 Open Access
Open Access
ARTICLE
Apolipoprotein C1 promotes tumor progression in gastric cancer
Department of General Surgery, The Second Affiliated Hospital of Nanjing Medical University, Nanjing, 210009, China
* Corresponding Author: Jianping Zhang,
(This article belongs to the Special Issue: Transcriptome Analysis in Tumor Microenvironment and Tumor Heterogeneity)
Oncology Research 2023, 31(3), 287-297. https://doi.org/10.32604/or.2023.028124
Received 30 November 2022; Accepted 15 February 2023; Issue published 22 May 2023
Abstract
Background: Gastric cancer (GC) is a malignancy with the worst prognosis that seriously threatens human health, especially in East Asia. Apolipoprotein C1 (apoc1) belongs to the apolipoprotein family. In addition, apoc1 has been associated with various tumors. However, its role in GC remains unclear. Methods: Firstly, we quantified its expression in GC and adjacent tumor tissues, using The Cancer Genome Atlas (TCGA). Next, we assessed cell invasion and migration abilities. Finally, we revealed the role of apoc1 in the tumor microenvironment (TME), immune cell infiltration and drug sensitivity. Results: Firstly, in TCGA database, it has been shown that elevated expression of apoc1 was identified in various cancers, including GC, then we found that high expression of apoc1 was significantly correlated with poor prognosis in GC. Histologically, apoc1 expression is proportional to grade, cancer stage, and T stage. The experimental results showed that apoc1 promoted cell invasion and migration. Then GO, KEGG, and GSEA pathway analyses indicated that apoc1 may be involved in the WNT pathway and immune regulation. Furthermore, we found out the tumor-infiltrating immune cells related to apoc1 in the tumor microenvironment (TME) using TIMER. Finally, we investigated the correlation between apoc1 expression and drug sensitivity, PD-1 and CTLA-4 therapy. Conclusions: These results suggest that apoc1 participates in the evolution of GC, and may represent a potential target for detection and immunotherapy in GC.Keywords
Gastric cancer (GC) threatens human health, especially in Asia. In 2020, GC was ranked fifth among new tumor cases and the leading cause of cancer death [1]. Risk factors for GC include smoking, Helicobacter pylori infection, alcohol consumption, high-salt diet, and familial inheritance [2]. Various kinds of radical gastric resection + lymph node dissection, combined with perioperative chemo-radiotherapy are currently commonly used therapies, and perioperative targeted drugs and immune checkpoint inhibitors are also potential therapies [3]. However, the therapeutic effects of GC have not progressed significantly. The median survival of patients with advanced GC is approximately 7–9 months, with a 2-year survival rate of only 10% [4]. Therefore, identifying new diagnostic and therapeutic targets is essential [5].
Apolipoprotein C1 (apoc1) belongs to the apolipoprotein family and has the smallest molecular weight among all the apolipoproteins. It participates in several biochemical processes including lipid transport and metabolism [6]. For example, it has been revealed that apoc1 may induce glomerulosclerosis (GS) by enhancing the cytokine response of macrophages, suggesting that apoc1 can serve as a promising new therapeutic target in GS [7]. In addition, apoc1 contributes to the non-invasive diagnosis of mycoplasma pneumonia in adolescents [8]. Gene polymorphisms of apoc1 are also associated with late-onset Alzheimer’s disease (LOAD) in China [9].
apoc1 also plays an integral role in tumor metastasis. It is distinctive from other members of the apolipoprotein family as it is a major driver of various aggressive behaviors in tumors, especially immune-regulatory processes, invasiveness, and metastasis. For example, high expression of apoc1 promotes the invasion and metastasis of breast cancer via the Epithelial Mesenchymal Transition (EMT) and JNK/MAPK pathways [10]. apoc1 is also upregulated in T4 and T3 compared with T2 in colorectal cancer (CRC) and is highly associated with immune cell infiltration, indicating that it can serve as an indicator of metastasis in CRC [11]. Similarly, apoc1 promotes renal cell carcinoma (RCC) proliferation, invasion, and metastasis by directly targeting Wnt family member 3A (wnt3a) [12]. In hepatocellular carcinoma (HCC), single-cell sequencing results have shown that apoc1 promotes the transformation of M2 macrophages into M1 macrophages, thus promoting the metastasis of HCC, reshaping the tumor immune microenvironment, and improving the immunotherapy effect of anti-PD-1 against HCC [13]. In GC, znf460 promotes apoc1 transcription by binding to its promoter, thus promoting GC metastasis through the EMT pathway. Thus, it can serve as a novel biomarker [14,15]. However, its role and specific mechanisms in GC progression remain unclear.
Gene profile and clinical information of GC and adjacent-tumor tissues
We downloaded data we need from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov), which includes 343 GC and 30 tumor-adjacent tissues. Expression data were converted using the R software (version 4.2.1) for subsequent analysis.
Analysis between clinical information and apoc1
After downloading clinical information, we evaluated the relationship between them and apoc1. Clinical information included age, sex, grade, and stage, T, N, M, and apoc1. After comparison, a relevant heat map was drawn based on previous results. “Limma” and “ggpubr” packages were included.
GC cell lines (AGS, MKN45) were purchased from the Type Culture Collection of the Chinese Academy of Science (Shanghai, China) and fed MKN45 into RPMI 1640 (Servicebio, Wuhan, China). AGS cells were cultured in F12K (Servicebio, Wuhan, China). All media were supplemented with fetal bovine serum (FBS) (Gibco, NY, USA) and a penicillin-streptomycin mixture. The cells were cultured in incubator with 5% CO2 at 37°C.
apoc1 protein expression was detected in GC and adjacent tissues by IHC. Formalin-fixed paraffin-embedded tissues sections were degreased in xylene and rehydrated in different concentrations of alcohol and distilled water before antigen retrieval, then followed by antigen retrieval. The fixed sections were washed three times in phosphate buffered saline (PBS) (pH 7.3), incubated in 3% H2O2 for 7 min, and then washed three times with PBS again. The sections were blocked with BSA (Servicebio, Wuhan, China) for 30 min, followed by incubation with Anti-apoc1 primary antibody (Abcam, USA) at 4°C overnight, followed by incubation with anti-rabbit antibodies (Proteintech Technology, Wuhan, China) for 50 min. Subsequently, the sections were incubated with diaminobenzidine as a chromogen for 5 min. The sections were then counterstained with hematoxylin and dehydrated using ethanol and xylene. The sections were viewed under an inverted microscope (Nikon Corporation, Japan).
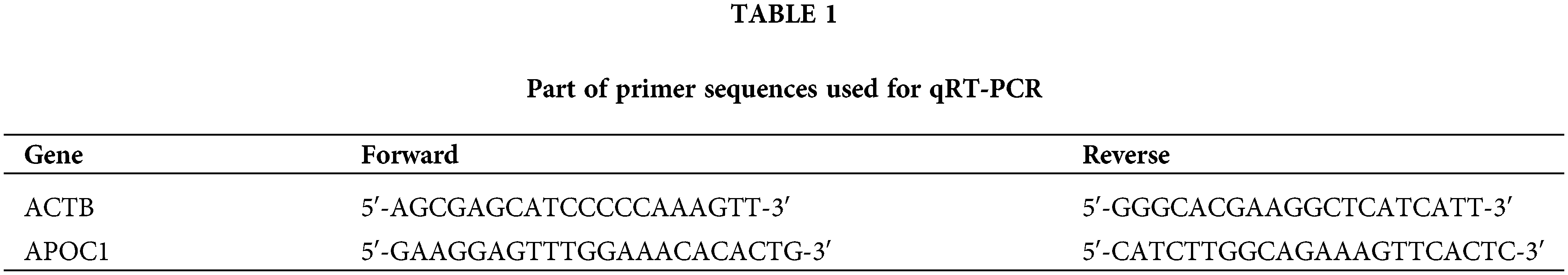
Total RNA was extracted from the GC cell lines using TRIzol regaent (Invitrogen, Carlsbad, USA). Hiscript III Reverse Transcriptase (Vazyme, Nanjing, China) was used for cDNA synthesis. cDNA was quantified using the ChamQ SYBR Color qPCR Master Mix (Vazyme, Nanjing, China) in StepOnePlus Real-Time PCR System (Applied Biosystems, Carlsbad, USA). Relative expression of apoc1 was calculated using the 2−ΔΔCT method. Primers used are listed in Table 1.
Small interfering RNA targeting apoc1 and a negative control were acquired from RiboBio (Guangzhou, China). Cells were cultured overnight in serum-free medium before transfection. The cells were then transfected with si-RNAs or si-NC using Lipofectamine 3000 (Invitrogen, Carlsbad, USA).
Total cell proteins were isolated in NP-40 buffer (Beyotime, Shanghai, China), and proteins were separated via SDS-PAGE, followed by transfer onto PVDF membranes (Invitrogen, Carlsbad, USA). The membranes were then blocked with BSA solution for 2 h. Finally, the membranes were incubated with the corresponding primary antibodies at 4°C overnight. After washing with TBST, the membranes were incubated with the corresponding secondary antibodies. The antibodies used in this assay were Anti-apoc1 (Abcam, Cambridge, UK) and Anti-gapdh (Proteintech Technology, Wuhan, China).
Cell migration and invasion assay
Cells were cultured in serum-free medium and seeded into the top of Transwell chambers (Corning, Shanghai Shang, China) with or without Matrigel-coated filters (BD, NY, USA). Each chamber contained 2 × 104 cells per well. A medium containing 10% FBS was added to the bottom of each chamber. 24 h after incubation, cells were stained with 1% crystal violet. Finally we counted the cell number using the ImageJ software.
First, cells were seeded into 6-well plates. When the density of cells reached at least 80% after transfection, a scratch was made in the middle of the well using a yellow pipette tip. The tip was maintained vertically at the bottom of the well. PBS was then adopted to wash the plate three times. The same scratch field was photographed at the indicated time (0, 24 and 48 h).
The animal work was approved by Nanjing Medical University. For the metastasis model, cells transfected with si-RNA or si-NC were injected into the caudal vein of anesthetized nude mice (three mice per group). 14 days later, the lungs of nude mice were removed to count metastatic foci.
Identification of relevant genes
We analyzed gene expression profile of GC patients using the ggpubr package, and relevant genes were defined with p < 0.05, along with corFilter ≥0.2, or ≤−0.2. After screening, scatter plots were drawn based on our results using the “ggExtra” package.
Identification of differential expression genes (DEGs)
We divided the patients with GC into two groups based on the mean value of apoc1. DEGs were defined as p < 0.05, and logFC ≥1, or ≤−1. Specific genes are shown in the heat map. Limma and pheatmap packages were included.
Functional enrichment analysis of DEGs
The “clusterProfiler” package was used for functional enrichment analysis. The threshold conditions were as follows: p < 0.05, q < 0.2.
TME and immune cell infiltration
We used the “estimate” package to calculate the TME scores of the GC samples. The TME score consists of two parts: stromal and immune. We then compared these differences based on the expression of apoc1. Finally, the “reshape2” and “ggpubr” packages were adopted for the violin plot.
To determine the relationship between apoc1 and immune cell infiltration, we first used CIBERSORT (http://cibersort.stanford.edu/) to calculate the percentage of immune cells. Next, the “reshape2” and “ggpubr” packages were also used in our study for box plot and scatter diagrams. Then, we drew the correlation circle diagram according to our previous results. Finally, we acquired a list of immune checkpoints and their expression profile data, and screened out checkpoints that are associated with apoc1.
Drug sensitivity and immunotherapy analysis
To calculate the sensitivity of the targeted drugs, we used the “pRRophetic” package. Then, apoc1 was analyzed and combined with the IC50 results to draw a boxplot. Similarly, we downloaded the immunotherapy scores from the TCIA website (https://tcia.at/) and analyzed the immunotherapy score and apoc1 expression.
Data are presented as the mean ± standard deviation, and analysis was conducted using GraphPad Prism 8.0 software (GraphPad Software Inc., La Jolla, CA, USA) and ImageJ (National Institutes of Health, USA). We also applied Student’s t-test for comparing the experimental groups and control groups. Statistical significance was set at p < 0.05.
Apoc1 is elevated in GC, indicating a poor prognosis
First, we conducted a differential analysis of the data of GC patients from the TCGA database, and filtered out 1621 differential genes, with |logFC| ≥ 2 and p < 0.05. These differential genes included 872 highly expressed genes and 749 genes with low expression. Based on our results, we drew volcano and heat maps (Figs. 1A and 1B). Among these differentially expressed genes, we chose apoc1 as the target of our study. We obtained its expression profile in 33 human tumors from the TIMER database (http://TIMER2.0 (cistrome.org)) and found that apoc1 is abnormally expressed in various types of human tumors, including GC, suggesting that apoc1 is a tumor-related gene (Fig. 1C). We compared the apoc1 expression profile between GC and adjacent tumor tissues and found that apoc1 was significantly elevated in GC (Figs. 1D and 1E). Immuno-histochemical studies also demonstrated significant upregulation of apoc1 in GC tissues compared with adjacent tissues (Fig. 1F). Finally, we conducted survival analysis based on survival time and the apoc1 expression profile from the KM plot database (http://kmplot.com/analysis/), and the results indicated that apoc1 expression was inversely proportional to patient prognosis (Fig. 1G). Overall, apoc1 expression was elevated in GC, indicating a poor prognosis.
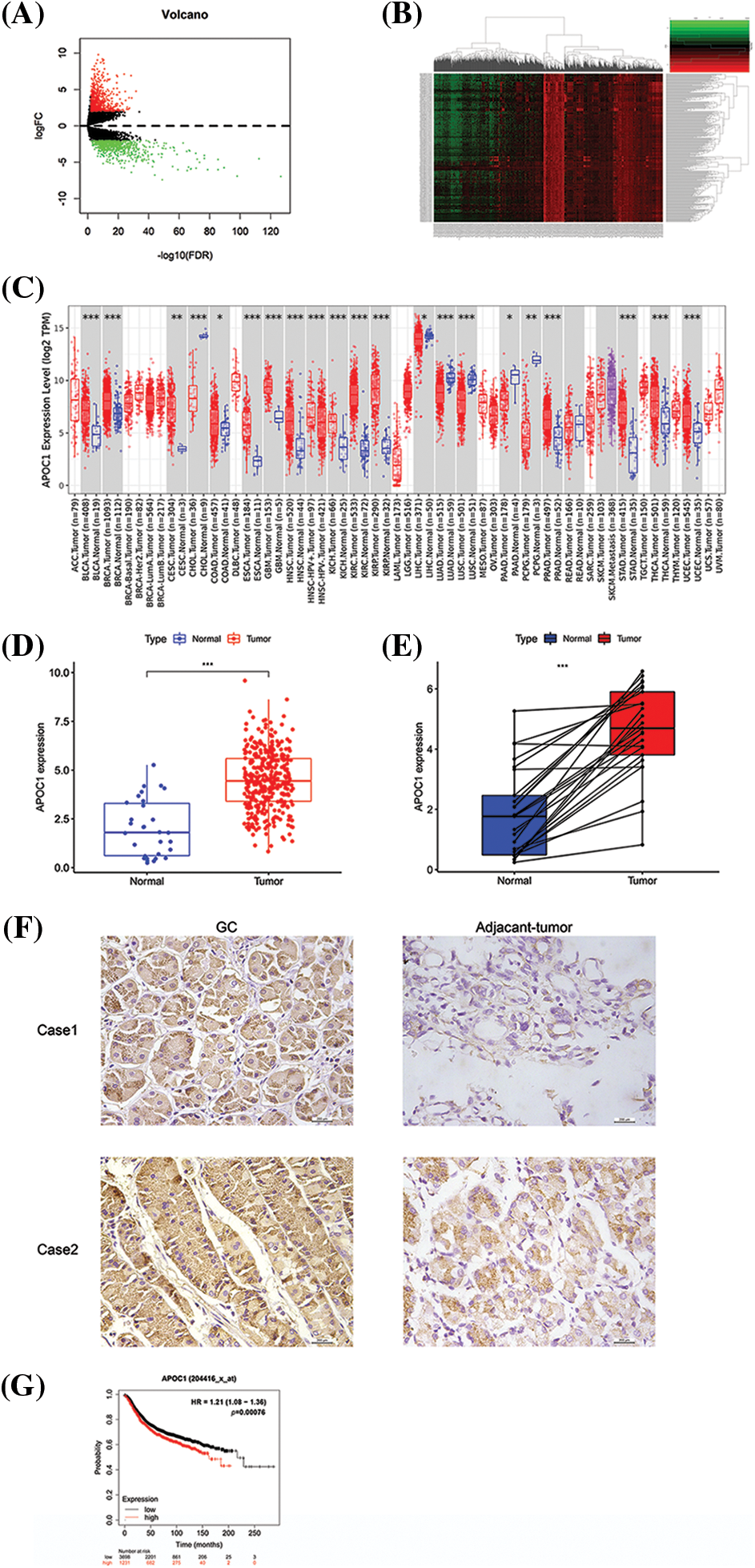
Figure 1: Apoc1 is elevated in GC, indicating a poor prognosis (A–B) Heat map and volcano map we adopted after differential analysis of TCGA GC data (C) Data of apoc1 expression in various kinds of cancers, including GC (D–E) Data of apoc1 expression levels in GC and adjacent-tumor tissues in TCGA dataset (F) Expression patterns in GC and para-carcinoma tissues revealed by immunohistochemistry analysis. (G) Survival analysis of apoc1 in GC patients TCGA, The Cancer Genome Atlas; GC, gastric cancer; *p < 0.05; **p < 0.01; ***p < 0.001.
Apoc1 is associated with tumor stage in GC
We first divided the samples into different groups based on their clinical and pathological features and compared the expression of apoc1. No statistical differences were observed in age (Fig. 2A), sex (Fig. 2B), grade (Fig. 2C), M (Fig. 2D) and N (Fig. 2G) in apoc1 expression. Compared with the T1 stage, apoc1 expression was significantly increased in the T2, T3 and T4 stages (Fig. 2F), and the same phenomenon was observed in Stages I, II, III, and IV (Fig. 2E). Finally, we created clinically relevant heat maps to confirm our conclusions (Fig. 2H).
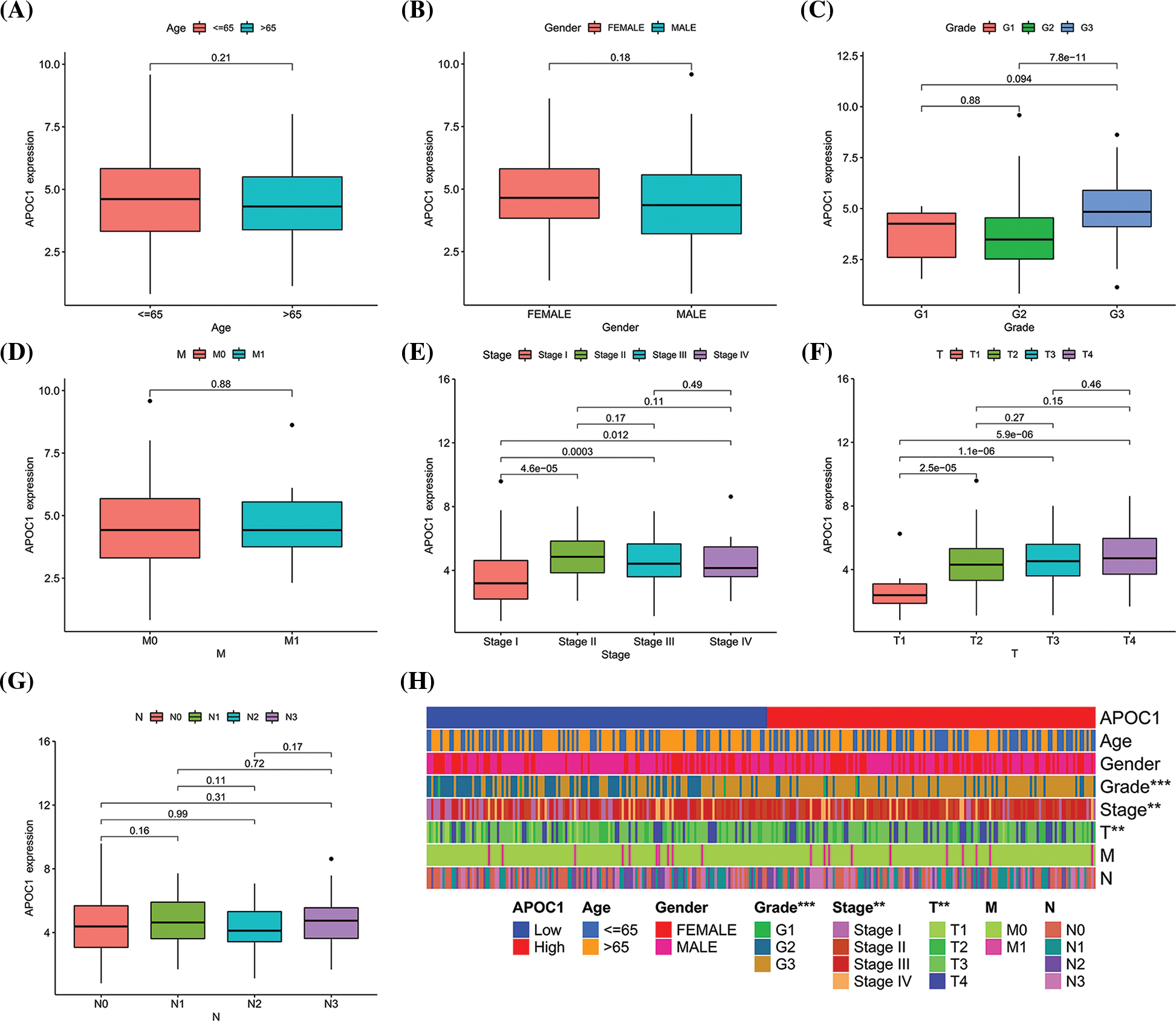
Figure 2: Apoc1 is associated with tumor in GC (A) age (B) gender (C) grade (D) M (E) stage (F) T and (G) N in GC patients from TCGA (H) clinic-pathological factors relevant heat map. **p < 0.01; ***p < 0.001.
Down-regulation of apoc1 inhibits abilities of migration and invasion
We obtained three siRNAs targeting apoc1 and a negative control for apoc1 knockdown. To confirm this efficiency, we performed qRT-PCR and western blotting (Figs. 3A and 3B). We then conducted cell migration and cell invasion assays, and the results showed that the previously mentioned abilities of GC cells transfected with siRNAs were reduced (Figs. 3C and 3D). Furthermore, this was confirmed by the results of the wound healing assay (Figs. 3E and 3F) and the model of pulmonary metastasis (Fig. 3G). Thus, we conclude that apoc1 plays a major role in cell migration and invasion.
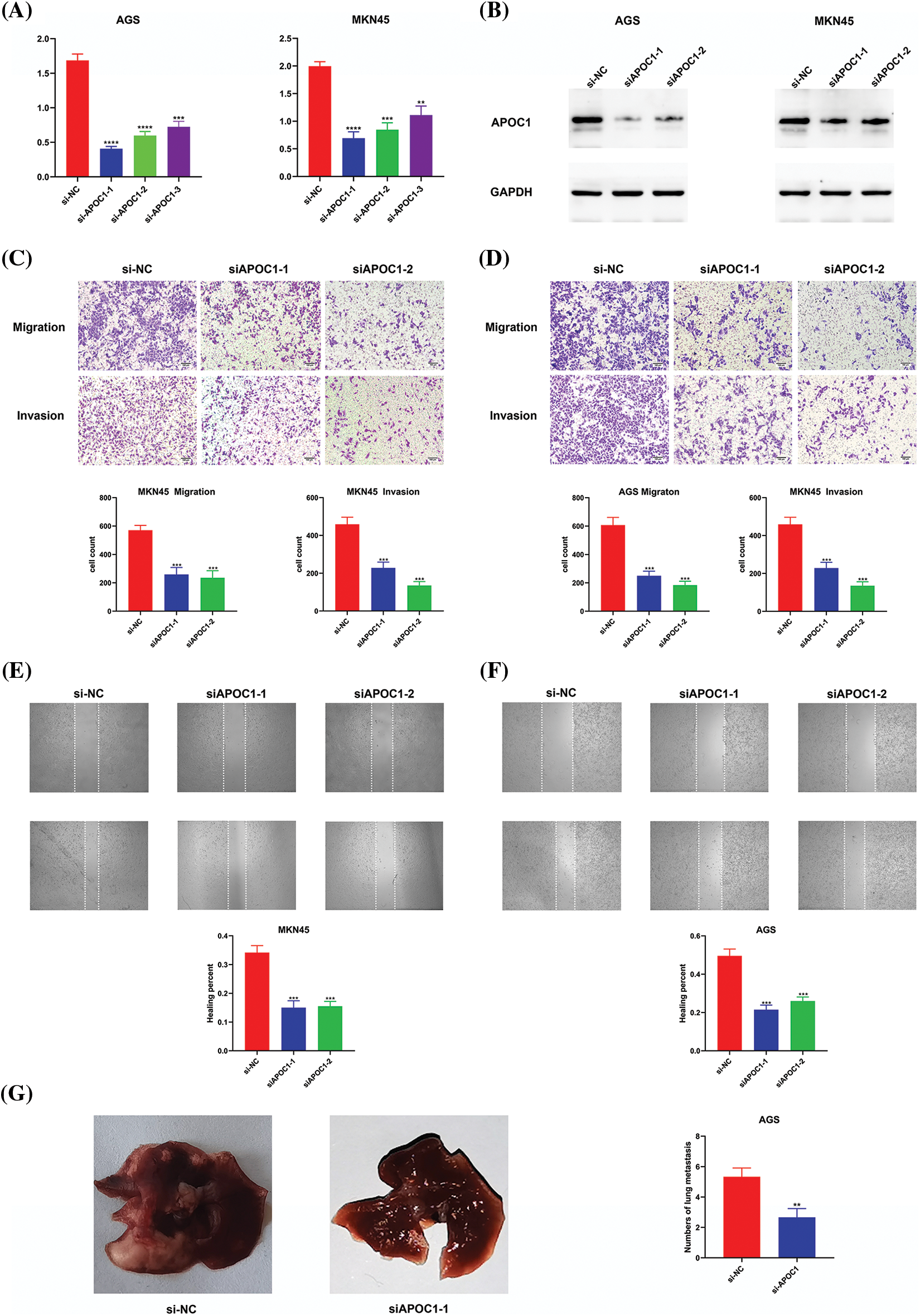
Figure 3: Down-regulation of apoc1 inhibits abilities of migration and invasion. (A) Efficiency of si-RNAs targeting apoc1 confirmed by qRT-PCR (B) Efficiency of si-RNAs targeting apoc1 confirmed by Western Blot (C–D) Transwell assay and (E–F) Wound healing assay were performed undergone different treatments (G) Model of pulmonary metastasis from nude mice **p < 0.01; ***p < 0.001; ****p < 0.0001.
Apoc1 may be involved in the Wnt pathway and immune regulation
To explore the mechanism by which apoc1 regulates the progression of GC, we conducted analysis to investigate the correlation between apoc1 and other genes in TCGA database. With |cor| ≥ 0.2 and p < 0.05, a total of 2752 correlated genes were identified and correlation circle maps were drawn (Fig. 4A).
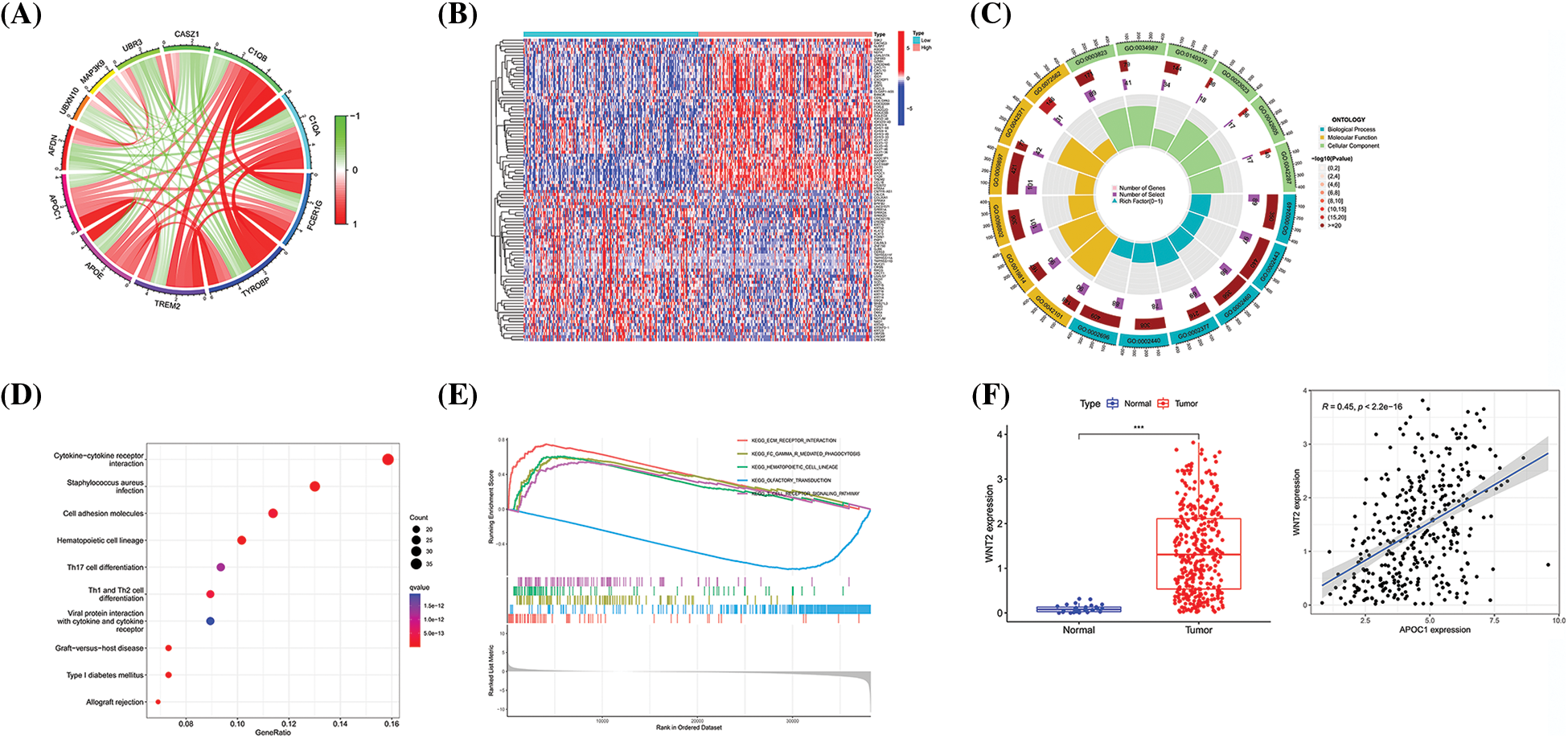
Figure 4: Apoc1 may be involved in the Wnt pathway and immune regulation (A) Circle diagram of genes which are strongly related to apoc1 (B) Heat map of differential genes (C) GO analysis (D) KEGG analysis and (E) GSEA analysis of differential genes apoc1 (F) Expression patterns of wnt2 in GC and Pearson coefficient between apoc1 and wnt2 ***p < 0.001.
Next, we analyzed and adopted 816 differentially expressed genes (DEGs) based on apoc1 expression, with |logFC| ≥ 1 and p < 0.05, as the standard and drew a heat map (Fig. 4B). GO, KEGG, and GSEA analyses of the DEGs showed that apoc1 was closely related to the Wnt pathway and immune regulation (Figs. 4C and 4E). We found that apoc1 may participate in Wnt signaling via wnt2 (Fig. 4F). Overall, apoc1 may participate in GC progression via the Wnt axis and immune activities.
Apoc1 relates to TME and immune cell infiltration
We further investigated its role in the TME. The results revealed that the stromal and immune scores of the apoc1 high expression group were higher (Fig. 5A).
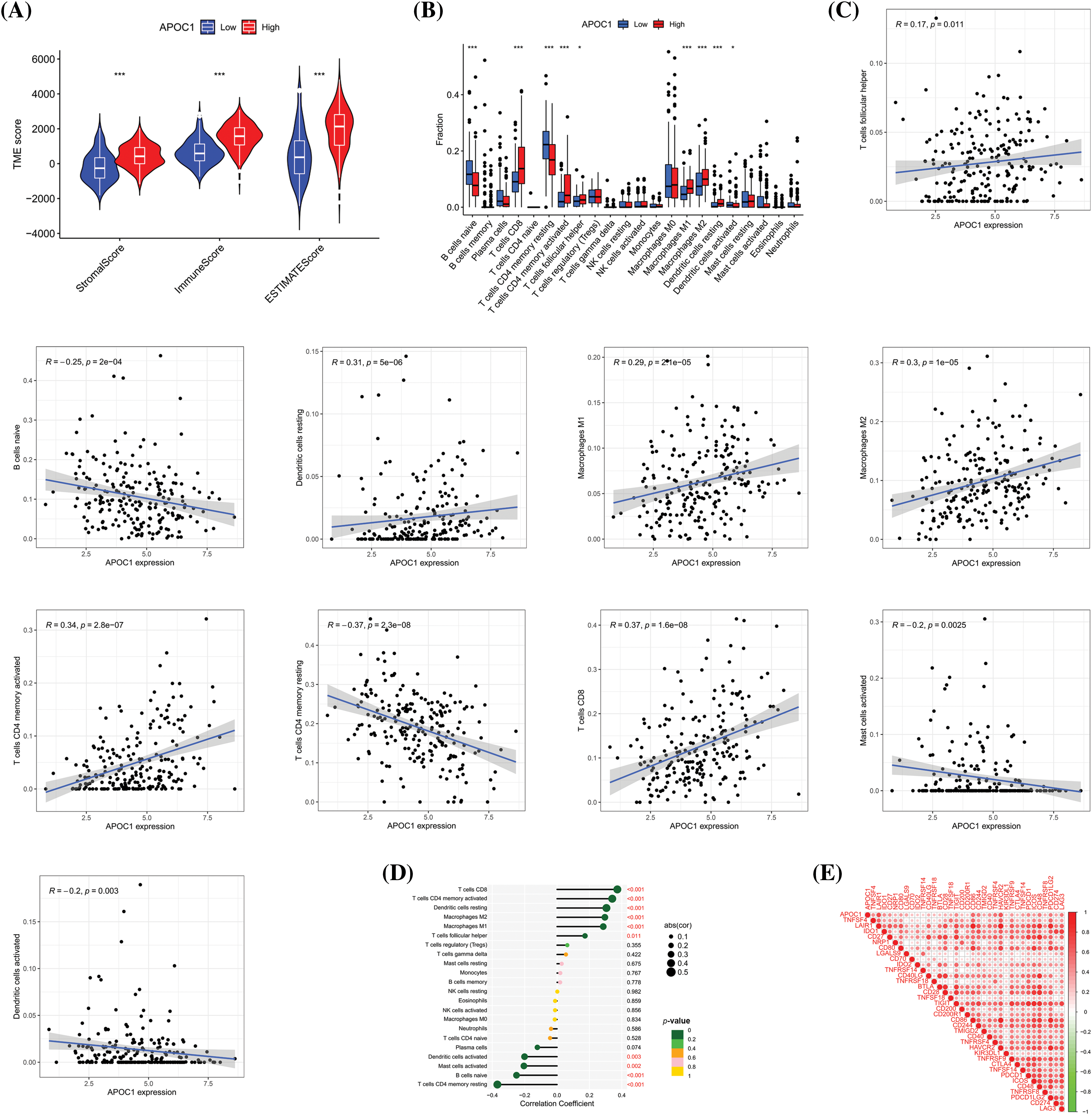
Figure 5: Apoc1 relates to TME and immune cell infiltration (A) Analysis of differences in TME scores (B) Analysis of differences in immune cells (C) Scatter plot of relevant immune cells (D) Correlation analysis of immune cells and (E) Immune checkpoint *p < 0.05, **p < 0.01, ***p < 0.001.
We then studied the relationship between the fraction of immune cells and the expression profile of apoc1 and found that its expression was related to the proportion of various immune cells (Fig. 5B). Next, we analyzed the correlation between the proportion of these immune cells and apoc1. The proportion of CD8+T cells, memory activated CD4+T cells, resting dendritic cells, Macrophages M1 and Macrophages M2 were positively proportional to apoc1, while the proportion of activated dendritic cells, activated mast cells, naïve B cells, and resting memory CD4+T cells were negatively proportional to their expression (Fig. 5C). Subsequent analysis of the immune cell correlation confirmed our conclusion (Fig. 5D). Finally, we obtained immune checkpoints related to apoc1 (Fig. 5E).
Drug sensitivity and immunotherapy analysis of apoc1
Targeted immunotherapy is a widely used treatment for advanced GC. To determine the relationship between apoc1 expression and the sensitivity of targeted drugs, we analyzed the expression and IC50 of various targeted drugs in TCGA GC samples, and found that the sensitivity of GC patients to Bexarotene, BEZ235, GNF-2, MG-132, OSI-930, Ruxolitinib, Sunitinib and Talazoparib was relevant to the expression of apoc1, and the IC50 of these targeted drugs in the apoc1-high group was lower (Fig. 6A), suggesting that doctors can decide which one to utilize based on the level of apoc1 in GC patients.
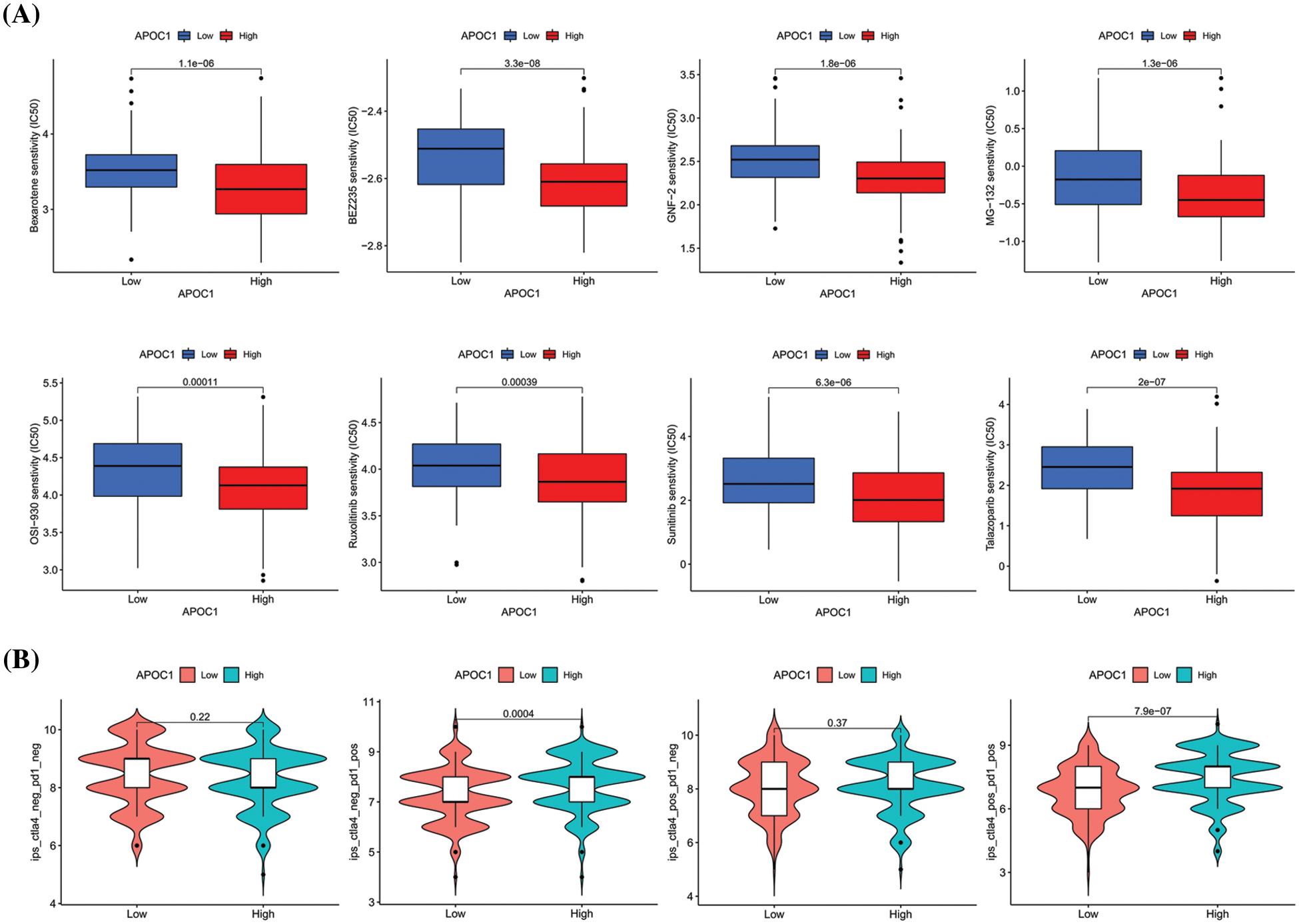
Figure 6: Drug sensitivity analysis of apoc1 (A) various kinds of small molecule drug whose IC50 are associated with APOC1 expression (B) Relationship between PD-1 and CTLA-4 effects and APOC1 expression.
Among the various immunotherapy regimens, PD-1 and CTLA-4 are the most commonly adopted. To study whether the expression of apoc1 affects the curative effect of PD-1 and CTLA-4, apoc1 expression and immunotherapy scores in GC patients were analyzed. The results indicated that the immune scores of the apoc1 highly group were higher than those in the low group, regardless of whether CTLA-4 was positive or not (Fig. 6B). However, whether CTLA-4 was positive or not and whether its curative effect was related to the expression of apoc1 was not significantly correlated with apoc1 expression. In summary, we found some targeted drugs that may have better efficacy in the apoc1 overexpression group, and apoc1 may play a major role in PD-1 therapy.
Apolipoprotein is a crucial structural component of lipoproteins and has been shown to be involved in their synthesis, secretion, and catabolism [16]. So far, eight human apolipoproteins (apoca1, apoca2, apoca4, apocb, apoc1, apoc2, apoc3, and apoce) have been identified. Apolipoprotein is associated with genetic susceptibility to atherosclerosis in humans, and investigation of the regulatory mechanism of apolipoprotein expression is helpful in developing new diagnostic and treatment tools for atherosclerosis [17]. For example, variation in Apolipoprotein E is one of the genetic factors that affect plasma lipoproteins and can be used to diagnose patients with type III hyperlipidemia and screen susceptible populations [18]. Similarly, apoa4 glycosylation (G-apoa4) is associated with coronary artery disease (CAD) in patients with type 2 diabetes mellitus (T2DM), and can induce atherosclerosis via nr4a3 [19]. apob is a key structural protein of atherosclerotic lipoproteins, and plasma apob concentration can be used as a measure of the amount of atherosclerotic lipoproteins [20]. apoc3 is a small protein that regulates TG metabolism and promotes atherosclerosis in humans by interfering with lipoprotein function and catabolism [21].
With further study of apolipoproteins, it was found that apolipoprotein family members also play significant roles in other physiological and biochemical processes, including different diseases [22]. For example, hereditary apoa1 amyloidosis, which is caused by pathogenic mutations in apoa1, can result in amyloid protein deposition in the heart, kidneys, nerves, larynx, and skin [23]. Another study has shown that low level of HDL-C and apoa1 indicates poor prognosis and recurrence in autoimmune encephalitis (AE): therefore, they may be utilized as biomarkers in the future [24]. An elevated apob/apoa1 ratio can be used to screen for metabolic syndrome (Met) and insulin resistance (IR) in patients with polycystic ovarian syndrome (PCOS) [25]. Decreased plasma levels of apob in patients with non-alcoholic fatty liver disease (NAFLD) impairs the anti-inflammatory ability, and the impaired anti-inflammatory activity is independently correlated with NAFLD [26]. Another study showed that high apob expression is associated with an increased incidence of CKD and metabolic dysfunction [27]. Fragments of apoe co-localize with neurofibrillary tangles and amyloid β (Aβ) plaques, which may lead to neurodegenerative changes that lead to Alzheimer’s disease (AD) [28].
Members of the apolipoprotein family have also been observed in human malignant tumors [29]. For instance, one study showed that the up-regulation of apoa1 in cervical squamous cell carcinoma cell lines can promote carboplatin resistance [30]. Plasma levels of HDL-C and apoa1 are up-regulated in non-small cell lung cancer patients with the epidermal growth factor receptor (EGFR) T790M mutation (NSCLC), suggesting that they have potential as markers in these patients [31]. In a retrospective analysis of apob, the preoperative apob/apoa1 ratio was used to diagnose osteosarcoma in juveniles [32]. In clear cell renal cell carcinoma (ccRCC), preoperative apob can be utilized to predict prognosis of patients [33]. In GC, apoc2 is elevated in peritoneal metastasis (PM) tissues and promotes PM via cd36-mediated PI3K/AKT/mTOR signaling [34]. One study suggested that apoe is elevated in patients with NPC and promotes tumor growth, migration, and invasion, which may serve as a potential biomarker for the diagnosis of NPC [35].
Apoc1 is an inhibitor of LDL and VLDL receptors and it can also inhibit the cholesterol ester transfer protein and uptake of fatty acids [36]. Four crystal structures of apoc1 have been identified, and these suggest a number of physiological functions [37], such as immunity, sepsis, diabetes, cancer, and viral infectivity. For example, it has been suggested that apoc1 may be used for diagnosis and prognosis of abdominal aortic aneurysm (AAA) [38]. Another study revealed that apoc1 can help detect metabolic abnormalities early in women with PCOS [39]. In sepsis, apoc1 may play a protective role against infection by regulating the response to LPS [40].
Apoc1 is also involved in tumor progression. In colorectal cancer (CRC), ZEB1-AS1 promotes liver metastasis via the miR-335-5p/APOC1 axis [41]. APOC1 expression is also upregulated in ccRCC and is significantly associated with clinico-pathological factors. apoc1 participates in cell growth and metastasis in ccRCC [42,43]. Knockdown of apoc1 also inhibits cell proliferation and induces apoptosis in pancreatic cancer [44].
Programmed cell death protein 1 (PD-1) is a well-known immune checkpoint receptor secreted by activated T cells. Antibodies, including PD-1 and PD-L1, are new drugs for tumor immunotherapy. Although it cannot kill tumor cells directly, it exerts anti-tumor effect by enhancing the patient’s own immune system [45]. PD-1/PD-L1 inhibitors may help to improve the prognosis of triple-negative breast cancer (TNBC) [46]. Similarly, another study suggested that approximately one-fifth of patients with anal squamous cell carcinoma (SCCA) benefit from PD-1 inhibitors [47]. Another meta-analysis showed that PD-1/PD-L1 inhibitors significantly improved the therapeutic efficacy of lung cancer patients with brain metastases compared to chemotherapy [48]. Anti PD-1 combined with SABR for metastatic NSCLC leads to a high response rate and prolongs the clinical benefits of immunotherapy and new systemic therapy by delaying further progression [49].
Antibodies against PD-1 and PD-L1 produce long-lasting responses in a large number of tumor patients; however, most of them eventually relapse due to acquired resistance [50]. For example, up-regulation of circhmgb2 in LUAD and LUSC can accelerate the progression of LUAD and LUSC by regulating the TME, indicating a new therapy for LUAD and LUSC [51]. ripk2 has also been reported to induce immunotherapy resistance by triggering cytotoxic T-lymphocyte dysfunction [52]. Multiple clinical trials have shown that LAG-3 blockade alleviates resistance to PD-1 inhibitors in pancreatic cancer. Therefore, the synchronous inhibition of LAG-3 and PD-1 may be a new strategy for alleviating tumor immunotherapy resistance [53]. PD-1 is a promising antitumor therapy; therefore, it is imperative to further study and eliminate acquired drug resistance in patients.
Acknowledgement: The authors thank the Laboratory of the Secondary Affiliated Hospital of Nanjing Medical University for providing financial support and technical guidance.
Funding Statement: This study was financed by the National Natural Science Foundation (Grant Number 81874058 to Jianping Zhang).
Author Contributions: JZ launched and designed an experimental roadmap for this study. QG performed the majority of the experiments. TZ and XG downloaded and analyzed data from the TCGA database. NL, GW and CL performed the TME, immune cell infiltration and drug sensitivity analyses online, respectively. LX and XG created the figures and edited the manuscript. All authors approved the submitted version.
Availability of Data and Materials: The datasets used and/or analyzed during the current study will be made available from the corresponding author on reasonable requests.
Ethics Approval: Animal experiments were performed and supervised by the Laboratory Animal Ethics Committee at Nanjing Medical University, and the number of the ethics approval is 2110011.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Siegel, R. L., Miller, K. D., Fuchs, H. E., Jemal, A. (2022). Cancer statistics, 2022. CA: A Cancer Journal for Clinicians, 72(1), 7–33. https://doi.org/10.3322/caac.21731 [Google Scholar] [CrossRef]
2. Thrumurthy, S. G., Chaudry, M. A., Hochhauser, D., Mughal, M. (2013). The diagnosis and management of gastric cancer. BMJ, 347, 1–9. https://doi.org/10.1136/bmj.f6367 [Google Scholar] [PubMed] [CrossRef]
3. Tung, I., Sahu, A. (2021). The treatment of resectable gastric cancer: A literature review of an evolving landscape. Journal of Gastrointestinal Oncology, 13(2), 871–884. https://doi.org/10.21037/jgo-21-721 [Google Scholar] [PubMed] [CrossRef]
4. Cervantes, A., Roselló, S., Roda, D., Rodríguez-Braun, E. (2008). The treatment of advanced gastric cancer: Current strategies and future perspectives. Annals of Oncology, 19(Supplement 5), 103–108. https://doi.org/10.1093/annonc/mdn321 [Google Scholar] [PubMed] [CrossRef]
5. Myer, N. M., Shitara, K., Chung, H. C., Lordick, F., Kelly, R. J. et al. (2022). Evolution of predictive and prognostic biomarkers in the treatment of advanced gastric cancer. Journal of Cancer Research Clinical Oncology, 148(8), 2023–2043. https://doi.org/10.1007/s00432-021-03902-1 [Google Scholar] [PubMed] [CrossRef]
6. Fuior, E. V., Gafencu, A. V. (2019). Apolipoprotein C1: Its pleiotropic effects in lipid metabolism and beyond. International Journal of Molecular Sciences, 20(23), 1–25. https://doi.org/10.3390/ijms20235939 [Google Scholar] [PubMed] [CrossRef]
7. Bus, P., Pierneef, L., Bor, R., Wolterbeek, R., van Es, L. A. et al. (2017). Apolipoprotein C-I plays a role in the pathogenesis of glomerulosclerosis. Journal of Pathology, 241(5), 589–599. https://doi.org/10.1002/path.4859 [Google Scholar] [PubMed] [CrossRef]
8. Li, J., Sun, L., Xu, F., Qi, H., Shen, C. et al. (2016). Screening and identification of APOC1 as a novel potential biomarker for differentiate of mycoplasma pneumoniae in children. Frontiers in Microbiology, 7, 1012. https://doi.org/10.3389/fmicb.2016.01961 [Google Scholar] [PubMed] [CrossRef]
9. Zhou, Q., Peng, D., Yuan, X., Lv, Z., Pang, S. et al. (2014). APOE and APOC1 gene polymorphisms are associated with cognitive impairment progression in Chinese patients with late-onset Alzheimer’s disease. Neural Regeneration Research, 9(6), 653–660. https://doi.org/10.4103/1673-5374.130117 [Google Scholar] [PubMed] [CrossRef]
10. Zhang, H., Wang, Y., Liu, C., Li, W., Zhou, F. et al. (2022). The Apolipoprotein C1 is involved in breast cancer progression via EMT and MAPK/JNK pathway. Pathology Research Practice, 229(2), 153746. https://doi.org/10.1016/j.prp.2021.153746 [Google Scholar] [PubMed] [CrossRef]
11. Shen, H. Y., Wei, F. Z., Liu, Q. (2021). Differential analysis revealing APOC1 to be a diagnostic and prognostic marker for liver metastases of colorectal cancer. World Journal of Clinical Cases, 9(16), 3880–3894. https://doi.org/10.12998/wjcc.v9.i16.3880 [Google Scholar] [PubMed] [CrossRef]
12. Jiang, H., Tang, J. Y., Xue, D., Chen, Y. M., Wu, T. C. et al. (2021). Apolipoprotein C1 stimulates the malignant process of renal cell carcinoma via the Wnt3a signaling. Cancer Cell International, 21(1), 41. https://doi.org/10.1186/s12935-020-01713-x [Google Scholar] [PubMed] [CrossRef]
13. Hao, X., Zheng, Z., Liu, H., Zhang, Y., Kang, J. et al. (2022). Inhibition of APOC1 promotes the transformation of M2 into M1 macrophages via the ferroptosis pathway and enhances anti-PD1 immunotherapy in hepatocellular carcinoma based on single-cell RNA sequencing. Redox Biology, 56, 102463. https://doi.org/10.1016/j.redox.2022.102463 [Google Scholar] [PubMed] [CrossRef]
14. Yi, J., Ren, L., Wu, J., Li, W., Zheng, X. et al. (2019). Apolipoprotein C1 (APOC1) as a novel diagnostic and prognostic biomarker for gastric cancer. Annals of Translational Medicine, 7(16), 380. https://doi.org/10.21037/atm.2019.07.59 [Google Scholar] [PubMed] [CrossRef]
15. An, L., Liu, Y. (2023). ZNF460 mediates epithelial-mesenchymal transition to promote gastric cancer progression by transactivating APOC1 expression. Experimental Cell Research, 422(2), 113452. https://doi.org/10.1016/j.yexcr.2022.113452 [Google Scholar] [PubMed] [CrossRef]
16. Breslow, J. L. (1985). Human apolipoprotein molecular biology and genetic variation. Annual Review of Biochemistry, 54(1), 699–727. https://doi.org/10.1146/annurev.bi.54.070185.003411 [Google Scholar] [PubMed] [CrossRef]
17. Breslow, J. L. (1988). Apolipoprotein genetic variation and human disease. Physiological Reviews, 68(1), 85–132. https://doi.org/10.1152/physrev.1988.68.1.85 [Google Scholar] [PubMed] [CrossRef]
18. Walden, C. C., Hegele, R. A. (1994). Apolipoprotein E in hyperlipidemia. Annals of Internal Medicine, 120(12), 1026–1036. https://doi.org/10.7326/0003-4819-120-12-199406150-00009 [Google Scholar] [PubMed] [CrossRef]
19. Dai, Y., Shen, Y., Li, Q. R., Ding, F. H., Wang, X. Q. et al. (2017). Glycated apolipoprotein A-IV induces atherogenesis in patients with CAD in type 2 diabetes. Journal of the American College of Cardiology, 70(16), 2006–2019. https://doi.org/10.1016/j.jacc.2017.08.053 [Google Scholar] [PubMed] [CrossRef]
20. Behbodikhah, J., Ahmed, S., Elyasi, A., Kasselman, L. J., de Leon, J. et al. (2021). Apolipoprotein B and cardiovascular disease: Biomarker and potential therapeutic target. Metabolites, 11(10), 690. https://doi.org/10.3390/metabo11100690 [Google Scholar] [PubMed] [CrossRef]
21. Dib, I., Khalil, A., Chouaib, R., El-Makhour, Y., Noureddine, H. (2021). Apolipoprotein C-III and cardiovascular diseases: When genetics meet molecular pathologies. Molecular Biology Reports, 48(1), 875–886. https://doi.org/10.1007/s11033-020-06071-5 [Google Scholar] [PubMed] [CrossRef]
22. Ceci, F. M., Ceccanti, M., Petrella, C., Vitali, M., Messina, M. P. et al. (2021). Alcohol drinking, apolipoprotein polymorphisms and the risk of cardiovascular diseases. Current Neurovascular Research, 18(1), 150–161. https://doi.org/10.2174/1567202618666210406123503 [Google Scholar] [PubMed] [CrossRef]
23. Cohen, O. C., Blakeney, I. J., Law, S., Ravichandran, S., Gilbertson, J. et al. (2022). The experience of hereditary apolipoprotein A-I amyloidosis at the UK National Amyloidosis Centre. Amyloid, 29(4), 237–244. https://doi.org/10.1080/13506129.2022.2070741 [Google Scholar] [PubMed] [CrossRef]
24. Liu, F., Huang, T., Wang, B., Wang, C., Guo, S. (2022). Low high-density lipoprotein cholesterol and apolipoprotein A-I levels are associated with poor outcome and relapse in autoimmune encephalitis. Neuroscience Letters, 775, 136546. https://doi.org/10.1016/j.neulet.2022.136546 [Google Scholar] [PubMed] [CrossRef]
25. He, H., Feng, J., Zhang, S., Wang, Y., Li, J. et al. (2022). The apolipoprotein B/A1 ratio is associated with metabolic syndrome components, insulin resistance, androgen hormones, and liver enzymes in women with polycystic ovary syndrome. Frontiers in Endocrinology, 12, 773781. https://doi.org/10.3389/fendo.2021.773781 [Google Scholar] [PubMed] [CrossRef]
26. Meng, H., Jiang, L., Song, Z., Wang, F. (2022). Causal associations of circulating lipids with osteoarthritis: A bidirectional mendelian randomization study. Nutrients, 14(7), 1327. https://doi.org/10.3390/nu14071327 [Google Scholar] [PubMed] [CrossRef]
27. Mazidi, M., Webb, R. J., Lip, G. Y. H., Kengne, A. P., Banach, M. et al. (2022). Discordance between LDL-C and apolipoprotein B levels and its association with renal dysfunction: Insights from a population-based study. Journal of Clinical Medicine, 11(2), 1–8. https://doi.org/10.3390/jcm11020313 [Google Scholar] [PubMed] [CrossRef]
28. Soiza, R. L., Donaldson, A. I. C., Myint, P. K. (2018). Vaccine against arteriosclerosis: An update. Therapeutic Advances in Vaccines, 9(6), 259–261. https://doi.org/10.1177/2051013617693753 [Google Scholar] [PubMed] [CrossRef]
29. Delk, S. C., Chattopadhyay, A., Escola-Gil, J. C., Fogelman, A. M., Reddy, S. T. (2021). Apolipoprotein mimetics in cancer. Seminars in Cancer Biology, 73(1), 158–168. https://doi.org/10.1016/j.semcancer.2020.11.002 [Google Scholar] [PubMed] [CrossRef]
30. He, Y., Han, S. B., Liu, Y., Zhang, J. J., Wu, Y. M. (2022). Role of APOA1 in the resistance to platinum-based chemotherapy in squamous cervical cancer. BMC Cancer, 22(1), 1–12. https://doi.org/10.1186/s12885-022-09528-x [Google Scholar] [PubMed] [CrossRef]
31. Ma, J., Bai, Y., Liu, M., Jiao, T., Chen, Y. et al. (2022). Pretreatment HDL-C and ApoA1 are predictive biomarkers of progression-free survival in patients with EGFR mutated advanced non-small cell lung cancer treated with TKI. Thoracic Cancer, 13(8), 1126–1135. https://doi.org/10.1111/1759-7714.14367 [Google Scholar] [PubMed] [CrossRef]
32. Ma, C., Li, R., Yu, R., Guo, J., Xu, J. et al. (2022). Predictive value of preoperative platelet-to-albumin ratio and apolipoprotein B-to-apolipoprotein A1 ratio for osteosarcoma in children and adolescents: A retrospective study of 118 cases. BMC Cancer, 22(1), 1–11. https://doi.org/10.1186/s12885-022-09223-x [Google Scholar] [PubMed] [CrossRef]
33. Wang, K., Wu, T., Chen, Y., Song, G., Chen, Z. (2020). Prognostic effect of preoperative apolipoprotein B level in surgical patients with clear cell renal cell carcinoma. Oncology Research and Treatment, 43(7–8), 340–345. https://doi.org/10.1159/000507964. [Google Scholar] [PubMed] [CrossRef]
34. Wang, C., Yang, Z., Xu, E., Shen, X., Wang, X. et al. (2021). Apolipoprotein C-II induces EMT to promote gastric cancer peritoneal metastasis via PI3K/AKT/mTOR pathway. Clinical and Translational Medicine, 11(8), e522. https://doi.org/10.1002/ctm2.522 [Google Scholar] [PubMed] [CrossRef]
35. Xue, Y., Huang, S., Huang, J., Li, S., Zhang, C. et al. (2020). Identification of apolipoprotein E as a potential diagnostic biomarker of nasopharyngeal carcinoma. Cancer Management and Research, 12, 8943–8950. https://doi.org/10.2147/CMAR.S239479 [Google Scholar] [PubMed] [CrossRef]
36. Shachter, N. S. (2001). Apolipoproteins C-I and C-III as important modulators of lipoprotein metabolism. Current Opinion in Lipidology, 12(3), 297–304. https://doi.org/10.1097/00041433-200106000-00009 [Google Scholar] [PubMed] [CrossRef]
37. McPherson, A., Larson, S. B. (2019). The structure of human apolipoprotein C-1 in four different crystal forms. Journal of Lipid Research, 60(2), 400–411. https://doi.org/10.1194/jlr.M089441 [Google Scholar] [PubMed] [CrossRef]
38. Moxon, J. V., Liu, D., Moran, C. S., Crossman, D. J., Krishna, S. M. et al. (2014). Proteomic and genomic analyses suggest the association of apolipoprotein C1 with abdominal aortic aneurysm. Proteomics—Clinical Applications, 8(9–10), 762–772. https://doi.org/10.1002/prca.201300119 [Google Scholar] [PubMed] [CrossRef]
39. Huang, S., Qiao, J., Li, R., Wang, L., Li, M. (2010). Can serum apolipoprotein C-I demonstrate metabolic abnormality early in women with polycystic ovary syndrome? Fertility and Sterility, 94(1), 205–210. https://doi.org/10.1016/j.fertnstert.2009.03.005 [Google Scholar] [PubMed] [CrossRef]
40. Berbée, J. F., van der Hoogt, C. C., Kleemann, R., Schippers, E. F., Kitchens, R. L. et al. (2006). Apolipoprotein CI stimulates the response to lipopolysaccharide and reduces mortality in Gram-negative sepsis. The FASEB Journal, 20(12), 2162–2164. https://doi.org/10.1096/fj.05-5639fje [Google Scholar] [PubMed] [CrossRef]
41. Lu, C., Luo, X., Xing, C., Mao, Y., Xu, Y. et al. (2021). Construction of a novel mRNA-miRNA-lncRNA network and identification of potential regulatory axis associated with prognosis in colorectal cancer liver metastases. Aging, 13(11), 14968–14988. https://doi.org/10.18632/aging.203049 [Google Scholar] [PubMed] [CrossRef]
42. Xiao, H., Xu, Y. (2020). Overexpression of apolipoprotein C1 (APOC1) in clear cell renal cell carcinoma and its prognostic significance. Medical Science Monitor, 27, 1–11. https://doi.org/10.1016/j.semcancer.2020.11.002 [Google Scholar] [CrossRef]
43. Wang, H. J., Ma, Y. X., Wang, A. H., Jiang, Y. S., Jiang, X. Z. (2021). Expression of apolipoprotein C1 in clear cell renal cell carcinoma: An oncogenic gene and a prognostic marker. Kaohsiung Journal of Medical Sciences, 37(5), 419–426. https://doi.org/10.1002/kjm2.12328 [Google Scholar] [PubMed] [CrossRef]
44. Takano, S., Yoshitomi, H., Togawa, A., Sogawa, K., Shida, T. et al. (2008). Apolipoprotein C-1 maintains cell survival by preventing from apoptosis in pancreatic cancer cells. Oncogene, 27(20), 2810–2822. https://doi.org/10.1038/sj.onc.1210951 [Google Scholar] [PubMed] [CrossRef]
45. Okazaki, T., Chikuma, S., Iwai, Y., Fagarasan, S., Honjo, T. (2013). A rheostat for immune responses: The unique properties of PD-1 and their advantages for clinical application. Nature Immunology, 14(12), 1212–1218. https://doi.org/10.1038/ni.2762 [Google Scholar] [PubMed] [CrossRef]
46. Liang, Y., Liu, X., Li, K., Li, H. (2022). Current situation of programmed cell death protein 1/programmed cell death ligand 1 inhibitors in advanced triple-negative breast cancer. Chinese Journal of Cancer Research, 34(2), 117–130. https://doi.org/10.21147/j.issn.1000-9604.2022.02.07 [Google Scholar] [PubMed] [CrossRef]
47. Jácome, A. A., Morris, V. K., Eng, C. (2022). The role of immunotherapy in the treatment of anal cancer and future strategies. Current Treatment Options in Oncology, 23(8), 1073–1085. https://doi.org/10.1007/s11864-022-00939-3 [Google Scholar] [PubMed] [CrossRef]
48. Yang, X., Zeng, Y., Tan, Q., Huang, Z., Jia, J. et al. (2022). Efficacy of PD-1/PD-L1 inhibitors versus chemotherapy in lung cancer with brain metastases: A systematic review and meta-analysis. Journal of Immunology Research, 2022(26), 1–14. https://doi.org/10.1155/2022/4518898 [Google Scholar] [PubMed] [CrossRef]
49. Chicas-Sett, R., Zafra, J., Rodriguez-Abreu, D., Castilla-Martinez, J., Benitez, G. et al. (2022). Combination of stereotactic ablative radiotherapy with Anti-PD-1 in oligoprogressive non-small-cell lung cancer and melanoma: Results of a prospective multicenter observational study. International Journal of Radiation Oncology Biology Physics, 114(4), 655–665. https://doi.org/10.1016/j.ijrobp.2022.05.013 [Google Scholar] [CrossRef]
50. Denis, M., Grasselly, C., Choffour, P. A., Wierinckx, A., Mathé, D. et al. (2022). In vivo syngeneic tumor models with acquired resistance to Anti-PD-1/PD-L1 therapies. Cancer Immunology Research, 10(8), 1013–1027. https://doi.org/10.1158/2326-6066.CIR-21-0802 [Google Scholar] [PubMed] [CrossRef]
51. Zhang, L. X., Gao, J., Long, X., Zhang, P. F., Yang, X. et al. (2022). The circular RNA circHMGB2 drives immunosuppression and anti-PD-1 resistance in lung adenocarcinomas and squamous cell carcinomas via the miR-181a-5p/CARM1 axis. Molecular Cancer, 21(1), 1–20. https://doi.org/10.1186/s12943-022-01586-w [Google Scholar] [PubMed] [CrossRef]
52. Song, J., Yang, R., Wei, R., Du, Y., He, P. et al. (2022). Pan‐cancer analysis reveals RIPK2 predicts prognosis and promotes immune therapy resistance via triggering cytotoxic T lymphocytes dysfunction. Molecular Medicine, 28(1), 47. https://doi.org/10.1186/s10020-022-00475-8 [Google Scholar] [PubMed] [CrossRef]
53. Wei, Y., Li, Z. (2022). LAG3-PD-1 combo overcome the disadvantage of drug resistance. Frontiers in Oncology, 12, 1–7. https://doi.org/10.3389/fonc.2022.831407 [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools