 Open Access
Open Access
ARTICLE
YWHAH activates the HMGA1/PI3K/AKT/mTOR signaling pathway by positively regulating Fra-1 to affect the proliferation of gastric cancer cells
1 NHC Key Laboratory of Carcinogenesis, Hunan Cancer Hospital and the Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Changsha, 410013, China
2 Cancer Research Institute, Basic School of Medicine, Central South University, Changsha, 410078, China
3 Hunan Key Laboratory of Cancer Metabolism, Hunan Cancer Hospital and the Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Changsha, 410013, China
* Corresponding Author: YANHONG ZHOU. Email:
Oncology Research 2023, 31(4), 615-630. https://doi.org/10.32604/or.2023.029698
Received 04 March 2023; Accepted 17 April 2023; Issue published 25 June 2023
Abstract
Fos-related antigen 1 (Fra-1) is a nuclear transcription factor that regulates cell growth, differentiation, and apoptosis. It is involved in the proliferation, invasion, apoptosis and epithelial mesenchymal transformation of malignant tumor cells. Fra-1 is highly expressed in gastric cancer (GC), affects the cycle distribution and apoptosis of GC cells, and participates in GC occurrence and development. However, the detailed mechanism of Fra-1 in GC is unclear, such as the identification of Fra-1-interacting proteins and their role in GC pathogenesis. In this study, we identified tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein eta (YWHAH) as a Fra-1-interacting protein in GC cells using co-immunoprecipitation combined with liquid chromatography-tandem mass spectrometry. Experiments showed that YWHAH positively regulated Fra-1 mRNA and protein expression, and affected GC cell proliferation. Whole proteome analysis showed that Fra-1 affected the activity of the high mobility group AT-hook 1 (HMGA1)/phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K)/protein kinase B (AKT)/mechanistic target of rapamycin (mTOR) signaling pathway in GC cells. Western blotting and flow cytometry confirmed that YWHAH activated HMGA1/PI3K/AKT/mTOR signaling pathway by positively regulating Fra-1 to affect GC cell proliferation. These results will help to discover new molecular targets for the early diagnosis, treatment, and prognosis prediction of GC.Keywords
Globally, gastric cancer (GC) is the fifth most common malignant tumor and the third leading cause of cancer-related mortality [1–5]. Its pathogenesis is a multi-factor and multi-step process, involving infection by microorganisms such as Helicobacter pylori, the activation of proto oncogenes such as FOSL1 (encoding Fos-related antigen 1, also known as Fra-1), BRD4 (encoding bromodomain containing 4), and THY1 (encoding Thy-1 cell surface antigen, also known as CD90), and the inactivation of tumor suppressor genes [6–12]. Helicobacter pylori is considered to be a major cause of GC, but not in all cases [12–16], and the mechanism of GC is not completely clear. Therefore, more experimental evidence is needed to clarify its pathogenesis and potential molecular targets.
Fra-1 is a member of the activator protein-1 (AP-1) transcription factor superfamily and Fos family of proteins, and its basic domain is highly homologous with c-Fos [17,18]. Fos family proteins encode leucine zipper proteins that can form dimers with JUN family proteins to form the transcription factor complex AP-1 [18]. Fra-1 is an important nuclear transcription factor that regulates the growth, differentiation, and apoptosis of normal cells. It is abnormally expressed in many cancer cells and tissues, and plays an important role in the tumorigenesis and progression or maintenance of many tumor types [6,7,19–21]. Fra-1 levels are frequently elevated in a variety of human cancers because of carcinogenic signal transduction, and Fra-1 is closely related to metastasis and poor prognosis [20–22]. Overexpression of Fra-1 can lead to morphological changes of fibroblasts and is related to mesenchymal characteristics and E-cadherin downregulation in cancer cells [23–25]. In the cell cycle, Fra-1 is recruited to the promoter of CCNA2 (encoding cyclin A2). Fra-1 induces the expression of Jun B, which interacts with the CCNA2 promoter to enhance cell proliferation [26,27]. Previous studies suggest that Fra-1 plays an important role in the occurrence and development of malignant tumors such as GC.
In a previous study, we used quantitative real-time polymerase chain reaction (qPCR), immunohistochemistry (IHC), and western blotting to show that Fra-1 is highly expressed in GC tissues. In vitro experiments confirmed that Fra-1 overexpression inhibited the apoptosis of GC cells, increased the proportion of S-phase cells, and was related to an imbalance of the phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K)/protein kinase B (AKT) signaling pathway [6]. However, the detailed mechanism of Fra-1 in GC has not been fully defined. In this study, we aimed to identify molecules that interact with Fra-1 in GC cells using co-immunoprecipitation (Co-IP) combined with liquid chromatography-tandem mass spectrometry (LC-MS/MS), and clarified their mutual regulation relationship. Then, western blotting and flow cytometry were used to confirm that the newly identified Fra-1 interacting molecule, tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein eta (YWHAH), could promote the proliferation of GC cells by positively regulating Fra-1 to activate the high mobility group AT-hook 1 (HMGA1)/PI3K/AKT/mechanistic target of rapamycin (mTOR) signaling pathway. Our results provide a new molecular target to explore the pathogenesis of GC.
Gastric cancer cells SGC7901 and AGS were preserved and provided by the Cancer Research Institute, Basic School of Medicine, Central South University. The cells were cultured via adherent growth in an incubator set at 37°C with 5% CO2 in Roswell Park Memorial Institute (RPMI) 1640 medium (Gibco-Life Technologies at Thermo Fisher Scientific, Waltham, MA, USA) with 10% fetal bovine serum (FBS; Gibco-Life Technologies) and 1% Penicillin-Streptomycin Solution.
The plasmids used in this study included: vector pLVX-mCMV-ZsGreen-PGK-Puro-Fra-1 (Fra-1 overexpression), vector pLVX-mCMV-ZsGreen-IRES-Puro-YWHAH (YWHAH overexpression), vector pLVX-mCMV-ZsGreen-PGK-Puro-Fra-1-D1 (expressing Fra-1 aa 1–107), vector pLVX-mCMV-ZsGreen-PGK-Puro-Fra-1-D2 (expressing Fra-1 aa 1–127), and vector pLVX-mCMV-ZsGreen-PGK-Puro-Fra-1-D3 vector (expressing Fra-1 aa 107–271). All plasmids were lentivirus expression vectors carrying an ampicillin resistance gene. Puromycin was used for screening.
After transfection, the cells were cultured in a 100 mm dish for 48 h and then lysed using IP lysis buffer containing a mixture of protease inhibitors and phosphatase inhibitors (Beyotime Biotechnology, P0013, Shanghai, China). Then, 1 µg of primary antibody was incubated with 30 µL of protein A+G beads on a rotator at room temperature for 2 h, the protein lysate was then added and incubated on the rotator at 4°C overnight. Next day, the beads and immune complexes were washed five times with IP lysis buffer by rotating at room temperature for five minutes at a speed (2000 rpm/min) of 20 s per round. The sample was added to the corresponding SDS loading buffer and heated at 95°C for 5 min. The immunoprecipitated samples were detected using western blotting.
The purified protein complex eluent was concentrated using IP beads, separated by SDS-PAGE, and stained with Coomassie brilliant blue (Beyotime Biotechnology). The corresponding protein band was excised from the gel, reduced, alkylated, and digested overnight using trypsin at 37°C (Thermo Fisher Scientific, Waltham, MA, USA). The digested peptides were dried and resuspended in MS compatible buffers, and the mixture was analyzed using an LTQ Orbitrap velos MS instrument (Thermo Fisher Scientific) in combination with a UltiMate RSLC Nano LC system (Dionex, Sunnyvale, CA, USA). Proteome Discoverer 1.4 software (Thermo Fisher Scientific) was used to identify proteins, import files, and search the UniProtKB/Swiss-Prot databases. The mass tolerances of precursors and fragments were set to 10 ppm and 0.8 Da, respectively. Peptide data with an error detection rate of <1% (p < 0.01) were discarded.
RNA extraction and quantitative real-time reverse transcription PCR (qRT-PCR)
The total RNA of GC cells was isolated using the TRIzol reagent (Invitrogen, Waltham, MA, USA), and cDNA synthesis was carried out using the RevertAid First Strand cDNA synthesis kit (CWBio) according to the manufacturer’s facturer’s recommendations. qRT‑PCR was carried out with GoTaq qPCR Master Mix (Promega, Fitchburg, WI, USA). For detection of Fra-1, YWHAH, AKT, PI3K, PDK1, MTOR, and HMGA1 mRNA expression levels, GAPDH was amplified in parallel as an internal control. The sequences of the primers used for qPCR were as follows: Fra-1 forward 5′-CAGTGGATGGTACAGCCTCATTTC-3′, reverse 5′-GCAGTCTCCTGTTCACAAGGC-3′; YWHAH forward 5′-TCAAGAAGGTGGTGAAGCAGG-3′, reverse 5′-TCAAAGGTGGAGGAGTGGGT-3′; AKT forward 5′-ACACCAGGTATTTTG ATGAGGAG-3′, reverse 5′-TCAGGCCGTGCCGCTGGCCGAGTAG-3′; PI3K forward 5′-AGCTGGTTTGGATCTTCGGA-3′, reverse 5′-CAGGTC ATCCCC AGAGTTGT-3′; PDK1 (encoding pyruvate dehydrogenase kinase 1) forward 5′-AGTTCATGTCACGCTGGGTA-3′, reverse 5′-CAGCTTCAGGTCTCCTTGGA-3′; MTOR forward 5′-GCAACCCTTCTTTGACAACATTTTT-3′, reverse 5′-ATTTCTTCTCTCAGACGCTCTCC-3′; HMGA1 forward 5′-TGCGAAGAAACTGGG AGAGA-3′, reverse 5′-TGCGAAGAAACTGGGAGAGA-3′; GAPDH (encoding glyceraldehyde-3-phosphate dehydr-ogenase) forward 5′-TCAAGAAGGTGGTGAAGCAGG-3′, reverse 5′-TCAAAGGTGGAGGAGT GGGT-3′. The expression of mRNA was assessed by evaluated threshold cycle (CT) values. The CT values were normalized to the expression levels of GAPDH and the relative amount of mRNA specific to each of the target genes was calculated using the 2−ΔΔCT method. qPCR was carried out using the Bio‑Rad CFK96™ Real‑Time System (Bio‑Rad, Hercules, CA, USA). The data were analyzed by Bio‑Rad CFK Manager software (Bio‑Rad).
To test the effect of silencing YWHAH and Fra-1, we selected three small interfering RNAs (siRNAs) targeting different regions of the YWHAH and Fra-1 genes. The three siRNA sequences for YWHAH were:
si-YWHAH-001 5′-GCUGGAGACAGUUUGCAAUTT-3′;
si-YWHAH-002 5′-CCAGAAUGCACCUGAGCAATT-3′;
si-YWHAH-003 5′-GCUGAGCUGGACACACUAATT-3′;
and the control si-NC 5′-ACGUGACACGUUCGG AGAATT-3′.
The three siRNA sequences for Fra-1 were:
si-Fra-1-001 5′-GTCGAAGGCCTTGTGAA-3′;
si-Fra-1-002 5′-GCTCATCGCAAGAGTAGCA-3′;
si-Fra-1-003 5′-GGAAGGAACTGACCGACTT-3′;
and the control si-NC 5′-ACGUGACACGUUC GGAGA ATT-3′.
For siRNA transfection, the riboFECT CP Transfection Kit (Guangzhou RiboBio Co., Ltd., Guangzhou, China) was used according to the manufacturer’s instructions, and then the silencing effect was detected. The most successful siRNA sequence for each gene was used for subsequent experiments.
5-ethynyl-2′-deoxyuridine (EdU) cell proliferation assay
Cells (1 × 105 to 3 × 106) were inoculated in 6-well plates and cultured at 37°C in a 5% CO2 incubator to the required cell density. Two hours before transfection, serum-free 1640 medium was used. The transfected cells were divided into groups according to the corresponding experiments. After 6 h, the mixed solution was sucked out and replaced with normal culture medium, and culture was continued for 48 h. Referring to the operating instructions of the EdU-647 cell proliferation test kit (Beyotime biotechnology, C0081S), the samples were processed before flow cytometry [28,29]. Finally, the samples were tested by flow cytometry at a 488 nm excitation wavelength and 520 nm emission wavelength. Cells were analyzed with Moflo XDP High-Performance Cell Sorter (Beckman Coulter). Data were acquired and analyzed with Summit v5.2 software.
Protein extraction and western blotting
Cells were digested with trypsin and then lysed with Radioimmunoprecipitation assay (RIPA) buffer (CWBio, Beijing, China). The sample was centrifuged at 12000 × g at 4°C for 15 min to remove insoluble matter. The protein concentration of the retained supernatant was detected, and protein electrophoresis was carried out using a 10% SDS-PAGE gel at 50 V for 40 min, and then at 120 V for 60 min (PowerPac Universal, Bio-Rad, Hercules, CA, USA). Then, the proteins were transferred to a polyvinylidene fluoride (PVDF) membrane (HyClone Laboratories, Logan, UT, USA) at 100 V for 90 min, and then incubated in phosphate-buffered saline (PBS)-Tween20 with 5% skimmed milk for 1–2 h. The membrane was then incubated with primary antibody at 4°C overnight.
The main antibodies used in this study include: Fra-1 (YM0122,Immunoway, Plano, TX, USA), YWHAH (ab206292, Abcam, Cambridge, MA, USA), HMGA1 (#7777, Cell Signaling Technology, Danvers, MA, USA), PI3K (AF6241, Affinity Biosciences, Cincinnati, OH, USA), AKT (10176-2-AP, Wuhan Sanying Biotechnology Co., Ltd., Wuhan, China), mTOR (ab32028, Abcam), phosphorylated (p)-mTOR (ab109268, Abcam), GAPDH (AB-P-R001, Hangzhou Xianzhi Biotechnology Co., Ltd., Hangzhou, China). The dilution ratio of the antibodies was 1:1000. Rabbit anti-β-tubulin antibody, mouse anti-β-actin, Rabbit anti-GAPDH, from Affinity Bioscience were used at a dilution of 1:5000; anti-rabbit secondary antibody and anti-mouse secondary antibody were purchased from Lianke company (Shanghai, China), and used at a dilution of 1:3000. The membranes were incubated with the secondary antibodies for 1 h and then the immunoreactive proteins were visualized using Molecular Imager Gel Dox XR System (Bio-Rad Laboratories).
GraphPad Prism 5 software (GraphPad Inc., La Jolla, CA, USA) was used perform the statistical analyses. T-tests were used to analyze whether the difference between any two groups was significant. One way analysis of variance (ANOVA) was used to analyze whether the data of more than two groups were significantly different. All experiments were set with three or more replicates, and p < 0.05 indicated a significant difference.
Identification of YWHAH as an interacting protein of Fra-1 in GC cells by Co-IP and LC-MS/MS technologies
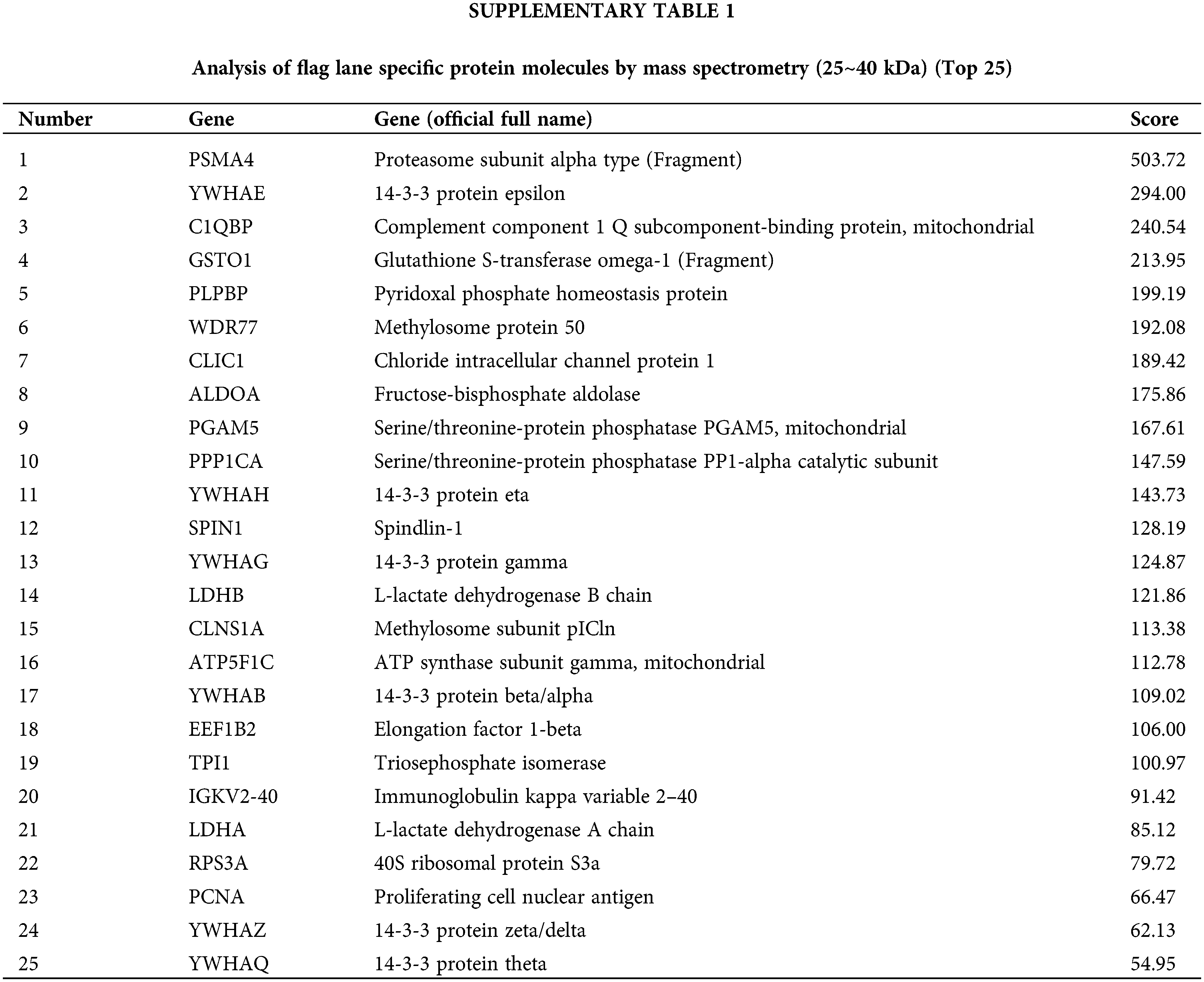
Our previous research results showed that Fra-1 was upregulated in GC tissue and is involved in its occurrence and development. To systematically explore the role and possible mechanism of Fra-1 in GC, we first used Co-IP combined with LC-MS/MS to identify proteins that interact with Fra-1 in GC cells. After the Fra-1 interaction proteins were separated by SDS-PAGE, stained with Coomassie blue, and decolorized, we observed obvious difference bands at 25~40, 40~70, and 70~180 kDa on the gel (Fig. 1A). The bands were then analyzed by endogel enzymolysis and mass spectrometry. Among them, 25 specific proteins of the FLAG lane were identified at 25~40 kDa, among which 6 protein molecules belonged to the 14-3-3 protein family, including YWHAE, YWHAH, YWHAG, YWHAZ, and YWHAQ, suggesting that the 14-3-3 protein family might interact with Fra-1 (Suppl. Table 1). To verify the interaction between the 14-3-3 protein family and Fra-1, we performed IP experiments for YWHAE, YWHAH, YWHAG, YWHAZ, and YWHAQ with Fra-1, respectively. The results showed that Fra-1 interacted only with YWHAH in SGC7901 GC cells (Fig. 1B). To further explore the interaction domain between YWHAH and Fra-1, we constructed vectors expressing Fra-1 3-segment domains D1 (1–107 aa), D2 (1–127 aa) and D3 (107–271 aa) with HA tags (Fig. 1C), and conducted IP experiments to verify the domain of interaction. The results showed that YWHAH interacted with full length Fra-1 and all three segment domains; however, the interaction between YWHAH and D2 (1–127 aa) was stronger (Fig. 1D), suggesting that the Bag domain of Fra-1 plays an important role in the interaction between the two proteins. In conclusion, we identified YWHAH as a interacting protein of Fra-1 in SGC7901 GC cells.
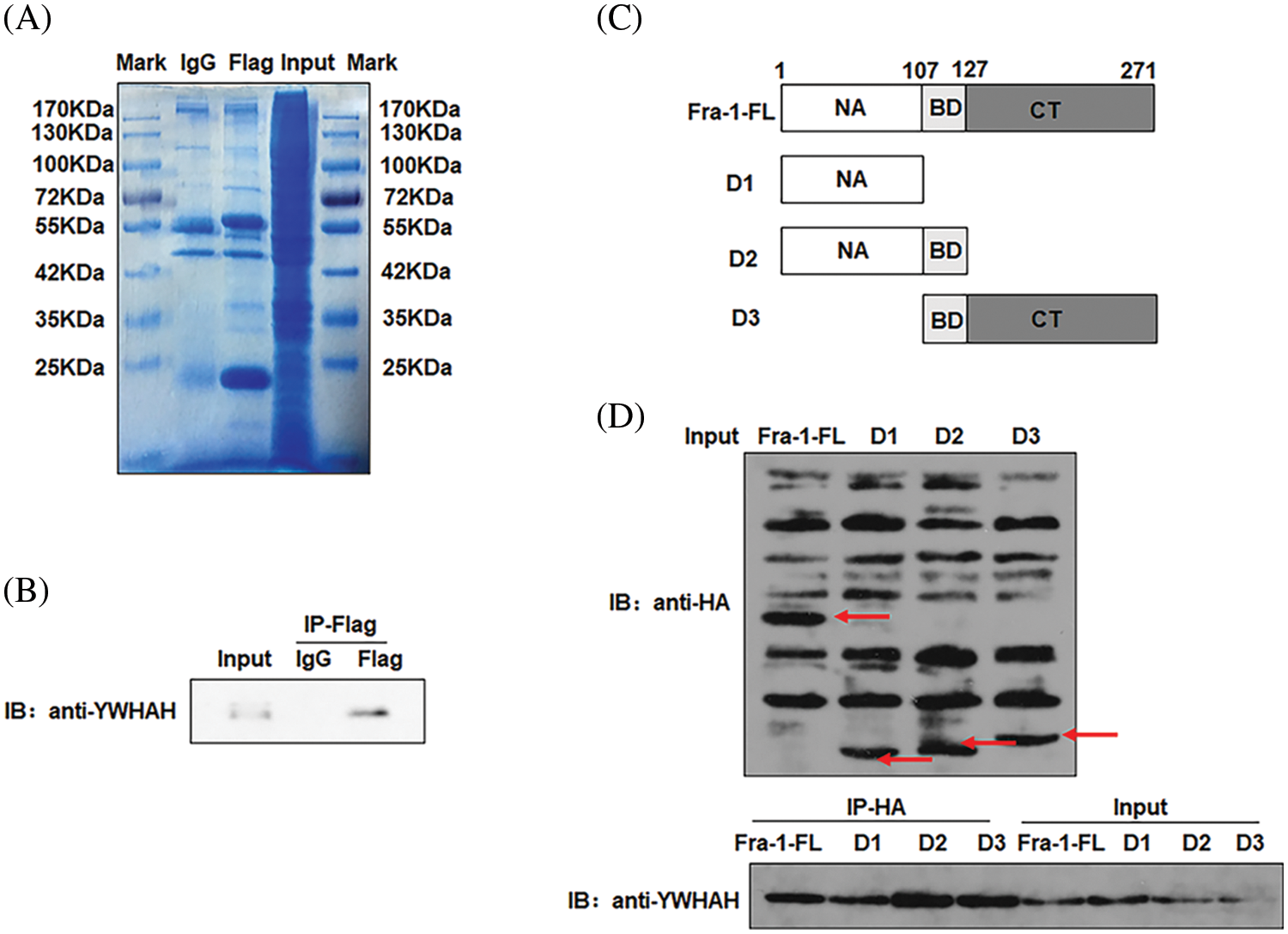
Figure 1: Identification of new interacting molecules of Fra-1 in SGC7901 GC cells. (A) Staining diagram of FLAG-Fra-1 precipitated using anti-FLAG affinity gel beads and its interacting proteins. Input lane: the whole cell protein as the positive control; IgG Lane: the protein bound to IgG as the negative control; FLAG lane: proteins binding to Fra-1. (B) Western blotting showing that there was an endogenous interaction between YWHAH and Fra-1 in SGC7901 GC cells. Input lane: the whole cell protein as the positive control; IgG Lane: the protein bound to IgG as the negative control; FLAG lane: proteins binding to Fra-1. (C) Schematic diagram of Fra-1 domains. Fra-1-FL: the full length amino acid sequence of Fra-1, D1 (1–107 aa), D2 (1–127 aa), and D3 (107–271 aa) domains of Fra-1. (D) The interaction domain between YWHAH and Fra-1 was detected by western blotting. Input Lane: the whole cell protein as the positive control; IP-HA: an anti-HA antibody was used to detect the domain interacting with YWHAH. The experiment was performed three times.
YWHAH positively regulates the mRNA and protein expression of Fra-1
To clarify the mutual regulation relationship between YWHAH and Fra-1, we first designed three siRNA sequences for the YWHAH gene, and transiently transfected the three siRNAs into SGC7901 GC cells. Western blotting showed that si-YWHAH-01 had the best effect in reducing the protein level of YWHAH (Suppl. Fig. 1); therefore, si-YWHAH-01 was used in subsequent experiments. Then, we overexpressed and silenced YWHAH, separately, in SGC7901 GC cells. Western blotting and qRT-PCR assays showed that the mRNA and protein levels of Fra-1 were upregulated after YWHAH overexpression (Figs. 2A and 2B) but downregulated after YWHAH silencing (Figs. 2B and 2C), suggesting that YWHAH further affected the translation level after regulating the transcription of Fra-1. To further verify our experimental results, we repeated the above experiments in AGS cells, and the results were consistent with those in SGC7901 cells (Figs. 2D–2F). The above results showed that YWHAH interacted with and positively regulated Fra-1, and affected its protein level by regulating Fra-1 transcription.
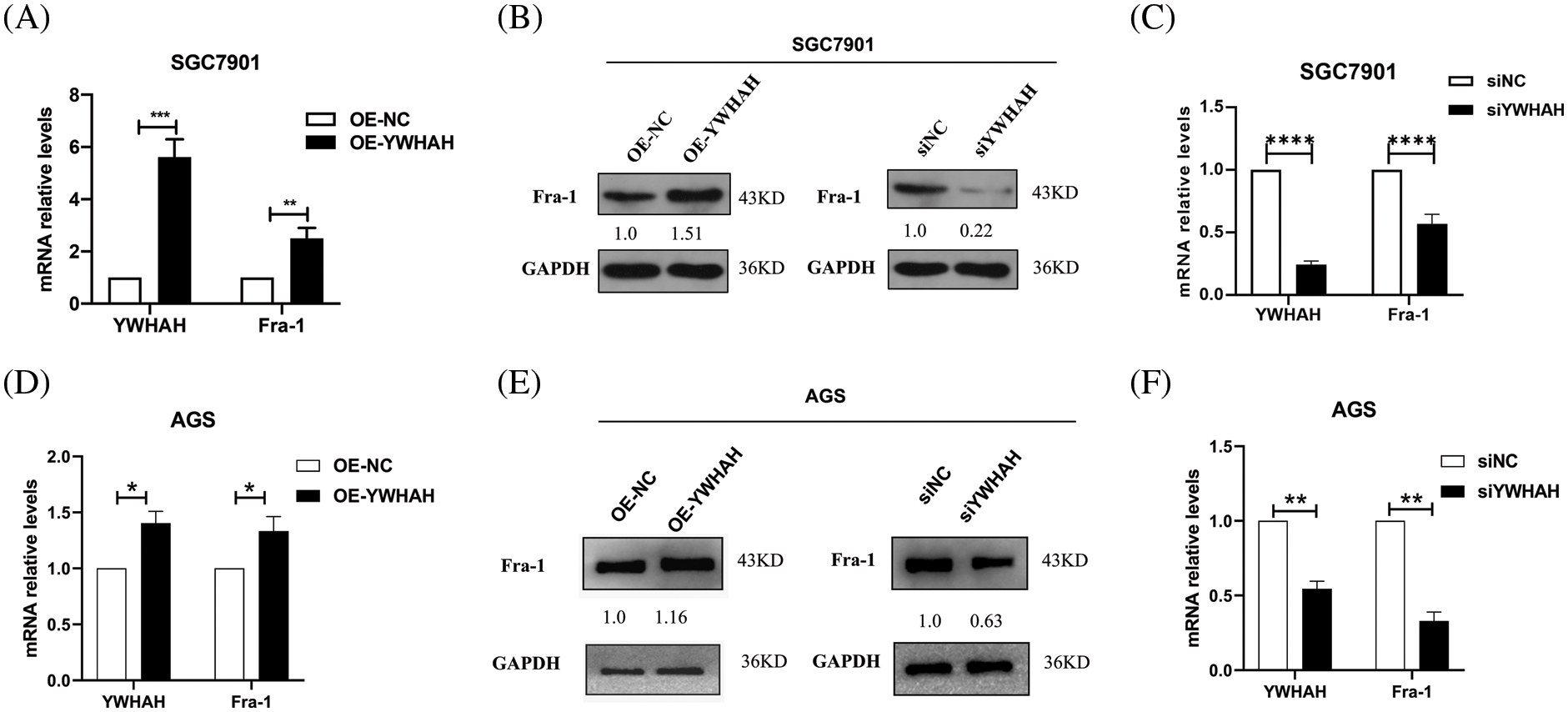
Figure 2: YWHAH positively regulates the mRNA and protein expression of Fra-1. (A) In SGC7901 GC cells, the effect of overexpression of YWHAH on the expression of Fra-1 mRNA was detected using qRT-PCR. (B) In SGC7901 GC cells, the effect of overexpression/interference of YWHAH on the level of the Fra-1 protein was detected using western blotting. (C) In SGC7901 GC cells, the mRNA level of Fra-1 after interference with YWHAH was detected using qRT-PCR. (D) In AGS GC cells, the effect of overexpression of YWHAH on the expression of Fra-1 mRNA was detected using qRT-PCR. (E) Western blotting was used to detect the effect of overexpression/interference of YWHAH on the level of the Fra-1 protein in AGS GC cells. (F) In AGS GC cells, the mRNA level of Fra-1 after interference with YWHAH was detected using qRT-PCR. GAPDH was the internal control, NS: not significant, **p < 0.01, ***p < 0.001. Three independent experiments were conducted.
YWHAH affects the proliferation of GC cells by positively regulating Fra-1
Based on the discovery that YWHAH interacts with and positively regulates the mRNA and protein levels of Fra-1, we assessed whether YWHAH could affect the proliferation of GC cells by regulating Fra-1. Cell proliferation assays combined with flow cytometry showed that the cell proliferation increased after the stable overexpression of YWHAH in SGC7901 GC cells. However, when Fra-1 was silenced in SGC7901 cells stably overexpressing YWHAH, cell proliferation was inhibited (Fig. 3A), suggesting that YWHAH might affect GC cell proliferation by regulating Fra-1. When we silenced with YWHAH in SGC7901 cells, their proliferation ability was inhibited. However, in cell silenced for YWHAH, Fra-1 overexpression restored cell proliferation (Fig. 3B). The above results showed that YWHAH affected the proliferation of GC cells by regulating Fra-1. To further verify our experimental results, we repeated the above experiments in AGS cells, and the results were consistent with those in SGC7901 cells (Figs. 3C and 3D).
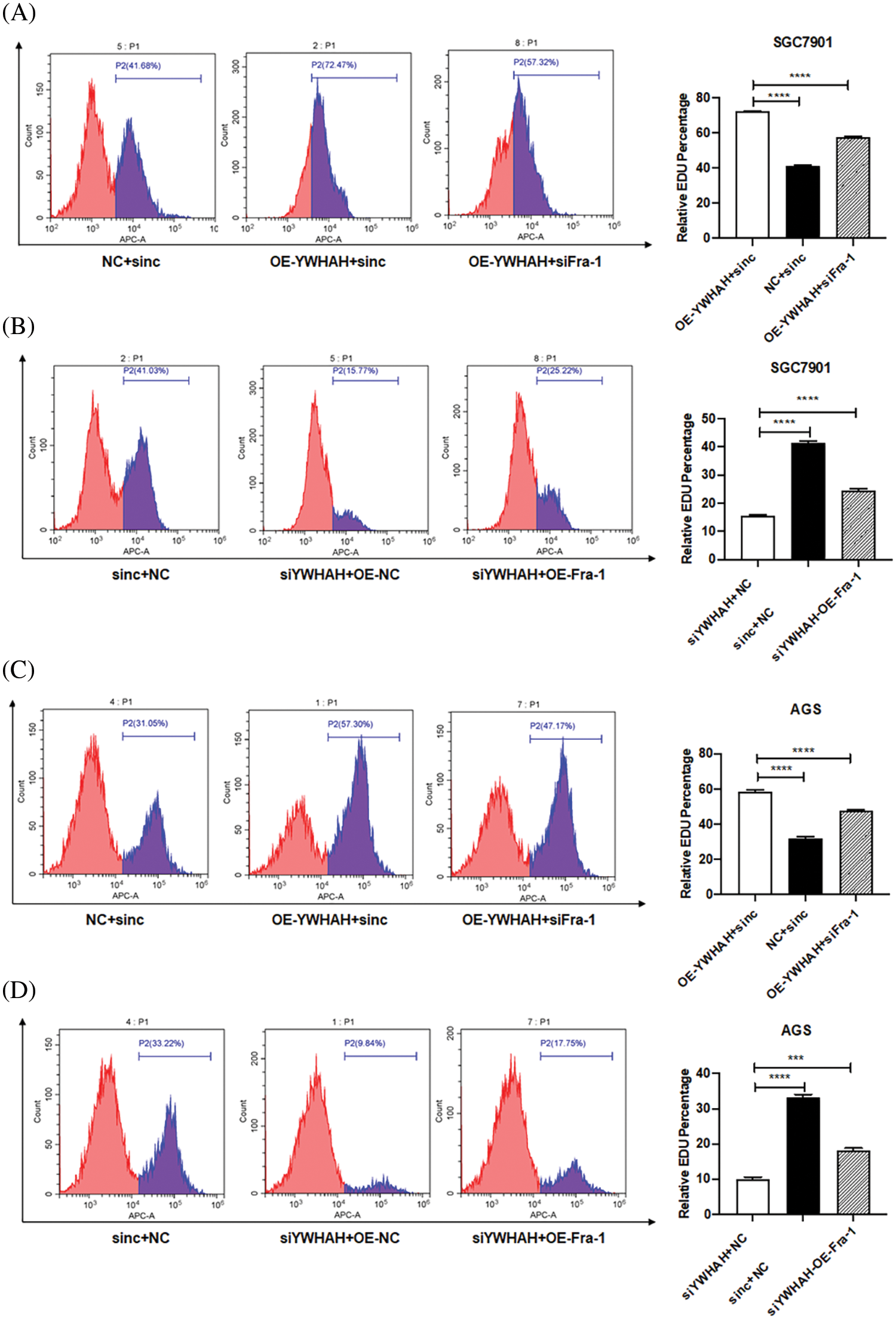
Figure 3: YWHAH affects the proliferation of GC cells by positively regulating Fra-1. (A) YWHAH+siNC, NC+siNC, and YWHAH+siFra-1 plasmids were transfected into SGC7901 GC cells, separately. Cell proliferation was detected using an EdU-647 cell proliferation assay combined with flow cytometry. (B) siYWHAH+ NC, siNC + NC, and siYWHAH+Fra-1 plasmids were transfected into SGC7901 GC cells, separately. Cell proliferation was detected using an EdU-647 cell proliferation assay combined with flow cytometry. (C) YWHAH+siNC, NC+siNC, and YWHAH+siFra-1 plasmids were transfected into AGS GC cells, separately. Cell proliferation was detected using an EdU-647 cell proliferation assay combined with flow cytometry. (D) siYWHAH+ NC, siNC + NC, and siYWHAH+Fra-1 plasmids were transfected into AGS GC cells, separately. Cell proliferation was detected using an EdU-647 cell proliferation assay combined with flow cytometry. ***p < 0.001. Three independent experiments were conducted.
Whole proteome analysis suggests that Fra-1 affects the activity of the PI3K/AKT/mTOR signal pathway in GC cells
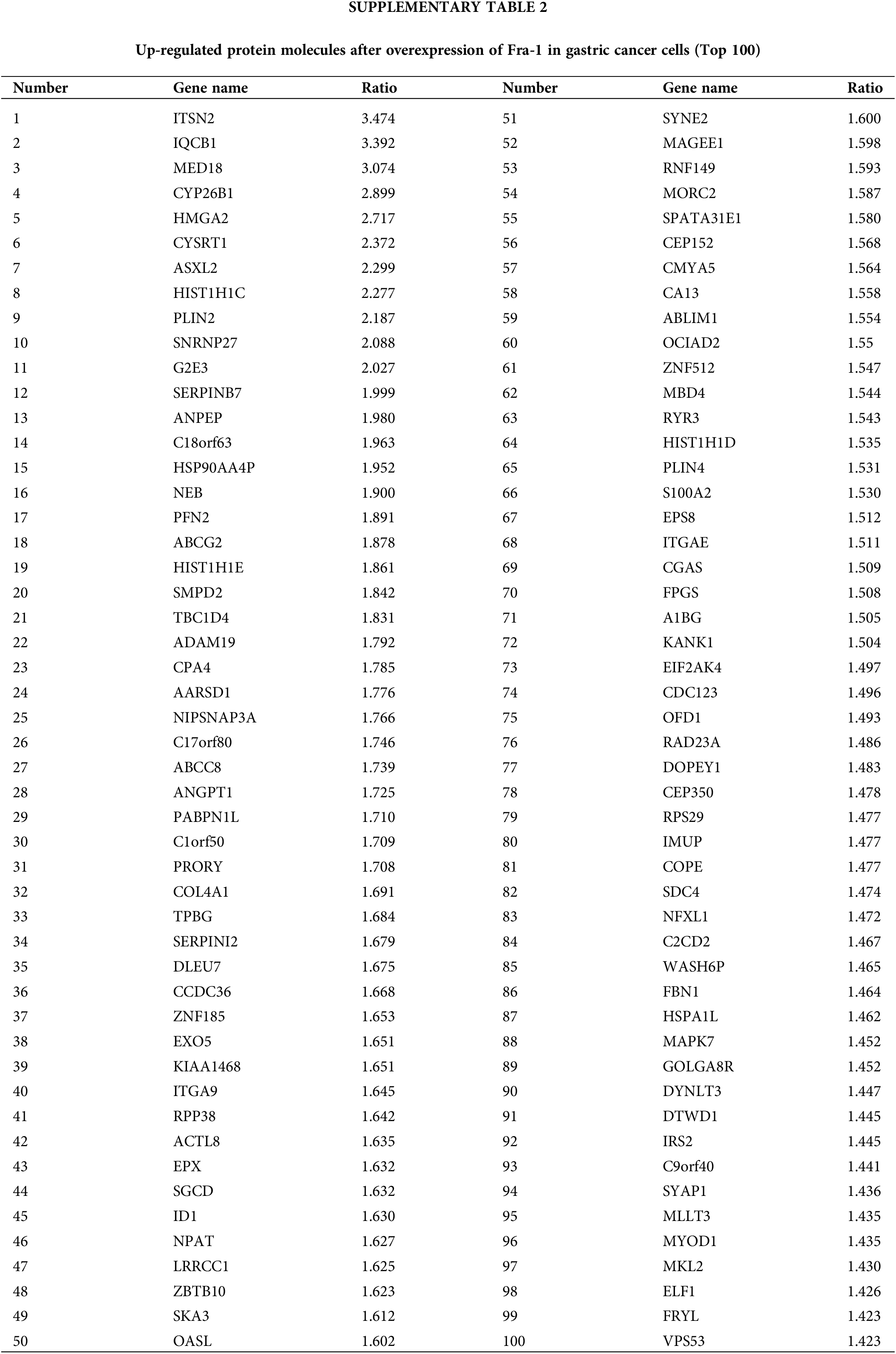
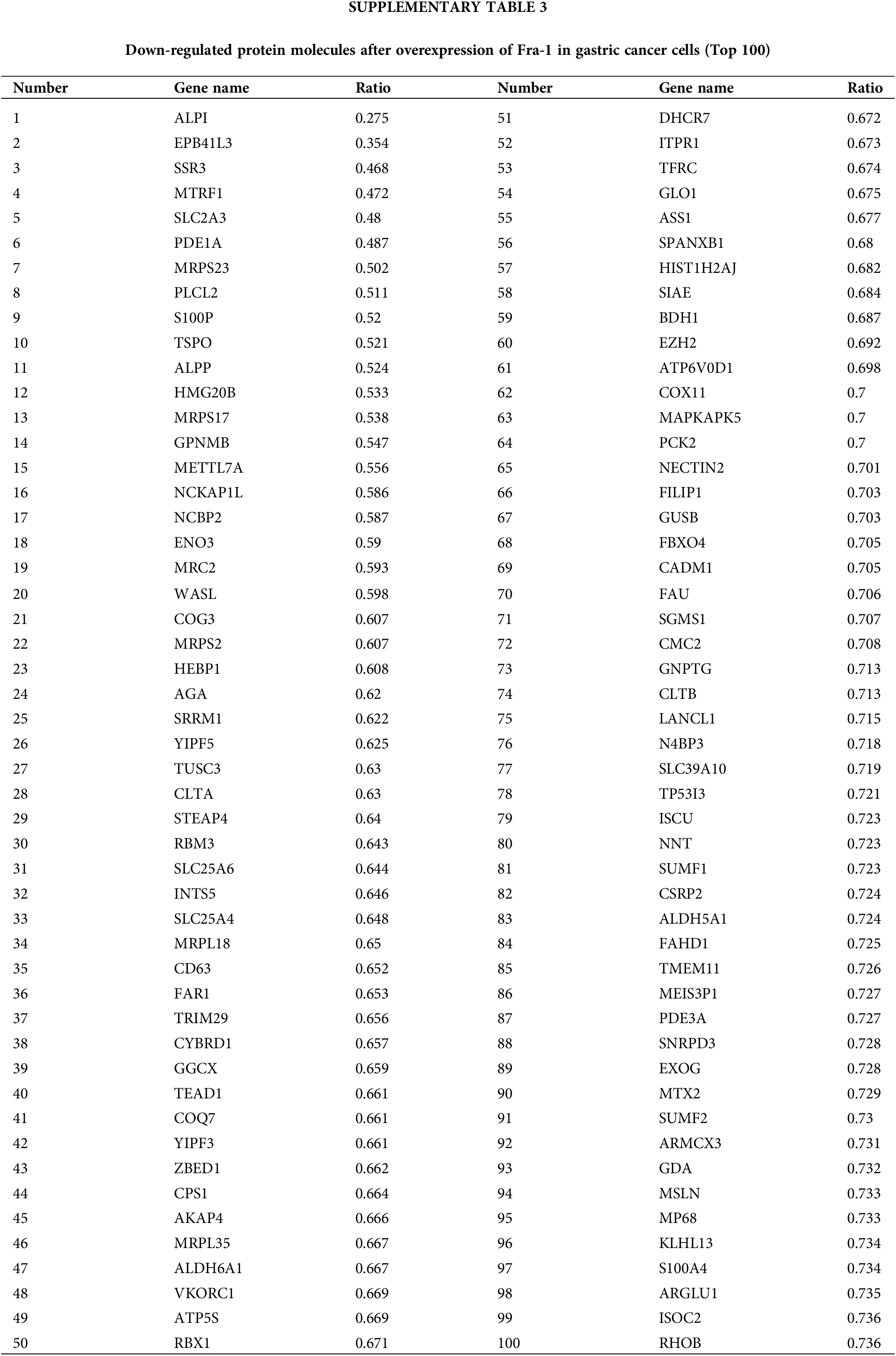
To explore the possible mechanism by which YWHAH regulates Fra-1 to affect the GC cell proliferation, the whole proteome was analyzed after vector (blank control) and Fra-1 plasmids were transfected into SGC7901 GC cells, separately. The results showed that 206 protein molecules were upregulated (Suppl. Table 2) and 146 proteins were downregulated after overexpression of Fra-1 in GC cells SGC7901 (Fig. 4A, Suppl. Table 3). Based on the analysis of these differentially abundant proteins, we performed functional classification, functional enrichment, and cluster analysis for all differentially abundant proteins. Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of upregulated proteins showed that the PI3K/AKT signaling pathway, the mitogen activated protein kinase (MAPK) signal transduction pathway, and the human papillomavirus infection signal transduction pathway were three main signal transduction pathways affected by Fra-1 (Fig. 4B). It has been reported that Fra-1 regulates the expression of HMGA1, and HMGA1 is a regulator of the PI3K/AKT/mTOR signaling pathway. Combined with our previous research results, we clarified whether YWHAH could promote the proliferation of GC cells by positively regulating Fra-1 to activate the HMGA1/PI3K/AKT/mTOR signaling pathway.
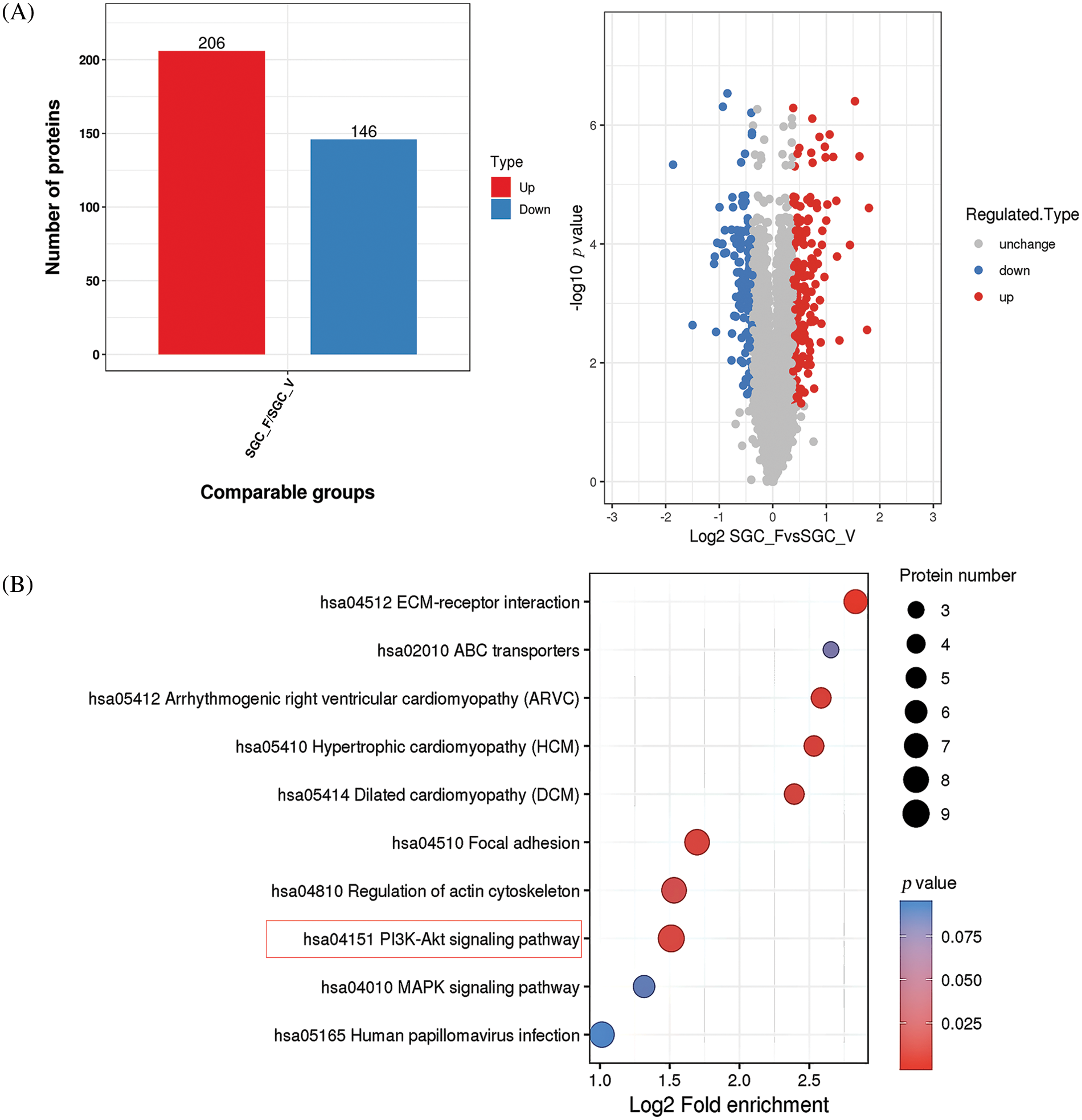
Figure 4: Whole proteome analysis suggests that Fra-1 affects the activity of the PI3K/AKT/mTOR signal pathway in GC cells. (A) Left: quantity distribution diagram of differentially abundant proteins between the Fra-1 overexpression group and the empty vector group of SGC7901 GC cells (column diagram); Right: quantitative volcano map of differentially abundant proteins between the Fra-1 overexpression group and empty vector group in SGC7901 GC cells. The upregulated differentially abundant proteins are represented by red dots in the map, and the downregulated differentially abundant proteins are represented by blue dots. (B) KEGG enrichment analysis of differentially abundant proteins: the vertical axis is the pathway, and the horizontal axis is the log2 converted value of the ratio of the differentially abundant protein in this functional type compared with the ratio of the identified protein. The color of the circle indicates the enrichment significance p value, and the size of the circle indicates the number of differentially abundant proteins in the functional class or pathway.
YWHAH activates the HMGA1/PI3K/AKT/mTOR signaling pathway by positively regulating Fra-1 to affect the proliferation of gastric cancer cells
To confirm whether YWHAH can affect the proliferation of GC cells by positively regulating Fra-1 to activate the HMGA1/PI3K/AKT/mTOR signaling pathway, we first transfected NC+siNC, OE-YWHAH+siNC, and OE-YWHAH+siFra-1 plasmids into SGC7901 GC cells, and detected the mRNA levels of PI3K/AKT/mTOR signal pathway related molecules (PI3K, AKT, PDK1, MTOR, etc.) using qRT-PCR. The results showed that the mRNA expression levels of PI3K, AKT, PDK1, and MTOR were upregulated after overexpression of YWHAH. However, after Fra-1 silencing in a background of YWHAH overexpression, upregulation of the mRNA expression levels of PI3K, AKT, PDK1, and MTOR molecules was inhibited (Fig. 5A). In addition, western blotting analysis also showed that the protein levels of PI3K and PDK1 were upregulated, as were the levels of phosphorylated (p)-AKT and p-mTOR after YWHAH overexpression. However, after Fra-1 silencing in a background of YWHAH overexpression, the upregulation of PI3K and PDK1 protein levels was inhibited, as were the upregulation of p-AKT and p-mTOR levels (Fig. 5B). The results suggest that YWHAH can activate the PI3K/AKT/mTOR signaling pathway through Fra-1.
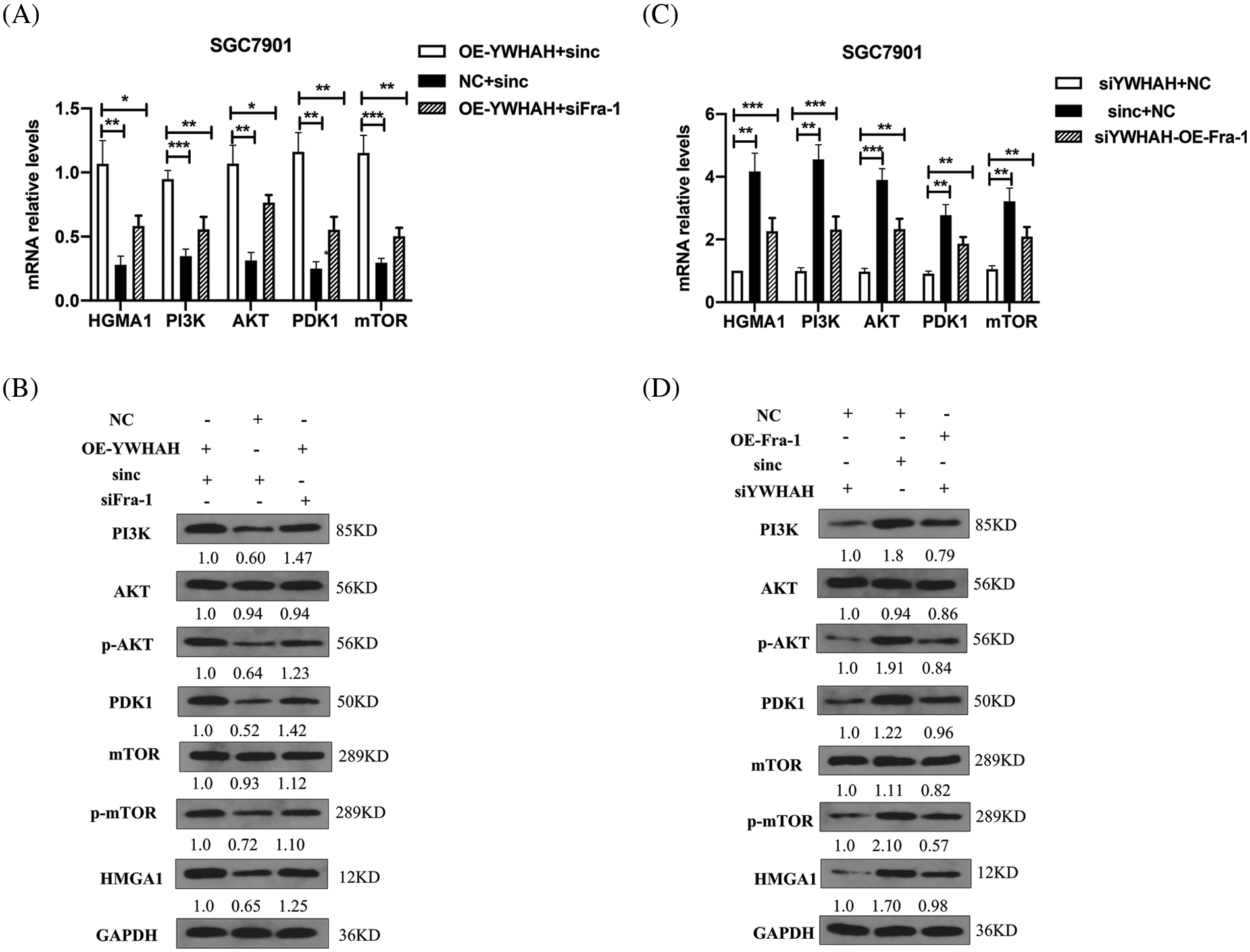
Figure 5: YWHAH regulates the activity of the HMGA1/PI3K/AKT/mTOR signaling pathway through Fra-1. (A and C) The mRNA levels of HMGA1/PI3K/AKT/mTOR signal pathway-related molecules were detected by qRT-PCR. (B and D) Western blotting detection of the protein level of HMGA1/PI3K/AKT/mTOR signaling pathway-related molecules. NS means: meaningless, *p < 0.05, **p < 0.01, ***p < 0.001, *** p < 0.0001. Internal reference: GAPDH, three independent repeated experiments.
At the same time, we silenced the expression of YWHAH, in which siNC+NC, siYWHAH+NC, and siYWHAH+OE-Fra-1 plasmids were transfected into SGC7901 GC cells, and the related molecules were detected using qRT-PCR. The results showed that the mRNA expression levels of PI3K, AKT, PDK1, and MTOR were downregulated after YWHAH silencing. However, overexpression of Fra-1 in a background of YWHAH silencing restored the mRNA expression levels of PI3K, AKT, PDK1, and MTOR (Fig. 5C). In addition, the protein levels of PI3K/AKT/mTOR signaling pathway-related molecules were detected by western blotting. The results showed that after YWHAH silencing, the protein levels of PI3K and PDK1 were downregulated, and the levels of p-AKT and p-mTOR were downregulated. Overexpression of Fra-1 in a background of YWHAH silencing restored the protein levels of PI3K and PDK1, as well as the levels of p-AKT and p-mTOR (Fig. 5D). Our results suggested that YWHAH can promote the proliferation of gastric cancer cells by activating the HMGA1/PI3K/AKT/mTOR signaling pathway through Fra-1.
To further confirm that YWHAH promotes gastric cancer cells proliferation by activating the PI3K/AKT/mTOR signaling pathway, we used rapamycin, an inhibitor of the PI3K/AKT/mTOR signaling pathway to treat gastric cancer cells. Firstly, we divided gastric cancer cells SGC7901 into three groups: OE-YWHAH+siNC+DMSO, OE-YWHAH+siFra-1+DMSO, OE-YWHAH+siFra-1+Rapamycin by transfecting the different plasmids and whether treating cells with rapamycin. Then, we tested the gastric cancer cells proliferation using an EDU proliferation detection kit and flow cytometry. Our results showed that compared to the OE-YWHAH+siNC+DMSO group, OE-YWHAH+siFra-1+DMSO group had a decrease in the proportion of proliferating cells, OE-YWHAH+siFra-1+Rapamycin group had a more significant reduction in the proportion of proliferating cells (Fig. 6A). To further confirm the effect of silencing YWHAH, we divided gastric cancer cells SGC7901 into three groups: siYWHAH+NC+DMSO, siYWHAH+OE-Fra-1+DMSO, siYWHAH+OE-Fra-1+Rapamycin by transfecting the different plasmids and whether treating cells with rapamycin. Then we tested the gastric cancer cells proliferation using an EDU proliferation detection kit and flow cytometry. Our results showed that compared to the siYWHAH+NC+DMSO group, the siYWHAH+OE-Fra-1+DMSO group had an increase in the proportion of proliferating cells. But, the siYWHAH+OE-Fra-1+Rapamycin group had a decrease in the proportion of proliferating cells (Fig. 6B). In AGS cells, we obtained results with the same trend (Figs. 6C and 6D). Above of all, our results suggested that YWHAH could promote the proliferation of gastric cancer cells by activating the PI3K/AKT/mTOR signaling pathway.
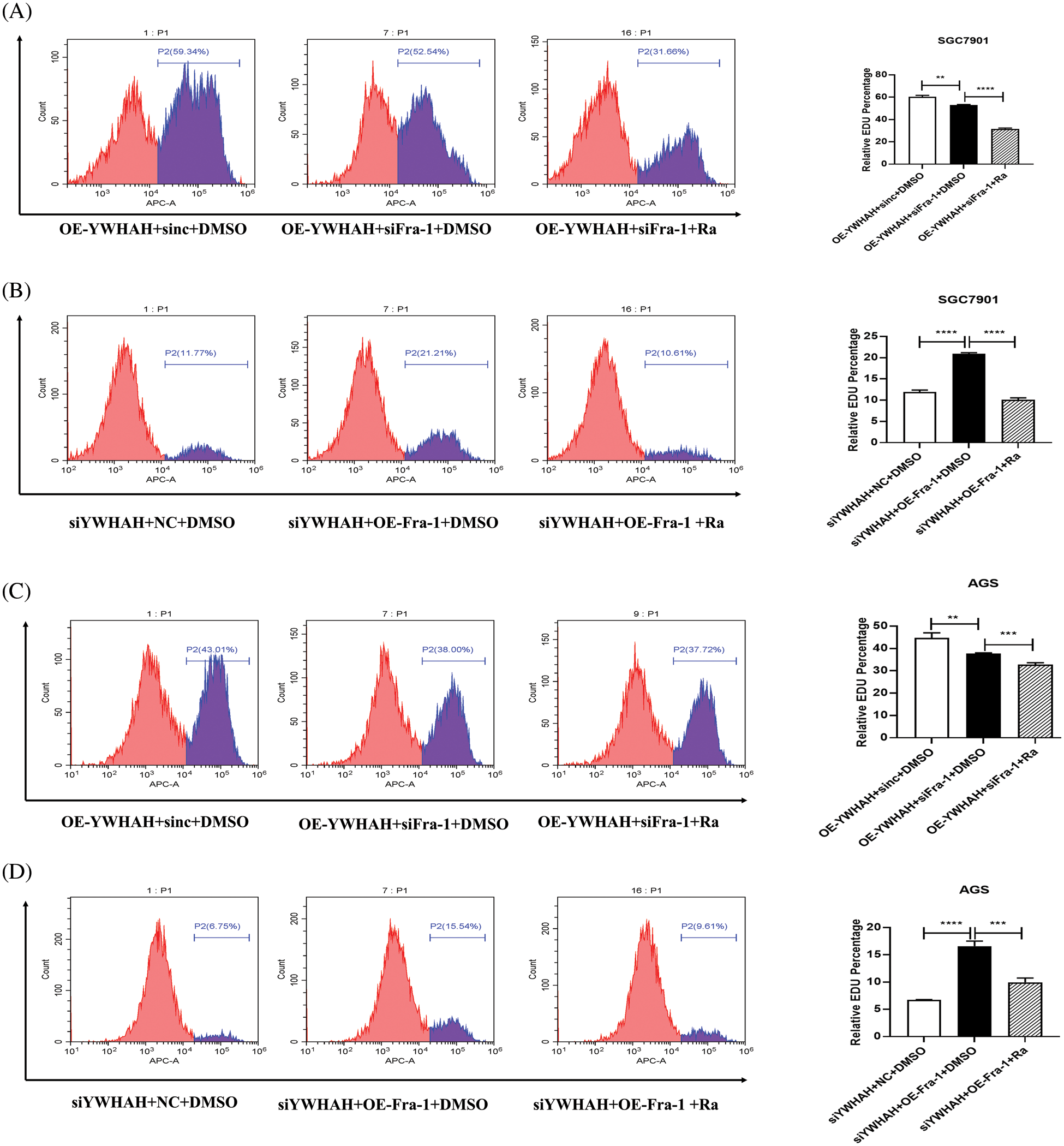
Figure 6: YWHAH promotes gastric cancer cell proliferation by activating the PI3K/AKT/mTOR signaling pathway. (A) Using EdU-647 cell proliferation assay combined with flow cytometry to detect the proliferation level of SGC7901 GC cells in three groups: OE-YWHAH+siNC+DMSO, OE-YWHAH+siFra-1+DMSO, OE-YWHAH+siFra-1+Rapamycin. (B) Using EdU-647 cell proliferation assay combined with flow cytometry to detect the proliferation level of SGC7901 GC cells in three groups: siYWHAH+NC+DMSO, siYWHAH+OE-Fra-1+DMSO, and siYWHAH+OE-Fra-1+Rapamycin. (C) Using EdU-647 cell proliferation assay combined with flow cytometry to detect the proliferation level of AGS GC cells in three groups: OE-YWHAH+siNC+DMSO, OE-YWHAH+siFra-1+DMSO, OE-YWHAH+siFra-1+Rapamycin. (D) Using EdU-647 cell proliferation assay combined with flow cytometry to detect the proliferation level of AGS GC cells in three groups: siYWHAH+NC+DMSO, siYWHAH+OE-Fra-1+DMSO, and siYWHAH+OE-Fra-1+Rapamycin. **p < 0.01, ***p < 0.001, ****p < 0.0001. Three independent experiments were conducted.
The pathogenesis of GC is complex. Although some progress has been made in its early diagnosis, surgical resection, and adjuvant chemotherapy, GC is still difficult to identify at the early stage of onset. The 5-year survival rate of patients with GC is not high. GC is one of the most aggressive cancers of the human digestive system [30–33]. Previous studies have shown that the pathogenesis of GC is closely related to Helicobacter pylori infection, proto oncogene activation, and tumor suppressor gene inactivation [6–8,12–14,34]; however, the detailed mechanism is unclear. More experimental evidence is needed to clarify its pathogenesis and potential molecular targets, to provide an experimental basis for early GC diagnosis, the identification of potential molecular therapeutic targets, and prognosis prediction.
Fra-1 is a member of the FOS family and is an important nuclear transcription factor that regulates the growth, differentiation, and apoptosis of normal cells [6,7,19–21]. Fra-1 is highly expressed in a variety of malignant tumors and plays an important role in cell transformation, proliferation, invasion, and metastasis [18–21,35]. Our previous study found that Fra-1 was highly expressed in GC tissues compared with that in adjacent non-cancerous tissues. In vitro experiments confirmed that Fra-1 overexpression inhibited GC cell apoptosis and increased the proportion of S-phase cells [6,36]. To further clarify the role and possible mechanism of Fra-1 in GC cell proliferation, we identified and confirmed YWHAH as a Fra-1-interacting protein in GC cells. Protein interaction is widely involved in cell-cell interactions, metabolic and developmental controls and other biological processes [37,38]. Protein interaction also plays a key role in predicting the function of target proteins [39,40]. We further studied the function of YWHAH, which provided a new perspective on the role and possible mechanism of Fra-1 in GC.
Based on the discovery that YWHAH interacts with Fra-1, further qRT-PCR and western blotting experiments showed that YWHAH positively regulates the transcription of Fra-1 and then affects the protein level of Fra-1. In addition, YWHAH overexpression in SGC7901 GC cells enhanced their proliferation. Compared with the vector group, cell proliferation was inhibited in the group transfected with YWHAH and siFra-1. The results of silencing YWHAH were opposite to those of YWHAH overexpression. These results suggested that YWHAH promotes GC cell proliferation by activating Fra-1. YWHAH is a 14-3-3 eta protein and a member of the 14-3-3 protein family [41,42]. A remarkable feature of 14-3-3 proteins is that they can bind to a variety of signal proteins with diverse functions, including kinases, phosphatases, and transmembrane receptors [43,44]. 14-3-3 proteins play an important role in biological processes, such as mitotic signal transduction, apoptotic cell death, and cell cycle control [45,46]. Wu et al. found that the miR-660-5p/YWHAH axis could activate the PI3K/AKT pathway to promote epithelial mesenchyme transition and the cell cycle process in hepatoma cells [47]. In thyroid cancer, the long noncoding RNA MAPKAPK5-AS1 promoted the proliferation and migration of thyroid cancer cell lines by targeting miR-519e-5p/YWHAH [48]. Our results provide new ideas and an experimental basis to clarify the role and possible mechanism of YWHAH in malignant tumors.
To further explore the specific mechanism by which YWHAH affects GC cell proliferation via Fra-1, we overexpressed Fra-1 in SGC7901 GC cells for whole proteome analysis. KEGG enrichment analysis of all differentially abundant proteins showed that the PI3K/AKT signaling pathway was abnormally activated. Then, we confirmed that YWHAH promoted GC cell proliferation by positively regulating Fra-1 to activate the HMGA1/PI3K/AKT/mTOR signaling pathway using western blotting, flow cytometry, and other technologies (Fig. 7). The PI3K/AKT signaling pathway plays an important role in basic intracellular functions, such as cell growth, apoptosis, translation, and cell metabolism [49–51]. It is important not only in carcinogenesis, but also in identifying potential new therapeutic targets [52–54]. The PI3K/AKT pathway is stimulated by receptor tyrosine kinases and by cytokine receptor activation. Tyrosine residues are subsequently phosphorylated and provide anchor sites for PI3K translocation to the membrane, thus participating in the transduction of various extracellular matrix molecules and cytokines [55,56]. Other studies have shown that changes in the activity of the PI3K/AKT/mTOR signaling pathway affect the proliferation of tumor cells [57–59]. Our results also confirmed that activation of the PI3K/AKT/mTOR signaling pathway promoted GC cell proliferation, which was consistent with the above related research results.
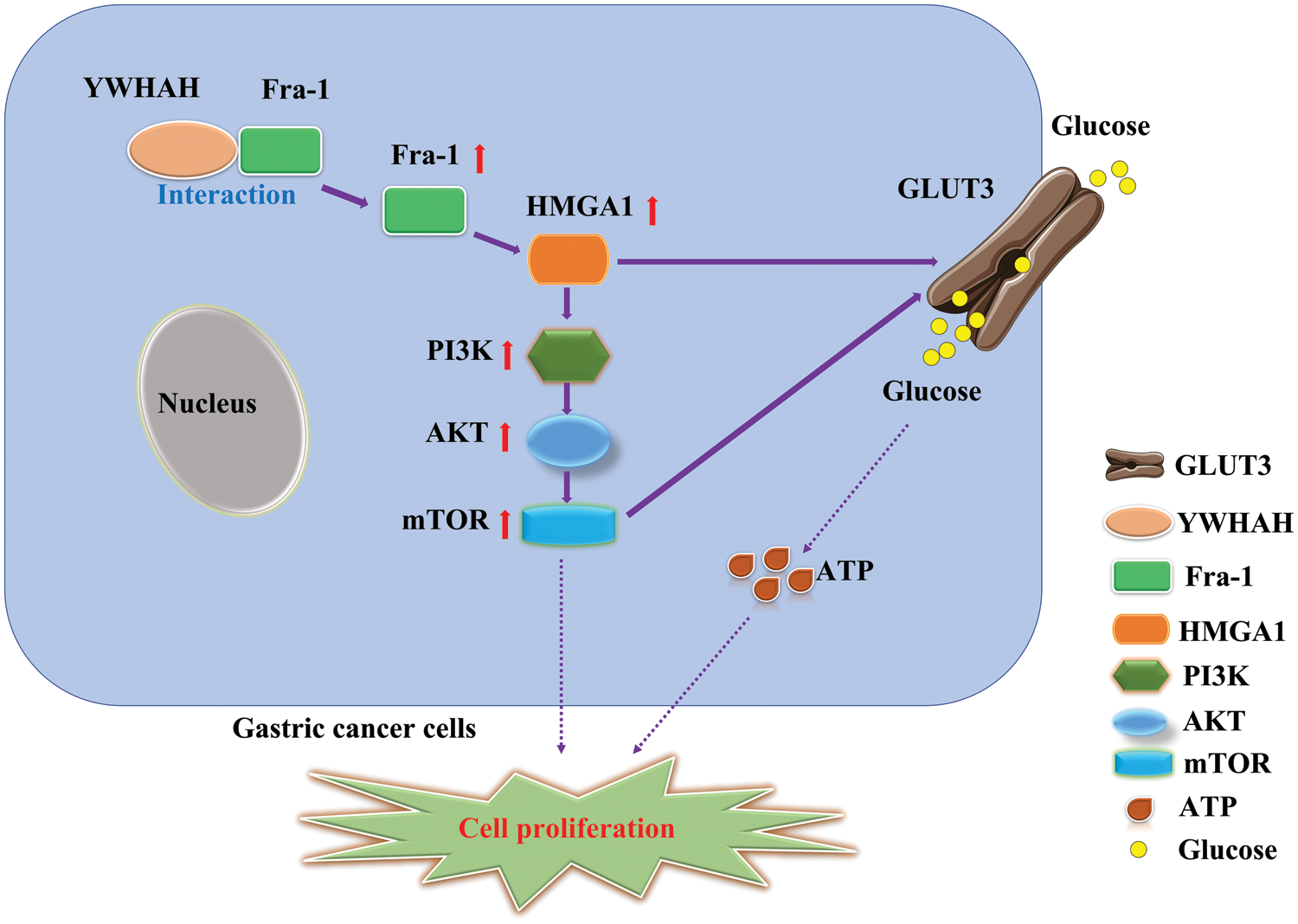
Figure 7: Schematic diagram of the effect of YWHAH interacting with Fra-1 on the proliferation of gastric cancer cells.
The experimental results provide new molecular targets to explore the pathogenesis of GC and provide an experimental basis to clarify the pathogenesis of GC.
Funding Statement: This work was supported by the Hunan Provincial Natural Science Foundation (2021JJ30915).
Author Contributions: JH, FZ and XJ performed the experiments. LL, MG and WL analyzed the data. GL and YZ designed the study and wrote the manuscript. All authors read and approved the final manuscript.
Availability of Data and Materials: The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Thrift, A. P., Nguyen, T. H. (2021). Gastric cancer epidemiology. Gastrointestinal Endoscopy Clinics of North America, 31(3), 425–439. https://doi.org/10.1016/j.giec.2021.03.001 [Google Scholar] [PubMed] [CrossRef]
2. Godeau, D., Petit, A., Richard, I., Roquelaure, Y., Descatha, A. (2021). Return-to-work, disabilities and occupational health in the age of COVID-19. Scandinavian Journal of Work Environment & Health, 47(5), 408–409. https://doi.org/10.5271/sjweh.3960 [Google Scholar] [PubMed] [CrossRef]
3. Petryszyn, P., Chapelle, N., Matysiak-Budnik, T. (2020). Gastric cancer: Where are we heading? Digestive Diseases, 38(4), 280–285. https://doi.org/10.1159/000506509 [Google Scholar] [PubMed] [CrossRef]
4. Zeng, Y., Jin, R. U. (2022). Molecular pathogenesis, targeted therapies, and future perspectives for gastric cancer. Seminars in Cancer Biology, 86(2), 566–582. https://doi.org/10.1016/j.semcancer.2021.12.004 [Google Scholar] [PubMed] [CrossRef]
5. Chen, Q., Cheng, C., Liu, Y., Guo, L., Xu, H. et al. (2022). Incidence and mortality of gastric cancer in 2018 and their trends from 2010 to 2018 in Henan province, China: Results from a provincial population-based cancer registry. Annals of Translational Medicine, 10(18), 1012. https://doi.org/10.21037/atm-22-4100 [Google Scholar] [PubMed] [CrossRef]
6. He, J., Zhu, G., Gao, L., Chen, P., Long, Y. et al. (2015). Fra-1 is upregulated in gastric cancer tissues and affects the PI3K/Akt and p53 signaling pathway in gastric cancer. International Journal of Oncology, 47(5), 1725–1734. https://doi.org/10.3892/ijo.2015.3146 [Google Scholar] [PubMed] [CrossRef]
7. Zeng, F., He, J., Jin, X., Liao, Q., Chen, Z. et al. (2022). FRA-1: A key factor regulating signal transduction of tumor cells and a potential target molecule for tumor therapy. Biomedicine & Pharmacotherapy, 150(1), 113037. https://doi.org/10.1016/j.biopha.2022.113037 [Google Scholar] [PubMed] [CrossRef]
8. Ba, M., Long, H., Yan, Z., Wang, S., Wu, Y. et al. (2018). BRD4 promotes gastric cancer progression through the transcriptional and epigenetic regulation of c-MYC. Journal of Cellular Biochemistry, 119(1), 973–982. https://doi.org/10.1002/jcb.26264 [Google Scholar] [PubMed] [CrossRef]
9. Zhu, G. C., Gao, L., He, J., Long, Y., Liao, S. et al. (2015). CD90 is upregulated in gastric cancer tissues and inhibits gastric cancer cell apoptosis by modulating the expression level of SPARC protein. Oncology Reports, 34(5), 2497–2506. https://doi.org/10.3892/or.2015.4243 [Google Scholar] [PubMed] [CrossRef]
10. Gao, L., Li, J., He, J., Liang, L., He, Z. et al. (2021). CD90 affects the biological behavior and energy metabolism level of gastric cancer cells by targeting the PI3K/AKT/HIF-1α signaling pathway. Oncology Letters, 21(3), 191. https://doi.org/10.3892/ol.2021.12451 [Google Scholar] [PubMed] [CrossRef]
11. Nie, K., Zheng, Z., Wen, Y., Shi, L., Xu, S. et al. (2020). Construction and validation of a TP53-associated immune prognostic model for gastric cancer. Genomics, 112(6), 4788–4795. https://doi.org/10.1016/j.ygeno.2020.08.026 [Google Scholar] [PubMed] [CrossRef]
12. Baj, J., Korona-Głowniak, I., Forma, A., Maani, A., Sitarz, E. et al. (2020). Mechanisms of the epithelial-mesenchymal transition and tumor microenvironment in helicobacter pylori-induced gastric cancer. Cells, 9(4), 1055. https://doi.org/10.3390/cells9041055 [Google Scholar] [PubMed] [CrossRef]
13. Ford, A. C., Yuan, Y., Moayyedi, P. (2020). Helicobacter pylori eradication therapy to prevent gastric cancer: systematic review and meta-analysis. Gut, 69(12), 2113–2121. https://doi.org/10.1136/gutjnl-2020-320839 [Google Scholar] [PubMed] [CrossRef]
14. Li, W. Q., Zhang, J. Y., Ma, J. L., Li, Z. X., Zhang, L. et al. (2019). Effects of Helicobacter pylori treatment and vitamin and garlic supplementation on gastric cancer incidence and mortality: Follow-up of a randomized intervention trial. BMJ (Clinical Research Edition), 366, l5016. https://doi.org/10.1136/bmj.l5016 [Google Scholar] [PubMed] [CrossRef]
15. Sokolova, O., Naumann, M. (2022). Matrix metalloproteinases in helicobacter pylori-associated gastritis and gastric cancer. International Journal of Molecular Sciences, 23(3), 1883. https://doi.org/10.3390/ijms23031883 [Google Scholar] [PubMed] [CrossRef]
16. Yan, L., Chen, Y., Chen, F., Tao, T., Hu, Z. et al. (2022). Effect of helicobacter pylori eradication on gastric cancer prevention: Updated report from a randomized controlled trial with 26.5 years of follow-up. Gastroenterology, 163(1), 154–162 e153. https://doi.org/10.1053/j.gastro.2022.03.039 [Google Scholar] [PubMed] [CrossRef]
17. Talotta, F., Casalino, L., Verde, P. (2020). The nuclear oncoprotein Fra-1: A transcription factor knocking on therapeutic applications’ door. Oncogene, 39(23), 4491–4506. https://doi.org/10.1038/s41388-020-1306-4 [Google Scholar] [PubMed] [CrossRef]
18. Casalino, L., Talotta, F., Cimmino, A., Verde, P. (2022). The Fra-1/AP-1 oncoprotein: From the “Undruggable” transcription factor to therapeutic targeting. Cancers, 14(6), 1480. https://doi.org/10.3390/cancers14061480 [Google Scholar] [PubMed] [CrossRef]
19. Hoang, V. T., Matossian, M. D., La, J., Hoang, K., Ucar, D. A. et al. (2021). Dual inhibition of MEK1/2 and MEK5 suppresses the EMT/migration axis in triple-negative breast cancer through FRA-1 regulation. Journal of Cellular Biochemistry, 122(8), 835–850. https://doi.org/10.1002/jcb.29916 [Google Scholar] [PubMed] [CrossRef]
20. Xu, H., Jin, X., Yuan, Y., Deng, P., Jiang, L. et al. (2017). Prognostic value from integrative analysis of transcription factors c-Jun and Fra-1 in oral squamous cell carcinoma: A multicenter cohort study. Scientific Reports, 7(1), 7522. https://doi.org/10.1038/s41598-017-05106-5 [Google Scholar] [PubMed] [CrossRef]
21. Toyozumi, T., Hoshino, I., Takahashi, M., Usui, A., Akutsu, Y. et al. (2017). Fra-1 regulates the expression of HMGA1, which is associated with a poor prognosis in human esophageal squamous cell carcinoma. Annals of Surgical Oncology, 24(11), 3446–3455. https://doi.org/10.1245/s10434-016-5666-5 [Google Scholar] [PubMed] [CrossRef]
22. Chen, Y., Dong, Y., Zhang, Z. L., Han, J., Chen, F. S. et al. (2023). Fra-1 induces apoptosis and neuroinflammation by targeting S100A8 to modulate TLR4 pathways in spinal cord ischemia/reperfusion injury. Brain Pathology, 33(1), e13113. https://doi.org/10.1111/bpa.13113 [Google Scholar] [PubMed] [CrossRef]
23. Kustikova, O., Kramerov, D., Grigorian, M., Berezin, V., Bock, E. et al. (1998). Fra-1 induces morphological transformation and increases in vitro invasiveness and motility of epithelioid adenocarcinoma cells. Molecular and Cellular Biology, 18(12), 7095–7105. https://doi.org/10.1128/MCB.18.12.7095 [Google Scholar] [PubMed] [CrossRef]
24. Lombaerts, M., van Wezel, T., Philippo, K., Dierssen, J. W., Zimmerman, R. M. et al. (2006). E-cadherin transcriptional downregulation by promoter methylation but not mutation is related to epithelial-to-mesenchymal transition in breast cancer cell lines. British Journal of Cancer, 94(5), 661–671. https://doi.org/10.1038/sj.bjc.6602996 [Google Scholar] [PubMed] [CrossRef]
25. Munmun, F., Mohiuddin, O. A., Hoang, V. T., Burow, M. E., Bunnell, B. A. et al. (2022). The role of MEK1/2 and MEK5 in melatonin-mediated actions on osteoblastogenesis, osteoclastogenesis, bone microarchitecture, biomechanics, and bone formation. Journal of Pineal Research, 73(2), e12814. https://doi.org/10.1111/jpi.12814 [Google Scholar] [PubMed] [CrossRef]
26. Casalino, L., Bakiri, L., Talotta, F., Weitzman, J. B., Fusco, A. et al. (2007). Fra-1 promotes growth and survival in RAS-transformed thyroid cells by controlling cyclin A transcription. Embo Journal, 26(7), 1878–1890. https://doi.org/10.1038/sj.emboj.7601617 [Google Scholar] [PubMed] [CrossRef]
27. Zhao, X., Miao, G., Zhang, L., Zhang, Y., Zhao, H. et al. (2022). Chlamydia pneumoniae infection induces vascular smooth muscle cell migration and atherosclerosis through mitochondrial reactive oxygen species-mediated JunB-Fra-1 activation. Frontiers in Cell and Developmental Biology, 10, 879023. https://doi.org/10.3389/fcell.2022.879023 [Google Scholar] [PubMed] [CrossRef]
28. He, T. G., Xiao, Z. Y., Xing, Y. Q., Yang, H. J., Qiu, H. et al. (2019). Tumor suppressor miR-184 enhances chemosensitivity by directly inhibiting SLC7A5 in retinoblastoma. Frontiers in Oncology, 9, 1163. https://doi.org/10.3389/fonc.2019.01163 [Google Scholar] [PubMed] [CrossRef]
29. Liao, S., Xiao, S., Chen, H., Zhang, M., Chen, Z. et al. (2017). The receptor for activated protein kinase C promotes cell growth, invasion and migration in cervical cancer. International Journal of Oncology, 51(5), 1497–1507. https://doi.org/10.3892/ijo.2017.4137 [Google Scholar] [PubMed] [CrossRef]
30. Joshi, S. S., Badgwell, B. D. (2021). Current treatment and recent progress in gastric cancer. CA: A Cancer Journal for Clinicians, 71(3), 264–279. https://doi.org/10.3322/caac.21657 [Google Scholar] [PubMed] [CrossRef]
31. Seeneevassen, L., Bessède, E., Mégraud, F., Lehours, P., Dubus, P. et al. (2021). Gastric cancer: Advances in carcinogenesis research and new therapeutic strategies. International Journal of Molecular Sciences, 22(7), 3418. https://doi.org/10.3390/ijms22073418 [Google Scholar] [PubMed] [CrossRef]
32. Necula, L., Matei, L., Dragu, D., Neagu, A. I., Mambet, C. et al. (2019). Recent advances in gastric cancer early diagnosis. World Journal of Gastroenterology, 25(17), 2029–2044. https://doi.org/10.3748/wjg.v25.i17.2029 [Google Scholar] [PubMed] [CrossRef]
33. Cao, F., Hu, C., Xu, Z. Y., Zhang, Y. Q., Huang, L. et al. (2022). Current treatments and outlook in adenocarcinoma of the esophagogastric junction: A narrative review. Annals of Translational Medicine, 10(6), 377. https://doi.org/10.21037/atm-22-1064 [Google Scholar] [PubMed] [CrossRef]
34. Yu, Z., Lan, J., Li, W., Jin, L., Qi, F. et al. (2022). Circular RNA hsa_circ_0002360 promotes proliferation and invasion and inhibits oxidative stress in gastric cancer by sponging miR-629-3p and regulating the PDLIM4 expression. Oxidative Medicine and Cellular Longevity, 2022(2), 2775433. https://doi.org/10.1155/2022/2775433 [Google Scholar] [PubMed] [CrossRef]
35. Ge, X., Lin, F., Wu, Z., Lin, Y., Tang, W. et al. (2022). Role of ROR2 in promoting gastric cancer metastasis by enhancing c-JUN-mediated MMP3 transcription. Annals of Translational Medicine, 10(20), 1117. https://doi.org/10.21037/atm-22-4583 [Google Scholar] [PubMed] [CrossRef]
36. He, Y. Y., Zhou, H. F., Chen, L., Wang, Y. T., Xie, W. L. et al. (2022). The Fra-1: Novel role in regulating extensive immune cell states and affecting inflammatory diseases. Frontiers in Immunology, 13, 954744. https://doi.org/10.3389/fimmu.2022.954744 [Google Scholar] [PubMed] [CrossRef]
37. Rabbani, G., Baig, M. H., Ahmad, K., Choi, I. (2018). Protein-protein interactions and their role in various diseases and their prediction techniques. Current Protein & Peptide Science, 19(10), 948–957. https://doi.org/10.2174/1389203718666170828122927 [Google Scholar] [PubMed] [CrossRef]
38. Liu, Q., Zheng, J., Sun, W., Huo, Y., Zhang, L. et al. (2018). A proximity-tagging system to identify membrane protein-protein interactions. Nature Methods, 15(9), 715–722. https://doi.org/10.1038/s41592-018-0100-5 [Google Scholar] [PubMed] [CrossRef]
39. Tanwar, H., George Priya Doss, C. (2018). Computational resources for predicting protein-protein interactions. Advances in Protein Chemistry and Structural Biology, 110(2), 251–275. https://doi.org/10.1016/bs.apcsb.2017.07.006 [Google Scholar] [PubMed] [CrossRef]
40. Athanasios, A., Charalampos, V., Vasileios, T., Ashraf, G. M. (2017). Protein-Protein Interaction (PPI) network: Recent advances in drug discovery. Current Drug Metabolism, 18(1), 5–10. https://doi.org/10.2174/138920021801170119204832 [Google Scholar] [PubMed] [CrossRef]
41. Wei, J. C., Leong, P. Y., Liu, G. Y. (2020). Chaperone/scaffolding/adaptor protein 14-3-3η (etaA diagnostic marker of rheumatoid arthritis. International Journal of Rheumatic Diseases, 23(11), 1439–1442. https://doi.org/10.1111/1756-185X.14004 [Google Scholar] [PubMed] [CrossRef]
42. Khorrami, A., Sharif Bagheri, M., Tavallaei, M., Gharechahi, J. (2017). The functional significance of 14-3-3 proteins in cancer: Focus on lung cancer. Hormone Molecular Biology and Clinical Investigation, 32(3), 1. https://doi.org/10.1515/hmbci-2017-0032 [Google Scholar] [PubMed] [CrossRef]
43. Stevers, L. M., Sijbesma, E., Botta, M., MacKintosh, C., Obsil, T. et al. (2018). Modulators of 14-3-3 protein-protein interactions. Journal of Medicinal Chemistry, 61(9), 3755–3778. https://doi.org/10.1021/acs.jmedchem.7b00574 [Google Scholar] [PubMed] [CrossRef]
44. Thompson, W. C., Goldspink, P. H. (2022). 14-3-3 protein regulation of excitation-contraction coupling. Pflugers Archiv-European Journal of Physiology, 474(3), 267–279. https://doi.org/10.1007/s00424-021-02635-x [Google Scholar] [PubMed] [CrossRef]
45. Yuan, R., Vos, H. R., van Es, R. M., Chen, J., Burgering, B. M. et al. (2018). Chk1 and 14-3-3 proteins inhibit atypical E2Fs to prevent a permanent cell cycle arrest. The EMBO Journal, 37(5), 2108. https://doi.org/10.15252/embj.201797877 [Google Scholar] [PubMed] [CrossRef]
46. Liu, J., Cao, S., Ding, G., Wang, B., Li, Y. et al. (2021). The role of 14-3-3 proteins in cell signalling pathways and virus infection. Journal of Cellular and Molecular Medicine, 25(9), 4173–4182. https://doi.org/10.1111/jcmm.16490 [Google Scholar] [PubMed] [CrossRef]
47. Wu, Y., Zhang, Y., Wang, F., Ni, Q., Li, M. (2020). MiR-660-5p promotes the progression of hepatocellular carcinoma by interaction with YWHAH via PI3K/Akt signaling pathway. Biochemical and Biophysical Research Communications, 531(4), 480–489. https://doi.org/10.1016/j.bbrc.2020.07.034 [Google Scholar] [PubMed] [CrossRef]
48. Zhou, Y., Liu, S., Luo, Y., Zhang, M., Jiang, X. et al. (2020). IncRNA MAPKAPK5-AS1 promotes proliferation and migration of thyroid cancer cell lines by targeting miR-519e-5p/YWHAH. European Journal of Histochemistry, 64(4), 3177. https://doi.org/10.4081/ejh.2020.3177 [Google Scholar] [PubMed] [CrossRef]
49. Hamzehzadeh, L., Atkin, S. L., Majeed, M., Butler, A. E., Sahebkar, A. (2018). The versatile role of curcumin in cancer prevention and treatment: A focus on PI3K/AKT pathway. Journal of Cellular Physiology, 233(10), 6530–6537. https://doi.org/10.1002/jcp.26620 [Google Scholar] [PubMed] [CrossRef]
50. Sheng, T., Mao, X. B., Zhang, S. H. (2020). CaMKKβ regulates proliferation, apoptosis, and glycolysis of hepatocellular carcinoma via PI3K/AKT pathway. Annals of Palliative Medicine, 9(6), 3857–3869. https://doi.org/10.21037/apm-20-1789 [Google Scholar] [PubMed] [CrossRef]
51. Hoxhaj, G., Manning, B. D. (2020). The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nature Reviews Cancer, 20(2), 74–88. https://doi.org/10.1038/s41568-019-0216-7 [Google Scholar] [PubMed] [CrossRef]
52. Evangelisti, C., Chiarini, F., Cappellini, A., Paganelli, F., Fini, M. et al. (2020). Targeting Wnt/β-catenin and PI3K/Akt/mTOR pathways in T-cell acute lymphoblastic leukemia. Journal of Cellular Physiology, 235(6), 5413–5428. https://doi.org/10.1002/jcp.29429 [Google Scholar] [PubMed] [CrossRef]
53. Liu, X., Sun, L., Zhang, S., Zhang, S., Li, W. (2020). GINS2 facilitates epithelial-to-mesenchymal transition in non-small-cell lung cancer through modulating PI3K/Akt and MEK/ERK signaling. Journal of Cellular Physiology, 235(11), 7747–7756. https://doi.org/10.1002/jcp.29381 [Google Scholar] [PubMed] [CrossRef]
54. Yang, Y., Zhang, J., Li, J. Y., Xu, L., Wang, S. N. et al. (2022). The ctDNA-based postoperative molecular residual disease status in different subtypes of early-stage breast cancer. Gland Surgery, 11(12), 1924–1935. https://doi.org/10.21037/gs-22-634 [Google Scholar] [PubMed] [CrossRef]
55. Davidson, C. D., Bolf, E. L., Gillis, N. E., Cozzens, L. M., Tomczak, J. A. et al. (2021). Thyroid hormone receptor beta inhibits PI3K-Akt-mTOR signaling axis in anaplastic thyroid cancer via genomic mechanisms. Journal of the Endocrine Society, 5(8), bvab102. https://doi.org/10.1210/jendso/bvab102 [Google Scholar] [PubMed] [CrossRef]
56. Han, S., Wang, P. F., Cai, H. Q., Wan, J. H., Li, S. W. et al. (2021). Alterations in the RTK/Ras/PI3K/AKT pathway serve as potential biomarkers for immunotherapy outcome of diffuse gliomas. Aging, 13(11), 15444–15458. https://doi.org/10.18632/aging.203102 [Google Scholar] [PubMed] [CrossRef]
57. Colín-Val, Z., López-Díazguerrero, N. E., López-Marure, R. (2021). DHEA inhibits proliferation, migration and alters mesenchymal-epithelial transition proteins through the PI3K/Akt pathway in MDA-MB-231 cells. Journal of Steroid Biochemistry and Molecular Biology, 208(109), 105818. https://doi.org/10.1016/j.jsbmb.2021.105818 [Google Scholar] [PubMed] [CrossRef]
58. Huang, X., Xu, X., Ke, H., Pan, X., Ai, J. et al. (2022). microRNA-16-5p suppresses cell proliferation and angiogenesis in colorectal cancer by negatively regulating forkhead box K1 to block the PI3K/Akt/mTOR pathway. European Journal of Histochemistry, 66(2), 3333. https://doi.org/10.4081/ejh.2022.3333 [Google Scholar] [PubMed] [CrossRef]
59. Kumar, S., Agnihotri, N. (2019). Piperlongumine, a piper alkaloid targets Ras/PI3K/Akt/mTOR signaling axis to inhibit tumor cell growth and proliferation in DMH/DSS induced experimental colon cancer. Biomedicine & Pharmacotherapy, 109, 1462–1477. https://doi.org/10.1016/j.biopha.2018.10.182 [Google Scholar] [PubMed] [CrossRef]

Supplementary Figure 1: Identification of gene knockout effect of YWHAH siRNA sequence. Three siRNA sequences were designed for the YWHAH gene and transiently transfected into SGC7901 cells. The knockout effects of the three YWHAH siRNA sequences were detected by western blot. The siRNA sequences of si-YWHAH-01, si-YWHHAH-02, and si-YWHHAH-03 were shown from left to right in the figure. Western blot analysis showed that si-YWHAH-01 had the best effect on reducing YWHAH protein level.
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools