 Open Access
Open Access
ARTICLE
Research on Clinical Effectiveness of Aspirin for Treating Breast Cancer and Cell Protein Biomarkers on Aspirin Treatment in Drug-Resistant Estrogen Receptor-Positive Breast Cancer Cells
1 Breast Disease Center, Peking University First Hospital, Peking University, Beijing, China
2 Institute of Precision Medicine, Peking University Shenzhen Hospital, Peking University, Shenzhen, China
* Corresponding Authors: Ruifang Pang. Email: ; Yinhua Liu. Email:
(This article belongs to the Special Issue: Biomarkers for Breast Cancer Diagnosis and Treatment Selection: from Basic Research to Practice)
Oncologie 2022, 24(4), 743-768. https://doi.org/10.32604/oncologie.2022.025419
Received 12 July 2022; Accepted 09 September 2022; Issue published 31 December 2022
Abstract
Background: Aspirin (ASA) has been reported to have an antitumor effect but the role of ASA in the prevention and treatment of breast cancer (BC) is still controversial. This study aimed to identify clinical effectiveness of ASA in the treatment of BC and explore the antitumor target proteins of ASA that may be involved in overcoming tamoxifen resistance in estrogen receptor (ER)-positive BC cells. Materials and Methods: Randomized controlled trials (RCTs) of ASA in the treatment of BC were queried from the databases, including PubMed, Web of Science, Cochrane Library, WanFang, and Chinese National Knowledge Infrastructure. According to the quality standard recommended in the Newcastle-Ottawa Scale (NOS), the outcome indexes were analyzed by RevMan 5.3 and Stata 12.0 software. Cell culture experiments were performed to explore the effect of tamoxifen combined with ASA on the proliferation of ER-positive BC cell lines MCF-7 and MCF-7/TAM. Cell cytotoxicity was determined by the 3-(4, 5-di-2-yl)-2, 5-ditetrazolium bromide (MTT) assay. A quantitative proteomic analysis was conducted between the control and experimental groups to identify differentially expressed proteins (DEPs). Subsequently, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were used for bioinformatic analysis of DEPs. The protein expression in patients of ER-positive BC was analyzed by immunochemistry (IHC). Results: Nine RCTs including 162,381 patients were selected for this study. The meta-analysis revealed that daily ASA use, as compared with its non-use, was associated with a decreased risk of BC death: relative risk (RR) = 0.83%, 95% CI [0.73, 0.94], Z = 2.89, P = 0.004 (P < 0.05). Cell culture experiments showed that tamoxifen combined with ASA can drastically inhibit the cell growth of MCF-7 and MCF-7/TAM cells more than the administration of tamoxifen alone (P < 0.05). Fifty-seven DEPs were up-regulated, while eighty-five DEPs were down-regulated in MCF-7/TAM cells after the ASA combination treatment. Several GO terms were significantly enriched, such as neutrophil degranulation, retinal metabolic process, sterol biosynthetic process, and prostaglandin metabolic process. KEGG pathway enrichment analysis also verified three associated pathways including metabolic pathways, chemical carcinogenesis-reactive oxygen species, and biosynthesis of amino acids. In ER-positive BC patients with Ki67 > 20%, the positive expression of one significantly DEP MYC was much higher than in BC patients with Ki67 ≤ 20% (40.91% vs. 12.50%, P < 0.05). There was no significant difference in MYC protein expression among the other subgroups (P > 0.05). Conclusions: Our results show that ASA has a clinical value in the treatment of BC. ASA could overcome tamoxifen resistance in ER-positive BC cells through some key proteins, which may be potential therapeutic targets for patients with tamoxifen resistance.Keywords
Around 12% of all cancer-related deaths worldwide occur due to breast cancer (BC), which is the leading cause of cancer-related death among women [1]. Depending on the clinical tumor subtypes of BC, therapeutic backbones include surgery, radiation therapy, endocrine therapy (ET), chemotherapy, and anti-HER2 targeting [2]. The most common type of BC is estrogen receptor (ER) positive, which accounts for about 60% of all BC cases [3]. These tumors are initially dependent on the steroid hormone estrogen acting through the ER. ET is a common treatment for ER-positive BC, which decreases the risk of recurrence and improves progression-free survival in a subset of patients with ER-positive BC. Most commonly, tamoxifen is used to treat BC for a 5-year treatment period at least because of its inhibitory effects on the transcriptional activity of ER [4]. Despite its initial therapeutic efficacy, resistance to tamoxifen limits its long-term benefit for patients with BC, which remains an important and challenging clinical problem [5,6]. Nearly 30% of patients with BC receiving tamoxifen eventually experience disease relapse, and secondary resistance to tamoxifen during treatment can lead tumor metastasis and death [7]. Molecular mechanisms of resistance to tamoxifen have not yet been determined. Some mechanisms of tamoxifen resistance are complex and include ER mutations [8], the metabolic pathway of tamoxifen [9], activation of alternative oncogenic signaling, and deregulation of oncogenic proteins [10]. As a result, there is a great need for more comprehensive research on novel treatment options to overcome the resistance of tamoxifen in ER-positive BC.
In recent years, several studies have demonstrated that aspirin (ASA) has anticancer effects in solid tumors, such as colorectal cancer, prostate cancer, and endometrial cancer [11–13]. However, previous studies of ASA in patients have produced controversial results. Some studies have shown that ASA can reduce the incidence of BC, while other studies were of the opposite opinion. The mechanism of the association between the anticancer effects and ASA is complex [14]. Multiple observational studies have reported an improved BC survival with regular ASA use [15,16]. As ASA has anti-tumor effects, we found it to play a novel role in tamoxifen resistance in ER-positive BC cells and proposed possible mechanisms in our previous study. Additionally, ASA combined with tamoxifen can inhibit cell cycle progression by downregulating cyclin D1, thus overcoming tamoxifen resistance [17]. In tamoxifen-resistant ER-positive BC cell lines, ASA might also inhibit the upstream regulators of MYC and cyclin D1 proteins, such as mTOR and NF-κB signaling pathways. These proteins and factors could upregulate MYC and cyclin D1 protein levels excessively, which is necessary for the survival of cancer cells. However, information regarding changes in proteins associated with tamoxifen resistance in ER-positive BC cells treated with the ASA and tamoxifen combination remains sparse.
Proteomics can help to identify and analyze a whole set of proteins in biological systems systematically and quantitatively. Proteomic studies can interpret not only changes in protein expression levels, but also posttranslational modifications that are essential for the regulation of protein functions [18,19]. In this study, based on evidence showing that ASA may contribute to the breakthrough of tamoxifen resistance, we further analysed tamoxifen-resistant MCF-7/TAM cells and identified altered protein expressions concerning ASA treatment using proteomic analysis techniques. We leveraged the data to better understand the effects of ASA treatment in overcoming tamoxifen resistance in BC.
2.1 Study Design and Research Sample
This study was designed according to the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) guidelines for literature search, study design, and data analysis. We identified eligible researches published up to 2021 on the treatment of BC with ASA from databases, including PubMed, Web of Science, Cochrane Library, Wanfang, and Chinese National Knowledge Infrastructure. Two experienced librarians helped develop a comprehensive search strategy including free text and MeSH terms. A number of search terms were used such as “breast cancer”, “breast neoplasms”, “ASA”, and “non-steroidal anti-inflammatory drugs (NSAIDs)”. Inclusion criteria included: (1) Published literature on aspirin in the treatment of BC; (2) The number of cases in each group exceeded 60 cases; (3) Follow-up data are available; (4) The data are detailed and reliable, and the results are clearly expressed. The relative risk (RR) and 95% confidence interval can be extracted from the relevant data. Exclusion criteria included: (1) Review, conference abstract and non-original literature; (2) Cannot provide data for statistical analysis; (3) Repeatability test; (4) Cell culture methods or animal studies; (5) Lack of follow-up data.
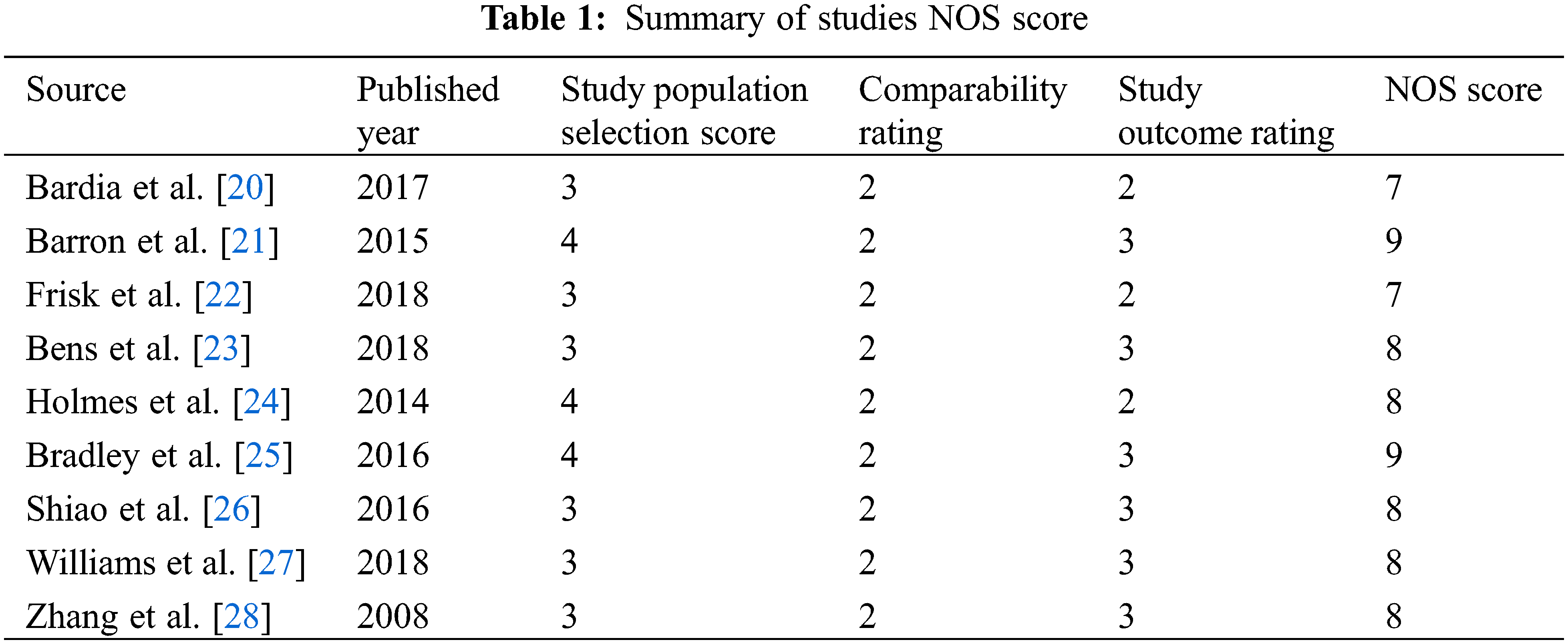
The design and quality of all studies were assessed using the Newcastle-Ottawa Scale (NOS). Two researchers independently read and evaluated the literature according to NOS scoring criteria, including patient selection, comparability, and study results. Studies with scores >5 were included in the follow-up studies.
4-hydroxy-tamoxifen (4-OHT) and ASA (Sigma-Aldrich, St. Louis, MO, USA) were dissolved in dimethyl sulphoxide (DMSO) (Sigma-Aldrich, St. Louis, MO, USA). Sodium deoxycholate, dithiothreitol, ammonium bicarbonate, and iodoacetamide were procured from Sigma (Sigma-Aldrich, St. Louis, MO, USA). Tris-(2-carboxyethyl) phosphine was procured from Thermo Scientific (Thermo Fisher Scientific, Waltham, MA, USA). Modified sequencing grade trypsin was procured from Promega (Promega Corporation, Wisconsin, USA). High-performance liquid chromatography-mass spectrometry (HPLC/MS) grade solvents were used to prepare all mobile phases and solutions, including water, acetonitrile, methanol, and formic acid (Thermo Fisher Scientific, Waltham, MA, USA). Anti-ER antibody (clone SP1), anti-PR antibody (clone 1E2), anti-HER2 antibody (clone 4B5), and anti-MYC antibody (clone Y69) (Roche, Switzerland) were used in accordance with the manufacturer’s instructions.
2.4 Cell Culture and Cell Viability Analysis
The human BC cell lines MCF-7 and MCF-7/TAM (tamoxifen-resistant) were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) (Gibco, USA) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin and 100 μg/ml streptomycin (Gibco, USA) in a 5% CO2 humidified incubator at 37°C. Cells were grown in 96 well culture plates and treated with both compounds and 4-OHT alone. The cytotoxicity of ASA and 4-OHT was evaluated by MTT assay according to the manufacturer’s instructions (Sigma-Aldrich, St. Louis, MO, USA). Each experiment was carried out three times. Optical density (OD) values at 450 nm wavelength were determined by enzyme-labeled instruments (Bio-Rad, USA).
2.5 Methods of Proteomics Experiments
Seeding of MCF-7 and MCF-7/TAM cells in 10 cm2 flasks was performed, and the cells were placed in a humidified incubator at 37°C and 5% CO2. At 70% confluency, treatments with 2 mM ASA and 3 μM 4-OHT (experimental group) or 4-OHT alone (control group) were carried out. Following six days of incubation, the cells in two groups were washed thrice with pre-cooled phosphate buffer saline (PBS) (Gibco, USA) and centrifuged to obtain cell pellets.
A total of 1 × 107 cells were lysed with 200 μL radioimmunoprecipitation assay (RIPA) lysis buffer (Thermo Fisher Scientific, Waltham, MA, USA) for 30 min at 4°C, and the debris was removed by centrifugation at 12,000 g for 30 min. Subsequently, the supernatant was collected and the protein concentration was determined by bicinchoninic acid protein assay (Thermo Fisher Scientific, Waltham, MA, USA).
Protein samples were reduced with a final concentration of 10 mM dithiothreitol at 25°C for 30 min on a shaking plate at 1,200 rpm. Afterward, samples were alkylated with 20 mM iodoacetamide (IAA) and incubated at 25°C for 30 min in the dark. Trichloroacetic acid (100%, 1/10 vol) was added to the samples, followed by incubation at 4°C for 30 min. The samples were washed with pre-chilled acetone and centrifuged at 16,000 g for 15 min at 4°C. This step was performed twice and the pellet was allowed to dry for 10 min. Finally, trypsin in 100 μL of 100 mM Tris/HCL (pH = 8.5) was added to each sample. The protein-to-enzyme ratio was 50:1. Samples were incubated overnight at 37°C to release peptides. Two microliters of 100% formic acid was added to the samples, followed by vortexing and centrifugation at 10,000 g for 30 s. Samples were desalted using an Oasis HLB extraction cartridge (Waters Corporation, Milford, MA, USA) according to the manufacturer’s instructions. After drying with a SpeedVac vacuum concentrator (Thermo Fisher Scientific, Waltham, MA, USA), peptides were dissolved in 0.1% formic acid for liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis.
For proteomics analysis, the samples were allowed to dissolve in 0.1% formic acid and then analyzed on a commercial C18 column (Acclaim PepMap RSLC, 75 μm × 50 cm, Thermo Scientific, Waltham, MA, USA) using an Ultra3000 UHPLC connected to Exploris 480 mass spectrometer (Thermo Scientific, Waltham, MA, USA). Peptides were separated at 300 nl/min for 105 min using a linear gradient of acetonitrile (ACN) 5% to 45% with 0.1% formic acid. The MS acquisition was set to the data-dependent acquisition (DDA) mode, including a full MS survey scan from m/z 350 to m/z 1,200 at a resolution of 60,000 full width at half maximum (FWHM) (at m/z 200) with accudenz gradient centrifugation (AGC) set to 5E6 (maximum injection time of 50 ms), followed by 20 MS/MS scans at a resolution of 15,000 FWHM with AGC set to 2E5 (maximum injection time of 100 ms). A total of twenty of the most intense precursors with an isolation width of m/z 1.6 were selected for fragmentation via high energy collision dissociation (HCD) with 30 normalized collision energy (NCE). An exclusion time of 60 s was set for dynamic exclusion.
2.5.5 Protein Identification and Quantification
In the UniProt human database, the SEQUEST searching engine was used to search MS/MS spectra in proteome discoverer (PD) 2.4 software. The search criteria were as follows: complete trypsin specificity was required; cleavages of up to two deletions were allowed; oxidation (M) and acetylation (N terms of protein) were set as the variable modification and carbamidomethyl (C) was set as fixed modification; precursor ion quality of tolerance was set to 10 ppm and used in orbitrap quality analyzer all MS; MS2 spectra of quality of fragment ions tolerance was set to 20 mmu. The percolator provided of PD was used to calculate peptide false discovery rate. A search in the reverse decoy database was performed to identify false findings based on peptide profile match. Peptides belonging only to a particular group of proteins were considered unique. The false discovery rate for protein identification was also set to 0.01. Label-free quantification was performed only for proteins with two or more unique peptide matches. Protein ratio was calculated as all belong to a protein peptide hit the median. Quantitative accuracy expressed protein ratio variability.
2.5.6 Study Population, Samples and Immunohistochemistry (IHC)
Patients with luminal-like invasive BC were included in the study who also had complete data on specimens analysis-based scores for ER, progesterone receptor (PR), human epidermal growth factor receptor 2 (HER2), Ki67, and MYC were included in the study. Formalin-fixed and paraffin-embedded (FFPE) tumor specimens were examined by IHC, anti-ER antibody, anti-PR antibody, anti-HER2 antibody, and anti-MYC antibody were used for the IHC analysis according to the manufacturer’s instructions. ER/PR-positivity was assessed as ≥1% of tumor cells in the tissue sample. HER2 was evaluated 0 or +1 as negative and +3 as positive. When the IHC test score was 2+, the gene amplification status was determined using fluorescence in situ hybridization (FISH), and MYC positivity was defined as ≥10% cells showing brown color. Ki67 index of ≤20% was considered low risk while >20% was considered high risk.
2.5.7 Bioinformatics and Statistical Analyses
Data were extracted from each observation in both the experimental and control groups. Review Manager 5.3 was utilized to assess the RR and 95% confidence interval (CI). Stata 12.0 software was used to conduct Begg’s test. A funnel plot and Begg’s test were used to visually evaluate the publication bias. A Principal component analysis (PCA) score plot and hierarchical clustering (Pearson correlation) based heat map were generated using PD 2.4 software. GO enrichment analysis and KEGG pathway enrichment analysis were performed using the DAVID web service (https://david.ncifcrf.gov/home.jsp) with the DEPs revealed by using the provided proteome data. SPSS 23.0 was utilized to detect the data, and P < 0.05 was considered statistically significant.
All studies were queried from domestic and foreign databases and nine studies were selected based on established standards [20–28] (Fig. 1). A total of 59,934 patients were enrolled in the ASA experimental group, while 102,447 patients were enrolled in the control group. ASA was administered at a low dose (75–150 mg/d), and and the follow-up time was in the range of 41.3–134 months. The NOS scores of nine screened studies were from 7 to 9. This suggests that the studies included in this research are of good quality and can be used for meta-analysis (Table 1).
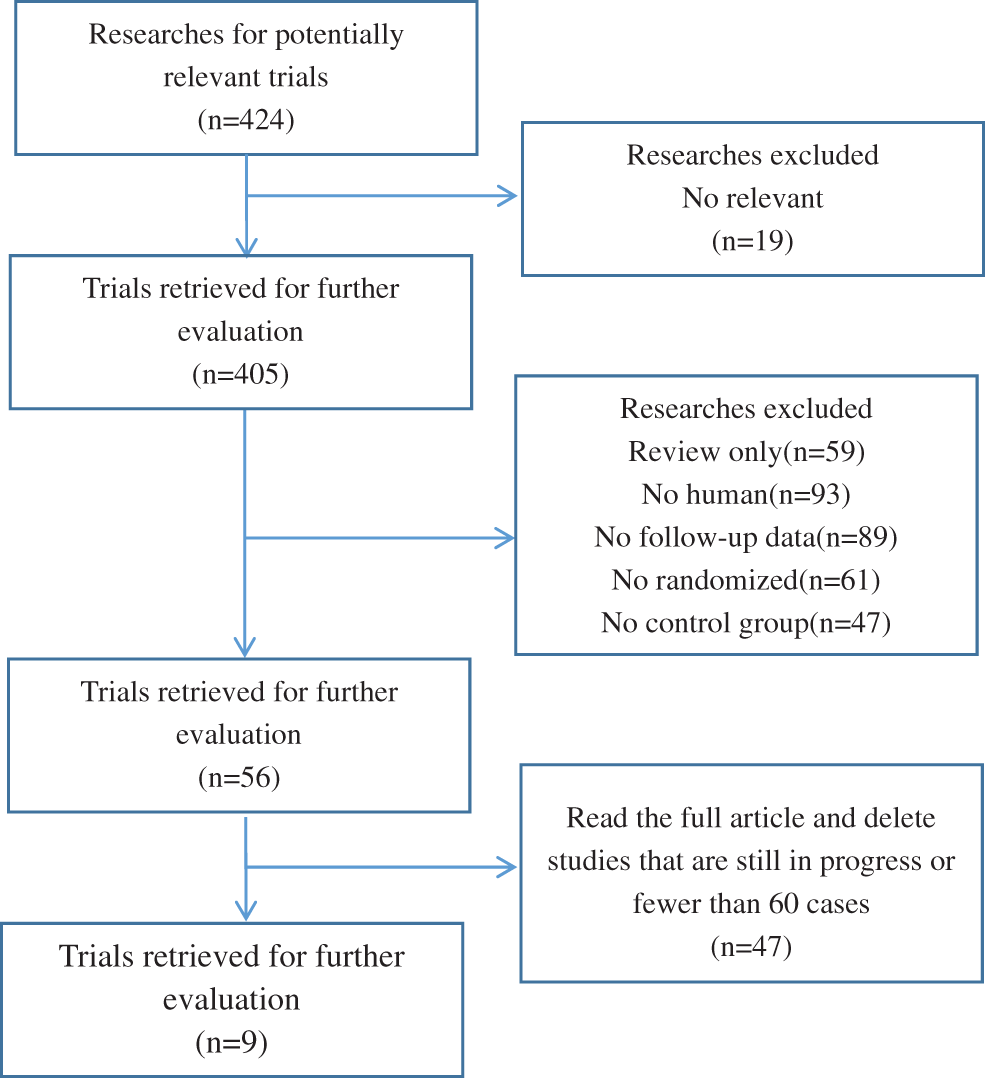
Figure 1: Flowchart of the selection of the clinical trials
3.2 Association between ASA use and Prognosis in Patients with BC
The combined analysis of the data of nine included studies showed that there was large heterogeneity among the studies (P < 0.0001, I2 = 75%). Therefore, the random effects model was adopted, with the combined effect size of RR being 0.83%, 95% CI [0.73, 0.94], Z = 2.89, P = 0.004 (P < 0.05). The difference was statistically significant. According to the forest plot (Fig. 2a), the survival rate of patients with BC was higher within the ASA group and the risk of death was less than that in the non-ASA group, thus suggesting that ASA helps improve the prognosis of patients with BC. For publication bias assessment, a funnel plot was constructed for all included studies (Fig. 2b). The results showed that the funnel plot was obviously symmetrical. The result of Begg’s test for publication bias was not statistically significant (P > 0.05), indicating that the results were less affected by publication bias and the conclusion was reliable.
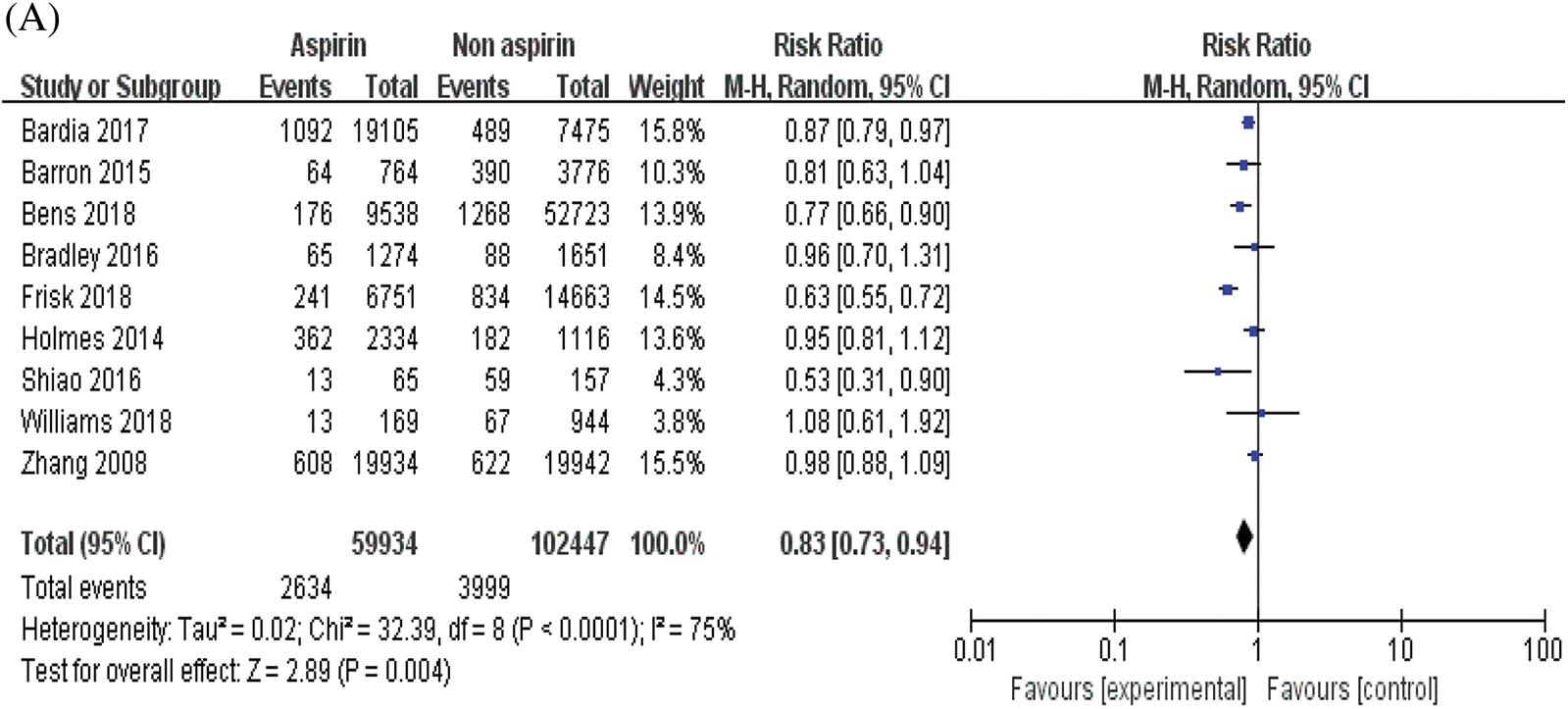
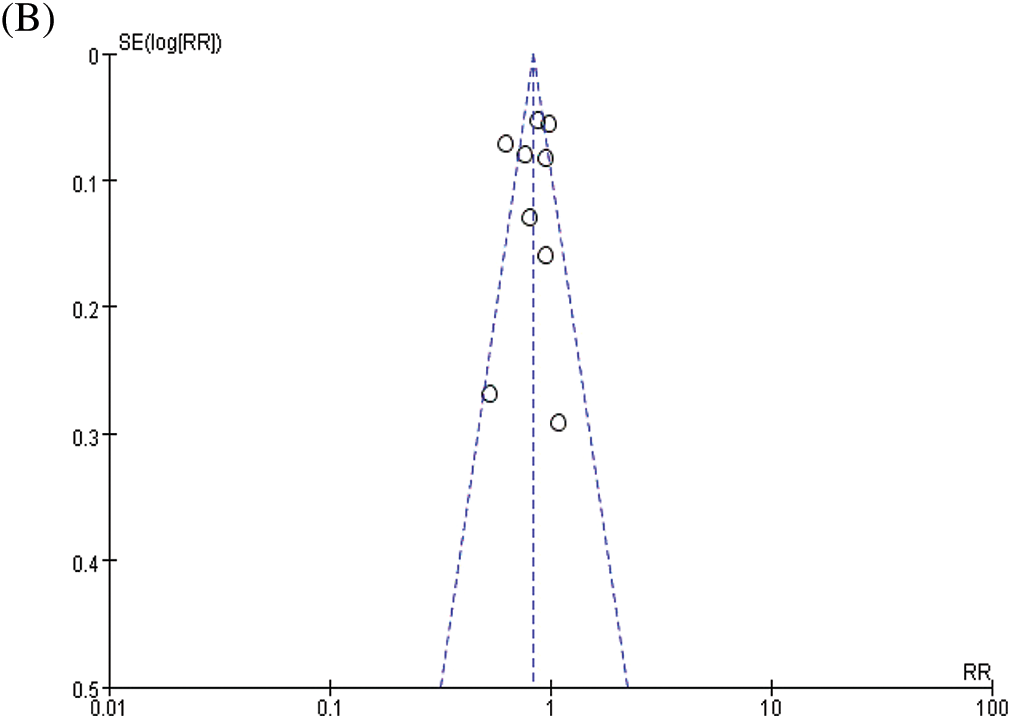
Figure 2: Meta-analyses of the included studies examining aspirin and overall survival in BC. (A) forest plot. (B) funnel plot
3.3 Effects of ASA Combined with 4-OHT on the Proliferation of MCF-7 and MCF-7/TAM Cells
MCF-7 and MCF-7/TAM cells were exposed to 4-OHT (3 μM) alone and 4-OHT (3 μM) combined with ASA (2 mM) for six days. After treatment, cell viability was determined by an MTT assay after treatment. The inhibitory effects of 4-OHT alone on MCF-7 and MCF-7/TAM cells were significantly different. Cell viability exhibited no significant decrease in MCF-7/TAM cells treated with 4-OHT alone. In contrast, cell viability of MCF-7/TAM cells in the 4-OHT combined with ASA groups (63.93 ± 5.98)% was significantly lower than in the 4-OHT alone groups (104.30 ± 5.67)% (P < 0.05) ( Fig. 3).
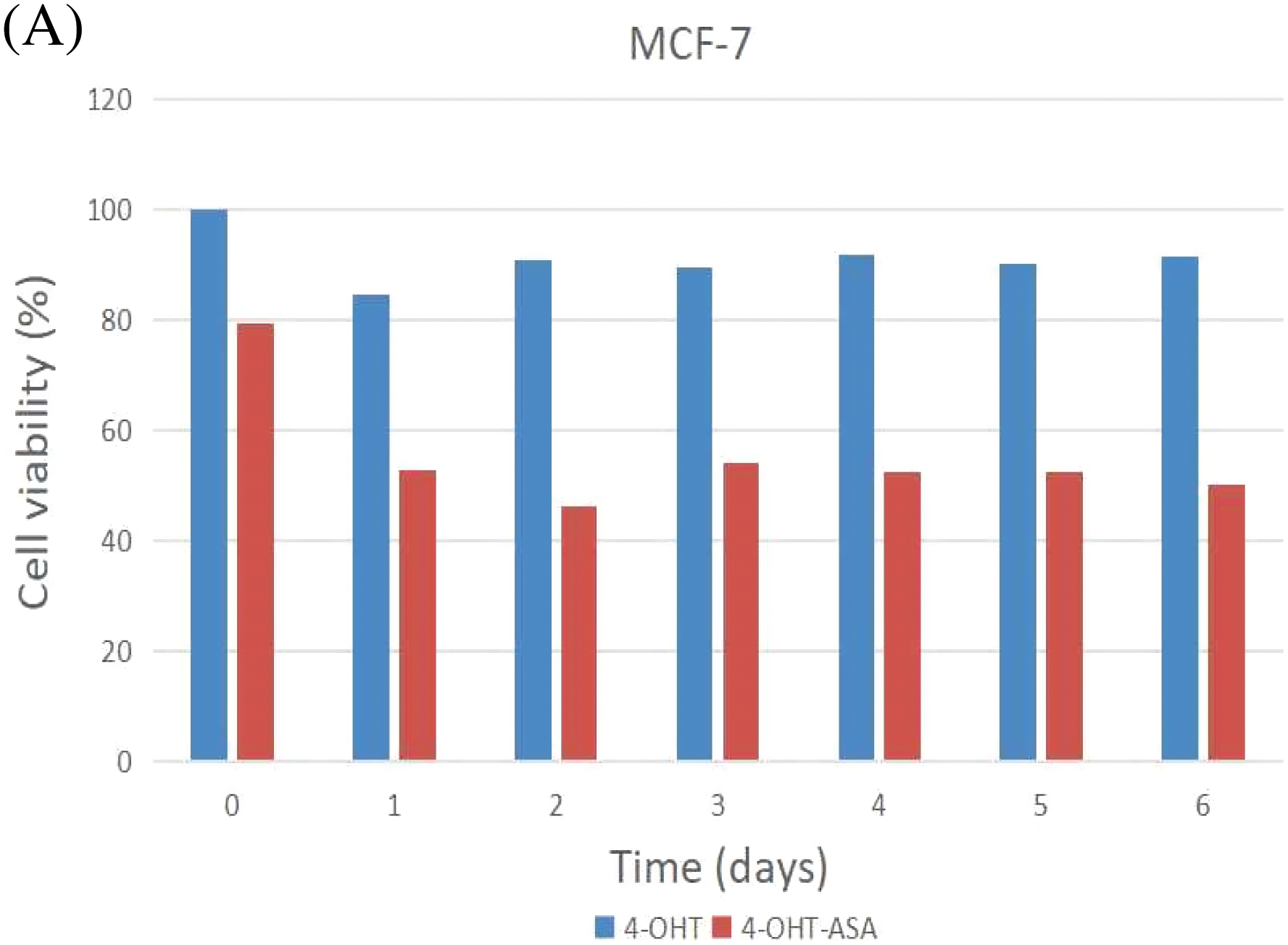
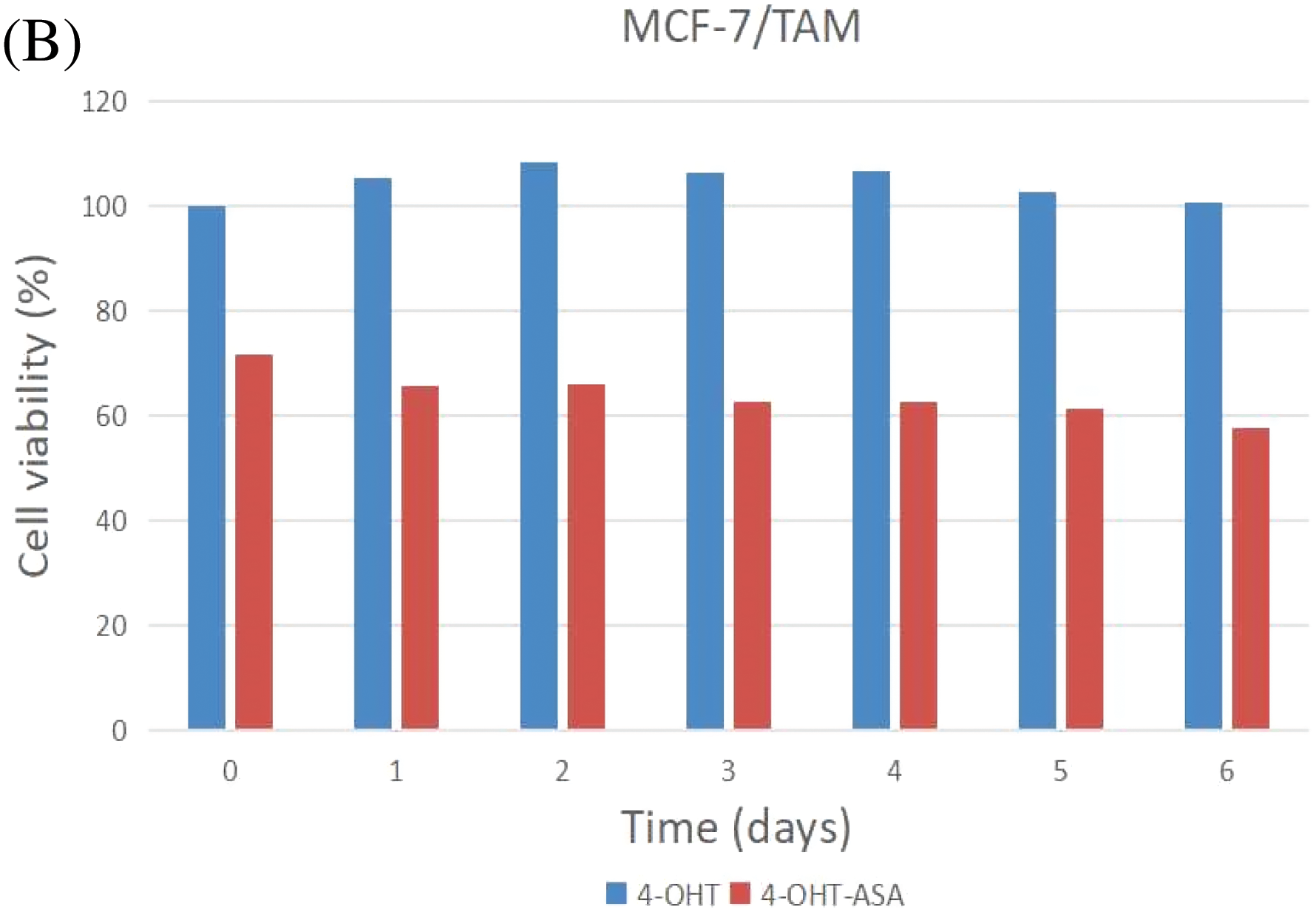
Figure 3: 4-OHT (3 μM) can inhibit cell growth in MCF-7 cells only and 4-OHT (3 μM) in combination with ASA (2 mM) can dramatically inhibit cell growth both in MCF-7 and MCF-7/TAM cells. (A) MCF-7 cells were treated with 4-OHT (3 μM) only and 4-OHT (3 μM) in combination with ASA (2 mM) for 0, 1, 2, 3, 4, 5, and 6 days. (B) MCF-7/TAM cells were treated with 4-OHT (3 μM) only and 4-OHT (3 μM) in combination with ASA (2 mM) for 0, 1, 2, 3, 4, 5, and 6 days
3.4 Overview of the Proteomic Results
To investigate the cell biological changes and mechanisms of ASA combined with 4-OHT, proteomic analysis was performed to find the difference in protein expression of MCF-7 and MCF-7/TAM cells before and after the ASA and 4-OHT combination treatment. 6,431 intracellular proteins were recognized with high reliability in all eight cell samples from LC-MS/MS (Fig. 4a). After filtration (the criteria were: found in all samples, FDR < 0.01 and unique peptides ≥ 2), 3,847 proteins were used to search for searching dysregulated proteins. PCA and hierarchical clustering of protein expression changes (Fig. 4b) showed a clear separation among each group.
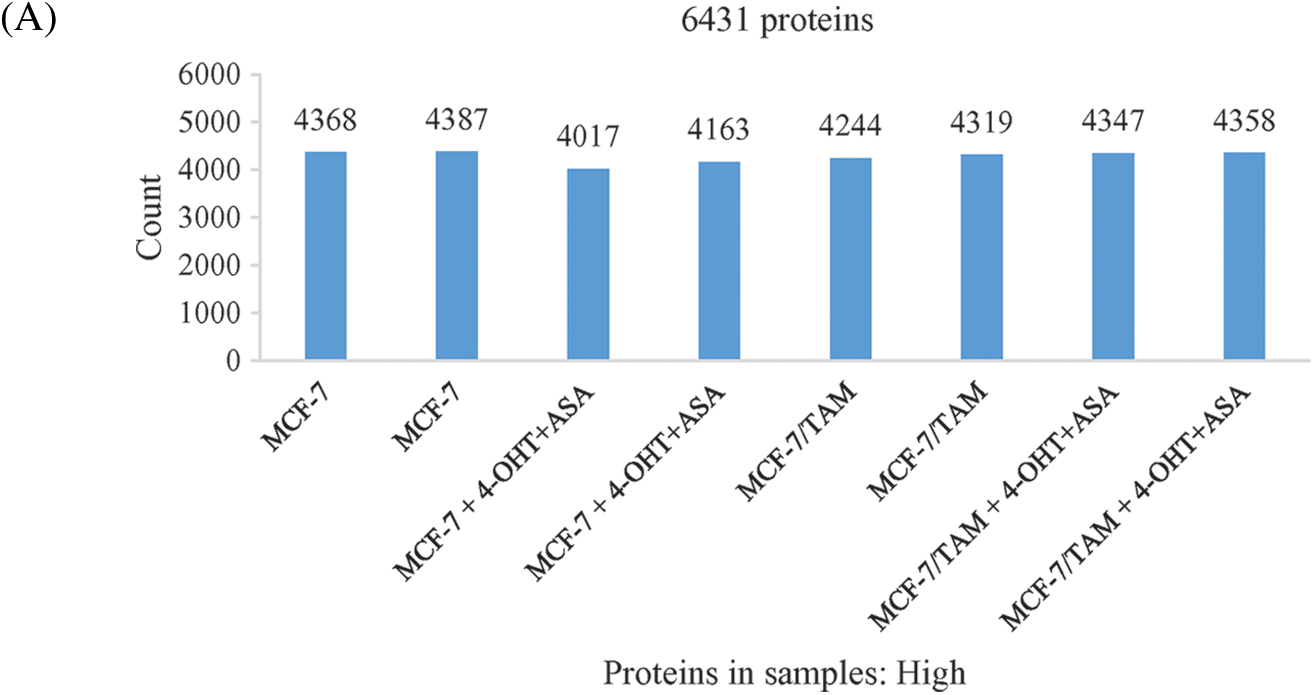
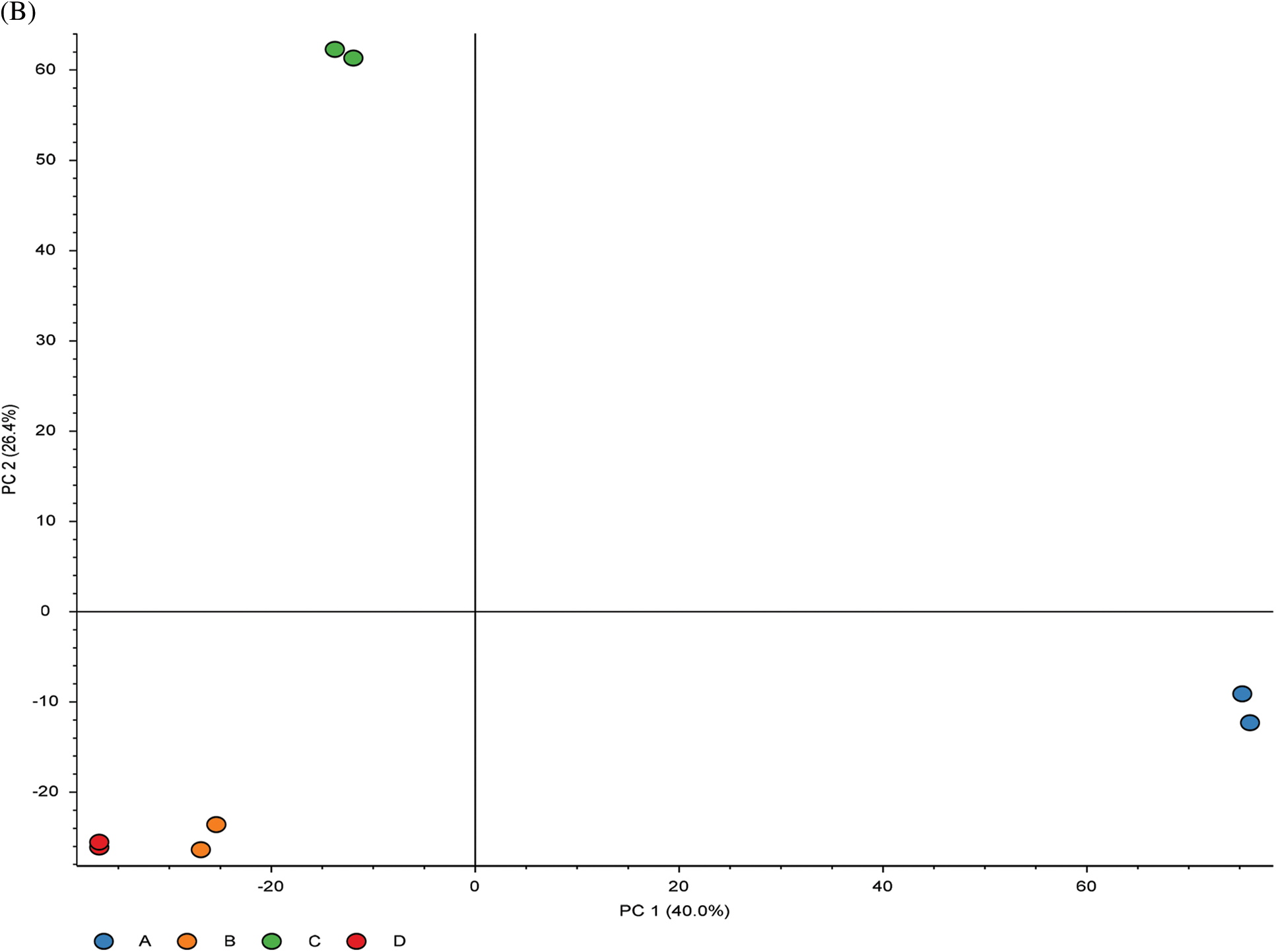
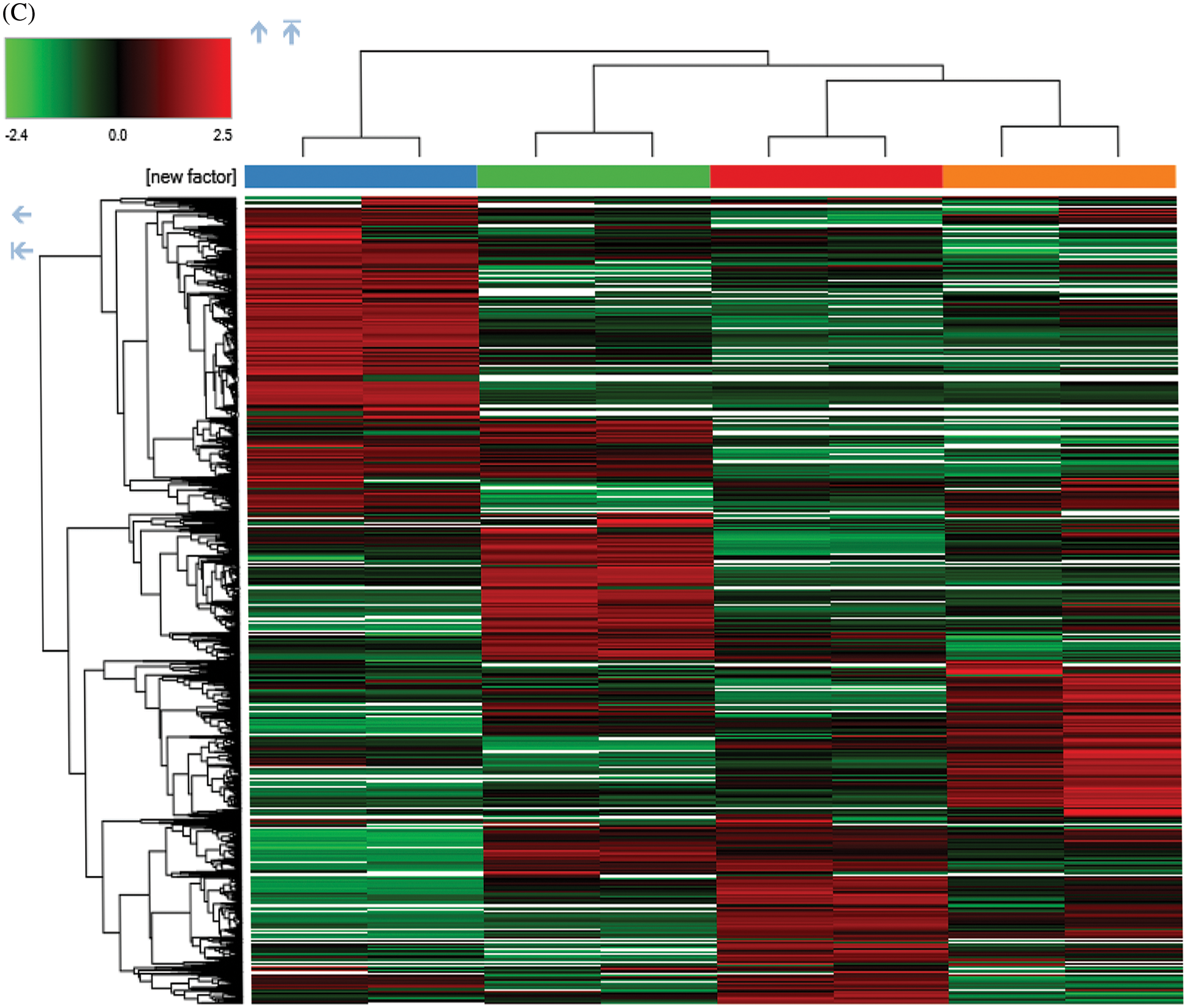
Figure 4: Overview of the proteomic results. (A) Proteins identified with high confidence in each cell sample. (B) Score plot of the PCA representing the distribution of samples in terms of protein expression profiles and hierarchical clustering heatmap representing the expression profiles of each sample. Sample groups are marked by colors. Blue represent MCF-7 cell samples; orange represent MCF-7/TAM cell samples; green represent MCF-7 cell samples in treatment of ASA and 4-OHT combination; red represent MCF-7/TAM cell samples in treatment of ASA and 4-OHT combination
3.5 Differentially Expressed Proteins (DEPs) of MCF-7/TAM Cells after Treatment with ASA and 4-OHT Combination
To identify the biological impact of ASA combined with 4-OHT on tamoxifen-resistant cells, we compared the proteomes of MCF-7/TAM cells before and after the ASA and 4-OHT combination treatment. A total of 57 DEPs were up-regulated after the combination treatment, while 85 DEPs were down-regulated (Fold change >2 or <0.5 and adjusted P < 0.05) (Fig. 5 and Table 2). GO analyses were then performed on these DEPs and the top-ranked GO terms were demonstrated in terms of biological process (Fig. 6a), cellular subtype (Fig. 6b), and molecular function (Fig. 6c). Several GO terms were significantly enriched, such as neutrophil degranulation, retinal metabolic process, sterol biosynthetic process, and prostaglandin metabolic process. KEGG pathway enrichment analysis also verified three associated pathways including metabolic pathways, chemical carcinogenesis-reactive oxygen species, and biosynthesis of amino acids (Fig. 6d).
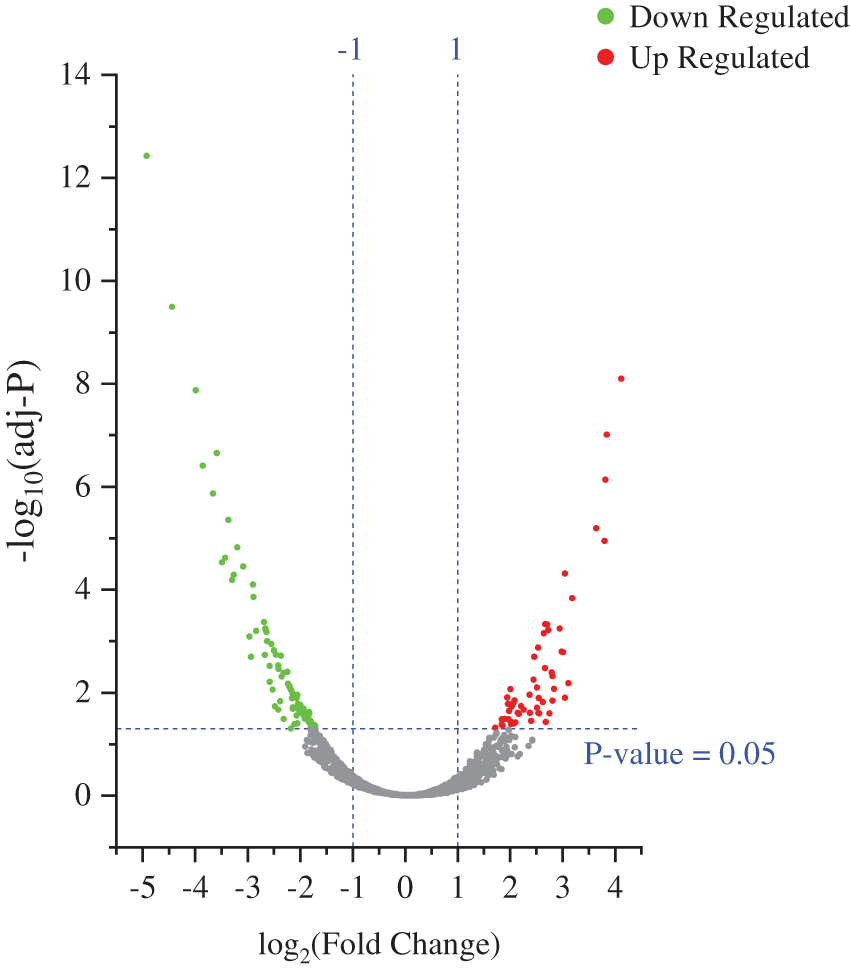
Figure 5: Volcano plot exhibit significantly DEPs
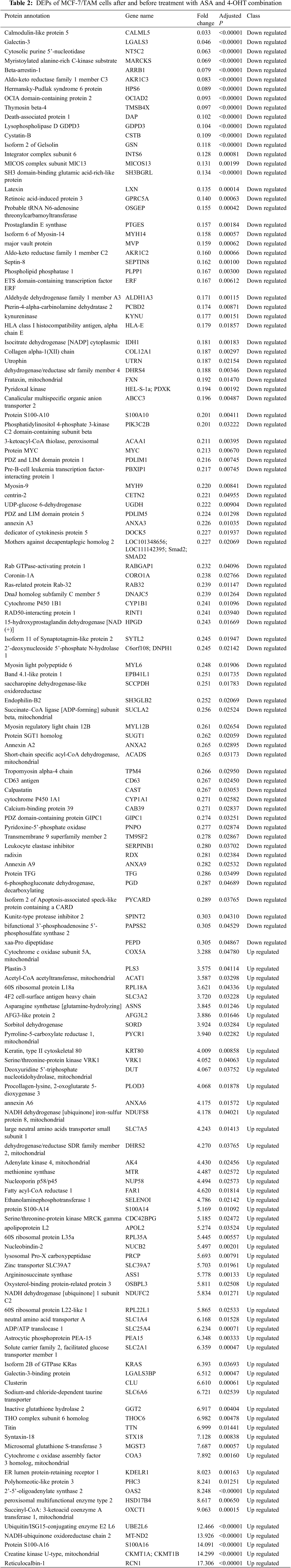
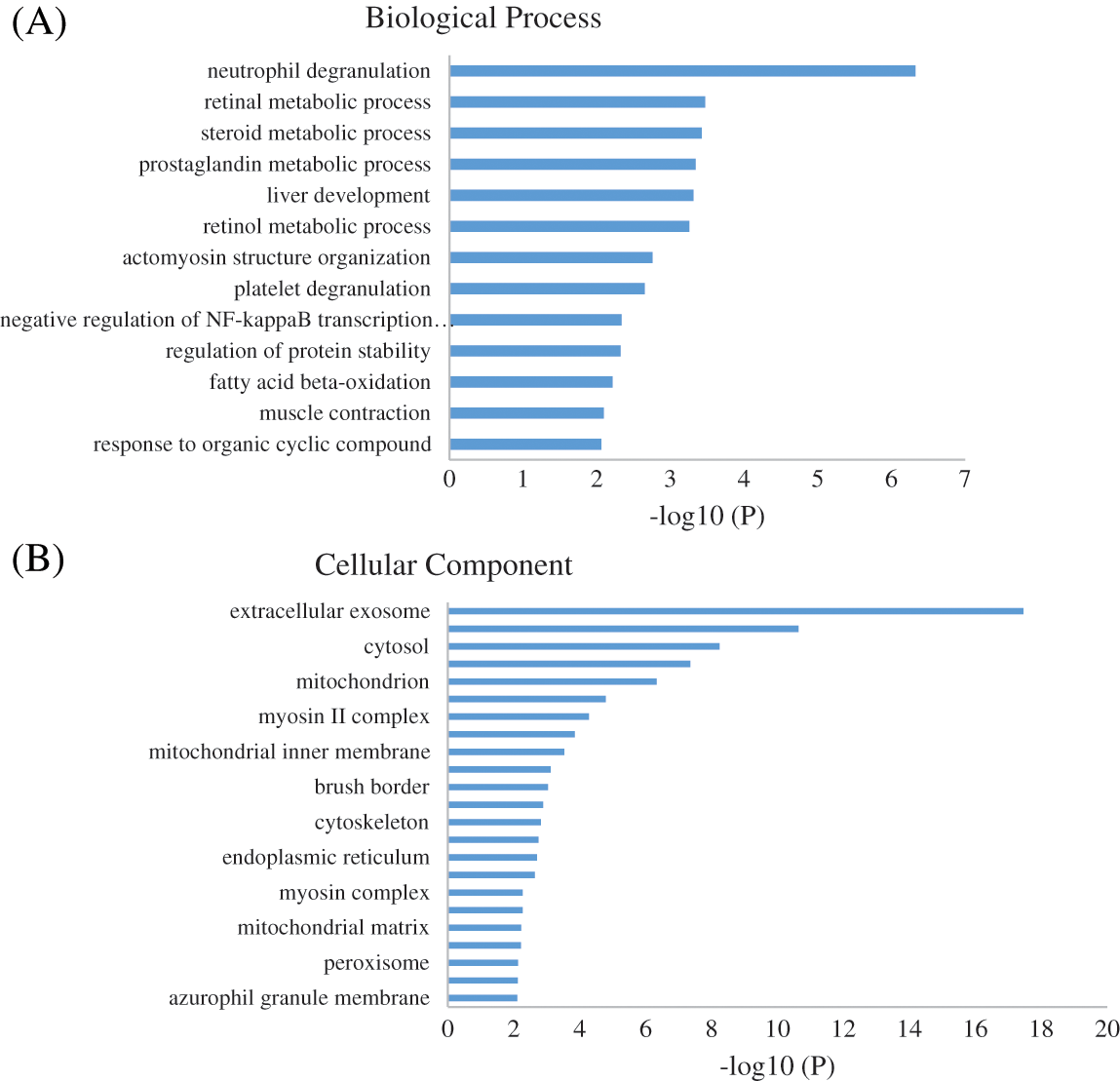
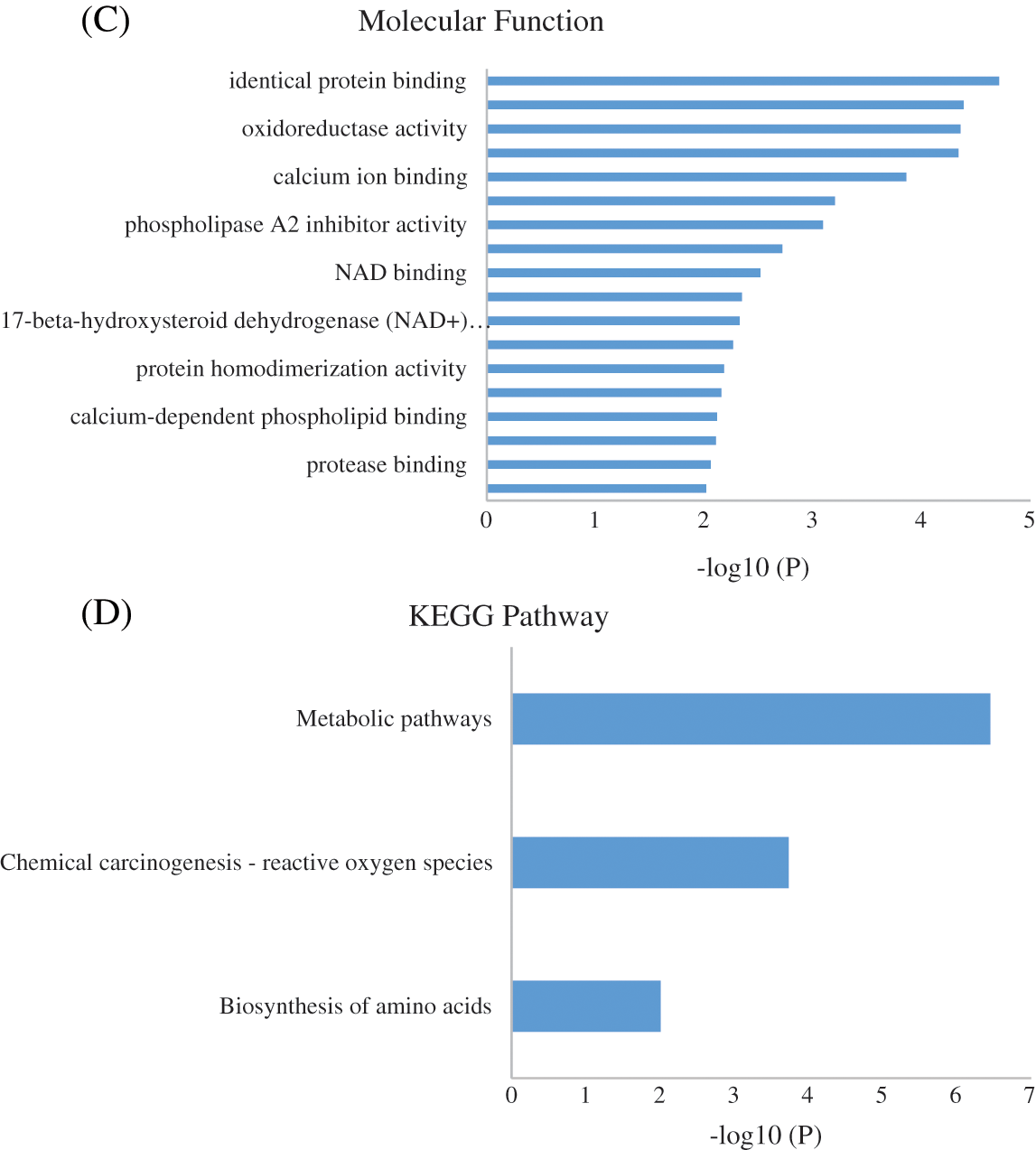
Figure 6: GO enrichment result of the DEPs of MCF-7/TAM Cells after Treatment with ASA and 4-OHT combination in terms of (A) biological process, (B) cellular component, and (C) molecular function (P < 0.01). (D) KEGG enrichment pathway result (P < 0.01)
3.6 Clinicopathological Characteristics of Patients
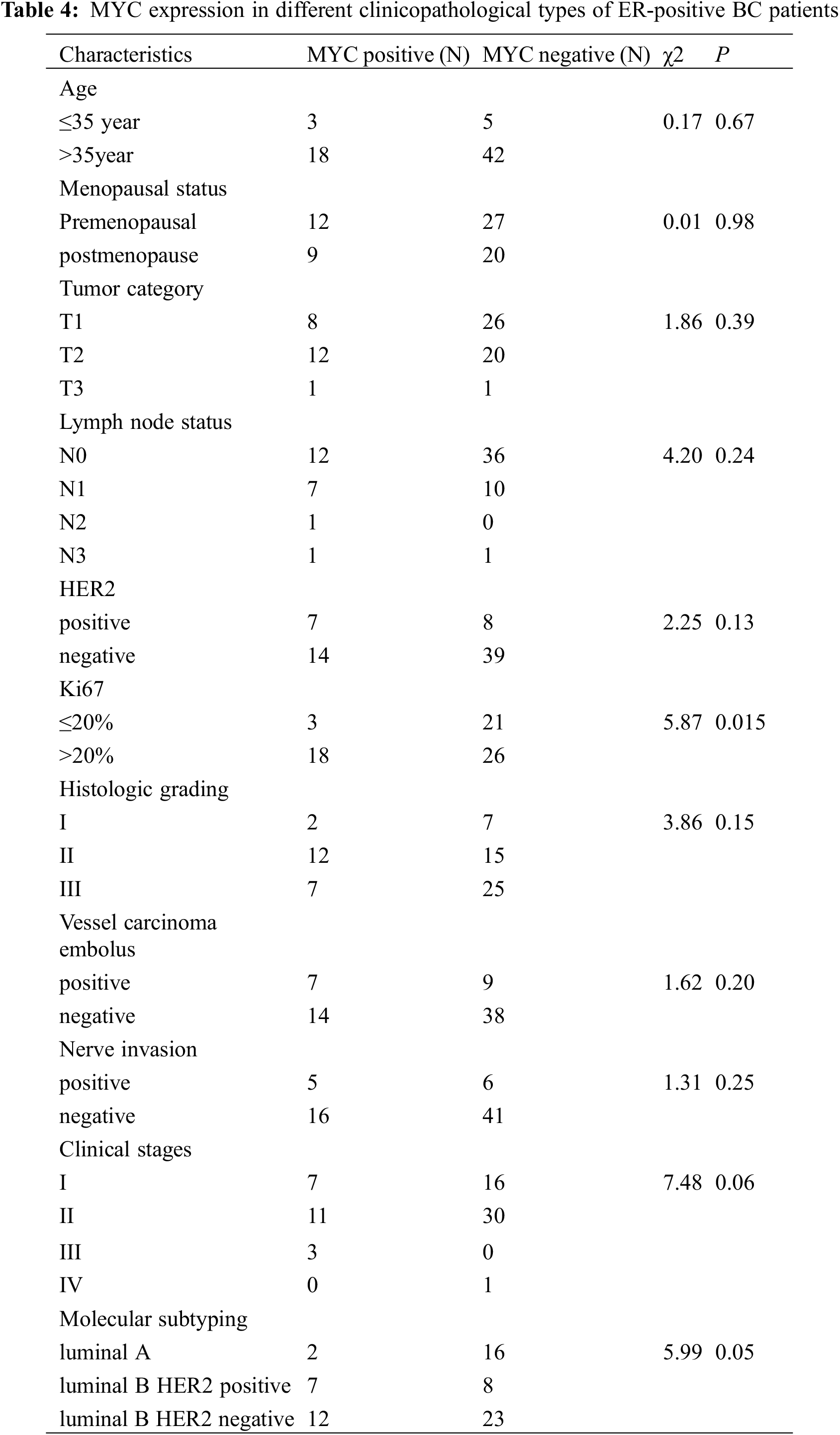
The proteomic analyses of MCF-7/TAM cells showed that MYC protein might be one of the most significantly therapeutic target protein for ASA combined with tamoxifen. Our previous work had demonstrated that MYC protein could enhance tamoxifen resistance in vitro experiment, we further analyzed the expression of MYC in tumor specimens of patients beginning or continuing long-term tamoxifen treatment. Between October 2019 to March 2022, sixty-eight patients were enrolled in Peking University Shenzhen Hospital. Data from patients within the an age range of 25–86 years (median age: 50.02 ± 13.58 years) were enrolled for this analysis. The pathological types were all invasive BC with ER positive. HER2 was positive in 14 (20.59%) of the 68 cases. There were 23 cases in clinical stage I (71.43%), 41 cases in clinical stage II (60.29%), three cases in clinical stage III (4.41%), and one case in clinical stage IV (1.48%). Also, 18 patients were luminal A (26.47%), 15 patients were luminal B HER2-positive (22.06%), and 35 patients were luminal B HER2-negative (51.47%). Of the 68 luminal-like invasive BC cases, 21 (30.88%) were positive for MYC protein expression, while 47 (69.12%) were negative for MYC protein (Table 3 and Fig. 7).
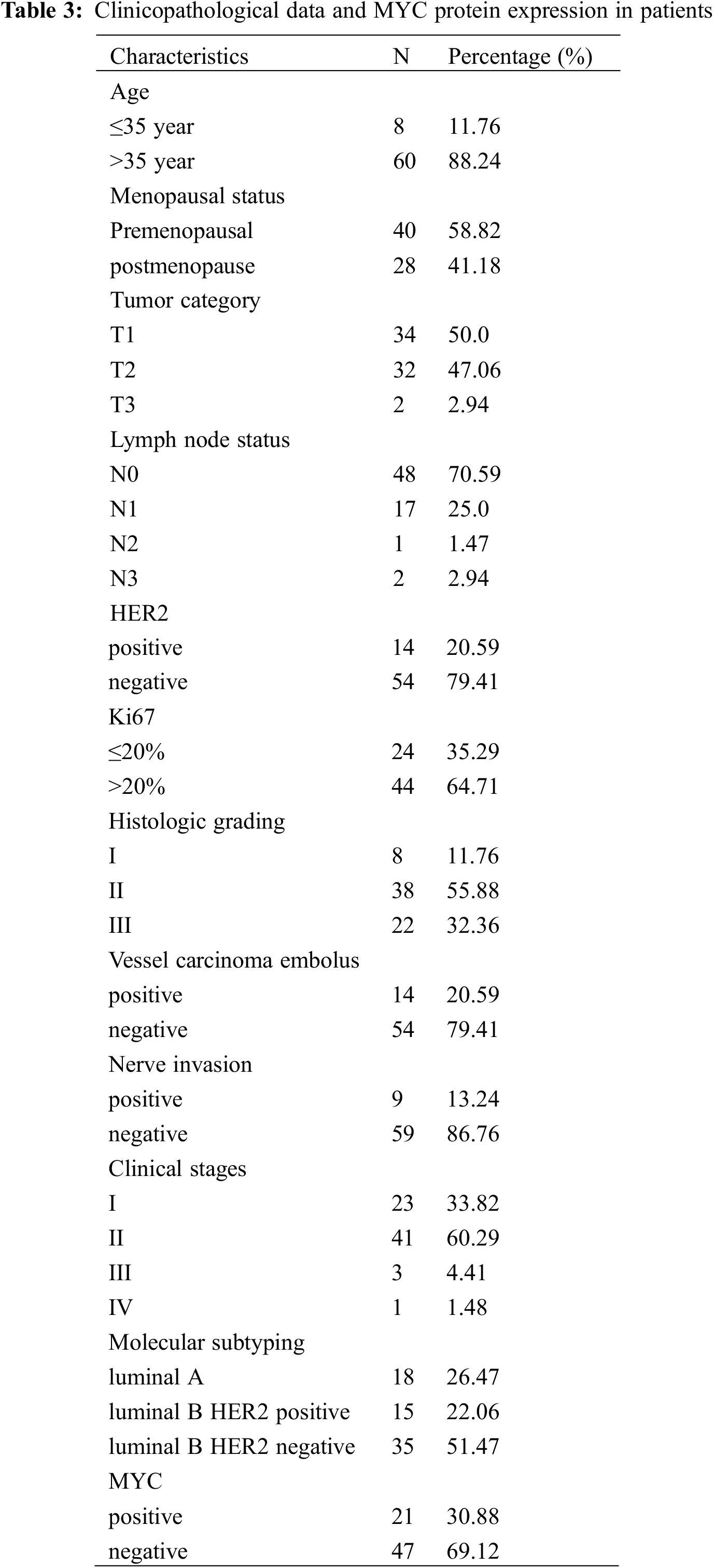
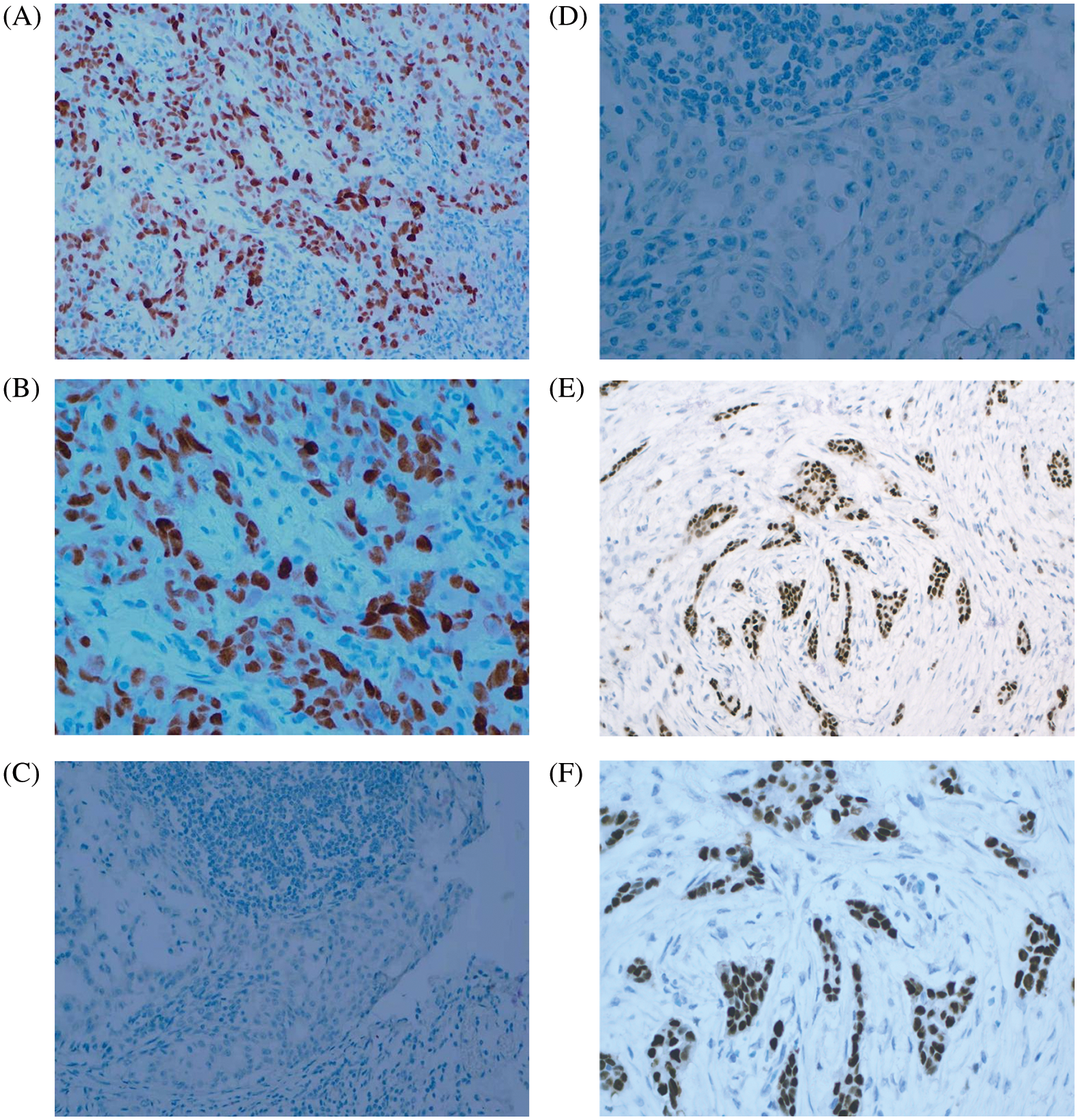
Figure 7: MYC and ER expression in BC tissues. (A) MYC positive (×200 magnification). (B) MYC positive (×400 magnification). (C) MYC negative (×200 magnification). (D) MYC negative (×400 magnification). (E) ER positive (×200 magnification). (F) ER positive (×400 magnification)
3.7 Associations between MYC and Clinical Markers
The MYC protein was expressed in different types of ER-positive BC tissues, with the highest expression in luminal B HER2-negative (57.14%), followed by luminal B HER2-positive BC (33.33%); whereas the lowest expression was in luminal A BC (9.53%). In patients with BC having Ki67 > 20% (40.91%), the proportion of MYC-positive cases was significantly higher (P < 0.05) than that of those having Ki67 ≤ 20% (12.50%). However, among the other subgroups, there was no significant difference in MYC protein expression (P > 0.05), as shown in Table 4.
In recent years, the continuous emergence of anti-cancer studies on ASA has enabled the traditional drug ASA to have important new functions. As a result, the potential application of ASA in colorectal cancer, BC, and other solid tumors has been developed [29,30]. Inflammation in the tumor microenvironment is one of the factors that promote tumor growth. A large number of epidemiological evidence suggests that ASA, which suppresses inflammation, reduces the risk of cancer [31]. ASA exerts its anti-inflammatory effects mainly by inhibiting cyclooxygenase (COX), which is a key enzyme in prostaglandin (PG) biosynthesis from arachidonic acid (AA). The two major isoforms of COX (COX-1 and COX-2) catalyze the conversion of AA to prostaglandins, which are metabolized into different prostaglandins by tissue-specific synthetase. ASA inhibits COX-1 and COX-2 activity, the latter of which is thought to be an important factor in the transition of inflammation to tumorigenesis. Another important effect of ASA is antiplatelet agglutination, which improves blood circulation by antagonizing platelet agglutination. A recent study found that ASA inhibits platelet aggregation, preventing cancer cells from taking advantage of platelet adhesion properties to combine with other cells, thus reducing the ability of tumors to metastasize. The clinical results of ASA in BC remain controversial. In some studies, low dose of ASA was associated with a reduced risk of BC, especially for ER-positive and HER2-negative subtypes, without an association with ER-negative BC [32,33]. This study included nine high-quality clinical trials. Due to the high heterogeneity of the studies, the conclusions are controversial. For instance, Zhang et al. [28] and Holmes et al. [24] thought ASA does not affect BC, whereas other researchers, such as Shiao et al. [26] made the opposite conclusion. The meta-analysis found that ASA could reduce the death rate of patients and improve the prognosis of patients compared with the non-ASA group. These results indicated that ASA had certain clinical effect on improving the prognosis of BC patients. Further experiments in vitro showed that ASA with 4-OHT could significantly inhibit the proliferation of tamoxifen resistance cells. Studies on the reversal of drug resistance of other tumors by ASA showed that cisplatin plus ASA significantly reduced the survival rate of cisplatin-resistant tumor cells, and ASA could inhibit the expression of tumor cell-related proteins, such as ALDH1, CD44, CD133, and P53 [34,35]. ASA reduced the growth and invasion of pancreatic ductal adenocarcinoma and significantly enhanced the therapeutic effect of gemcitabine. These findings were confirmed in tissue samples from patients who had taken ASA or not before surgery [36]. Therefore, combined with the previous experimental results, this study further explored the changes in cellular proteins related to the tumor-sensitizing effect of ASA to tamoxifen treatment.
Proteomic analysis is important in the comprehensive study of human biology, and the biological processes involved in complex diseases, including tumors, can be identified by combining proteomic and bioinformatics tools [37,38]. Systems biology tools for data analysis are often used to interpret results to define the biological significance of differentially abundant proteins. Quantitative proteomics technology combined with bioinformatics was used to identify tamoxifen resistance-associated proteins. According to the proteomic profiling of the BC cell lines understudied, 60 proteins were up-regulated and 34 proteins were down-regulated in MCF-7/TAM cells. To further analyze the effect and related mechanism of ASA on tamoxifen resistance in BC, the proteins differentially expressed after ASA and 4-OHT combined treatment were further identified from the dysregulated proteins described above. It was found that the expression of proteins, including CALML5, ARRB1, ACAA1, OCIAD2, DNAJC5, PLPP1, and MYC, were upregulated in MCF-7/TAM cells and depressed after the ASA and 4-OHT combined treatment. In our previous research, the MYC gene and protein levels in tamoxifen resistant cells were knocked down using shRNA gene interference technology, which also increased the sensitivity of resistant BC cells to tamoxifen again [17]. The high expression of the MYC protein also brings a high risk to patients with BC, and the MYC protein has been reported as a tumor marker of BC [39]. Based on the Kaplan-Meier Plotter database, researchers conducted survival correlation analysis on 3,951 patients with BC and the results showed that the MYC protein was positively correlated with the survival risk of BC. MYC is also involved in the process of endocrine resistance in BC. Compared with non-endocrine drug-resistant cells, it was found that the MYC protein was overexpressed in all drug-resistant cell lines and that inhibition of the MYC protein resulted in differential blocking of the non-estrogen-dependent proliferation of drug-resistant cells [40]. Among the patients enrolled in this study, the MYC protein was expressed in the highest expression ratio in luminal B HER2 negative patients, while patients with luminal B HER2 negative were the largest proportion of patients receiving endocrine therapy. Therefore, the occurrence of drug resistance should be monitored in long-term endocrine therapy. MYC protein is significantly associated with tumor histological type. MYC protein expression level was higher in medullary carcinoma and lower in lobular carcinoma [41]. We have further analyzed the difference in MYC protein expression among BC tissues with different clinicopathological indicators and found that the positive proportion of the MYC protein was higher in BC tissues with high Ki67 expression. Among the BC tissues with positive MYC protein expression, the proportion of Ki67 > 20% was as high as 85.71%, which was much higher than that of the MYC protein negative group, thus indicating that the MYC protein expression was closely related to tumor cell proliferation. Salicylic acid is the main metabolite of ASA, and it could reduce the MYC protein levels in human colon cancer cells [42]. Human platelets enhance colon cancer cell proliferation by upregulating and activating the MYC protein, and the upregulation of the MYC protein and proliferation of cancer cells are both reversed by ASA against the platelet concentration, thus suggesting that ASA inhibition of platelets may affect cancer cell proliferation by regulating the MYC protein [43].
In this study, here we also found that some proteins as ASS1, OXCT1, RCN1, DUT, and GGT2 were downregulated in MCF-7/TAM cells and recovered after ASA and 4-OHT combined treatment. ASS1 is a key enzyme in arginine biosynthesis, and its abundance is reduced in many tumors. In a random sample of 149 BCs, ASS1 was low or undetected in more than 60% of the patients independent molecular subtype [44]. In former researches, ASS1 had been reported as a tumor suppressor, and a lack of ASS1 in cancer was found to induce arginine auxotrophy [45–47]. In this study, the combination of ASA and 4-OHT increased the expression level of ASS1. Genetic manipulation studies of ASS1 had confirmed that ASS1 expression inhibited fibroblast proliferation, migration, and invasion. OXCT1 was also recovered after ASA and 4-OHT combined treatment in MCF-7/TAM cells. Epigenetic silencing of OXCT1 in ovarian cancer was confirmed to be associated with cisplatin resistance. Overexpression of OXCT1 restored sensitivity to cisplatin chemotherapy, suggesting that OXCT1 is a resistant repressor in cancer [48].
In conclusion, our data provide the necessary evidence that ASA may have a certain effect on improving the prognosis of BC patients, and tamoxifen resistant of ER-positive BC cells can be reversed through ASA and 4-OHT combined treatment. Several proteins were downregulated or activated involved in ASA and 4-OHT combined treatment, which might be identified as potential targets for the treatment of ER-positive BC with tamoxifen resistance. However, the limitations of this study may be the limited data from animal intervention studies and the long-term follow-up of participants, and further research is needed in the future.
Ethical Approval and Informed Consent Statement: Written informed consents were obtained from all participants, and this study was permitted by the Ethics Committee of Peking University Shenzhen Hospital (No. 2021-037. Approval Date: 2021.03.04).
Author Contributions: All authors contributed to the study design. Y.L. conceptualized and designed this study. J.C. and M.L. performed the cell culture, sample preparation experiments and immunohistochemistry. J.C. and R.P. performed the meta-analysis, proteomic experiments and analyzed the data. J.C. and R.P. wrote the draft of the manuscript. M.L. and Y.L. revised the manuscript. All author have read and approved the final manuscript.
Availability of Data and Materials: The dataset supporting the conclusions of this article is included within this article and is available from the corresponding author upon request.
Funding Statement: This work was supported by Shenzhen Key Medical Discipline Construction Fund (No. SZXK017), Shenzhen High-Level Hospital Construction Fund.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Leon-Ferre, R. A., Polley, M. Y., Liu, H., Gilbert, J. A., Cafourek, V. et al. (2018). Impact of histopathology, tumor-infiltrating lymphocytes, and adjuvant chemotherapy on prognosis of triple-negative breast cancer. Breast Cancer Research and Treatment, 167(1), 89–99. DOI 10.1007/s10549-017-4499-7. [Google Scholar] [CrossRef]
2. Harbeck, N., Gnant, M. (2017). Breast cancer. Lancet, 389(10074), 1134–1150. DOI 10.1016/S0140-6736(16)31891-8. [Google Scholar] [CrossRef]
3. Li, Y., Kong, X., Xuan, L., Wang, Z., Huang, Y. H. (2021). Prolactin and endocrine therapy resistance in breast cancer: The next potential hope for breast cancer treatment. Journal of Cellular and Molecular Medicine, 25(22), 10327–10348. DOI 10.1111/jcmm.16946. [Google Scholar] [CrossRef]
4. Paganini-Hill, A., Clark, L. J. (2000). Preliminary assessment of cognitive function in breast cancer patients treated with tamoxifen. Breast Cancer Research and Treatment, 64(2), 165–176. DOI 10.1023/A:1006426132338. [Google Scholar] [CrossRef]
5. Araki, K., Miyoshi, Y. (2018). Mechanism of resistance to endocrine therapy in breast cancer: The important role of PI3K/Akt/mTOR in estrogen receptor-positive, HER2-negative breast cancer. Breast Cancer, 25(4), 392–401. DOI 10.1007/s12282-017-0812-x. [Google Scholar] [CrossRef]
6. Caffa, I., Spagnolo, V., Vernieri, C., Valdemarin, F., Becherini, P. et al. (2020). Fasting-mimicking diet and hormone therapy induce breast cancer regression. Nature, 583(7817), 620–624. DOI 10.1038/s41586-020-2502-7. [Google Scholar] [CrossRef]
7. Bekele, R. T., Venkatraman, G., Liu, R. Z., Tang, X., Mi, S. et al. (2016). Oxidative stress contributes to the tamoxifen-induced killing of breast cancer cells: Implications for tamoxifen therapy and resistance. Scientific Reports, 6, 21164. DOI 10.1038/srep21164. [Google Scholar] [CrossRef]
8. Toy, W., Weir, H., Razavi, P., Lawson, M., Goeppert, A. U. et al. (2017). Activating ESR1 mutations differentially affect the efficacy of ER antagonists. Cancer Discovery, 7(3), 277–287. DOI 10.1158/2159-8290.CD-15-1523. [Google Scholar] [CrossRef]
9. Cronin-Fenton, D. P., Damkier, P. (2018). Tamoxifen and CYP2D6: A controversy in pharmacogenetics. Advances in Pharmacology, 83, 65–91. DOI 10.1016/bs.apha.2018.03.001. [Google Scholar] [CrossRef]
10. Tryfonidis, K., Zardavas, D., Katzenellenbogen, B. S., Piccart, M. (2016). Endocrine treatment in breast cancer: Cure, resistance and beyond. Cancer Treatment Reviews, 50, 68–81. DOI 10.1016/j.ctrv.2016.08.008. [Google Scholar] [CrossRef]
11. Burn, J., Sheth, H., Elliott, F., Reed, L., Macrae, F. et al. (2020). Cancer prevention with aspirin in hereditary colorectal cancer (Lynch syndrome10-year follow-up and registry-based 20-year data in the CAPP2 study: A double-blind, randomised, placebo-controlled trial. Lancet, 395(10240), 1855–1863. DOI 10.1016/S0140-6736(20)30366-4. [Google Scholar] [CrossRef]
12. Assayag, J., Pollak, M. N., Azoulay, L. (2015). The use of aspirin and the risk of mortality in patients with prostate cancer. The Journal of Urology, 193(4), 1220–1225. DOI 10.1016/j.juro.2014.11.018. [Google Scholar] [CrossRef]
13. Matsuo, K., Cahoon, S. S., Yoshihara, K., Shida, M., Kakuda, M. et al. (2016). Association of low-dose aspirin and survival of women with endometrial cancer. Obstetrics and Gynecology, 128(1), 127–137. DOI 10.1097/AOG.0000000000001491. [Google Scholar] [CrossRef]
14. Xie, S., Wang, Y., Huang, Y., Yang, B. (2021). Mechanisms of the antiangiogenic effects of aspirin in cancer. European Journal of Pharmacology, 898, 173989. DOI 10.1016/j.ejphar.2021.173989. [Google Scholar] [CrossRef]
15. Chen, W. Y., Holmes, M. D. (2017). Role of aspirin in breast cancer survival. Current Oncology Reports, 19(7), 48. DOI 10.1007/s11912-017-0605-6. [Google Scholar] [CrossRef]
16. Ma, S., Guo, C., Sun, C., Han, T., Zhang, H. et al. (2021). Aspirin use and risk of breast cancer: A meta-analysis of observational studies from 1989 to 2019. Clinical Breast Cancer, 21(6), 552–565. DOI 10.1016/j.clbc.2021.02.005. [Google Scholar] [CrossRef]
17. Cheng, R., Liu, Y. J., Cui, J. W., Yang, M., Liu, X. L. et al. (2017). Aspirin regulation of MYC and cyclind1 proteins to overcome tamoxifen resistance in estrogen receptor-positive breast cancer cells. Oncotarget, 8(18), 30252–30264. DOI 10.18632/oncotarget.16325. [Google Scholar] [CrossRef]
18. Wang, D., Eraslan, B., Wieland, T., Hallström, B., Hopf, T. et al. (2019). A deep proteome and transcriptome abundance atlas of 29 healthy human tissues. Molecular Systems Biology, 15(2), e8503. DOI 10.15252/msb.20188503. [Google Scholar] [CrossRef]
19. Martínez-Rodríguez, F., Limones-González, J. E., Mendoza-Almanza, B., Esparza-Ibarra, E. L., Gallegos-Flores, P. I. et al. (2021). Understanding cervical cancer through proteomics. Cells, 10(8), 1854. DOI 10.3390/cells10081854. [Google Scholar] [CrossRef]
20. Bardia, A., Keenan, T. E., Ebbert, J. O., Lazovich, D., Wang, A. H. et al. (2016). Personalizing aspirin use for targeted breast cancer chemoprevention in postmenopausal women. Mayo Clinic Proceedings, 91(1), 71–80. DOI 10.1016/j.mayocp.2015.10.018. [Google Scholar] [CrossRef]
21. Barron, T. I., Murphy, L. M., Brown, C., Bennett, K., Visvanathan, K. et al. (2015). De novo post-diagnosis aspirin use and mortality in women with stage I-III breast cancer. Cancer Epidemiology, Biomarkers & Prevention, 24(6), 898–904. DOI 10.1158/1055-9965.EPI-14-1415. [Google Scholar] [CrossRef]
22. Frisk, G., Ekberg, S., Lidbrink, E., Eloranta, S., Sund, M. et al. (2018). No association between low-dose aspirin use and breast cancer outcomes overall: A Swedish population-based study. Breast Cancer Research, 20(1), 142. DOI 10.1186/s13058-018-1065-0. [Google Scholar] [CrossRef]
23. Bens, A., Friis, S., Dehlendorff, C., Jensen, M. B., Ejlertsen, B. et al. (2018). Low-dose aspirin use and risk of contralateral breast cancer: A danish nationwide cohort study. Preventive Medicine, 116, 186–193. DOI 10.1016/j.ypmed.2018.09.015. [Google Scholar] [CrossRef]
24. Holmes, M. D., Olsson, H., Pawitan, Y., Holm, J., Lundholm, C. et al. (2014). Aspirin intake and breast cancer survival-A nation-wide study using prospectively recorded data in Sweden. BMC Cancer, 14, 391. DOI 10.1186/1471-2407-14-391. [Google Scholar] [CrossRef]
25. Bradley, M. C., Black, A., Freedman, A. N., Barron, T. I. (2016). Prediagnostic aspirin use and mortality in women with stage I to III breast cancer: A cohort study in the prostate, lung, colorectal, and ovarian cancer screening trial. Cancer, 122(13), 2067–2075. DOI 10.1002/cncr.30004. [Google Scholar] [CrossRef]
26. Shiao, J., Thomas, K. M., Rahimi, A. S., Rao, R., Yan, J. et al. (2017). Aspirin/antiplatelet agent use improves disease-free survival and reduces the risk of distant metastases in stage II and III triple-negative breast cancer patients. Breast Cancer Research and Treatment, 161(3), 463–471. DOI 10.1007/s10549-016-4081-8. [Google Scholar] [CrossRef]
27. Williams, A. D., Li, Y. R., So, A., Steel, L., Carrigan, E. et al. (2018). The impact of aspirin use on breast cancer subtype and clinical course. The Journal of Surgical Research, 230, 71–79. DOI 10.1016/j.jss.2018.04.040. [Google Scholar] [CrossRef]
28. Zhang, S. M., Cook, N. R., Manson, J. E., Lee, I. M., Buring, J. E. (2008). Low-dose aspirin and breast cancer risk: Results by tumour characteristics from a randomised trial. British Journal of Cancer, 98(5), 989–991. DOI 10.1038/sj.bjc.6604240. [Google Scholar] [CrossRef]
29. Yang, L., Lv, Z., Xia, W., Zhang, W., Xin, Y. et al. (2018). The effect of aspirin on circulating tumor cells in metastatic colorectal and breast cancer patients: A phase II trial study. Clinical & Translational Oncology, 20(7), 912–921. DOI 10.1007/s12094-017-1806-z. [Google Scholar] [CrossRef]
30. Coyle, C., Cafferty, F. H., Rowley, S., MacKenzie, M., Berkman, L. et al. (2016). ADD-ASPIRIN: A phase III, double-blind, placebo controlled, randomised trial assessing the effects of aspirin on disease recurrence and survival after primary therapy in common non-metastatic solid tumours. Contemporary Clinical Trials, 51, 56–64. DOI 10.1016/j.cct.2016.10.004. [Google Scholar] [CrossRef]
31. Gilligan, M. M., Gartung, A., Sulciner, M. L., Norris, P. C., Sukhatme, V. P. et al. (2019). Aspirin-triggered proresolving mediators stimulate resolution in cancer. Proceedings of the National Academy of Sciences, 116(13), 6292–6297. DOI 10.1073/pnas.1804000116. [Google Scholar] [CrossRef]
32. Clarke, C. A., Canchola, A. J., Moy, L. M., Neuhausen, S. L., Chung, N. T. et al. (2017). Regular and low-dose aspirin, other non-steroidal anti-inflammatory medications and prospective risk of HER2-defined breast cancer: The california teachers study. Breast Cancer Research, 19(1), 52. DOI 10.1186/s13058-017-0840-7. [Google Scholar] [CrossRef]
33. Bardia, A., Olson, J. E., Vachon, C. M., Lazovich, D., Vierkant, R. A. et al. (2011). Effect of aspirin and other NSAIDs on postmenopausal breast cancer incidence by hormone receptor status: Results from a prospective cohort study. Breast Cancer Research and Treatment, 126(1), 149–155. DOI 10.1007/s10549-010-1074-x. [Google Scholar] [CrossRef]
34. Zhao, M., Wang, T., Hui, Z. (2020). Aspirin overcomes cisplatin resistance in lung cancer by inhibiting cancer cell stemness. Thoracic Cancer, 11(11), 3117–3125. DOI 10.1111/1759-7714.13619. [Google Scholar] [CrossRef]
35. Guo, J., Zhu, Y., Yu, L., Li, Y., Guo, J. et al. (2021). Aspirin inhibits tumor progression and enhances cisplatin sensitivity in epithelial ovarian cancer. PeerJ, 9, e11591. DOI 10.7717/peerj.11591. [Google Scholar] [CrossRef]
36. Zhang, Y., Liu, L., Fan, P., Bauer, N., Gladkich, J. et al. (2015). Aspirin counteracts cancer stem cell features, desmoplasia and gemcitabine resistance in pancreatic cancer. Oncotarget, 6(12), 9999–10015. DOI 10.18632/oncotarget.3171. [Google Scholar] [CrossRef]
37. Aslam, B., Basit, M., Nisar, M. A., Khurshid, M., Rasool, M. H. (2017). Proteomics: Technologies and their applications. Journal of Chromatographic Science, 55(2), 182–196. DOI 10.1093/chromsci/bmw167. [Google Scholar] [CrossRef]
38. Nusinow, D. P., Szpyt, J., Ghandi, M., Rose, C. M., McDonald, E. R. et al. (2020). Quantitative proteomics of the cancer cell line encyclopedia. Cell, 180(2), 387–402.e16. DOI 10.1016/j.cell.2019.12.023. [Google Scholar] [CrossRef]
39. Constantinou, C., Papadopoulos, S., Karyda, E., Alexopoulos, A., Agnanti, N. et al. (2018). Expression and clinical significance of claudin-7, PDL-1, PTEN, c-kit, c-met, MYC, ALK, CK5/6, CK17, p53, EGFR, ki67, p63 in triple-negative breast cancer-A single centre prospective observational study. In Vivo, 32(2), 303–311. [Google Scholar]
40. Sengupta, S., Biarnes, M. C., Jordan, V. C. (2014). Cyclin dependent kinase-9 mediated transcriptional de-regulation of cMYC as a critical determinant of endocrine-therapy resistance in breast cancers. Breast Cancer Research and Treatment, 143(1), 113–124. DOI 10.1007/s10549-013-2789-2. [Google Scholar] [CrossRef]
41. Green, A. R., Aleskandarany, M. A., Agarwal, D., Elsheikh, S., Nolan, C. C. et al. (2016). MYC functions are specific in biological subtypes of breast cancer and confers resistance to endocrine therapy in luminal tumours. British Journal of Cancer, 114(8), 917–928. [Google Scholar]
42. Ai, G., Dachineni, R., Muley, P., Tummala, H., Bhat, G. J. (2016). Aspirin and salicylic acid decrease MYC expression in cancer cells: A potential role in chemoprevention. Tumour Biology, 37(2), 1727–1738. [Google Scholar]
43. Mitrugno, A., Sylman, J. L., Ngo, A. T., Pang, J., Sears, R. C. et al. (2017). Aspirin therapy reduces the ability of platelets to promote colon and pancreatic cancer cell proliferation: Implications for the oncoprotein MYC. American Journal of Physiology. Cell Physiology, 312(2), C176–C189. [Google Scholar]
44. Qiu, F., Chen, Y. R., Liu, X., Chu, C. Y., Shen, L. J. et al. (2014). Arginine starvation impairs mitochondrial respiratory function in ASS1-deficient breast cancer cells. Science Signaling, 7(319), ra31. [Google Scholar]
45. Li, J. M., Yang, D. C., Oldham, J., Linderholm, A., Zhang, J. et al. (2021). Therapeutic targeting of argininosuccinate synthase 1 (ASS1)-deficient pulmonary fibrosis. Molecular Therapy, 29(4), 1487–1500. [Google Scholar]
46. Trott, J. F., Hwang, V. J., Ishimaru, T., Chmiel, K. J., Zhou, J. X. et al. (2018). Arginine reprogramming in ADPKD results in arginine-dependent cystogenesis. American Journal of Physiology. Renal Physiology, 315(6), F1855–F1868. [Google Scholar]
47. Patil, M. D., Bhaumik, J., Babykutty, S., Banerjee, U. C., Fukumura, D. (2016). Arginine dependence of tumor cells: Targeting a chink in cancer’s armor. Oncogene, 35(38), 4957–4972. [Google Scholar]
48. Yang, S. D., Ahn, S. H., Kim, J. I. (2018). 3-Oxoacid CoA transferase 1 as a therapeutic target gene for cisplatin-resistant ovarian cancer. Oncology Letters, 15(2), 2611–2618. [Google Scholar]
Cite This Article
 Copyright © 2022 The Author(s). Published by Tech Science Press.
Copyright © 2022 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.

 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools