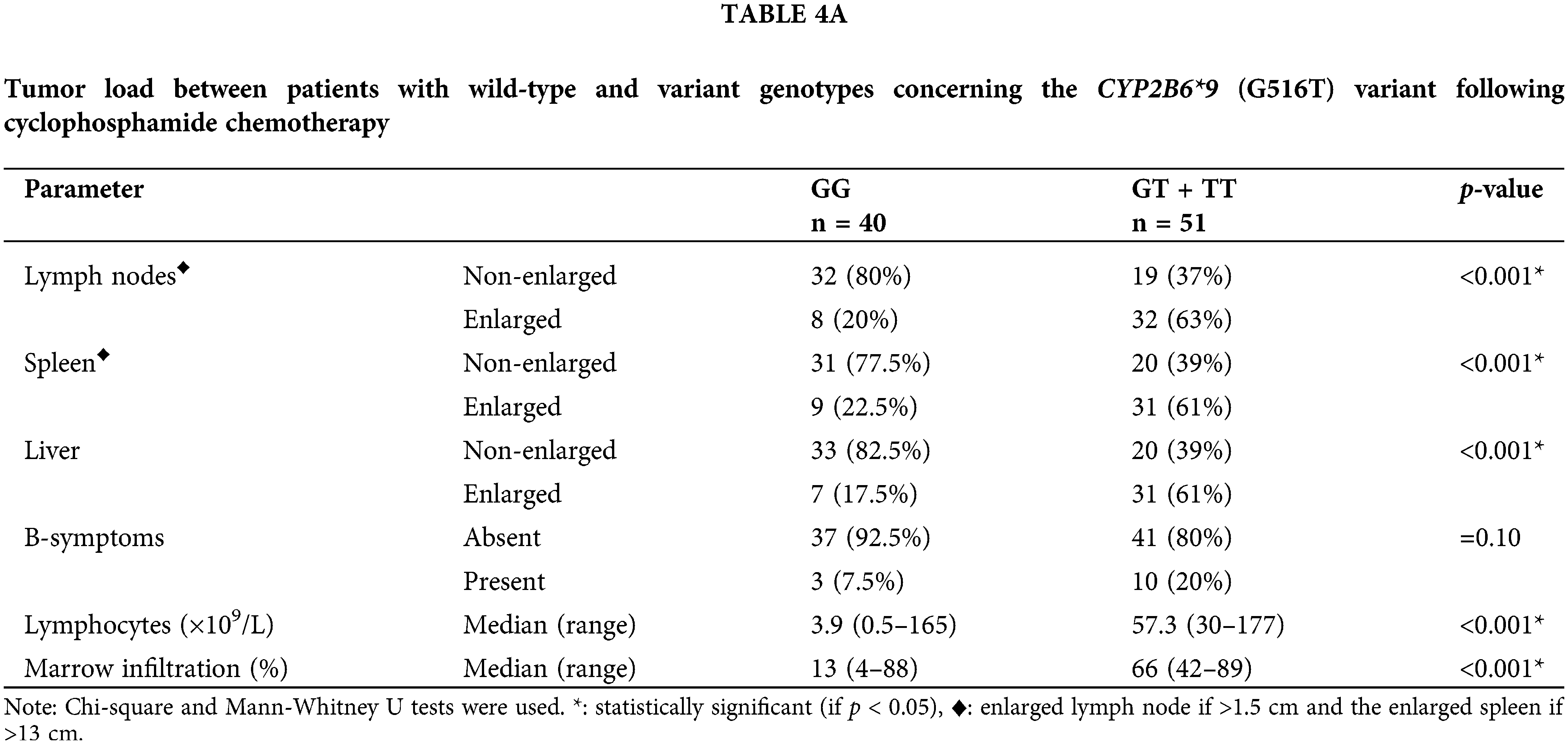
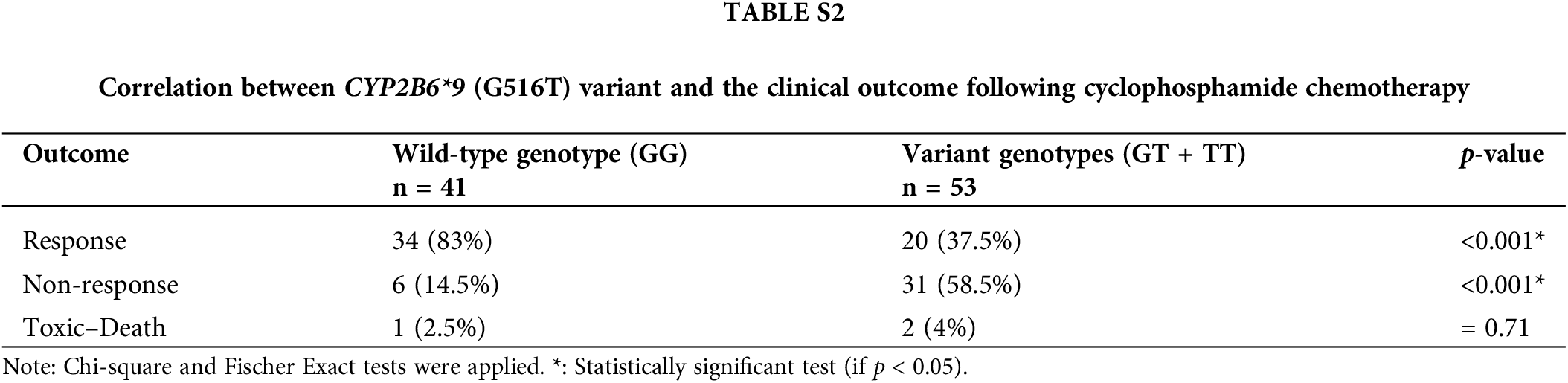
We analyzed the genotypes of the wild-type (GG) and variant (GT + TT) to see if the CYP2B6 (G516T) polymorphism had an impact on the clinical outcome after cyclophosphamide treatment. IW-CLL was used to evaluate the clinical outcome based on the tumor load and bone marrow function recovery. In contrast to patients with the GG genotype, who were classified as 83% responders, 14.5% non-responders, and 2.5%) no responders, patients carrying variant genotypes (GT + TT) were classified as (37.5% responders, 58.5% non-responders, and 4% toxic death). Based on the data in Table S2, we found that there was a significant (p < 0.001) association between the variant genotypes and response failure.
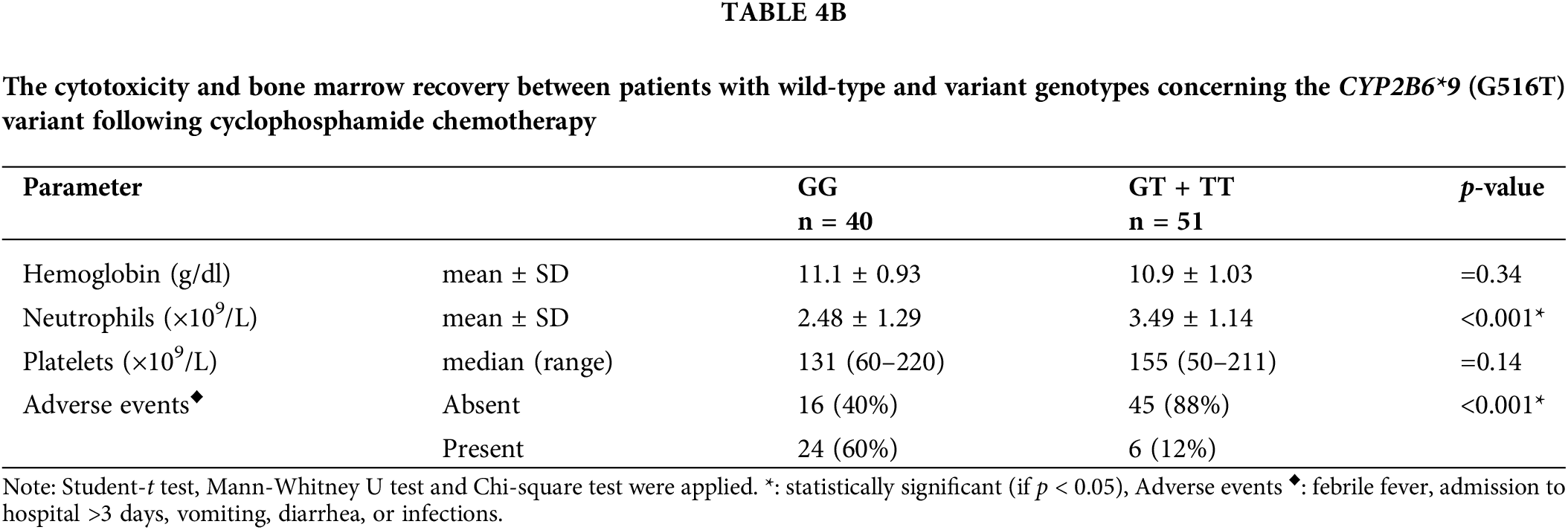
Results in Table 4A demonstrated that, with respect to the tumor load, the (GT + TT) group exhibited significantly higher mean percentages of lymphocytes infiltrating the bone marrow, absolute lymphocyte counts, and percentages of patients presenting with enlarged lymph nodes, liver, and spleen in comparison with the (GG) group (p < 0.001 for each). The cytotoxicity and bone marrow recovery results in Table 4B showed that the (GG) group’s absolute neutrophil count was significantly less than the (GT + TT) group’s. Furthermore, it was shown that patients in the (GG) group had more adverse events (p < 0.001) following chemotherapy than patients in the (GT + TT) group. These adverse events included fever episodes, hospitalization for longer than three days, infections, vomiting, and diarrhea.
B-chronic lymphocytic leukemia (B-CLL) is a chronic lymphoproliferative disorder characterized by monoclonal B-cell proliferation [26,27]. B-CLL can manifest as an aggressive, fatal form of the disease or as an indolent variant that does not need to be treated for many years. Although the currently existing clinical staging techniques for B-CLL are simple and inexpensive, they are not accurate enough to forecast the course of the disease and a patient’s prognosis [28].
Chemotherapy responses across and within B-CLL patients vary widely, and this variability is unknown for any given patient. It has been proposed that variations in chemotherapy metabolism may be involved in controlling responses over time, both between and within patients [25]. Genetic polymorphisms in important drug-metabolizing enzymes such as cytochromes p450 (CYPs) have been emphasized as one of the main causes of variability in drug responses. As the liver is the main site of drugs’ metabolism, changes in the activities or expressions of drug metabolizing enzymes will have the potential to alter hepatic clearance and drug bioavailability [29].
This study aimed to investigate the potential correlation between the CYP2B6*9 (G516T, rs3745274) SNP and B-CLL susceptibility, as well as the effect of this connection on the clinical outcome after cyclophosphamide treatment among Egyptians. Our population is a perfect genetic model for conducting this kind of research because of its relative genetic homogeneity without inter-ethnic variations. The process through which populations of organisms alter over many generations is known as evolution. Genetic variations underlie these changes. Genetic variations can result from normal processes such as genetic recombination, which is the rearranging of genetic material as a cell prepares to divide, or from gene variants, often known as mutations. Different traits can be introduced into an organism by genetic variations that change the function of proteins or gene activity. Natural selection refers to the process wherein a genetic variant is more likely to be passed down to the following generation if the characteristic is favorable and helps the individual survive and reproduce. As generations of individuals with the trait continue to reproduce, the advantageous trait becomes more prevalent in a population over time, making the population different from an ancestral one. We particularly chose to investigate the CYP2B6*9 variant allele because of its high frequency and its recognized effect on the decrease in CYP2B6 expression and function.
The B-CLL diagnosis was performed based on the diagnostic criteria of NCCN guidelines for B-chronic lymphocytic leukemia and the World Health Organization (WHO) classification. A typical immunophenotyping of monoclonal B-cells with a B-CLL score (4–5), <55% prolymphocytes-like cells in the peripheral blood smear and >5 × 109/L absolute mature lymphocytes were the main characteristics of the patients [30,17]. B-CLL cells highly express CD5, CD19, and CD23 in addition to a single light chain expression (κ or λ), supporting the monoclonality of B-cells. Additional markers may also have been identified, such as CD10 and CD200, which help in the differential identification of suspicious cases, and CD38 and CD49d, which have prognostic value in B-CLL [31]. Using flow cytometry, we discovered that the expression of (CD5, CD19, CD20, CD23, CD10, CD22, FMC7, CD38, CD49d, sIg, ƛ-light chain and ƙ-light chain) was present in (97%, 100%, 100%, 95%, 7%, 10%, 6%, 35%, 42%, 6%, 43% and 57%, respectively). Expression of CD38 and CD49d is thought to be a poor prognostic factor as they help leukemia cells migrate and homing to secondary lymphoid organs and stimulate B-CLL cell proliferation in tandem with BCR signaling [32].
Cytogenetic analysis is useful for identifying certain chromosomal abnormalities that affect the disease’s prognosis and could affect the choice of treatment [33]. We observed that 1%, 18%, 4% and 1% of patients, respectively, had 13q14 del, 11q23 del, 17p13 del and trisomy. Only one patient had unmutated (IGHV-UM). B-cells with IGHV-UM are generally thought to be poor prognostic markers since they are highly susceptible to antigenic stimulation and are correlated with a lower survival time [34]. Leukemic B-cells are thought to constitutively secrete β2 microglobulin (β2M), and the tumor burden is correlated with the serum levels of β2M. A serum level of β2M is used in determining the risk rank of B-CLL patients, and β2M >3.5 mg/L is also considered a bad prognostic marker [25].
Leukemic cells invade the lymphoid organs and bone marrow as the disease worsens. Before and after starting cytotoxic agent treatment, bone marrow examination gives oncologists the information they need to determine if cytopenias are caused by leukemic infiltration or treatment-related myelo-suppression. The lab results showed that the patients had considerably greater WBCs and absolute lymphocyte counts than the controls (p < 0.001 for both). Absolute neutrophil and platelet counts in the patients were substantially lower than in the controls (both with p < 0.001). Likewise, the patients’ Hb level was considerably lower than that of the control group (p < 0.001). This may be explained by the fact that megakaryocytes, myeloid precursors, and erythroid precursors are all reduced concurrently with lymphocytic infiltration of the bone marrow [18]. Serum LDH levels, which indicate the disease’s proliferative load, were higher in patients than in controls (p < 0.001) which could be explained by the related tumor lysis syndrome, which is brought on by the disintegration of leukemic cells [35].
Analysis of CYP2B6*9 polymorphism showed that the patients displayed greater percentages of (GT) and (TT) genotypes than controls (p < 0.001 for both), carrying a 4- and 13-fold risk of developing B-CLL [(OR = 4.2, 95% CI 1.9–9.3) and (OR = 12.8, 95% CI 3.5–45.6), respectively]. Additionally, patients had a variant T-allele frequency that was substantially greater than that of controls (p < 0.001), with an estimated 5-fold increased risk of B-CLL (OR = 4.8, 95% CI 2.8–7.9).
Several studies provide evidence for the pathogenic role of the CYP2B6 (G516T) polymorphism in the susceptibility to hematological malignancies. Yuan et al. have reported that the T-allele could be one of the risk factors for ALL and AML predispositions. They found that GT and GT + TT genotype frequencies were greater in ALL (37.5% and 42.7%, respectively, p < 0.01), and AML (37.2% and 40.9%, respectively, p < 0.01) than in control (23.9% and 25.9%, respectively), suggesting that the inherited defective function of the CYP2B6 detoxification pathway may be an important genetic determinant of leukemia risk [16]. Daraki et al. also informed us that the T-allele was related to AML; higher frequencies of the variant genotypes (GT and TT) were found in cases compared to controls (GT: 38.8% vs. 29.8% and TT: 9.3% vs. 5.3%, respectively, p = 0.001) [36]. Berköz et al. informed us that the risk of acute leukemia development was 2.48-fold for GT genotype carriers and 1.92-fold for T-allele carriers [37]. In Egypt, Botros et al. reported that the CYP2B6 gene polymorphism carries a 3-fold risk of AML [22]. No study has examined the impact of the CYP2B6 (G516T) polymorphism on the incidence of CLL; therefore, it is quite demanding to anticipate the effective role of these genotypes in CLL risk.
When age at disease diagnosis was considered in our study, it was found that the B-CLL patients were distributed in all age groups (30–39, 40–49, 50–59, 60–69 and 70–79 years), but the highest frequency of patients (46%) was found in the (50–59 years) group. It was previously known that the risk of B-CLL development rises with advanced age, with a median age at disease onset of 70 years old [38].
Upon examining the association of the CYP2B6*9 (G516T) polymorphism with B-CLL, we observed that the median age of the disease onset was significantly lower in the (GT) and (TT) groups than the (GG) group (p < 0.001). Individuals with variant genotypes (GT) and (TT) have less metabolic activity of the CYP2B6 enzyme. Absence or lowering the CYP2B6 enzyme activity leads to improper metabolism, resulting in a carcinogenic effect, which might be responsible for an earlier onset of disease.
Given that men are more likely than women to acquire CLL, we investigated if there might be a relationship between gender and the CYP2B6 (G516T) polymorphism. We discovered that the percentage of the (GT) genotype’s carrier was higher in males than females (p = 0.009). The regulatory region of CYP2B6 has an estrogen-responsive element, and it has recently been shown that females have 1.7 times higher enzyme activity than males. This implies that males may be more susceptible than females to the development of such lymphoproliferative disorders due to the CYP2B6 enzyme’s declining activity [39].
Oxidative stress products induce CYP2B6, which is a defense mechanism against DNA damage that may aid in leukemogenesis. The T-allele is associated with decreased enzymatic activity as well as a decreased capacity to metabolize and render numerous carcinogenic substances inactive, including naphthalene, aflatoxin B1, benzene metabolites, trichloroethylene, and alkylating agents. This could imply that those who are homozygous or heterozygous (GT or TT) are unable to metabolize genotoxic substances effectively, which causes cellular damage to accumulate. When considering the aforementioned information along with the higher frequency of (GT) and (TT) genotypes observed in patients with chromosomal aberrations (p < 0.001), one could hypothesize that CYP2B6 enzyme deficiency could influence a person’s susceptibility to heamatotoxic exposure to leukemogens and could raise the risk of developing B-CLL.
For individuals with B-CLL, chemotherapy remains the cornerstone of current frontline treatment. Alkylating agents, like chlorambucil and cyclophosphamide, and purine analogues, like fludarabine, are the two main groups of chemotherapeutic medications employed [40]. For B-CLL patients lacking a Tp53 mutation or 17p deletion, cyclophosphamide (CPA) therapy remains the optimal option [41]. According to the patient’s age and fitness, CPA is typically used in combination with other medications like FC (fludarabine + cyclophosphamide) and CVP (cyclophosphamide + vincristine + prednisone). Despite the fact that most patients benefit after the first treatment, B-CLL is still an incurable condition with a clinical course marked by recurrent episodes that eventually lead to chemotherapy resistance. Individual patients differ significantly in their growth dynamics, response to treatment, and tendency for disease acceleration and transformation. Some biological characteristics have been identified as being linked to unfavorable outcomes. These variables include advanced disease stage, male gender, older age, β2 microglobulin level, IGHV gene status, TP53 mutation or deletion, and 11q chromosomal deletion [25]. TP53 continues to be the most highly correlated factor with chemoresistance; yet, not all patients who did not experience a response may be explained by this factor. As such, we ought to take into account other factors that could influence the pharmacokinetics or pharmacodynamics of the chemotherapeutic drugs.
CYP2B6 is an essential enzyme involved in the conversion of CPA to an active form responsible for its anticancer activity. CPA is a prodrug that requires metabolic activation to produce its cytotoxic metabolite. The oxidation of CPA to create 4-hydroxy-CPA (4-OH-CPA), which is catalyzed by numerous members of the cytochrome P450 enzyme superfamily, including CYP2B6, is a crucial step in the bioactivation of CPA. This unstable intermediate binds to erythrocytes before being delivered to the tumor tissue, where it undergoes a spontaneous process of elimination to produce phosphoramide mustard and acrolein. The cytotoxic effects of phosphoramide mustard are caused by the cross-linking of DNA strands that occurs when it spontaneously cyclizes into aziridinium ions [42]. The most striking finding in our work is the relationship between the CYP2B6*9 (G516T) polymorphism and the failure to respond to CPA treatment. We found that the presence of the T-allele was associated with a significantly lower response rate in treated patients, unlike patients who had the normal G-allele (p < 0.001). Patients with the wild-type genotype (GG) were (83% responders, 14.5% non-responders and 2.5% toxic deaths), in contrast to the patients with variant genotypes (GT + TT), who were (37.5% responders, 58.5% non-responders, and 4% toxic deaths). This observation could be explained by the fact that the presence of the T-allele is associated with lower CYP2B6 gene expression and enzyme activity [12].
The restoration of hemopoietic function is one of the criteria used to define remission. Therefore, rather than reflecting worse cytoreduction, it is probable that the decreased response rate seen in patients with the variant genotypes actually reflects higher hematopoietic toxicity; care was taken to ensure that this impact was frank. This option was ruled out by the greater post-treatment neutrophil count and higher residual tumor burden (lymphocytosis, lymphadenopathy, and hepatosplenomegaly) seen in the T-allele patients (p < 0.001 for each). It was of interest to determine whether this effect extended to treatment toxicity as evidenced by documented adverse events that are indicative of the toxicity of alkylating agents, including days in the hospital, febrile episodes, nausea and vomiting and diarrhea.
To address this question, patients with wild-type and variant genotypes were compared for the frequency of these adverse events determined after receiving chemotherapy, and we found that patients with the variant genotypes had fewer adverse events (p < 0.001). All of these data point to a strong possibility that the T-allele reduces the effectiveness and toxicity of CPA by interfering with its conversion to its active form.
Our results were in concordance with the Shu et al. findings [43]. They showed that CYP2B6*9 encoded an enzyme with low metabolic activity in CPA 4-hydroxylation in vivo and in vitro. The in-vitro findings confirmed that the T-allele confers lower mRNA expression and slower CPA 4-hydroxylation, indicating that its carriers are poor CPA metabolizers. Furthermore, Johnson et al. found that individuals with at least one *6 allele had a lower chance of achieving a full response to FC; this suggests that the CYP2B6*6 allele is an independent predictor of a worse response to FC. A higher probability of not responding to the FC protocol was associated with the *6 allele (OR = 4.375, 95% CI: 1.046–18.265, p = 0.042). According to their findings, CYP2B6*6 is associated with reduced CPA efficacy, fewer treatment-related side effects, and a higher chance of FC chemotherapeutic response failure in CLL patients [44]. To our knowledge, though, this SNP has not been examined in CLL patients in any other reports outside of the Johnson et al.’s study.
The study’s primary constraint is the comparatively small sample size of the patient and control groups. Even though the findings might have therapeutic implications, more instances in the study are needed to validate our data. To validate our results and determine the prevalence of this SNP in other populations while taking ethnic heterogeneity into account, more research is advised. Furthermore, to fully comprehend the potential of the CYP2B6 candidate gene for cancer susceptibility, additional research, including large sample sizes, gene-gene, and gene-environment interactions, is needed.
In conclusion, our research indicates that the CYP2B6*9 polymorphism plays a pathogenic role in the predisposition to B-CLL. This implies that inherited malfunction of the CYP2B6 detoxification pathway could be a crucial genetic factor influencing the likelihood of developing B-CLL. Furthermore, the findings point to CYP2B6 genotyping as a desirable candidate marker for routine clinical application and a step toward personalized medicine to get the most effective course of treatment.
Acknowledgement: The authors are very thankful to colleagues at the Oncology Center Mansoura University (OCMU) for supporting the selection of the study participants involved in this work.
Funding Statement: The authors declare that they have not any source of funding for this study.
Author Contributions: Menna Al-Adl contributed to project administration, investigation, and the performance of the statistical analysis. Magdy M. Youssef contributed to methodology, formal analysis, and supervision. Ahmed El-Sebaie was involved in validation, writing, and editing. Sherif Refaat contributed to software development, data curation, and formal analysis. Afaf El-Said was involved in formal analysis, methodology, and visualization.
Availability of Data and Materials: The dataset utilized in the preparation of this study will be available from the corresponding author upon reasonable request.
Ethics Approval: All procedures accomplished in this work, including human subjects, were in accordance with the ethical guidelines of the Mansoura University Faculty of Medicine’s Ethics Committee (Code No. MD.21.03.62) and with the 1964 Helsinki declaration and its adjustments. Informed consent was obtained from all participants in this work.
Conflicts of Interest: The authors declare that they have no funding, financial relationships, or potential conflicts of interest on the subject of this study.
References
1. Arruga, F., Gyau, B. B., Iannello, A., Vitale, N., Vaisitti, T. et al. (2020). Immune response dysfunction in chronic lymphocytic leukemia: Dissecting molecular mechanisms and microenvironmental conditions. International Journal of Molecular Sciences, 21(5), 1825. https://doi.org/10.3390/ijms21051825 [Google Scholar] [PubMed] [CrossRef]
2. Ou, Y., Long, Y., Ji, L., Zhan, Y., Qiao, T. et al. (2022). Trends in disease burden of chronic lymphocytic leukemia at the global, regional, and national levels from 1990 to 2019, and projections until 2030: A population-based epidemiologic study. Frontiers in Oncology, 12, 840616. https://doi.org/10.3389/fonc.2022.840616 [Google Scholar] [PubMed] [CrossRef]
3. Hallek, M., Al-Sawaf, O. (2021). Chronic lymphocytic leukemia: 2022 update on diagnostic and therapeutic procedures. American Journal of Hematology, 96(12), 1679–1705. https://doi.org/10.1002/ajh.26367 [Google Scholar] [PubMed] [CrossRef]
4. Hwa Yun, B., Guo, J., Bellamri, M., Turesky, R. J. (2020). DNA adducts: Formation, biological effects, and new biospecimens for mass spectrometric measurements in humans. Mass Spectrometry Reviews, 39(1–2), 55–82. https://doi.org/10.1200/JCO.2010.32.3865 [Google Scholar] [CrossRef]
5. Pankov, K. V., McArthur, A. G., Gold, D. A., Nelson, D. R., Goldstone, J. V. et al. (2021). The cytochrome P450 (CYP) superfamily in cnidarians. Scientific Reports, 11(1), 9834. https://doi.org/10.1038/s41598-021-88700-y [Google Scholar] [PubMed] [CrossRef]
6. Acharya, P., Chouhan, K., Weiskirchen, S., Weiskirchen, R. (2021). Cellular mechanisms of liver fibrosis. Frontiers in Pharmacology, 12, 671640. https://doi.org/10.3389/fphar.2021.671640 [Google Scholar] [PubMed] [CrossRef]
7. Kuban, W., Daniel, W. A. (2021). Cytochrome P450 expression and regulation in the brain. Drug Metabolism Reviews, 53(1), 1–29. https://doi.org/10.1080/03602532.2020.1858856 [Google Scholar] [PubMed] [CrossRef]
8. Olack, E. M., Heintz, M. M., Baldwin, W. S. (2022). Dataset of endo- and xenobiotic inhibition of CYP2B6: Comparison to CYP3A4. Data in Brief, 41, 108013. https://doi.org/10.1016/j.dib.2022.108013 [Google Scholar] [PubMed] [CrossRef]
9. Langmia, I. M., Just, K. S., Yamoune, S., Brockmöller, J., Masimirembwa, C. et al. (2021). CYP2B6 functional variability in drug metabolism and exposure across populations-implication for drug safety, dosing, and individualized therapy. Frontiers in Genetics, 12, 692234. https://doi.org/10.3389/fgene.2021.692234 [Google Scholar] [PubMed] [CrossRef]
10. Mangó, K., Fekete, F., Kiss, Á.F., Erdős, R., Fekete, J. T. et al. (2023). Association between CYP2B6 genetic variability and cyclophosphamide therapy in pediatric patients with neuroblastoma. Scientific Reports, 13(1), 11770. https://doi.org/10.1038/s41598-023-38983-0 [Google Scholar] [PubMed] [CrossRef]
11. Bunu, S. J., Owaba, A. D. C., Vaikosen, E. N., Ebeshi, B. U. (2022). The Cyp2b6 gene polymorphism and phenotypic correlation of efavirenz-based combination therapy among the niger delta ethnic population: Implications in modern pharmacogenomics. Pharmacogenomics and Personalized Medicine, 15, 45–54. https://doi.org/10.2147/PGPM.S345038 [Google Scholar] [PubMed] [CrossRef]
12. Kassogue, Y., Diakite, B., Kassogue, O., Konate, I., Tamboura, K. et al. (2021). Distribution of alleles, genotypes and haplotypes of the CYP2B6 (rs3745274; rs2279343) and CYP3A4 (rs2740574) genes in the Malian population: Implication for pharmacogenetics. Medicine, 100(29), e26614. https://doi.org/10.1097/MD.0000000000026614 [Google Scholar] [PubMed] [CrossRef]
13. Carvalho Henriques, B., Yang, E. H., Lapetina, D., Carr, M. S., Yavorskyy, V. et al. (2020). How can drug metabolism and transporter genetics inform psychotropic prescribing? Frontiers in Genetics, 11, 491895. https://doi.org/10.3389/fgene.2020.491895 [Google Scholar] [PubMed] [CrossRef]
14. Mangó, K., Kiss, A. F., Fekete, F., Erdős, R., Monostory, K. (2022). CYP2B6 allelic variants and non-genetic factors influence CYP2B6 enzyme function. Scientific Reports, 12(1), 2984. https://doi.org/10.1038/s41598-022-07022-9 [Google Scholar] [PubMed] [CrossRef]
15. Justenhoven, C., Pentimalli, D., Rabstein, S., Harth, V., Lotz, A. et al. (2014). CYP2B6*6 is associated with increased breast cancer risk. International Journal of Cancer, 134(2), 426–430. https://doi.org/10.1002/ijc.28356 [Google Scholar] [PubMed] [CrossRef]
16. Yuan, Z. H., Liu, Q., Zhang, Y., Liu, H. X., Zhao, J. et al. (2011). CYP2B6 gene single nucleotide polymorphisms and leukemia susceptibility. Annals of Hematology, 90(3), 293–299. https://doi.org/10.1007/s00277-010-1085-z [Google Scholar] [PubMed] [CrossRef]
17. Wierda, W. G., Byrd, J. C., Abramson, J. S., Bilgrami, S. F., Bociek, G. et al. (2020). Chronic lymphocytic leukemia/small lymphocytic lymphoma, version 4.2020, NCCN clinical practice guidelines in oncology. Journal of the National Comprehensive Cancer Network, 18(2), 185–217. https://doi.org/10.6004/jnccn.2020.0006 [Google Scholar] [PubMed] [CrossRef]
18. Hallek, M., Cheson, B. D., Catovsky, D., Caligaris-Cappio, F., Dighiero, G. et al. (2018). iwCLL guidelines for diagnosis, indications for treatment, response assessment, and supportive management of CLL. Blood, 131(25), 2745–2760. https://doi.org/10.1182/blood-2017-09-806398 [Google Scholar] [PubMed] [CrossRef]
19. Haas, D. W., Ribaudo, H. J., Kim, R. B., Tierney, C., Wilkinson, G. R. et al. (2004). Pharmacogenetics of efavirenz and central nervous system side effects: An adult AIDS clinical trials group study. AIDS, 18(18), 2391–2400. [Google Scholar] [PubMed]
20. Müller, T. E., Ellwanger, J. H., Michita, R. T., Matte, M. C. C., Renner, J. D. P. (2017). CYP2B6 516 G>T polymorphism and side effects of the central nervous system in HIV-positive individuals under Efavirenz treatment: Study of a sample from southern Brazil. Anais da Academia Brasileira de Ciencias, 89(1 Suppl), 497–504. https://doi.org/10.1590/0001-3765201720160355 [Google Scholar] [PubMed] [CrossRef]
21. Malcikova, J., Tausch, E., Rossi, D., Sutton, L. A., Soussi, T. et al. (2018). ERIC recommendations for TP53 mutation analysis in chronic lymphocytic leukemia-update on methodological approaches and results interpretation. Leukemia, 32(5), 1070–1080. https://doi.org/10.1038/s41375-017-0007-7 [Google Scholar] [PubMed] [CrossRef]
22. Botros, S. K. A., El Gharbawi, N., Shahin, G., Al Lithy, H., El Sherbiny, M. (2021). Impact of Cytochromes P450 3A4 and 2B6 gene polymorphisms on predisposition and prognosis of acute myeloid leukemia: An Egyptian case-control study. Egypt Journal Medical. Egyptian Journal of Medical Human Genetics, 22(38), 1–10. https://doi.org/10.1186/s43042-021-00145-0 [Google Scholar] [CrossRef]
23. Rai, K. R., Sawitsky, A., Cronkite, E. P., Chanana, A. D., Levy, R. N. et al. (1975). Clinical staging of chronic lymphocytic leukemia. Blood, 46(2), 219–234. https://doi.org/10.1182/blood-2016-08-737650 [Google Scholar] [CrossRef]
24. Binet, J. L., Auquier, A., Dighiero, G., Chastang, C., Piguet, H. et al. (1981). A new prognostic classification of chronic lymphocytic leukemia derived from a multivariate survival analysis. Cancer, 48(1), 198–206. https://doi.org/10.1002/1097-0142(19810701)48:1<198::aid-cncr2820480131>3.0.co;2-v [Google Scholar] [PubMed] [CrossRef]
25. Eichhorst, B., Robak, T., Montserrat, E., Ghia, P., Niemann, C. U. et al. (2021). Chronic lymphocytic leukaemia: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Annals of Oncology, 32(1), 23–33. https://doi.org/10.1016/j.annonc.2020.09.019 [Google Scholar] [PubMed] [CrossRef]
26. Shadman, M. (2023). Diagnosis and treatment of chronic lymphocytic leukemia: A review. JAMA, 329(11), 918–932. https://doi.org/10.1001/jama.2023.1946 [Google Scholar] [PubMed] [CrossRef]
27. Kikushige, Y. (2020). Pathogenesis of chronic lymphocytic leukemia and the development of novel therapeutic strategies. Journal of Clinical and Experimental Hematopathology, 60(4), 146–158. https://doi.org/10.3960/jslrt.20036 [Google Scholar] [PubMed] [CrossRef]
28. Hallek, M. (2019). Chronic lymphocytic leukemia: 2020 update on diagnosis, risk stratification and treatment. American Journal of Hematology, 94(11), 1266–1287. https://doi.org/10.1002/ajh.25595 [Google Scholar] [PubMed] [CrossRef]
29. Vaja, R., Rana, M. (2020). Drugs and the liver. Anaesthesia and Intensive Care Medicine, 21(10), 517–523. https://doi.org/10.1016/j.mpaic.2020.07.001 [Google Scholar] [CrossRef]
30. Alaggio, R., Amador, C., Anagnostopoulos, I., Attygalle, A. D., Araujo, I. B. O. et al. (2022). The 5th edition of the world health organization classification of haematolymphoid tumours: Lymphoid neoplasms. Leukemia, 36(7), 1720–1748. https://doi.org/10.1038/s41375-022-01620-2 [Google Scholar] [PubMed] [CrossRef]
31. Darwiche, W., Gubler, B., Marolleau, J. P., Ghamlouch, H. (2018). Chronic lymphocytic leukemia B-cell normal cellular counterpart: Clues from a functional perspective. Frontiers in Immunology, 9, 683. https://doi.org/10.3389/fimmu.2018.00683 [Google Scholar] [PubMed] [CrossRef]
32. Giudice, V., Serio, B., Bertolini, A., Mettivier, L., D'Alto, F. et al. (2022). Implementation of international prognostic index with flow cytometry immunophenotyping for better risk stratification of chronic lymphocytic leukemia. European Journal of Haematology, 109(5), 483–493. https://doi.org/10.1111/ejh.13833 [Google Scholar] [CrossRef]
33. Wan Mohamad Zamri, W. N., Mohd Yunus, N., Abdul Aziz, A. A., Zulkipli, N. N., Sulong, S. (2023). Perspectives on the application of cytogenomic approaches in chronic lymphocytic leukaemia. Diagnostics, 13(5), 964. https://doi.org/10.3390/diagnostics13050964 [Google Scholar] [PubMed] [CrossRef]
34. Visentin, A., Facco, M., Gurrieri, C., Pagnin, E., Martini, V. et al. (2019). Prognostic and predictive effect of IGHV mutational status and load in chronic lymphocytic leukemia: Focus on FCR and BR treatments. Clinical Lymphoma, Myeloma Leukemia, 19(10), 678–685.E4. https://doi.org/10.1016/j.clml.2019.03.002 [Google Scholar] [PubMed] [CrossRef]
35. Autore, F., Strati, P., Innocenti, I., Corrente, F., Trentin, L. et al. (2019). Elevated lactate dehydrogenase has prognostic relevance in treatment-naïve patients affected by chronic lymphocytic leukemia with trisomy 12. Cancers, 11(7), 896. https://doi.org/10.3390/cancers11070896 [Google Scholar] [PubMed] [CrossRef]
36. Daraki, A., Zachaki, S., Koromila, T., Diamantopoulou, P., Pantelias, G. E. et al. (2014). The G516T CYP2B6 germline polymorphism affects the risk of acute myeloid leukemia and is associated with specific chromosomal abnormalities. PLoS One, 9(2), e88879. https://doi.org/10.1371/journal.pone.0088879 [Google Scholar] [PubMed] [CrossRef]
37. Berköz, M., Yalin, S. (2009). Association of CYP2B6 G15631T polymorphism with acute leukemia susceptibility. Leukemia Research, 33(7), 919–923. https://doi.org/10.1016/j.leukres.2008.11.014 [Google Scholar] [PubMed] [CrossRef]
38. Hallek, M. (2023). First line therapy of CLL. Hematological Oncology, 41(S1), 129–135. https://doi.org/10.1002/hon.3145 [Google Scholar] [PubMed] [CrossRef]
39. Desta, Z. (2021). PharmVar GeneFocus: CYP2B6. Clinical Pharmacology and Therapeutics, 110, 82–97. https://doi.org/10.1002/cpt.2166 [Google Scholar] [PubMed] [CrossRef]
40. Cirillo, M., Tan, P., Sturm, M., Cole, C. (2018). Cellular immunotherapy for hematologic malignancies: Beyond bone marrow transplantation. Biology of Blood and Marrow Transplantation, 24(3), 433–442. https://doi.org/10.1016/j.bbmt.2017.10.035 [Google Scholar] [PubMed] [CrossRef]
41. Bewarder, M., Stilgenbauer, S., Thurner, L., Kaddu-Mulindwa, D. (2021). Current treatment options in CLL. Cancers, 13(10), 2468. https://doi.org/10.3390/cancers13102468 [Google Scholar] [PubMed] [CrossRef]
42. Krüger-Genge, A., Köhler, S., Laube, M., Haileka, V., Lemm, S. et al. (2023). Anti-cancer prodrug cyclophosphamide exerts thrombogenic effects on human venous endothelial cells independent of CYP450 activation-relevance to thrombosis. Cells, 12(15), 1965. https://doi.org/10.3390/cells12151965 [Google Scholar] [PubMed] [CrossRef]
43. Shu, W., Chen, L., Hu, X., Zhang, M., Chen, W. et al. (2017). Cytochrome P450 genetic variations can predict mRNA expression, cyclophosphamide 4-hydroxylation, and treatment outcomes in Chinese patients with non-Hodgkin’s lymphoma. Journal of Clinical Pharmacology, 57(7), 886–898. https://doi.org/10.1002/jcph.878 [Google Scholar] [PubMed] [CrossRef]
44. Johnson, G. G., Lin, K., Cox, T. F., Oates, M., Sibson, D. R. et al. (2013). CYP2B6*6 is an independent determinant of inferior response to fludarabine plus cyclophosphamide in chronic lymphocytic leukemia. Blood, 122(26), 4253–4258. https://doi.org/10.1182/blood-2013-07-516666 [Google Scholar] [PubMed] [CrossRef]
Supplementary Materials
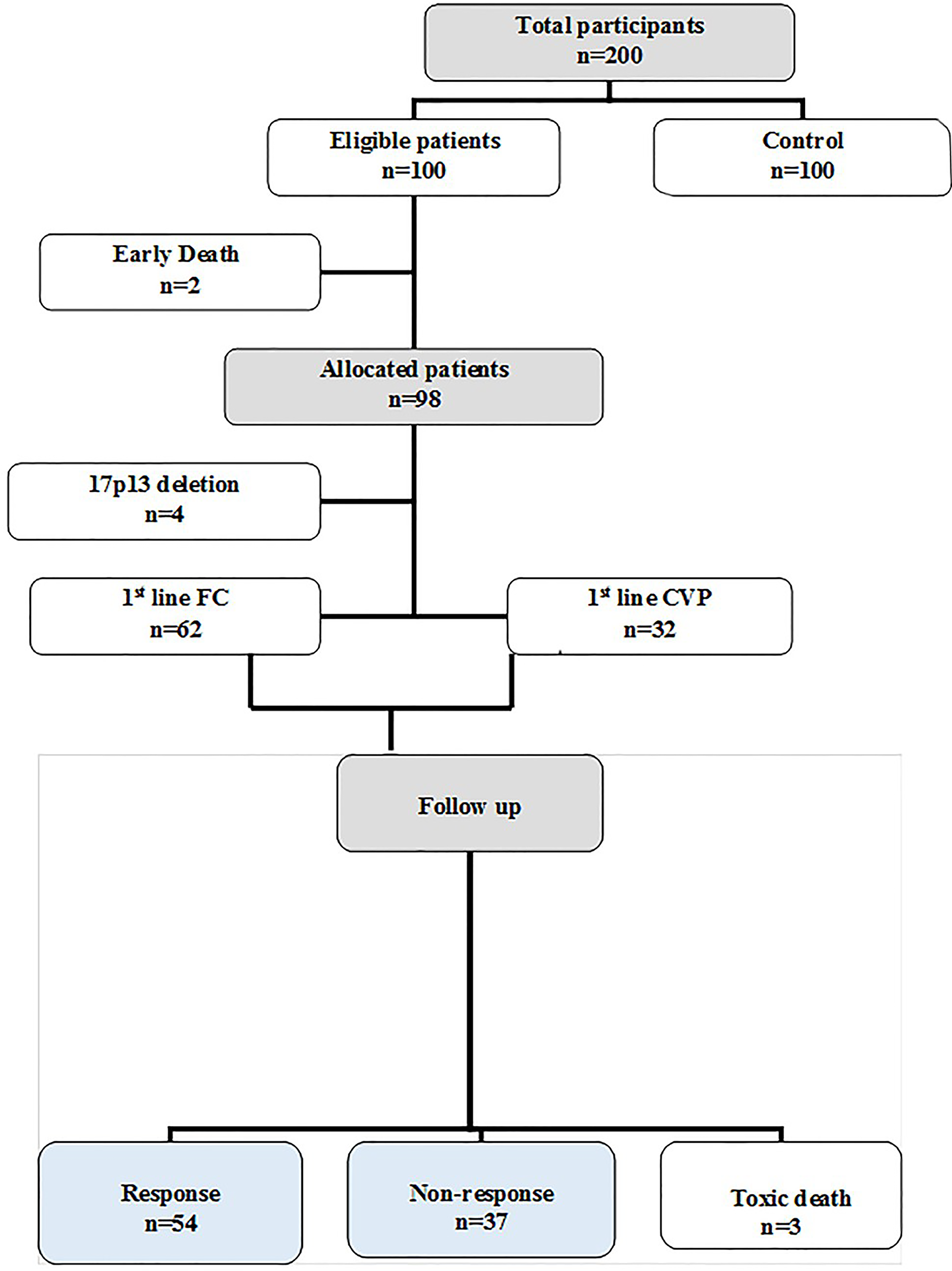
Figure S1: Flow diagram of the studied groups.
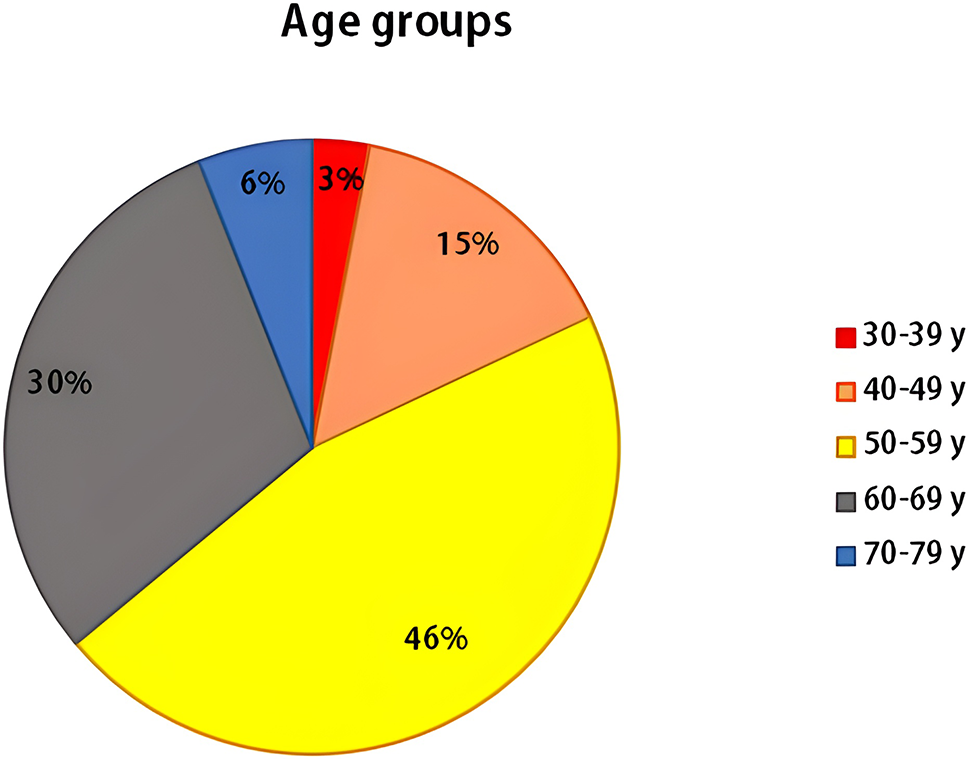
Figure S2: Spreading of B-CLL cases among various age groups.
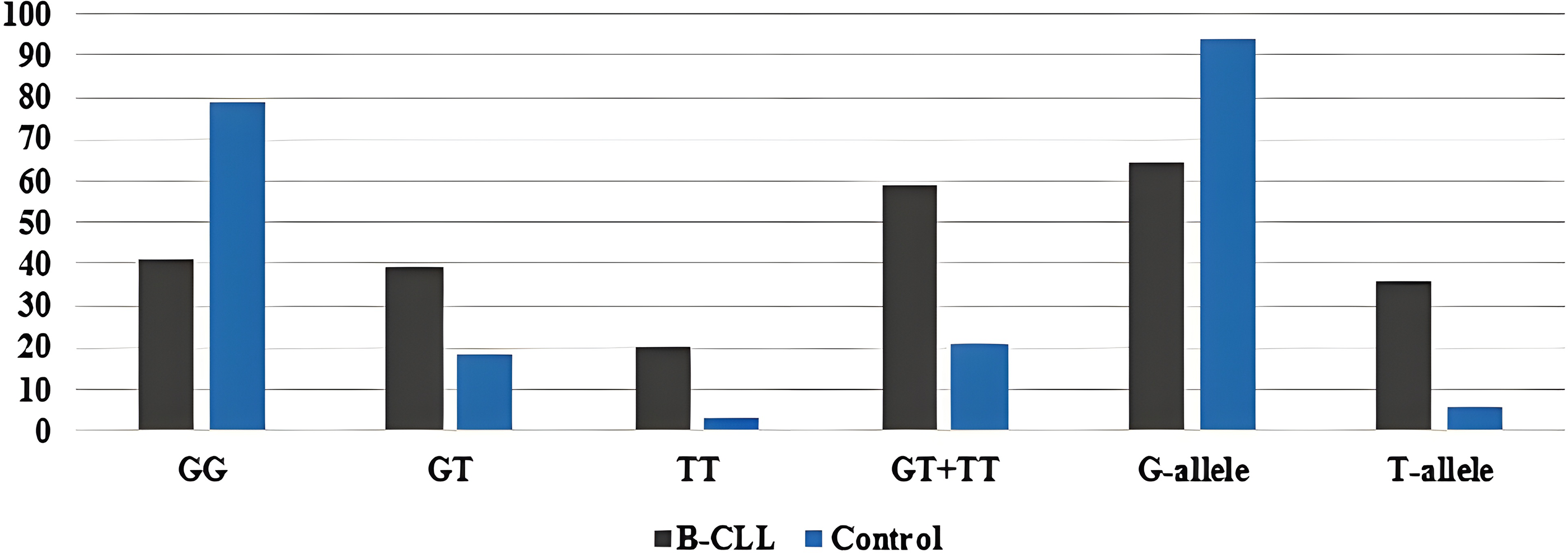
Figure S3: Genotypic and allelic frequencies of the CYP2B6*9 (G561T) SNP among the patients and controls.
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools