 Open Access
Open Access
ARTICLE
An inflammatory-related genes signature based model for prognosis prediction in breast cancer
Jiangsu Breast Disease Center, The First Affiliated Hospital with Nanjing Medical University, Nanjing, China
* Corresponding Author: TIANSONG XIA. Email:
Oncology Research 2023, 31(2), 157-167. https://doi.org/10.32604/or.2023.027972
Received 24 November 2022; Accepted 14 February 2023; Issue published 10 April 2023
Abstract
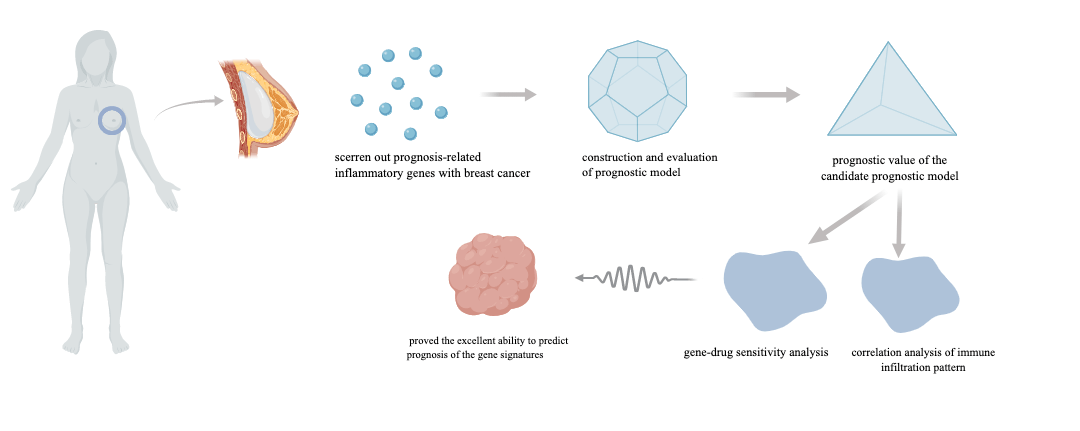
Background: Breast cancer has become the most common malignant tumor in the world. It is vital to discover novel prognostic biomarkers despite the fact that the majority of breast cancer patients have a good prognosis because of the high heterogeneity of breast cancer, which causes the disparity in prognosis. Recently, inflammatory-related genes have been proven to play an important role in the development and progression of breast cancer, so we set out to investigate the predictive usefulness of inflammatory-related genes in breast malignancies. Methods: We assessed the connection between Inflammatory-Related Genes (IRGs) and breast cancer by studying the TCGA database. Following differential and univariate Cox regression analysis, prognosis-related differentially expressed inflammatory genes were estimated. The prognostic model was constructed through the Least Absolute Shrinkage and Selector Operation (LASSO) regression based on the IRGs. The accuracy of the prognostic model was then evaluated using the Kaplan-Meier and Receiver Operating Characteristic (ROC) curves. The nomogram model was established to predict the survival rate of breast cancer patients clinically. Based on the prognostic expression, we also looked at immune cell infiltration and the function of immune-related pathways. The CellMiner database was used to research drug sensitivity. Results: In this study, 7 IRGs were selected to construct a prognostic risk model. Further research revealed a negative relationship between the risk score and the prognosis of breast cancer patients. The ROC curve proved the accuracy of the prognostic model, and the nomogram accurately predicted survival rate. The scores of tumor-infiltrating immune cells and immune-related pathways were utilized to calculate the differences between the low- and high-risk groups, and then explored the relationship between drug susceptibility and the genes that were included in the model. Conclusion: These findings contributed to a better understanding of the function of inflammatory-related genes in breast cancer, and the prognostic risk model provides a potentially promising prognostic strategy for breast cancer.Graphic Abstract
Keywords
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools