
 | Oncologie |  |
DOI: 10.32604/oncologie.2021.018713
ARTICLE
Circular RNA hsa_circ_0016863 Regulates the Proliferation, Migration, Invasion and Apoptosis of Hepatocellular Carcinoma
1Department of Laboratory Medicine, Nantong City No.1 People’s Hospital and Second Affiliated Hospital of Nantong University, Nantong, China
2Department of Laboratory Medicine, Affiliated Hospital of Nantong University, Nantong, China
3Department of Laboratory Medicine, School of Public Health, Nantong University, Nantong, China
*Corresponding Authors: Xiang Chen. Email: ntchenx0520@163.com; Feng Wang. Email: richardwangf@163.com
#Ping Zhao and Yao Xu contributed equally to this work
Received: 12 August 2021; Accepted: 21 October 2021
Abstract: Background: Although there has been a part of research that the expression of circular RNAs (circRNAs) is bound up with the occurrence, development and prognosis of multiple tumors, what roles circRNAs have in hepatocellular carcinoma (HCC) remain to be explored. Methods: Hsa_circ_0016863 showed a trend of high expression in HCC tissues by Microarray GSE97332. The expression of hsa_circ_0016863 in HCC and adjacent non-cancerous tissues was detected by qRT-PCR. The role of hsa_circ_0016863 in HCC was investigated by colony formation assay, cell proliferation assay, Transwell assay and flow cytometry. Results: Hsa_circ_0016863 exhibited a trend of high expression in HCC. Cell experiments showed that upregulated hsa_circ_0016863 promoted HCC cell proliferation, migration and invasion, whereas it inhibited cell apoptosis, while silencing hsa_circ_0016863 showed the opposite results. Conclusions: Hsa_circ_0016863 shows a trend of high expression in HCC, which may be related to the development of HCC.
Keywords: Proliferation; migration; invasion; apoptosis; HCC; hsa_circ_0016863
Due to various reasons, such as virus infection, heavy drinking and fatty liver caused by obesity, the incidence of hepatocellular carcinoma (HCC) is increasing year by year [1]. HCC is the sixth most common cancer in the world with a high incidence. It also has a high mortality rate and ranks fourth in cancer [2,3]. Liver resection, liver transplantation, and local ablative therapies, among others, are several of the most commonly used methods to treat HCC [4]. According to the stage of HCC, patients with early-stage HCC can obtain better treatment outcomes, with 5-year survival rates as high as 75%, whereas the treatment outcomes for patients with advanced stage or those who have entered advanced stage are not very good, with 1-year survival rates less than 10 [5]. In recent years, much effort has been devoted to the study of molecular mechanisms of HCC, which in turn has led to new insights into the occurrence and development of HCC.
Circular RNAs (circRNAs) are non-coding RNAs that lack 3′-poly (A) tails and 5′-cap, which form closed circular structures by covalent bonds [6]. At present, it has been found that circRNAs has many biological functions. CircRNAs may act as sponges for microRNAs (miRNAs), for example, circZNF566 promotes HCC progression by sponging miR-4738-3p [7]. CircRNA-5692 adsorbs miR-328-5p to decrease its expression, which in turn enhances the expression of DAB2IP, ultimately inhibiting HCC development [8]. CircRNAs can directly bind to proteins, for example, circRHOT1, which is upregulated in HCC, can initiate NR2F6 by recruiting TIP60, resulting in the inhibition of HCC development and progression [9]. In addition, circRNAs were also found to translate proteins, and circβ-catenin, whose expression levels were obviously up-regulated in HCC, could encode a novel protein β-catenin-370aa. The protein activates the Wnt/β-catenin pathway, which promotes cancer development [10]. There have been numerous studies showing that circRNAs are associated with multiple diseases, including tumors, and the role of circRNAs in HCC needs further exploration, thereby providing ideas for treatment.
Here, we screened a novel circRNA, hsa_circ_0016863, in Microarray GSE97332. Hsa_circ_0016863 was obviously up-regulated in HCC. Upon overexpression of hsa_circ_0016863, the ability of HCC cells to proliferate, migrate and invade increases and apoptosis decreases, suggesting that hsa_circ_0016863 may contribute to HCC development.
2.1 Collection of Tissue Samples
50 pairs of HCC specimens were gathered in the Affiliated Hospital of Nantong University from January 2018 to December 2019. All patients did not receive any treatment preoperatively. After the tissues were removed during surgery, they were immediately snap frozen and later stored in a −80°C freezer. This study was approved by the ethics committee of the Affiliated Hospital of Nantong University. All participating patients gave written informed consent.
Cell lines required for experiments, including normal liver cell line LO2 and HCC cell lines (BEL-7404, HCCLM3, SMMC-7721, SK-Hep1 and MHCC-97H) were purchased from the Cell Resources Center of the Chinese Academy of Science. Cells were grown in DMEM containing 10% FBS and 1% penicillin-streptomycin and placed in an incubator at 37°C and 5% CO2.
2.3 Total RNA Extraction and qRT-PCR Analysis
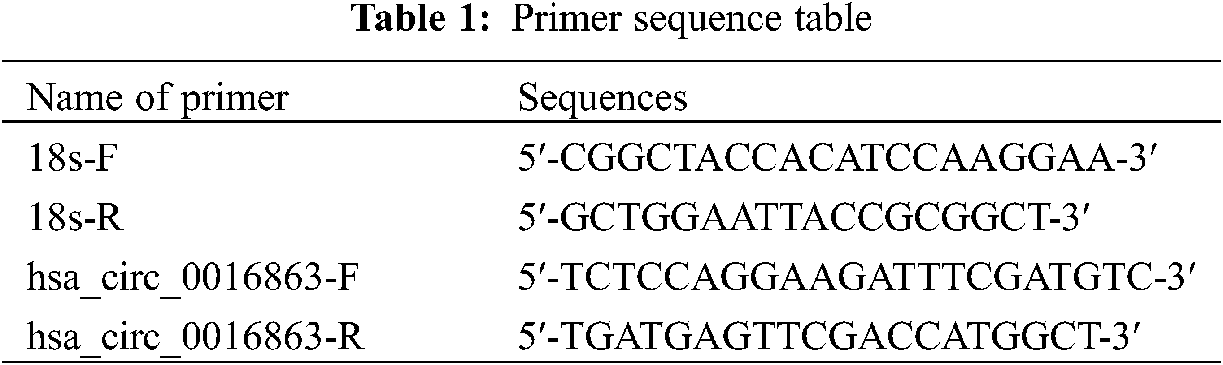
Total RNA was extracted from tissues and cells using reagents such as Trizol (Thermo Fisher Scientific, USA), chloroform, isopropyl alcohol and 75% ethanol. The resulting RNA is then reverse transcribed into cDNA. Samples were loaded on ice in the dark and qRT-PCR was performed using a Roche LightCycler 480 (Roche, Germany). 18S rRNA served as an internal reference. Primer sequence as shown in Table 1.
Total RNA (10 μg) was incubated with 3–4 U/μg RNase R for 45 min at 37°C. Then, the RNA expression levels of 18sRNA and hsa_circ_0016863 were detected by qRT-PCR.
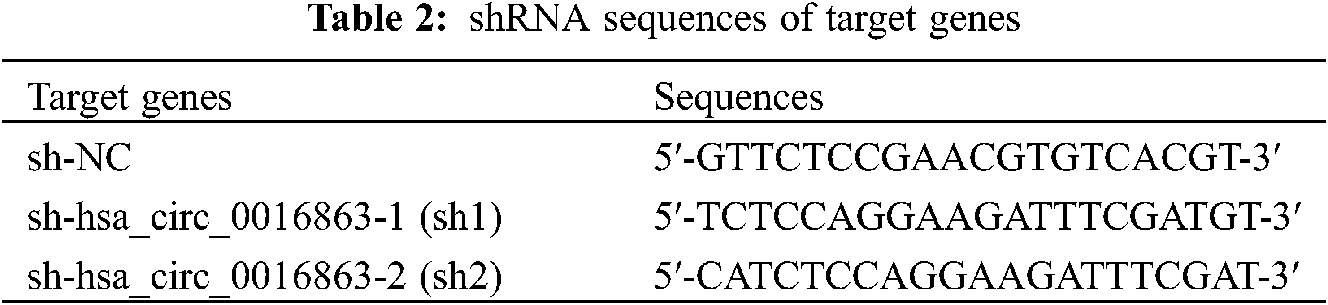
HCC cell lines were plated in 6 or 12 well plates, and cell transfection was performed when the cell amount reached 60% to 80%. Overexpression plasmid (ov) of hsa_circ_0016863 and its corresponding negative control (nc), interference plasmid (sh-circ) and its corresponding negative control (sh-NC) were fully mixed with Lipofectamine 3000 in serum-free medium. The mixture was transferred to the cells after standing for 20 min and allowed to distribute evenly with gentle shaking. The cells were put into an incubator and 6h later changed to complete medium containing 10% FBS. QRT-PCR was performed to determine transfection efficiency and cell function experiments after continuing the incubation for 24–72 h. ShRNA sequences of targrt genes as shown in Table 2.
The transfected cells with good growth trend were cultured in 6 well plates at a certain density, and the medium was carefully changed around one week. After approximately 2 weeks of incubation in the incubator, then stop the culture. After fixation with 4% paraformaldehyde, the cells were stained with crystal violet, and finally the cell clone rate was calculated.
A certain number of cells transfected with shRNA or overexpressed vectors were implanted into 96 well plates. Then add 10 μl cck-8 reagent (Dojindo, Japan) to each well at 24, 48, 72, 96, 120 h, respectively. After incubation for 2 h, the OD value of each well was determined at the wavelength of 450 nm.
The upper chamber of Transwell with Matrigel added was used for invasion experiments, while the upper chamber without Matrigel was used for migration experiments. Cells were inoculated into the upper compartment, followed by another 500 ml of DMEM without serum, and DMEM with 10% serum was added in the lower compartment of the Transwell. The incubation was continued for 24 h, the upper chamber was removed, and after washing, fixing, and staining, cell invasion and metastasis functions were evaluated.
Cell cycle and apoptosis were detected with a flow cytometer (BD, USA). After 48 h of cell transfection, cells were collected and washed with PBS. The cell cycle was measured after treatment with RNase A and PI dye. Cell apoptosis was analyzed after treatment with Annexin V-Kfluor647 and PI dye.
The data of the present experiments were analyzed using GraphPad prism version 8.0 (GraphPad Software, Inc., La Jolla, CA, USA). The t-test was used for comparison between the two groups, and P < 0.05 was considered statistically significant.
3.1 Hsa_circ_0016863 is Overexpressed in HCC Tissues and Its Characteristics in HCC
To screen candidate circRNAs in HCC patients, we discovered that hsa_circ_0016863 was up-regulated in HCC in the GSE97332 database. We subsequently validated by qRT-PCR, demonstrating that hsa_circ_0016863 indeed exhibits a trend of high expression in 50 pairs of HCC tissues (Figs. 1a and 1b).
Hsa_circ_0016863 is composed of exon 2 to exon 8 of the COG2 gene with a span of 827nt (Fig. 1c). Then, we treated it with RNase R and observed the stability of hsa_circ_0016863. In this experiment, 18sRNA was a linear RNA and served as the control group. The qRT-PCR showed that the expression of 18sRNA after RNase R treatment decreased more obviously than that without treatment, while the relative expression decline of hsa_circ_0016863 was not obvious. This indicates that hsa_circ_0016863 is not digested by RNase R and is more stable (Fig. 1d). Next, the band recovered after qRT-PCR was Sanger sequenced, verifying the circularization site junction of hsa_circ_0016863 (Fig. 1e). The above results indicate that hsa_circ_0016863 has an intact circular structure that exists stably.
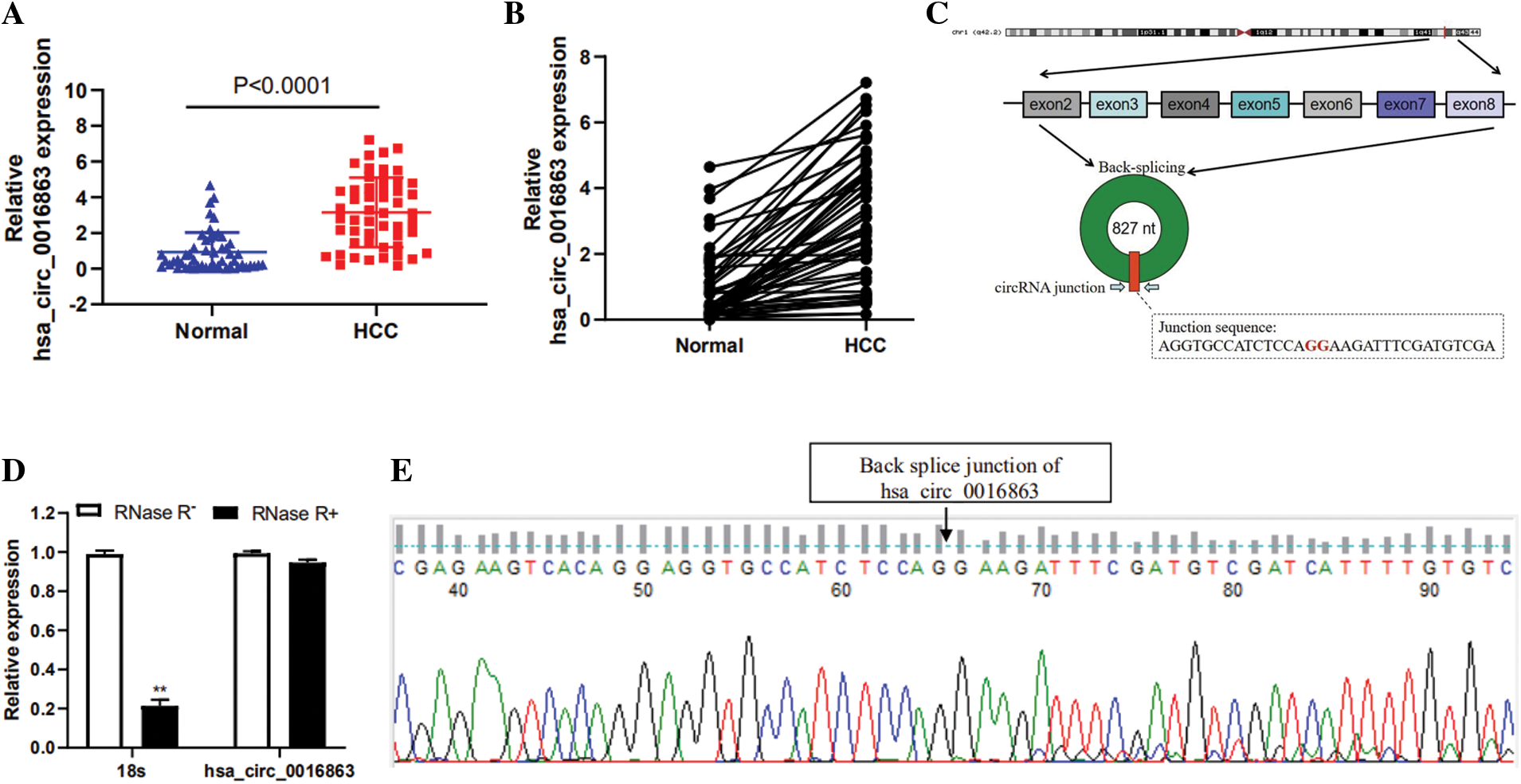
Figure 1: Relative expression levels and properties of hsa_circ_0016863 in HCC tissues. (A, B) qRT-PCR was performed to detect the expression of hsa_circ_0016863 in 50 pairs of HCC tissues and their corresponding para-carcinoma tissues. (C) Hsa_circ_0016863 results from exon 2 to exon 8 of the COG2 gene. (D) The expression of hsa_circ_0016863 and 18s rRNA was analyzed by qRT-PCR after RNase R treatment and untreated, and hsa_circ_0016863 was found to be resistant to RNase R. (E) The results of Sanger sequencing of the qRT-PCR product of hsa_circ_0016863 showed that there is a circularization site for hsa_circ_0016863. **: P < 0.01
3.2 Hsa_circ_0016863 is Overexpressed in HCC Cells
We tested the expression of hsa_circ_0016863 in five HCC cell lines (BEL-7404, HCCLM3, SMMC-7721, SK-Hep-1, MHCC-97H) by qRT-PCR (Fig. 2a). The expression level of hsa_circ_0016863 in HCC cell lines was visibly higher than that of LO2. Among them, hsa_circ_0016863 showed markedly higher expression in HCCLM3 than the other four cells, while the expression in SMMC-7721 was the lowest. Therefore, we selected SMMC-7721 cell line for overexpression experiment and HCCLM3 cell line for interference experiment.
To understand the effect of hsa_circ_0016863 on HCC, the growth of HCC cells was observed after overexpression and interference of hsa_circ_0016863. Transfection was performed in SMMC-7721 cells with an overexpression plasmid that results in a significant increase in hsa_circ_0016863 expression levels compared to the control (Fig. 2B). Subsequently, two RNA interference vectors, sh1 and sh2, against hsa_circ_0016863 were constructed, and stable transfection of shRNAs was performed in HCCLM3 cells (Fig. 2C). Although both sh1 and sh2 can inhibit the expression of hsa_circ_0016863, the inhibitory effect of sh1 is more obvious. Therefore, we chose sh1 in the subsequent interference experiments.
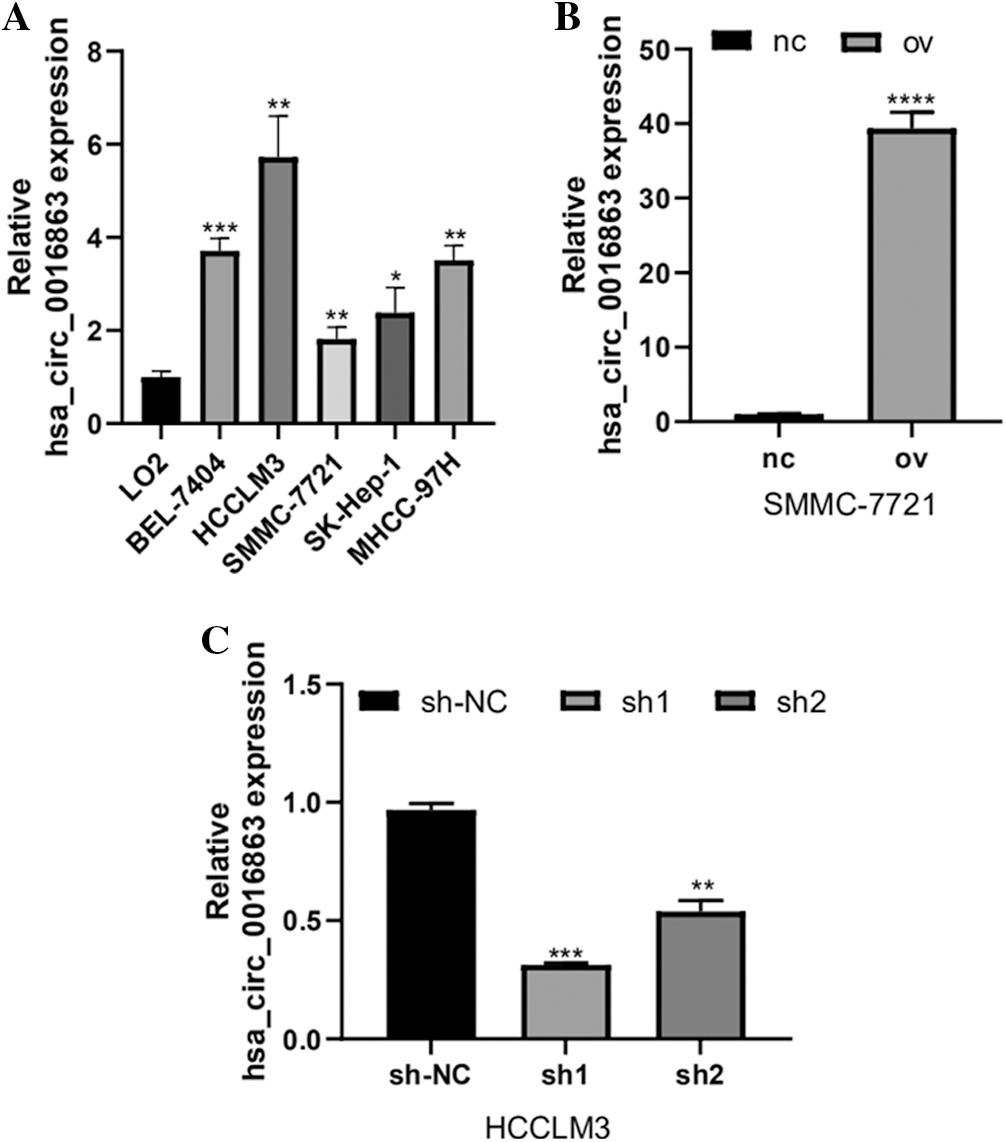
Figure 2: The expression level and transfection efficiency of hsa_circ_0016863 in HCC cells. (A) qRT-PCR was performed to detect the expression of hsa_circ_0016863 in normal liver cell lines and five HCC cell lines. (B) qRT-PCR was performed to detect the expression level of hsa_circ_0016863 after transfection of overexpression plasmid in SMMC-7721 cell line. (C) The interference efficiency of shRNA against hsa_circ_0016863 in HCCLM3 cell line was detected by qRT-PCR. *: P < 0.05, **: P < 0.01, ***: P < 0.001, ****: P < 0.0001
3.3 Hsa_circ_0016863 Promotes Proliferation and Growth Levels of HCC Cells
After transfection of overexpression plasmid and negative control plasmid in SMMC-7721 cells, CCK-8 assay, colony formation assay as well as cell cycle detection were performed. CCK-8 results showed that the proliferation activity of SMMC-7721 cells in the negative control was considerably lower than that in the overexpression groups (Fig. 3A). Results from colony formation assay revealed that the cell clonogenic rate after hsa_circ_0016863 overexpression was also higher than that of the negative control (Fig. 3C). In a cell cycle assay, we found that cells in the G0/G1 phase decreased and cells in the S + G2/M phase increased in the overexpression groups compared with the negative control (Fig. 3E).
After that, we performed transfection of interference plasmids in HCCLM3 and downregulation of hsa_circ_0016863 produced opposite effects. In cell proliferation assay and cell cloning assay, the cell proliferation activity and clonogenic rate of sh-circ treatment were lower than those of sh-NC (Figs. 3B and 3D). In cell cycle experiments, cells in G0/G1 phase increased and cells in S + G2/M phase decreased in sh-circ treated group (Fig. 3F).
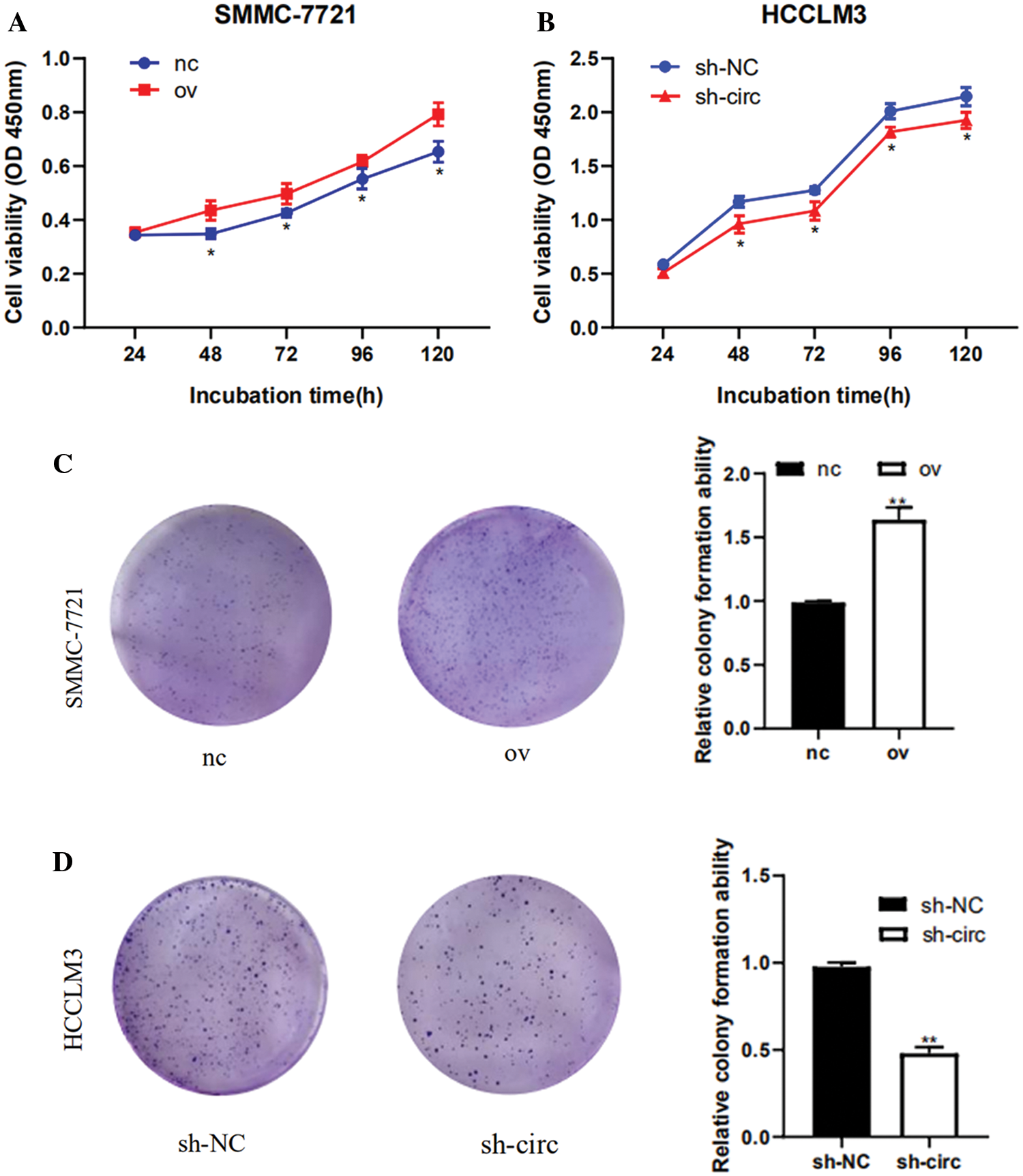
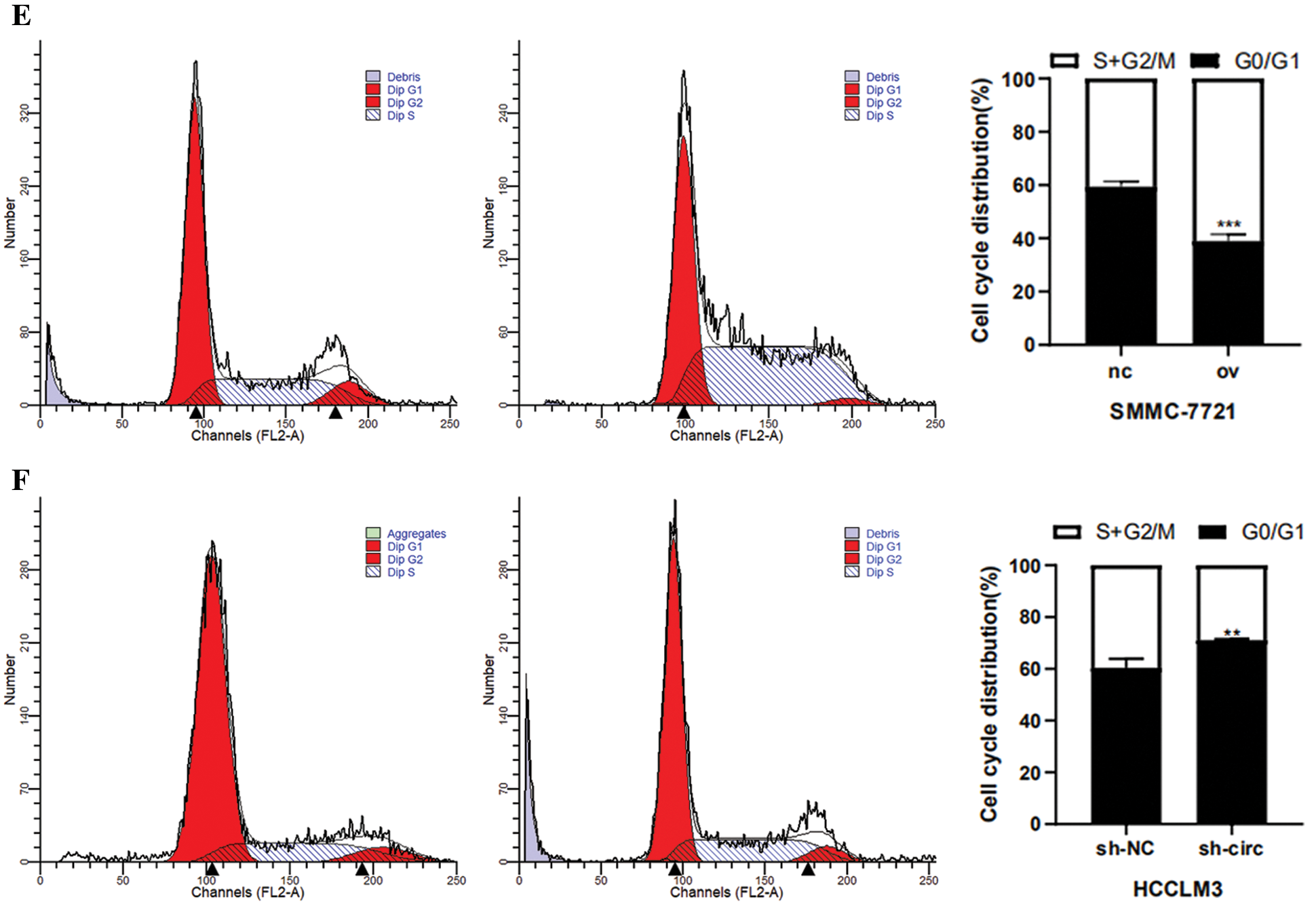
Figure 3: The proliferation and cell cycle of HCC cells were observed after transfection with overexpression plasmids and shRNA. (A, B) The proliferation of HCC cells was detected with CCK-8. (C, D) Results of colony formation assay. (E, F) Cell cycle was detected by flow technique. *: P < 0.05, **: P < 0.01, ***: P < 0.001
3.4 Hsa_circ_0016863 Promotes Migration and Invasion of HCC Cells
The effect of hsa_circ_0016863 on invasion and migration in HCC cells was investigated by Transwell experiment. The results showed that the migration and invasion abilities of HCC cells after hsa_circ_0016863 upregulation were markedly higher than those of the control group (Figs. 4A and 4C). In contrast, the experimental results obtained after sh-circ treatment revealed that the migration and invasion abilities of HCC cells were sharply attenuated (Figs. 4B and 4D). Taken together, from the transwell assay to analyze, hsa_circ_0016863 may promote metastasis of hepatoma cells with carcinogenic effect.
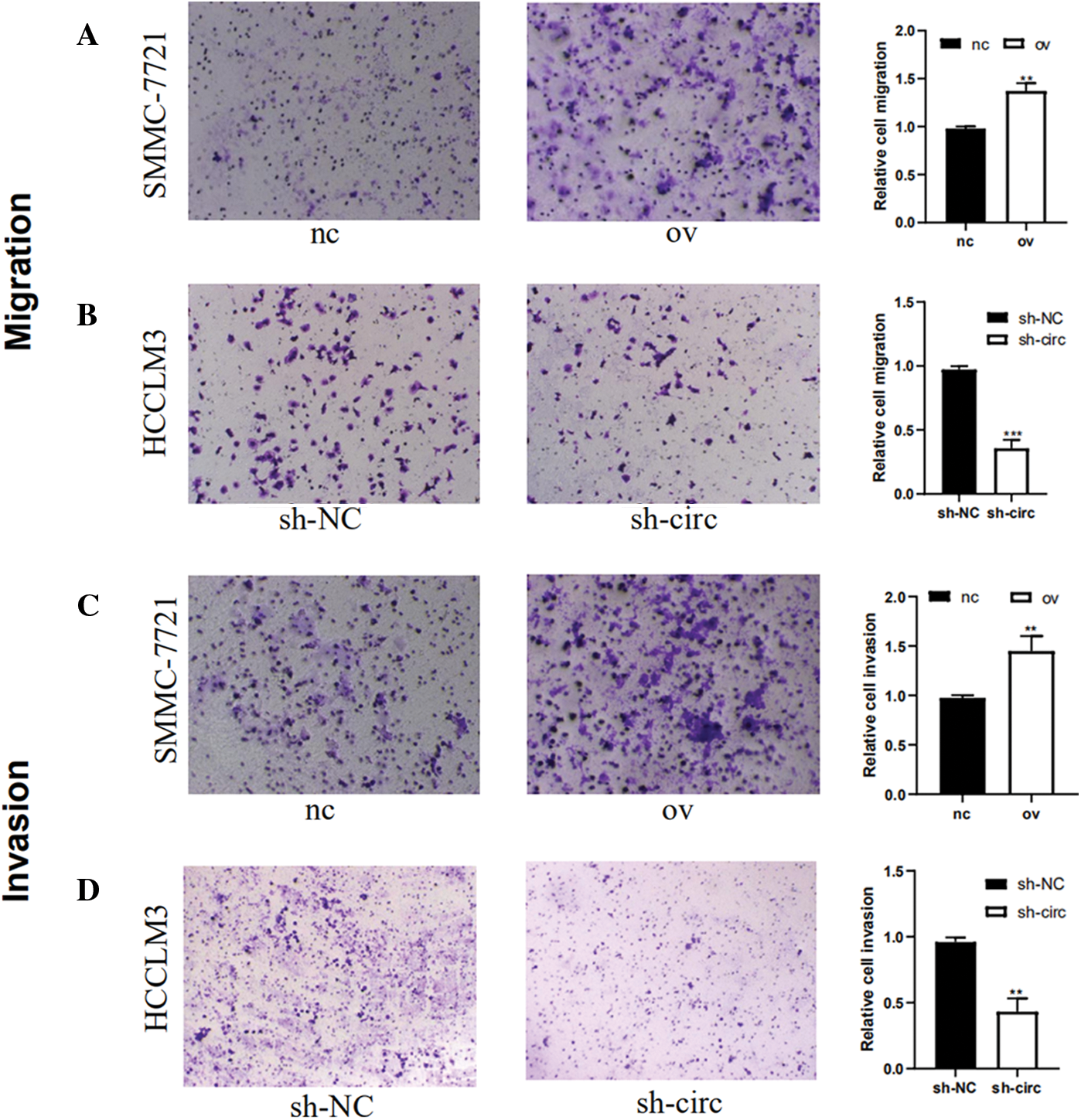
Figure 4: The changes of invasion and migration of HCC cells were observed after transfection with overexpression plasmid and shRNA. (A, B) Results of cell migration experiments. (C, D) Results of cell invasion assay. **: P < 0.01, ***: P < 0.001
3.5 Hsa_circ_0016863 Inhibits Apoptosis in HCC Cells
Next, the influence relationship of hsa_circ_0016863 on HCC cell apoptosis was accomplished by flow cytometry. In the overexpression experiments, there were significantly fewer apoptotic cells in the overexpression group (Fig. 5A); while in interference experiments, the amount of apoptosis of cells after sh-circ treatment was drastically more than that of sh-NC, indicating that hsa_circ_0016863 may inhibit the apoptosis of HCC cells (Fig. 5B).
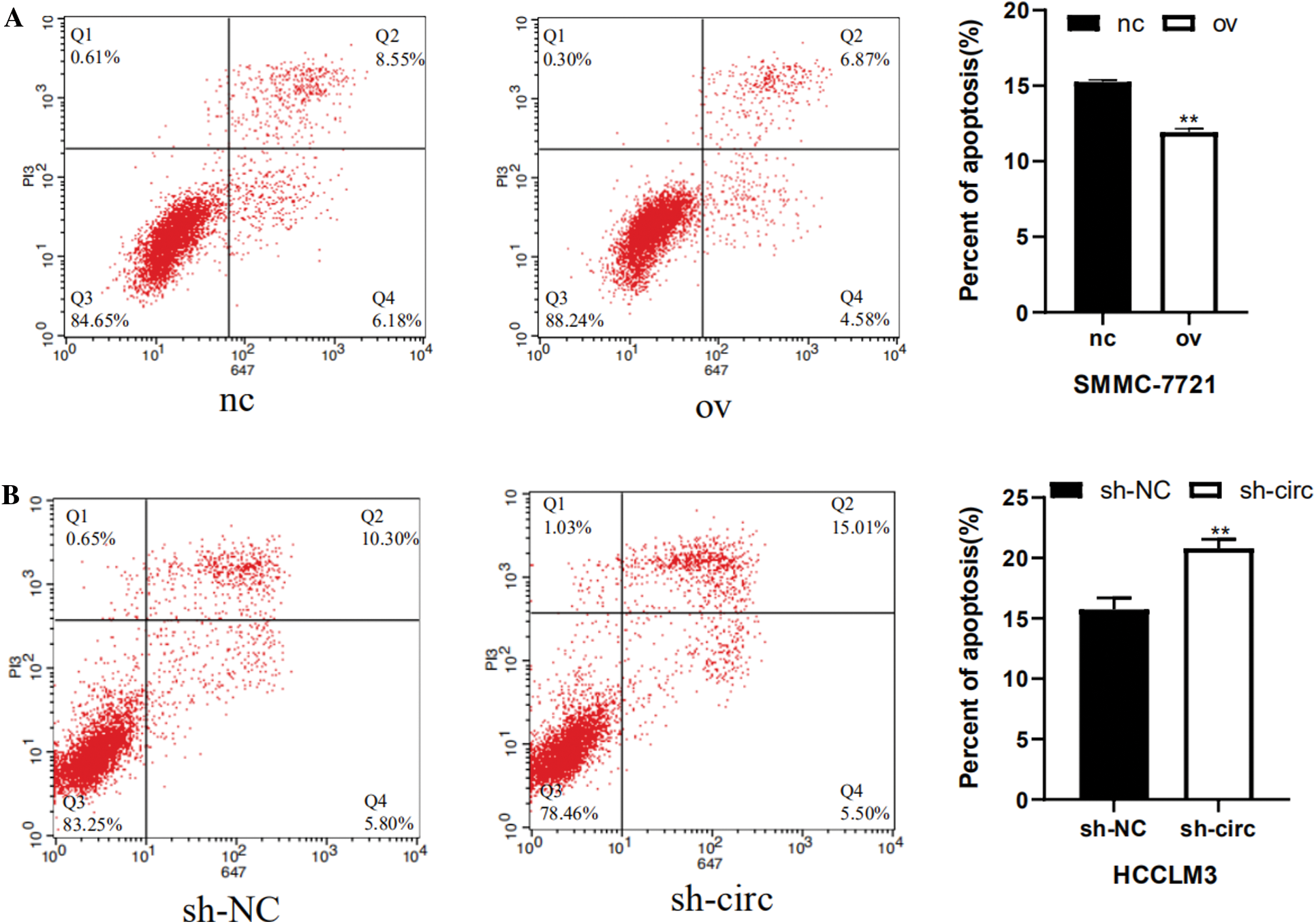
Figure 5: Effects of hsa_circ_0016863 on apoptosis in HCC cells. (A) After the overexpression plasmids were transfected in SMMC-7721 cells, flow cytometry assay revealed that the apoptotic cells decreased. (B) Increased apoptotic cells were detected by flow cytometry after shRNA transfection in HCCLM3 cells. **: P < 0.01
3.6 Exploration of Downstream Regulatory Networks of Hsa_circ_0016863 in HCC
Next, we used bioinformatics analysis to predict the possible circRNA-miRNA-mRNA regulatory pathway of hsa_circ_0016863 in HCC. As shown in Fig. 6, six miRNAs (hsa-miR-665, hsa-miR-568, hsa-miR-643, hsa-miR-769-5p, hsa-miR-384 and hsa-miR-583) and their corresponding target mRNAs were mapped, which may provide a new direction for exploring the downstream regulatory pathways of hsa_circ_0016863 in HCC afterwards.
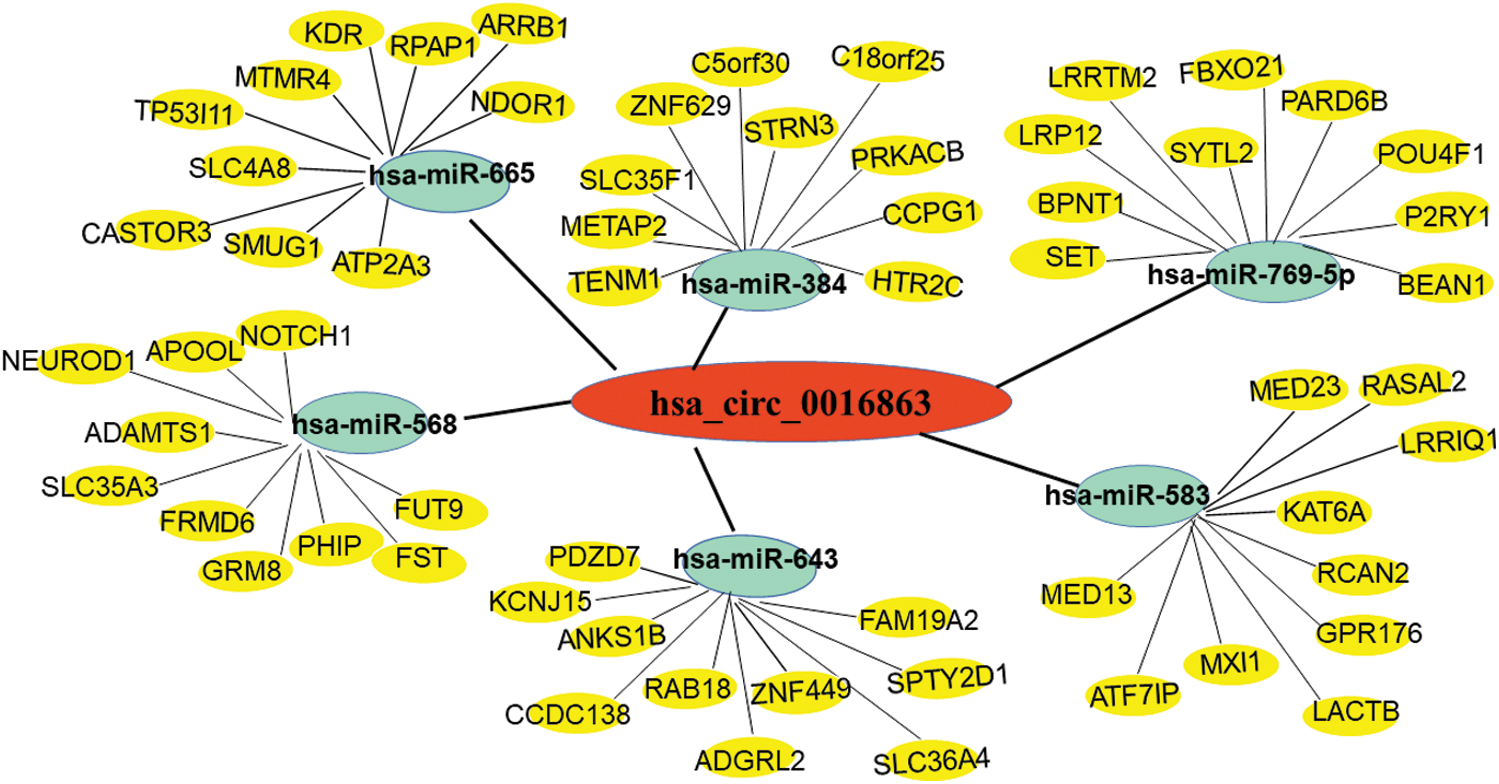
Figure 6: Prediction of circRNA-miRNA-mRNA network regulatory diagram. Red represents hsa_circ_0016863; Green represents 6 miRNAs that can bind to hsa_circ_0016863; Yellow represents target genes downstream of miRNAs
Up to now, massive studies have confirmed the abnormal expression of circRNAs in a variety of tumors, such as liver, lung, bladder, colorectal, breast, stomach, pancreatic, prostate, esophageal, cervical, ovarian, glioma and so on [11–15]. Currently, generous evidence has indicated that circRNAs are differentially expressed in HCC. Several circRNAs are up-regulated in HCC and related to patient prognosis, such as circUHRF1 [16], circRNA-SORE [17], hsa_circ_0082002 [18], and their high expression may promote HCC progression. There is also a portion that is down-regulated in HCC, such as circRNA-ITCH [19], circSMYD4 [20], hsa_circ_0004018 [21], thereby exerting a tumor suppressive effect. So, circRNAs not only affect the development of tumors, but also some circRNAs are associated with tumor diagnosis and prognostic factors, suggesting that circRNAs have great potential in the study of tumor mechanisms.
In this study, we picked a new molecule of circRNAs, hsa_circ_0016863, in the published Microarray GSE97332. Hsa_circ_0016863 was overexpressed in HCC tissues as verified by qRT-PCR. In cellular experiments, we discovered that knockdown of hsa_circ_0016863 impaired the propagation and metastasis of HCC cells, but also arrested cell cycle progression and increased apoptosis, whereas elevating the expression of hsa_circ_0016863 resulted in the contrary effect.
This experiment mainly studied the expression and function of hsa_circ_0016863 in HCC, and the mechanism of action of hsa_circ_0016863 in HCC remains to be further explored. A growing number of researches have indicated that there are binding sites for miRNAs in circRNAs, and miRNAs can inhibit protein production by affecting the stability of mRNAs or downregulating the translation of mRNAs [22]. Thus, circRNAs can regulate mRNA by acting as decoys for miRNAs [23]. In this study, we mapped the circRNA-miRNA-mRNA regulatory axis of HCC by bioinformatics prediction and found that hsa_circ_0016863 may interact with hsa-miR-665, hsa-miR-568, hsa-miR-643, hsa-miR-769-5p, hsa-miR-384 and hsa-miR-583. Furthermore, we found that in lung adenocarcinoma (LUAD), circ-TSPAN4 could upregulate ZEB1 by sponging miR-665, thereby promoting tumor metastasis [24]. CircPAPPA, which is lowly expressed in preeclampsia (PE) trophoblasts, can bind miR-384, thereby reducing the expression of STAT3, ultimately inhibiting trophoblast cell proliferation and invasion [25]. In acute myeloid leukemia (AML), circRAD18 acts as a sponge for miR-206, while PRKACB is a miR-206 downstream target gene. In AML, upregulated circRAD18 promotes AML progression by decreasing miR-206 and elevating PRKACB, providing a reference direction for the treatment of AML [26]. NOTCH1 has been implicated in the progression of multiple tumors. Many studies have shown that NOTCH1 is involved in the development and progression of squamous cell carcinoma [27,28]. In renal cell carcinoma (RCC), highly expressed circPDK1 promotes malignant progression of RCC cells through the circPDK1-miR-377-3P-NOTCH1 axis [29].
Therefore, we speculate that hsa_circ_0016863 can promote proliferation, metastasis and inhibit apoptosis of HCC through the circRNA-miRNA-mRNA regulatory axis. Hsa_circ_0016863 may own the function of sponge to regulate the signaling pathway of HCC cell proliferation and growth by absorbing a certain miRNA, and finally promote the progression of HCC.
Collectively, the above various experimental results imply that hsa_circ_0016863 is closely associated with the progression of HCC and has great potential in clinical applications.
Ethical Approval and Informed Consent Statement: Written informed consents were obtained from all participants, and this study was permitted by the Ethics Committee of Nantong City No.1 People’s Hospital (IRB No. 2021KT020, Approval Date: 2021.04.16).
Availability of Data and Materials: The dataset supporting the conclusions of this article is included within this article and is available from the corresponding author upon request.
Authors’ Contributions: P.Z. and Y.X. jointly collected the paper and wrote the manuscript. A.W., Y. H., A.C. and X.W. revised the manuscript. X.C. and F.W. participated in the review and helped revise the manuscript. All authors read and approved the final manuscript.
Funding Statement: This work was financially supported by the National Natural Science Foundation of China (81873978), the Key Project of Social Development in Jiangsu Province (BE2019691), the Sixth Talent Peaks Project of Jiangsu Province (2018-WSW-068), Chinese Postdoctoral Science Foundation (2018M642298), Jiangsu Postdoctoral Research Fund Project (2021K012A), Nantong Science and Technology Project (MS22021001).
Conflicts of Interest: As an Editorial board member of Oncologie, the author Feng Wang did not participate in the review process of this manuscript. Other authors declare that they have no conflicts of interest to report regarding the present study.
1. Kulik, L., El-Serag, H. B. (2019). Epidemiology and management of hepatocellular carcinoma. Gastroenterology, 156(2), 477–491. DOI 10.1053/j.gastro.2018.08.065. [Google Scholar] [CrossRef]
2. Mak, L. Y., Cruz-Ramón, V., Chinchilla-López, P., Torres, H. A., LoConte, N. K. et al. (2018). Global epidemiology, prevention, and management of hepatocellular carcinoma. American Society of Clinical Oncology Education Book, 38(38), 262–279. DOI 10.1200/EDBK_200939. [Google Scholar] [CrossRef]
3. Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A. et al. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. A Cancer Journal for Clinicians, 68(6), 394–424. DOI 10.3322/caac.21492. [Google Scholar] [CrossRef]
4. Bang, A., Dawson, L. A. (2019). Radiotherapy for HCC: Ready for prime time? JHEP Reports: Innovation in Hepatology, 1(2), 131–137. DOI 10.1016/j.jhepr.2019.05.004. [Google Scholar] [CrossRef]
5. El-Serag, H. B., Marrero, J. A., Rudolph, L., Reddy, K. R. (2008). Diagnosis and treatment of hepatocellular carcinoma. Gastroenterology, 134(6), 1752–1763. DOI 10.1053/j.gastro.2008.02.090. [Google Scholar] [CrossRef]
6. Cheng, D., Wang, J., Dong, Z., Li, X. (2021). Cancer-related circular RNA: Diverse biological functions. Cancer Cell International, 21(1), 11. DOI 10.1186/s12935-020-01703-z. [Google Scholar] [CrossRef]
7. Li, S., Weng, J., Song, F., Li, L., Xiao, C. et al. (2020). Circular RNA circZNF566 promotes hepatocellular carcinoma progression by sponging miR-4738-3p and regulating TDO2 expression. Cell Death and Disease, 11(6), 452. DOI 10.1038/s41419-020-2616-8. [Google Scholar] [CrossRef]
8. Liu, Z., Yu, Y., Huang, Z., Kong, Y., Hu, X. et al. (2019). CircRNA-5692 inhibits the progression of hepatocellular carcinoma by sponging miR-328-5p to enhance DAB2IP expression. Cell Death And Disease, 10(12), 900. DOI 10.1038/s41419-019-2089-9. [Google Scholar] [CrossRef]
9. Wang, L., Long, H., Zheng, Q., Bo, X., Xiao, X. et al. (2019). Circular RNA circRHOT1 promotes hepatocellular carcinoma progression by initiation of NR2F6 expression. Molecular Cancer, 18(1), 119. DOI 10.1186/s12943-019-1046-7. [Google Scholar] [CrossRef]
10. Liang, W. C., Wong, C. W., Liang, P. P., Shi, M., Cao, Y. et al. (2019). Translation of the circular RNA circβ-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biology, 20(1), 84. DOI 10.1186/s13059-019-1685-4. [Google Scholar] [CrossRef]
11. Chen, J., Chen, T., Zhu, Y., Li, Y., Zhang, Y. et al. (2019). circPTN sponges miR-145-5p/miR-330-5p to promote proliferation and stemness in glioma. Journal of Experimental and Clinical Cancer Research: CR, 38(1), 398. DOI 10.1186/s13046-019-1376-8. [Google Scholar] [CrossRef]
12. Kong, Y., Li, Y., Luo, Y., Zhu, J., Zheng, H. et al. (2020). circNFIB1 inhibits lymphangiogenesis and lymphatic metastasis via the miR-486-5p/PIK3R1/VEGF-C axis in pancreatic cancer. Molecular Cancer, 19(1), 82. DOI 10.1186/s12943-020-01205-6. [Google Scholar] [CrossRef]
13. Chen, R. X., Liu, H. L., Yang, L. L., Kang, F. H., Xin, L. P. et al. (2019). Circular RNA circRNA_0000285 promotes cervical cancer development by regulating FUS. European Review For Medical Pharmacological Sciences, 23(20), 8771–8778. DOI 10.26355/eurrev_201910_19271. [Google Scholar] [CrossRef]
14. Liu, J., Xue, N., Guo, Y., Niu, K., Gao, L. et al. (2019). CircRNA_100367 regulated the radiation sensitivity of esophageal squamous cell carcinomas through miR-217/Wnt3 pathway. Aging (Albany NY), 11(24), 12412–12427. DOI 10.18632/aging.102580. [Google Scholar] [CrossRef]
15. Kristensen, L. S., Hansen, T. B., Venø, M. T., Kjems, J. (2018). Circular RNAs in cancer: Opportunities and challenges in the field. Oncogene, 37(5), 555–565. DOI 10.1038/onc.2017.361. [Google Scholar] [CrossRef]
16. Zhang, P. F., Gao, C., Huang, X. Y., Lu, J. C., Guo, X. J. et al. (2020). Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Molecular Cancer, 19(1), 110. DOI 10.1186/s12943-020-01222-5. [Google Scholar] [CrossRef]
17. Xu, J., Ji, L., Liang, Y., Wan, Z., Zheng, W. et al. (2020). CircRNA-SORE mediates sorafenib resistance in hepatocellular carcinoma by stabilizing YBX1. Signal Transduction and Targeted Therapy, 5(1), 298. DOI 10.1038/s41392-020-00375-5. [Google Scholar] [CrossRef]
18. Huang, X. Y., Zhang, P. F., Wei, C. Y., Peng, R., Lu, J. C. et al. (2020). Circular RNA circMET drives immunosuppression and anti-PD1 therapy resistance in hepatocellular carcinoma via the miR-30-5p/snail/DPP4 axis. Molecular Cancer, 19(1), 92. DOI 10.1186/s12943-020-01213-6. [Google Scholar] [CrossRef]
19. Yang, B., Zhao, J., Huo, T., Zhang, M., Wu, X. (2020). Effects of CircRNA-ITCH on proliferation and apoptosis of hepatocellular carcinoma cells through inhibiting Wnt/β-catenin signaling pathway. Official Journal of the Balkan Union of Oncology, 25(3), 1368–1374. [Google Scholar]
20. Zhang, Y., Wang, H., Li, C., Gao, L., Zheng, Y. et al. (2020). CircSMYD4 regulates proliferation, migration and apoptosis of hepatocellular carcinoma cells by sponging miR-584-5p. Cancer Cell International, 20(1), 556. DOI 10.1186/s12935-020-01648-3. [Google Scholar] [CrossRef]
21. Zhu, P., Liang, H., Huang, X., Zeng, Q., Liu, Y. et al. (2020). Circular RNA Hsa_circ_0004018 inhibits Wnt/β-Catenin signaling pathway by targeting microRNA-626/DKK3 in hepatocellular carcinoma. Onco Targets and Therapy, 13, 9351–9364. DOI 10.2147/OTT.S254997. [Google Scholar] [CrossRef]
22. Shukla, G. C., Singh, J., Barik, S. (2011). MicroRNAs: Processing, maturation, target recognition and regulatory functions. Molecular and Cellular Pharmacology, 3(3), 83–92. [Google Scholar]
23. Verduci, L., Strano, S., Yarden, Y., Blandino, G. (2019). The circRNA-microRNA code: Emerging implications for cancer diagnosis and treatment. Molecular Oncology, 13(4), 669–680. DOI 10.1002/1878-0261.12468. [Google Scholar] [CrossRef]
24. Ying, X., Zhu, J., Zhang, Y. (2019). Circular RNA circ-TSPAN4 promotes lung adenocarcinoma metastasis by upregulating ZEB1 via sponging miR-665. Molecular Genetics and Genomic Medicine, 7(12), e991. DOI 10.1002/mgg3.991. [Google Scholar] [CrossRef]
25. Zhou, W., Wang, H., Yang, J., Long, W., Zhang, B. et al. (2019). Down-regulated circPAPPA suppresses the proliferation and invasion of trophoblast cells via the miR-384/STAT3 pathway. Bioscience Reports, 39(9), BSR20191965. DOI 10.1042/BSR20191965. [Google Scholar] [CrossRef]
26. Wang, Y., Guo, T., Liu, Q., Xie, X. (2020). CircRAD18 accelerates the progression of acute myeloid leukemia by modulation of miR-206/PRKACB axis. Cancer Management and Research, 12, 10887–10896. DOI 10.2147/CMAR.S277432. [Google Scholar] [CrossRef]
27. Inamura, N., Kimura, T., Wang, L., Yanagi, H., Tsuda, M. et al. (2017). Notch1 regulates invasion and metastasis of head and neck squamous cell carcinoma by inducing EMT through c-Myc. Auris Nasus Larynx, 44(4), 447–457. DOI 10.1016/j.anl.2016.08.003. [Google Scholar] [CrossRef]
28. Natsuizaka, M., Whelan, K. A., Kagawa, S., Tanaka, K., Giroux, V. et al. (2017). Interplay between Notch1 and Notch3 promotes EMT and tumor initiation in squamous cell carcinoma. Nature Communications, 8(1), 1758. DOI 10.1038/s41467-017-01500-9. [Google Scholar] [CrossRef]
29. Huang, Z., Ding, Y., Zhang, L., He, S., Jia, Z. et al. (2020). Upregulated circPDK1 promotes RCC cell migration and invasion by regulating the miR-377-3P-NOTCH1 axis in renal cell carcinoma. OncoTargets and Therapy, 13, 11237–11252. DOI 10.2147/OTT.S280434. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |