
 | Oncologie |  |
DOI: 10.32604/oncologie.2022.021863
REVIEW
Gene Editing in Non-Small Cell Lung Cancer: Current Application and Future Perspective
1Clinical Medical College, Guizhou Medical University, Guiyang, 550004, China
2Second Clinical Medical College of Lanzhou University, Lanzhou, 730000, China
3School of Nursing, Guizhou Medical University, Guiyang, 550025, China
4Department of Pathophysiology, Guizhou Medical University, Guiyang, 550025, China
5Guizhou Provincial Key Laboratory of Pathogenesis and Drug Research on Common Chronic Diseases, Guizhou Medical University, Guiyang, 550025, China
*Corresponding Authors: Huimei Zou. Email: huimeizou@yeah.net; Fan Zhang. Email: zfan1985@yeah.net
#Equal Contribution: Hangxing Wang, Jingyun Fang and Yujiao Wang contributed to this work equally and should be regarded as co-first authors
Received: 10 February 2022; Accepted: 06 March 2022
Abstract: Lung cancer is the most common malignant tumor with the highest morbidity and mortality in the world, and non-small cell lung cancer (NSCLC) accounts for the vast majority of cases. At present, its main treatment methods are still traditional surgery, radiotherapy and chemotherapy, with disadvantages such as a high recurrence rate and limited effectiveness. Therefore, a new, better treatment method is urgently needed. Gene editing technology, as a new genetic engineering approach, has shown great potential in gene research, gene therapy and genetic improvement. It has also emerged as a promising treatment for lung cancer. This paper reviews the current research and applications of gene editing technology in NSCLC and other aspects of NSCLC, the therapeutic principle and mechanism of action, and the existing problems and prospects. This review aims to provide a basis for the prevention and treatment of NSCLC in the future and improve the survival rate of lung cancer patients.
Keywords: Non-small cell lung cancer; gene editing; CRISPR/Cas9
Lung cancer is one of the most common malignant tumors and accounts for 20% of all cancer-related deaths globally [1,2]. It was estimated that there were 2.1 million new cases and 1.8 million deaths in 2018 [3]. Lung cancer can be divided into small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC). NSCLC accounts for 85% of lung cancers, and the 5-year overall survival (OS) rate is approximately 15% [4]. NSCLC has multiple pathological types, including lung adenocarcinoma, lung squamous cell carcinoma, large cell carcinoma and others. It is a group of highly heterogeneous and highly mutated tumor types, which has been a great challenge for cancer treatment and a research hotspot in recent years. As one of the most common cancer types in the world, it has important clinical significance, and this article will give a general overview of NSCLC.
Although new treatments such as targeted therapy, immunotherapy, and gene therapy are being researched, the current treatment of non-small cell lung cancer mainly relies on traditional surgery, radiotherapy and chemotherapy. However, different treatment regimens are urgently needed because traditional treatment methods cannot completely overcome the side effects of metastasis, recurrence and resistance of cancer cells.
Gene editing, as a newer and more accurate genetic engineering technology that can modify specific target genes in the genome of organisms, has shown great potential in gene research, gene therapy and genetic improvement because of its high efficiency of site-specific editing. Currently, it is a promising method for tumor therapy. This article reviews the principles and mechanisms of gene editing technology in the treatment of NSCLC, other applications related to NSCLC, and the existing problems and prospects to provide new ideas and references for the treatment and prevention of lung cancer.
2 The Role of Gene Editing Technology in the Treatment of Non-Small Cell Lung Cancer
2.1 The Principle of Gene Editing Technology
Gene editing involves the removal of a nucleotide sequence or even a single nucleotide from a target gene in a cell’s genome and the addition or insertion of an exogenous DNA sequence [5–7]. Homologous recombination (HR), zinc finger nuclease (ZFN), transcriptional activator (TALEN) and regularly spaced short harmonic sequence repeats (CRISPR) are some of the most widely used techniques. The common feature of various gene editing technologies is that they assume that the DNA of the target gene will undergo a specific double strand breakage (DSB). However, there are two ways to repair a broken DNA double-strand [8]. One is nonhomologous end-joining (NHEJ), which does not need a homologous template chain. Gene mutation and gene knockout can be performed by inserting or deleting bases followed by NHEJ repair. The other is homology-directed repair (HDR) with template DNA, which realizes gene mutations, gene markers and target gene insertion through the template provided. The core principles and techniques of gene editing are briefly introduced as follows.
HR, the earliest gene editing technique, involves the exchange of genetic information (recombination) between two similar (homologous) strands of DNA. By producing and isolating DNA fragments with genome sequences similar to those of the genome to be edited, followed by injecting these fragments into monocytes or by making the cells absorb them with special chemicals, these fragments, once inside the cell, can be recombined with the cell’s DNA to replace the target part of the genome. However, due to its low frequency of recombination, high error rate, time consumption, need for extensive labor [9] and the difficulty of applying it to embryonic stem cells, the application of this technology is limited to specific model organisms, such as flies and mice [10–12].
ZFN is an artificially modified nuclease obtained by fusion of a zinc finger DNA binding domain and a DNA cutting domain from a nuclease [13]. The design of the zinc finger domain can achieve targeted cutting of a specific DNA sequence in the target gene [14], which also enables ZFN to locate the unique target sequence in a complex genome. By incorporating endogenous DNA repair mechanisms, ZFN can be utilized to accurately modify the genomes of higher organisms. However, more effective techniques for gene editing are urgently needed as a result of the high cost and lengthy production time required to design ZFNs. Compared with traditional gene manipulation techniques, this technique has obvious advantages, especially the breakthrough of achieving site-specific integration, but its main disadvantages are cytotoxicity, a high miss rate and a need for multiple screening steps.
TALEN is a restriction enzyme that has been genetically engineered to cut precise DNA sequences. The actual process is to fuse a TAL effector DNA binding domain with a DNA cleavage domain from a nuclease. Terence can be designed to bind to any desired DNA sequence, so the DNA can be cut at a specific location. Talen has unique advantages over traditional ZFN technologies, namely, a simpler design and higher specificity. The main disadvantages include a certain cytotoxicity and cumbersome module assembly processes.
The CRISPR/Cas system was first discovered in bacteria and archaea, where it functions as a form of adaptive immunity against viruses [15–20]. Overall, Cas proteins typically recognize a small protospacer-adjacent motif (PAM) present in invading DNA [21]. After PAM recognition, a downstream stretch of DNA (approximately 20 bp), called the protospacer, is copied from the exogenous DNA and entered into the CRISPR array for transcription into short CRISPR RNA (crRNA) [22]. These then anneal to the transactivating crRNAs (tracrRNAs) already present in the cell, forming stem–loop structures to allow Cas enzymes to bind [22]. The RNA complex then acts as a guide for the Cas endonuclease, initiating sequence-specific cleavage of the foreign DNA, generating DSBs and silencing the pathogen [22]. Once CRISPR/Cas forms DSBs in DNA, cells typically undergo two major repair mechanisms. One, NHEJ, is a DNA ligase that joins two broken DNA ends together to rejoin the sequence. During this process, one or more nucleotides are typically inserted and/or deleted at the DSB site. Thus, NHEJ can result in DNA frameshifts or nonsense mutated DNA that alter the amino acid sequence. Repair by homology-directed repair (HDR) is also possible. Second, HDR is an error-free repair pathway in which a homologous DNA template, usually obtained from a gene copy or homologous gene, is inserted to populate the DSB and recombine with the original DNA sequence [23], allowing insertion of specific DNA sequences. This facilitates overexpression studies as it allows targeted gene insertion at precise DNA locations. Therefore, CRISPR/Cas can effectively induce gene mutations and is widely used in the functional study of single genes, and it can also allow precise insertion of targeted genes into DNA, which is helpful for the study of gene expression. In conclusion, CRISPR, as an efficient, fast, simple and inexpensive intracellular knockout technology, has higher specificity, higher efficiency and simpler operation in binding to DNA target sequences than previous gene editing technologies [24], easier access to homozygous mutants and allowing for the simultaneous introduction of multiple mutants at different sites (Fig. 1).
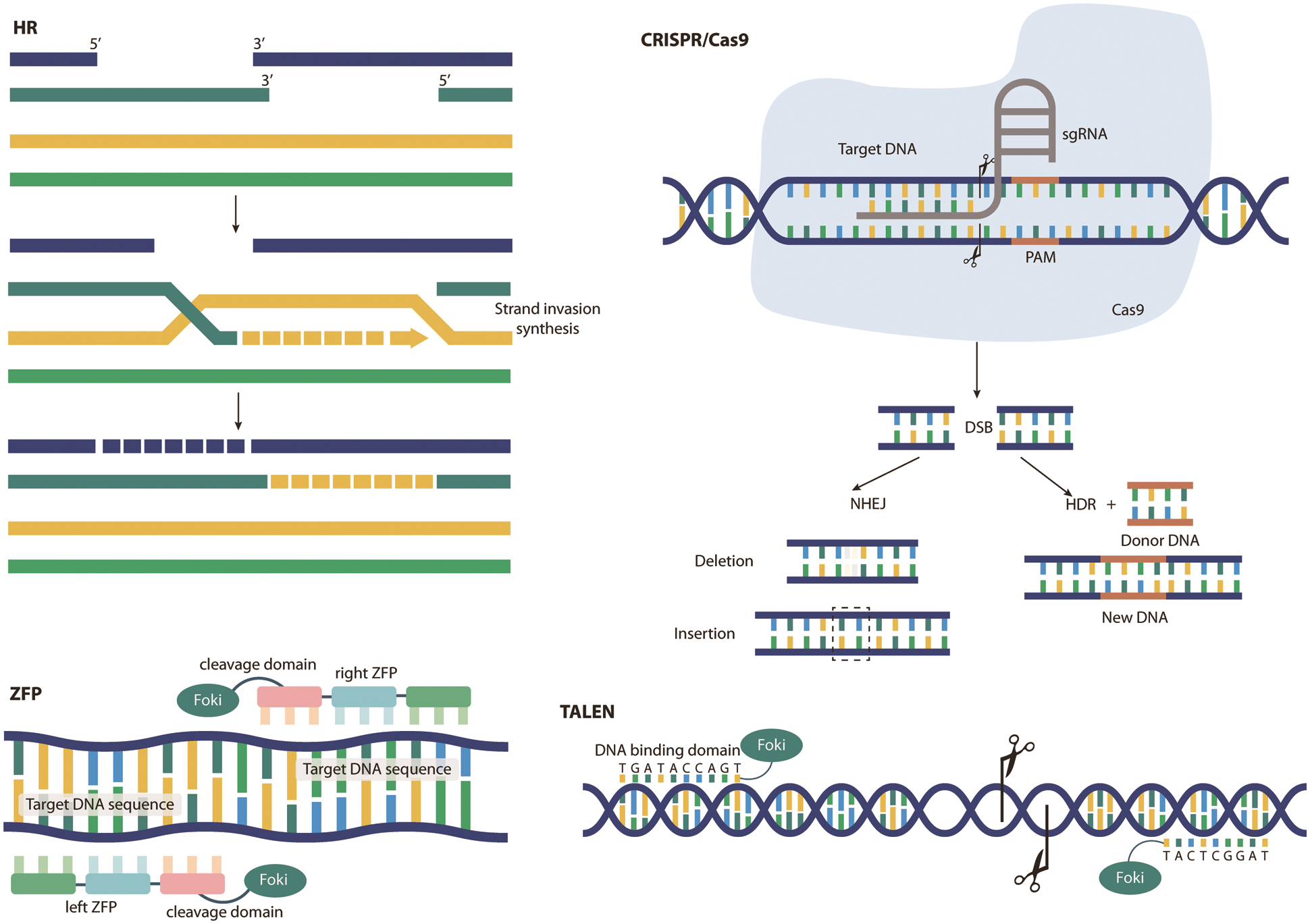
Figure 1: Principles of different gene editing techniques. As an emerging genetic engineering technology that can modify specific target genes of an organism’s genome, gene editing technology mainly includes homologous recombination (HR), zinc finger nucleases (ZFN), transcription activators (TALEN) and regular spacing short harmonic sequence repeats (CRISPR). The principles of these techniques are shown in this figure
Although gene modification-mediated gene therapy still faces many problems, such as off-target effects, the development of this technology has undoubtedly brought hope for the treatment of many diseases. In recent years, researchers have used gene editing for molecular targeted intervention therapy of tumors [25], which shows the great potential of these gene editing tools for the treatment of diseases.
2.2 Mechanism of Gene Editing in Non-Small Cell Lung Cancer Treatment
At present, gene editing technology has been implemented in the clinical treatment of NSCLC. Depending on the different types of lung cancer, different editing strategies have been adopted. Currently, its participation in the treatment of NSCLC mainly includes the following three methods: gene knockout, special targets in the regulatory signaling pathways and combined effects with other therapeutic methods. Currently, gene editing technology can be used to knock out the PD-1 gene of T cells to enhance their ability to attack tumor cells or by inhibiting a specific target in a signaling pathway related to the occurrence of NSCLC to achieve a therapeutic effect against NSCLC. However, gene editing technology is currently most often used in clinical practice in combination with chemotherapy, medicinal plant therapy or other therapeutic methods to achieve therapeutic effects.
2.2.1 Oncogenes/Suppressor Genes Are Subjected to Gene Knockout
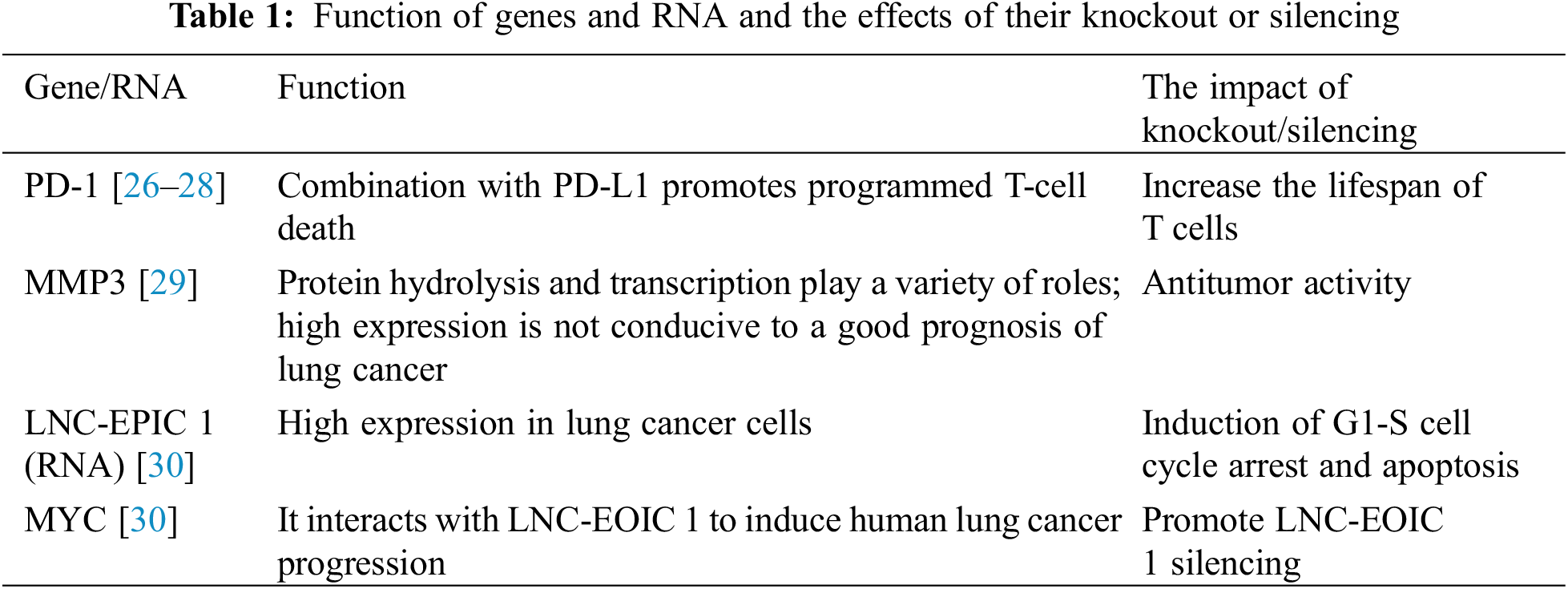
To date, in a clinical trial conducted by Sichuan University, CRISPR/Cas9 technology has been shown to be an effective intervention for lung cancer [26], and the effect of CRISPR–Cas9 PD-1-edited T cells in patients with advanced non-small cell lung cancer have been tested. The results of the first-in-human phase I clinical trial proved that it is generally safe and feasible in clinical application [27]. The approach is to isolate T cells, use CRISPR–Cas9 technology to knock out the PD-1 gene in the T cells, expand the T cells in vitro, and then return the T cells to the patients. Since PD-1 is a gene expressed in T cells and its encoded protein can promote T-cell programmed cell death by binding to the corresponding receptor (PD-L1), knocking out the PD-1 gene increases the lifespan of T cells and enhances their ability to attack tumor cells to treat lung cancer [28].
Matrix metalloproteinase 3 (MMP3) plays multiple roles in extracellular proteolysis and intracellular transcription. At present, studies have found that CRISPR/Cas9-mediated MMP3 gene knockout has an obvious antitumor effect, inhibiting the migration and invasion of tumor cells and reducing the activity of the CCN2/CTGF promoter and causing in vitro fragmentation. At the same time, experimental studies have proven that the high expression of MMP3 or CCN2/CTGF mRNA is detrimental to the prognosis of patients with lung cancer [29]. Therefore, knocking out the MMP gene will be a promising new route for the treatment of lung cancer.
Studies have shown that MYC gene knockout using CRISPR/Cas9 gene editing can block the silencing or overexpression of long noncoding RNA EPIC1 (LNC-EPIC1) to induce the death of human lung cancer cells [30]. LNC-EPIC1 is an lncRNA that interacts with MYC, and its expression in human lung cancer/primary lung cancer cells is significantly higher than that in human lung epithelial cells. By silencing LNC-EPIC1 with targeted siRNA, LNC-EPIC1 significantly inhibited the growth, survival and proliferation of human lung cancer cells and induced G1-S cell cycle arrest and apoptosis. Moreover, the targets of MYC, such as cyclin A1, CD 20 and CD 45, can be downregulated by LNC-EPIC1 siRNA. The experimental results suggest that Lnc-EPIC1 may promote the growth of human lung cancer cells by targeting MYC, which provides an approach to lung cancer treatment (Table 1).
2.2.2 Suppression of Specific Targets in the Key Signaling Pathways
In addition, gene editing technology can be employed to regulate specific targets in the corresponding signaling pathways to participate in the treatment of lung cancer. In the PTEN/Akt signaling pathway, the expression of miR-92a inhibited the expression of PTEN, which regulated the activity of the PI3K/Akt pathway. In the WNT/β-catenin (CTNNB1) pathway, MUC1-C silencing was achieved by genome editing to inhibit MYC and its target genes. In addition, in the GRM8/MAPK signaling pathway, suppressing the transmission of TGF-β signaling by gene editing can inhibit the occurrence and spread of non-small cell lung cancer (NSCLC) and reduce the resistance of NSCLC to targeted drugs and anticancer drugs so they can play a better role in the treatment progress.
The PTEN/Akt Signaling Pathway
PTEN, as a member of the protein tyrosine phosphatases (PTP) gene family, is an important antioxidant that negatively regulates the activity of the PI3K/Akt signaling pathway, thus affecting the proliferation, apoptosis, autophagy and other biological activities of tumor cells. Zhao et al. [31] reported that an anticancer drug could induce autophagy and apoptosis in lung cancer cells by blocking the activity of this pathway, thereby inhibiting the growth of tumors. Further studies have confirmed [32,33] that blocking this pathway can promote autophagy in tumor cells and activate this pathway to inhibit autophagy in tumor cells. Overexpression of miR-92a can inhibit PTEN expression, thereby regulating PI3K/Akt signaling activity and promoting NSCLC cell migration, invasion and tumor growth [34].
WNT/β-catenin (CTNNB1) Pathway
Wnt signaling pathways consist of three types: the Wnt/Ca2+ signaling pathway, the cell plane polarity pathway, and the classical Wnt signaling pathway. The classical Wnt signaling pathway is closely related to tumor formation, and its regulation is mediated by the phosphorylation/degradation of cytoplasmic catenin [35]. In the absence of a Wnt signal, catenin remains in the cytoplasm at a low level and cannot activate nuclear targets. In the presence of a Wnt signal, catenin could not continue to be phosphorylated, and it could accumulate in the cytoplasm and then enter the nucleus to participate in tumor epithelial-mesenchymal transition (EMT) and metastasis.
Dysregulation of MYC expression is a hallmark of cancer. Studies have shown that MUC1-C is a common cancer transmembrane protein and can induce the expression of MYC in KRAS mutant NSCLC. This effect can be a result of targeting MUC1-C by shRNA silencing, CRISPR editing or GO-203 pharmacological inhibition. MUC1-C activates the WNT/CTNNB1 pathway [36] and promotes the expression of the MUC1-C/catenin/TCF4 complex of MYC promoters. At the same time, MUC1-C also promotes the recruitment of P300 histone acetylase (EP300), thus inducing histone H3 acetylation and MYC gene transcription activation. In addition, studies have found that targeting MUC1-C also reduces the expression of key MYC target genes crucial for the growth and survival of NSCLC cells (such as TERT and CDK4) [37]. In human KRAS mutant NSCLC tumors, the expression of MUC1 is substantially correlated with MYC and its target genes. Therefore, MUC1-C silencing by CRISPR genome editing can inhibit the expression of MYC and its target genes in A549 and H460 cells. Thus, the purpose of treating lung cancer is achieved [38].
Other Pathways
The TGF-β signaling pathway plays a major role in the occurrence and development of NSCLC. It promotes the invasion and metastasis of NSCLC by promoting epithelial-mesenchymal transformation and angiogenesis and is closely linked to the drug resistance of NSCLC. EMT induced by TGF-β plays a major role in targeted therapy and chemotherapy against NSCLC. Asakura et al. [39] showed that TGF-β signaling could positively regulate ZEB1 and negatively regulate E-cadherin expression. This causes EMT in EGFR-mutated lung cancer cell lines and antagonizes the effect of some chemotherapy drugs. Existing studies have shown that miRNAs can affect resistance to targeted inhibitors by regulating the TGF-β-EMT axis. The inhibitory effect of miR-181b on TβR-I antagonizes both classical and nonclassical downstream signaling of TGF-β and promotes chemical sensitization [40]. In addition, Sato et al. [41] showed that overexpression of miR-200 could restore the sensitivity of NSCLC cell lines to EGFR inhibitors and was associated with the reduction of ZEB1 mediated by miR-200. Gene regulation of the TGF-β signaling pathway can inhibit the spread of NSCLC and alter the drug resistance of NSCLC, thereby playing a more effective role in treatment progression. Therefore, targeted inhibition of the TGF-β signaling pathway can play an important role in the treatment of lung cancer and has promising clinical applications (Fig. 2).
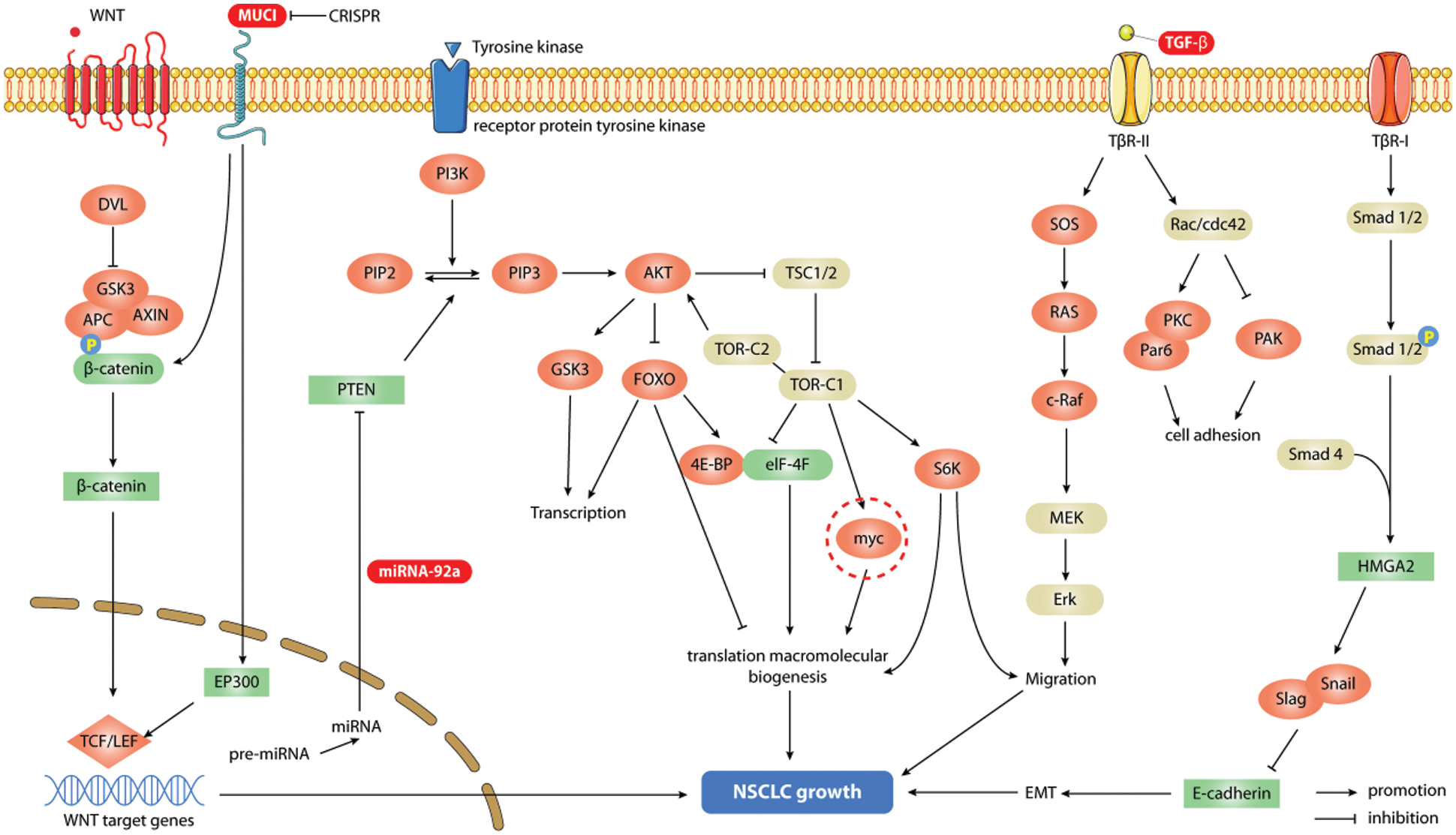
Figure 2: Signaling pathways associated with gene editing therapy for lung cancer. Gene editing technology can regulate specific targets in corresponding signaling pathways to participate in the treatment of lung cancer. In the PI3K/AKt signaling pathway, the overexpression of miR-92a by gene editing technology can inhibit the expression of PTEN, which regulates the activity of the pathway, and promotes the migration, invasion and tumor growth of NSCLC cells; in the WNT/β-catenin (CTNNB1) pathway, the gene editing silencing of MUC1-C can inhibit the expression of MYC and its target genes to achieve the purpose of treating NSCLC; while the inhibition of TGF-β signaling by gene editing can inhibit the cascade reaction of its pathway, which can eventually lead to the spread of NSCLC
2.3 Gene Editing and Other Combinations to Treat Non-Small Cell Lung Cancer
Compared with gene knockout and regulation of special targets, the current clinical use of gene editing technology is usually via combining it with other treatments. The most commonly used combination is gene editing plus chemotherapy, which can enhance the drug sensitivity of lung cancer cells and improve the antitumor effect. Compared with the former, BRM270, as a medicinal plant, in combination with gene editing can greatly reduce the toxicity and side effects of treatment. The combination of gene editing and immunotherapy is the most rapidly emerging curative method in recent years. It has concentrated targeting and can specifically enhance the immune response. In addition, gene editing could be associated with a variety of other therapies to improve survival in patients with certain types of non-small cell lung cancer (Table 2).
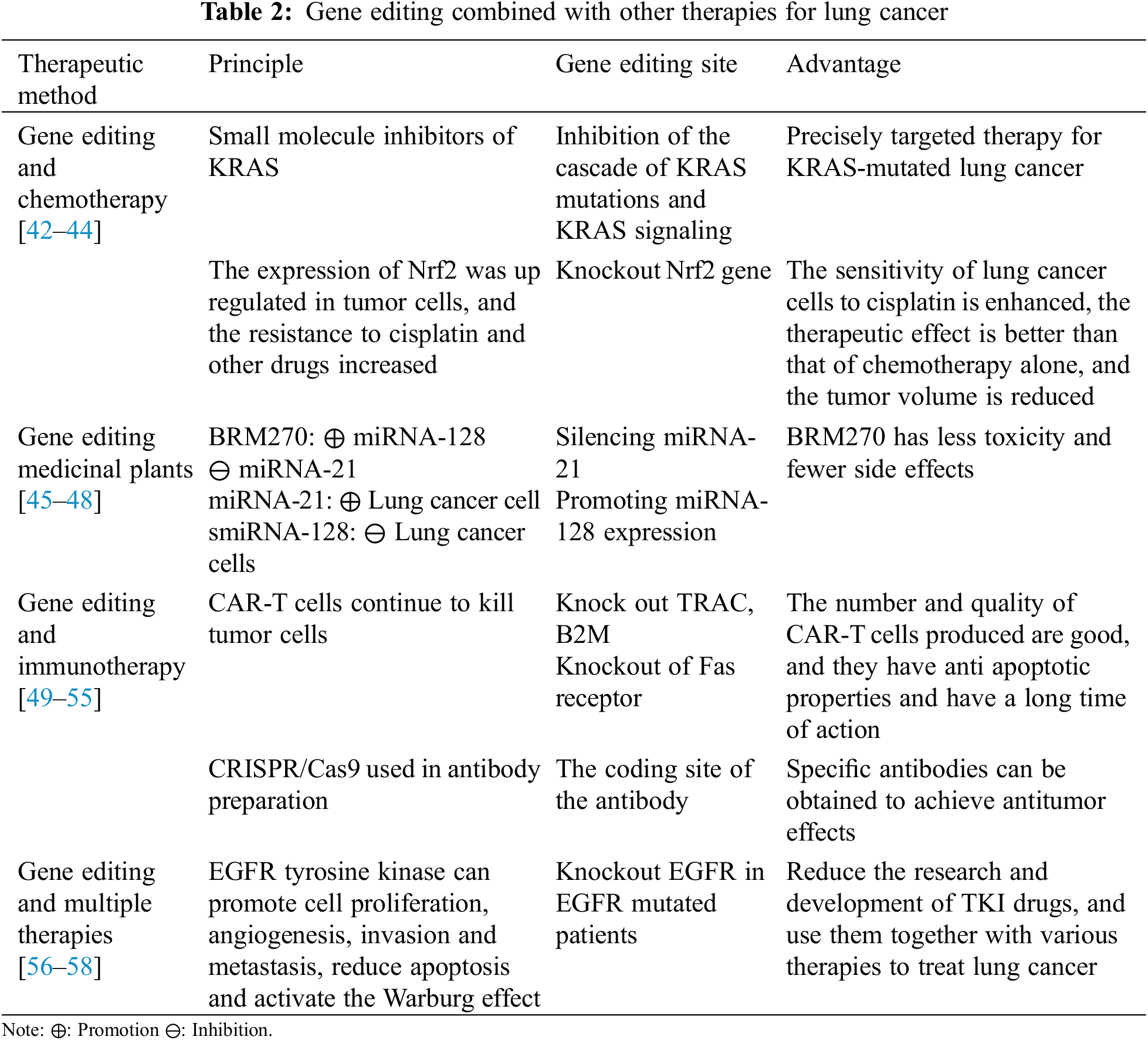
2.3.1 Combination Therapy of Gene Editing and Chemotherapy
Currently, chemotherapy remains an important treatment for lung cancer. KRAS mutation is one of the most common gene mutations in NSCLC, and inhibitors have been used to inhibit mutated KRAS and oncogenic KRAS signaling in recent decades. Today, KRAS small molecule inhibitors are entering the clinic. Their tight integration with gene editing technology enhances the precise targeting of cancer cell therapy as a way to combat KRAS-driven refractory cancers [42,43].
The body’s resistance to chemotherapy drugs may increase over time. A recent study found that nuclear factor red pigment 2-related factor (Nrf2) is an important transcription factor regulating the cellular oxidative stress response and is an important regulator involved in maintaining internal REDOX homeostasis. Chemotherapeutic drugs have been shown to activate the transcriptional activity of the Nrf2 gene and upregulate Nrf2 expression. This increases the resistance of cells to chemotherapeutic drugs. CRISPR/Cas9 technology was used to knock out the Nrf2 gene in lung cancer cells, which inhibited their expression of functional proteins. At the same time, the knockout lung cancer cells were more sensitive to chemotherapy drugs such as display, carboxylates, and chlororubin [44]. Therefore, the combination therapy of Nrf2 gene knockout in lung cancer cells and chemotherapeutic drugs can be regarded as a promising lung cancer treatment method.
2.3.2 Gene Editing in Combination with Medicinal Plant Therapy
The upregulated expression of miRNA-21 in NSCLC can promote the growth and invasion of lung cancer cells. However, the expression of miRNA-128 in lung cancer is inhibited due to its inhibitory effects on lung cancer cells [45,46]. BRM270 is an extract from seven Asian medicinal plants [47] and is a nontoxic alternative to chemotherapy, which has been shown to inhibit the growth of various tumor cells. It can enhance the expression of miRNA-128 in lung adenocarcinoma A456 cells and inhibit the expression of miRNA-21 [48]. Moreover, BRM270 was also found to regulate the self-renewal and tumor initiation of cancer stem cells through positive regulation of miRNA-128. Therefore, the combination of BRM270 and miRNA-128 can achieve the purpose of treating NSCLC more effectively, which will play an important role in clinical applications.
2.3.3 Gene Editing in Combination with Immunotherapy
Car-T therapy (chimeric antigen receptor-T-cell therapy) is immunotherapy currently approved for the treatment of acute lymphoblastic leukemia and non-Hodgkin’s lymphoma [49,50]. Car-T cells are autologous T cells that carry receptors for genetically modified antigens. Traditional Car-T-cell production for clinical trials has certain limitations, and high-quality T cells from healthy donors are urgently needed to manufacture Car T cells. CRISPR/Cas9 technology provides multiple knockout of TRAC and β2 M for such T cells. Technical support and an operation method for microglobulin (an important subunit of HLA) [51] are needed. Additionally, CRISPR/Cas9 was utilized to knockout the anti-apoptotic Fas receptors Car-T cells can produce, thus prolonging their action time and increasing their function [52]. Therefore, a combination of gene editing technology and CAR-T-cell therapy can be invoked as a therapeutic means to treat tumors [53].
The study found that CRISPR/Cas9 plays a role in the preparation of antibodies. Some researchers have used CRISPR/Cas9 technology to achieve IgM-IgG-IgA antibody type conversion [54]. Therefore, we propose that this technology can be used to encode specific antibody genes of immune cells to obtain specific types of antibodies to achieve antitumor activity [55].
2.3.4 Gene Editing in Combination with Other Treatments
Under the premise of the existing technology, for individual EGFR-mutated NSCLC patients, the virus-transmitted CRISPR/Cas system can be used to repair or destroy the EGFR-mutated gene to achieve therapeutic purposes. The most common real-life cases of primary and secondary EGFR mutations demonstrate the feasibility of this approach [56]. This personalized molecular surgery utilizing gene editing technology directly targets the etiology of the disease and is often combined with traditional surgery, radiation therapy or chemotherapy/targeted therapy to achieve the therapeutic effect of “1 + 1 > 2” [57,58].
3 Gene Editing in Other Approaches to Non-Small Cell Lung Cancer
In addition to clarifying the pathogenesis of non-small cell lung cancer and playing a role in its treatment, gene editing technology can also be used to establish animal models of NSCLC, diagnose and screen for early clinical NSCLC, monitor the effects after treatment, explore the mechanism of action of drugs and promote the development of new anticancer drugs.
3.1 Gene Editing Can Be Used to Construct Animal Models of Non-Small Cell Lung Cancer
Gene editing technology can be used to construct experimental animal NSCLC models, which provides more convenience for NSCLC-related experiments. The establishment of an experimental animal NSCLC model is conducive to further study of the etiology and pathogenesis of NSCLC to explore therapeutic methods. Meanwhile, it is also conducive to the study of the effectiveness of certain therapeutic means or therapeutic drugs to obtain the best treatment plan. At present, there are three main methods that have been used to apply gene editing technology to the construction of animal models of NSCLC.
The first one was a mouse model of the conditional expression of Cas9 constructed by Zhang Feng and his team [59]. The researchers selected KRAS, P53 and LKBl genes with the most significant mutation effects in lung adenocarcinoma, introduced sgDNA (single guided DNA) of P53 and LKBl genes into Cas9 mice using AAV vectors, and introduced Kras (Gl2D) gene mutations using homologous recombination-mediated repair. After the mutation, tumor nodules appear in the lungs of mice, and they increase over time and finally achieve the purpose of building a mouse lung cancer model [60]. This effectively overcomes the difficulty of introducing Cas9 into adult mice, simplifies the steps of gene editing in vivo, and improves the editing efficiency.
The second was to fuse the EML4 and ALK genes by using gene translocation caused by chromosome 2 breakage of tumor cells in NSCLC and applied to study the mechanism of the potential antigenic effect of the fused genes [61]. The research team of Maddalo et al. [62] adopted CRISPR editing technology to fuse EML4-ALK and express it in tumor cells, thus building an in vitro model of NSCLC, which is conducive to experimental studies on the pathogenesis of NSCLC and the efficacy of relevant drugs [62]. This technique is cheaper and faster than genetic targeting to build tumor models in experimental animals.
The third was constructed by Sanchez-Rivera et al. [63] Who injected CRISPR into the lung tissues of mice with retrovirus as the vector and achieved the construction of a mouse lung cancer model carrying the Kras oncogene. The animal model was constructed to facilitate the study of Kras-mutated lung cancer, which will allow for subsequent research on the treatment of this lung cancer.
Traditionally, to study the tumor suppressor or carcinogenic role of a protein in lung cancer, laboratories have had to generate new strains of mice for each protein, an extremely time-consuming and laborious task. Therefore, the introduction of CRISPR gene targeting enables the realization of the 3R principles of in vivo experiments: it can significantly reduce the total number of experimental animals required and the overall cost of animal generation, optimize existing experimental animal modeling methods, and expand the application of CRISPR to various model systems. These organ model systems can replace in vivo tumor models in the future.
3.2 Application of Gene Editing in the Screening/Diagnosis of Non-Small Cell Lung Cancer
Gene editing technology also plays a major role in the screening and diagnosis of lung cancer. Ubiquitin is a protein modifier and can play a role in posttranslational modification. Ubiquitin-specific peptidase 24 (USP24) was found by gene editing to be highly expressed in tumor-enhanced cell lines and advanced lung cancer tissues, especially the expression level of USP24-930T/USP24-7656C, which was significantly increased [47,64]. By analyzing the variations of USP24, it can be found that its mutation rate in patients with lung cancer is higher than that in normal people, so USP24-930T/USP24-7656C can be used as a predictor of a lung cancer diagnosis, which suggests it may have use in comprehensive screening for lung cancer or have utility in monitoring people at high risk of lung cancer and early lung cancer patients to provide more timely treatment, extend their length of life and improve their quality of life.
3.3 Gene Editing Can Predict Prognosis
The use of gene editing technology to determine the prognosis of patients with lung cancer is convenient for clinicians to more accurately grasp the patient’s physical condition and provide more information for subsequent treatment and program selection [65,66]. Currently, the zygote arrest protein 1 (ZAR1) methylation level and changes in microRNA editing can be used as critical indicators of the prognosis of lung cancer.
ZAR1 is known to play a role in lung cancer through epigenetic inactivation. ZAR1 promoter methylation can occur throughout CpG islands, and its hypermethylation is closely associated with its decreased expression in cancer. Researchers used CRISPR/Cas9 to reactivate ZAR1 to study the role of ZAR1 in human cancer. It was found that ZAR1 is a novel lung cancer biomarker that can be silenced by DNA methylation in various cancers. In addition, ZAR1 exerts its tumor-suppressive function through p53 and zinc finger domains [67]. Therefore, the prognosis of lung cancer patients can be judged by detecting the level of ZAR1 methylation.
Adenosine to inosine (A to I) microRNA editing is well known to be associated with tumor phenotypes in various cancer types. By comparing the small RNA in-depth sequencing data of 74 publicly released lung adenocarcinoma (AD) and corresponding normal corresponding (NC) specimens with the latest analysis of the Cancer Genome Atlas (TCGA) dataset, researchers confirmed that changes in the editing level of microRNAs in lung adenocarcinoma could be used as a potential biomarker [68]. Then, by improving the quantification of the editing level, some researchers clarified the relationship between the deletion of miR-99a-5p in cancer tissues and the poor prognosis of surgically resected lung adenocarcinoma specimens; that is, in the existing patient cohort, its deletion was associated with a poor prognosis after complete resection of lung adenocarcinoma [49]. Therefore, changes in microRNA editing in lung adenocarcinoma can be utilized as a potential biomarker to predict the outcomes of patients [69,70].
Glutamate receptor 8 (GRM8) is one of the G-protein-coupled receptors of the glutamate family, which can bind with various intracellular second messengers to regulate the excitability and development of neurons. Studies have found through gene editing technology that GRM8 mediates a reduction of intracellular cAMP concentration by inhibiting the activity of adenylate cyclase, and the amplification of GRM8 can promote the progression of lung squamous cell carcinoma (LUSC) [71]. The experimental results showed that the activation of GRM8 significantly reduced the intracellular cAMP concentration and PKA activity, and the activated cAMP pathway could block the activation of the MAPK pathway, which is the main pathway for intracellular transmission of proliferation and survival signals [72]. They also showed that the phosphorylation levels of MEK and ERK, key molecules in the MAPK pathway, were significantly enhanced after GRM8 activation. Therefore, transcriptional activation of GRM8 promotes LUSC tumor cell proliferation and survival by inhibiting the cAMP pathway and activating the MAPK pathway. Amplification of GRM8 enhances the transcription level of GRM8. This enables GRM8 to act as an activated variant and promote the development of LUSC. Through gene editing technology, the pathogenesis of lung cancer needs to be further explored to identify more preventive measures against lung cancer in the future (Fig. 3).
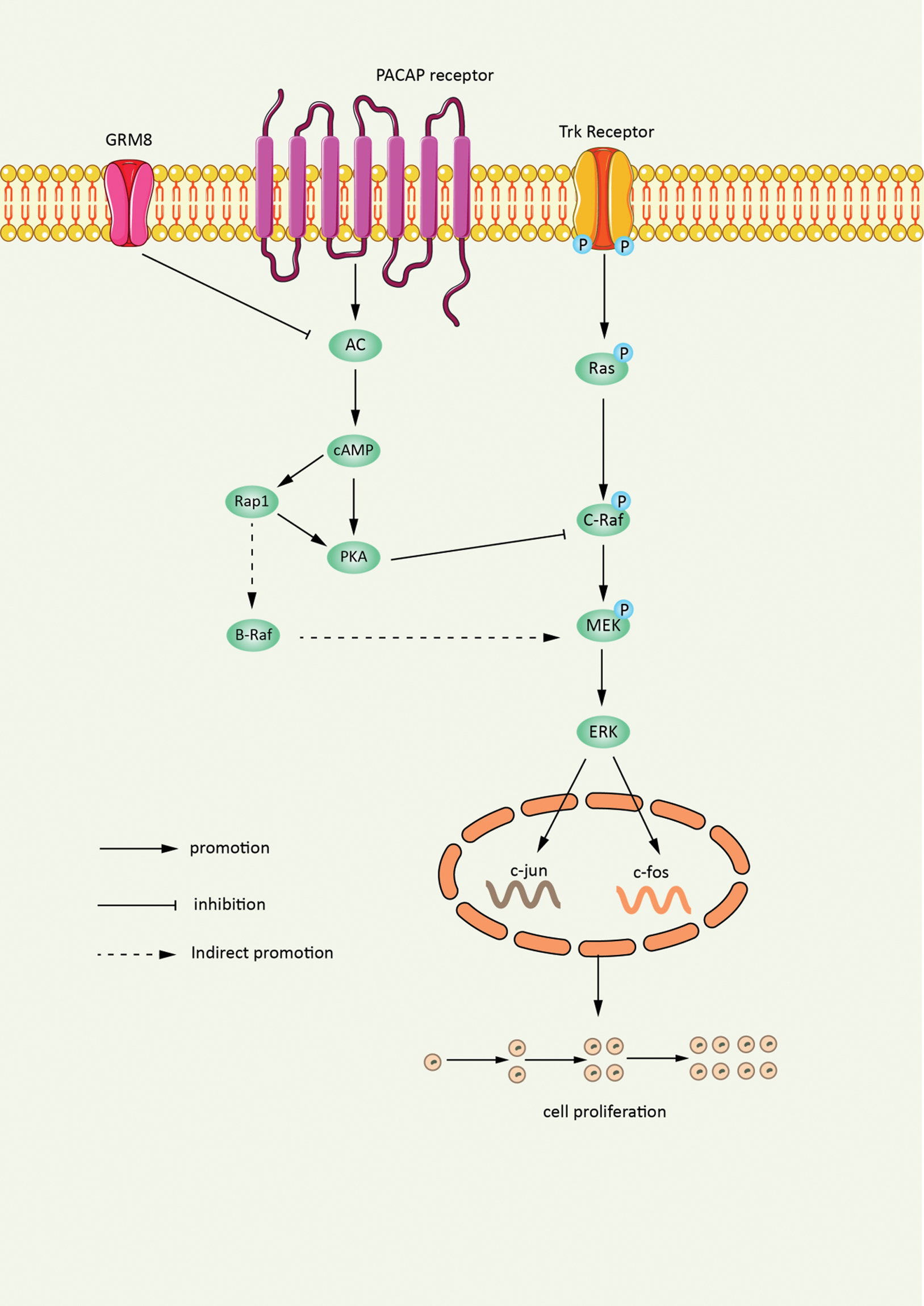
Figure 3: Gene editing amplification of GRM8 promotes tumor proliferation. Gene editing technology enables transcriptional amplification of GRM8. The cAMP pathway is inhibited while the MAPK pathway is activated, which ultimately promote the proliferation and survival of LUSC tumor cells
Gene editing technology can also be employed to study the drug resistance mechanism of NSCLC. Researchers using CRISPR/Cas9 technology inserted the EGFR T790 M mutation into the drug-resistant pc9 human lung cancer cell line and found that its drug resistance was increased [73]. Therefore, CRISPR/Cas9 can be used to endow all kinds of cancer cell lines with specific mutations, which is of profound significance for research on the mechanism of anticancer drug resistance and drug efficacy.
In addition, gene editing techniques can be used to explore tumor development. Compared with NSCLC, small cell lung cancer (SCLC), another highly aggressive subtype of lung cancer, is one of the deadliest solid tumors [74], with rapid initial growth and extensive metastasis, so most SCLC patients often present with systemic disease at the time of diagnosis [75]. The researchers used the CRISPR/Cas9 system to establish a good SCLC model to rapidly detect functional missing mutations in candidate genes. In-depth studies found that the deletion of P107 (also known as Rbl1) repeatedly occurs in SCLC, which can significantly promote the development of tumors, and the deletion of P107 can be used to simulate the occurrence and development of tumors [76].
Gene editing technology is also commonly used to create specific cell lines for different types of NSCLC. For example, CRISPR/Cas9 gene editing technology was used to construct A549 cell lines with ubiquity C-terminal hydrolase L1 (UCHL1) gene knockout to explore the effect of UCHL1 on A549 cells in NSCLC [77] (Fig. 4).
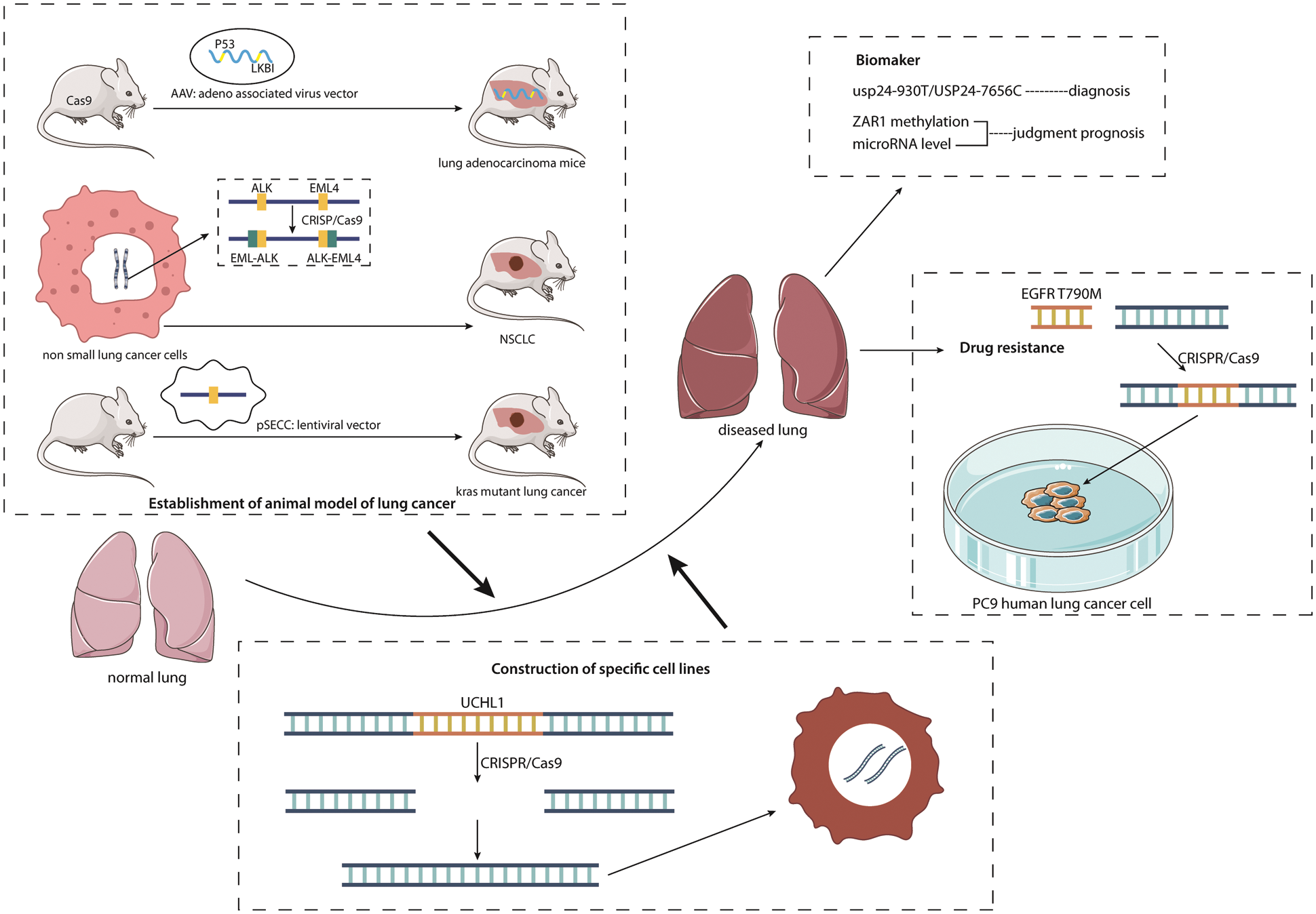
Figure 4: Multiple applications of gene editing in lung cancer. The figure lists the different applications of gene editing technology in NSCLC. It can be seen that there are three methods for gene editing technology to establish experimental animal NSCLC models; gene editing is used to discover new biomarkers and apply them to NSCLC screening, diagnosis and prognosis; gene editing technology is also used for NSCLC drug resistance research and the construction of specific cell lines for different types of NSCLC
In recent years, the morbidity and lethality of NSCLC have been increasing, and it has become one of the most common cancer types, posing a major threat to people’s lives and health. Currently, gene editing is one of the hottest fields in cancer research and has important application prospects in the diagnosis, screening and treatment of lung cancer. The occurrence and development of cancer are closely related to genetic changes. Compared with traditional cancer treatment, gene therapy can treat cancer more thoroughly and fundamentally. Its targeting is restricted. At the same time, it also has the advantages of fewer side effects and an obvious effect of improving patient prognosis. In addition, targeted modification of endogenous loci using gene editing technology can edit mutated genes directly or regulate signaling pathways, resulting in better therapeutic effects.
Nevertheless, there are some drawbacks of gene editing. The first is the difficulty of design and the cost. Even though the use of ZFNS and TALEN simplifies the work of gene editing, there are still difficult, time-consuming and laborious problems [78]. Second, there is a nontarget effect. Due to the similarity between target sites and nontarget sites, ZFNS often recognizes nontarget sites [79]. Numerous nontarget loci of TALEN were also found [80]. CRISPR/Cas9 can induce higher mutation rates in nontarget cells [81]. Third, gene editing technology has improved significantly in recent years in terms of efficiency and delivery. CRISPR/Cas9 has become the method of choice for generating knockout and knockin mutants in a wide variety of species, and the technology has also been successfully applied to a variety of cell lines but is limited due to poor dissemination of primary cells and difficulties in transfection. Its application in primary cells is currently limited [82]. Early studies using CRISPR/Cas9 for gene editing in human primary T cells have attempted viral delivery using Cas9 and gRNA [83] or by electroporation of gRNA/Cas9 expression constructs for transfection [84]. These methods all show low targeting efficiency, and electroporation of DNA is also highly toxic to T cells. In recent years, the electroporation method of the complex of Cas9 ribonucleoprotein (RNP), recombinant Cas9 and in vitro transcribed or synthesized single guide RNA (sgRNA) has been used to transfect activated human T cells, and the targeted transfection rate has been improved. However, the scope of application cannot be expanded, and there are many limitations. Therefore, the effect of gene editing and therapy is currently limited [85]. Moreover, the low rate of homologous directed recombination (HDR) limits the CRISPR/Cas9 system [86].
Finally, there is the issue of the ethics and the regulation of gene editing. On the one hand, gene editing technology offers a viable prospect for the treatment and prevention of human diseases. However, the International Bioethics Committee (IBC) of UNESCO points out that the application of gene editing technology should be restricted to prevention, diagnosis and treatment procedures and should not be used to alter human embryos. At the same time, scientists recognize that the use of CRISPR/Cas9 in human diseases is not entirely safe and effective [87]. These problems have brought challenges to the wider application of gene editing technology in future development. One of the greatest challenges of gene editing technology at present is to deliver genome editing molecules directly, efficiently and safely to target only specific cells [87]. The problems mentioned above should be avoided as far as possible in animal experiments or clinical applications, but the author believes that more in-depth research is needed in order to solve these problems and challenges to obtain more effective gene editing techniques.
In addition, most of the drugs used to treat non-small-cell lung cancer are administered intravenously and work in patients but are affected by poor pharmacokinetics. There are some limitations to their role. Studies are currently being reported of the development of CRISPR/Cas9 inhaled therapeutics [88], which are expected to be used to treat lung diseases. Its advantage is that it can be delivered directly and rapidly to target cells in the lung at high concentrations, thereby reducing systemic exposure [89]. However, due to the complexity of the treatment process, the specific clinical application remains to be studied, but this provides a new development direction for gene editing technology in the treatment of non-small-cell lung cancer [90].
With the continual development of gene editing technology, it is believed that it will play a more significant role in the prevention and treatment of non-small cell lung cancer in the future.
Authorship: Hangxing Wang, Jingyun Fang and Yujiao Wang wrote the paper. Shuo Li and Zirui Wang edited the figures. Wei He, Nan Wang and Shuang Luo edited the tables and revised the manuscript accordingly. Huimei Zou and Fan Zhang obtained the funding support and managed the project. The authors declare that all data were generated in-house and that no paper mill was used.
Ethics Approval and Informed Consent: Not applicable.
Funding Statement: The current work was supported by the Academic Seedling Project of National Natural Science Foundation of China (19NSP031), the Natural Science Foundation of Guizhou Province, China (QianKeHeJiChu-ZK[2022] General 409), and the Youth Talent Development Project of Guizhou Education Department (Qian Jiao He [2021]177).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Cao, M., Chen, W. (2019). Epidemiology of lung cancer in China. Thoracic Cancer, 10(1), 3–7. DOI 10.1111/1759-7714.12916. [Google Scholar] [CrossRef]
2. Qiu, Y., Li, H., Xie, J., Qiao, X., Wu, J. (2021). Identification of ABCC5 among ATP-binding cassette transporter family as a new biomarker for hepatocellular carcinoma based on bioinformatics analysis. International Journal of General Medicine, 7(14), 7235–7246. DOI 10.2147/IJGM.S333904. [Google Scholar] [CrossRef]
3. Bade, B. C., Dela Cruz, C. S. (2020). Lung cancer 2020: Epidemiology, etiology, and prevention. Clinics in Chest Medicine, 41(1), 1–24. DOI 10.1016/j.ccm.2019.10.001. [Google Scholar] [CrossRef]
4. Remark, R., Becker, C., Gomez, J. E., Damotte, D., Dieu-Nosjean, M. C. et al. (2015). The non-small cell lung cancer immune contexture. A major determinant of tumor characteristics and patient outcome. American Journal of Respiratory and Critical Care Medicine, 191(4), 377–390. DOI 10.1164/rccm.201409-1671PP. [Google Scholar] [CrossRef]
5. Zou, X., Wang, L., Li, Z., Luo, J., Wang, Y. et al. (2018). Genome engineering and modification toward synthetic biology for the production of antibiotics. Medicinal Research Reviews, 38(1), 229–260. DOI 10.1002/med.21439. [Google Scholar] [CrossRef]
6. Bak, R. O., Gomez-Ospina, N., Porteus, M. H. (2018). Gene editing on center stage. Trends in Genetics, 34(8), 600–611. DOI 10.1016/j.tig.2018.05.004. [Google Scholar] [CrossRef]
7. Tan, W. S., Carlson, D. F., Walton, M. W., Fahrenkrug, S. C., Hackett, P. B. (2012). Precision editing of large animal genomes. Advances in Genetics, 80, 37–97. DOI 10.1016/B978-0-12-404742-6.00002-8. [Google Scholar] [CrossRef]
8. Janssen, J. M., Chen, X., Liu, J. (2019). The chromatin structure of CRISPR Cas9 target DNA controls the balance between mutagenic and homology-directed gene-editing events. Molecular Therapy. Nucleic Acids, 16, 141–154. DOI 10.1016/j.omtn.2019.02.009. [Google Scholar] [CrossRef]
9. Puchta, H., Fauser, F. (2013). Gene targeting in plants: 25 years later. The International Journal of Developmental Biology, 57, 629–637. DOI 10.1387/ijdb.130194hp. [Google Scholar] [CrossRef]
10. Golic, K. G., Golic, M. M. (1996). Engineering the drosophila genome: Chromosome rearrangements by design. Genetics, 144(4), 1693–1711. DOI 10.1093/genetics/144.4.1693. [Google Scholar] [CrossRef]
11. Koller, B. H., Smithies, O. (1989). Inactivating the beta 2-microglobulin locus in mouse embryonic stem cells by homologous recombination. Proceedings of the National Academy of Sciences of the United States of America, 86(22), 8932–8935. DOI 10.1073/pnas.86.22.8932. [Google Scholar] [CrossRef]
12. Thomas, K. R., Capecchi, M. R. (1987). Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell, 51(3), 503–512. DOI 10.1016/0092-8674(87)90646-5. [Google Scholar] [CrossRef]
13. Carroll, D. (2011). Genome engineering with zinc-finger nucleases. Genetics, 188, 773–782. DOI 10.1534/genetics.111.131433. [Google Scholar] [CrossRef]
14. Pei, D., Corey, D. R., Schultz, P. G. (1990). Site-specific cleavage of duplex DNA by a semisynthetic nuclease via triple-helix formation. Proceedings of the National Academy of Sciences of the United States of America, 87, 9858–9862. DOI 10.1073/pnas.87.24.9858. [Google Scholar] [CrossRef]
15. Ishino, Y., Shinagawa, H., Makino, K., Amemura, M., Nakata, A. (1987). Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. Journal of Bacteriology, 169(12), 5429–5433. DOI 10.1128/jb.169.12.5429-5433.1987. [Google Scholar] [CrossRef]
16. Nakata, A., Amemura, M., Makino, K. (1989). Unusual nucleotide arrangement with repeated sequences in the Escherichia coli K-12 chromosome. Journal of Bacteriology, 171(6), 3553–3556. DOI 10.1128/jb.171.6.3553-3556.1989. [Google Scholar] [CrossRef]
17. Hermans, P. W., van Soolingen, D., Bik, E. M., de Haas, P. E., Dale, J. W. et al. (1991). Insertion element IS987 from mycobacterium bovis BCG is located in a hot-spot integration region for insertion elements in mycobacterium tuberculosis complex strains. Infection and Immunity, 59(8), 2695–2705. DOI 10.1128/iai.59.8.2695-2705.1991. [Google Scholar] [CrossRef]
18. Mojica, F. J., Díez-Villaseñor, C., García-Martínez, J., Soria, E. (2005). Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. Journal of Molecular Evolution, 60(2), 174–182. DOI 10.1007/s00239-004-0046-3. [Google Scholar] [CrossRef]
19. Jansen, R., Embden, J. D., Gaastra, W., Schouls, L. M. (2002). Identification of genes that are associated with DNA repeats in prokaryotes. Molecular Microbiology, 43(6), 1565–1575. DOI 10.1046/j.1365-2958.2002.02839.x. [Google Scholar] [CrossRef]
20. Mojica, F. J., Juez, G., Rodríguez-Valera, F. (1993). Transcription at different salinities of haloferax mediterranei sequences adjacent to partially modified PstI sites. Molecular Microbiology, 9(3), 613–621. DOI 10.1111/j.1365-2958.1993.tb01721.x. [Google Scholar] [CrossRef]
21. Gleditzsch, D., Pausch, P., Müller-Esparza, H., Özcan, A., Guo, X. et al. (2019). PAM identification by CRISPR-cas effector complexes: Diversified mechanisms and structures. RNA Biology, 16(4), 504–517. DOI 10.1080/15476286.2018.1504546. [Google Scholar] [CrossRef]
22. Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A. et al. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science, 337(6096), 816–821. DOI 10.1126/science.1225829. [Google Scholar] [CrossRef]
23. Heyer, W. D., Ehmsen, K. T., Liu, J. (2010). Regulation of homologous recombination in eukaryotes. Annual Review of Genetics, 44, 113–139. DOI 10.1146/annurev-genet-051710-150955. [Google Scholar] [CrossRef]
24. Gaj, T., Gersbach, C. A., Barbas III., C. F. (2013). ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends in Biotechnology, 31, 397–405. DOI 10.1016/j.tibtech.2013.04.004. [Google Scholar] [CrossRef]
25. Sachdeva, M., Sachdeva, N., Pal, M., Gupta, N., Khan, I. A. et al. (2015). CRISPR/Cas9: Molecular tool for gene therapy to target genome and epigenome in the treatment of lung cancer. Cancer Gene Therapy, 22(11), 509–517. DOI 10.1038/cgt.2015.54. [Google Scholar] [CrossRef]
26. Cyranoski, D. (2016). CRISPR gene-editing tested in a person for the first time. Nature, 539(7630), 479. DOI 10.1038/nature.2016.20988. [Google Scholar] [CrossRef]
27. Lu, Y., Xue, J., Deng, T., Zhou, X., Yu, K. et al. (2020). Safety and feasibility of CRISPR-edited T cells in patients with refractory non-small-cell lung cancer. Nature Medicine, 26(5), 732–740. DOI 10.1038/s41591-020-0840-5. [Google Scholar] [CrossRef]
28. Zhang, Y., Huang, S., Gong, D., Qin, Y., Shen, Q. (2010). Programmed death-1 upregulation is correlated with dysfunction of tumor-infiltrating CD8+ T lymphocytes in human non-small cell lung cancer. Cellular & Molecular Immunology, 7(5), 389–395. DOI 10.1038/cmi.2010.28. [Google Scholar] [CrossRef]
29. Okusha, Y., Eguchi, T., Tran, M. T., Sogawa, C., Yoshida, K. et al. (2020). Extracellular vesicles enriched with moonlighting metalloproteinase Are highly transmissive, pro-tumorigenic, and trans-activates cellular communication network factor (CCN2/CTGFCRISPR against cancer. Cancers, 12(4), 881. DOI 10.3390/cancers12040881. [Google Scholar] [CrossRef]
30. Zhang, B., Lu, H. Y., Xia, Y. H., Jiang, A. G., Lv, Y. X. (2018). Long non-coding RNA EPIC1 promotes human lung cancer cell growth. Biochemical and Biophysical Research Communications, 503(3), 1342–1348. DOI 10.1016/j.bbrc.2018.07.046. [Google Scholar] [CrossRef]
31. Zhao, Z. Q., Yu, Z. Y., Li, J., Ouyang, X. N. (2016). Gefitinib induces lung cancer cell autophagy and apoptosis via blockade of the PI3K/AKT/mTOR pathway. Oncology Letters, 12(1), 63–68. DOI 10.3892/ol.2016.4606. [Google Scholar] [CrossRef]
32. Datta, K., Suman, S., Fornace, A. J. Jr. (2014). Radiation persistently promoted oxidative stress, activated mTOR via PI3K/Akt, and downregulated autophagy pathway in mouse intestine. The International Journal of Biochemistry & Cell Biology, 57, 167–176. DOI 10.1016/j.biocel.2014.10.022. [Google Scholar] [CrossRef]
33. Wang, C., Zhang, X., Teng, Z., Zhang, T., Li, Y. (2014). Downregulation of PI3K/Akt/mTOR signaling pathway in curcumin-induced autophagy in APP/PS1 double transgenic mice. European Journal of Pharmacology, 740, 312–320. DOI 10.1016/j.ejphar.2014.06.051. [Google Scholar] [CrossRef]
34. Lu, C., Shan, Z., Hong, J., Yang, L. (2017). MicroRNA-92a promotes epithelial-mesenchymal transition through activation of PTEN/PI3K/AKT signaling pathway in non-small cell lung cancer metastasis. International Journal of Oncology, 51(1), 235–244. DOI 10.3892/ijo.2017.3999. [Google Scholar] [CrossRef]
35. Zhan, T., Rindtorff, N., Boutros, M. (2017). Wnt signaling in cancer. Oncogene, 36(11), 1461–1473. DOI 10.1038/onc.2016.304. [Google Scholar] [CrossRef]
36. Yamamoto, M., Bharti, A., Li, Y., Kufe, D. (1997). Interaction of the DF3/MUC1 breast carcinoma- associated antigen and β-catenin in cell adhesion. Journal of Biological Chemistry, 272, 12492–12494. DOI 10.1074/jbc.272.19.12492. [Google Scholar] [CrossRef]
37. Bouillez, A., Rajabi, H., Pitroda, S., Jin, C., Alam, M. (2016). Inhibition of MUC1-C suppresses MYC expression and attenuates malignant growth in KRAS mutant lung adenocarcinomas. Cancer Research, 76(6), 1538–1548. DOI 10.1158/0008-5472.CAN-15-1804. [Google Scholar] [CrossRef]
38. Zhao, R., Gao, S., He, H., Zhang, J., Zhang, G. et al. (2021). Evaluation on the distribution of EGFR, KRAS and BRAF genes and the expression of PD-l1 in different types of lung cancer. International Journal of General Medicine, 14, 5615–5620. DOI 10.2147/IJGM.S316151. [Google Scholar] [CrossRef]
39. Asakura, T., Yamaguchi, N., Ohkawa, K., Yoshida, K. (2015). Proteasome inhibitor-resistant cells cause EMT-induction via suppression of E-cadherin by miR-200 and ZEB1. International Journal of Oncology, 46(5), 2251–2260. DOI 10.3892/ijo.2015.2916. [Google Scholar] [CrossRef]
40. Wang, X., Chen, X., Meng, Q., Jing, H., Lu, H. et al. (2015). MiR-181b regulates cisplatin chemosensitivity and metastasis by targeting TGFpR1/Smad signaling pathway in NSCLC. Scientific Reports, 5, 17618. DOI 10.1038/srep17618. [Google Scholar] [CrossRef]
41. Sato, H., Shien, K., Tomida, S., Okayasu, K., Suzawa, K. et al. (2017). Targeting the miR-200c/LIN28B axis in acquired EGFR-TKI resistance non-small cell lung cancer cells harboring EMT features. Scientific Reports, 7, 40847. DOI 10.1038/srep40847. [Google Scholar] [CrossRef]
42. Asimgil, H., Ertetik, U., Çevik, N. C., Ekizce, M., Doğruöz, A. et al. (2022). Targeting the undruggable oncogenic KRAS: The dawn of hope. JCI Insight, 7(1), e153688. DOI 10.1172/jci.insight.153688. [Google Scholar] [CrossRef]
43. Hao, F. (2021). Systemic profiling of KDM5 subfamily signature in non-small-cell lung cancer. International Journal of General Medicine, 14, 7259–7275. DOI 10.2147/IJGM.S329733. [Google Scholar] [CrossRef]
44. Bialk, P., Wang, Y., Banas, K., Kmiec, E. B. (2018). Functional gene knockout of NRF2 increases chemosensitivity of human lung cancer A549 cells in vitro and in a xenograft mouse model. Molecular Therapy Oncolytics, 11, 75–89. DOI 10.1016/j.omto.2018.10.002. [Google Scholar] [CrossRef]
45. Hu, J., Cheng, Y., Li, Y., Jin, Z., Pan, Y. et al. (2014). MicroRNA-128 plays a critical role in human non-small cell lung cancer tumourigenesis, angiogenesis and lymphangiogenesis by directly targeting vascular endothelial growth factor-C. European Journal of Cancer, 50(13), 2336–2350. DOI 10.1016/j.ejca.2014.06.005. [Google Scholar] [CrossRef]
46. Yanaihara, N., Caplen, N., Bowman, E., Seike, M., Kumamoto, K. et al. (2006). Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell, 9(3), 189–198. DOI 10.1016/j.ccr.2006.01.025. [Google Scholar] [CrossRef]
47. Mongre, R. K., Sodhi, S. S., Ghosh, M., Kim, J. H., Kim, N. et al. (2015). The novel inhibitor BRM270 downregulates tumorigenesis by suppression of NF-κB signaling cascade in MDR-induced stem like cancer-initiating cells. International Journal of Oncology, 46(6), 2573–2585. DOI 10.3892/ijo.2015.2961. [Google Scholar] [CrossRef]
48. Kwon, T., Chandimali, N., Huynh, D. L., Zhang, J. J., Kim, N. et al. (2018). BRM270 inhibits cancer stem cell maintenance via microRNA regulation in chemoresistant A549 lung adenocarcinoma cells. Cell Death & Disease, 9(2), 244. DOI 10.1038/s41419-018-0277-7. [Google Scholar] [CrossRef]
49. Porter, D. L., Levine, B. L., Kalos, M., Bagg, A., June, C. H. (2011). Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. The New England Journal of Medicine, 365(8), 725–733. DOI 10.1056/NEJMoa1103849. [Google Scholar] [CrossRef]
50. Kochenderfer, J. N., Wilson, W. H., Janik, J. E., Dudley, M. E., Stetler-Stevenson, M. et al. (2010). Eradication of B-lineage cells and regression of lymphoma in a patient treated with autologous T cells genetically engineered to recognize CD19. Blood, 116(20), 4099–4102. DOI 10.1182/blood-2010-04-281931. [Google Scholar] [CrossRef]
51. Liu, X., Zhang, Y., Cheng, C., Cheng, A. W., Zhang, X. et al. (2017). CRISPR-Cas9-mediated multiplex gene editing in CAR-T cells. Cell Research, 27(1), 154–157. DOI 10.1038/cr.2016.142. [Google Scholar] [CrossRef]
52. Ren, J., Zhang, X., Liu, X., Fang, C., Jiang, S. et al. (2017). A versatile system for rapid multiplex genome-edited CAR T cell generation. Oncotarget, 8(10), 17002–17011. DOI 10.18632/oncotarget.15218. [Google Scholar] [CrossRef]
53. Mollanoori, H., Shahraki, H., Rahmati, Y., Teimourian, S. (2018). CRISPR/Cas9 and CAR-T cell, collaboration of two revolutionary technologies in cancer immunotherapy, an instruction for successful cancer treatment. Human Immunology, 79(12), 876–882. DOI 10.1016/j.humimm.2018.09.007. [Google Scholar] [CrossRef]
54. Anguille, S., Smits, E. L., Bryant, C., van Acker, H. H., Goossens, H. et al. (2015). Dendritic cells as pharmacological tools for cancer immunotherapy. Pharmacological Reviews, 67(4), 731–753. DOI 10.1124/pr.114.009456. [Google Scholar] [CrossRef]
55. Xie, J., Li, H., Chen, L., Cao, Y., Hu, Y. et al. (2021). A novel pyroptosis-related lncRNA signature for predicting the prognosis of skin cutaneous melanoma. International Journal of General Medicine, 14, 6517–6527. DOI 10.2147/IJGM.S335396. [Google Scholar] [CrossRef]
56. da Cunha Santos, G., Shepherd, F. A., Tsao, M. S. (2011). EGFR mutations and lung cancer. Annual Review of Pathology, 6, 49–69. DOI 10.1146/annurev-pathol-011110-130206. [Google Scholar] [CrossRef]
57. Huang, M., Ma, Y., Lv, C., Li, S., Lu, F. et al. (2021). Aneuploid circulating tumor cells as a predictor of response to neoadjuvant chemotherapy in non-small cell lung cancer. International Journal of General Medicine, 14, 6609–6620. DOI 10.2147/IJGM.S330361. [Google Scholar] [CrossRef]
58. Tang, H., Shrager, J. B. (2016). CRISPR/Cas-mediated genome editing to treat EGFR-mutant lung cancer: A personalized molecular surgical therapy. EMBO Molecular Medicine, 8(2), 83–85. DOI 10.15252/emmm.201506006. [Google Scholar] [CrossRef]
59. Platt, R. J., Chen, S., Zhou, Y., Yim, M. J., Swiech, L. et al. (2014). CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell, 159(2), 440–455. DOI 10.1016/j.cell.2014.09.014. [Google Scholar] [CrossRef]
60. Hartmann, O., Reissland, M., Maier, C. R., Fischer, T., Prieto-Garcia, C. (2021). Implementation of CRISPR/Cas9 genome editing to generate murine lung cancer models that depict the mutational landscape of human disease. Frontiers in Cell and Developmental Biology, 9, 641618. DOI 10.3389/fcell.2021.641618. [Google Scholar] [CrossRef]
61. Soda, M., Choi, Y. L., Enomoto, M., Takada, S., Yamashita, Y. et al. (2007). Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature, 448(7153), 561–566. DOI 10.1038/nature05945. [Google Scholar] [CrossRef]
62. Maddalo, D., Manchado, E., Concepcion, C. P., Bonetti, C., Vidigal, J. A. et al. (2015). In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature, 516(7531), 423–427. DOI 10.1038/nature13902. [Google Scholar] [CrossRef]
63. Sánchez-Rivera, F. J., Papagiannakopoulos, T., Romero, R., Tammela, T., Bauer, M. R. et al. (2014). Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature, 516(7531), 428–431. DOI 10.1038/nature13906. [Google Scholar] [CrossRef]
64. Wang, Y. C., Wang, S. A., Chen, P. H., Hsu, T. I., Yang, W. B. et al. (2016). Variants of ubiquitin-specific peptidase 24 play a crucial role in lung cancer malignancy. Oncogene, 35(28), 3669–3680. DOI 10.1038/onc.2015.432. [Google Scholar] [CrossRef]
65. Guo, L., Song, B., Xiao, J., Lin, H., Chen, J. et al. (2021). The prognostic value of biomarkers on detecting non-small cell lung cancer in a Chinese elderly population. International Journal of General Medicine, 14, 5279–5286. DOI 10.2147/IJGM.S331311. [Google Scholar] [CrossRef]
66. Xie, H., Zhang, J. F., Li, Q. (2021). Development of a prognostic nomogram for patients with lung adenocarcinoma in the stages I, II, and III based on immune scores. International Journal of General Medicine, 14, 8677–8688. DOI 10.2147/IJGM.S337934. [Google Scholar] [CrossRef]
67. Deutschmeyer, V., Breuer, J., Walesch, S. K., Sokol, A. M., Graumann, J. et al. (2019). Epigenetic therapy of novel tumour suppressor ZAR1 and its cancer biomarker function. Clinical Epigenetics, 11(1), 182. DOI 10.1186/s13148-019-0774-2. [Google Scholar] [CrossRef]
68. Du, J., Qian, J., Zheng, B., Xu, G., Chen, H. et al. (2021). MiR-21-5p is a biomarker for predicting prognosis of lung adenocarcinoma by regulating PIK3R1 expression. International Journal of General Medicine, 14, 8873–8880. DOI 10.2147/IJGM.S337149. [Google Scholar] [CrossRef]
69. Liu, B., Yang, S. (2021). A five autophagy-related long non-coding RNA prognostic model for patients with lung adenocarcinoma. International Journal of General Medicine, 14, 7145–7158. DOI 10.2147/IJGM.S334601. [Google Scholar] [CrossRef]
70. Wu, G. J., Ren, K., He, M., Xu, J. X., Li, Z. Q. et al. (2021). SNX20 expression correlates with immune cell infiltration and can predict prognosis in lung adenocarcinoma. International Journal of General Medicine, 14, 7599–7611. DOI 10.2147/IJGM.S337198. [Google Scholar] [CrossRef]
71. Zhang, P., Kang, B., Xie, G., Li, S., Gu, Y. et al. (2021). Genomic sequencing and editing revealed the GRM8 signaling pathway as potential therapeutic targets of squamous cell lung cancer. Cancer Letters, 442, 53–67. DOI 10.1016/j.canlet.2018.10.035. [Google Scholar] [CrossRef]
72. Stork, P. J., Schmitt, J. M. (2002). Crosstalk between cAMP and MAP kinase signaling in the regulation of cell proliferation. Trends in Cell Biology, 12(6), 258–266. DOI 10.1016/S0962-8924(02)02294-8. [Google Scholar] [CrossRef]
73. Park, M. Y., Jung, M. H., Eo, E. Y., Kim, S., Lee, S. H. et al. (2017). Generation of lung cancer cell lines harboring EGFR T790M mutation by CRISPR/Cas9-mediated genome editing. Oncotarget, 8(22), 36331–36338. DOI 10.18632/oncotarget.16752. [Google Scholar] [CrossRef]
74. Govindan, R., Page, N., Morgensztern, D., Read, W., Tierney, R. et al. (2006). Changing epidemiology of small-cell lung cancer in the United States over the last 30 years: Analysis of the surveillance, epidemiologic, and end results database. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology, 24(28), 4539–4544. DOI 10.1200/JCO.2005.04.4859. [Google Scholar] [CrossRef]
75. Califano, R., Abidin, A. Z., Peck, R., Faivre-Finn, C., Lorigan, P. (2012). Management of small cell lung cancer: Recent developments for optimal care. Drugs, 72(4), 471–490. DOI 10.2165/11597640-000000000-00000. [Google Scholar] [CrossRef]
76. Ng, S. R., Rideout III, W. M., Akama-Garren, E. H., Bhutkar, A., Mercer, K. L. et al. (2020). CRISPR-mediated modeling and functional validation of candidate tumor suppressor genes in small cell lung cancer. Proceedings of the National Academy of Sciences of the United States of America, 117(1), 513–521. DOI 10.1073/pnas.1821893117. [Google Scholar] [CrossRef]
77. Zhang, Y., Shi, Y., Song, D., Zhang, X., Yu, L. et al. (2018). Establishment and characterization of A549 tumor monoclonal cell line with UCHL1 gene deletion. Sheng Li Xue Bao, 70(2), 184–192. DOI 10.13294/j.aps.2018.0032. [Google Scholar] [CrossRef]
78. El-Kenawy, A., Benarba, B., Neves, A. F., de Araujo, T. G., Tan, B. L. et al. (2019). Gene surgery: Potential applications for human diseases. EXCLI Journal, 18, 908–930. DOI 10.17179/excli2019-1833. [Google Scholar] [CrossRef]
79. Li, M., Suzuki, K., Kim, N. Y., Liu, G. H., Izpisua Belmonte, J. C. (2014). A cut above the rest: Targeted genome editing technologies in human pluripotent stem cells. The Journal of Biological Chemistry, 289(8), 4594–4599. DOI 10.1074/jbc.R113.488247. [Google Scholar] [CrossRef]
80. Grau, J., Boch, J., Posch, S. (2013). TALENoffer: Genome-wide TALEN off-target prediction. Bioinformatics, 29(22), 2931–2932. DOI 10.1093/bioinformatics/btt501. [Google Scholar] [CrossRef]
81. Fu, Y., Foden, J. A., Khayter, C., Maeder, M. L., Reyon, D. et al. (2013). High-frequency off-target mutagenesis induced by CRISPR-cas nucleases in human cells. Nature Biotechnology, 31(9), 822–826. DOI 10.1038/nbt.2623. [Google Scholar] [CrossRef]
82. Seki, A., Rutz, S. (2018). Optimized RNP transfection for highly efficient CRISPR/Cas9-mediated gene knockout in primary T cells. The Journal of Experimental Medicine, 215, 985–997. DOI 10.1084/jem.20171626. [Google Scholar] [CrossRef]
83. Wang, W., Ye, C., Liu, J., Zhang, D., Kimata, J. T. et al. (2014). CCR5 gene disruption via lentiviral vectors expressing Cas9 and single guided RNA renders cells resistant to HIV-1 infection. PLoS One, 9(12), e115987. DOI 10.1371/journal.pone.0115987. [Google Scholar] [CrossRef]
84. Li, C., Guan, X., Du, T., Jin, W., Wu, B. et al. (2015). Inhibition of HIV-1 infection of primary CD4+ T-cells by gene editing of CCR5 using adenovirus-delivered CRISPR/Cas9. The Journal of General Virology, 96(8), 2381–2393. DOI 10.1099/vir.0.000139. [Google Scholar] [CrossRef]
85. Liang, X., Potter, J., Kumar, S., Zou, Y., Quintanilla, R. et al. (2015). Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. Journal of Biotechnology, 208, 44–53. DOI 10.1016/j.jbiotec.2015.04.024. [Google Scholar] [CrossRef]
86. Aird, E. J., Lovendahl, K. N., Martin, A. S., Harris, R. S., Gordon, W. R. (2018). Increasing Cas9-mediated homology-directed repair efficiency through covalent tethering of DNA repair template. Communications Biology, 1(1), 54. DOI 10.1038/s42003-018-0054-2. [Google Scholar] [CrossRef]
87. Carroll, D. (2016). Genome editing: Progress and challenges for medical applications. Genome Medicine, 8, 120. DOI 10.1186/s13073-016-0378-9. [Google Scholar] [CrossRef]
88. Zhang, H., Bahamondez-Canas, T. F., Zhang, Y., Leal, J., Smyth, H. D. C. (2018). PEGylated chitosan for nonviral aerosol and mucosal delivery of the CRISPR/Cas9 system in vitro. Molecular Pharmaceutics, 15(11), 4814–4826. DOI 10.1021/acs.molpharmaceut.8b00434. [Google Scholar] [CrossRef]
89. Bennett, W. D., Brown, J. S., Zeman, K. L., Hu, S. C., Scheuch, G. (2002). Targeting delivery of aerosols to different lung regions. Journal of Aerosol Medicine: The Official Journal of the International Society for Aerosols in Medicine, 15(2), 179–188. DOI 10.1089/089426802320282301. [Google Scholar] [CrossRef]
90. Chow, M. Y. T., Chang, R. Y. K., Chan, H. K. (2021). Inhalation delivery technology for genome-editing of respiratory diseases. Advanced Drug Delivery Reviews, 168, 217–228. DOI 10.1016/j.addr.2020.06.001. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |