

 | Oncologie |  |
DOI: 10.32604/Oncologie.2021.018869
REVIEW
Epigenetics of Sirtuins: Relevance to Hepatocellular Carcinoma
1National Drug Clinical Trial Center, The First Affiliated Hospital of Bengbu Medical College, Bengbu, 233000, China
2School of Pharmacy, Bengbu Medical College, Bengbu, 233000, China
3School of Design, Hefei University, Hefei, 230601, China
4The Affiliated Anqing Hospital of Anhui Medical University, Anqing, 230022, China
5School of Pharmacy, Anhui Medical University, Hefei, 230032, China
*Corresponding Authors: Jing Xie. Email: byyfyxj@126.com; Huan Zhou. Email: zhouhuanbest@126.com
#Xingyu Zhu, Yongjun Wang and Shuang Chang contributed equally to this work
Received: 23 August 2021; Accepted: 19 October 2021
Abstract: Sirtuins (SIRTs), members of the enzyme family found in yeast cells, are related to silent information regulator (SIR) 2 homologous to its gene family. SIRTs play an important role in many physiological functions from overexpression of gene silencing at the molecular level to the expression of related proteins and RNA to apoptosis. Studies have indicated that SIRTs may be related to the occurrence, development, and metastasis of cancer. However, the current mechanism of action of SIRTs in various diseases and the principle of molecular biology are not fully understood. Therefore, the present article discusses the main regulatory role and function of SIRTs in HCC and evaluates SIRTs as new targets for HCC treatment.
Keywords: Sirtuins (SIRTs); hepatocellular carcinoma (HCC); deacetylation; microRNA (miRNA); methylation
| Abbreviations | |
| AFP: | α-fetoprotein; |
| ART: | ADP-ribosyltransferase; |
| AS lncRNAs: | antisense long noncoding RNAs; |
| AKT: | p-protein kinase B; |
| Bax: | BCL2-associated X protein; |
| DOX: | doxorubicin; |
| EMT: | epithelial-to-mesenchymal transition; |
| FASN: | fatty acid synthase; |
| FOXO: | forkhead box O; |
| GPC3: | glypican-3; |
| GSK-3β: | glycogen synthase kinase 3β; |
| HDAC: | histone deacetylases; |
| HIF-1α: | hypoxia-inducible factor 1-α; |
| HPV16: | human papillomaviruses 16; |
| MiRNAs: | microRNAs; |
| NAD+: | nicotinamide adenine dinucleotide; |
| NAFLD: | nonalcoholic fatty liver disease; |
| PARP: | poly ADP-ribose polymerase; |
| PI3K: | phosphoinositide-3-kinase; |
| PTEN: | phosphatase and tensin homolog deleted on chromosome ten; |
| PTM: | posttranslational modification; |
| RES: | resveratrol; |
| ROS: | reactive oxygen species; |
| SIR2: | silent information regulator 2; |
| SIRTs: | sirtuins; |
| SOD2: | superoxide dismutase 2; |
| SREBP: | sterol regulatory element binding protein; |
| TCA: | tricarboxylic acid; |
| TERT: | telomerase reverse transcriptase; |
| USP22: | ubiquitin-specific peptidase 22; |
| YAP2: | yes-associated protein 2; |
| ZEB2: | zinc finger E-box binding homeobox 2 |
Hepatocellular carcinoma (HCC) is one of the most common fatal tumors and affects the physical and mental health of high-risk groups. A previous study issued a report stating that the global incidence of HCC ranks among all cancers of all tumors, and its death ranks third (8.3% of all cancer deaths) [1,2]. East Asia, especially China, has the most HCC patients. However, patients with early HCC have no obvious symptoms, and they often miss the opportunity for the best treatment. Most patients are usually diagnosed when HCC enters the advanced stages. At this time, the survival time generally does not exceed one year [3]. The current main treatment options include surgical resection, liver transplantation, local radiotherapy, chemotherapy, combination therapy, and targeted therapy [4]. Despite the continuous improvement of treatment methods, the prognosis of patients with advanced liver cancer is still poor [5]. The latest targeted therapy drugs significantly prolong the median survival time of patients with advanced HCC, but drug resistance gradually appears in the later period of treatment [6]. Therefore, the clinically urgent problem is to obtain high-precision identification of early- and mid-stage HCC tumor biomarkers, block the phenomenon of immune escape, and solve the problems of drug resistance, disease recurrence, and metastasis. The occurrence and development of HCC is an extremely complex pathological process caused by a variety of factors, such as aflatoxin B1 intake, alcohol consumption, HBV, and HCV, and its molecular mechanism is still unclear. Studies have shown that various liver inflammatory diseases are the main reason for the progression of HCC [7]. Among them, repeated mutations of HCC chromatin regulatory factors have been confirmed by whole-genome sequencing, and the activity and structure of chromatin are deeply affected by these mutations. Histones undergo posttranslational modification after chromatin regulator methylation, acetylation, and ubiquitination to regulate gene expression [8]. Among them, Class I and Class II histone deacetylases (HDACs) are currently new strategies for the treatment of HCC with compounds, such as pabirestat. Sirtuins (SIRTs) constitute Class III HDACs and, therefore, may be another representative of potential targeted chromatin modulators [9]. Currently, studies have shown that the abnormal expression of SIRTs in HCC may be related to the occurrence, development, metastasis, and prognosis of tumors. SIRT proteins, including seven subtypes (SIRT1-SIRT7), are highly conserved nicotinamide adenine dinucleotide (NAD+)-dependent deacetylases, and their structures contain a conserved catalytic core domain. SIRTs are localized in different cellular compartments, and they have differences in functionality [10]. In mammals, each member has a unique subcellular localization, function, and substrate specificity [11,12]. The dependence of SIRTs on NAD+ indicate that the activity of SIRTs can be used as a sensor for the cytoplasmic ratio of NAD+/NADH, thereby directly linking the activity of SIRTs with cell metabolism and cellular energy status. Since the discovery of the enzyme activity of SIRTs, various physiological functions of SIRTs in the body have also been discovered [13]. In this review, we summarize the functions and roles of SIRTs in HCC and provide references for future research.
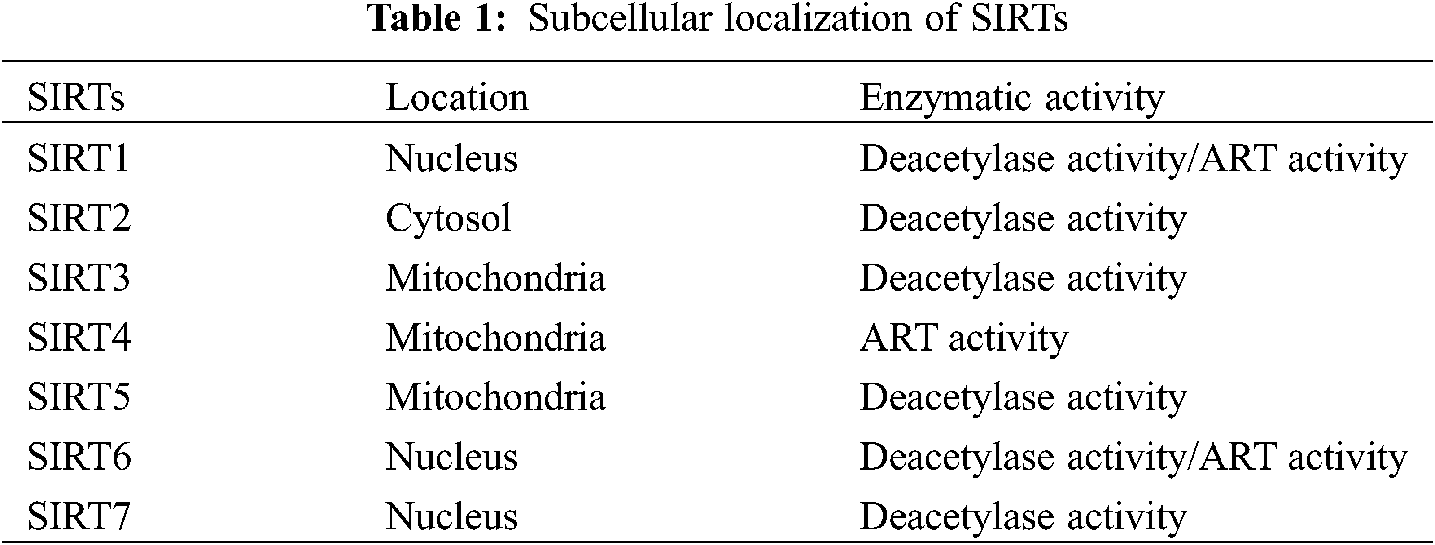
A previous study has shown that SIRT1 is related to the stability of genome structure, chromatin silencing, and histone deacetylase in simple eukaryotes [14]. SIRTs are Class III highly conserved histone deacetylases that have homology with yeast SIR2 [15]. SIR2 was first discovered as a yeast ortholog, and SIRTs have also been found in prokaryotes and multicellular animals. Recent research has shown that SIRTs are activated in different organs and tissues. In addition, SIRT proteins have different localizations according to classification, such as the nucleus (SIRT1, SIRT6, and SIRT7) [11,12], cytoplasm (SIRT2), and mitochondria (SIRT3-SIRT5) [16,17]. Due to the different localizations, SIRTs are exposed to different environments, resulting in various physiological and potential pathological changes. SIRTs have NAD+-dependent deacetylase activity. SIRT1 and SIRT6 not only have deacetylase activity but also have weak ADP-ribosyltransferase (ART) activity [18,19]. However, SIRT3 has only ART activity [20] (Table 1).
Sirtuins are proteases that are found in a variety of cancers and play different roles at different stages of cancer. For example, SIRT1 enhances the DNA damage response, changes the repair mechanism function of DNA in cells, and promotes the occurrence of cancer [21]. In addition, SIRT1 is located at the promoters of a considerable number of abnormally silenced tumor suppressor genes. The DNA of these genes is highly methylated, which prevents the normal expression of tumor suppressor genes and promotes the development of cancer [22,23]. Furthermore, HepG2 and SMMC-7721 cellular studies have revealed that SIRT1 decreases the expression of E-cadherin and increases the expression of vimentin, Snail, and Twist1 in cells. Moreover, SIRT1 promotes the expression of epithelial-mesenchymal transition (EMT) and enhances invasion and metastasis ability, thereby promoting metastasis [24,25]. Moreover, sirtuins play an indispensable role in the mechanism of resistance. A previous study has shown that autophagy plays an important role in tumor resistance [26–28]. A previous study has shown that sorafenib, a small molecule-targeted drug for the treatment of HCC, also exhibits drug resistance in the later stage of treatment. The last study showed that miR-425 regulates SIRT1 in HCC. When miR-425 is inhibited, SIRT1 promotes fat autophagy by mediating the autophagy process and promoting sorafenib resistance [29]. Additionally, SIRT1 promotes gene mutations to induce drug resistance in cancer cells [30].
Overall, sirtuins play an important role in the occurrence, development, transfer, and drug resistance of HCC. The exploration of sirtuins in the pathogenesis of HCC will provide a new treatment for HCC.
3 Expression and Functional Roles of SIRTs in HCC
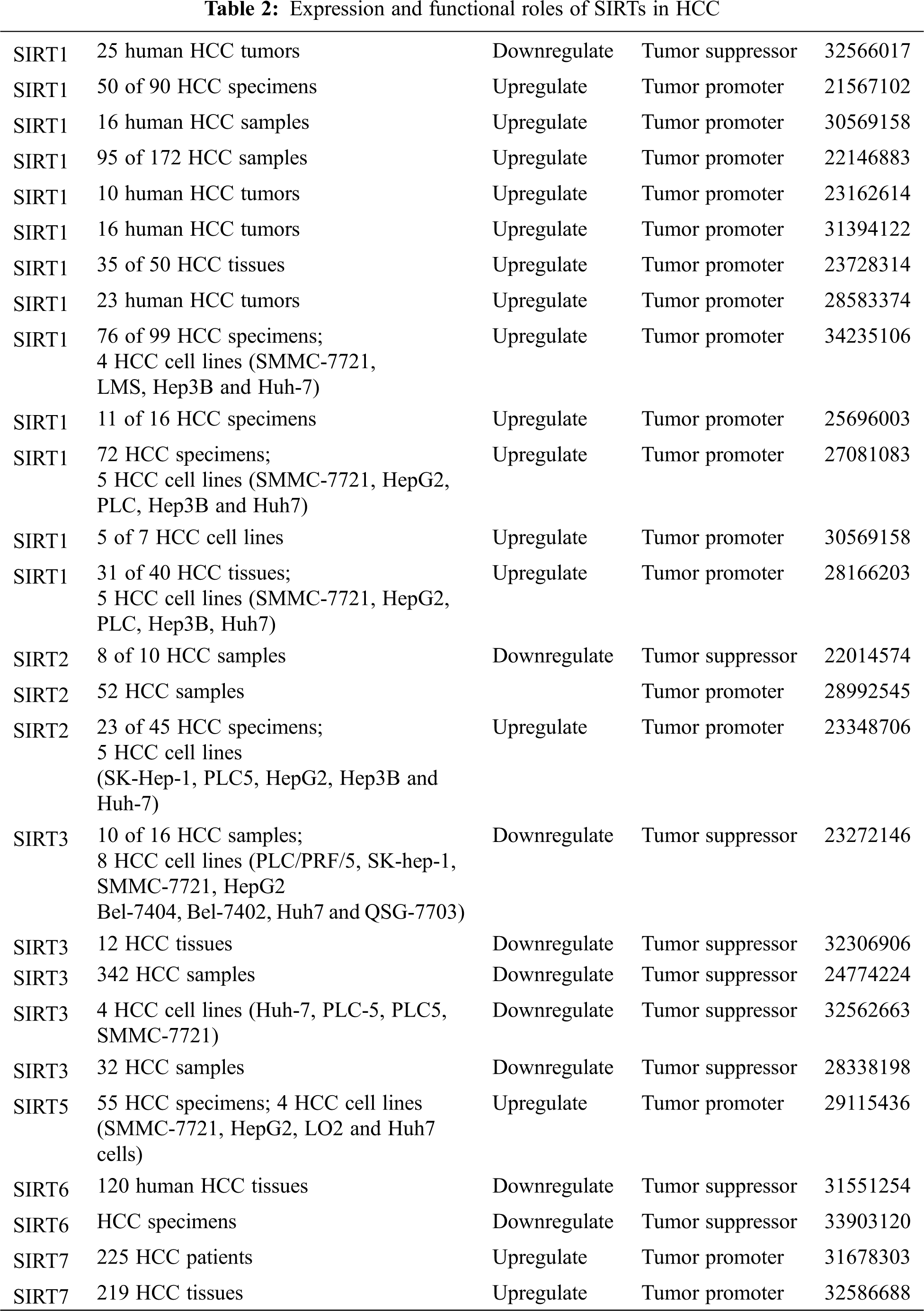
Sirtuins are involved in all stages of cancer development. Different sirtuin family members play different roles in the mechanisms of cells. This section summarizes the current reports about the regulatory effects of different SIRTs in HCC (Table 2).
SIRT1 is an NAD+-dependent histone deacetylase. SIRT1 protects cells from harsh environments by deacetylating key cell cycle molecules and apoptosis regulatory proteins [31,32]. Scientists have different views on the regulatory role of SIRT1 in tumors. On the one hand, SIRT1 is overexpressed in HCC, and the expression level of SIRT1 is positively correlated with tumor progression and grade [33,34]. Portman et al. observed that SIRT1 promotes the development of HCC cells and accelerates the formation of tumors in orthotopic xenograft models [35]. Multiple groups have also reported similar findings. HepG2 and HLF cell growth and cell proliferation are significantly inhibited after SIRT1-siRNA transduction [36,37]. The expression of SIRT1 is negatively correlated with the proliferation of HepG2 cells [37]. On the other hand, Wang et al. found that SIRT1 may play a tumor suppressor role in HCC samples and cell lines [14]. Consequently, the type of cancer and the complicated factors of upstream and downstream genes induced by SIRT1 may play different roles in cancer [38,39]. Recent studies have found that SIRT1 is overexpressed in HCC tissues and may be involved in tumor development [35].
In another study, Hao et al. found that SIRT1 protein is overexpressed in HCC cell lines, which causes epithelial-mesenchymal transition (EMT) in HCC, thereby promoting cancer migration and invasion [24]. EMT is a sequence event that converts adherent epithelial cells into migratory cells that invade the extracellular matrix, which is related to the metastasis of tumor cells [40]. The study of SIRT1 in liver cancer stem cells (CSCs) has further revealed that overexpression and continuous self-renewal of such cancer cells are related to their carcinogenic potential in the HCC cell subset with stem cell-like characteristics [41]. SIRT1 silencing causes telomere dysfunction and nuclear abnormalities, which correlate with the downregulation of telomerase reversed transcriptase (TERT) and PTOP expression, and ectopic expression of TERT or PTOP significantly restores cell proliferation in SIRT1-depleted cells. Additionally, a previous study has indicated that SIRT1 knockdown inhibits the development of HCC and causes further depletion of PTOP and TERT [42]. Additionally, Zhou et al. found that SIRT1 is an essential regulator of the immune reaction that inhibits the migration and growth of malignant HCC cells, suggesting that macrophage SIRT1 is an innovative target to treat HCC [43]. Moreover, resveratrol ameliorates oxidative stress in the kidneys of HCC rats in a catalase-dependent and glutathione peroxidase-independent manner [44].
Overall, the mechanism of SIRT1 in cancer is complex and changeable. The overexpression of SIRT1 in the cytoplasm indicates a longer overall survival rate, but the high expression of SIRT1 in the nucleus indicates a lower overall survival rate. Therefore, it is necessary to redefine the role of SIRT1 in HCC [45,46].
SIRT2 is a histone deacetylase that regulates the expression of DNA and RNA in cells, maintains the stability of gene structure, and mediates cell apoptosis as a chromatin silencer. SIRT2 exists in the cytoplasm and colocalizes with microtubules. Some substrates of SIRT1 are the focal point of SIRT2, such as H4K16, p53, and p65. Furthermore, recent studies have shown that the cytoplasmic localization, role in tubulin dynamics, and neuronal movement regulation of SIRT2 may have different functions compared with SIRT1 [47,48]. Other studies have found that SIRT2 plays a dual role in tumorigenesis and development as the expression of SIRT2 in gliomas is significantly reduced [49] but is elevated in other cancers [50]. Consequently, there is still controversy about SIRT2-regulated metabolism and molecular mechanisms in HCC cells [51].
Studies have shown that SIRT2 regulates epigenetic changes associated with the biological activities of malignant tumors [52]. Inhibiting the expression of SIRT2 induces tumor occurrence. In addition, the reduction rate of SIRT2 has been shown to be 80% in 10 HCC samples, suggesting that SIRT2 may inhibit human HCC [16,51].
Additionally, siRNA-mediated silencing of SIRT2 expression has revealed a significant reduction in movement and invasion in HCC cell lines. The results have shown that the expression of SIRT2 is negatively correlated with the expression of EMT markers [52,53]. Additionally, the mechanism of SIRT2-induced HCC in HBV has also been studied, demonstrating that HBV stimulates the expression of SIRT2. Moreover, hepatitis B virus X (HBx) activates the SIRT2 promoter and upregulates the expression levels of SIRT2 mRNA and protein in HepG2 and Huh-7 cells [54]. Another study has shown that HBV transcription and replication are promoted by SIRT2 overexpression. Previous studies have found that SIRT2 may play some potential roles in HBV and HBV-mediated HCC, and these effects may be related to HBx [54].
In summary, these findings indicate that the migration and invasion of human HCC cells are correspondingly weakened when the expression of SIRT2 is reduced. Therefore, inhibiting SIRT2 may be an effective treatment for HCC [51].
SIRT3, as a mitochondrial SIRT, has deacetylase activity and regulates metabolism, including fatty acid oxidation, amino acid metabolism, redox balance, and tricarboxylic acid (TCA) cycle [55]. Mitochondrial redox has also been demonstrated to be regulated by SIRT3. In addition, SIRT3 plays a dual role in cancer as it is reduced in some cancers but increased in others [56–58].
Recently, a study has shown that SIRT3 is significantly reduced in HCC [59]. Additionally, SIRT3 is a prognostic cytokine for tumors as it has different expression levels in different cancer stages [60,61]. The growth of HepG2 and HuH-7 cells is inhibited by SIRT3 overexpression in vitro. Furthermore, SIRT3 overexpression causes HCC cell membrane damage and apoptosis. Song et al. found that SIRT3 induces apoptosis as a tumor suppressor in HCC [62]. In summary, SIRT3 is considered to be the most important tumor suppressor in HCC [63]. In addition, the expression of hypoxia-inducible factor 1-α (HIF-1α) is increased in HCC cells, and SIRT3 inhibits tumor development by inhibiting HIF-1α and mitochondrial reactive oxygen species (ROS) [64,65]. As a carcinogen, ROS are produced by mitochondria, which play an important role in the process of apoptosis [65]. Recent studies have indicated that ROS regulates the progression, metastasis, DNA damage, and mitochondrial pathway in HCC [65,66]. Moreover, SIRT3 promotes the sensitivity of hepatocellular carcinoma cells to regorafenib by accelerating mitochondrial dysfunction [67,68]. Overall, the occurrence and development of HCC decrease the expression of SIRT3, thereby suggesting a new method to treat HCC. However, further research is required to verify these relationships.
4 Epigenetic Role of SIRTs in HCC
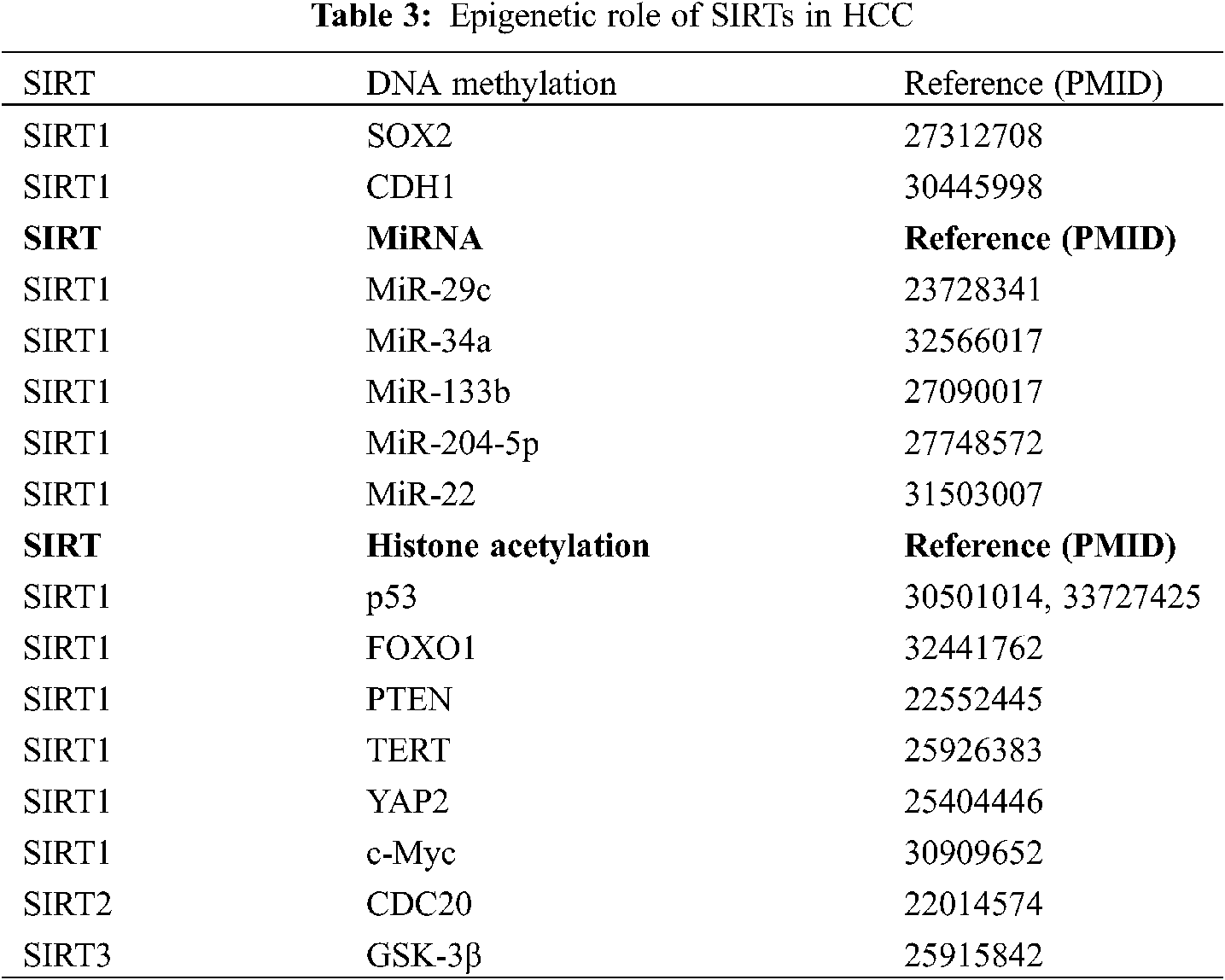
Epigenetic changes are observed in the occurrence, development, and metastasis of all cancers, and these changes include DNA methylation defects and abnormal covalent histone modifications, which are involved in the entire tumor formation process [69]. A study has shown that epigenetic modifiers inhibit HCC metastasis and proliferation in vivo and in vitro, suggesting that they are important targets for the treatment of HCC [70]. Thus, there is a relationship between epigenetic modifications and changes associated with SIRTs and the development of HCC (Table 3).
DNA Methylation
DNA hypermethylation of multiple tumor suppressor genes plays an important role in the pathogenesis of hepatocellular carcinoma. Moreover, this epigenetic defect is observed in the noncancerous liver tissues of HCC patients. Therefore, methylation-induced silencing plays a regulatory role in HCC [71].
Qi et al. found that the self-renewal of cancer stem cells is regulated by the involvement of sex-determining area Y-box 2 (SOX2). In addition, SIRT1 regulates the self-renewal and tumorigenicity of liver cancer stem cells [72]. A previous study has indicated that SIRT1 is the upstream regulator of SOX2, which mediates the self-renewal and pathogenicity of liver cancer cells by regulating the expression of SOX2. Molecularly, DNA methylation plays an important role in the regulation of SOX2 gene transcription by SIRT1 through epigenetic changes in chromatin [41]. When the E-cadherin (CDH1) promoter is hypermethylated, CDH1 is downregulated in cell models expressing the genotype 1b HCV core protein. In addition, the expression of SIRT1 increases as the HCV core protein of genotype 1b is upregulated. When SIRT1 is inhibited by its inhibitor, the methylation level of the CDH1 promoter is also downregulated. Studies have shown that SIRT1 participates in CDH1 promoter methylation in HCV-HCC [71]. Furthermore, the role of DNA methylation in the occurrence, development, and metastasis of HCC needs to be further investgated, especially experiments focusing on histone modification, methylation, and chromatin remodeling.
MiRNA
MicroRNAs (miRNAs) are evolutionarily conserved short noncoding RNAs that play a role in protein synthesis and many tumors. MiRNAs combine with target mRNAs and regulate mRNA expression after transcription, which further regulates mRNA translation or inhibits gene silencing. When the miRNA is paired with the 3′-nontranslated region (3′-UTR), a partial complementary base of the target mRNA is obtained. The biological functions of cells, such as differentiation, may be inhibited. In addition, cancer, cardiovascular diseases, and many other diseases and disorders are related to miRNAs [73]. Additionally, miRNAs regulate the expression of SIRT1 in various tissues. For example, Zia et al. [74] found that miR-9, miR-204, miR-199b, and miR-135a modulate SIRT1 expression in aging and age-associated diseases. Recently, a study has demonstrated that miRNAs participate in the occurrence and development of HCC by regulating SIRT1 (Fig. 1).
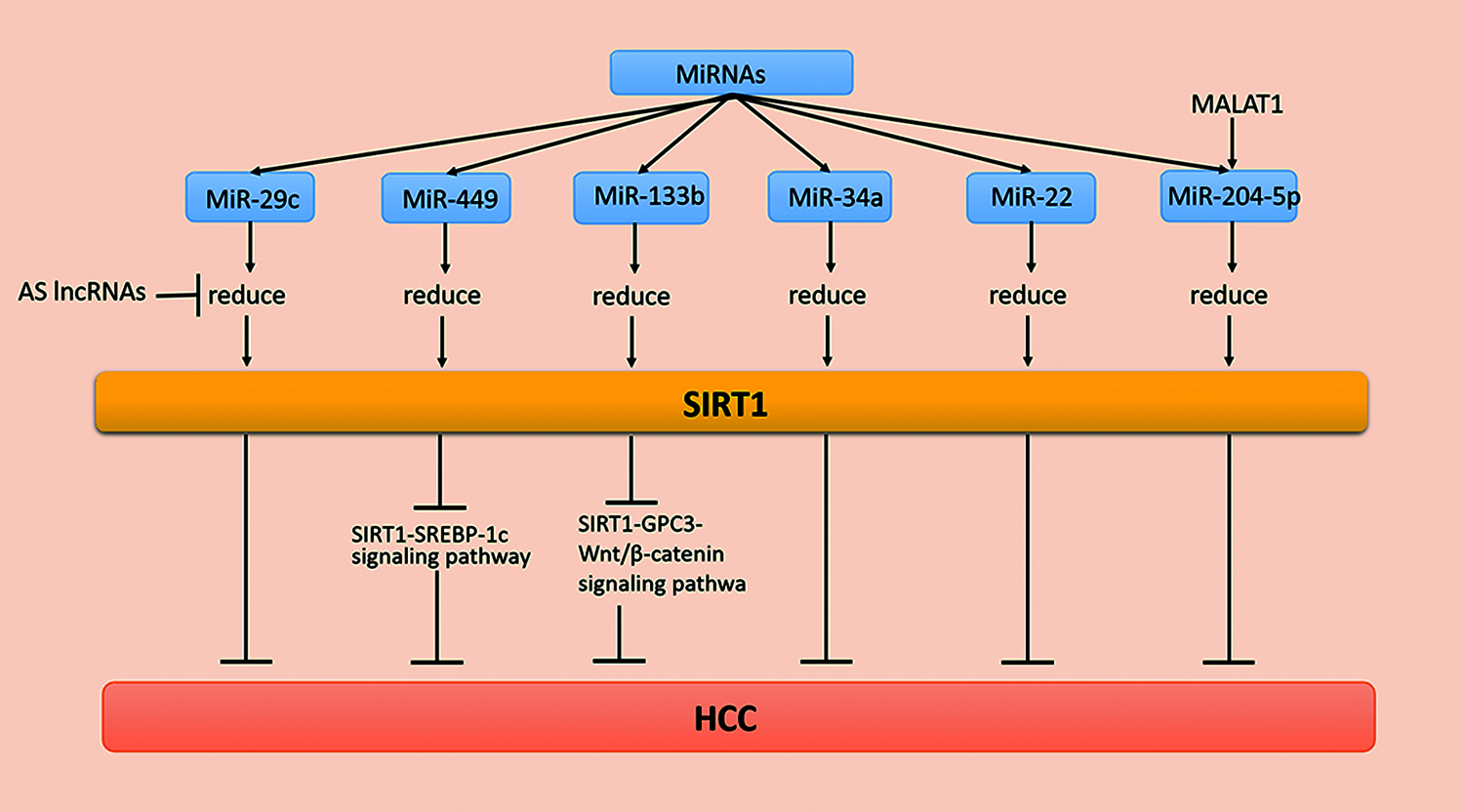
Figure 1: MiRNA regulation of SIRT1 in HCC
The low survival rate of HCC is related to miR-29c, a member of the miR-29 family [75]. Studies have shown that overexpression of miR-29c suppresses SIRT1 levels and reduces the growth and proliferation of HCC cells. It is worth noting that the decreased expression of miR-29c in HCC patients may cause a poor prognosis for HCC patients.
MiR-29c inhibits the biofunction of hepatoma cells by binding carcinogenic SIRT1 with the 3′-UTR of SIRT1 mRNA. For example, miR-29c suppresses SIRT1 in HCC as a tumor inhibitor [76]. In addition, most antisense long noncoding RNAs (AS lncRNAs) are found in natural sense transcription (NAT). Moreover, several AS lncRNAs are disordered and overexpressed in cancer tissues during the development of tumorigenesis. When SIRT1-AS binds to SIRT1 mRNA at the 3′-UTR, it competitively inhibits miR-29c from binding at the binding site, which inhibits SIRT1 protein levels, causing the malignant proliferation of HCC cells [77].
MiR-34a, as a tumor suppressor, inhibits tumor growth. For example, miR-34a arrests cells in the G1 phase by inhibiting the expression of cell cycle regulatory genes and other oncogenes. Additionally, lower expression levels of miR-34a causes poor prognosis in patients with HCC [78]. The 3′-UTR of SIRT1 is the direct target of miR-34a, while miR-34a inhibits the expression of tumor suppressor genes and antiapoptotic genes by acting on SIRT1 mRNAs. These studies provide multipathway selection for HCC treatment [78]. Furthermore, miR-133b is significantly reduced in various cancer types as a tumor suppressor. A previous study has indicated that miR-133b is involved in the progression of HCC. In addition, the upregulation of SIRT1 induces the expression of glypican-3 (GPC3) and then reverses the effects of transcription factor β-catenin cytoplasm accumulation and nuclear translocation [79]. In conclusion, miR-133b suppresses the occurrence and development of cancer by reducing the expression of the SIRT1/GPC3/Wnt/β-catenin signaling pathway and induces the death of cancer cells. These findings provide new ideas and new targets for treating patients with HCC [80]. Additionally, miR-204-5p inhibits HCC metastasis and vascular infiltration, while miR-204-5p expression is downregulated in the late (III and IV) subgroups. MiR-204-5p in BEL-7405 cells reduces the expression of the 3′-UTR of SIRT1. Studies have indicated that miR-204-5p acts directly on SIRT1. In vivo experiments have shown that miR-204-5p downregulates the HCC cell cycle and metastasis through SIRT1. In summary, these findings indicate that miR-204 may become a new strategy for the treatment of HCC metastasis as an inhibitor of SIRT1 [81,82]. Interestingly, another study has shown that miR-22, which binds the 3′-UTR of SIRT1, is underexpressed in human HCC tissues and acts on the target SIRT1. MiR-22 reduces SIRT1, p-protein kinase B (AKT), and β-catenin expression levels but increases glycogen synthase kinase 3β (GSK-3β) and phosphatase and tensin homologs on chromosome 10 (PTEN) expression. Additionally, miR-22 inhibits the occurrence, development, and metastasis of cancer cells as a tumor suppressor gene. Therefore, inhibiting the expression of miR-22 may be a new strategy for the treatment of HCC [83,84]. The above studies suggest that the expression of SIRT1 at the protein level deserves more attention, and the various relationships between SIRT1 and miRNA expression provide novel ideas for the treatment of HCC [84].
Histone Acetylation
Histones and nonhistone proteins regulate cell signal transduction and gene expression by acetylation. Previous studies have confirmed that SIRT1 has a deacetylation function, which silences the expression of genes through histone deacetylation. In addition, SIRT1 also regulates the expression of tumor suppressor factors, such as FOXO protein and p53 [85] (Fig. 2).
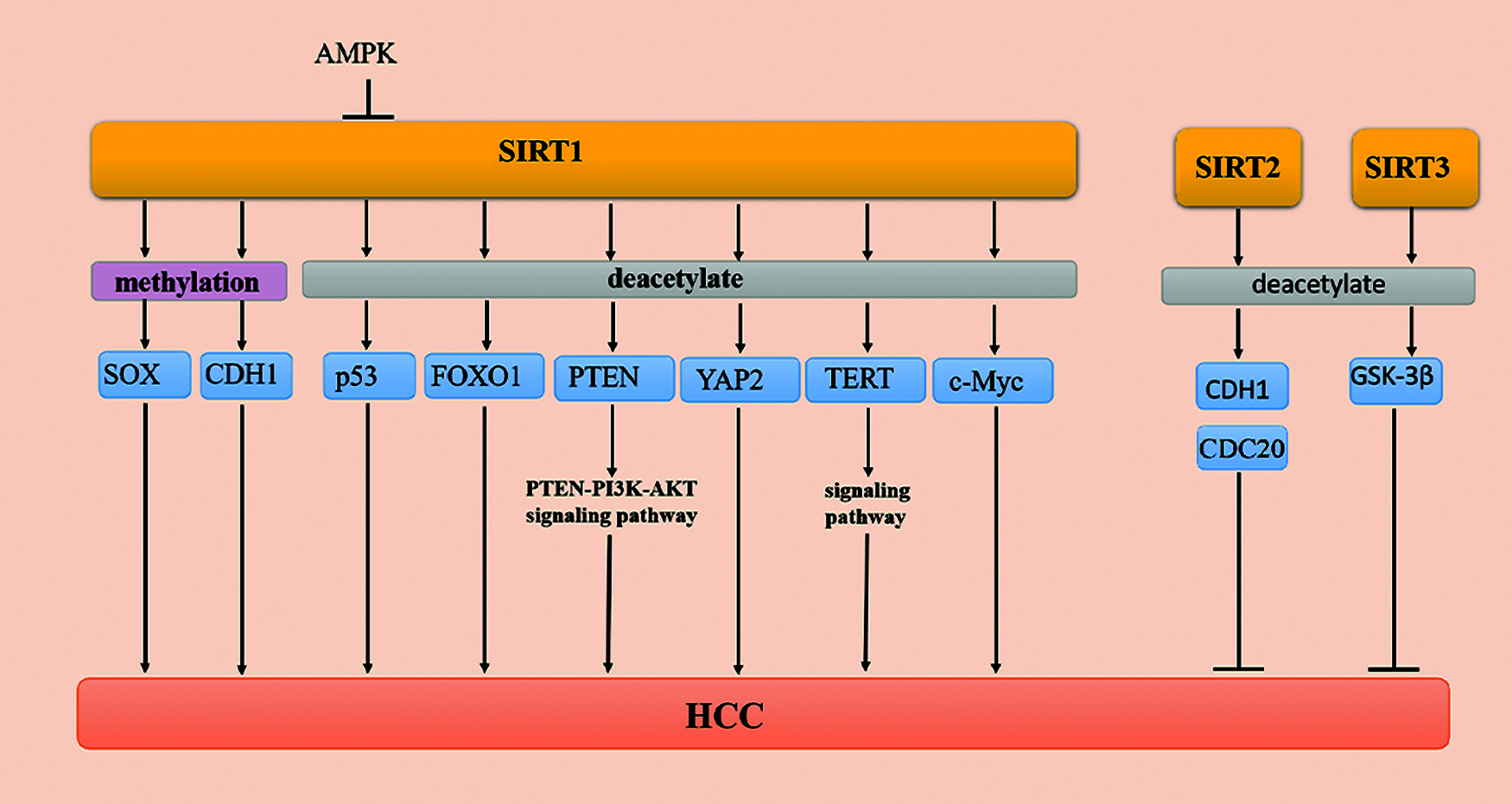
Figure 2: Methylation and acetylation of SIRTs in HCC
p53 is involved in the change of the chromatin structure of the core promoter and has a negative correlation with the expression of the AFP gene [33]. There is no clear conclusion about the relationship among the production, expression, and role of SIRT1 and AFP in HCC. A previous study has indicated that p53 inhibits the activity of the AFP promoter, while SIRT1 deacetylates the p53 protein to reduce its transcriptional activity and then increases the activity of the AFP promoter in HepG2 cells [86].
The AMP-activated protein kinase (AMPK) signaling pathway is a serine/threonine-protein kinase that maintains energy homeostasis balance in response to metabolic stress as sensors in cells. Notably, the activation of AMPK reduces a variety of cancer symptoms, including HCC [87]. Recent studies have shown that AMPK directly phosphorylates SIRT1 at Thr344 to reduce SIRT1 deacetylase activity on p53 and then inhibits the development of cancer by activating the p53-dependent apoptosis pathway [88]. These results demonstrate that the AMPK signaling pathway inhibits the occurrence and migration of HCC [89]. It has been shown that the AMPK/SIRT1/p53 signaling pathway plays an integral-regulatory role in HCC.
FOXO family members are considered to be important transcription factors involved in the regulation of a variety of cancers, including HCC [90,91]. The EMT program is reversed by FOXO1 as the overexpression of FOXO1 reduces the expression of mesenchymal markers. FOXO1 silencing increases the level of epithelial markers and inhibits the proliferation of human HCC cells [92,93]. The level of EMT inducers have an inverse relationship with the level of FOXO1 and positively correlates with the binding of the zinc finger E-box to homeobox 2 (ZEB2) in HCC cells [93]. The decrease in the acetylation level of the FOXO1 substrate of SIRT1 is related to the increase in SIRT1, indicating that the expression and activity of SIRT1 are consistent with the decrease [94]. Studies have shown that FOXO1 overexpression may be a new strategy in the treatment of HCC.
PTEN is a member of the tumor suppressor family, and it participates in various activities of the human body, including regulating gene expression, cell cycle progression, and apoptosis. Additionally, PTEN is downregulated in cancer as a tyrosine phosphatase [95,96]. Studies have shown that SIRT1 plays a negative regulatory role in the phosphoinositide 3-kinase (PI3K)/AKT signaling pathway and promotes the proliferation of cancer cells [97]. When PTEN is acetylated, the activity of SIRT is reduced, thereby activating the AKT signaling pathway [98,99]. The phosphorylation level of PI3K/AKT is significantly reduced when SIRT1 is overexpressed in HepG2 and L0-2 cells. Furthermore, the silencing of SIRT1 increases the level of acetylated PTEN and promotes cancer cell apoptosis. A recent review has suggested that the PTEN/PI3K/AKT signaling pathway interacts with SIRT1 and participates in the various stages of HCC tumorigenesis, development, and migration [100].
Telomerase is a unique ribonucleoprotein polymerase with reverse transcription activity that is composed of an RNA subunit (hTERC) and a protein catalytic subunit (hTERT). Telomerase functions to synthesize telomere DNA in the cell, maintain the length of the telomere, increase the number of cell divisions, and induce cells to proliferate indefinitely. Additionally, telomeres play a dual regulatory role in HCC tissue. On the one hand, chromosomal instability, telomere shortening, and cell cycle arrest occur in the early stages of liver cancer. On the other hand, long telomeres and telomerase activity are upregulated in advanced HCC. In addition, SIRT1 not only regulates the CpG methylation pattern of the TERT promoter but also regulates the stability of the 3′-UTR of TERT mRNA in the normal biological activity mechanism [101]. Zhang et al. found that TERT is upregulated in 40 pairs of HCC samples. In conclusion, these studies indicate that SIRT1/TERT may be regulated by regulatory axes in vivo [102]. Moreover, overexpression of SIRT1 reduces the level of histone H3-K9 acetylation but induces H3-K9 trimethylation at the TERT promoter. These studies indicate that reducing the expression of SIRT1 also induces TERT activation during the treatment of HCC, which is a challenge that still needs to be overcome [103].
YAP is a downstream regulator of the Hippo pathway that participates in the regulation of cell proliferation and apoptosis in a variety of cancers by regulating yeast transcription factors. The YAP signaling pathway plays a role in the regulation of cell proliferation and apoptosis in cancer. Zhang et al. found that excessive bile acids and cytokines induce the activation of YAP in HCC [104]. The expression of the YAP2 signaling pathway in HCC is positively correlated with the expression of SIRT1 [105]. SIRT1 is a type III deacetylase, and YAP2 is an acetylated protein. Mao et al. confirmed that when SIRT1 is silenced, the acetylation level of the YAP2 protein is significantly increased. In summary, these studies indicate that SIRT1 is the deacetylase of the YAP2 protein, and the YAP2 signaling pathway inhibits cell apoptosis in cancer cells [105].
C-Myc is a downstream signaling pathway of Wnt/β-catenin that is upregulated in a variety of cancers. The half-life of c-Myc is prolonged when c-Myc is deacetylated by SIRT1, which promotes c-Myc binding to Max, promotes cell proliferation, and inhibits cell apoptosis. Additionally, the expression of c-Myc increases when the expression of SIRT1 is upregulated. The expression of SIRT1 and c-Myc in HCC cells plays a positive regulatory role in promoting the proliferation of cancer cells [105]. Thus, SIRT1 and c-Myc regulate each other in the occurrence and development of liver tumors, and they can also be used as clinical indicators for the prognosis of HCC [106].
Studies have shown that SIRT1, as a deacetylase, regulates cellular activity. SIRT1 inhibits the PTEN/PI3K/AKT signaling pathway by cooperating with the c-Myc signaling pathway, affects the lipid metabolism of normal liver tissues, induces the expression of oncogenes, and promotes growth, migration, and drug resistance. SIRT1 participates in multilink signaling pathways and multi regulatory factors as a target for the treatment of HCC [106]. However, the molecular therapy mechanism of SIRT1 is complex, and the precise gene regulatory network is still unclear. The regulatory mechanism of SIRT1 in HCC needs to be explored by further study. Integration of the pathophysiology of HCC into SIRT1 biology will aid in further understanding of the relevant regulatory mechanisms of HCC, thereby providing new ideas for efficient, rapid treatment, and a specific diagnosis.
Studies have shown that SIRT2 (a histone deacetylase) plays a dynamic regulatory role in cancer cells [51]. Cell division cycle 20 (CDC20) overexpression has been reported in various malignancies and plays a role in tumorigenesis and tumor progression [107]. Kim et al. [16] found that SIRT2 plays a critical role in maintaining mitosis by modulating the activity of anaphase-promoting complex/cyclosome (APC/C) by deacetylation of the coactivator proteins, CDH1 and CDC20. Furthermore, SIRT2 ensures normal mitotic progression, maintains genome integrity, and suppresses HCC as a positive regulator of APC/C activity [16] (Fig. 2). However, there have been few studies on the deacetylation of SIRT2 in recent years. However, some progress has been made in understanding the regulatory role of SIRT2 in HCC.
SIRT3 is a mitochondrial deacetylase, and it regulates gene expression, cell death, energy metabolism, and carcinogenesis. The mechanism by which SIRT3 mediates tumor regulators has been elucidated. GSK-3β is a common serine/threonine kinase that is involved in glycogen metabolism and transcriptional regulation [108]. Cancer progression is suppressed when the expression of SIRT3 increases in HCC. A previous study has shown that the expression and activity of the GSK-3β signaling pathway are consistent with the expression of SIRT3. The deacetylation of GSK-3β at lysine 205 depends on the regulation of SIRT3. The GSK-3β signaling pathway is activated by SIRT3 deacetylation, which induces the expression of the proapoptotic protein, Bax, and mitochondrial translocation to promote cell apoptosis. In summary, the SIRT3/GSK-3β/Bax signaling pathway plays an important role in the occurrence, development, and metastasis of HCC, and it provides a new potential target for HCC treatment [62] (Fig. 2).
5 Potential Therapeutic Substances for HCC-Targeting SIRTs
Chen et al. discovered the potential role of SIRT1 in the treatment of HCC and determined that it enhances the efficacy of the DNA damage drug, doxorubicin (DOX) [42,109]. It is worth noting that K-Hep-1 and PLC5 cells become more sensitive to DOX when SIRT1 expression is decreased, thereby promoting the process of DOX-induced apoptosis.
Butyrate is a short-chain fatty acid produced by the fermentation of cellulose by specific bacteria in the intestine. Butyrate has been demonstrated to be used as an HDAC inhibitor for the treatment of diabetes. A recent study has demonstrated that butyrate induces ROS-mediated apoptosis by modulating miR-22-SIRT1 in HCC [83]. Another study has reported that butyrate has anticancer efficacy by inhibiting SIRT3 [110].
Metformin is widely used as a hypoglycemic agent in the treatment of type 2 diabetes. Bhalla et al. found that metformin has a positive effect on the treatment of HCC by promoting the senescence of liver cancer cells through the AMPK/SIRT1 pathway [87].
Another study has shown that MIAM inhibits the growth of HCC in drug-resistant variants of the Bel-7402 cell line by activating the SIRT3/SOD2 and SIRT3/p53/p2 signaling pathways, demonstrating that MIAM has an intervening effect on the development of HCC. In addition, potential quinazoline derivatives, as tumor-targeting drugs, target multiple receptors. A previous study has demonstrated that a newly synthesized phenylchloromethine-quinazoline derivative with SIRT1 as its target effectively promotes the apoptosis of hepatocellular carcinoma in vivo and in vitro [111].
Furthermore, some natural products show a certain antitumor effect. For example, quercetin induces HCC (HepG2 and Huh7 cells) growth inhibition. Additionally, when HepG2 cells are stimulated by quercetin, the expression level of miR-34a increases and the sensitivity of HCC cells to quercetin decreases [112]. In addition, a new experiment has shown that Dulcitol, a natural product extracted from Euonymus, not only downregulates the protein expression of SIRT1 and Bcl-2 but also inhibits the proliferation of liver cancer cells. Research results have shown that dulcitol induces apoptosis and inhibits the proliferation of HepG2 cells as a SIRT1 inhibitor [113]. Gallotannin is a hydrolyzable tannic acid that also promotes apoptosis and senescence of HCC cells [114]. Resveratrol (RES) is a polyphenol compound rich in grapes and red wine, and it is an anticancer mechanism that inhibits SIRT1 to induce HCC cell apoptosis [115].
Chronic hepatitis will develop into liver cirrhosis and HCC without intervention. Therefore, it is necessary to clarify the mechanism of SIRTs in HCC. The above research results all demonstrate the mechanism by which SIRT1 participates in the body, and it may be possible to make a breakthrough in the treatment of HCC in the future [116]. Additionally, many studies have provided insight into the effects of the clustered regularly spaced short palindrome repeats/CRISPR-associated protein 9 system (CRISPR/Cas9) on SIRT biology, and SIRT editing has shown its relevance to tumors in the latest research. For example, human papillomavirus 16 (HPV16) uses its E1 and E2 proteins to bind the host genome, and the SIRT1 acetylase participates in viral DNA replication. Importantly, the replication of the virus is decreased by knocking out SIRT1. Therefore, enzyme-inhibited SIRT1 may become a new method to prevent or treat HPV [117].
CRISPR/Cas9-mediated gene knockout (KO) provides a theoretical basis for the development of novel therapies for HCC. Aspartate β-hydroxylase (ASPH) expression is involved in HCC growth and progression, and its protein level is higher in HepG2 cells. ASPH knockout significantly reduces cell proliferation, thereby retarding HCC progression [118]. In addition, a new type of nuclease crisp/Cas13 specifically recognizes RNA sequences and cuts RNA. In the future, Cas13 will be used to destroy lncRNAs that increase SIRT1 in HCC. Furthermore, the ability of Cas13 to recognize specific RNAs has been demonstrated in cancer diagnosis [119,120]. Additionally, many RNAs involved in the regulation of HCC will become important targets for the diagnosis and treatment of HCC. All these data show the promising application prospects of SIRT editing in HCC.
SIRTs are promising in the treatment of diseases, and the related research is advancing. It is necessary to identify the targets of SIRTs in diseases, especially in HCC, and to clarify their mechanism of action.
Ethics Approval and Informed Consent Statement: Not Applicable.
Availability of Data and Materials: The data supporting the article is available within the article.
Author Contributions: XY-Z, YJ-W and S-C drafted and revised the manuscript by reviewing the literature. Y-S, CX-H, S-H, MH-Z and YZ-D participated in the discussion. NN-R and QZ-W drew the chart. The corresponding author J-X and H-Z conducted the entire manuscript. All authors have read and approved the final manuscript.
Funding Statement: 512 Talent Cultivation Plan of Bengbu Medical College (by51201313) Natural Science Foundation of Anhui Province (2008085QH401).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I. et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. A Cancer Journal for Clinicians, 71(3), 209–249. DOI 10.3322/caac.21660. [Google Scholar] [CrossRef]
2. Feng, S., Li, M., Zhang, J., Liu, S., Wang, Q. et al. (2015). Regulation of HepG2 cell apoptosis by hepatitis C virus (HCV) core protein via the sirt1-p53-bax pathway. Virus Genes, 51(3), 338–346. DOI 10.1007/s11262-015-1253-2. [Google Scholar] [CrossRef]
3. Hollebecque, A., Malka, D., Ferte, C., Ducreux, M., Boige, V. (2015). Systemic treatment of advanced hepatocellular carcinoma: From disillusions to new horizons. European Journal of Cancer, 51(3), 327–339. DOI 10.1016/j.ejca.2014.12.005. [Google Scholar] [CrossRef]
4. Longerich, T. (2020). Hepatocellular carcinoma. Pathologe, 41(5), 478–487. DOI 10.1007/s00292-020-00801-z. [Google Scholar] [CrossRef]
5. Cainap, C., Qin, S., Huang, W. T., Chung, I. J., Pan, H. et al. (2015). Linifanib versus Sorafenib in patients with advanced hepatocellular carcinoma: Results of a randomized phase III trial. Journal of Clinical Oncology, 33(2), 172–179. DOI 10.1200/JCO.2013.54.3298. [Google Scholar] [CrossRef]
6. Kuczynski, E. A., Lee, C. R., Man, S., Chen, E., Kerbel, R. S. (2015). Effects of sorafenib dose on acquired reversible resistance and toxicity in hepatocellular carcinoma. Cancer Research, 75(12), 2510–2519. DOI 10.1158/0008-5472.CAN-14-3687. [Google Scholar] [CrossRef]
7. Berretta, M., Cavaliere, C., Alessandrini, L., Stanzione, B., Facchini, G. et al. (2017). Serum and tissue markers in hepatocellular carcinoma and cholangiocarcinoma: Clinical and prognostic implications. Oncotarget, 8(8), 14192–14220. DOI 10.18632/oncotarget.13929. [Google Scholar] [CrossRef]
8. Fu, W., Gao, L., Huang, C., Yao, J., Lin, Y. et al. (2019). Mechanisms and importance of histone modification enzymes in targeted therapy for hepatobiliary cancers. Discovery Medicine, 28(151), 17–28. [Google Scholar]
9. Chelladurai, P., Boucherat, O., Stenmark, K., Kracht, M., Seeger, W. et al. (2021). Targeting histone acetylation in pulmonary hypertension and right ventricular hypertrophy. British Journal of Pharmacology, 178(1), 54–71. DOI 10.1111/bph.14932. [Google Scholar] [CrossRef]
10. Laemmle, A., Lechleiter, A., Roh, V., Schwarz, C., Portmann, S. et al. (2012). Inhibition of SIRT1 impairs the accumulation and transcriptional activity of HIF-1alpha protein under hypoxic conditions. PLoS One, 7(3), e33433. DOI 10.1371/journal.pone.0033433. [Google Scholar] [CrossRef]
11. Kim, J. K., Noh, J. H., Jung, K. H., Eun, J. W., Bae, H. J. et al. (2013). Sirtuin7 oncogenic potential in human hepatocellular carcinoma and its regulation by the tumor suppressors MiR-125a-5p and MiR-125b. Hepatology, 57(3), 1055–1067. DOI 10.1002/hep.26101. [Google Scholar] [CrossRef]
12. Zhang, B., Qin, L., Zhou, C. J., Liu, Y. L., Qian, H. X. et al. (2013). SIRT3 expression in hepatocellular carcinoma and its impact on proliferation and invasion of hepatoma cells. Asian Pacific Journal of Tropical Medicine, 6(8), 649–652. DOI 10.1016/S1995-7645(13)60112-1. [Google Scholar] [CrossRef]
13. Liu, L., Su, X., Quinn, W. J., III, Hui, S., Krukenberg, K. et al. (2018). Quantitative analysis of NAD synthesis-breakdown fluxes. Cell Metabolism, 27(5), 1067–1080.e5. DOI 10.1016/j.cmet.2018.03.018. [Google Scholar] [CrossRef]
14. Wang, R. H., Sengupta, K., Li, C., Kim, H. S., Cao, L. et al. (2008). Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell, 14(4), 312–323. DOI 10.1016/j.ccr.2008.09.001. [Google Scholar] [CrossRef]
15. Pantazi, E., Zaouali, M. A., Bejaoui, M., Folch-Puy, E., Ben Abdennebi, H. et al. (2013). Role of sirtuins in ischemia-reperfusion injury. World Journal of Gastroenterology, 19(43), 7594–7602. DOI 10.3748/wjg.v19.i43.7594. [Google Scholar] [CrossRef]
16. Kim, H. S., Vassilopoulos, A., Wang, R. H., Lahusen, T., Xiao, Z. et al. (2011). SIRT2 maintains genome integrity and suppresses tumorigenesis through regulating APC/C activity. Cancer Cell, 20(4), 487–499. DOI 10.1016/j.ccr.2011.09.004. [Google Scholar] [CrossRef]
17. Kratz, E. M., Solkiewicz, K., Kubis-Kubiak, A., Piwowar, A. (2021). Sirtuins as important factors in pathological states and the role of their molecular activity modulators. International Journal of Molecular Sciences, 22(2), 630. DOI 10.3390/ijms22020630. [Google Scholar] [CrossRef]
18. Zhang, C., Yu, Y., Huang, Q., Tang, K. (2019). SIRT6 regulates the proliferation and apoptosis of hepatocellular carcinoma via the ERK1/2 signaling pathway. Molecular Medicine Reports, 20(2), 1575–1582. DOI 10.3892/mmr.2019.10398. [Google Scholar] [CrossRef]
19. Hui, X., Zhang, M., Gu, P., Li, K., Gao, Y. et al. (2017). Adipocyte SIRT1 controls systemic insulin sensitivity by modulating macrophages in adipose tissue. EMBO Reports, 18(4), 645–657. DOI 10.15252/embr.201643184. [Google Scholar] [CrossRef]
20. Zhang, Y. Y., Zhou, L. M. (2012). Sirt3 inhibits hepatocellular carcinoma cell growth through reducing Mdm2-mediated p53 degradation. Biochemical and Biophysical Research Communications, 423(1), 26–31. DOI 10.1016/j.bbrc.2012.05.053. [Google Scholar] [CrossRef]
21. Choi, H. N., Bae, J. S., Jamiyandorj, U., Noh, S. J., Park, H. S. et al. (2011). Expression and role of SIRT1 in hepatocellular carcinoma. Oncology Reports, 26(2), 503–510. DOI 10.3892/or.2011.1301. [Google Scholar] [CrossRef]
22. Voelter-Mahlknecht, S., Mahlknecht, U. (2010). The sirtuins in the pathogenesis of cancer. Clinical Epigenetics, 1(3–4), 71–83. DOI 10.1007/s13148-010-0008-0. [Google Scholar] [CrossRef]
23. Chen, H. C., Jeng, Y. M., Yuan, R. H., Hsu, H. C., Chen, Y. L. (2012). SIRT1 promotes tumorigenesis and resistance to chemotherapy in hepatocellular carcinoma and its expression predicts poor prognosis. Annals of Surgical Oncology, 19(6), 2011–2019. DOI 10.1245/s10434-011-2159-4. [Google Scholar] [CrossRef]
24. Hao, C., Zhu, P. X., Yang, X., Han, Z. P., Jiang, J. H. et al. (2014). Overexpression of SIRT1 promotes metastasis through epithelial-mesenchymal transition in hepatocellular carcinoma. BMC Cancer, 14(1), 69. DOI 10.1186/1471-2407-14-978. [Google Scholar] [CrossRef]
25. Li, Y., Xu, S., Li, J., Zheng, L., Feng, M. et al. (2016). SIRT1 facilitates hepatocellular carcinoma metastasis by promoting PGC-1α-mediated mitochondrial biogenesis. Oncotarget, 7(20), 29255–29274. DOI 10.18632/oncotarget.8711. [Google Scholar] [CrossRef]
26. Tang, H. X., Wang, M. Y., Xiao, W., Wen, J. W. (2019). SIRT2-reverses drug-resistance of HL-60/A through autophagy mechanism. Zhongguo Shi Yan Xue Ye Xue Za Zhi, 27(2), 409–414. DOI 10.19746/j.cnki.issn.1009-2137.2019.02.016. [Google Scholar] [CrossRef]
27. Ling, S., Li, J., Shan, Q., Dai, H., Lu, D. et al. (2017). USP22 mediates the multidrug resistance of hepatocellular carcinoma via the SIRT1/AKT/MRP1 signaling pathway. Molecular Oncology, 11(6), 682–695. DOI 10.1002/1878-0261.12067. [Google Scholar] [CrossRef]
28. Xiong, H., Ni, Z., He, J., Jiang, S., Li, X. et al. (2017). LncRNA HULC triggers autophagy via stabilizing Sirt1 and attenuates the chemosensitivity of HCC cells. Oncogene, 36(25), 3528–3540. DOI 10.1038/onc.2016.521. [Google Scholar] [CrossRef]
29. Sun, G., Yang, L., Wei, S., Jin, H., Li, B. et al. (2021). miR-425 regulates lipophagy via SIRT1 to promote sorafenib resistance in liver cancer. Oncology Letters, 22(4), 695. DOI 10.3892/ol.2021.12956. [Google Scholar] [CrossRef]
30. Wang, Z., Yuan, H., Roth, M., Stark, J. M., Bhatia, R. et al. (2013). SIRT1 deacetylase promotes acquisition of genetic mutations for drug resistance in CML cells. Oncogene, 32(5), 589–598. DOI 10.1038/onc.2012.83. [Google Scholar] [CrossRef]
31. Molla, M. D., Dessie, G., Akalu, Y., Ayelign, B. (2020). Hepatocellular expression of SIRT1 and its effect on hepatocellular carcinoma progression: A future therapeutic perspective. International Journal of Hepatology, 2020(1), 1–10. DOI 10.1155/2020/2374615. [Google Scholar] [CrossRef]
32. Chen, C., Zhou, M., Ge, Y., Wang, X. (2020). SIRT1 and aging related signaling pathways. Mechanisms of Ageing and Development, 187(6), 111215. DOI 10.1016/j.mad.2020.111215. [Google Scholar] [CrossRef]
33. Garten, A., Grohmann, T., Kluckova, K., Lavery, G. G., Kiess, W. et al. (2019). Sorafenib-induced apoptosis in hepatocellular carcinoma is reversed by SIRT1. International Journal of Molecular Sciences, 20(16), 4048. DOI 10.3390/ijms20164048. [Google Scholar] [CrossRef]
34. Farcas, M., Gavrea, A. A., Gulei, D., Ionescu, C., Irimie, A. et al. (2019). SIRT1 in the development and treatment of hepatocellular carcinoma. Frontiers in Nutrition, 6, 1485. DOI 10.3389/fnut.2019.00148. [Google Scholar] [CrossRef]
35. Portmann, S., Fahrner, R., Lechleiter, A., Keogh, A., Overney, S. et al. (2013). Antitumor effect of SIRT1 inhibition in human HCC tumor models in vitro and in vivo. Molecular Cancer Therapeutics, 12(4), 499–508. DOI 10.1158/1535-7163.MCT-12-0700. [Google Scholar] [CrossRef]
36. Wang, H., Liu, H., Chen, K., Xiao, J., He, K. et al. (2012). SIRT1 promotes tumorigenesis of hepatocellular carcinoma through PI3K/PTEN/AKT signaling. Oncology Reports, 28(1), 311–318. DOI 10.3892/or.2012.1788. [Google Scholar] [CrossRef]
37. Zang, Y., Fan, L., Chen, J., Huang, R., Qin, H. (2018). Improvement of lipid and glucose metabolism by capsiate in palmitic acid-treated HepG2 cells via activation of the AMPK/SIRT1 signaling pathway. Journal of Agricultural and Food Chemistry, 66(26), 6772–6781. DOI 10.1021/acs.jafc.8b01831. [Google Scholar] [CrossRef]
38. Ma, F., Wu, J., Jiang, Z., Huang, W., Jia, Y. et al. (2019). P53/NRF2 mediates SIRT1’s protective effect on diabetic nephropathy. Biochimica et Biophysica Acta (BBA)–Molecular Cell Research, 1866(8), 1272–1281. DOI 10.1016/j.bbamcr.2019.04.006. [Google Scholar] [CrossRef]
39. Alves-Fernandes, D. K., Jasiulionis, M. G. (2019). The role of SIRT1 on DNA damage response and epigenetic alterations in cancer. International Journal of Molecular Sciences, 20(13), 3153. DOI 10.3390/ijms20133153. [Google Scholar] [CrossRef]
40. Liu, Q., Tong, D., Liu, G., Xu, J., Do, K. et al. (2017). Metformin reverses prostate cancer resistance to enzalutamide by targeting TGF-beta1/STAT3 axis-regulated EMT. Cell Death & Disease, 8(8), e3007. DOI 10.1038/cddis.2017.417. [Google Scholar] [CrossRef]
41. Liu, L., Liu, C., Zhang, Q., Shen, J., Zhang, H. et al. (2016). SIRT1-mediated transcriptional regulation of SOX2 is important for self-renewal of liver cancer stem cells. Hepatology, 64(3), 814–827. DOI 10.1002/hep.28690. [Google Scholar] [CrossRef]
42. Chen, J., Zhang, B., Wong, N., Lo, A. W., To, K. F. et al. (2011). Sirtuin 1 is upregulated in a subset of hepatocellular carcinomas where it is essential for telomere maintenance and tumor cell growth. Cancer Research, 71(12), 4138–4149. DOI 10.1158/0008-5472.CAN-10-4274. [Google Scholar] [CrossRef]
43. Zhou, B., Yang, Y., Li, C. (2019). SIRT1 inhibits hepatocellular carcinoma metastasis by promoting M1 macrophage polarization via NF-kappaB pathway. OncoTargets and Therapy, 12, 2519–2529. DOI 10.2147/OTT. [Google Scholar] [CrossRef]
44. Rawat, D., Shrivastava, S., Naik, R. A., Chhonker, S. K., Koiri, R. K. (2020). SIRT1-mediated amelioration of oxidative stress in kidney of alcohol-aflatoxin-B1-induced hepatocellular carcinoma by resveratrol is catalase dependent and GPx independent. Journal of Biochemical and Molecular Toxicology, 34(11), 1408. DOI 10.1002/jbt.22576. [Google Scholar] [CrossRef]
45. Ceballos, M. P., Angel, A., Delprato, C. B., Livore, V. I., Ferretti, A. C. et al. (2021). Sirtuin 1 and 2 inhibitors enhance the inhibitory effect of sorafenib in hepatocellular carcinoma cells. European Journal of Pharmacology, 892, 173736. DOI 10.1016/j.ejphar.2020.173736. [Google Scholar] [CrossRef]
46. Song, S., Luo, M., Song, Y., Liu, T., Zhang, H. et al. (2014). Prognostic role of SIRT1 in hepatocellular carcinoma. Journal of the College of Physicians and Surgeons Pakistan, 24(11), 849–854. [Google Scholar]
47. Tang, F., Pan, M. H., Wan, X., Lu, Y., Zhang, Y. et al. (2018). Kif18a regulates Sirt2-mediated tubulin acetylation for spindle organization during mouse oocyte meiosis. Cell Division, 13(1), 32302. DOI 10.1186/s13008-018-0042-4. [Google Scholar] [CrossRef]
48. Jeon, H. J., Oh, J. S. (2020). RASSF1A regulates spindle organization by modulating tubulin acetylation via SIRT2 and HDAC6 in mouse Oocytes. Frontiers in Cell and Developmental Biology, 8, 83. DOI 10.3389/fcell.2020.601972. [Google Scholar] [CrossRef]
49. Shi, L., Zhang, Y., Zhang, J., Gao, Y., Liu, J. et al. (2021). MiR-339 is a potential biomarker of coronary heart disease to aggravate oxidative stress through Nrf2/FOXO3 targeting Sirt2. Annals of Palliative Medicine, 10(3), 2596–2609. DOI 10.21037/apm-20-603. [Google Scholar] [CrossRef]
50. Igci, M., Kalender, M. E., Borazan, E., Bozgeyik, I., Bayraktar, R. et al. (2016). High-throughput screening of Sirtuin family of genes in breast cancer. Gene, 586(1), 123–128. DOI 10.1016/j.gene.2016.04.023. [Google Scholar] [CrossRef]
51. Huang, S., Zhao, Z., Tang, D., Zhou, Q., Li, Y. et al. (2017). Downregulation of SIRT2 inhibits invasion of hepatocellular carcinoma by inhibiting energy metabolism. Translational Oncology, 10(6), 917–927. DOI 10.1016/j.tranon.2017.09.006. [Google Scholar] [CrossRef]
52. Chen, G., Huang, P., Hu, C. (2020). The role of SIRT2 in cancer: A novel therapeutic target. International Journal of Cancer, 147(12), 3297–3304. DOI 10.1002/ijc.33118. [Google Scholar] [CrossRef]
53. Piracha, Z. Z., Kwon, H., Saeed, U., Kim, J., Jung, J. et al. (2018). Sirtuin 2 isoform 1 enhances hepatitis B virus RNA transcription and DNA synthesis through the AKT/GSK-3beta/beta-Catenin signaling pathway. Journal of Virology, 92(21), e00955–18. DOI 10.1128/JVI.00955-18. [Google Scholar] [CrossRef]
54. Cheng, S. T., Ren, J. H., Cai, X. F., Jiang, H., Chen, J. (2018). HBx-elevated SIRT2 promotes HBV replication and hepatocarcinogenesis. Biochemical and Biophysical Research Communications, 496(3), 904–910. DOI 10.1016/j.bbrc.2018.01.127. [Google Scholar] [CrossRef]
55. Yang, W., Nagasawa, K., Munch, C., Xu, Y., Satterstrom, K. et al. (2016). Mitochondrial sirtuin network reveals dynamic SIRT3-dependent deacetylation in response to membrane depolarization. Cell, 167(4), 985–1000.e21. DOI 10.1016/j.cell.2016.10.016. [Google Scholar] [CrossRef]
56. Zhang, J., Zhu, Y., Hu, L., Yan, F., Chen, J. (2019). miR-494 induces EndMT and promotes the development of HCC (Hepatocellular Carcinoma) by targeting SIRT3/TGF-beta/SMAD signaling pathway. Scientific Reports, 9(1), 753. DOI 10.1038/s41598-019-43731-4. [Google Scholar] [CrossRef]
57. Alhazzazi, T. Y., Kamarajan, P., Joo, N., Huang, J. Y., Verdin, E. et al. (2011). Sirtuin-3 (SIRT3a novel potential therapeutic target for oral cancer. Cancer, 117(8), 1670–1678. DOI 10.1002/cncr.25676. [Google Scholar] [CrossRef]
58. Kim, H. S., Patel, K., Muldoon-Jacobs, K., Bisht, K. S., Aykin-Burns, N. et al. (2010). SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell, 17(1), 41–52. DOI 10.1016/j.ccr.2009.11.023. [Google Scholar] [CrossRef]
59. Zhang, C. Z., Liu, L., Cai, M., Pan, Y., Fu, J. et al. (2012). Low SIRT3 expression correlates with poor differentiation and unfavorable prognosis in primary hepatocellular carcinoma. PLoS One, 7(12), e51703. DOI 10.1371/journal.pone.0051703. [Google Scholar] [CrossRef]
60. Wang, J. X., Yi, Y., Li, Y. W., Cai, X. Y., He, H. W. et al. (2014). Down-regulation of sirtuin 3 is associated with poor prognosis in hepatocellular carcinoma after resection. BMC Cancer, 14(1), 1245. DOI 10.1186/1471-2407-14-297. [Google Scholar] [CrossRef]
61. Liu, Y., Liu, Y. L., Cheng, W., Yin, X. M., Jiang, B. (2017). The expression of SIRT3 in primary hepatocellular carcinoma and the mechanism of its tumor suppressing effects. European Review for Medical and Pharmacological Sciences, 21(5), 978–998. [Google Scholar]
62. Song, C. L., Tang, H., Ran, L. K., Ko, B. C., Zhang, Z. Z. et al. (2016). Sirtuin 3 inhibits hepatocellular carcinoma growth through the glycogen synthase kinase-3beta/BCL2-associated X protein-dependent apoptotic pathway. Oncogene, 35(5), 631–641. DOI 10.1038/onc.2015.121. [Google Scholar] [CrossRef]
63. Li, Y., Wang, W., Xu, X., Sun, S., Qu, X. J. (2015). {2-[1-(3-Methoxycarbonylmethyl-1H-indol-2-yl)-1-methyl-ethyl]-1H-indol-3-yl}-acetic acid methyl ester (MIAM) inhibited human hepatocellular carcinoma growth through upregulation of Sirtuin-3 (SIRT3). Biomedicine & Pharmacotherapy, 69, 125–132. DOI 10.1016/j.biopha.2014.11.005. [Google Scholar] [CrossRef]
64. de Matteis, S., Scarpi, E., Granato, A. M., Vespasiani-Gentilucci, U., La Barba, G. et al. (2019). Role of SIRT-3, p-mTOR and HIF-1alpha in hepatocellular carcinoma patients affected by metabolic dysfunctions and in chronic treatment with metformin. International Journal of Molecular Sciences, 20(6), 1503. DOI 10.3390/ijms20061503. [Google Scholar] [CrossRef]
65. Feng, W., Xue, T., Huang, S., Shi, Q., Tang, C. et al. (2018). HIF-1alpha promotes the migration and invasion of hepatocellular carcinoma cells via the IL-8-NF-kappaB axis. Cellular & Molecular Biology Letters, 23(1), 533. DOI 10.1186/s11658-018-0077-1. [Google Scholar] [CrossRef]
66. Rizza, S., di Leo, L., Mandatori, S., de Zio, D., Filomeni, G. (2020). Mitophagy contributes to alpha-tocopheryl succinate toxicity in GSNOR-deficient hepatocellular carcinoma. Biochemical Pharmacology, 176(15), 113885. DOI 10.1016/j.bcp.2020.113885. [Google Scholar] [CrossRef]
67. Jo, H., Park, Y., Kim, T., Kim, J., Lee, J. S. et al. (2020). Modulation of SIRT3 expression through CDK4/6 enhances the anti-cancer effect of sorafenib in hepatocellular carcinoma cells. BMC Cancer, 20(1), 394. DOI 10.1186/s12885-020-06822-4. [Google Scholar] [CrossRef]
68. Wang, R., Liu, Y., Mi, X., Chen, Q., Jiang, P. et al. (2020). Sirt3 promotes hepatocellular carcinoma cells sensitivity to regorafenib through the acceleration of mitochondrial dysfunction. Archives of Biochemistry and Biophysics, 689(1), 108415. DOI 10.1016/j.abb.2020.108415. [Google Scholar] [CrossRef]
69. Werner, R. J., Kelly, A. D., Issa, J. J. (2017). Epigenetics and precision oncology. Cancer Journal, 23(5), 262–269. DOI 10.1097/PPO.0000000000000281. [Google Scholar] [CrossRef]
70. Han, T. S., Ban, H. S., Hur, K., Cho, H. S. (2018). The epigenetic regulation of HCC metastasis. International Journal of Molecular Sciences, 19(12), 3978. DOI 10.3390/ijms19123978. [Google Scholar] [CrossRef]
71. Ripoli, M., Barbano, R., Balsamo, T., Piccoli, C., Brunetti, V. et al. (2011). Hypermethylated levels of E-cadherin promoter in Huh-7 cells expressing the HCV core protein. Virus Research, 160(1–2), 74–81. DOI 10.1016/j.virusres.2011.05.014. [Google Scholar] [CrossRef]
72. Qi, W., Ren, D., Wang, P., Song, Z., Wu, H. et al. (2020). Upregulation of Sirt1 by tyrosol suppresses apoptosis and inflammation and modulates extracellular matrix remodeling in interleukin-1beta-stimulated human nucleus pulposus cells through activation of PI3K/Akt pathway. International Immunopharmacology, 88(2), 106904. DOI 10.1016/j.intimp.2020.106904. [Google Scholar] [CrossRef]
73. Xu, T., Li, L., Hu, H. Q., Meng, X. M., Huang, C. et al. (2019). MicroRNAs in alcoholic liver disease: Recent advances and future applications. Journal of Cellular Physiology, 234(1), 382–394. DOI 10.1002/jcp.26938. [Google Scholar] [CrossRef]
74. Zia, A., Sahebdel, F., Farkhondeh, T., Ashrafizadeh, M., Zarrabi, A. et al. (2021). A review study on the modulation of SIRT1 expression by miRNAs in aging and age-associated diseases. International Journal of Biological Macromolecules, 188(1), 52–61. DOI 10.1016/j.ijbiomac.2021.08.013. [Google Scholar] [CrossRef]
75. Karbasforooshan, H., Hayes, A. W., Mohammadzadeh, N., Zirak, M. R., Karimi, G. (2020). The possible role of Sirtuins and microRNAs in hepatocellular carcinoma therapy. Cell Cycle, 19(23), 3209–3221. DOI 10.1080/15384101.2020.1843813. [Google Scholar] [CrossRef]
76. Bae, H. J., Noh, J. H., Kim, J. K., Eun, J. W., Jung, K. H. et al. (2014). MicroRNA-29c functions as a tumor suppressor by direct targeting oncogenic SIRT1 in hepatocellular carcinoma. Oncogene, 33(20), 2557–2567. DOI 10.1038/onc.2013.216. [Google Scholar] [CrossRef]
77. Liu, J., Wu, W., Jin, J. (2015). A novel mutation in SIRT1-AS leading to a decreased risk of HCC. Oncology Reports, 34(5), 2343–2350. DOI 10.3892/or.2015.4205. [Google Scholar] [CrossRef]
78. Sharma, P., Dando, I., Strippoli, R., Kumar, S., Somoza, A. et al. (2020). Nanomaterials for autophagy-related miRNA-34a delivery in cancer treatment. Frontiers in Pharmacology, 11, 219. DOI 10.3389/fphar.2020.01141. [Google Scholar] [CrossRef]
79. Zheng, C. G., Chen, B. Y., Sun, R. H., Mou, X. Z., Han, F. et al. (2019). miR-133b downregulation reduces vulnerable plaque formation in mice with AS through inhibiting macrophage immune responses. Molecular Therapy–Nucleic Acids, 16, 745–757. DOI 10.1016/j.omtn.2019.04.024. [Google Scholar] [CrossRef]
80. Tian, Z., Jiang, H., Liu, Y., Huang, Y., Xiong, X. et al. (2016). MicroRNA-133b inhibits hepatocellular carcinoma cell progression by targeting Sirt1. Experimental Cell Research, 343(2), 135–147. DOI 10.1016/j.yexcr.2016.03.027. [Google Scholar] [CrossRef]
81. Chen, K., Hou, Y., Liao, R., Li, Y., Yang, H. et al. (2021). LncRNA SNHG6 promotes G1/S-phase transition in hepatocellular carcinoma by impairing miR-204-5p-mediated inhibition of E2F1. Oncogene, 40(18), 3217–3230. DOI 10.1038/s41388-021-01671-2. [Google Scholar] [CrossRef]
82. Wu, H. Y., Cai, K. T., Ma, J., Chen, G., Dang, Y. W. et al. (2019). Evaluation of miR-302b-5p expression and molecular mechanism in hepatocellular carcinoma: Findings based on RT-qPCR and in silico analysis. Pathology–Research and Practice, 215(7), 152424. DOI 10.1016/j.prp.2019.04.016. [Google Scholar] [CrossRef]
83. Pant, K., Yadav, A. K., Gupta, P., Islam, R., Saraya, A. et al. (2017). Butyrate induces ROS-mediated apoptosis by modulating miR-22/SIRT-1 pathway in hepatic cancer cells. Redox Biology, 12, 340–349. DOI 10.1016/j.redox.2017.03.006. [Google Scholar] [CrossRef]
84. Hou, Z., Xu, X., Zhou, L., Fu, X., Tao, S. et al. (2017). The long non-coding RNA MALAT1 promotes the migration and invasion of hepatocellular carcinoma by sponging miR-204 and releasing SIRT1. Tumor Biology, 39(7), 1010428317718135. DOI 10.1177/1010428317718135. [Google Scholar] [CrossRef]
85. Gomes, A. R., Yong, J. S., Kiew, K. C., Aydin, E., Khongkow, M. et al. (2016). Sirtuin1 (SIRT1) in the acetylation of downstream target proteins. Methods in Molecular Biology, 1436, 169–188. DOI 10.1007/978-1-4939-3667-0. [Google Scholar] [CrossRef]
86. Nakamura, K., Zhang, M., Kageyama, S., Ke, B., Fujii, T. et al. (2017). Macrophage heme oxygenase-1-SIRT1-p53 axis regulates sterile inflammation in liver ischemia-reperfusion injury. Journal of Hepatology, 67(6), 1232–1242. DOI 10.1016/j.jhep.2017.08.010. [Google Scholar] [CrossRef]
87. Bhalla, K., Hwang, B. J., Dewi, R. E., Twaddel, W., Goloubeva, O. G. et al. (2012). Metformin prevents liver tumorigenesis by inhibiting pathways driving hepatic lipogenesis. Cancer Prevention Research, 5(4), 544–552. DOI 10.1158/1940-6207.CAPR-11-0228. [Google Scholar] [CrossRef]
88. Park, S. A., Joo, N. R., Park, J. H., Oh, S. M. (2020). Role of the SIRT1/p53 regulatory axis in oxidative stressmediated granulosa cell apoptosis. Molecular Medicine Reports, 23(1), 1. DOI 10.3892/mmr.2020.11658. [Google Scholar] [CrossRef]
89. Lee, C. W., Wong, L. L., Tse, E. Y., Liu, H. F., Leong, V. Y. et al. (2012). AMPK promotes p53 acetylation via phosphorylation and inactivation of SIRT1 in liver cancer cells. Cancer Research, 72(17), 4394–4404. DOI 10.1158/0008-5472.CAN-12-0429. [Google Scholar] [CrossRef]
90. Jiang, S., Yang, Z., di, S., Hu, W., Ma, Z. et al. (2018). Novel role of forkhead box O 4 transcription factor in cancer: Bringing out the good or the bad. Seminars in Cancer Biology, 50(24), 1–12. DOI 10.1016/j.semcancer.2018.04.007. [Google Scholar] [CrossRef]
91. Jiramongkol, Y., Lam, E. W. (2020). FOXO transcription factor family in cancer and metastasis. Cancer and Metastasis Reviews, 39(3), 681–709. DOI 10.1007/s10555-020-09883-w. [Google Scholar] [CrossRef]
92. Jia, G., Tang, Y., Deng, G., Fang, D., Xie, J. et al. (2019). miR-590-5p promotes liver cancer growth and chemotherapy resistance through directly targeting FOXO1. American Journal of Translational Research, 11(4), 2181–2193. [Google Scholar]
93. Dong, T., Zhang, Y., Chen, Y., Liu, P., An, T. et al. (2017). FOXO1 inhibits the invasion and metastasis of hepatocellular carcinoma by reversing ZEB2-induced epithelial-mesenchymal transition. Oncotarget, 8(1), 1703–1713. DOI 10.18632/oncotarget.13786. [Google Scholar] [CrossRef]
94. Kim, H. N., Han, L., Iyer, S., de Cabo, R., Zhao, H. et al. (2015). Sirtuin1 suppresses osteoclastogenesis by deacetylating FoxOs. Molecular Endocrinology, 29(10), 1498–1509. DOI 10.1210/me.2015-1133. [Google Scholar] [CrossRef]
95. Liu, L., Long, H., Wu, Y., Li, H., Dong, L. et al. (2018). HRD1-mediated PTEN degradation promotes cell proliferation and hepatocellular carcinoma progression. Cellular Signalling, 50(2), 90–99. DOI 10.1016/j.cellsig.2018.06.011. [Google Scholar] [CrossRef]
96. Jane, P., Gogl, G., Kostmann, C., Bich, G., Girault, V. et al. (2020). Interactomic affinity profiling by holdup assay: Acetylation and distal residues impact the PDZome-binding specificity of PTEN phosphatase. PLoS One, 15(12), e0244613. DOI 10.1371/journal.pone.0244613. [Google Scholar] [CrossRef]
97. Datta, S. R., Brunet, A., Greenberg, M. E. (1999). Cellular survival: A play in three Akts. Genes & Development, 13(22), 2905–2927. DOI 10.1101/gad.13.22.2905. [Google Scholar] [CrossRef]
98. Zhang, H., Xu, H. B., Kurban, E., Luo, H. W. (2020). LncRNA SNHG14 promotes hepatocellular carcinoma progression via H3K27 acetylation activated PABPC1 by PTEN signaling. Cell Death & Disease, 11(8), 380. DOI 10.1038/s41419-020-02808-z. [Google Scholar] [CrossRef]
99. Li, Y., Tsang, C. K., Wang, S., Li, X. X., Yang, Y. et al. (2016). MAF1 suppresses AKT-mTOR signaling and liver cancer through activation of PTEN transcription. Hepatology, 63(6), 1928–1942. DOI 10.1002/hep.28507. [Google Scholar] [CrossRef]
100. Xin, X., Wu, M., Meng, Q., Wang, C., Lu, Y. et al. (2018). Long noncoding RNA HULC accelerates liver cancer by inhibiting PTEN via autophagy cooperation to miR15a. Molecular Cancer, 17(1), e3118. DOI 10.1186/s12943-018-0843-8. [Google Scholar] [CrossRef]
101. Yang, W., Zhang, G., Jiang, F., Zeng, Y., Zou, P. et al. (2019). BPDE and B[a]P induce mitochondrial compromise by ROS-mediated suppression of the SIRT1/TERT/PGC-1alpha pathway in spermatogenic cells both in vitro and in vivo. Toxicology and Applied Pharmacology, 376(5666), 17–37. DOI 10.1016/j.taap.2019.05.004. [Google Scholar] [CrossRef]
102. Zhang, B., Chen, J., Cheng, A. S., Ko, B. C. (2014). Depletion of sirtuin 1 (SIRT1) leads to epigenetic modifications of telomerase (TERT) gene in hepatocellular carcinoma cells. PLoS One, 9(1), e84931. DOI 10.1371/journal.pone.0084931. [Google Scholar] [CrossRef]
103. Billard, P., Poncet, D. A. (2019). Replication stress at telomeric and mitochondrial DNA: Common origins and consequences on ageing. International Journal of Molecular Sciences, 20(19), 4959. DOI 10.3390/ijms20194959. [Google Scholar] [CrossRef]
104. Zhang, X., Li, Y., Ma, Y., Yang, L., Wang, T. et al. (2018). Yes-associated protein (YAP) binds to HIF-1alpha and sustains HIF-1alpha protein stability to promote hepatocellular carcinoma cell glycolysis under hypoxic stress. Journal of Experimental & Clinical Cancer Research, 37(1), 115. DOI 10.1186/s13046-018-0892-2. [Google Scholar] [CrossRef]
105. Mao, B., Hu, F., Cheng, J., Wang, P., Xu, M. et al. (2014). SIRT1 regulates YAP2-mediated cell proliferation and chemoresistance in hepatocellular carcinoma. Oncogene, 33(11), 1468–1474. DOI 10.1038/onc.2013.88. [Google Scholar] [CrossRef]
106. Jang, K. Y., Noh, S. J., Lehwald, N., Tao, G. Z., Bellovin, D. I. et al. (2012). SIRT1 and c-Myc promote liver tumor cell survival and predict poor survival of human hepatocellular carcinomas. PLoS One, 7(9), e45119. DOI 10.1371/journal.pone.0045119. [Google Scholar] [CrossRef]
107. Gao, Y., Zhang, B., Wang, Y., Shang, G. (2018). Cdc20 inhibitor apcin inhibits the growth and invasion of osteosarcoma cells. Oncology Reports, 40(2), 841–848. DOI 10.3892/or.2018.6467. [Google Scholar] [CrossRef]
108. Zhang, N., Liu, X., Liu, L., Deng, Z., Zeng, Q. et al. (2018). Glycogen synthase kinase-3beta inhibition promotes lysosome-dependent degradation of c-FLIPL in hepatocellular carcinoma. Cell Death & Disease, 9(2), 19. DOI 10.1038/s41419-018-0309-3. [Google Scholar] [CrossRef]
109. Pan, J. X., Chen, G., Li, J. J., Zhu, Q. D., Li, J. J. et al. (2018). Isocorydine suppresses doxorubicin-induced epithelial-mesenchymal transition via inhibition of ERK signaling pathways in hepatocellular carcinoma. American Journal of Cancer Research, 8(1), 154–164. [Google Scholar]
110. Xu, S., Liu, C. X., Xu, W., Huang, L., Zhao, J. Y. et al. (2017). Butyrate induces apoptosis by activating PDC and inhibiting complex I through SIRT3 inactivation. Signal Transduction and Targeted Therapy, 2(1), 1111. DOI 10.1038/sigtrans.2016.35. [Google Scholar] [CrossRef]
111. Lv, J. J., Song, W. T., Li, X. M., Gao, J. M., Yuan, Z. L. (2020). Synthesis of a new phenyl chlormethine-quinazoline derivative, a potential anti-cancer agent, induced apoptosis in hepatocellular carcinoma through mediating Sirt1/Caspase 3 signaling pathway. Frontiers in Pharmacology, 11, 903. DOI 10.3389/fphar.2020.00911. [Google Scholar] [CrossRef]
112. Lou, G., Liu, Y., Wu, S., Xue, J., Yang, F. et al. (2015). The p53/miR-34a/SIRT1 positive feedback loop in quercetin-induced apoptosis. Cellular Physiology and Biochemistry, 35(6), 2192–2202. DOI 10.1159/000374024. [Google Scholar] [CrossRef]
113. Lin, X. L., Li, K., Yang, Z., Chen, B., Zhang, T. (2020). Dulcitol suppresses proliferation and migration of hepatocellular carcinoma via regulating SIRT1/p53 pathway. Phytomedicine, 66(9), 153112. DOI 10.1016/j.phymed.2019.153112. [Google Scholar] [CrossRef]
114. Kwon, H. Y., Kim, J. H., Kim, B., Srivastava, S. K., Kim, S. H. (2018). Correction to: Regulation of SIRT1/AMPK axis is critically involved in gallotannin-induced senescence and impaired autophagy leading to cell death in hepatocellular carcinoma cells. Archives of Toxicology, 92(9), 2979. DOI 10.1007/s00204-018-2262-4. [Google Scholar] [CrossRef]
115. Chai, R., Fu, H., Zheng, Z., Liu, T., Ji, S. et al. (2017). Resveratrol inhibits proliferation and migration through SIRT1 mediated posttranslational modification of PI3K/AKT signaling in hepatocellular carcinoma cells. Molecular Medicine Reports, 16(6), 8037–8044. DOI 10.3892/mmr.2017.7612. [Google Scholar] [CrossRef]
116. Wang, W., Liu, F., Xu, C., Liu, Z., Ma, J. et al. (2021). Lactobacillus plantarum 69-2 combined with galacto-oligosaccharides alleviates d-galactose-induced aging by regulating the AMPK/SIRT1 signaling pathway and gut microbiota in mice. Journal of Agricultural and Food Chemistry, 69(9), 2745–2757. DOI 10.1021/acs.jafc.0c06730. [Google Scholar] [CrossRef]
117. Das, D., Smith, N., Wang, X., Morgan, I. M. (2017). The deacetylase SIRT1 regulates the replication properties of human papillomavirus 16 E1 and E2. Journal of Virology, 91(10), 2639. DOI 10.1128/JVI.00102-17. [Google Scholar] [CrossRef]
118. Zhang, B. C., Luo, B. Y., Zou, J. J., Wu, P. Y., Jiang, J. L. et al. (2020). Co-delivery of sorafenib and CRISPR/Cas9 based on targeted core-shell hollow mesoporous organosilica nanoparticles for synergistic HCC therapy. ACS Applied Materials & Interfaces, 12(51), 57362–57372. DOI 10.1021/acsami.0c17660. [Google Scholar] [CrossRef]
119. Zhou, T., Huang, R., Huang, M., Shen, J., Shan, Y. et al. (2020). CRISPR/Cas13a powered portable electrochemiluminescence chip for ultrasensitive and specific MiRNA detection. Advanced Science, 7(13), 1903661. DOI 10.1002/advs.201903661. [Google Scholar] [CrossRef]
120. Bruch, R., Johnston, M., Kling, A., Mattmüller, T., Baaske, J. et al. (2021). CRISPR-powered electrochemical microfluidic multiplexed biosensor for target amplification-free miRNA diagnostics. Biosensors and Bioelectronics, 177(333), 112887. DOI 10.1016/j.bios.2020.112887. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |