 Open Access
Open Access
ARTICLE
Heparanase/Syndecan-1 Axis Regulates the Grade of Liver Cancer and Proliferative Ability of Hepatocellular Carcinoma Cells
Institute of Molecular Medicine, Medical College of Eastern Liaoning University, Dandong, 118003, China
* Corresponding Authors: Yu Jiang. Email: ; Lijuan Lin. Email:
Oncologie 2022, 24(3), 539-551. https://doi.org/10.32604/oncologie.2022.024882
Received 12 June 2022; Accepted 10 August 2022; Issue published 19 September 2022
Abstract
Background: Although heparanase/syndecan-1 axis is involved in malignant progression of many cancers, its significance in liver cancer is not well understood. In this study, we explored the value of heparanase/syndecan-1 axis expression in liver cancer and the intervention mechanisms that target this axis by inhibiting the proliferation of hepatocellular carcinoma cells. Materials and Methods: We conducted tissue microarray analysis that included 90 primary liver cancer and their corresponding adjacent samples to evaluate the expression of heparanase and syndecan-1 and their correlation with clinicopathologic characteristics. RNA interference and western blot assays were performed to analyze the effect of heparanase on syndecan-1 expression in human hepatocellular carcinoma cells MHCC97-H. Cell proliferative capability, Erk and Akt signaling molecules were respectively detected with CCK- 8, cell colony formation and western blot assays after the treatment of low molecular weight heparin (LMWH) and syndecan-1 monoclonal antibody. Results: Heparanase showed a high positive rates (75.56%) in liver cancer patients. The cellular positivity for membrane-located syndecan-1 was less in liver cancer (36.67%) compared with adjacent tissue (57.78%, P < 0.05). Heparanase expression was positively correlated with tumor grade, stages, and Ki- 67 expression (P < 0.05). In contradistinction, both cytoplasmic and membranal syndecan-1 expression exhibited negative correlations with tumor grade, stages, and Ki-67 expression (P < 0.05). Transfection with a heparanase small-interfering RNA resulted in a significant rise in membrane syndecan-1 expression but a marked decline in shed syndecan-1. LMWH and syndecan-1 monoclonal antibody notably inhibited cellular proliferation. Further studies revealed that LMWH and syndecan-1 monoclonal antibody significantly down-regulated Erk and Akt phosphorylation. Conclusions: Heparanase and syndecan-1 present a contrary correlation with the malignant capability of liver cancer, but work together to facilitate the proliferation of hepatocellular carcinoma cells that can be attenuated by LMWH and syndecan-1 monoclonal antibody by inhibiting Erk and Akt signals.Keywords
Liver cancer constitutes one of the most common malignant tumors and comprises the third- and second-most common causes of malignancy-related death in Asia and the rest of the world, respectively [1]. In addition, the aberrant proliferation of tumor cells remains the critical regulatory step in the multistep process of tumorigenesis and development. Therefore, exploring the expression patterns of the tumor proliferation-related molecules and interventional targets involved may provide a novel strategy for slowing cancer procession and even curing cancer ultimately.
Syndecan-1 (SDC-1) is a member of the heparan sulfate proteoglycan (HSPG) family and is known to be an adhesion molecule expressed on cell membranes. There is increasing evidence that SDC-1 regulates the interaction of various signaling molecules and their receptors in tumor cell growth, apoptosis, adhesion, invasion, and angiogenesis [2,3]. SDC-1 is also involved in the formation of the Wnt signalosome and drives aberrant Wnt/β-catenin signaling and cellular growth in multiple myeloma [4]. Alan et al. reported that SDC-1 was necessary in organizing a ternary receptor complex composed of SDC-1, insulin-like growth factor-1 receptor, and integrin αVβ3 during vascular endothelium-cadherin-dependent endothelial cell invasion [5].
Heparanase (HPSE) is an endo-β-D-glucuronidase, with numerous analyses revealing high expression of HPSE RNA and protein in cancers including myeloma, bladder, colon, breast, and liver [6]. It has also been confirmed that HPSE promotes tumorigenesis and tumor development by regulating tumor growth, apoptosis, cellular invasion, and angiogenesis [7,8]. Recent studies have suggested that HPSE and SDC-1 coordinate with one another to portray critical roles in pathologic events such as inflammation, hypoplasia, and tumor progression [9,10]. Investigators have additionally demonstrated augmented serum levels of HPSE and SDC-1 in patients undergoing open or endovascular thoracoabdominal aortic aneurysm repair, and that these proteins also induced systemic inflammatory responses in mice [11]. Kero et al. [12] found that SDC-1 and its interactors (including HPSE) were involved in regulating the epithelial-mesenchymal interaction in human odontogenesis, and Cassinelli et al. [13] posited that the HPSE/HSPG axis may constitute a potential target for sarcoma treatment. However, the significance of HPSE/SDC-1 axis expression and its effect on liver cancer progression are poorly understood.
In this study, we confirmed that cytoplasm-localized HPSE was positively correlated with tumor grade, stages, and Ki-67 expression, but membrane-localized SDC-1 was negatively correlated with tumor grade, stages, and Ki-67 expression in liver cancer patients. Additionally, HPSE-induced shedding of membrane SDC-1 facilitated the proliferation of the human hepatocellular carcinoma cell (HCC) line MHCC97-H, which was subsequently attenuated by SDC-1 monoclonal antibody and low molecular weight heparin (an inhibitor of HPSE activity) acting via the inhibition of Erk and Akt signals. Therefore, the present study will provide a therapeutic strategy by targeting the HPSE/SDC-1 axis so as to intervene in the progression of liver cancer.
2.1 Tissue Microarray Analysis and Immunohistochemical Staining
The tissue microarray (TMA) slides were purchased from Shanghai Outdo Biotech Company (China) (no. JSW-11-01, Shanghai, China) and consisted of 90 liver cancer samples and corresponding adjacent samples. Surgical resections were performed between January 2007 and November 2009 for all patients, and tumors were staged according to the 7th Edition of the American Joint Committee on Cancer. Of the 90 liver cancer tissue samples, 38 were determined to be early stage (I/II) and 52 as late stage (III/IV). We then conducted immunohistochemical (IHC) staining to ascertain HPSE and SDC-1 protein expression. Briefly, TMA slides were treated in citrate buffer (pH 6.0) for 30 min at room temperature and incubated with anti-HPSE (no. sc-293205, Santa Cruz, CA, USA) or anti-SDC-1 (no. ab128936, Abcam, Cambridge, UK) antibody overnight at 4°C, followed by incubation with HRP-labeled secondary antibody (no. KGAA37, no. KGAA35, KeyGen Biotech, Nanjing, China) for 30 min at room temperature. The immunostaining for HPSE or SDC-1 was semi-quantitatively scored using “−”, “+”, “++” and “+++” to indicate the percentages of less than 5%, 5%–25%, 26%–50%, and greater than 50% of positive cells.
Experiments were approved by the Ethics Committee of Shanghai Outdo Biotech Company (China) (no. YBM-05-02), and were conducted in compliance with the Declaration of Helsinki. Each patient signed her/his consent form.
Human HCCs (MHCC97-H) possessing a high metastatic potential were purchased from Cell Biological Institute (no. BFN608006110, Shanghai, China). The cells were grown in Dulbecco’s modified Eagle’s medium containing 4.5 g/L of glucose (Gibco, USA) supplemented with 10% heat-inactivated fetal bovine serum (Gibco), and incubated at 37°C in a humidified atmosphere of 5% CO2 in compressed air.
2.4 Transfection of Small-Interfering RNAs (siRNAs)
The siRNAs for HPSE (siHPSE) (sc-40685) and the negative control (siNC) were purchased from Santa Cruz Biotech Inc. (USA) and transfected into HCCs according to the manufacturer’s recommendation using Lipofectamine™ 2000 (Invitrogen).
2.5 Cell Treatment of Low Molecular Weight Heparin (LMWH)
The HCCs were treated with LMWH (20 IU/mL or 40 IU/mL, Pharmion) for 24 h, and the untreated cells were used as a negative control.
Real-time PCR was performed with SYBR Premix DimerEraser kit (TaKaRa, Dalian, China) using an ABI PRISM 7900 detection system. We performed the cDNA synthesis using a PrimeScript RT Reagent kit (TaKaRa, Dalian, China) according to the manufacturer’s instructions. Specific primers for SDC-1 and β-actin were purchased from GenePharma (Shanghai, China), with sequences as follows: SDC-1 forward, 5′-CTGGAGGCTACAGCTGCCTC-3′ and reverse, 5′-CTGCTGCTGGGAGCTGACTG-3′; and β-actin forward, 5′-CATGTACGTTGCTATCCAGG-3′ and reverse, 5′-GCTCATTGCCAATGGTGATG-3′. Relative changes in mRNA expression were normalized to β-actin and calculated using the 2−ΔΔCt method.
Protein was extracted from total cells, cellular membranes, cytoplasmic compartments, and nuclei with a protein extraction kit (Sigma-Aldrich Corp., St. Louis, MO, USA), and quantified using a BCA assay kit (Pierce, Rockford, IL, USA). Equal amounts of denatured proteins were subsequently subjected to 10% SDS-PAGE and blotted onto PVDF membranes (Pall Corporation, Port 5 Washington, NY, USA). We used antibodies generated against HPSE (no. sc-293205, Santa Cruz, CA, USA), SDC-1 (no. ab128936, Abcam, Cambridge, UK), Erk1/2 (no. BS65610, Bioworld Technology, Inc., Louis Park, MN, USA), p-Erk1/2 (no. BS94044, Bioworld Technology, Inc., Louis Park, MN, USA), Akt (no. 4691, Cell Signaling Technology, Inc., Danvers, MA, USA), p-Akt (no. 4060, Cell Signaling Technology, Inc., Danvers, MA, USA), and β-actin (no. sc-81178, Santa Cruz, CA, USA) as primary antibodies. Antibody detection was implemented using an ECL kit (Amersham Biosciences Corp., Piscataway, New Jersey, USA) according to the manufacturer’s instructions.
After the indicated treatments, cell culture supernatants were collected and then evaluated using an ELISA kit for SDC-1 (Diaclone, Besancon Cedex, France) according to the manufacturer’s instructions.
2.9 Immunodepletion (IP) Treatment
MHCC97-H cells were incubated with SDC-1 monoclonal antibody (2 μg/mL, no. ab128936, Abcam, Cambridge, UK) for 8 h to deplete the SDC-1 from the cell culture supernatants, and we adopted rabbit IgG (2 μg/mL, no. ab172730, Abcam, Cambridge, UK) as the isotype control.
2.10 Cell Colony Formation Assay
Cells that underwent different treatments were seeded into six-well plates (Corning, NY, USA) at a density of 200 cells per well and incubated at 37°C in 5% CO2 for 14 days. After three washes with PBS, cells were fixed with 4% paraformaldehyde for 15 min and stained with 0.1% crystal violet for 30 min. The clones were imaged, and the number of clones >50 cells was counted using a light optical microscope.
Cells that underwent different treatments were seeded into 96-well plates (Corning, NY, USA) and measured using CCK-8 assay kits (Cell Signaling Technology, USA) following the manufacturer’s protocol.
We applied SPSS 17.0 software for statistical analysis. A one-way analysis of variance with the post hoc Tukey’s test and Student’s t-test were adopted for the comparisons of continuous variables that involved more than two groups and two groups, respectively. We executed the Chi-squared test to compare categorical variables. Statistical significance was defined as P < 0.05.
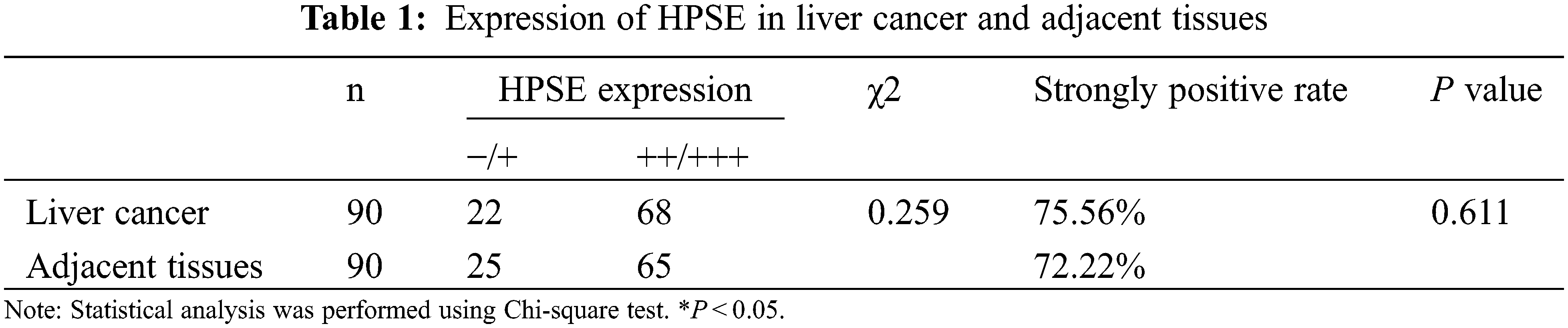
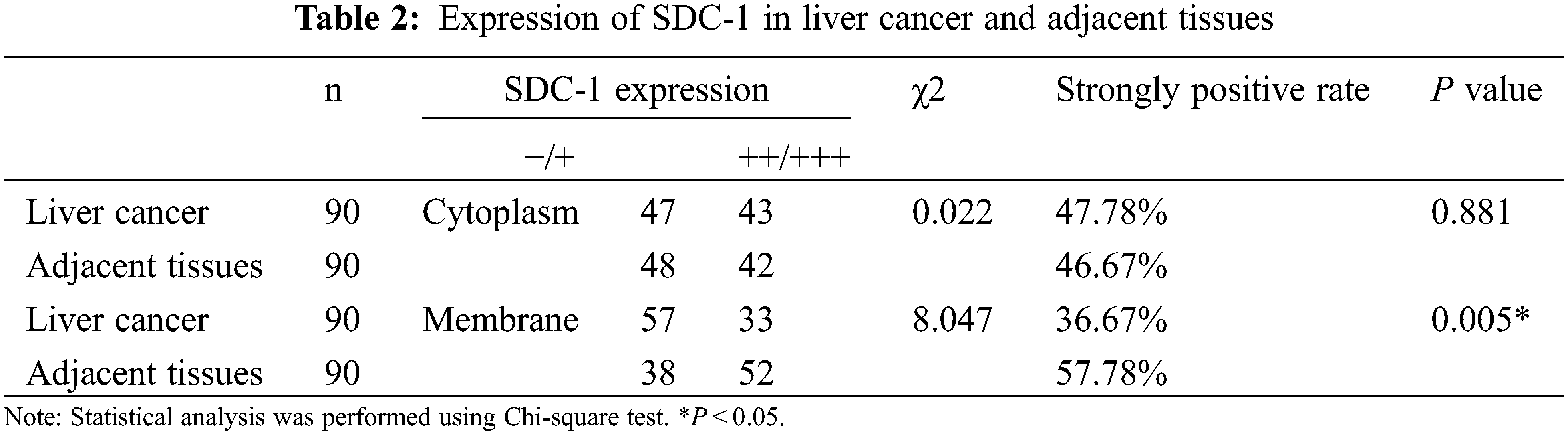
3.1 The Expression Analyses of HPSE and SDC-1 in the Human Liver Cancer Tissue Microarray
IHC staining was executed to analyze HPSE (Fig. 1A) and SDC-1 (Fig. 1B) protein expression in the TMA that encompassed 90 liver cancer samples and corresponding adjacent samples. HPSE protein showed robust cytoplasmic staining but displayed no significant differences in staining intensities (Figs. 1C and 1D) and strongly positive rate (Table 1) between liver cancer and corresponding adjacent tissues. Positive SDC-1 expression reflected cytoplasmic and membranal staining in liver cancer (Fig. 1E) and corresponding adjacent tissue (Fig. 1F); and while cytoplasm-localized SDC-1 manifested no significant differences, membrane-localized SDC-1 was markedly less in liver cancer than in the corresponding adjacent tissue. Though the strongly positive rate of cytoplasm-localized SDC-1 protein was not different from the corresponding adjacent tissues, that of membrane-localized SDC-1 protein was notably reduced in liver cancer (36.67% vs. 57.78%, respectively; P < 0.05) (Table 2). These data suggested that membrane SDC-1 constituted a target of HPSE due to the high expression of HPSE and the reduced localization of membrane SDC-1 in liver cancer.
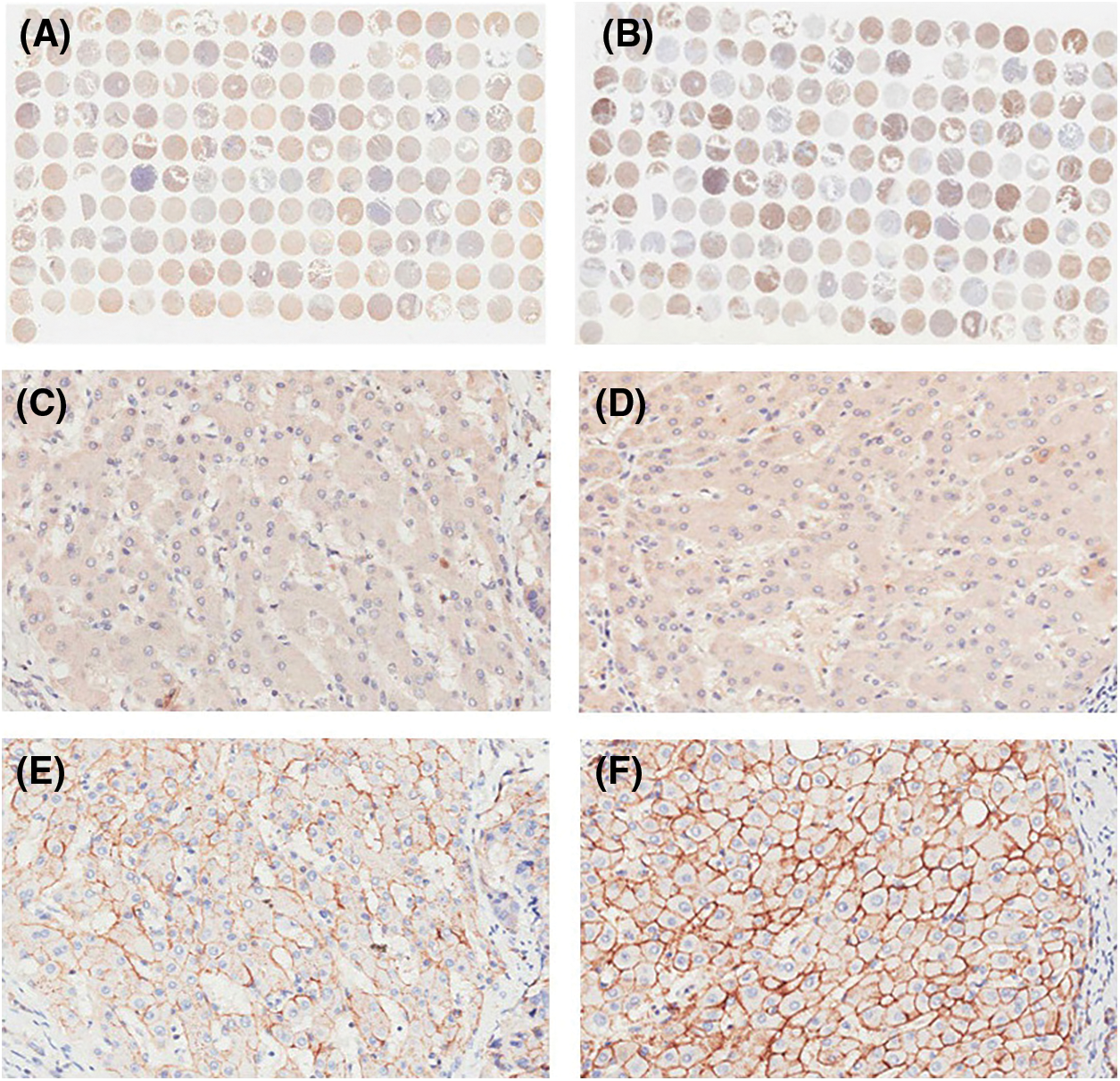
Figure 1: IHC staining for HPSE and SDC-1 in TMA. Overview of HPSE (A) and SDC-1 (B) staining. Positive HPSE staining was not significantly different between liver cancer (C) and corresponding adjacent tissues (D). Membrane SDC-1 staining was weaker in liver cancer (E) than in corresponding adjacent tissues (F), while cytoplasmic SDC-1 staining was not altered (original magnification, ×200)
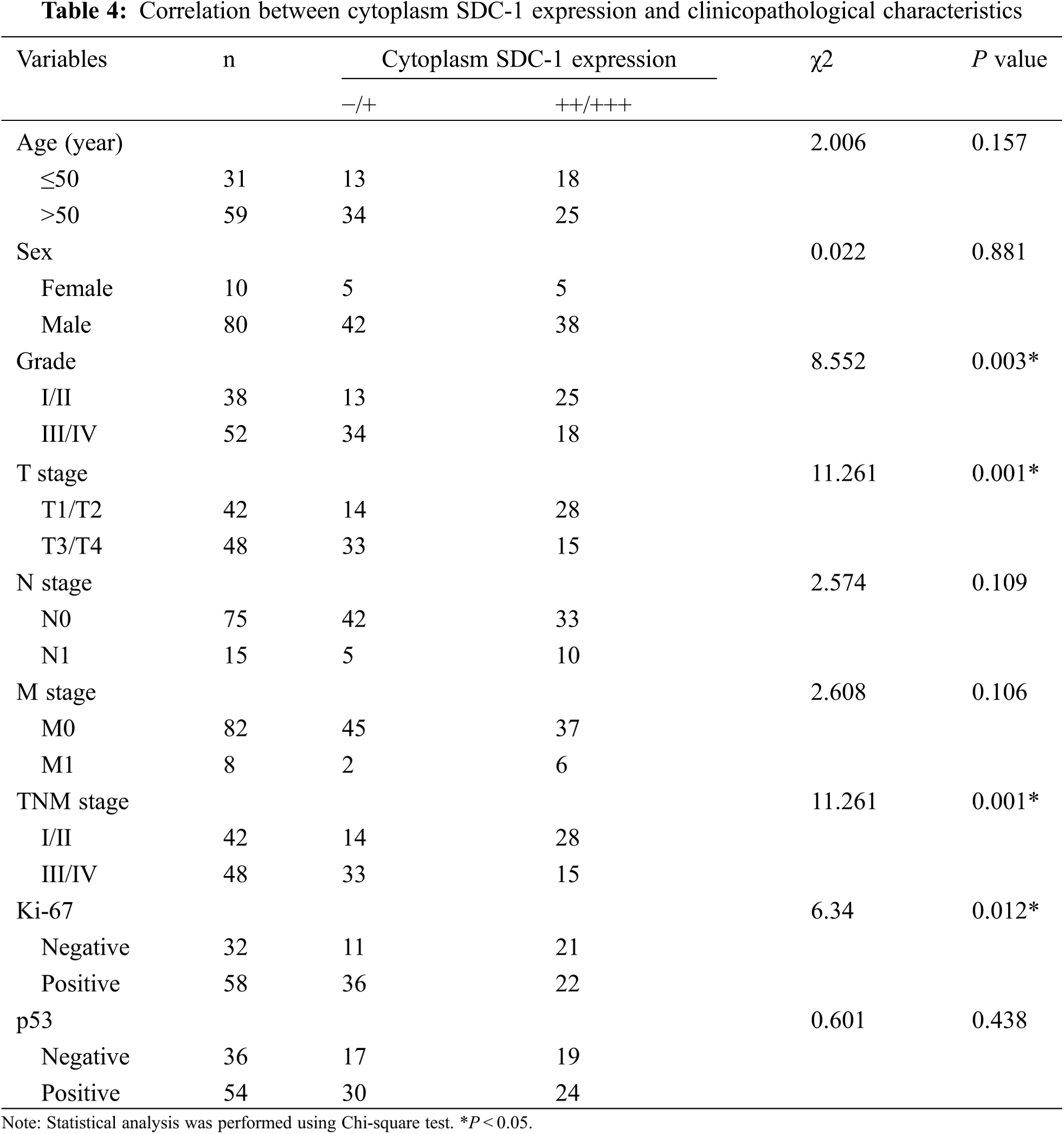
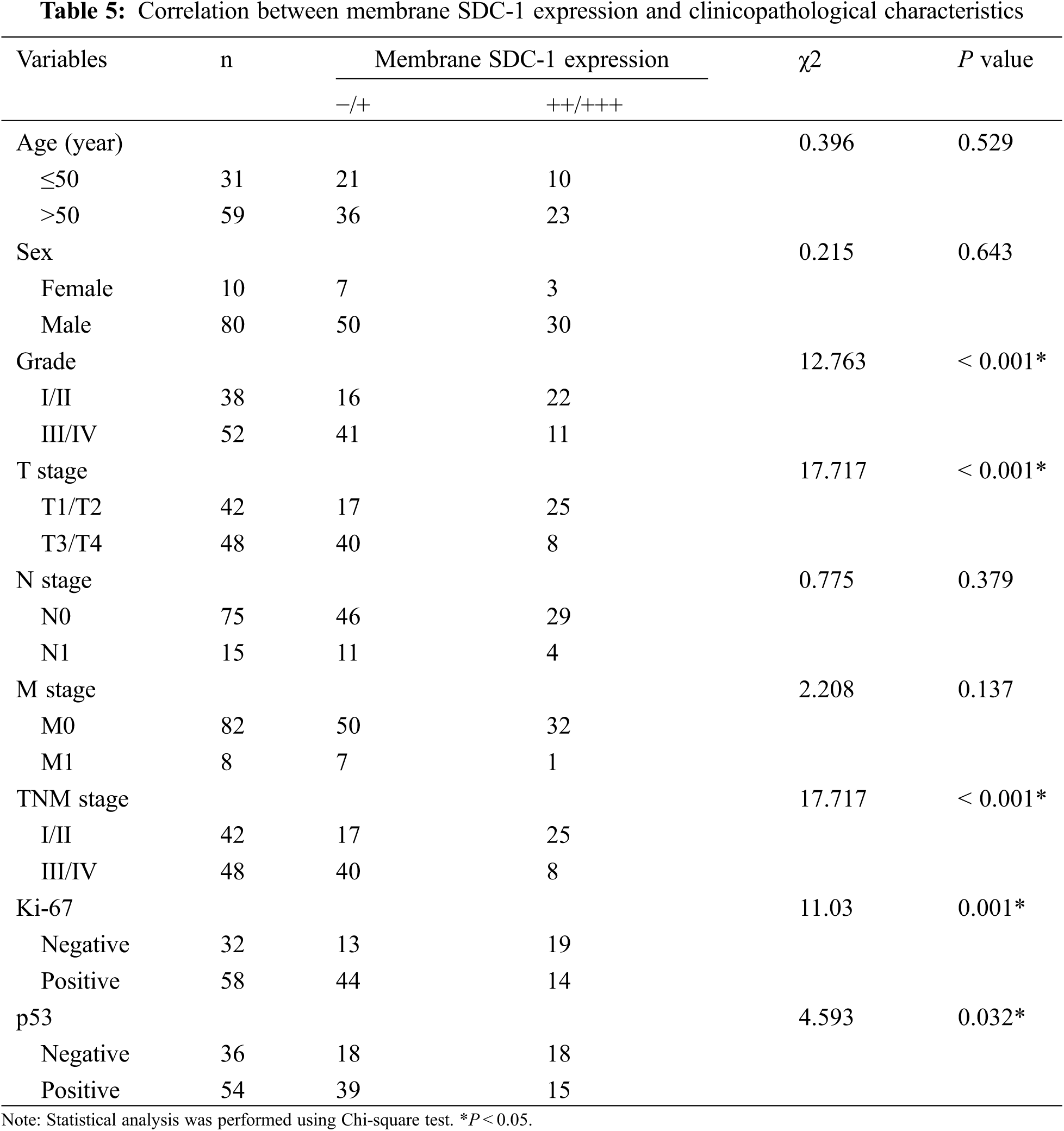
3.2 The Correlation between HPSE or SDC-1 Expression and Clinicopathologic Characteristics
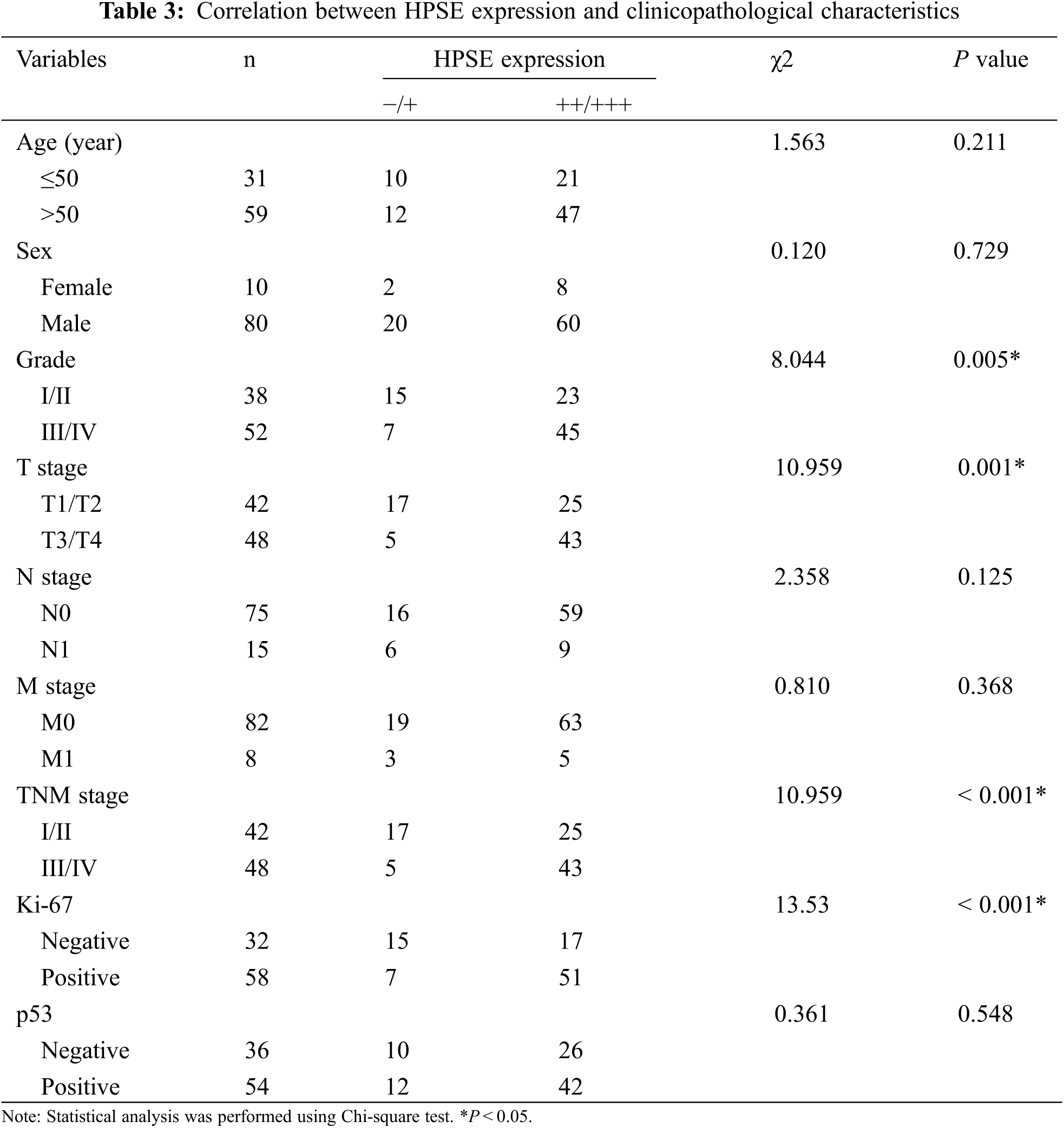
To evaluate the role of heparanase/syndecan-1 axis in the malignant progression of liver cancer, we analyzed the correlation between HPSE or SDC-1 protein and major clinicopathologic characteristics of liver cancer. Our results showed that HPSE expression was positively correlated with tumor grade, stages, and Ki-67 expression (P < 0.05) (Table 3); that cytoplasmic SDC-1 expression revealed a negative association with tumor grade, stages, and Ki-67 expression (P < 0.05) (Table 4); and that membrane SDC-1 expression was negatively related to tumor grade, stages, and the expression of p53 and Ki-67 (P< 0.05) (Table 5). These data indicated that the expression of HPSE and membrane SDC-1 presented a contrary correlation with clinicopathologic characteristics in liver cancer patients.
3.3 HPSE Induces the Shedding of SDC-1 from MHCC97-H Cells
To investigate the regulatory pattern of heparanase/syndecan-1 axis expression in HCCs, a HPSE small-interfering RNA (siHPSE) and LMWH (an inhibitor of HPSE activity) were applied to disrupt the roles of HPSE in the human HCC line MHCC97-H. The transfection of siHPSE considerably inhibited the protein expression of HPSE compared with that of the negative control small-interfering RNA, and LMWH exerted no effect on HPSE expression (Fig. 2A). The levels of SDC-1 mRNA were not different between the siHPSE-transfected and negative control-transfected cells (Fig. 2B). However, treatment with siHPSE or LMWH significantly up-regulated the protein levels of total and membrane SDC-1 without an effect on cytoplasmic SDC-1 compared with the treatment with negative control small-interfering RNA or untreated MHCC97-H cells (Fig. 2C). We further measured the levels of SDC-1 in the medium aspirated from different HCC incubates using ELISA, and noted that the shedding levels of SDC-1 were notably diminished in siHPSE-transfected and LMWH-treated cells relative to levels from negative control-transfected or untreated cells (Fig. 2D). These results suggested that HPSE drove the shedding of SDC-1 from MHCC97-H cells.
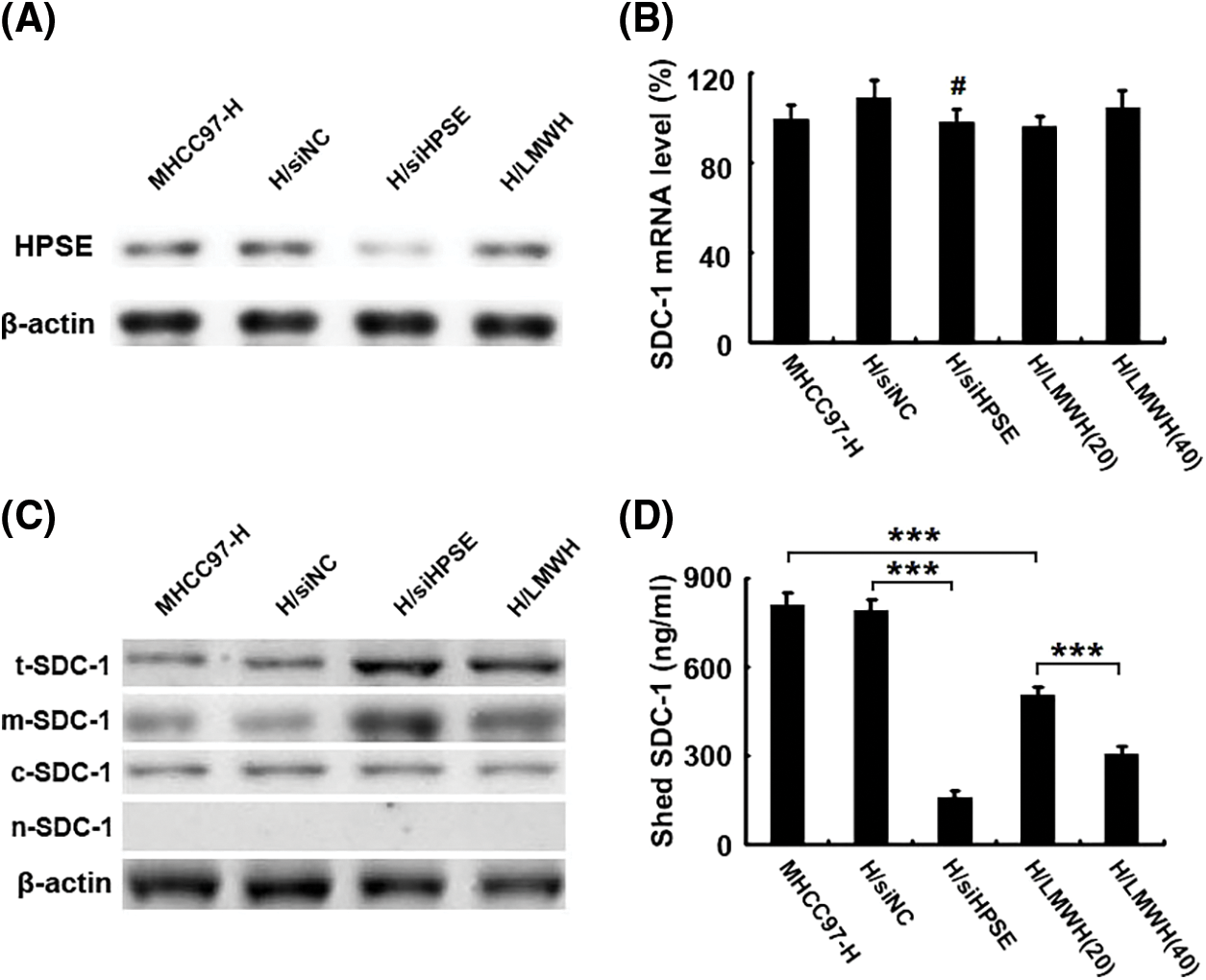
Figure 2: HPSE induces the shedding of SDC-1 from MHCC97-H cells. (A) Western blot results showed that the transfection of a HPSE small-interfering RNA into MHCC97-H cells (H/siHPSE) markedly inhibited HPSE protein expression compared with that of the negative control small-interfering RNA (H/siNC). However, the treatment with LMWH (H/LMWH) did not exert an effect on HPSE expression (β-actin blotting served as a control). (B) Real-time PCR results displayed levels of SDC-1 mRNA that were not significantly different between the H/siHPSE-and H/siNC-transfected cells. (C) Western blot results showed that treatment with a small-interfering RNA for HPSE (H/siHPSE) and LMWH (H/LMWH) significantly up-regulated the protein levels of total and membrane SDC-1 (i.e., t-SDC-1 and m-SDC-1). However, they did not alter cytoplasmic SDC-1 (c-SDC-1). There was no detection signal in the nucleus SDC-1 (i.e., n-SDC-1) (β-actin blotting served as a control). (D) As shown in ELISA results, the shed levels of SDC-1 were notably reduced in the medium aspirated from H/siHPSE cells and 20 IU/mL or 40 IU/mL LMWH treated cells compared to levels from H/siNC cells or those from untreated MHCC97-H cells (***P < 0.001, #P > 0.05)
3.4 HPSE-Induced Shedding of SDC-1 Facilitates the Proliferation of MHCC97-H Cells, Which Can be Attenuated by Treatment with LMWH and SDC-1 Monoclonal Antibody by Inhibiting Erk and Akt Signals
To investigate the role of HPSE-induced shedding of SDC-1 in the proliferation of HCCs and its underlying interventional mechanisms, we used a monoclonal antibody to SDC-1 to deplete soluble SDC-1 from the medium in which the MHCC97-H cells were incubated; and colony formation (Fig. 3A) and CCK-8 (Fig. 3B) assays demonstrated that cellular growth was obviously inhibited after immunodepletion of soluble SDC-1 compared with the isotype control IgG, and that treatment with siHPSE and LMWH also resulted in significant declines in the proliferative capability of the HCCs relative to treatment with negative control or to untreated cells. To further explore the anti-proliferative mechanisms underlying the actions of the SDC-1 monoclonal antibody and LMWH, we determined the activational levels of Erk and Akt signaling molecules by western blot analysis. As shown in Fig. 3C, SDC-1 immunodepletion and LMWH significantly down-regulated the levels of Erk and Akt phosphorylation. These data suggested that LMWH and SDC-1 monoclonal antibody effectively prevented HPSE/SDC-1 axis-mediated proliferation of MHCC97-H cells via inhibition of Erk and Akt signals.
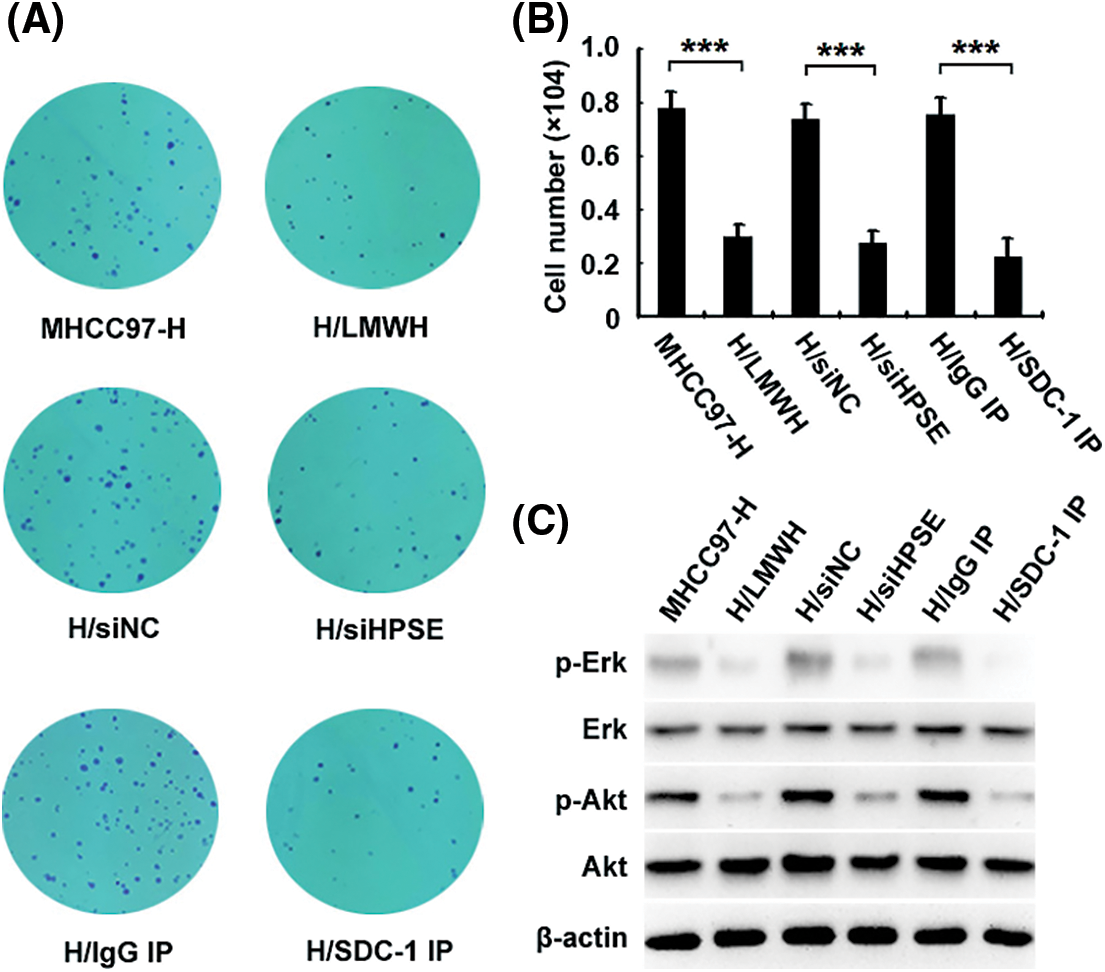
Figure 3: HPSE-induced shedding of SDC-1 facilitates the proliferation of MHCC97-H cells, which can be attenuated by LMWH and SDC-1 monoclonal antibody via inhibition of Erk and Akt signals. Colony formation (A) and CCK-8 (B) assays revealed that the growth of H/siHPSE-transfected, 40 IU/mL LMWH-treated (H/LMWH), and SDC-1 monoclonal antibody-immunodepleted (H/SDC-1 IP) cells was notably inhibited relative to the cells transfected with H/siNC, untreated, or that were IgG immunodepleted (H/IgG IP). (C) Western blot results showed that SDC-1 immunodepletion and LMWH significantly down-regulated the levels of phosphorylated Erk and Akt (***P < 0.001)
SDC-1 is an important transmembrane proteoglycan that participates in regulating cellular behaviors such as adhesion, differentiation, and proliferation [2]. Several studies have indicated that HPSE and SDC-1 work together to mediate pathologic progression that includes that of cancer [9]. Duplancic et al. reported that the expression of SDC-1 and HPSE was correlated with the presence of inflammatory infiltrate in diseased periodontal tissue [14]. The up-regulation of HPSE also promoted infection with the herpes simplex virus by acting on SDC-1 [15], and HPSE-induced shedding of SDC-1 activated the VEGFR2-dependent invasion of myeloma and endothelial cells [16]. Both HPSE and SDC-1 show positive expression in liver cancer tissue [17,18] and suggest a relationship between them. However, any significant correlations with clinicopathologic characteristics in liver cancer patients are still poorly elucidated. In the present study, tissue microarray data revealed that cytoplasm-localized HPSE protein was highly expressed, with cell positivity rates of 75.56% in liver cancer patients; and that it was positively correlated with tumor grade, stages, and Ki-67 expression. However, protein expression of membrane-localized SDC-1 was attenuated in liver cancer compared with the corresponding adjacent tissue, and the cellular positivity rate for membrane-localized SDC-1 was 36.67% in liver cancer significantly lower than that observed for the corresponding adjacent tissue. Furthermore, membrane-localized SDC-1 reflected a negative correlation with tumor grade, stages, and Ki-67 expression. We speculate that membrane SDC-1 may be a target of HPSE according to its expression features and divergent correlations with clinicopathologic characteristics in liver cancer.
Metwaly et al. [19] found that the higher levels of shed SDC-1 detected in the serum of liver cancer patients were accompanied by a worse prognosis. Our previous work demonstrated that HPSE resulted in the dissociation of SDC-1/VEGF-C complex in the mouse HCC line Hca-F and drove the proliferation of lymphatic endothelial cells [20]. Yasuo et al. [18] even predicted that SDC-1 might be a diagnostic marker because of the increase in the levels of shed SDC-1 in the serum of patients with liver cancer. Regardless, discerning whether the shedding of SDC-1 is related to the high expression of HPSE in liver cancer deserves additional intensive study. To further investigate the regulatory patterns associated with HPSE and SDC-1 in liver cancer, we employed an HPSE small-interfering RNA (siHPSE) and LMWH (an inhibitor of HPSE activity) to evaluate the effect of HPSE on the expression of SDC-1 in the HCC line MHCC97-H. We noted that siHPSE notably down-regulated HPSE protein expression but did not change SDC-1 mRNA expression. Importantly, intervention in HPSE by siHPSE and LMWH resulted in a significant elevation in the levels of membrane-localized SDC-1 and a remarkable decline in the levels of soluble SDC-1 in cellular incubates. This suggested that HPSE induced the shedding of SDC-1 from MHCC97-H cells.
Due to the generally critical roles of HPSE and HSPGs in tumor progression, more attention has been allocated to both clinical and basic research on anti-tumor correlates of the HPSE/HSPG axis. The heparan sulfate mimetic PI-88 exhibited anti-metastatic activity through inhibition of HPSE and heparan sulfate-dependent growth factors, and was recently included in phase III clinical evaluation for liver cancer therapy [21–23]. Multiple authors have confirmed that blocking the collaborative effect between HPSE and SDC-1 may comprise a novel strategy for treating multiple myeloma and pancreatic cancer [24,25]. Several reports indicated that the SDC-1-mimetic peptide synstatins prevented HPSE/SDC-1 axis-mediated tumor invasion, tumorigenesis, and angiogenesis; and activated shedding SDC-1-suppressed tumor cell immune [16,26,27]. We herein found that immunodepletion of soluble SDC-1 from the medium of incubated MHCC97-H cells using SDC-1 monoclonal antibody significantly inhibited cellular proliferation, and that LMWH achieved the same blocking effect. Our further studies then revealed that SDC-1 immunodepletion and LMWH markedly down-regulated the levels of phosphorylated Erk and Akt proteins. Rangarajan et al. demonstrated that heparanase is able to translocate to the nucleus where it regulates the expression of multiple genes such as vascular endothelial growth factor (VEGF) and hepatocyte growth factor (HGF) [28]. Other evidence has indicated that mitochondria promoted the survival of tumor cells in tumor microenvironment, and facilitated immune evasion, progression, and treatment resistance [29]. In addition, heparanase also increased the resistance to stress-induced apoptosis of glioma cells by maintaining mitochondrial membrane potential [30]. Although we herein used only one HCC line (MHCC97-H), we expect to further explore the effects of the HPSE/SDC-1 axis on malignant behaviors and to ascertain the underlying mechanisms involved by employing other types of human HCCs.
In summary, HPSE and SDC-1, particularly membrane SDC-1, occupy important roles in the progression of liver cancer. However, their protein expression in the present study depicted a divergent correlation with clinicopathologic characteristics in liver cancer patients. HPSE drove the shedding of SDC-1 from human HCCs that facilitated cellular proliferation, and the HPSE/SDC-1 axis-induced proliferation was then prevented by LMWH and SDC-1 monoclonal antibody through the inhibition of Erk and Akt signals. This study provides new insights into the significance of HPSE and SDC-1 expression in liver cancer, and into the regulatory patterns involving HPSE/SDC-1 axis expression and its underlying interventional mechanisms in the malignant progression of human HCCs.
Authorship: The authors confirm contribution to the paper as follows: study conception and design: Shengjin Yu, Yu Jiang, Lijuan Lin; data collection: Shengjin Yu, Huiming Lv, Jinhui Zhang; analysis and interpretation of results: Shengjin Yu, He Zhang, Weiwei Ju; draft manuscript preparation: Shengjin Yu. All authors reviewed the results and approved the final version of the manuscript.
Ethics Approval and Informed Consent Statement: This study was approved by the Ethics Committee of Shanghai Outdo Biotech Company (No. YBM-05-02) and was conducted in compliance with the Helsinki Declaration. Each patient signed the consent form.
Availability of Data and Materials: The data and materials are available from the corresponding author upon reasonable request.
Funding Statement: This work was supported by grants from the Project of Education Department in Liaoning Province (Nos. LNSJYT201917, LNSJYT201910, LJKZ1133).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. McGlynn, K. A., Petrick, J. L., London, W. T. (2015). Global epidemiology of hepatocellular carcinoma: An emphasis on demographic and regional variability. Clinics in Liver Disease, 19(2), 223–238. DOI 10.1016/j.cld.2015.01.001. [Google Scholar] [CrossRef]
2. Agere, S. A., Kim, E. Y., Akhtar, N., Ahmed, S. (2018). Syndecans in chronic inflammatory and autoimmune diseases: Pathological insights and therapeutic opportunities. Journal of Cellular Physiology, 233(9), 6346–6358. DOI 10.1002/jcp.26388. [Google Scholar] [CrossRef]
3. Gharbaran, R. (2015). Advances in the molecular functions of syndecan-1 (SDC1/CD138) in the pathogenesis of malignancies. Critical Reviews in Oncology/Hematology, 94(1), 1–17. DOI 10.1016/j.critrevonc.2014.12.003. [Google Scholar] [CrossRef]
4. Ren, Z., van Andel, H., de Lau, W., Hartholt, R. B., Maurice, M. M. et al. (2018). Syndecan-1 promotes Wnt/β-catenin signaling in multiple myeloma by presenting Wnts and R-spondins. Blood, 131(9), 982–994. DOI 10.1182/blood-2017-07-797050. [Google Scholar] [CrossRef]
5. Rapraeger, A. C., Ell, B. J., Roy, M., Li, X. H., Morrison, O. R. et al. (2013). VE-cadherin stimulates syndecan-1-coupled IGF1R and cross-talk between αVβ3 integrin and VEGFR2 during the onset of endothelial cell dissemination in angiogenesis. The FEBS Journal, 280(10), 2194–2206. DOI 10.1111/febs.12134. [Google Scholar] [CrossRef]
6. Vlodavsky, I., Gross-Cohen, M., Weissmann, M., Ilan, N., Sanderson, R. D. (2018). Opposing functions of heparanase-1 and heparanase-2 in cancer progression. Trends in Biochemical Sciences, 43(1), 18–31. DOI 10.1016/j.tibs.2017.10.007. [Google Scholar] [CrossRef]
7. Heyman, B., Yang, Y. (2016). Mechanisms of heparanase inhibitors in cancer therapy. Experimental Hematology, 44(11), 1002–1012. DOI 10.1016/j.exphem.2016.08.006. [Google Scholar] [CrossRef]
8. Sanderson, R. D., Elkin, M., Rapraeger, A. C., Ilan, N., Vlodavsky, I. (2017). Heparanase regulation of cancer, autophagy and inflammation: New mechanisms and targets for therapy. The FEBS Journal, 284(1), 42–55. DOI 10.1111/febs.13932. [Google Scholar] [CrossRef]
9. Karamanos, N. K. (2017). Matrix pathobiology-central roles for proteoglycans and heparanase in health and disease. The FEBS Journal, 284(1), 7–9. DOI 10.1111/febs.13945. [Google Scholar] [CrossRef]
10. Ramani, V. C., Purushothaman, A., Stewart, M. D. (2013). The heparanase/syndecan-1 axis in cancer: Mechanisms and therapies. The FEBS Journal, 280(10), 2294–2306. DOI 10.1111/febs.12168. [Google Scholar] [CrossRef]
11. Martin, L., Gombert, A., Chen, J., Liebens, J., Verleger, J. et al. (2017). The β-d-endoglucuronidase heparanase is a danger molecule that drives systemic inflammation and correlates with clinical course after open and endovascular thoracoabdominal aortic aneurysm repair: Lessons learnt from mice and men. Frontiers in Immunology, 8, 681. DOI 10.3389/fimmu.2017.00681. [Google Scholar] [CrossRef]
12. Kero, D., Bilandzija, T. S., Arapovic, L. L., Vukojevic, K., Saraga-Babic, M. (2018). Syndecans and enzymes involved in heparan sulfate biosynthesis and degradation are differentially expressed during human odontogenesis. Frontiers in Physiology, 9, 732. DOI 10.3389/fphys.2018.00732. [Google Scholar] [CrossRef]
13. Cassinelli, G., Zaffaroni, N., Lanzi, C. (2016). The heparanase/heparan sulfate proteoglycan axis: A potential new therapeutic target in sarcomas. Cancer Letters, 382(2), 245–254. DOI 10.1016/j.canlet.2016.09.004. [Google Scholar] [CrossRef]
14. Duplancic, R., Roguljic, M., Puhar, I., Vecek, N., Dragun, R. et al. (2019). Syndecans and enzymes for heparan sulfate biosynthesis and modification differentially correlate with presence of inflammatory infiltrate in periodontitis. Frontiers in Physiology, 10, 1248. DOI 10.3389/fphys.2019.01248. [Google Scholar] [CrossRef]
15. Hadigal, S., Koganti, R., Yadavalli, T., Agelidis, A., Suryawanshi, R. et al. (2020). Heparanase-regulated syndecan-1 shedding facilitates herpes simplex virus 1 egress. Journal of Virology, 94(6), e01672–19. DOI 10.1128/JVI.01672-19. [Google Scholar] [CrossRef]
16. Jung, O., Trapp-Stamborski, V., Purushothaman, A., Jin, H., Wang, H. et al. (2016). Heparanase-induced shedding of syndecan-1/CD138 in myeloma and endothelial cells activates VEGFR2 and an invasive phenotype: Prevention by novel synstatins. Oncogenesis, 5(2), e202. DOI 10.1038/oncsis.2016.5. [Google Scholar] [CrossRef]
17. Dong, S., Wu, X. Z. (2010). Heparanase and hepatocellular carcinoma: Promoter or inhibitor? World Journal of Gastroenterology, 16(3), 306–311. DOI 10.3748/wjg.v16.i3.306. [Google Scholar] [CrossRef]
18. Yasuo, T., Ryosuke, T., Kazuhiko, K. (2018). Proteoglycans are attractive biomarkers and therapeutic targets in hepatocellular carcinoma. International Journal of Molecular Sciences, 19(10), 3070. DOI 10.3390/ijms19103070. [Google Scholar] [CrossRef]
19. Metwaly, H. A., Al-Gayyar, M. M., Eletreby, S., Ebrahim, M. A., El-Shishtawy, M. M. (2012). Relevance of serum levels of interleukin-6 and syndecan-1 in patients with hepatocellular carcinoma. Scientia Pharmaceutica, 80(1), 179–188. DOI 10.3797/(ISSN)0036-8709. [Google Scholar] [CrossRef]
20. Yu, S., Lv, H., Zhang, H., Jiang, Y., Hong, Y. et al. (2017). Heparanase-1-induced shedding of heparan sulfate from syndecan-1 in hepatocarcinoma cell facilitates lymphatic endothelial cell proliferation via VEGF-C/ERK pathway. Biochemical and Biophysical Research Communications, 485(2), 432–439. DOI 10.1016/j.bbrc.2017.02.060. [Google Scholar] [CrossRef]
21. Elli, S., Stancanelli, E., Handley, P. N., Carroll, A., Urso, E. et al. (2018). Structural and conformational studies of the heparin sulfate mimetic PI-88. Glycobiology, 28(10), 731–740. DOI 10.1093/glycob/cwy068. [Google Scholar] [CrossRef]
22. Liu, C. J., Chang, J., Lee, P. H., Lin, D. Y., Wu, C. C. et al. (2014). Adjuvant heparanase inhibitor PI-88 therapy for hepatocellular carcinoma recurrence. World Journal of Gastroenterology, 20(32), 11384–11393. DOI 10.3748/wjg.v20.i32.11384. [Google Scholar] [CrossRef]
23. Ferro, V., Dredge, K., Liu, L., Hammond, E., Bytheway, I. et al. (2007). PI-88 and novel heparan sulfate mimetics inhibit angiogenesis. Seminars in Thrombosis and Hemostasis, 33(5), 557–568. DOI 10.1055/s-2007-982088. [Google Scholar] [CrossRef]
24. Bertrand, J., Bollmann, M. (2019). Soluble syndecans: Biomarkers for diseases and therapeutic options. British Journal of Pharmacology, 176(1), 67–81. DOI 10.1111/bph.14397. [Google Scholar] [CrossRef]
25. Vlodavsky, I., Singh, P., Boyango, I., Gutter-Kapon, L., Elkin, M. et al. (2016). Heparanase: From basic research to therapeutic applications in cancer and inflammation. Drug Resistance Updates, 29, 54–75. DOI 10.1016/j.drup.2016.10.001. [Google Scholar] [CrossRef]
26. Rapraeger, A. C. (2013). Synstatin: A selective inhibitor of the syndecan-1-coupled IGF1R-αvβ3 integrin complex in tumorigenesis and angiogenesis. The FEBS Journal, 280(10), 2207–2215. DOI 10.1111/febs.12160. [Google Scholar] [CrossRef]
27. Jung, O., Beauvais, D. M., Adams, K. M., Rapraeger, A. C. (2019). VLA-4 phosphorylation during tumor and immune cell migration relies on its coupling to VEGFR2 and CXCR4 by syndecan-1. Journal of Cell Science, 132(20), jcs232645. DOI 10.1242/jcs.232645. [Google Scholar] [CrossRef]
28. Rangarajan, S., Richter, J. R., Richter, R. P., Bandari, S. K., Tripathi, K. et al. (2020). Heparanase-enhanced shedding of syndecan-1 and its role in driving disease pathogenesis and progression. Journal of Histochemistry & Cytochemistry, 68(12), 823–840. DOI 10.1369/0022155420937087. [Google Scholar] [CrossRef]
29. Taghizadeh-Hesary, F., Behnam, B., Akbari, H., Bahadori, M. (2022). Targeted anti-mitochondrial therapy: The future of oncology. https://www.preprints.org/manuscript/202201.0171/v3. [Google Scholar]
30. Riaz, A., Ilan, N., Vlodavsky, I., Li, J. P., Johansson, S. (2013). Characterization of heparanase-induced phosphatidylinositol 3-kinase-AKT activation and its integrin dependence. The Journal of Biological Chemistry, 288(17), 12366–12375. DOI 10.1074/jbc.M112.435172. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2022 The Author(s). Published by Tech Science Press.
Copyright © 2022 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.

 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools