 Open Access
Open Access
ARTICLE
Ampelopsin Inhibits Breast Cancer Glucose Metabolism Reprogramming Based on Network Pharmacology and Molecular Docking
1
Hebei Key Laboratory of Chinese Medicine Research on Cardiocerebrovascular Disease, Hebei University of Chinese Medicine,
Shijiazhuang, 050200, China
2
Department of Biochemistry and Molecular Biology, College of Basic Medicine, Hebei University of Chinese Medicine,
Shijiazhuang, 050200, China
3
College of Pharmacy, Hebei University of Chinese Medicine, Shijiazhuang, 050200, China
* Corresponding Author: Yunfeng Li. Email:
# These authors contributed equally to this work
Oncologie 2022, 24(3), 483-498. https://doi.org/10.32604/oncologie.2022.025148
Received 24 June 2022; Accepted 02 August 2022; Issue published 19 September 2022
Abstract
Background: Breast cancer (BC) is the most frequent type of gynecology tumors with high morbidity and mortality. Ampelopsin, the main active compound of Ampelopsis grossedentata, exerts an anti-tumor effect on a variety of cancers. However, the anti-cancer role of ampelopsin in BC remains unclear. The aim of this study is to explore the mechanism of ampelopsin against breast cancer. Materials and Methods: The target genes of ampelopsin in the treatment of breast cancer were determined and analyzed by network pharmacology and molecular docking. Cytoscape software was used to identify the core target genes and construct a protein–protein interaction (PPI) network. Discovery Studio software was used to perform the molecular docking of ampelopsin and core genes and glycolytic metabolic enzymes. Results: In total, 25 potential target genes of ampelopsin were screened out. The core target genes of ampelopsin against breast cancer were AKT1, ESR1, ESR2, NCOA1, HSP90AA1, NCOA2, BECN1, COMT, HMOX1, and CDK6, with AKT1, ESR1 and ESR2 considered as the key target proteins. Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis showed that ampelopsin inhibited breast cancer via modulating the estrogen signaling pathway, apoptosis regulation, carbohydrate metabolism, and inflammation. Molecular docking analysis showed that ampelopsin possessed a stable binding ability to regulate the three target proteins and glycolytic metabolic enzymes such as ALDOA and LDHA. Conclusions: Ampelopsin may inhibit the proliferation of breast cancer cells by acting on AKT and estrogen-related glucose metabolic pathways and inhibiting the enzymes involved in glycolysis and oxidative phosphorylation.Keywords
Nomenclature
| ALDOA | aldolase |
| BC | breast cancer |
| COX7RP | cytochrome C oxidase subunit 7α-related polypeptide |
| CTD | comparative toxicogenomics database |
| ECM | extracellular matrix |
| ER | estrogen receptor |
| ETCM | Encyclopedia of Traditional Chinese Medicine |
| GO | Gene Ontology |
| HK2 | hexokinase 2 |
| HR | hormone receptor |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| LDHA | lactate dehydrogenase A |
| MCT | monocarboxylic acid transporter |
| OXPHOS | oxidative phosphorylation |
| PFKL | liver phosphofructokinase |
| PK | pyruvate kinase |
| PPI | protein–protein interaction |
| PR | progesterone receptor |
| TCA | tricarboxylic acid |
| TNBC | triple-negative breast cancer |
Breast cancer (BC) is the most common type of cancers and the leading cause of cancer-related deaths among women [1]. Clinical treatments for BC fall under the radiotherapy, surgical resection, chemotherapy, immunotherapy, and hormonal therapy [2]. Most breast cancers express hormone receptors (HR) such as the estrogen receptor α (ERα) and progesterone receptor (PR), whereas others express human epidermal growth factor receptor 2 (HER2). HR- and HER2-positive cancers can be treated with endocrine therapy and anti-HER2 molecules (such as Trastuzumab), respectively [3]. Triple-negative breast cancer (TNBC) is a subtype of BC that lacks ERα, PR, and HER2 expression, accounting for 15%–20% of BC cases. Owing to the lack of relevant receptors, TNBC is often unresponsive to targeted therapy. Currently, conventional chemotherapy and surgical resection are the mainstays for the treatment of TNBC. However, inherent or acquired drug resistance in all BC types leads to treatment failure [4,5]. Thus, there is an urgent need to develop new and alternative BC therapies.
Cancer cells undergo metabolic changes to satisfy their energy requirements for increased proliferation, migration, and invasion. Multiple metabolic pathways in cancer cells, such as glycolysis, oxidative phosphorylation (OXPHOS), tricarboxylic acid (TCA) cycle, and amino acid and lipid metabolism, are reprogrammed as they undergo adaptation [6]. Although a hypoxic microenvironment generally increases glucose consumption and glycolysis, cancer cells differ from normal cells in which they prefer glycolysis even in the presence of sufficient oxygen. This phenomenon is called the Warburg effect [7]. MCF-7 (HR-positive), SKBR3 (HER2-positive), and MAD-MB-231 (TNBC) BC cells rely on glycolysis to meet approximately 20% [8,9], 50% [10], and 70% [11] of their ATP requirements, respectively. Therefore, by targeting glycolysis and related processes, researchers can discover new strategies for treating TNBC.
Ampelopsin, also known as dihydromyricetin, is the main active flavonoid in Ampelopsis grossedentata [12]. Modern pharmacological studies have shown that ampelopsin exhibits anti-inflammatory, antimicrobial, antioxidant, and hypoglycemic activitie [13]. Several studies have explored the anti-tumor effects of ampelopsin. For example, ampelopsin promotes apoptosis in lung adenocarcinoma [14], liver cancer [15], breast cancer [16], and ovarian cancer cells [17]. Moreover, ampelopsin could inhibit the proliferation of cholangiocarcinoma cells via regulating miR-21 [18] and promotes apoptosis of nasopharyngeal cancer cells via inhibiting the activation of NF-κB [19]. In vivo studies have demonstrated that ampelopsin inhibits the progression of tumors through the Wnt/β-catenin signaling pathway [15,20]. Besides, ampelopsin can restrain the proliferation of BC stem cells by interfering with the OXPHOS in TNBC cells through the TNFα/NF-κB signaling pathway [21]. In addition, ampelopsin can inhibit the occurrence and development of TNBC by enhancing mitochondrial autophagy and endoplasmic reticulum stress through the mammalian target of rapamycin (mTOR) signaling pathway [16]. However, it is not clear whether ampelopsin inhibits the progression by regulating glycolysis.
In this study, we investigated the targets of ampelopsin against BC using network pharmacology and molecular docking to determine the potential mechanism of ampelopsin against BC and whether ampelopsin can prevent against the proliferation of tumor cells by regulating glycolysis.
Network pharmacology and molecular docking could provide an effective way to identify the mechanisms underlying the effects of traditional Chinese medicine and active compounds [22]. Traditional network pharmacology and molecular docking combine multiple database resources to identify core genes that can be targeted by drugs or active compounds. Thus, network pharmacology and molecular docking were employed in this study to discover the role of ampelopsin in the treatment of human breast cancer.
2.1 Target Screening of Ampelopsin
The Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMCP, https://www.tcmsp-e.com/), HERB (http://herb.ac.cn/), and Encyclopedia of Traditional Chinese Medicine (ETCM, http://www.tcmip.cn/ETCM/) databases were used to obtain the necessary information of the target genes of ampelopsin.
2.2 Potential Therapeutic Targets of Ampelopsin in BC
Information related to BC markers and target genes was collected from GeneCards (https://www.genecards.org/), MalaCards (https://www.malacards.org/), and Comparative Toxicogenomics Database (CTD, http://ctdbase.org/), using the keyword “BC”. The potential targets of ampelopsin in the treatment of BC were identified by analyzing the intersection targets of ampelopsin and BC online (https://bioinfogp.cnb.csic.es/tools/venny/index.html).
2.3 Potential Protein–Protein Interactions (PPI) Network Construction and Analysis
Analysis was performed using the STRING database (https://string-db.org/) [23], and the species selected to assess PPI was “Homo sapiens” with a confidence degree of 0.9. The network visualization software Cytoscape [24] was used to construct the ampelopsin–BC target network, and the MCODE plug-in of Cytoscape was employed to screen the core targets. The clusters containing molecules in the network with the highest scores were considered core genes.
2.4 Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Enrichment Analyses
The ClueGO plug-in of Cytoscape (version 3.8.2) and the Database for Annotation, Visualization and Integrated Discovery (DAVID) software (https://david.ncifcrf.gov/) were used to analyze the core target genes in the PPI network. The core targets were imported into Kyoto Encyclopedia of Genes and Genomes (KEGG) for pathway visualization in order to screen reliable biological processes, molecular functions, and signaling pathways. Results with P < 0.05 and Q < 0.05 were considered statistically significant.
Ampelopsin was used as the ligand, and the core target proteins related to glucose metabolism were used as receptors. Molecular docking verification was performed using Discovery Studio 2019 software. Discovery Studio is a life science prediction software developed by BIOVIA company that integrates classic and advanced algorithms as well as high-quality graphical interfaces from the current molecular simulation field mainly used for binding and interaction between proteins and small molecule compounds [25]. The 3D structure of the human target protein was downloaded from the Protein Data Bank (PDB) database (https://www.rcsb.org/). PDB database provides archive-information about the 3D shapes of proteins, nucleic acids, and complex assemblies [26]. The PubChem database (https://pubchem.ncbi.nlm.nih.gov/) was used to determine the 3D structure of ampelopsin and the core target proteins [26]. Molecular Operating Environment software (MEO v2019.0102), a drug discovery software platform that integrates visualization, modeling simulation, and method development, was used to optimize the target protein structure [27]. RSMD (RSMD ≤ 4 A is reliable and RSMD ≤ 2 A is accurate), representing the accuracy parameter of the molecular docking model, and S, referring to the binding free energy [28], were used to verify the binding of ampelopsin to the target proteins and enzymes. The potential binding modes of ampelopsin, glycolytic enzymes and target proteins were predicted and analyzed using molecular docking. The interaction energy of the ligand and receptor was calculated, and the optimal receptor–ligand complex was selected for mapping.
3.1 Potential Therapeutic Target Genes of Ampelopsin in BC
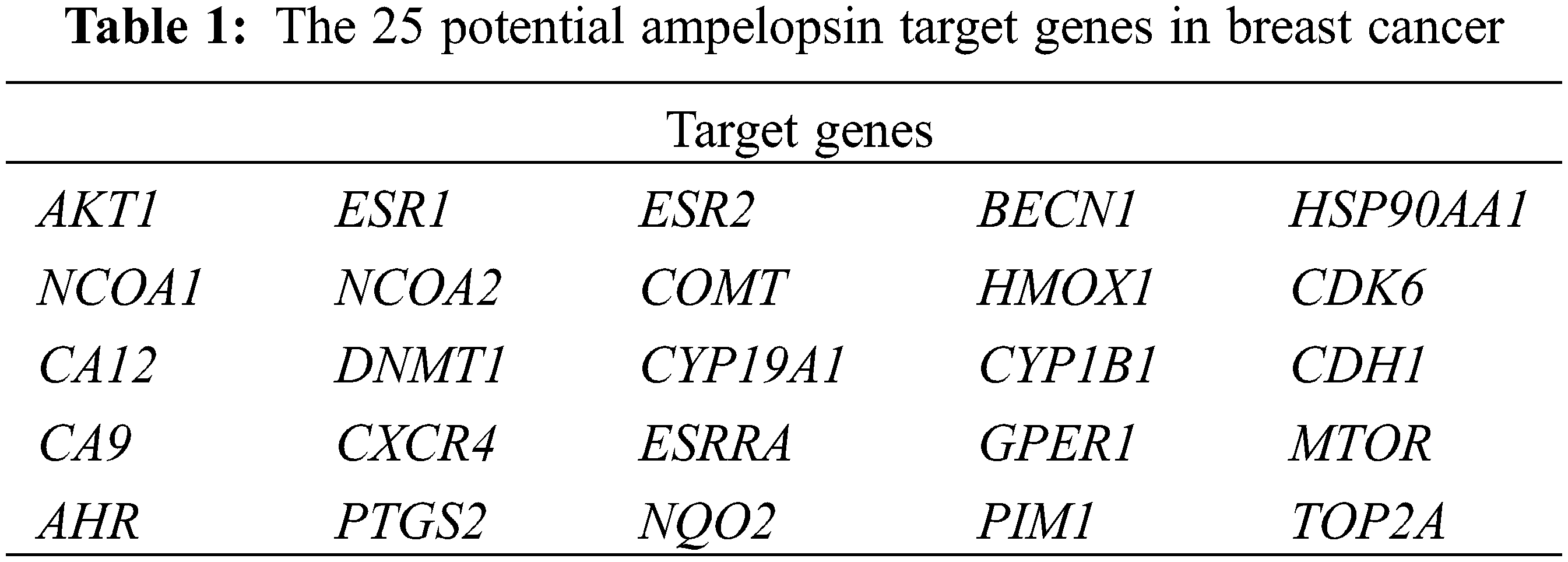
A total of 85 target genes of ampelopsin were identified in the three TCM databases, including protein molecules involved in various pathways such as glucose and lipid metabolism, cell proliferation, inflammation, and tumor microenvironment. A total of 622 BC markers and genes were retrieved from the CTD, GeneCards, and MalaCards databases. Veeny was used to analyze and compare the target genes of ampelopsin and BC (Fig. 1). Twenty-five potential intersection targets were identified and are shown in Table 1.
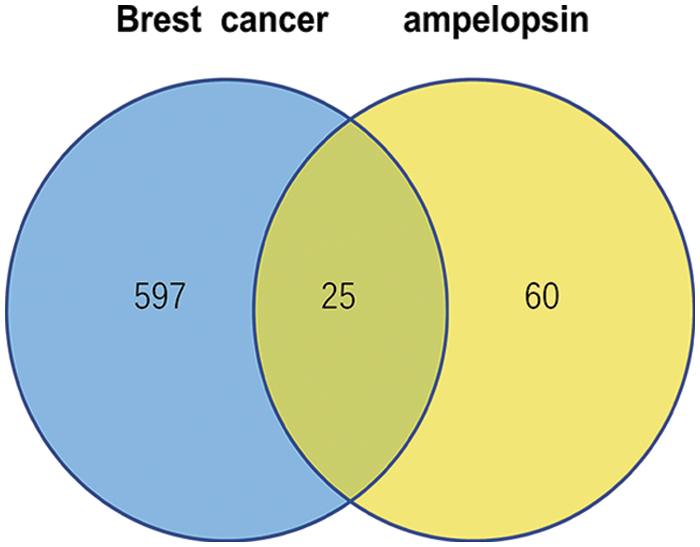
Figure 1: Ampelopsin-breast cancer target intersection
3.2 Construction and Analysis of a PPI Network and the Central Node
The 25 intersection targets were uploaded to the STRING database. Multiple proteins were selected with a confidence interval > 0.4 and a protein-protein interaction (PPI) network was constructed. The PPI network had 25 nodes (for the target genes in Table 1) and 102 links, with an average node degree of 8.16 (Fig. 2). The MCODE plug-in of Cytoscape was used for the cluster analysis, and the results showed ten important genes which were AKT1, ESR1, ESR2, NCOA1, HSP90AA1, NCOA2, BECN1, COMT, HMOX1, and CDK6. Cluster 1 consisted of 30 nodes and 363 edges, with an average node degree of 25.034; cluster 2 consisted of 18 nodes and 32 edges, with an average node degree of 3.592; cluster 3 consisted of 4 nodes and 5 edges, with an average node degree of 3.333. Genes such as AKT1, ESR1 and ESR2 were selected by degree to be the core targets (Fig. 3).
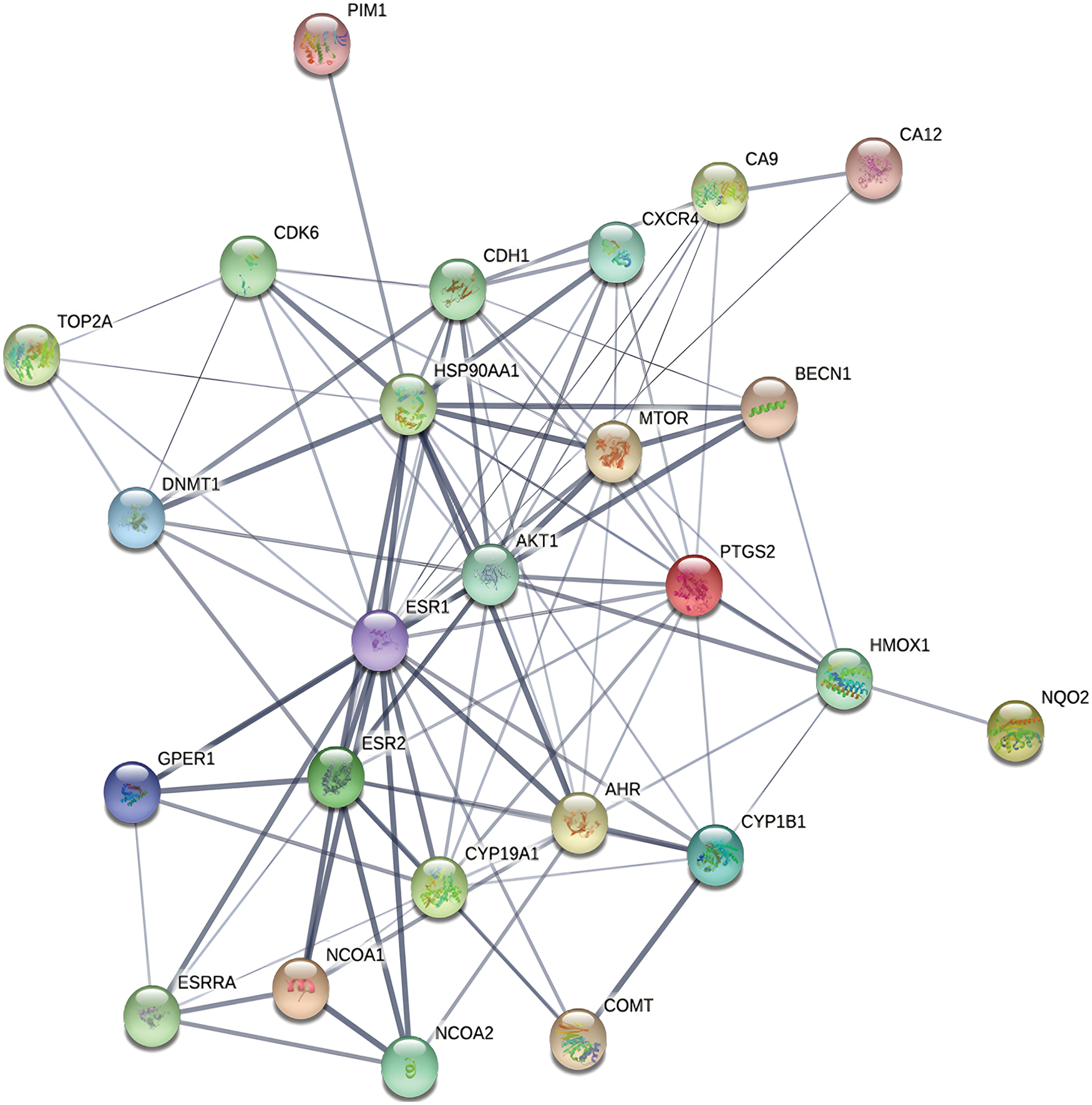
Figure 2: Protein–protein interaction network of the intersection targets
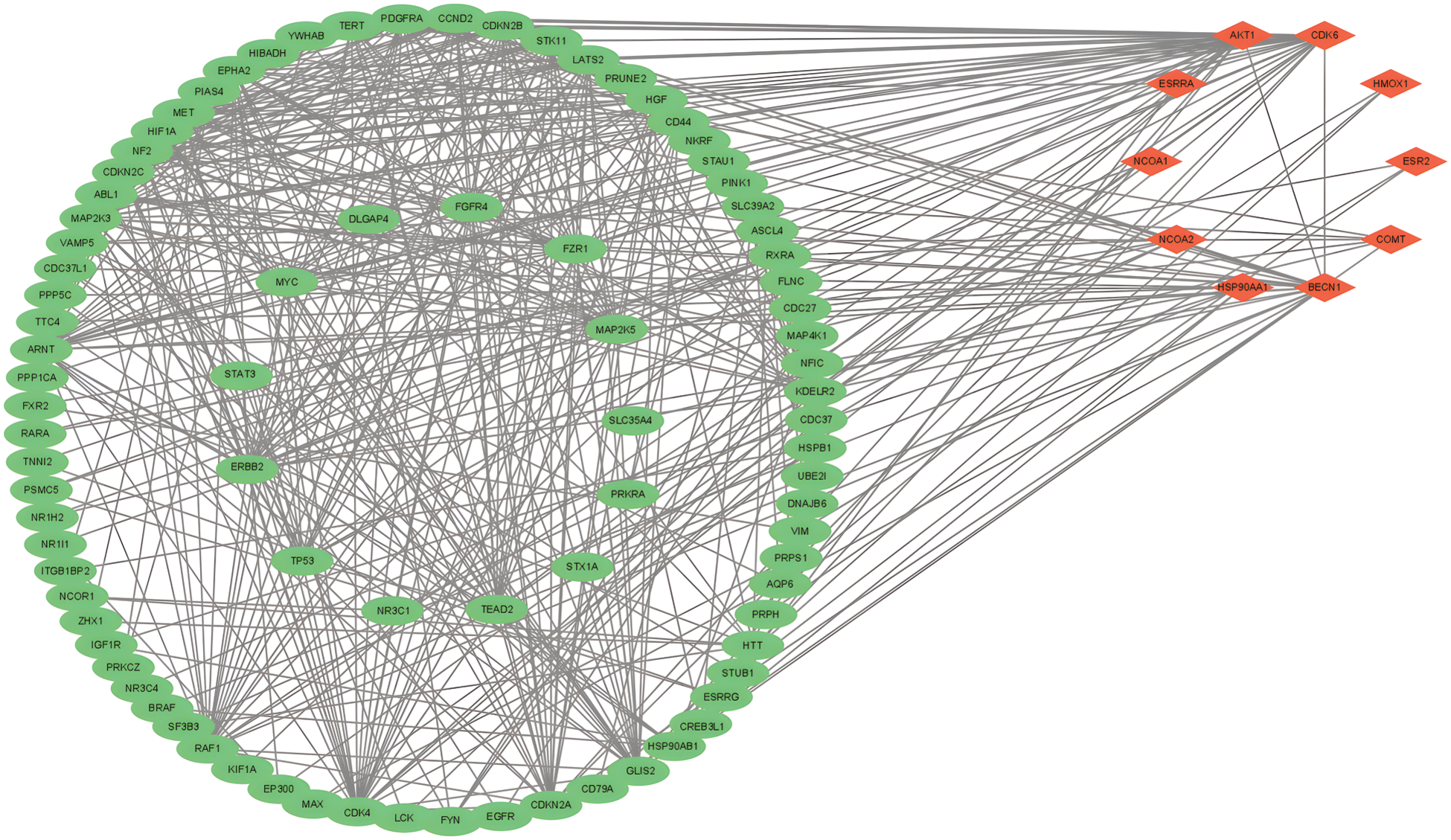
Figure 3: Cluster of MCODE cluster analysis
3.3 GO and KEGG Enrichment Analyses
A total of 34 biological processes of the intersection targets were identified using the GO enrichment analysis. As shown in Fig. 4, the biological processes associated with intersection target genes included cell migration in angiogenesis, activity of steroid HR, negative regulation of autophagy, and cell proliferation.
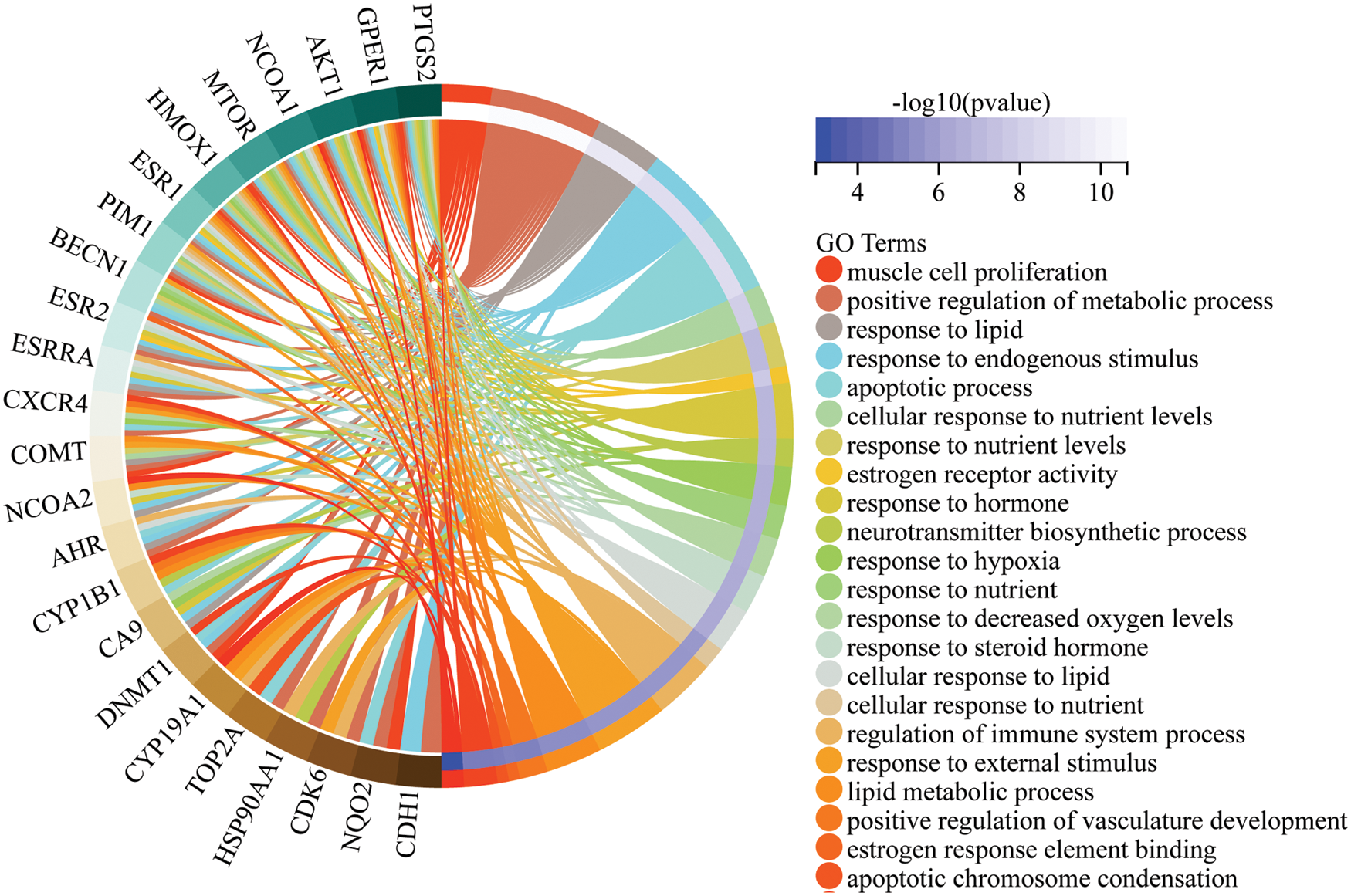
Figure 4: Gene Ontology (GO) enrichment analysis of the intersection targets
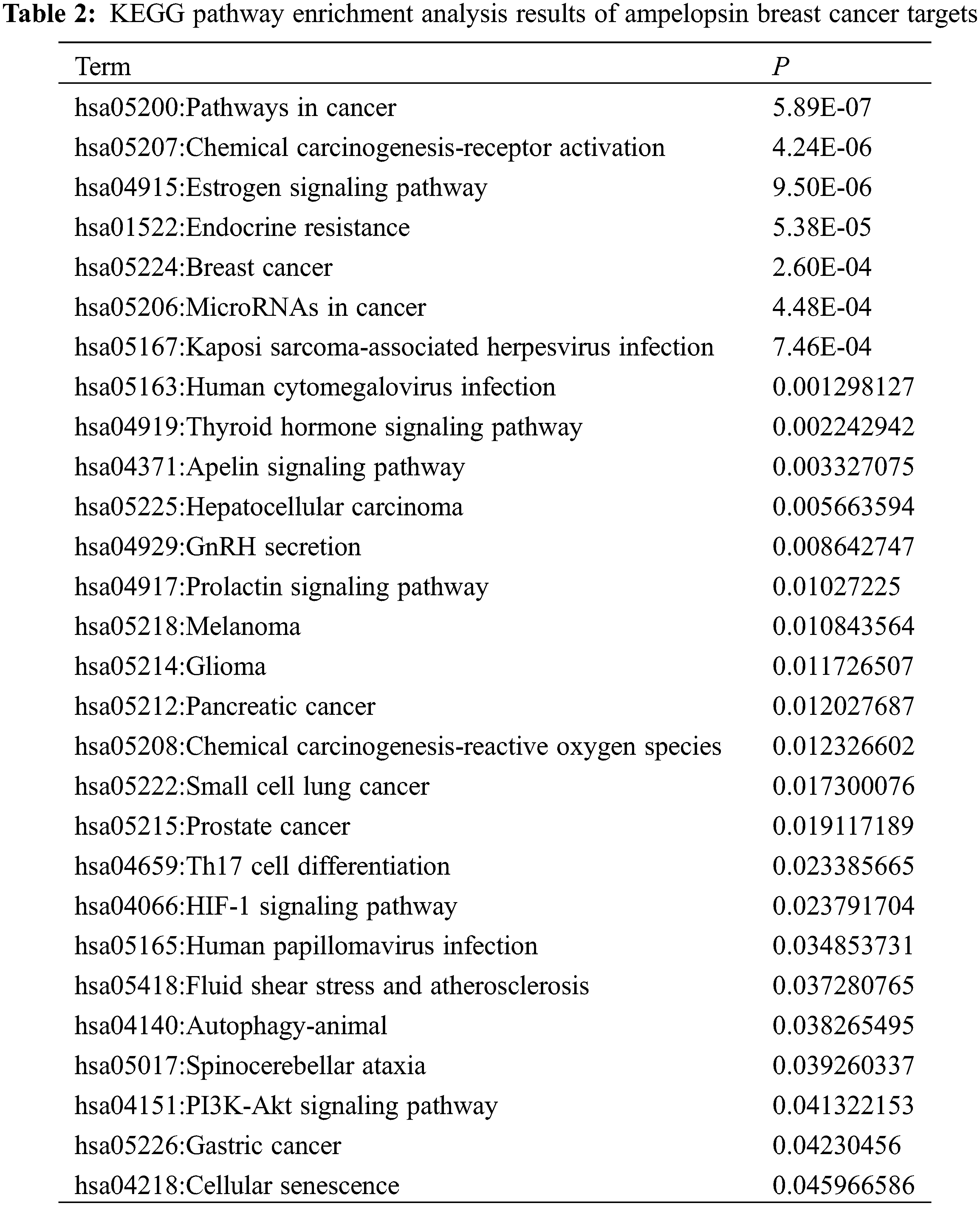
The KEGG enrichment analysis revealed several tumor-related endocrine pathways, indicating that ampelopsin could inhibit the occurrence and development of BC through the hormone-receptor pathway. The hormone signaling pathways included endocrine resistance, estrogen signaling, prolactin signaling, thyroid hormone signaling, and other pathways such as autophagy (Fig. 5 and Table 2).
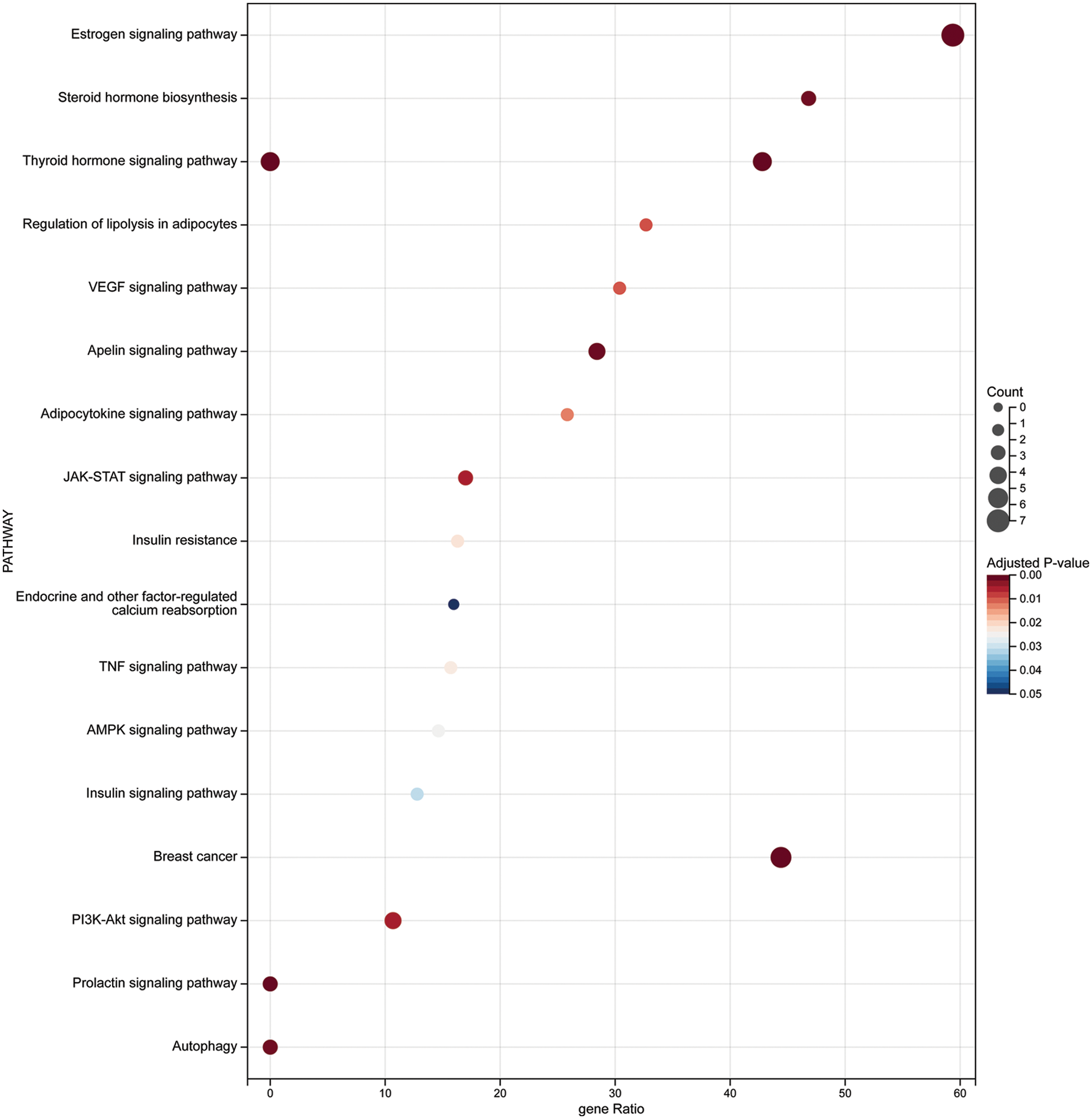
Figure 5: KEGG enrichment analysis of the intersection targets
Through KEGG pathway enrichment, we concluded that metabolic enzymes in the glycolytic pathway were regulated by hormone signalings. Thus, ampelopsin might regulate glycolysis through PI3K/AKT and promote lipid metabolism through the apelin signaling pathway, which plays an important role in the regulation of autophagy in BC (Figs. 6–8).
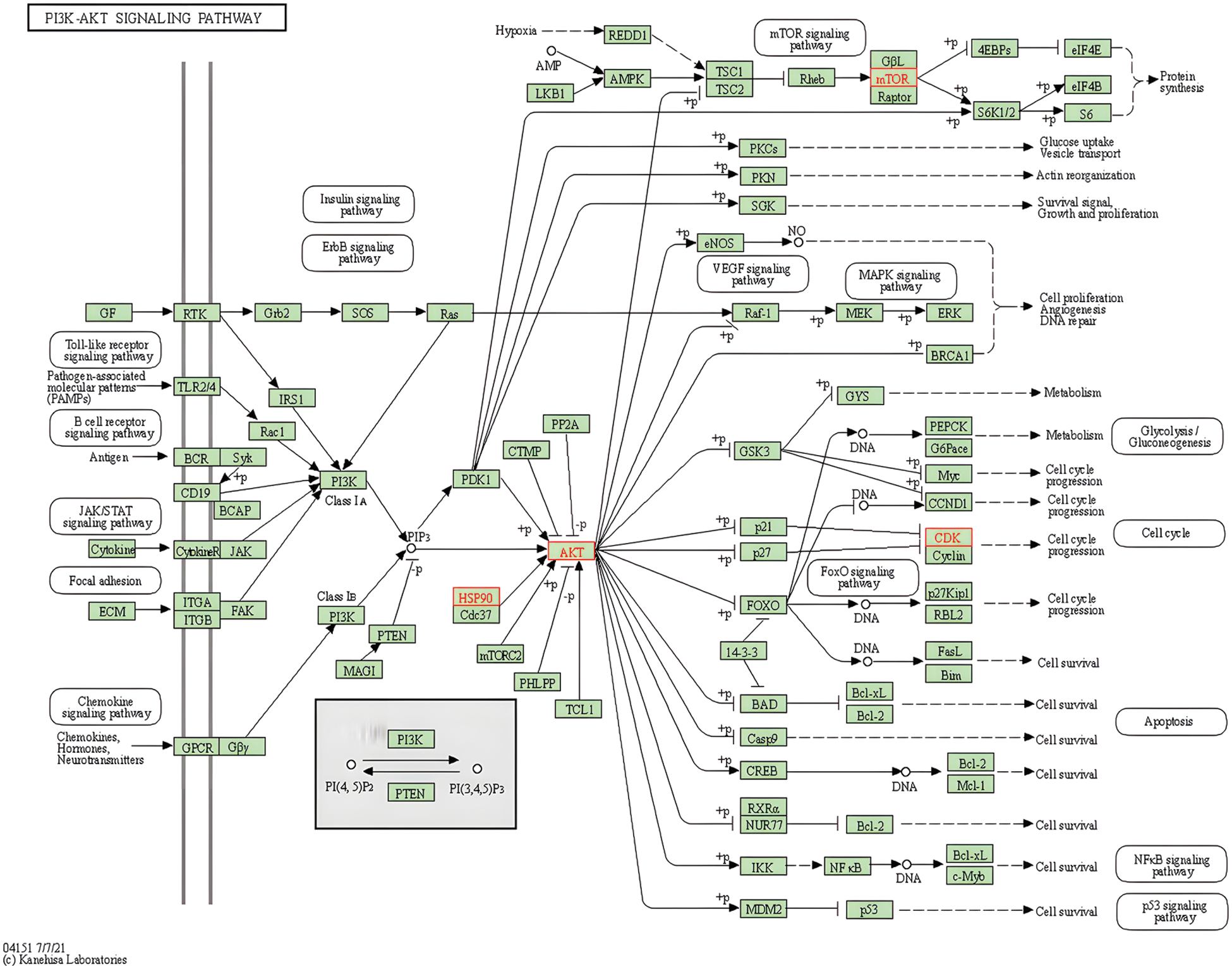
Figure 6: Potential role of ampelopsin in the PI3K/AKT signaling pathway
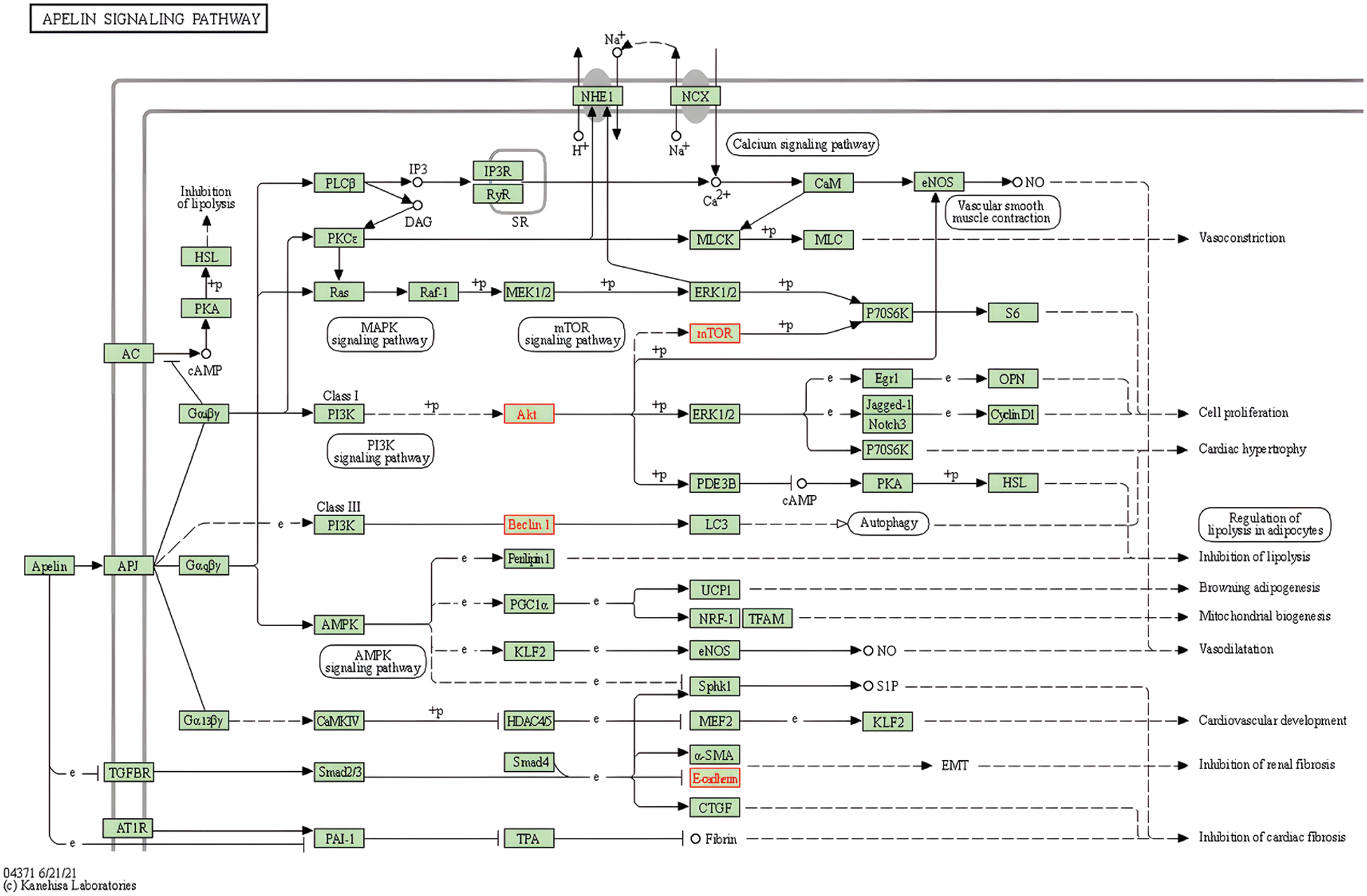
Figure 7: Potential role of ampelopsin in the apelin signaling pathway
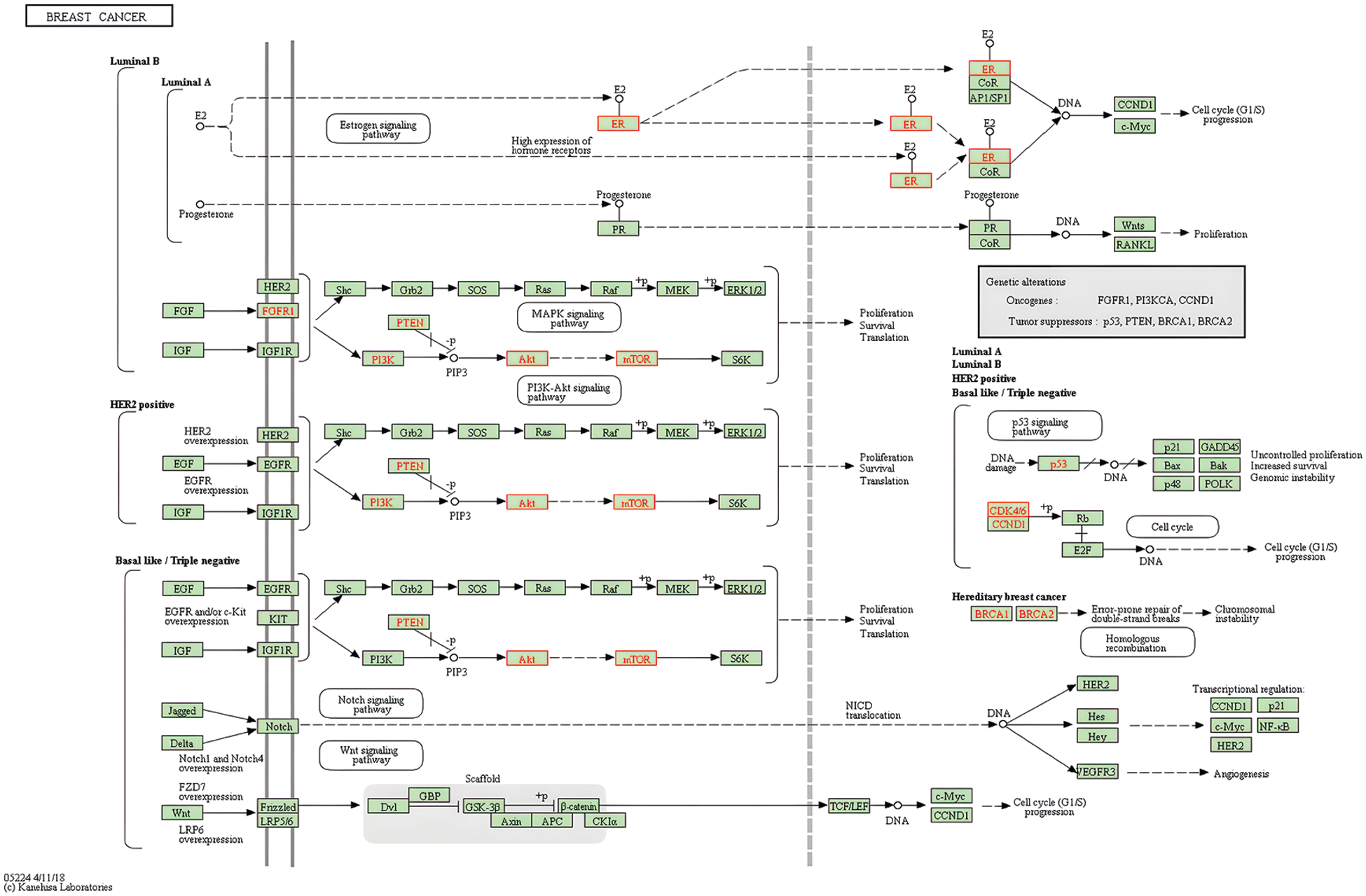
Figure 8: Potential role of the PI3K/AKT/mTOR signaling pathway in breast cancer
As shown in Figs. 2–5, ampelopsin might bind to AKT1 and other key target proteins to regulate AKT downstream signaling molecules. Glycolytic pathway is regulated by the PI3K/AKT signaling pathway. Therefore, we analyzed the effect of ampelopsin on the target proteins and metabolic enzymes involved in glycolysis. We performed molecular docking analysis using ampelopsin as the ligand and AKT1, ESR1, ESR2, and glycolytic metabolic enzymes such as hexokinase 2 (HK2), liver phosphofructokinase (PFKL), aldolase (ALDOA), pyruvate kinase (PK), lactate dehydrogenase A (LDHA), and cytochrome C oxidase subunit 7α-related polypeptide (COX7RP) as the receptors.
The binding of ampelopsin to the core target proteins and enzymes related to glycolytic pathway was determined (Table 3 and Fig. 9). Our results indicated that ampelopsin had a high affinity for its targets, such as ESR1 and ESR2 (S < −6 kcal/mol and RMSD < 2), and a moderate affinity for AKT1. Docking with enzymes related to glycolytic pathway showed that the binding of ampelopsin to ALDOA and LDHA was stable and strong (S < −6 kcal/mol and RMSD < 2) (Table 3 and Fig. 9). However, there was a moderate affinity between ampelopsin and some other enzymes such as HK2, PFKL, PK and COX7RP (Table 3).
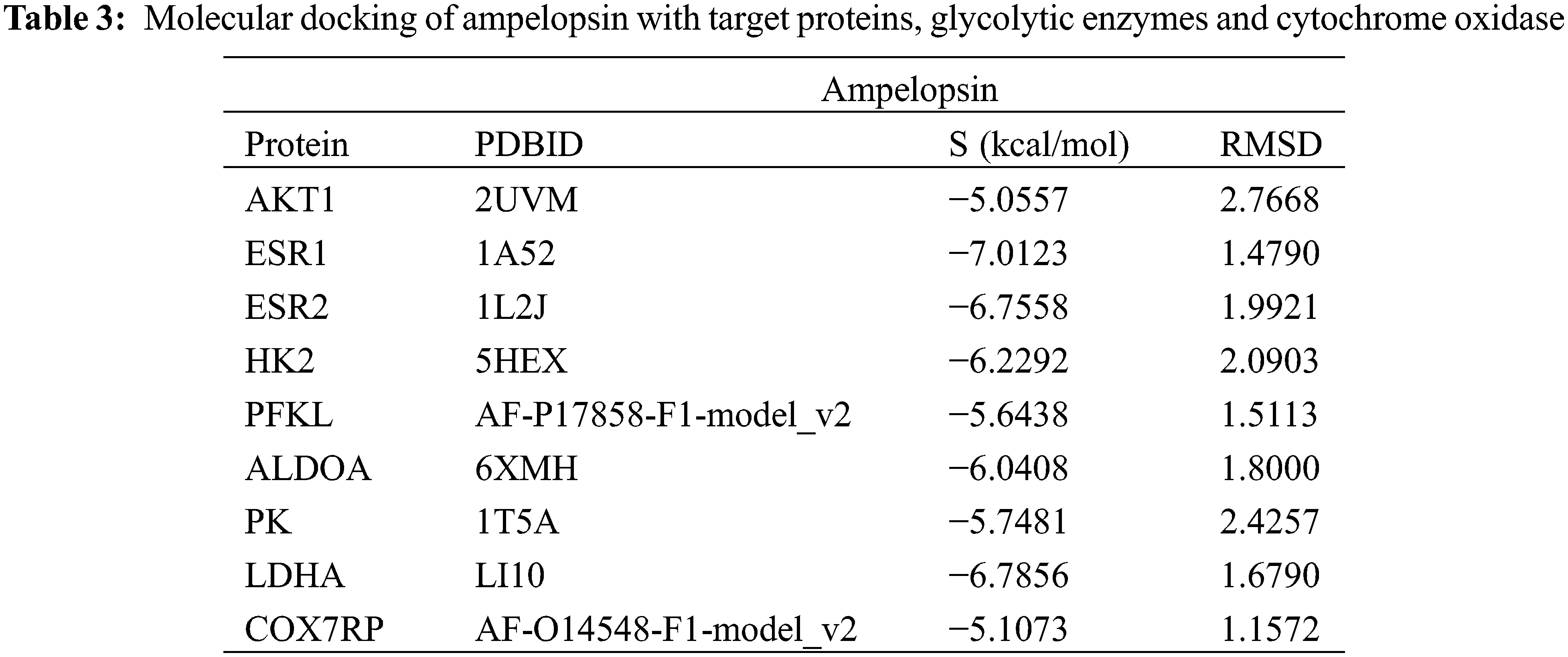
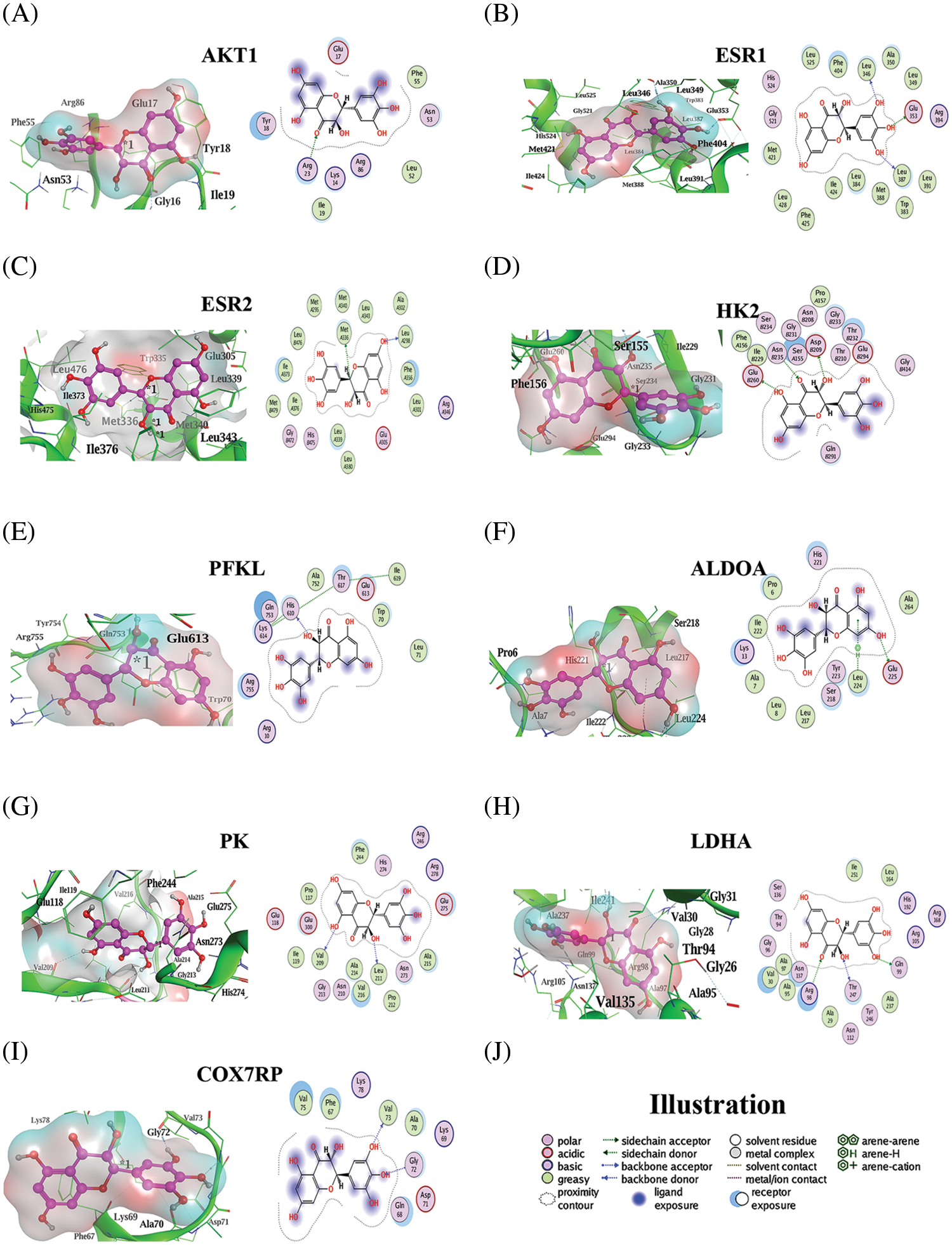
Figure 9: Molecular docking of ampelopsin with target genes, glycolytic enzymes, and cytochrome oxidase. (A–I) Molecular docking pattern of ampelopsin with AKT1 (A), ESR1 (B), ESR2 (C), HK2 (D), PFKL (E), ALDOA (F), PK (G), LDHA (H), and COX7RP (I), (J) Illustration of the ligand-receptor binding pattern. AKT1 (PKB1), Protein kinase B1; ESR, estrogen receptor gene; HK2, hexokinase 2; PFKL, liver phosphofructokinase; ALDOA, aldolase; PK, pyruvate kinase; LDHA, lactate dehydrogenase A; COX7RP, cytochrome C oxidase subunit 7α-related polypeptide
In this study, we have evaluated the therapeutic role of ampelopsin in the treatment of BC. Our results indicated the following: (1) there were 25 potential targets of ampelopsin in BC; (2) the target genes of ampelopsin were AKT1, ESR1, ESR2, NCOA1, HSP90AA1, NCOA2, BECN1, COMT, HMOX1, and CDK6, with AKT1, ESR1, and ESR2 considered as the core target proteins; (3) ampelopsin inhibits BC by regulating the estrogen signaling pathway, apoptosis, carbohydrate metabolism, and inflammation; (4) ampelopsin possesses a stable binding ability to regulate target proteins such as AKT1, ESR1, and ESR2 and the glycolytic enzymes ALDOA and LDHA.
OXPHOS and mitochondrial super-complexes exhibit an essential effect on promoting cell proliferation. Although intracellular ATP production is measured differently, the proportion of energy produced via glycolytic pathway in MAD-MB-231 cells is among 40%–70%, suggesting that glycolysis exhibits an essential effect on TNBC cells [11,29]. Previous studies have indicated that tumor cells showing the “Warburg effect” exhibit specific metabolic profiles at different times; glycolysis is the main metabolic pathway during the day. At night, however, tumor cells revert to the mitochondrial aerobic oxidation and OXPHOS pattern of normal healthy cells [30,31]. Owing to this circadian rhythm, these tumors can become cancerous at some point in time [31–33]. During the day, tumor cells undergo glycolysis in the environment even in presence of sufficient oxygen, and the overproduced lactic acid flows out of the cells through the monocarboxylic acid transporters [34]. Lactic acid changes the pH of the tumor microenvironment and promotes the decomposition of the extracellular matrix as well as the migration and invasion of tumor cells [35]. Additionally, an acidic microenvironment can increase angiogenesis and promote tumor metastasis. Hence, glycolysis can improve the proliferation, migration, and invasion of tumor cells. Therefore, the regulation of the glycolytic pathway may be an important strategy for the treatment of breast cancer.
In tumor cells, multiple metabolic processes are reprogrammed to provide energy. For instance, OXPHOS is facilitated by the formation of the mitochondrial respiratory chain supercomplex (complexes I, III, and IV) [36]. Ampelopsin can promote mitochondrial autophagy in BC cells and inhibit the activity of mitochondrial supercomplexes through the PI3K/AKT/mTOR signaling pathway [21]. Glycolysis plays an important role in promoting tumor proliferation, migration, and invasion. Therefore, it is important to elucidate the role of ampelopsin in regulating glycolysis.
In the present study, 25 potential target genes of ampelopsin were identified against BC (Table 1). The PPI network analysis suggested that AKT1, ESR1, ESR2, NCOA1, HSP90AA1, NCOA2, BECN1, COMT, HMOX1, and CDK6 were the core target genes of ampelopsin against BC (Figs. 1–3 and Table 1). AKT1 is a well-known oncogene involved in the regulation of the metabolism and proliferation of tumor cells. The estrogen receptors ESR1 and ESR2 activate the second messengers in the cells by binding to estrogen and regulating signaling pathways such as the PI3K/AKT/mTOR [37], MAPK/ERK [38] and cAMP/PKA pathways [39]. The KEGG enrichment analysis results suggested that ampelopsin could affect the development of BC by regulating the estrogen signaling pathway, cell cycle, and metabolism (Figs. 4–8 and Table 2).
HK2, the first key enzyme in the glycolytic pathway, is inhibited by the negative feedback of glucose-6-phosphate, whereas PFKL is the second rate-limiting enzyme in glycolysis and the most important rate-limiting enzyme regulating glycolysis. PFKL catalyzes the conversion of fructose-6-phosphate to fructose-1,6-diphosphate. Incidentally, fructose-2,6-bisphosphate synthesized by 6-phosphate fructokinase 2/fructose-2,6-bisphosphatase (PFK2) is the strongest allosteric activator of PFKL. The PFKL activity is significantly increased in TNBC, and it may contribute to the increased lymph node infiltration and metastasis [40]. By regulating the PI3K/AKT and cAMP/PKA signaling pathways activated by ESR1 and ESR2, ampelopsin can promote the phosphorylation of PFK2 and the hydrolysis of fructose-2,6-bisphosphate, reduce the production of fructose-2,6-bisphosphate, and inhibit the glycolytic rate-limiting enzyme PFKL [41]. As a result, ampelopsin is a potent inhibitor of TNBC cell proliferation and metastasis.
As a downstream enzyme of PFKL, ALDOA is regulated by the PI3K/AKT signaling pathway and is associated with cancer progression [42]. PK is the third key enzyme in glycolysis, and its activity is regulated by the cAMP/PKA signaling pathway. Ampelopsin activates cAMP/PKA through the estrogen signaling pathway, promoting PK phosphorylation and inactivation. LDHA catalyzes the final step of aerobic glycolysis, generating lactic acid. LDHA expression is associated with the invasion and metastasis of tumor cells [43]. Hence, ampelopsin can effectively bind to enzymes such as HK2, PFKL, ALDOA, PK, LDHA, and COX7RP, especially ALDOA and LDHA, to inhibit their activity and reduce lactic acid production and energy generation (Fig. 9 and Table 3). These results suggest that ampelopsin could inhibit the progression of BC through glycolysis regulation.
The molecular docking results confirmed the possibility of ampelopsin binding to BC core target genes (AKT1, ESR1 and ESR2) and enzymes involved in glycolysis (HK2, PFK1, ALDOA, PK and LDHA). The findings suggested a strong binding between ampelopsin and the target proteins ESR1 and ESR2 (S < −6 kcal/mol and RMSD < 2) (Fig. 9 and Table 3). Moreover, ampelopsin inhibited ALDOA and LDHA by strongly binding to them (S < −6 kcal/mol and RMSD < 2). Furthermore, ampelopsin regulated glycolysis and OXPHOS by binding to HK2, PFK1, PK, and COX7RP (Fig. 9 and Table 3).
In this study, we used network pharmacology and molecular docking analysis to confirm that ampelopsin may inhibit BC by acting with three core genes and five glucose-metabolism regulatory enzymes. However, our study had some limitations. First, some key targets and pathways may not have been adopted in this study because of the limitations of setting screening conditions and application databases. Second, this study did not consider the influence of ampelopsin concentration on its anti-cancer activity. Third, the effects of ampelopsin on other glucose metabolism processes such as the pentose phosphate pathway, OXPHOS, glycogen synthesis and decomposition, and gluconeogenesis were not discussed. Thus, further studies are needed to determine the role of ampelopsin in regulating glycolysis and inhibiting the occurrence and development of TNBC.
In conclusion, our study provides evidence that ampelopsin inhibits glycolysis in BC cells by regulating the PI3K/AKT and cAMP/PKA signaling pathways and suppresses the activities of enzymes involved in glycolysis, thereby providing a theoretical basis and experimental support for ampelopsin treatment in breast cancer.
Authorship Statement: The authors confirm contribution to the paper as follows: Conceptualization, Lin Liu, Yunfeng Li, and Jingshan Zhao; Data curation, Lin Liu, Rong Zeng, and Wenmei Zhang; Formal analysis, Lin Liu and Rong Zeng; Funding acquisition, Lin Liu, Jingshan Zhao, Guohong Zhang and Yunfeng Li; Investigation, Jingshan Zhao and Yunfeng Li; Methodology, Lin Liu; Supervision, Jingshan Zhao and Yunfeng Li; Writing–original draft, Lin Liu; Writing–review & editing, Rong Zeng, Jingshan Zhao, Guohong Zhang and Yunfeng Li. All authors reviewed the results and approved the final version of the manuscript.
Ethics Approval and Informed Consent Statement: Not applicable.
Availability of Data and Materials: The data generated and/or analyzed in this study can be found at: TCMSP (https://old.tcmsp-e.com/tcmsp.php), HERB (http://herb.ac.cn/), ETCM (http://www.tcmip.cn/ETCM/), GeneCards (https://www.genecards.org), MALACards (https://www.malacards.org), CTD (http://ctdbase.org/), Cytoscape software (version 3.8.2), STRING database (https://string-db.org/), DiscoveryStudio, PDH database (http://www.rcsb.org/), and PubChem database (https://pubchem.ncbi.nlm.nih.gov/).
Funding Statement: This research was funded by Medical Science Research Project of Hebei Provincial Health Commission 20221478 to Lin Liu; National Natural Science Foundation 81971434 to Yunfeng Li; Hebei Traditional Chinese Medicine Administration 2022080 to Lin Liu, 2017010 to Guohong Zhang and 2017009 to Jingshan Zhao; Science and Technology Project of Hebei Education Department QN2022092 to Lin Liu; Doctor Fund of Hebei University of Chinese Medicine BSZ2021007 to Lin Liu; Natural Science Foundation of Hebei Province H2020423061 and Key Program of Scientific and Technological Research Projects of Colleges and Universities in Hebei Province ZD2017056 to Jingshan Zhao.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I. et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer Journal for Clinicians, 71(3), 209–249. DOI 10.3322/caac.21660. [Google Scholar] [CrossRef]
2. Wang, B., Xing, Z., Wang, F., Yuan, X., Zhang, Y. (2017). Fangchinoline inhibits migration and causes apoptosis of human breast cancer MDA-MB-231 cells. Oncology Letters, 14(5), 5307–5312. DOI 10.3892/ol.2017.6831. [Google Scholar] [CrossRef]
3. von Minckwitz, G.,Huang, C. S., Mano, M. S., Loibl, S., Mamounas, E. P. et al. (2019). Trastuzumab emtansine for residual invasive HER2-positive breast cancer. The New England Journal of Medicine, 380(7), 617–628. DOI 10.1056/NEJMoa1814017. [Google Scholar] [CrossRef]
4. Ali, S., Coombes, R. C. (2002). Endocrine-responsive breast cancer and strategies for combating resistance. Nature Reviews Cancer, 2(2), 101–112. DOI 10.1038/nrc721. [Google Scholar] [CrossRef]
5. Nahta, R., Yu, D., Hung, M. C., Hortobagyi, G. N., Esteva, F. J. (2006). Mechanisms of disease: Understanding resistance to HER2-targeted therapy in human breast cancer. Nature Clinical Practice Oncology, 3(5), 269–280. DOI 10.1038/ncponc0509. [Google Scholar] [CrossRef]
6. Sobanski, T., Rose, M., Suraweera, A., O’Byrne, K., Richard, D. J. et al. (2021). Cell metabolism and DNA repair pathways: Implications for cancer therapy. Frontiers in Cell and Developmental Biology, 9, 633305. DOI 10.3389/fcell.2021.633305. [Google Scholar] [CrossRef]
7. Warburg, O. (1956). On the origin of cancer cells. Science, 123(3191), 309–314. DOI 10.1126/science.123.3191.309. [Google Scholar] [CrossRef]
8. Guppy, M., Leedman, P., Zu, X., Russell, V. (2002). Contribution by different fuels and metabolic pathways to the total ATP turnover of proliferating MCF-7 breast cancer cells. The Biochemical Journal, 364, 309–315. DOI 10.1042/bj3640309. [Google Scholar] [CrossRef]
9. Louie, M. C., Ton, J., Brady, M. L., Le, D. T., Mar, J. N. et al. (2020). Total cellular ATP production changes with primary substrate in MCF7 breast cancer cells. Frontiers in Oncology, 10, 1703. [Google Scholar]
10. Wu, H., Ying, M., Hu, X. (2016). Lactic acidosis switches cancer cells from aerobic glycolysis back to dominant oxidative phosphorylation. Oncotarget, 7(26), 40621–40629. DOI 10.18632/oncotarget.9746. [Google Scholar] [CrossRef]
11. Klepinin, A., Miller, S., Reile, I., Puurand, M., Rebane-Klemm, E. et al. (2022). Stable isotope tracing uncovers reduced γ/β-ATP turnover and metabolic flux through mitochondrial-linked phosphotransfer circuits in aggressive breast cancer cells. Frontiers in Oncology, 12, 892195. DOI 10.3389/fonc.2022.892195. [Google Scholar] [CrossRef]
12. Xin, Q., Yuan, M., Li, H., Lu, J., Song, X. et al. (2019). In vitro efficacy of ampelopsin against Echinococcus granulosus and Echinococcus multilocularis. The Journal of Veterinary Medical Science , 81(12), 1853–1858. DOI 10.1292/jvms.19-0347. [Google Scholar] [CrossRef]
13. Tong, Q., Hou, X., Fang, J., Wang, W., Xiong, W. et al. (2015). Determination of dihydromyricetin in rat plasma by LC-MS/MS and its application to a pharmacokinetic study. Journal of Pharmaceutical and Biomedical Analysis, 114, 455–461. DOI 10.1016/j.jpba.2015.06.030. [Google Scholar] [CrossRef]
14. Kao, S. J., Lee, W. J., Chang, J. H., Chow, J. W., Chung, C. L. et al. (2017). Suppression of reactive oxygen species-mediated ERK and JNK activation sensitizes dihydromyricetin-induced mitochondrial apoptosis in human non-small cell lung cancer. Environmental Toxicology, 32(4), 1426–1438. DOI 10.1002/tox.22336. [Google Scholar] [CrossRef]
15. Jiang, L., Ye, W. C., Li, Z., Yang, Y., Dai, W. et al. (2021). Anticancer effects of dihydromyricetin on the proliferation, migration, apoptosis and in vivo tumorigenicity of human hepatocellular carcinoma Hep3B cells. BMC Complementary Medicine and Therapies, 21(1), 194. DOI 10.1186/s12906-021-03356-5. [Google Scholar] [CrossRef]
16. Li, Y., Zhou, Y., Wang, M., Lin, X., Zhang, Y. et al. (2021). Ampelopsin inhibits breast cancer cell growth through mitochondrial apoptosis pathway. Biological & Pharmaceutical Bulletin, 44(11), 1738–1745. DOI 10.1248/bpb.b21-00470. [Google Scholar] [CrossRef]
17. Xu, Y., Wang, S., Chan, H. F., Lu, H., Lin, Z. et al. (2017). Dihydromyricetin induces apoptosis and reverses drug resistance in ovarian cancer cells by p53-mediated downregulation of survivin. Scientific Reports, 7(1), 46060. DOI 10.1038/srep46060. [Google Scholar] [CrossRef]
18. Chen, L., Yang, Z. S., Zhou, Y. Z., Deng, Y., Jiang, P. et al. (2020). Dihydromyricetin inhibits cell proliferation, migration, invasion and promotes apoptosis regulating miR-21 in Human Cholangiocarcinoma cells. Journal of Cancer, 11(19), 5689–5699. DOI 10.7150/jca.45970. [Google Scholar] [CrossRef]
19. Li, C. H., Ding, H., Shi, J. L., Huang, B., Ding, H. et al. (2021). Dihydromyricetin promotes apoptosis, suppresses proliferation and tumor necrosis factor-α-mediated nuclear factor kappa-B activation in nasopharyngeal carcinoma CNE-2 cell. Journal of Traditional Chinese Medicine, 41(3), 367–375. [Google Scholar]
20. Ye, L., Yin, G., Jiang, M., Tu, B., Li, Z. et al. (2021). Dihydromyricetin exhibits antitumor activity in nasopharyngeal cancer cell through antagonizing Wnt/β-catenin signaling. Integrative Cancer Therapies. DOI 10.1177/1534735421991217. [Google Scholar] [CrossRef]
21. Truong, V. N., Nguyen, Y. T., Cho, S. K. (2021). Ampelopsin suppresses stem cell properties accompanied by attenuation of oxidative phosphorylation in chemo- and radio-resistant MDA-MB-231 breast cancer cells. Pharmaceuticals, 14(8), 794. DOI 10.3390/ph14080794. [Google Scholar] [CrossRef]
22. Sakle, N. S., More, S. A., Mokale, S. N. (2020). A network pharmacology-based approach to explore potential targets of Caesalpinia pulcherima: An updated prototype in drug discovery. Scientific Reports, 10(1), 17217. DOI 10.1038/s41598-020-74251-1. [Google Scholar] [CrossRef]
23. Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S. et al. (2019). STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Research, 47(D1), D607–D613. DOI 10.1093/nar/gky1131. [Google Scholar] [CrossRef]
24. Otasek, D., Morris, J. H., Bouças, J., Pico, A. R., Demchak, B. (2019). Cytoscape automation: Empowering workflow-based network analysis. Genome Biology, 20(1), 185. DOI 10.1186/s13059-019-1758-4. [Google Scholar] [CrossRef]
25. Pinzi, L., Rastelli, G. (2019). Molecular docking: Shifting paradigms in drug discovery. International Journal of Molecular Sciences, 20(18), 4331. DOI 10.3390/ijms20184331. [Google Scholar] [CrossRef]
26. Burley, S. K., Berman, H. M., Kleywegt, G. J., Markley, J. L., Nakamura, H. et al. (2019). Protein data bank: The single global archive for 3D macromolecular structure data. Nucleic Acids Research, 47(D1), D520–D528. DOI 10.1093/nar/gky949. [Google Scholar] [CrossRef]
27. Rampogu, S., Baek, A., Son, M., Park, C., Yoon, S. et al. (2020). Discovery of lonafarnib-like compounds: Pharmacophore modeling and molecular dynamics studies. ACS Omega, 5(4), 1773–1781. DOI 10.1021/acsomega.9b02263. [Google Scholar] [CrossRef]
28. Gioia, D., Bertazzo, M., Recanatini, M., Masetti, M., Cavalli, A. (2017). Dynamic docking: A paradigm shift in computational drug discovery. Molecules, 22(11), 2029. DOI 10.3390/molecules22112029. [Google Scholar] [CrossRef]
29. Pacheco-Velázquez, S. C., Robledo-Cadena, D. X., Hernández-Reséndiz, I., Gallardo-Pérez, J. C., Moreno-Sánchez, R. et al. (2018). Energy metabolism drugs block triple negative breast metastatic cancer cell phenotype. Molecular Pharmaceutics, 15(6), 2151–2164. DOI 10.1021/acs.molpharmaceut.8b00015. [Google Scholar] [CrossRef]
30. Li, M., Hao, B., Zhang, M., Reiter, R. J., Lin, S. et al. (2021). Melatonin enhances radiofrequency-induced NK antitumor immunity, causing cancer metabolism reprogramming and inhibition of multiple pulmonary tumor development. Signal Transduction and Targeted Therapy, 6(1), 330. DOI 10.1038/s41392-021-00745-7. [Google Scholar] [CrossRef]
31. Reiter, R. J., Sharma, R., Pires, D. C. Z. D., de Almeida, C. L., Manucha, W. et al. (2021). Melatonin synthesis in and uptake by mitochondria: Implications for diseased cells with dysfunctional mitochondria. Future Medicinal Chemistry, 13(4), 335–339. DOI 10.4155/fmc-2020-0326. [Google Scholar] [CrossRef]
32. Reiter, R. J., Sharma, R., Ma, Q. (2021). Switching diseased cells from cytosolic aerobic glycolysis to mitochondrial oxidative phosphorylation: A metabolic rhythm regulated by melatonin? Journal of Pineal Research, 70(1), e12677. DOI 10.1111/jpi.12677. [Google Scholar] [CrossRef]
33. Reiter, R. J., Sharma, R., Ma, Q., Rorsales-Corral, S., de Almeida Chuffa, C. L. (2020). Melatonin inhibits Warburg-dependent cancer by redirecting glucose oxidation to the mitochondria: A mechanistic hypothesis. Cellular and Molecular Life Sciences, 77(13), 2527–2542. DOI 10.1007/s00018-019-03438-1. [Google Scholar] [CrossRef]
34. Jha, M. K., Morrison, B. M. (2020). Lactate transporters mediate glia-neuron metabolic crosstalk in homeostasis and disease. Frontiers in Cellular Neuroscience, 14, 589582. DOI 10.3389/fncel.2020.589582. [Google Scholar] [CrossRef]
35. Pérez-Tomás, R., Pérez-Guillén, I. (2020). Lactate in the tumor microenvironment: An essential molecule in cancer progression and treatment. Cancers, 12(11), 3244. DOI 10.3390/cancers12113244. [Google Scholar] [CrossRef]
36. Ikeda, K., Horie-Inoue, K., Suzuki, T., Hobo, R., Nakasato, N. et al. (2019). Mitochondrial supercomplex assembly promotes breast and endometrial tumorigenesis by metabolic alterations and enhanced hypoxia tolerance. Nature Communications, 10(1), 4108. DOI 10.1038/s41467-019-12124-6. [Google Scholar] [CrossRef]
37. Ciruelos, G. E. (2014). Targeting the PI3K/AKT/mTOR pathway in estrogen receptor-positive breast cancer. Cancer Treatment Reviews, 40(7), 862–871. DOI 10.1016/j.ctrv.2014.03.004. [Google Scholar] [CrossRef]
38. Sun, Q., Liang, Y., Zhang, T., Wang, K., Yang, X. (2017). ER-α36 mediates estrogen-stimulated MAPK/ERK activation and regulates migration, invasion, proliferation in cervical cancer cells. Biochemical and Biophysical Research Communications, 487(3), 625–632. DOI 10.1016/j.bbrc.2017.04.105. [Google Scholar] [CrossRef]
39. Szego, E. M., Barabás, K., Balog, J., Szilágyi, N., Korach, K. S. et al. (2006). Estrogen induces estrogen receptor alpha-dependent cAMP response element-binding protein phosphorylation via mitogen activated protein kinase pathway in basal forebrain cholinergic neurons in vivo. Journal of Neuroscience, 26(15), 4104–4110. DOI 10.1523/JNEUROSCI.0222-06.2006. [Google Scholar] [CrossRef]
40. Coelho, R. G., Calaça, I. C., Celestrini, D. M., Correia-Carneiro, A. H., Costa, M. M. et al. (2015). Hexokinase and phosphofructokinase activity and intracellular distribution correlate with aggressiveness and invasiveness of human breast carcinoma. Oncotarget, 6(30), 29375–29387. DOI 10.18632/oncotarget.4910. [Google Scholar] [CrossRef]
41. Lee, J. H., Liu, R., Li, J., Wang, Y., Tan, L. et al. (2018). EGFR-phosphorylated platelet isoform of phosphofructokinase 1 promotes PI3K activation. Molecular Cell, 70(2), 197–210.e7. DOI 10.1016/j.molcel.2018.03.018. [Google Scholar] [CrossRef]
42. Ji, S., Zhang, B., Liu, J., Qin, Y., Liang, C. et al. (2016). ALDOA functions as an oncogene in the highly metastatic pancreatic cancer. Cancer Letters, 374(1), 127–135. DOI 10.1016/j.canlet.2016.01.054. [Google Scholar] [CrossRef]
43. Yang, Y., Su, D., Zhao, L., Zhang, D., Xu, J. et al. (2014). Different effects of LDH-A inhibition by oxamate in non-small cell lung cancer cells. Oncotarget, 5(23), 11886–11896. DOI 10.18632/oncotarget.2620. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2022 The Author(s). Published by Tech Science Press.
Copyright © 2022 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.

 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools