 Open Access
Open Access
ARTICLE
MiR-145-5p Suppresses Hepatocellular Carcinoma Progression by Targeting ABHD17C
1 Department of Hepatobiliary and Pancreatic Surgery, The Second Affiliated Hospital of Fujian Medical University, Quanzhou, 362000, China
2 Department of Health Medicine, The 910th Hospital of People’s Liberation Army, Quanzhou, 362000, China
3 Department of General Surgery, The Second Affiliated Hospital of Fujian Medical University, Quanzhou, 362000, China
* Corresponding Authors: Wei Wang. Email: ; Shaojian Chen. Email:
# These authors contributed equally in the study
Oncologie 2022, 24(4), 897-912. https://doi.org/10.32604/oncologie.2022.025693
Received 26 July 2022; Accepted 05 October 2022; Issue published 31 December 2022
Abstract
Background: MicroRNA-145-5p (miR-145-5p) reportedly inhibits hepatocellular carcinoma (HCC) by targeting ARF6, SPATS2, CDCA3, KLF5, and NRAS, indicating that miR-145-5p plays an important role in the occurrence and development of HCC by regulating the expression of various genes. In this study, we aimed to explore novel downstream targets of miR-145-5p and elucidate the potential mechanism of miR-145-5p in HCC. Materials and Methods: A bioinformatics analysis was performed to determine the clinical significance of miR-145-5p and alpha/beta hydrolase domain-containing protein 17C (ABHD17C) in patients with HCC. The ability of Hep3B cells to proliferate, migrate, and invade was examined after overexpression of miR-145-5p and ABHD17C or knockdown of ABHD17C. Tumorigenesis of Hep3B cells overexpressing miR-145 was detected using in vivo experiments. Results: miR-145-5p was downregulated in HCC tissues, and this was associated with poor prognosis in patients with HCC. Based on the bioinformatics analysis, miR-145-5p was predicted to target ABHD17C, as demonstrated by a luciferase reporter assay. ABHD17C downregulation inhibited cell viability, migration, and invasion and arrested the cell cycle. Overexpression of miR-145-5p significantly reduced the expression of ABHD17C. Moreover, ABHD17C expression was elevated in HCC tissues, which was associated with an unfavorable prognosis. Re-expressing ABHD17C into HCC cells rescued the suppressed cell viability, migration, and invasion mediated by ectopic expression of miR-145-5p. Importantly, miR-145-5p suppressed tumor growth in mice and downregulated the levels of Ki67 and ABHD17C in tumor. Conclusion: miR-145-5p could attenuate HCC progression via suppressing ABHD17C.Keywords
Hepatocellular carcinoma (HCC) is a leading cause of cancer-related deaths worldwide [1]. The five-year survival of patients with HCC remains poor, although multiple therapeutic advances have been achieved in recent decades [2]. Aberrant genetic events can result in the mutagenesis of critical driver genes, such as CTNNB1 and P53, resulting in HCC initiation and progression [3]. Multiple predictive biomarkers, including alpha-fetoprotein (AFP), have been validated for early diagnosis and subsequent prognosis of HCC [4,5]. However, specific biomarkers for improving the early detection of HCC need to be established. Moreover, prognostic biomarkers with superior effectiveness could benefit patients with HCC for clinical stratification. Chemotherapeutic strategies, such as sorafenib therapy, were found to be an effective treatment strategy in patients with HCC; nonetheless, resistance to chemotherapeutic drugs markedly undermines their benefits to patients [6]. Therefore, additional druggable targets need to be explored for adjuvant therapies.
As a family of small non-coding RNAs, microRNAs (miRNAs) suppress target gene expression via sequence matching, thereby modulating cellular processes [7,8]. Certain miRNAs are preferentially expressed in liver tissues or during HCC, affording ideal predictive biomarkers and/or promising targets for therapeutic purposes [9]. For instance, miR-122 accounts for more than 70% of all miRNAs in the liver, and loss of miR-122 reportedly results in HCC development [10]. As a miRNA whose tumor-suppressive effects have been validated in multiple cancer types, miR-145-5p was shown to exert tumor-inhibitory effects in HCC by mitigating the expression of several target genes, including ARF6, SPATS2, CDCA3, KLF5, and NRAS [11–15], indicating that miR-145-5p plays an important role in the occurrence and development of HCC. Therefore, the systematic identification of critical miR-145-5p targets in HCC cells can aid in comprehensively elucidating the mechanism through which miR-145-5p modulates HCC progression.
Alpha/beta hydrolase domain-containing protein 17 (ABHD17) has been identified as a protein acyl thioesterase that selectively eliminates palmitoylation of stress-regulated exon linkers to regulate channel gating [16]. ABHD17 is divided into ABHD17A, ABHD17B, and ABHD17C, which function as protein depalmitoylases [17]. Reportedly, ABHD17C can hydrolyze fatty acids from S-acylated cysteine residues in proteins and exert depalmitoylation activity against NRAS and DLG4/PSD95 [18]. In addition, ABHD17C was shown to excise the potassium channel lipidome precisely [19]. Although ABHD17C is highly expressed in breast tissue tumor xenografts and patients with breast cancer [20], its expression pattern and role in HCC remain unexplored.
In this study, we confirmed the clinical significance of miR-145-5p as a deregulated miRNA in patients with HCC. In-depth in silico screening using coupling prediction results from diverse platforms can offer multiple target gene hits, among which oncogenic ABHD17C was confirmed as a pivotal target gene of miR-145-5p. Notably, ABHD17C re-expression into HCC cells mitigated the tumor-suppressive role of miR-145-5p. Finally, the results from animal experiments confirmed the negative effects of miR-145-5p on HCC growth in vivo. Our study coupled bioinformatic approaches and experimental analyses to comprehensively reveal the clinical significance and biological functions of miR-145-5p.
Raw transcriptome analysis data of normal liver and HCC tissues from patients with HCC were downloaded from The Cancer Genome Atlas Liver Hepatocellular Carcinoma (TCGA-LIHC) (https://portal.gdc.cancer.gov/). Subsequently, the R edgeR package (Version 3.30.3) was used to normalize the bioinformatics data, and a comparison was conducted. The Kaplan-Meier curve R survival package (version 3.1 12) was used to analyze patient survival. For screening miR-145-5p target genes, analysis was performed on six online platforms (TargetScan [21], MicroT-CDS [22], miRDB [23], miTarBase [24], starBase [25], and mirDIP [26]).
HEK293T and Hep3B cells were obtained from IMMOCELL (Xiamen, China) and cultured in Dulbecco’s modified Eagle’s medium (Gibco, China) containing 10% fetal bovine serum (FBS; Gibco, China) at 37°C in a humidified incubator with 5% CO2.
2.3 Plasmids, Transfections, and Lentiviral Transduction
miR-145 (pri-miR-145-5p) or ABHD17C was amplified and inserted into a pCDH-EF1A-MCS-T2A vector to generate plasmids encoding miR-145 and ABHD17C, named miR-145 OE and ABHD17C OE, respectively. The short hairpin RNA of ABHD17C was designed and inserted into a pLKO.1-puro vector to generate plasmids encoding the short hairpin RNA of ABHD17C, namely shABHD17C. The negative control (NC) of shABHD17C was named shNC. The wild-type (WT) 3’UTR of ABHD17C (ABHD17C 3’UTR WT) and mutant 3’UTR of ABHD17C (ABHD17C 3’UTR MUT) were also PCR-amplified and ligated into the pmirGLO vector. The primers used to construct plasmids are as follows. ABHD17C forward primer: 5′-CTAGAGAATTCGGATCCATGCCCGAGCCAGGCCCCAGGATGAAC-3′, ABHD17C reverse primer: 5′-AGCTTCCATGGCTCGAGGGAATTAGGAAGTTCGTGAG-3′, pri-miR-145-5p forward primer: 5′-ATTCACGCGTGCGGCCGCCGGCGGCCTTGGCGCTGAAG-3′, pri-miR-145-5p reverse primer: 5′-TAGGGATCCGGGCCCGGGGTGGGAAGGAGGCAAATCC-3′, ABHD17C 3’UTR WT forward primer: 5′-AGCTCGCTAGCCTCGAGATGGGAATGAGAGCTGAATG-3′, ABHD17C 3’UTR WT reverse primer: 5′-CATGCCTGCAGGTCGACTCTATGCACAATGTGATTGC-3′, ABHD17C 3’UTR MUT-1 forward primer: 5′-AGTGTTTGACCTAAGCTAGTGTGGTGAAAATTC-3′, ABHD17C 3’UTR MUT-1 reverse primer: 5′-ACTAGCTTAGGTCAAACACTATCACAGCTTCAC-3′, ABHD17C 3’UTR MUT-2 forward primer: 5′-TATAAATATTTGACCTATATTCTTAAACAAAAAG-3′, ABHD17C 3’UTR MUT-2 reverse primer: 5′-ATATAGGTCAAATATTTATATGGTTACAG-3′, shABHD17C forward primer: 5′-CCGGAAGAUGUUGCAGUUGAUGCGGCTCGAGCCGCATCAACTGCAACATCTTTTTTT-3′, shABHD17C reverse primer: 5′-AATTAAAAAAAGAUGUUGCAGUUGAUGCGGCTCGAGCCGCATCAACTGCAACATCTT-3′, shNC forward primer: 5′-CCGGTTCTCCGAACGTGTCACGTCTCGAGACGTGACACGTTCGGAGAATTTTT-3′, shNC reverse primer: 5′-AATTAAAAATTCTCCGAACGTGTCACGTCTCGAGACGTGACACGTTCGGAGAA-3′.
miR-145-5p mimic, NC of miR-145-5p mimic (mimic NC), miR-145-5p inhibitor, NC of miR-145-5p (inhibitor NC) (RiboBio, Guangzhou, China), shABHD17C, shNC, and ABHD17C OE were transfected into Hep3B cells using Lipofectamine™ 2000 (11668500, Invitrogen, China) to increase the level of miR-145-5p, inhibit miR-145-5p, decrease the expression of ABHD17C, and overexpress ABHD17C, respectively. The cells were analyzed two days post-transfection. Plasmids expressing pri-miR-145-5p were transfected into HEK293T cells along with pMD2G and pspax2 plasmids. After 48 h, the lentivirus-containing supernatant was collected; lentivirus was enriched, and the titer was determined as described previously [27]. The sequence of miR-145-5p mimic: 5′-GUCCAGUUUUCCCAGGAAUCCCU-3′; sequence of mimic NC: 5′-UCACAACCUCCUAGAAAGAGUAGA-3′; sequence of inhibitor NC: 5′-CAGUACUUUUGUGUAGUACAA-3′; sequence of miR-145-5p inhibitor: 5′-AGGGAUUCCUGGGAAAACUGGAC-3′.
2.4 Construction of Cell Line Stably Expressing pri-miR-145-5p
In the presence of 5 μg/mL polypropylene, the lentivirus was transduced into Hep3B cells at a multiplicity of infection of 10. After 48 h, the medium was replaced with fresh medium, and puromycin was added at a final concentration of 1.5 μg/mL. After 72 h, the cells were collected to analyze miR-145-5p expression.
2.5 Quantitative Reverse Transcription-PCR (qPCR)
Total RNA was isolated using the TRIzol reagent (15596018, Invitrogen, USA). The same amount of RNA was transcribed into cDNA using a Mir-X miRNA first-strand synthesis kit (638315, Takara, China) with specific primers for miRNAs or using the Superscript III First-Strand Synthesis System (18080051, Invitrogen, China) with random primers for other genes, according to the manufacturer’s instructions. qPCR was performed using Mir-X miRNA qPCR TB Green® Kit (638314, Takara, China) for miRNAs and U6, or TB Green® Fast qPCR Mix (RR430A, Takara, China) for other genes, and Thermal Cycler Dice™ Real Time System III with PC (TP970, Takara, China), according to the manufacturer’s instructions. The cDNAs of miRNA and U6 were pre-denatured at 95°C for 10 s, denatured at 95°C for 5 s, annealed, and extended at 60°C for 20 s. Denaturation, annealing, and extension were performed for 40 cycles. Finally, the melting curve was obtained. The melting curve program consisted of 95°C for 60 s, 55°C for 30 s, and 95°C for 30 s. The thermal cycling conditions for qPCR of the other genes were 95°C for 30 s, 40 cycles of 95°C for 5 s, and 60°C for 15 s, followed by melting curves. Data for miRNAs and other genes were normalized using the 2−ΔΔCq method according to U6 and 18S rRNA levels, respectively. The primers used for qPCR are as follows: U6 reverse transcription primer: 5′-AACGCTTCACGAATTTGCGT-3′, U6 forward primer: 5′-CTCGCTTCGGCAGCACA-3′, U6 reverse primer: 5′-AACGCTTCACGAATTTGCGT-3′, miR-145-5p reverse transcription primer: 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGGGAT-3′, miR-145-5p forward primer:5′-CGGTCCAGTTTTCCCAGGA-3′, miR-145-5p reverse primer: 5′-AGTGCAGGGTCCGAGGTATT-3′, 18S rRNA forward primer: 5′-AGGCGCGCAAATTACCCAATCC-3′, 18S rRNA reverse primer: 5′-GCCCTCCAATTGTTCCTCGTTAAG-3′, ABHD17C forward primer: 5′-TACTGCTTTGATGCTTTCC-3′, ABHD17C reverse primer: 5′-ATCCTCTGTACCATGAATGA-3′, ANGPT2 forward primer: 5′-GTGGCTAATGAAGGTATT-3′, ANGPT2 reverse primer: 5′-TTATTGACTGTAGTTGGAT-3′, CTNNBIP1 forward primer: 5′-TGACCAACAGAAACCTTT-3′, CTNNBIP1 reverse primer: 5′-AATCAGACCTCTTCACATT-3′, PDGFD forward primer: 5′-GGATTAGAGGAAGCAGAA-3′, PDGFD reverse primer: 5′-TCGGACTTGAATGTGATT-3′, PODXL forward primer: 5′-AAGATAAGTGCGGCATAC-3′, PODXL reverse primer: 5′-GCTTAGTGTGAATAGTGATT-3′, SMAD3 forward primer: 5′-GCACATAATAACTTGGACCT-3′, SMAD3 reverse primer: 5′-GCTCGTAGTAGGAGATGG-3′, SMAD5 forward primer: 5′-AAGCCGTTGGATATTTGT-3′, SMAD5 reverse primer: 5′-AGGTAAGACTGGACTCTC-3′, SRGAP1 forward primer: 5′-ATCAATCTCTATGGTCTTCA-3′, SRGAP1 reverse primer: 5′-ATGGTTACTCTGGTCATC-3′.
Briefly, ice-cold RIPA buffer (P0013C, Beyotime, Shanghai, China) supplemented with phenylmethanesulfonyl fluoride (PMSF; ST506, Beyotime) was used to lyse cells or tumor tissues. Proteins (20 μg) were separated via sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) after determining protein concentration using a BCA Protein Concentration Assay Kit (P0010S, Beyotime). After transferring the separated proteins onto a polyvinylidene fluoride membrane, which was then blocked with 5% skimmed milk at 26°C for 2 h, the membrane was incubated with diluted ABHD17C (PA5-61831, Thermo Fisher Scientific, USA) antibody, β-actin antibody (20536-1-AP, Proteintech, China) and GAPDH antibody (10494-1-AP, Proteintech, China) at 26°C for 2 h. Subsequently, horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (SA00001-2, Proteintech, China) and an ECL substrate (Bio-Rad, China) were used to visualize the proteins. Densiometric analysis was performed using ImageJ v1.48 (National Institutes of Health).
Briefly, 1,000 Hep3B cells exhibiting upregulated miR-145-5p and ABHD17C or downregulated ABHD17C were plated on 96-well plates. The following day, cell viability was quantified using the CyQUANT™ MTT Cell Viability Assay kit (V13154, Invitrogen, China) according to manufacturer’s instruction.
2.8 Invasion and Migration Assays
Briefly, 1 × 105 Hep3B cells, exhibiting upregulated miR-145-5p and ABHD17C or downregulated ABHD17C, were suspended in serum-free medium and then added to the upper Transwell inserts in the absence (for migration assays) or presence (for invasion assays) of Matrigel. 500 μL medium containing 10% FBS were added to the lower chamber. After incubation for 48 h, cells in the lower phase of the upper chamber were fixed with 4% paraformaldehyde and stained with 0.5% crystal violet. Migrated or invasive cells were photographed and counted under a microscope (Motic Incorporation, Ltd., China).
ABHD17C 3’UTR WT or ABHD17C 3’UTR MUT and pRL-TK vector (D2760, Beyotime) were co-transfected with miR-145-5p mimic or mimic NC into HEK293T cells. After 2 days, the cells were harvested and treated using a dual luciferase reporter assay kit (E1910, Promega, China), according to the manufacturer’s instructions. Subsequently, firefly luciferase and Renilla luciferase activities were detected using a multi-mode microplate reader (Synergy Neo2, BioTek, USA). Data were normalized by dividing firefly luciferase activity by that of Renilla luciferase.
2.10 Cell Cycle Quantification
For cell cycle analysis, Hep3B cells with upregulated miR-145-5p and ABHD17C or downregulated ABHD17C were fixed with pre-cooled 75% ethanol at 4°C for 4 h, permeated with 0.2% Triton X-100 in the presence of RNase A (Beyotime, Shanghai, China) for 30 min, and stained with propidium iodide (PI; 50 μg/mL) for 0.5 h at 26°C. Subsequently, stained cells were measured using a FACScan flow cytometer (Beckman Coulter, USA), and the resulting data were analyzed using Cell Quest software.
Experiments were performed under a project license granted by the ethics board of Fujian Medical University in compliance with the institutional guidelines for the care and use of animals. Briefly, 1 × 106 Hep3B cells stably overexpressing miR-145 were subcutaneously injected into six-week male BALB/c nude mice, procured from the Shanghai Laboratory Animal Research Center. Tumors were monitored, and all mice were sacrificed on day 37 post-injection. Obtained tumors were subjected to biological analyses, including qPCR, western blotting, and immunohistochemistry (IHC) assays.
Tumor samples were sliced, fixed, and blocked, as previously described. Subsequently, blocked slices were incubated with diluted Ki67 antibody (catalog number: 27309-1-AP; Proteintech) for 12 h at 4°C. After incubation with HRP-conjugated goat anti-rabbit IgG (catalog number: SA00001-2; Proteintech), sections were incubated with DAB for color development. Subsequently, the slices were treated with hematoxylin to stain nuclei and dehydrated. Finally, the sections were sealed with transparent resin and observed under an Olympus light microscope (BX51, Olympus Corporation, Tokyo, Japan).
Data were analyzed using GraphPad Prism 8 software (GraphPad Software, Inc., La Jolla, CA, USA). The Shapiro–Wilk test was performed to determine whether data followed a normal distribution. Levene’s test was used to ensure homogeneity of variance. The Mann–Whitney U test was performed to compare differences between two groups of nonparametric data. Survival curves were calculated using the Kaplan-Meier method, and significance was determined using the log-rank test. The Student’s t test was performed to compare differences between the two groups of parametric data. Analysis of variance (ANOVA) followed by Tukey’s post-hoc test was used for multiple comparisons among three groups of parametric data. Statistical significance was set at P < 0.05.
3.1 MiR-145-5p Is Enriched as an HCC-Suppressive miRNA and Correlates with a Better Prognosis
To evaluate the clinical significance of miR-145-5p, we analyzed miRNA transcriptome data from normal non-tumor (NT) and HCC tissues. Notably, miR-145-5p was significantly downregulated in HCC tissues compared with that in NT tissues (Fig. 1A). We then determined whether miR-145-5p expression could be employed as a candidate predictive biomarker. Analysis of survival data of patients with HCC revealed that patients with higher miR-145-5p expression exhibited longer overall survival (OS) and disease-free intervals (DFI; Figs. 1B and 1C). Collectively, miR-145-5p expression correlated with a favorable prognosis in patients with HCC.
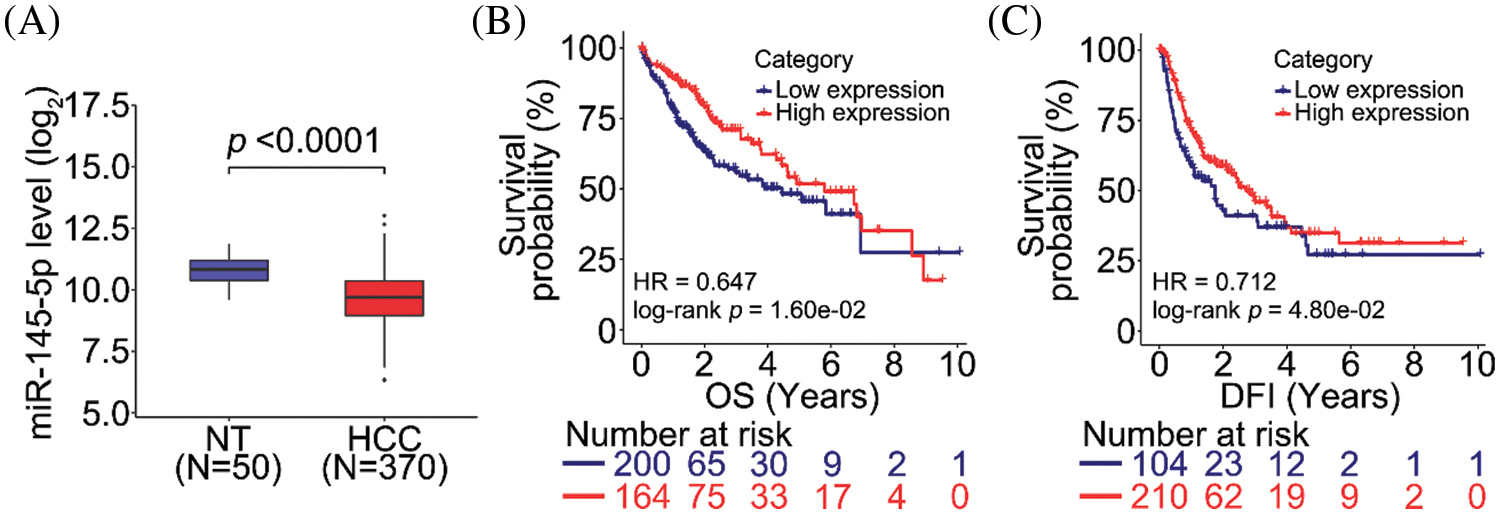
Figure 1: miR-145-5p correlates with a favorable prognosis in patients with HCC. (A) miR-145-5p expression is downregulated in HCC tissues when compared with that in normal non-tumor (NT) tissues. (B, C) miR-145-5p expression correlates with better overall survival (OS; B) and disease-free interval (DFI; C) in patients with HCC. Data analyses were performed using the Mann–Whitney test (A) and log-rank test (B and C). HCC, hepatocellular carcinoma
3.2 ABHD17C is a Target of miR-145-5p
Given that a miRNA may target a panel of mRNAs for specific suppression, the six most prevalent target prediction platforms were applied to identify actual miR-145-5p targets. Ultimately, 12 genes were identified as hits with the highest possibility of miR-145-5p interaction (Fig. 2). Further analysis was conducted by overlapping 12 putative miR-145-5p targeting genes with genes that were upregulated in patients with HCC from TCGA-LIHC (Fig. 3A). Eight genes were identified in the intersecting compartment (Fig. 3A). To experimentally validate these predictive results, miR-145-5p mimic or a control non-targeting miRNA mimic was transfected into the commonly used HCC cell line, Hep3B (Fig. 3B). qPCR analysis revealed that the expression of six of eight genes was significantly suppressed following ectopic expression of miR-145-5p (Fig. 3C). Among these six genes, ABHD17C expression was markedly inhibited by miR-145-5p, suggesting that it may be a vital target gene for mediating the biological effects of miR-145-5p (Fig. 3C). Dual luciferase reporter assays indicated that miR-145-5p directly bound to ABHD17C 3′-UTR (WT; Figs. 3D and 3E). Nevertheless, the interaction was suppressed when the sequence of the identified interaction site was mutated (Figs. 3D and 3E). Downregulation of ABHD17C had no significant effect on miR-145-5p levels (Figs. 3F and 3G). Moreover, increasing miR-145-5p expression significantly decreased the expression of ABHD17C, whereas inhibiting miR-145-5p expression significantly enhanced ABHD17C expression (Figs. 3H and 3I). Collectively, miR-145-5p specifically targeted ABHD17C to inhibit its expression.
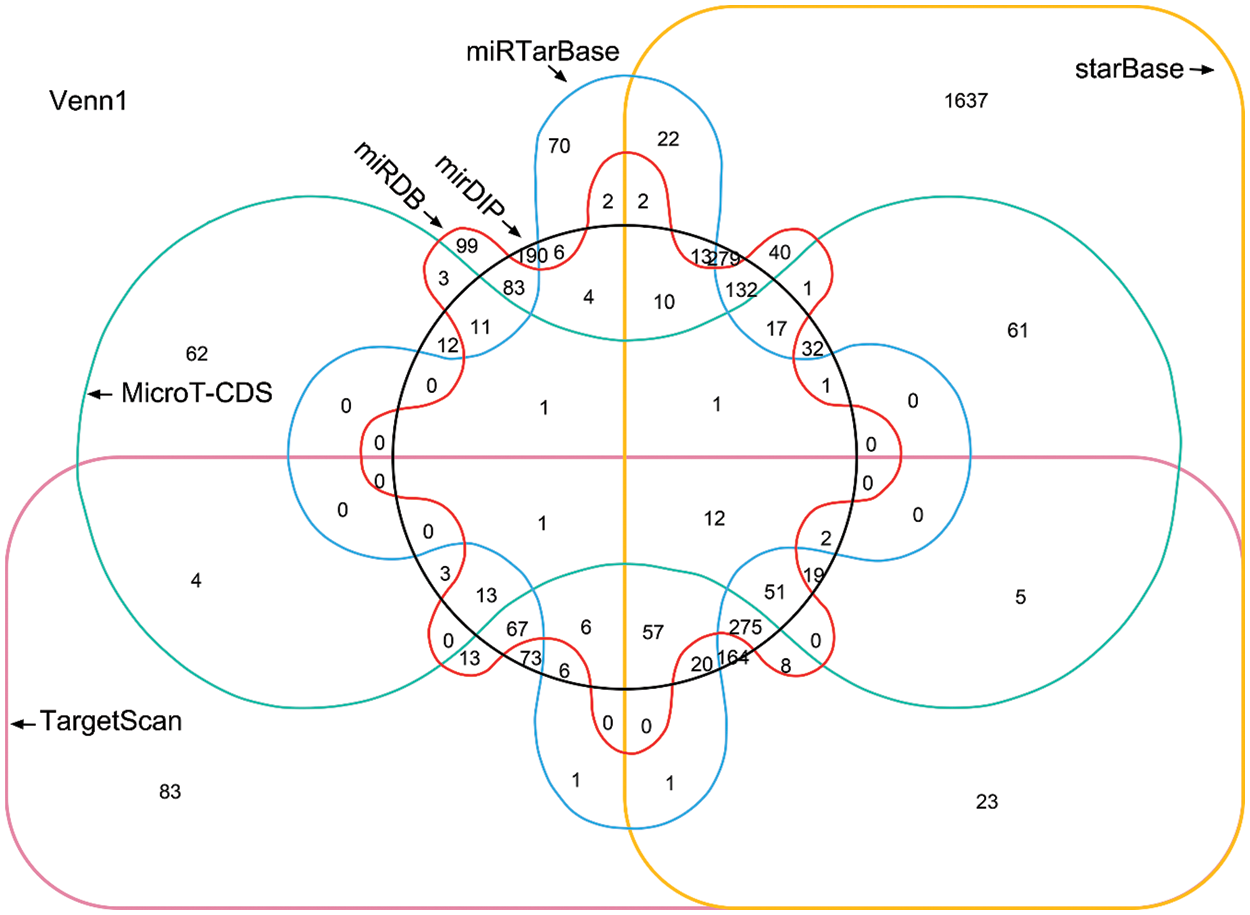
Figure 2: Venn diagram showing the prediction results of miR-145-5p target genes from six platforms
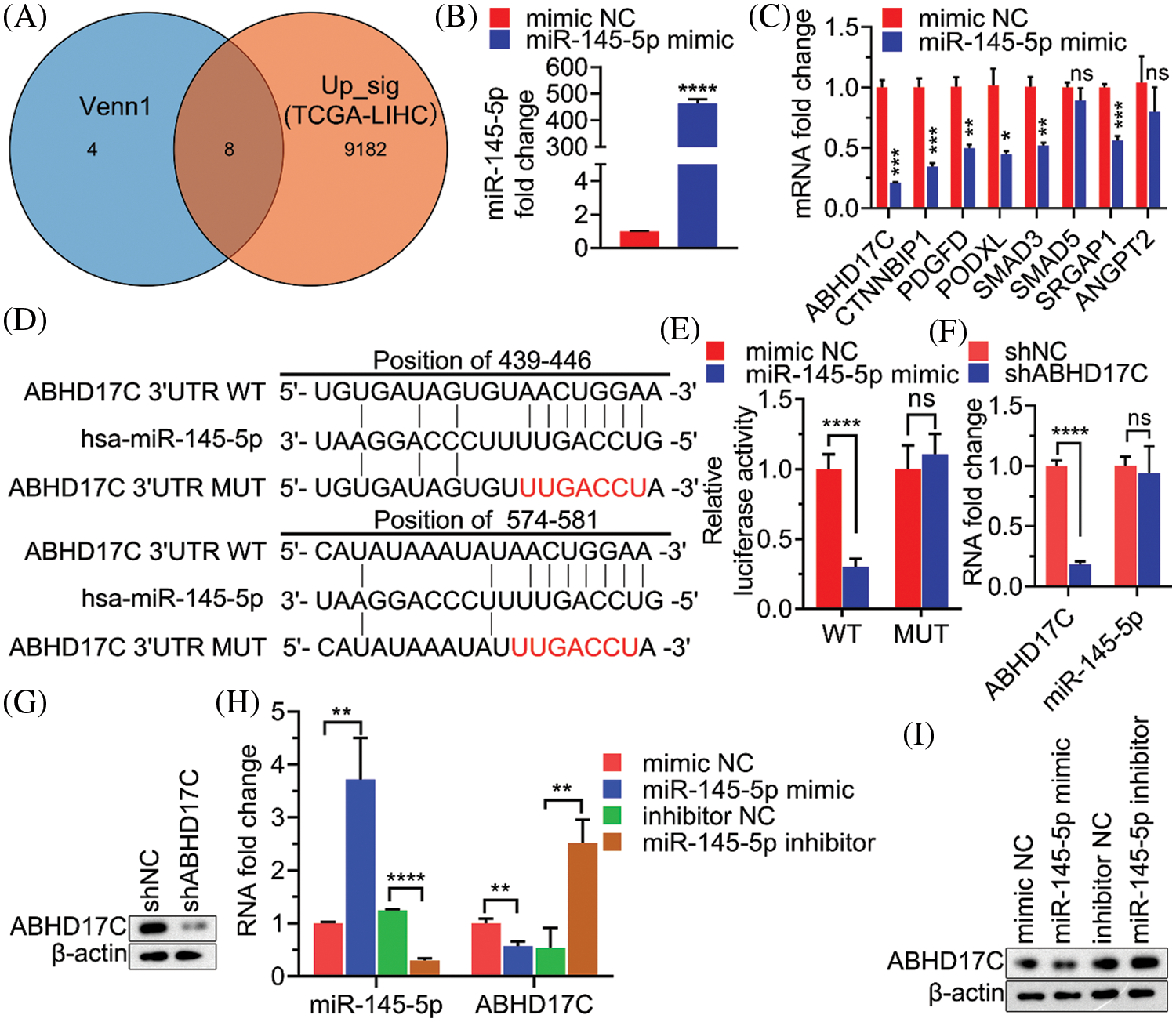
Figure 3: ABHD17C is a target gene of miR-145-5p. (A) Venn diagram showing the overlap between the gene list from Fig. 2 and genes upregulated in patients with HCC from TCGA-LIHC. (B) qPCR results validating miR-145-5p overexpression in Hep3B cells. (C) Expression of miR-145-5p target gene candidates in miR-145-5p-overexpressing Hep3B cells. (D) A schematic panel illustrating the matching sequences between ABHD17C and miR-145-5p wild-type (WT) or mutated (MUT). (E) Luciferase reporter assays assessing the interaction between ABHD17C and miR-145-5p WT or MUT. (F, G) Expression of miR-145-5p (F) and ABHD17C (F and G) in Hep3B cells, transfected with shNC or shABHD17C. (H and I) Expression of miR-145-5p (H) and ABHD17C (H and I) in Hep3B cells, transfected with miR-145-5p mimic, mimic negative control (NC), miR-145-5p inhibitor, and inhibitor NC. Data are presented as mean ± standard deviation (SD) and were analyzed using Student’s t test (B, C, E, F, and I). ABHD17C, alpha/beta hydrolase domain-containing protein 17C; HCC, hepatocellular carcinoma, ns: not significant. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001
3.3 ABHD17C Expression Is Associated with an Unfavorable Prognosis in Patients with HCC
To evaluate the clinical significance of ABHD17C, the same clinical data of patients with HCC was used to analyze ABHD17C expression. In contrast to miR-145-5p expression, ABHD17C expression levels were enhanced in HCC tissue samples when compared with those in NT tissues (Fig. 4A). ABHD17C levels were not significantly associated with better or worse OS (Fig. 4B), but high ABHD17C expression correlates with poor DFI (Fig. 4C).
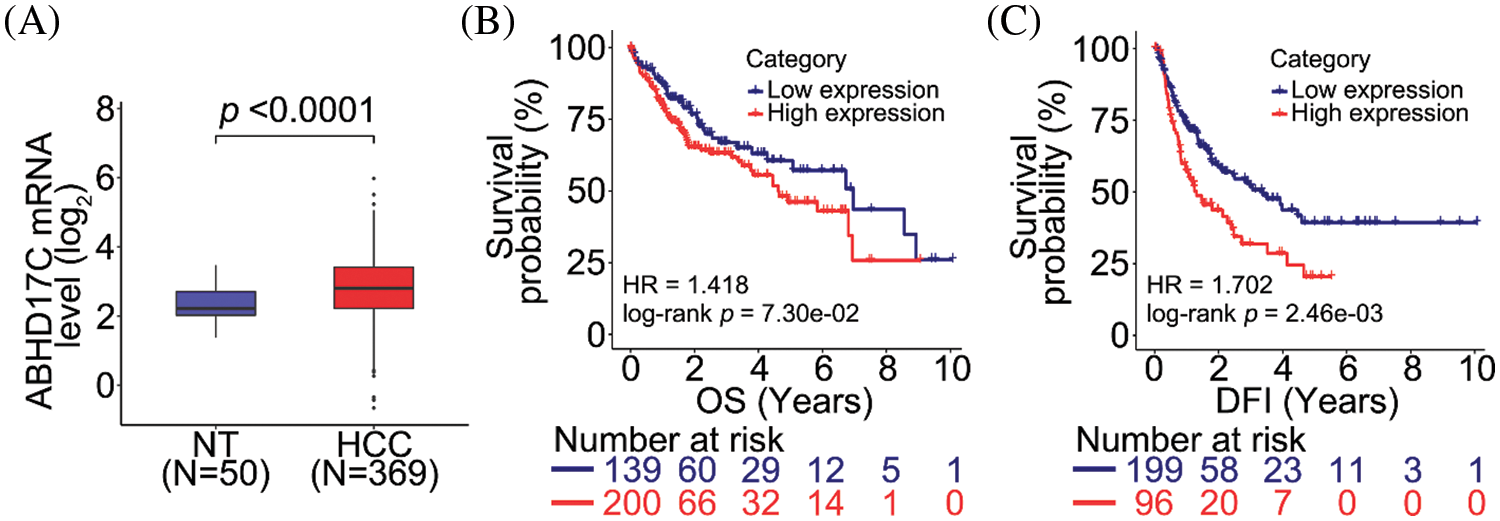
Figure 4: ABHD17C correlates with unfavorable prognosis in patients with HCC. (A) ABHD17C expression is upregulated in HCC tissues when compared with that in normal non-tumor (NT) tissues. (B, C) Correlation between ABHD17C level and overall survival (OS; B) or disease-free interval (DFI; C) in patients with HCC. Data were analyzed using the Mann–Whitney test (A) and log-rank test (B and C). ABHD17C, alpha/beta hydrolase domain-containing protein 17C; HCC, hepatocellular carcinoma
3.4 Downregulation of ABHD17C Expression Inhibits Cell Viability, Migration, and Invasion and Arrests Cell Cycle
To determine the role of ABHD17C in the biological processes of HCC cells, we knocked down ABHD17C in Hep3B cells. Downregulation of ABHD17C expression significantly reduced cell viability (Fig. 5A) and arrested the cell cycle in the G0/G1 phase (Fig. 5B). Moreover, deleting ABHD17C expression could suppress the migratory and invasive abilities of these cells (Fig. 5C). These findings suggested that ABHD17C knockdown significantly suppressed the malignant behavior of Hep3B cells.
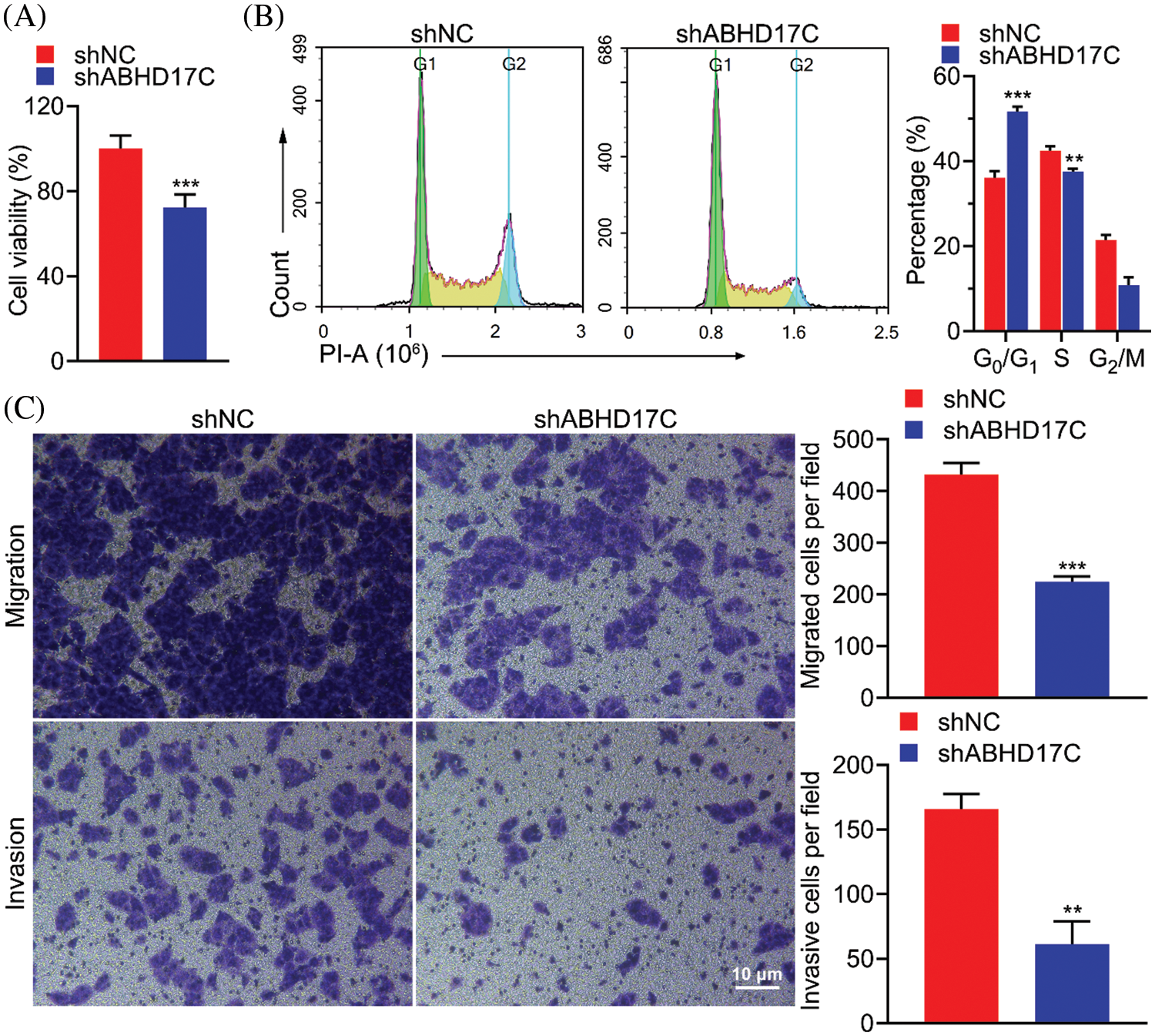
Figure 5: Downregulation of ABHD17C expression inhibits cell viability, migration, and invasion and arrests cell cycle. (A) MTT assays were performed to determine the viability of Hep3B cells exhibiting downregulated ABHD17C expression. (B) Flow cytometric analysis and quantification of Hep3B cells exhibiting downregulated ABHD17C expression to quantify the cell cycle distribution. (C) Transwell assays were performed to evaluate the migratory (upper) and invasive (lower) abilities of Hep3B cells exhibiting downregulated ABHD17C expression. Data are presented as mean ± SD and were analyzed using Student’s t test (A, B, and C). ABHD17C, alpha/beta hydrolase domain-containing protein 17C. **P < 0.01, ***P < 0.001
3.5 Mitigation of Cell Viability, Migration, and Invasion Mediated by miR-145-5p Is ABHD17C Dependent
To determine whether ABHD17C is biologically indispensable to mediate the effect of miR-145-5p in HCC cells, ABHD17C was re-expressed in Hep3B cells by transient transfection. qPCR and western blotting confirmed that ABHD17C was efficiently re-transduced into Hep3B cells overexpressing miR-145-5p (Figs. 6A and 6B). Next, we examined the biological parameters of Hep3B cells. As shown in Fig. 6C, miR-145-5p alleviated Hep3B cell viability, whereas ABHD17C re-expression significantly restored this effect (Fig. 6C). Consistently, the HCC cell cycle was dramatically arrested in the G0/G1 phase in miR-145-5p-overexpressing cells. However, re-expression of ABHD17C in Hep3B cells substantially attenuated miR-145-5p-directed cell cycle arrest (Figs. 6D and 6E). Given that gain of migratory and invasive properties is a hallmark of cancer progression [28], we examined the effects of miR-145-5p and ABHD17C expression on HCC cell migration and invasion. Consistent with the cell viability results, the suppressive effects of miR-145-5p on Hep3B cell migration and invasion were rescued following ABHD17C re-expression (Figs. 7A and 7B). In conclusion, ABHD17C was experimentally confirmed as a critical target of miR-145-5p for suppressing malignant HCC cells.
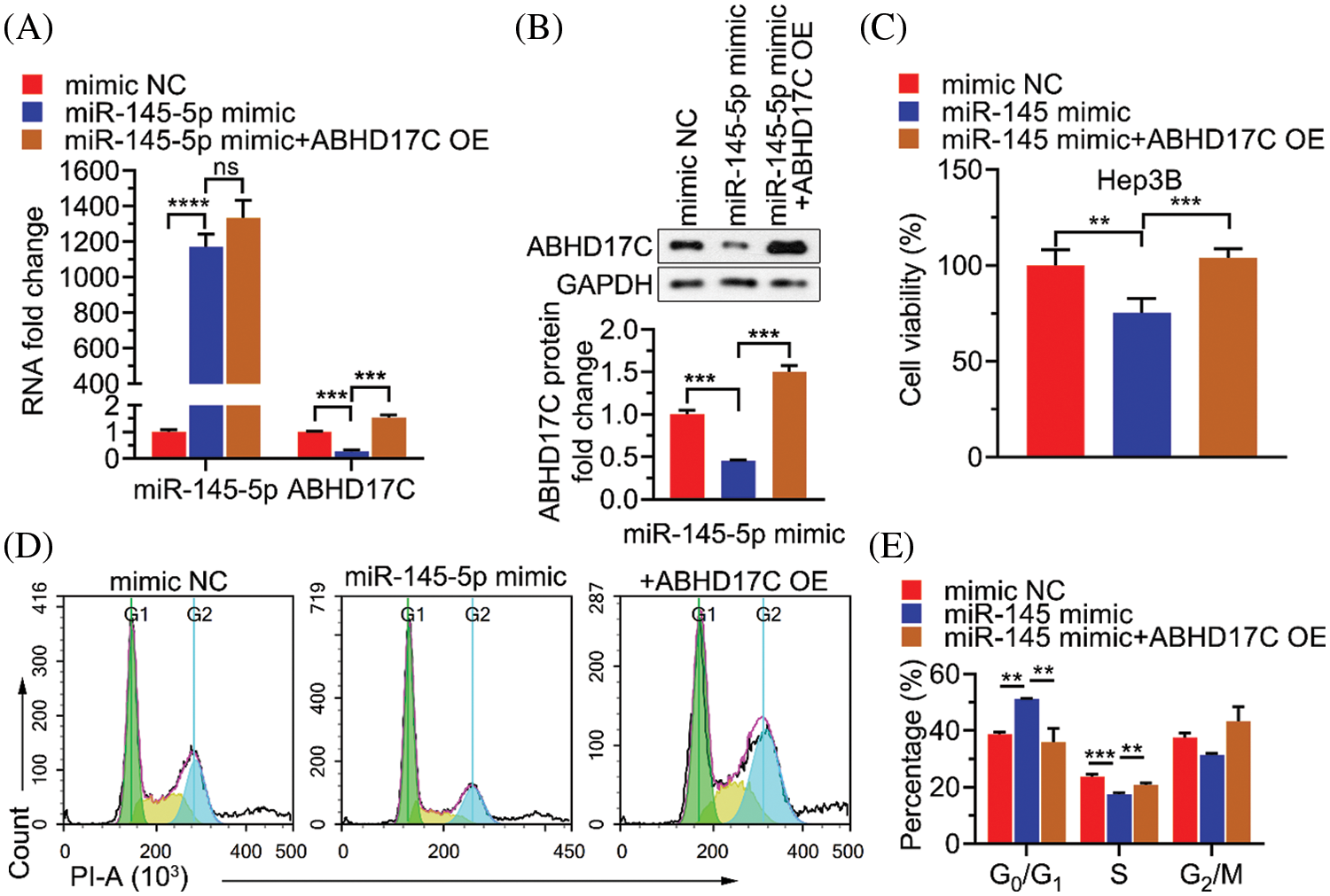
Figure 6: Inhibitory effects of miR-145-5p on HCC cell viability depend on the suppressed ABHD17C expression. (A) qPCR analysis of miR-145-5p or ABHD17C expression in Hep3B cells upon miR-145-5p mimic or/and ABHD17C transfection. (B) Western blotting results (upper) and quantification (lower) of ABHD17C expression in Hep3B cells following miR-145-5p mimic or/and ABHD17C transfection. (C) MTT assays were performed to determine the viability of Hep3B cells exhibiting miR-145-5p and ABHD17C misexpression. (D, E) Flow cytometric analysis (D) and quantification (E) on Hep3B cells exhibiting miR-145-5p and ABHD17C misexpression to quantify cell cycle distribution. Data are presented as mean ± SD and were analyzed using analysis of variance (ANOVA), followed by Tukey’s post-hoc test (A, B, C, and E). ABHD17C, alpha/beta hydrolase domain-containing protein 17C; HCC, hepatocellular carcinoma, ns: not significant. **P < 0.01, ***P < 0.001, ****P < 0.0001
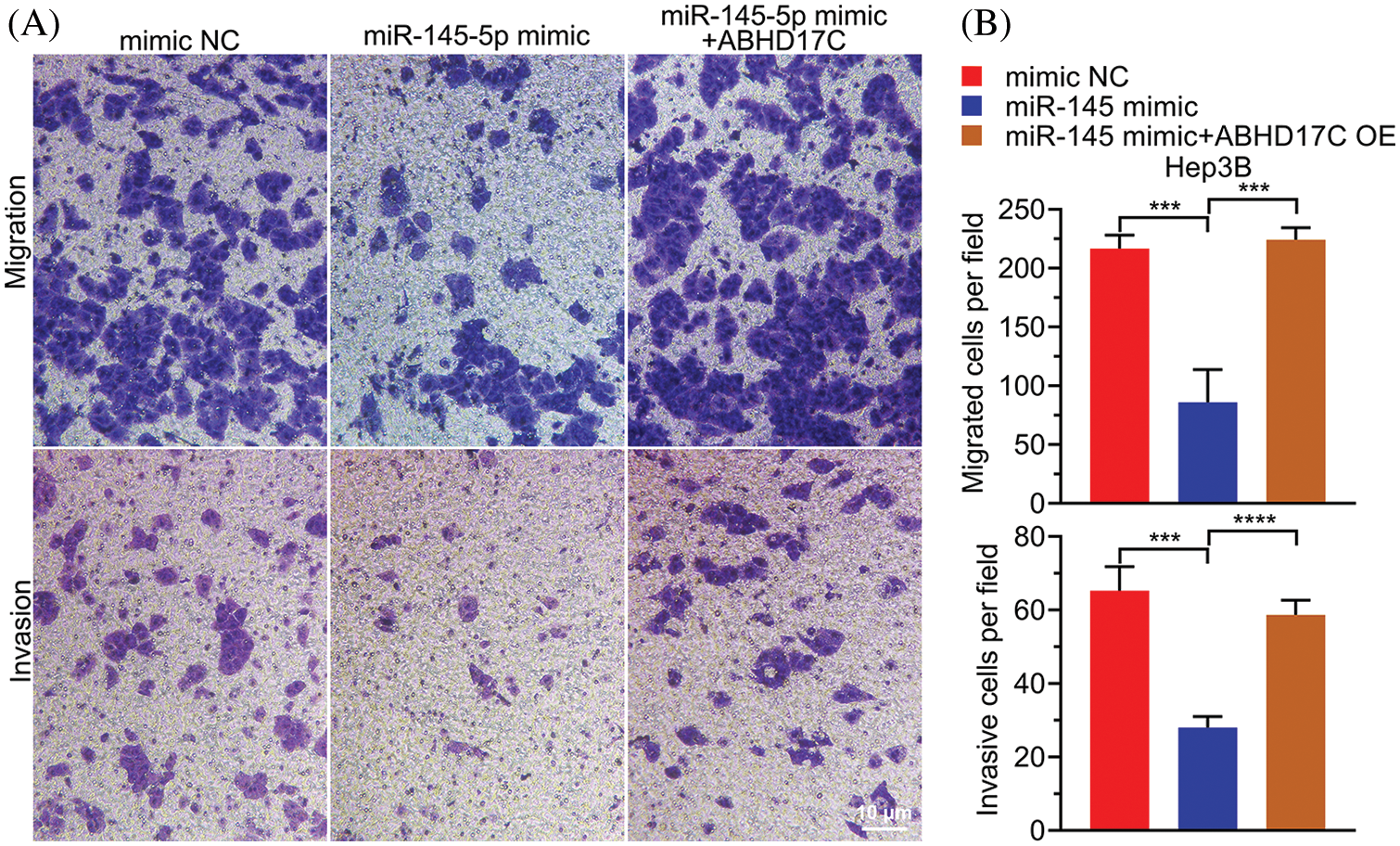
Figure 7: miR-145-5p-mediated suppressive impacts on HCC cell migration and invasion are ABHD17C dependent. (A) Transwell assays were performed to evaluate the migratory (upper) and invasive (lower)abilities of Hep3B cells exhibiting miR-145-5p and ABHD17C misexpression. (B) Statistical histogram of migrated and invasive cells. Data are presented as mean ± SD and were analyzed using analysis of variance (ANOVA), followed by Tukey’s post-hoc test (B). ABHD17C, alpha/beta hydrolase domain-containing protein 17C; HCC, hepatocellular carcinoma. ***P < 0.001, ****P < 0.0001
3.6 MiR-145-5p Suppresses HCC Tumor Growth in Vivo
Finally, we used a mouse xenograft model to confirm whether in vitro suppressive effects of miR-145-5p could be observed in vivo. Notably, miR-145-5p overexpression markedly reduced the volume and weight of Hep3B-derived tumors (Figs. 8A–8C). Ki67 levels were significantly reduced in Hep3B-derived tumors overexpressing miR-145-5p (Figs. 8D and 8E). In addition, miR-145-5p expression was retained in tumors derived from miR-145-5p-overexpressing cells (Fig. 8F). Consistent with the in vitro results, evaluation of the Hep3B-derived tumors confirmed that ABHD17C expression was significantly attenuated on transducing Hep3B cells with miR-145-5p, which were further grown in mice (Figs. 8G–8I). Taken together, our results indicated that miR-145-5p could inhibit HCC tumor growth and ABHD17C expression levels in mice.
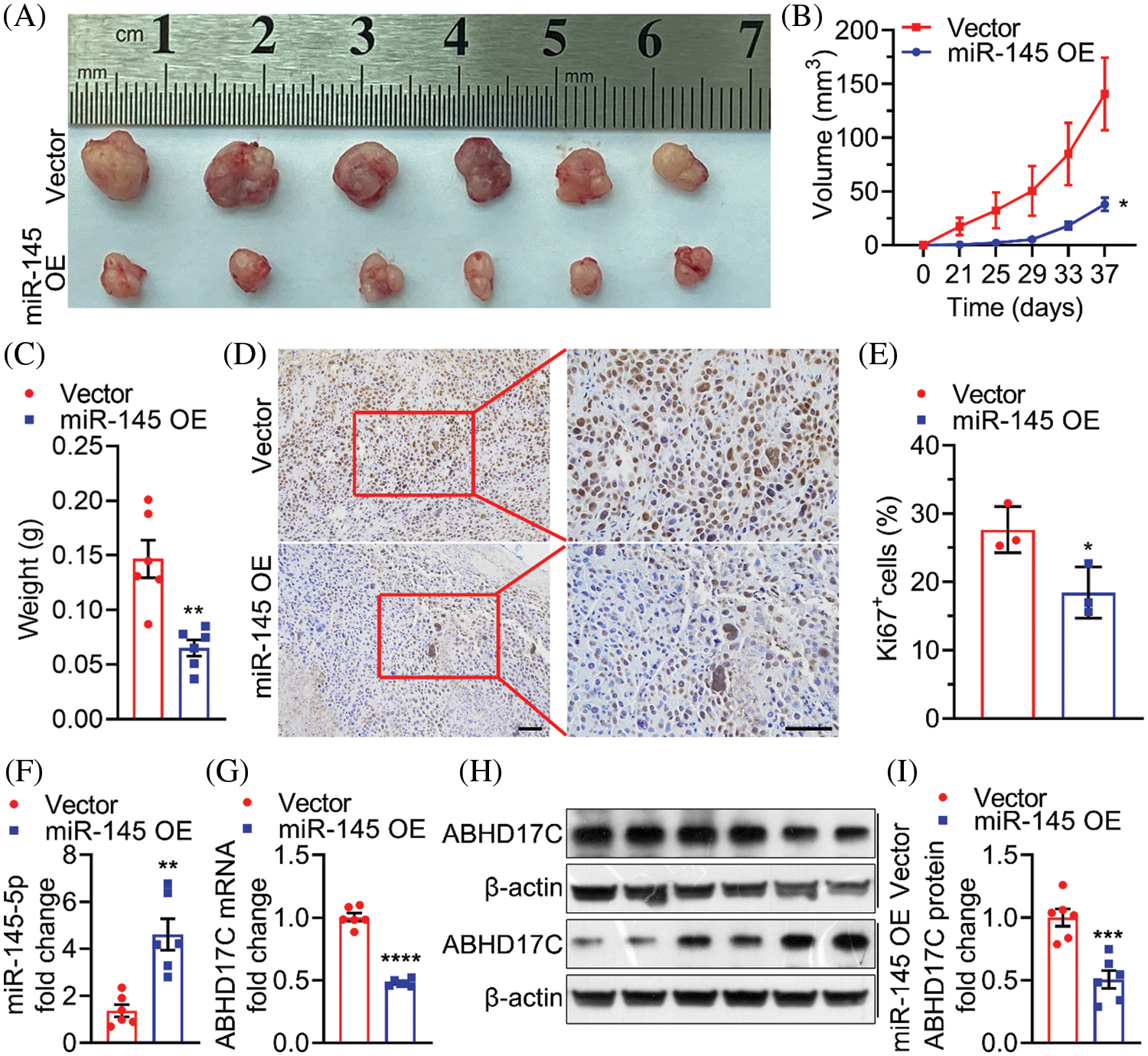
Figure 8: miR-145-5p alleviates the growth of HCC cells in mice. (A–C) Representative images (A), volume (B), and weight (C) of tumors formed by Hep3B cells upon ectopic miR-145-5p expression. (D and E) Ki67 was tested using IHC. Scale bar: 100 μm. (F and G) qPCR detection of miR-145-5p (F) and ABHD17C (G) expression in tumors with or without miR-145-5p overexpression. (H, I) Western blotting representative images (H) and quantification (I) of ABHD17C expression in tumors with or without miR-145-5p overexpression. Data are presented as mean ± SD and were analyzed using Student’s t test (B, C, E, F, G, and I). ABHD17C, alpha/beta hydrolase domain-containing protein 17C; HCC, hepatocellular carcinoma. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001
HCC, which accounts for 80% of all cases of primary liver cancer, is one of the primary causes of cancer-related deaths worldwide [29]. To date, numerous risk factors, including viral infection, alcoholism, nonalcoholic fatty liver disease, and cirrhosis, have been associated with the onset and progression of HCC [29–31]. Furthermore, the accumulation of genetic and epigenetic changes can contribute to HCC development [32]. In the present study, we explored the role of miR-145-5p in HCC and examined its underlying mechanism.
The importance of miRNAs in modulating cellular processes highlights their potential role as vital candidate biomarkers and promising therapeutic agents [33]. miR-145-5p, a member of the p53 tumor suppressor network, is known to be involved in the post-transcriptional regulation of several cancer-related genes and has been shown to suppress glioma, breast cancer, and bladder cancer [29]. Moreover, miR-145-5p is reportedly downregulated in HCC [14]. Consistently, our bioinformatics analysis revealed that miR-145-5p was downregulated in HCC tissues, and high levels of miR-145-5p were associated with better prognosis in patients with HCC. In colorectal cancer, miR-145-5p expression was found to be downregulated, and increasing miR-145-5p expression could inhibit cell viability, migration, invasion, and epithelial-mesenchymal transition [34]. Similarly, we showed that upregulating miR-145-5p expression could suppress HCC cell invasion and migration, induce cell cycle arrest, and inhibit tumor growth.
It has been reported that miR-145-5p inhibits breast cancer cell proliferation by downregulating SOX2 expression [35] and can suppress gastric cancer cell proliferation and promote gastric cancer cell differentiation by negatively regulating KLF5 expression [36]. Furthermore, miR-145-5p can inhibit bladder cancer cell proliferation and migration by reducing TAGLN2 expression [37], suppress the malignancy of colon cancer cells by downregulating expression levels of CXCL1 and ITGA2 [29], and increase the sensitivity of acquired gefitinib-resistant cells to gefitinib in non-small cell lung cancer cells by inhibiting NRAS and MEST expression [38]. Moreover, miR-145-5p suppresses ovarian cancer cell proliferation and promotes cell apoptosis by downregulating ARK5 expression [39]. In HCC cells, miR-145-5p significantly increases apoptosis, decreases cell proliferation, and inhibits HCC cell migration by downregulating PKM2 and HDAC11 expression [40,41]. Although these studies have identified and validated multiple targets of miR-145-5p in HCC cells, the complexity of miR-145-5p and its downstream target networks remain poorly explored. In the present study, we found that miR-145-5p negatively regulated ABHD17C expression, which, in turn, arrested the cell cycle in the G0/G1 phase and inhibited cell migration, invasion, and tumor growth.
ABHD17C, a member of ABHD17, is highly expressed in breast cancer tissues, indicating that ABHD17C could be involved in regulating tumorigenesis and breast cancer development [17,20]. In the present study, we found that ABHD17C was upregulated in HCC tissues, and patients with HCC exhibiting high ABHD17C levels had worse OS when compared with those presenting low ABHD17C levels. Furthermore, ABHD17C knockdown suppressed the malignant behavior of Hep3B cells. These data suggest that ABHD17C could potentially regulate HCC development.
To better understand the complexity and unbiased screening of miR-145-5p targets, six platforms and a dataset of upregulated genes in patients with HCC were employed, resulting in the enrichment of eight candidate genes. Importantly, miR-145-5p could downregulate six of the eight candidate genes, confirming the robustness of our screening methodology. However, none of the miR-145-5p target genes identified in previous studies were identified in the present analysis. Therefore, it is unsurprising that one miRNA may target multiple genes for degradation. Notably, a few genes (CTNNBIP1 and SMAD3) that belong to critical cancer-driving pathways, such as transforming growth factor (TGF)-β/SMAD and Wnt/CTNNB1, were enriched, although the decreased expression mediated by miR-145-5p was not as prominent as afforded by ABHD17C. We cannot exclude the possibility that miR-145-5p may tightly control these genes, thereby fine-tuning the aforementioned cancer-related signaling and affecting HCC progression. Therefore, future studies could focus on determining the contribution of these genes to miR-145-5p-mediated negative effects on HCC cell malignancy.
In this study, we revealed the direct interaction between ABHD17C 3′UTR and miR-145-5p via dual luciferase reporter analysis. Notably, the biological role of ABHD17C has been poorly investigated. Patient-derived data revealed a negative correlation between ABHD17C expression and survival of patients with HCC. Furthermore, rescue experiments showed that miR-145-5p depended on ABHD17C to exert its tumor-inhibitory function, strengthening the importance of ABHD17C expression in promoting HCC progression. Thus, in-depth investigations should be conducted to uncover the functions of ABHD17C expression in the progression of HCC and other cancer types. The mechanistic details of how ABHD17C mediates its biological effects warrant further investigation.
There are certain limitations to this study. First, the present study mainly explored the roles of miR-145-5p and ABHD17C in HCC by upregulating the expression levels of miR-145-5p and ABHD17C, which were not further verified by inhibiting miR-145-5p. Second, we did not explore the therapeutic effects of targeting miR-145-5p in HCC, which should be explored in future investigations. Third, the possible utilization of miR-145-5p in preclinical models was not further determined using a complementary experiment, patient-derived xenograft models or organoid systems.
In the study, the tumor-suppressive effects of miR-145-5p were validated in vitro and vivo experiments. Therefore, this study demonstrated that targeting miR-145-5p has the potential to treat HCC, laying a foundation for the development of targeted therapeutic drugs for HCC.
Author Contributions: (I) Conception and design: Linpei Wang, Xiaoqiu Ma, Wei Wang, and Shaojian Chen; (II) Administrative support: Wei Wang and Shaojian Chen; (III); Provision of study materials and patients: Linpei Wang, Youqi Chen, and Shaojian Chen; (IV) Data collection and assembly: Linpei Wang, Xiaoqiu Ma, Youqi Chen, Jiahui Zhang, Jiawei Zhang, and Wei Wang; (V) Data analysis and interpretation: Linpei Wang, Xiaoqiu Ma, and Shaojian Chen; (VI) Manuscript writing: All authors; (VII) Final approval of the manuscript: All authors.
Availability of Data and Materials: The authors declare that all data supporting the findings of the present study are available within the article and its supplementary information.
Ethical Statement: The authors are accountable for all aspects of the work, ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Experiments were performed under a project license granted by the Ethics Board of the Second Affiliated Hospital of Fujian Medical University (Approval No. 2020 No. 15) in compliance with the institutional guidelines for the care and use of animals.
Funding Statement: This work was supported by the Educational Research Project for Young and Middle-Aged Teachers of Fujian Educational Committee (JAT190217), the Natural Science Foundation of Fujian Province, China (2020J01208), and the Quanzhou High-Level Talents Project (2021C048R).
Conflicts of Interest: All authors declare that there is no conflict of interest exists in the submission of this manuscript and approve the manuscript for publication.
References
1. Villanueva, A. (2019). Hepatocellular carcinoma. The New England Journal of Medicine, 380(15), 1450–1462. DOI 10.1056/NEJMra1713263. [Google Scholar] [CrossRef]
2. Zheng, R., Qu, C., Zhang, S., Zeng, H., Sun, K. et al. (2018). Liver cancer incidence and mortality in China: Temporal trends and projections to 2030. Chinese Journal of Cancer Research, 30(6), 571–579. DOI 10.21147/j.issn.1000-9604.2018.06.01. [Google Scholar] [CrossRef]
3. Yim, S. Y., Lee, J. S. (2021). An overview of the genomic characterization of hepatocellular carcinoma. Journal of Hepatocellular Carcinoma, 8, 1077–1088. DOI 10.2147/JHC.S270533. [Google Scholar] [CrossRef]
4. Wong, R. J., Ahmed, A., Gish, R. G. (2015). Elevated alpha-fetoprotein: Differential diagnosis-hepatocellular carcinoma and other disorders. Clinics in Liver Disease, 19(2), 309–323. DOI 10.1016/j.cld.2015.01.005. [Google Scholar] [CrossRef]
5. Piñero, F., Dirchwolf, M., Pessôa, M. G. (2020). Biomarkers in hepatocellular carcinoma: Diagnosis, prognosis and treatment response assessment. Cells, 9(6), 1370. DOI 10.3390/cells9061370. [Google Scholar] [CrossRef]
6. Wei, L., Lee, D., Law, C. T., Zhang, M. S., Shen, J. et al. (2019). Genome-wide CRISPR/Cas9 library screening identified PHGDH as a critical driver for sorafenib resistance in HCC. Nature Communications, 10(1), 4681. DOI 10.1038/s41467-019-12606-7. [Google Scholar] [CrossRef]
7. Adams, B. D., Kasinski, A. L., Slack, F. J. (2014). Aberrant regulation and function of microRNAs in cancer. Current Biology, 24(16), R762–R776. DOI 10.1016/j.cub.2014.06.043. [Google Scholar] [CrossRef]
8. Pencheva, N., Tavazoie, S. F. (2013). Control of metastatic progression by microRNA regulatory networks. Nature Cell Biology, 15(6), 546–554. DOI 10.1038/ncb2769. [Google Scholar] [CrossRef]
9. Wang, X., He, Y., Mackowiak, B., Gao, B. (2021). MicroRNAs as regulators, biomarkers and therapeutic targets in liver diseases. Gut, 70(4), 784–795. DOI 10.1136/gutjnl-2020-322526. [Google Scholar] [CrossRef]
10. Girard, M., Jacquemin, E., Munnich, A., Lyonnet, S., Henrion-Caude, A. (2008). miR-122, a paradigm for the role of microRNAs in the liver. Journal of Hepatology, 48(4), 648–656. DOI 10.1016/j.jhep.2008.01.019. [Google Scholar] [CrossRef]
11. Wang, S., Wang, T., Gu, P. (2021). microRNA-145-5p inhibits migration, invasion, and metastasis in hepatocellular carcinoma by inhibiting ARF6. Cancer Management and Research, 13, 3473–3484. DOI 10.2147/CMAR.S300678. [Google Scholar] [CrossRef]
12. Dong, G., Zhang, S., Shen, S., Sun, L., Wang, X. et al. (2020). SPATS2, negatively regulated by miR-145-5p, promotes hepatocellular carcinoma progression through regulating cell cycle. Cell Death & Disease, 11(10), 837. DOI 10.1038/s41419-020-03039-y. [Google Scholar] [CrossRef]
13. Liang, H., Sun, H., Yang, J., Yi, C. (2018). miR-145-5p reduces proliferation and migration of hepatocellular carcinoma by targeting KLF5. Molecular Medicine Reports, 17(6), 8332–8338. DOI 10.3892/mmr.2018.8880. [Google Scholar] [CrossRef]
14. Ding, B., Fan, W., Lou, W. (2020). hsa_circ_0001955 enhances in vitro proliferation, migration, and invasion of HCC cells through miR-145-5p/NRAS axis. Molecular Therapy. Nucleic Acids, 22, 445–455. DOI 10.1016/j.omtn.2020.09.007. [Google Scholar] [CrossRef]
15. Gu, X., Zhang, J., Ran, Y., Pan, H., Jia, J. et al. (2021). Circular RNA hsa_circ_101555 promotes hepatocellular carcinoma cell proliferation and migration by sponging miR-145-5p and regulating CDCA3 expression. Cell Death & Disease, 12(4), 356. DOI 10.1038/s41419-021-03626-7. [Google Scholar] [CrossRef]
16. McClafferty, H., Runciman, H., Shipston, M. J. (2020). Site-specific deacylation by ABHD17a controls BK channel splice variant activity. The Journal of Biological Chemistry, 295(49), 16487–16496. DOI 10.1074/jbc.RA120.015349. [Google Scholar] [CrossRef]
17. Lin, D. T. S., Davis, N. G., Conibear, E. (2017). Targeting the Ras palmitoylation/depalmitoylation cycle in cancer. Biochemical Society Transactions, 45(4), 913–921. DOI 10.1042/BST20160303. [Google Scholar] [CrossRef]
18. Lin, D. T., Conibear, E. (2015). ABHD17 proteins are novel protein depalmitoylases that regulate N-Ras palmitate turnover and subcellular localization. eLife, 4, e11306. DOI 10.7554/eLife.11306. [Google Scholar] [CrossRef]
19. del Rivero Morfin, P. J., Ben-Johny, M. (2020). Cutting out the fat: Site-specific deacylation of an ion channel. The Journal of Biological Chemistry, 295(49), 16497–16498. DOI 10.1074/jbc.H120.016490. [Google Scholar] [CrossRef]
20. Chen, L., Pan, X., Zhang, Y. H., Hu, X., Feng, K. et al. (2019). Primary tumor site specificity is preserved in patient-derived tumor xenograft models. Frontiers in Genetics, 10, 738. DOI 10.3389/fgene.2019.00738. [Google Scholar] [CrossRef]
21. McGeary, S. E., Lin, K. S., Shi, C. Y., Pham, T. M., Bisaria, N. et al. (2019). The biochemical basis of microRNA targeting efficacy. Science, 366(6472), eaav1741. DOI 10.1126/science.aav1741. [Google Scholar] [CrossRef]
22. Paraskevopoulou, M. D., Georgakilas, G., Kostoulas, N., Vlachos, I. S., Vergoulis, T. et al. (2013). DIANA-microT web server v5.0: Service integration into miRNA functional analysis workflows. Nucleic Acids Research, 41(W1), W169–W173. DOI 10.1093/nar/gkt393. [Google Scholar] [CrossRef]
23. Chen, Y., Wang, X. (2020). miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Research, 48(D1), D127–D131. DOI 10.1093/nar/gkz757. [Google Scholar] [CrossRef]
24. Huang, H. Y., Lin, Y. C., Li, J., Huang, K. Y., Shrestha, S. et al. (2020). miRTarBase 2020: Updates to the experimentally validated microRNA-target interaction database. Nucleic Acids Research, 48(D1), D148–D154. [Google Scholar]
25. Li, J. H., Liu, S., Zhou, H., Qu, L. H., Yang, J. H. (2014). starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-seq data. Nucleic Acids Research, 42(D1), D92–D97. DOI 10.1093/nar/gkt1248. [Google Scholar] [CrossRef]
26. Tokar, T., Pastrello, C., Rossos, A. E. M., Abovsky, M., Hauschild, A. C. et al. (2018). mirDIP 4.1-integrative database of human microRNA target predictions. Nucleic Acids Research, 46(D1), D360–D370. DOI 10.1093/nar/gkx1144. [Google Scholar] [CrossRef]
27. Li, J., Hu, L., Liu, Y., Huang, L., Mu, Y. et al. (2015). DDX19A senses viral RNA and mediates NLRP3-dependent inflammasome activation. Journal of Immunology, 195(12), 5732–5749. DOI 10.4049/jimmunol.1501606. [Google Scholar] [CrossRef]
28. Hanahan, D. (2022). Hallmarks of cancer: New dimensions. Cancer Discovery, 12(1), 31–46. DOI 10.1158/2159-8290.CD-21-1059. [Google Scholar] [CrossRef]
29. Zhuang, W., Niu, T., Li, Z. (2021). MicroRNA miR-145-5p regulates cell proliferation and cell migration in colon cancer by inhibiting chemokine (C-X-C motif) ligand 1 and integrin α2. Bioengineered, 12(2), 9909–9917. DOI 10.1080/21655979.2021.2000243. [Google Scholar] [CrossRef]
30. Lou, W., Liu, J., Ding, B., Chen, D., Xu, L. et al. (2019). Identification of potential miRNA-mRNA regulatory network contributing to pathogenesis of HBV-related HCC. Journal of Translational Medicine, 17(1), 7. DOI 10.1186/s12967-018-1761-7. [Google Scholar] [CrossRef]
31. Ding, B., Lou, W., Liu, J., Li, R., Chen, J. et al. (2019). In silico analysis excavates potential biomarkers by constructing miRNA-mRNA networks between non-cirrhotic HCC and cirrhotic HCC. Cancer Cell International, 19, 186. DOI 10.1186/s12935-019-0901-3. [Google Scholar] [CrossRef]
32. Cai, C., Wang, W., Tu, Z. (2019). Aberrantly DNA methylated-differentially expressed genes and pathways in hepatocellular carcinoma. Journal of Cancer, 10(2), 355–366. DOI 10.7150/jca.27832. [Google Scholar] [CrossRef]
33. Hayes, J., Peruzzi, P. P., Lawler, S. (2014). MicroRNAs in cancer: Biomarkers, functions and therapy. Trends in Molecular Medicine, 20(8), 460–469. DOI 10.1016/j.molmed.2014.06.005. [Google Scholar] [CrossRef]
34. Chen, Q., Zhou, L., Ye, X., Tao, M., Wu, J. (2020). miR-145-5p suppresses proliferation, metastasis and EMT of colorectal cancer by targeting CDCA3. Pathology, Research and Practice, 216(4), 152872. DOI 10.1016/j.prp.2020.152872. [Google Scholar] [CrossRef]
35. Tang, W., Zhang, X., Tan, W., Gao, J., Pan, L. et al. (2019). miR-145-5p suppresses breast cancer progression by inhibiting SOX2. The Journal of Surgical Research, 236, 278–287. DOI 10.1016/j.jss.2018.11.030. [Google Scholar] [CrossRef]
36. Zhou, T., Chen, S., Mao, X. (2019). miR-145-5p affects the differentiation of gastric cancer by targeting KLF5 directly. Journal of Cellular Physiology, 234(5), 7634–7644. DOI 10.1002/jcp.27525. [Google Scholar] [CrossRef]
37. Zhang, H., Jiang, M., Liu, Q., Han, Z., Zhao, Y. et al. (2018). miR-145-5p inhibits the proliferation and migration of bladder cancer cells by targeting TAGLN2. Oncology Letters, 16(5), 6355–6360. DOI 10.3892/ol.2018.9436. [Google Scholar] [CrossRef]
38. Yu, C., Li, B., Wang, J., Zhang, Z., Li, S. et al. (2021). miR-145-5p modulates gefitinib resistance by targeting NRAS and MEST in non-small cell lung cancer. Annals of Clinical and Laboratory Science, 51(5), 625–637. [Google Scholar]
39. Wu, L., Zhou, W. Q., Yuan, L. N., Li, J., Pei, M. L. (2021). Role of miR-145-5p targeting ARK5 in regulating the proliferation and apoptosis of human epithelial ovarian cancer cells. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. Acta Academiae Medicinae Sinicae, 43(5), 669–676. [Google Scholar]
40. Jia, G., Wang, Y., Lin, C., Lai, S., Dai, H. et al. (2021). LNCAROD enhances hepatocellular carcinoma malignancy by activating glycolysis through induction of pyruvate kinase isoform PKM2. Journal of Experimental & Clinical Cancer Research, 40(1), 299. DOI 10.1186/s13046-021-02090-7. [Google Scholar] [CrossRef]
41. Wang, W., Ding, B., Lou, W., Lin, S. (2020). Promoter hypomethylation and miR-145-5p downregulation-mediated HDAC11 overexpression promotes sorafenib resistance and metastasis of hepatocellular carcinoma cells. Frontiers in Cell and Developmental Biology, 8, 724. DOI 10.3389/fcell.2020.00724. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2022 The Author(s). Published by Tech Science Press.
Copyright © 2022 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.

 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools