 Open Access
Open Access
ARTICLE
Identification of M2 macrophage-related genes for establishing a prognostic model in pancreatic cancer: FCGR3A as key gene
1 Department of Hepatopancreatobiliary Surgery, XuZhou Central Hospital Affiliated to Medical School of Southeast University, Xuzhou, 221000, China
2 Department of Surgery, School of Medicine, Southeast University, Nanjing, 210000, China
3 Department of Oncology, Albert Einstein College of Medicine, Bronx, NY 10461, USA
* Corresponding Author: DUNFENG QI. Email:
(This article belongs to the Special Issue: Novel Biomarkers and Treatment Strategies in Solid Tumor Diagnosis, Progression, and Prognosis)
Oncology Research 2024, 32(12), 1851-1866. https://doi.org/10.32604/or.2024.055286
Received 22 June 2024; Accepted 13 September 2024; Issue published 13 November 2024
Abstract
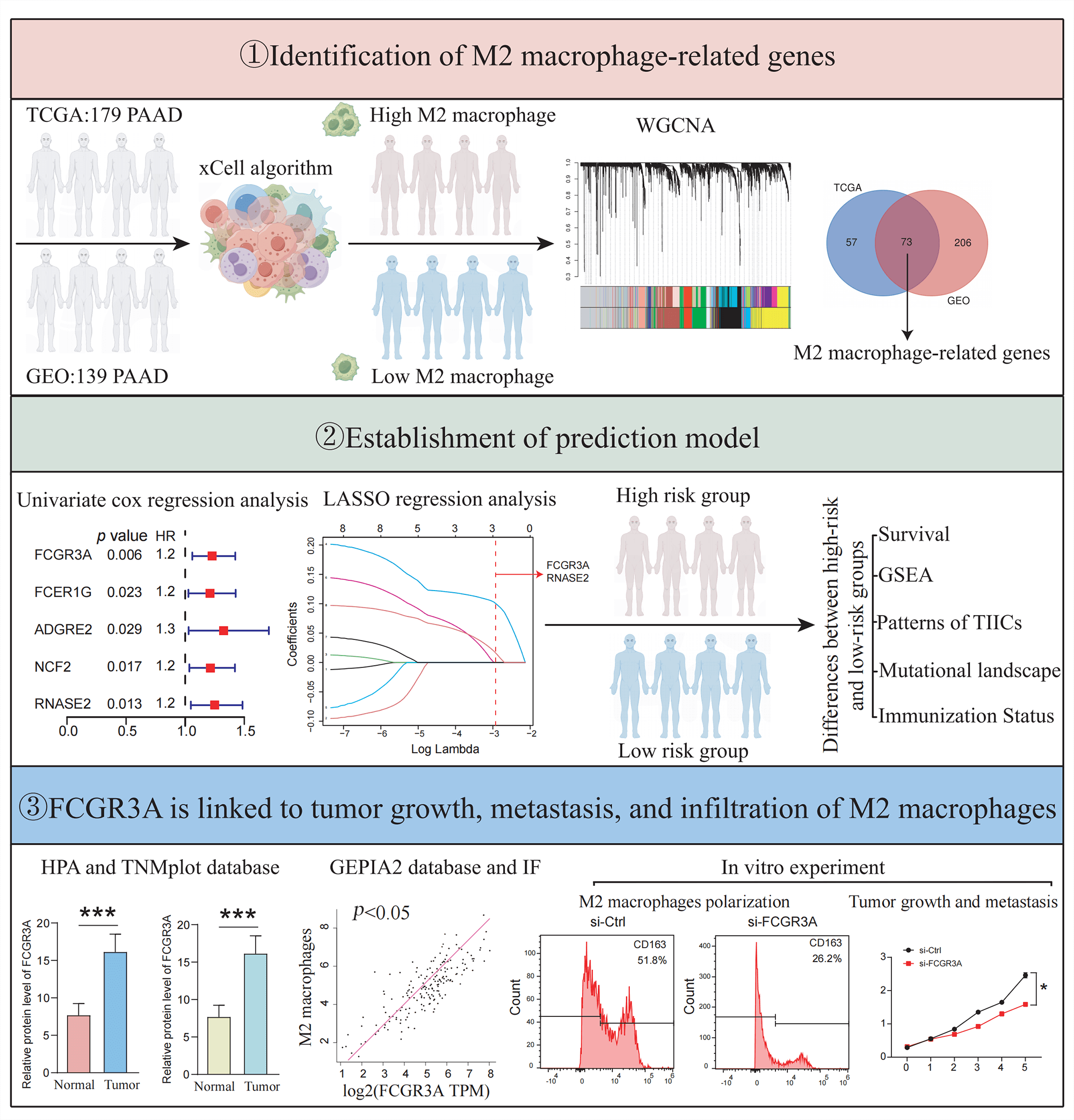
Background: Pancreatic ductal adenocarcinoma (PDAC) has a rich and complex tumor immune microenvironment (TIME). M2 macrophages are among the most extensively infiltrated immune cells in the TIME and are necessary for the growth and migration of cancers. However, the mechanisms and targets mediating M2 macrophage infiltration in pancreatic cancer remain elusive. Methods: The M2 macrophage infiltration score of patients was assessed using the xCell algorithm. Using weighted gene co-expression network analysis (WGCNA), module genes associated with M2 macrophages were identified, and a predictive model was designed. The variations in immunological cell patterns, cancer mutations, and enrichment pathways between the cohorts with the high- and low-risk were examined. Additionally, the expression of FCGR3A and RNASE2, as well as their association with M2 macrophages were evaluated using the HPA, TNMplot, and GEPIA2 databases and verified by tissue immunofluorescence staining. Moreover, in vitro cell experiments were conducted, where FCGR3A was knocked down in pancreatic cancer cells using siRNA to analyze its effects on M2 macrophage infiltration, tumor proliferation, and metastasis. Results: The prognosis of patients in high-risk and low-risk groups was successfully distinguished using a prognostic risk score model of M2 macrophage-related genes (p = 0.024). Between the high- and low-risk cohorts, there have been notable variations in immune cell infiltration patterns, tumor mutations, and biological functions. The risk score was linked to the manifestation of prevalent immunological checkpoints, immunological scores, and stroma values (all p < 0.05). In vitro experiments and tissue immunofluorescence staining revealed that FCGR3A can promote the infiltration or polarization of M2 macrophages and enhance tumor proliferation and migration. Conclusions: In this study, an M2 macrophage-related pancreatic cancer risk score model was established, and found that FCGR3A was correlated with tumor formation, metastasis, and M2 macrophage infiltration.Graphic Abstract
Keywords
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools