
 | Oncologie |  |
DOI: 10.32604/oncologie.2021.018514
REVIEW
Circulating circRNAs as Potential Biomarkers for Cancers
1Department of Laboratory Medicine, Affiliated Hospital of Nantong University, Nantong, China
2Department of Laboratory Medicine, School of Public Health, Nantong University, Nantong, China
3Department of Gastroenterology and Laboratory Medicine, Nantong Third Hospital Affiliated to Nantong University, Nantong, China
4Department of Laboratory Medicine, Hai’an People’s Hospital Affiliated to Nantong University, Nantong, China
5Department of Laboratory Medicine, Taizhou Jiangyan Hospital of Traditional Chinese Medicine, Taizhou, China
*Corresponding Authors: Feng Wang. Email: richardwangf@163.com; Henggui Xu. Email: jyzyyjyk@163.com
#Co-first authors
Received: 30 July 2021; Accepted: 03 September 2021
Abstract: Cancers are diseases with a high mortality rate worldwide. In order to better diagnose and improve the survival rate, many studies have been conducted. In recent years, the role of non-coding RNAs in cancers has been confirmed, and circular RNAs (circRNAs) have attracted much attention. CircRNAs are involved in the occurrence and development of cancers with high stability. Experiments have shown that they can exist stably in peripheral blood. Therefore, the expression of circulating circRNAs can be detected to help diagnose cancers and reflect tumor progression. In this review, we summarized the role of circulating circRNAs in cancers and discussed their potential as biomarkers.
Keywords: Circulating circRNAs; plasma; serum; diagnosis biomarkers; cancers
Abbreviations:
| circRNAs: | Circular RNAs |
| miRNAs: | MicroRNAs |
| lncRNAs: | Long non-coding RNAs |
| RBP: | RNA binding protein |
| ceRNA: | Competitive endogenous RNA |
| RBM3: | RNA binding protein 3 |
| GC: | Gastric cancer |
| Pol II: | Polymerase II |
| IRES: | Internal ribosome entry sites |
| ORFs: | Open reading frames |
| NSCLC: | Non-small cell lung cancer |
| LUAD: | Lung adenocarcinoma |
| ROC: | Receiver operating characteristic |
| OS: | Overall survival |
| NPC: | Nasopharyngeal carcinoma |
| AUC: | Area under the receiver operating characteristic curve |
| CEA: | Carcinoembryonic antigen |
| CA199: | Carbohydrate antigen199 |
| CA724: | Carbohydrateantigen724 |
| EGC: | Early gastric cancer |
| HCC: | Hepatocellular carcinoma |
| AFP: | Alpha-fetoprotein |
| CRC: | Colorectal cancer |
| ESCC: | Esophageal squamous cell carcinoma |
| TNM: | Tumor-node-metastasis |
| PDAC: | Pancreatic ductal adenocarcinoma |
| CA153: | Cancer antigen-153 |
| OC: | Ovarian cancer |
| SOC: | Serous ovarian cancers |
| EOC: | Epithelial ovarian cancer |
| PCa: | Prostate cancer |
| PSA: | Prostate-specific antigen |
| BPH: | Benign prostatic hyperplasia |
| CLL: | Chronic lymphocytic leukemia |
| CML: | Chronic myeloid leukemia |
| MM: | Multiple myeloma |
| DLBCL: | Diffused large B-cell lymphomaa |
Cancer, resulted from uncontrollable cell proliferation and differentiation, is one of the most lethal diseases suffered by a large number of people worldwide, leading to spike in death cases each year [1,2]. Today there are a lot of ongoing researches on diagnosis and treatment for cancers. Early detection and diagnosis of cancers may help increase the survival rate of patients, but the detection methods currently used in clinical are not sufficiently sensitive and specific to early diagnosis, which makes it imperative to find new biomarkers as soon as possible [3]. In recent years, many studies have confirmed that non-coding RNAs that mainly include microRNAs (miRNAs), long noncoding RNAs (lncRNAs) and circRNAs, etc., play an important role in tumor diagnosis [4] (Fig. 1).
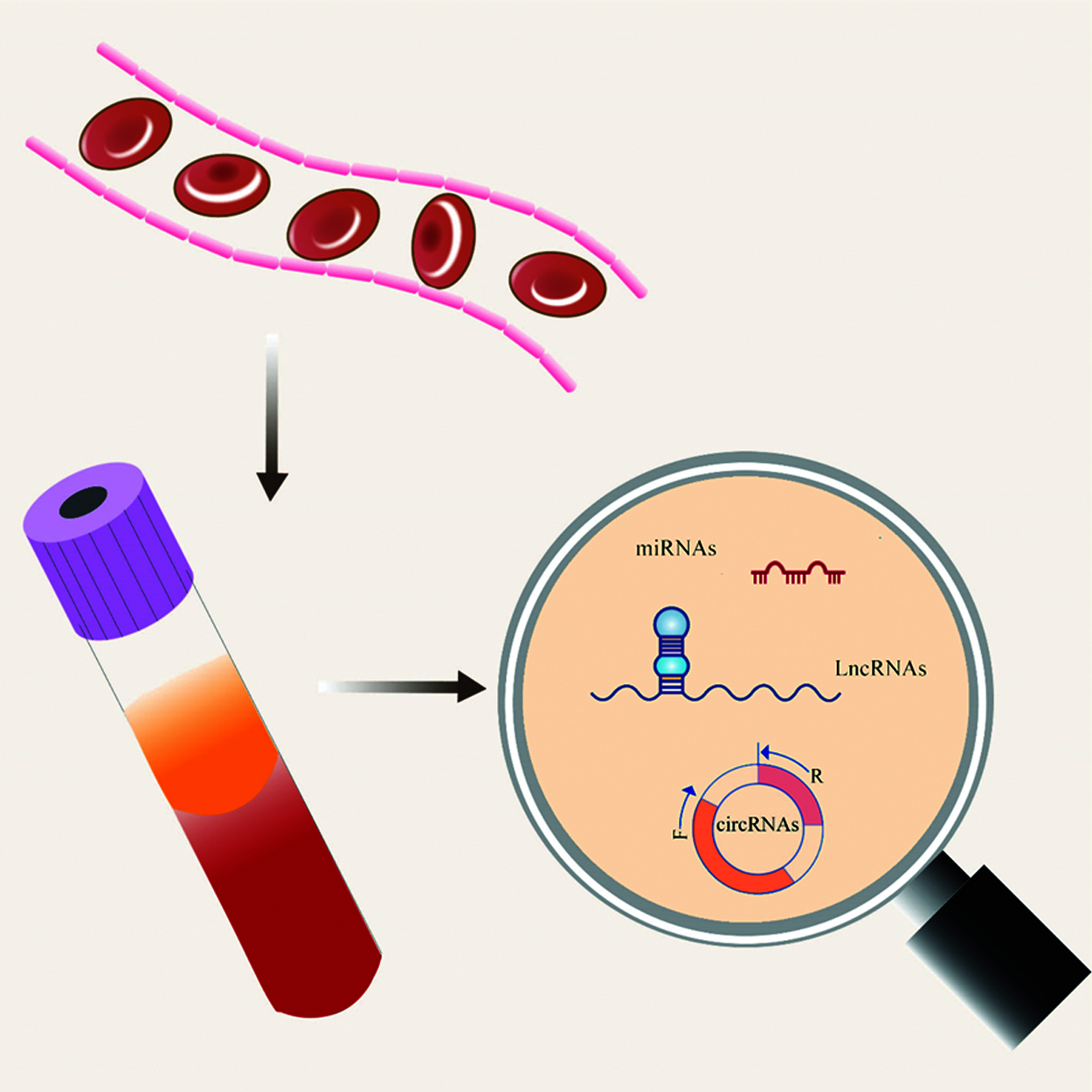
Figure 1: Non-coding RNA that can be detected in peripheral blood, including microRNA, lncRNA and circRNA
CircRNAs refer to endogenous non-coding RNAs with a wide expression in the mammalian genome [5]. Initially observed in RNA viruses, circRNAs were deemed as a by-product of splicing-mediated splicing errors because of their low expression [6]. CircRNAs vary in types, and thousands of circRNAs have been identified in different categories of human cell [7]. CircRNAs have a covalent closed-loop structure without 5’-end cap and 3’-end poly (A) tail, which makes them more stable than linear RNA and less susceptible to degradation resulted from RNA exonuclease [8–10]. The function of circRNAs can be divided into five aspects: they can act as miRNA sponge, interact with RNA binding protein (RBP), act as autophagy regulators, regulate the transcription process and encode proteins [11–16]. Referring to the in-depth study on circRNAs, speculations of the correlation between abnormal function and the development of human diseases are made. Mounting evidence has shown that circRNAs have a close correlation with cancers [17,18]. Since circRNAs evolve slowly with long half-life, and become highly stable on account of its circular structure, so they can stably exist in plasma, saliva, and other surrounding tissues as potential biomarkers with minimal invasiveness [19]. Studies have shown that circRNAs have half-lifes of more than 48 h, longer than lncRNA [20]. In this review, the functions of circRNAs and the roles of circulating circRNAs in cancers were inducted, and the feasibility of circulating circRNAs as biomarkers of cancers were discussed.
2 The Biological Functions of circRNAs
This is a well-studied function of circRNAs, which may act as miRNA sponges by indirectly regulating gene expression through binding of competing miRNAs [21]. CircRNAs regulate their activity by binding to miRNAs and then reduce their ability to target mRNA [22,23]. Compared with other competitive endogenous RNA (ceRNA), they have a higher binding capacity to miRNAs [24,25]. For instance, Chen et al. found that the upregulation of circ-MALAT1 increased JAK2, and it is known that circRNA acts as a miRNA sponge to regulate target gene expression [26]. CircRHOBTB3 is considered to be the sponge of miR-654-3p, which inhibits the growth of gastric cancers by activating the p21 signaling pathway [27] (Fig. 2a).
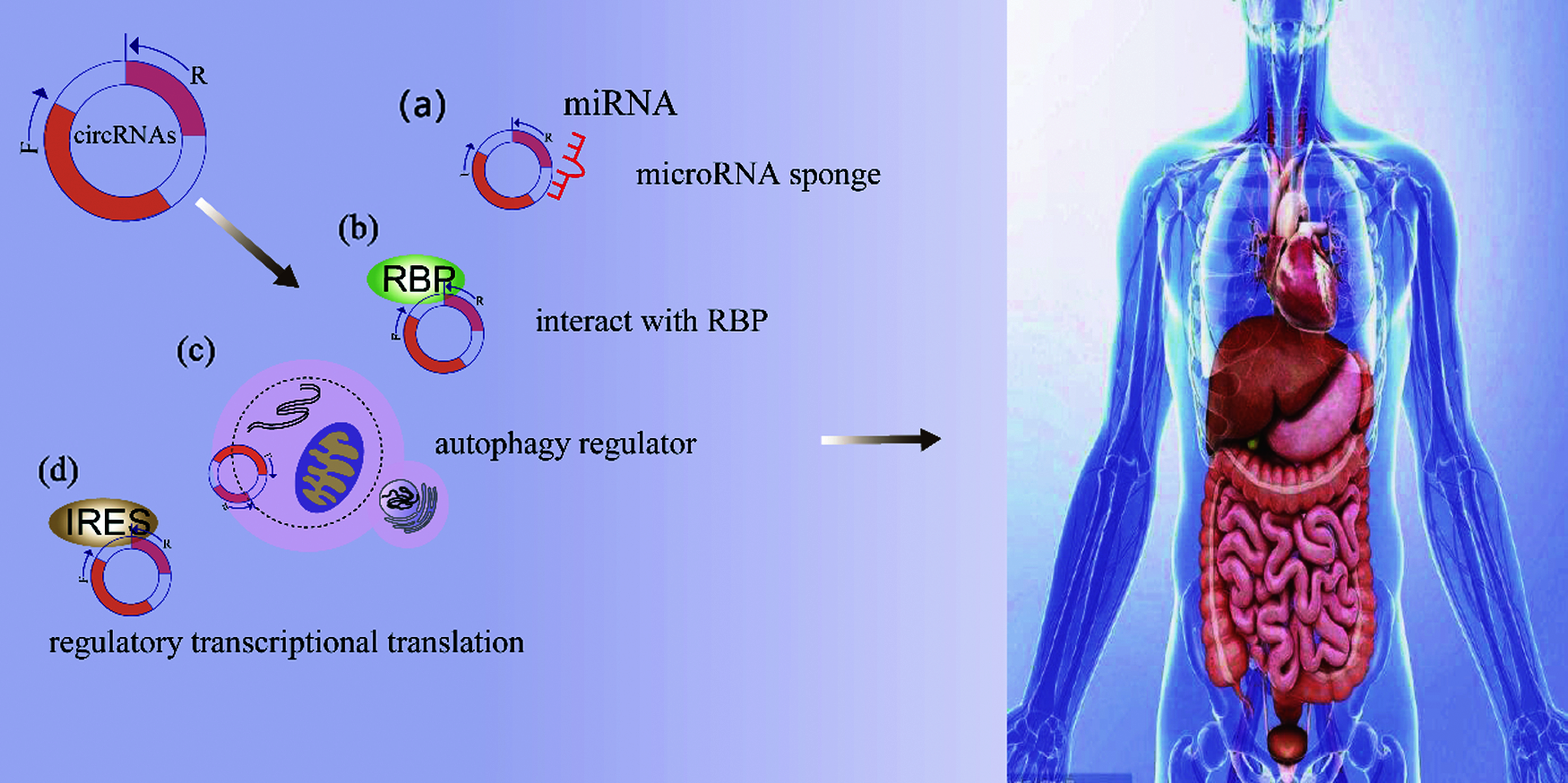
Figure 2: The biological function of circRNA (a) As microRNA sponges: circRNAs may act as miRNA sponges by competing for miRNAs binding and indirectly regulating gene expression (b) Interact with RBP: circRNAs interact with RBP and participate in the regulation of gene expression (c) As autophagy regulator: circRNA-mediated autophagy can promote the proliferation and invasion of cancer cells (d) Regulate the transcription and translation: (1) CircRNAs can regulate gene expression transcription or post-transcription level. (2) CircRNAs can encode regulatory peptides, and there may be hidden proteomes encoded by circRNAs
RBP plays a vital role in a variety of cellular processes, such as cell function, transport, and localization, especially in the post-transcriptional regulation of RNA [28]. RBP can be used as a trans-acting factor that regulates circRNAs biogenesis, and can also isolate, store, and classify RBP, thereby controlling intracellular positioning [2,29]. There is increasing evidence that circRNAs interact with RBP and participate in the regulation of gene expression. For example, the production of SCD-circRNA2 is dynamically regulated by RNA binding protein 3 (RBM3). By regulating the level of RBM3 or SCD-circRNA2, it is found that RBM3 relies on SCD-circRNA2 to promote the proliferation of HCC cells [30] (Fig. 2b).
Autophagy is a highly conservative and continuous self-degradation process that plays an important role in cellular stress response and survival [31]. This process usually occurs during tumorigenesis, progression, metastasis, and chemotherapy, leading to drug resistance in cancers treatment [32]. CircRNA-mediated autophagy can promote the proliferation and invasion of cancers cells [33]. Some studies have found that circRNAs are involved in cancer autophagy, affecting the occurrence and development of human cancers. For example, the over-expression of circ_0032821 inhibits autophagy of human Gastric cancer (GC) cells in vitro [13]. The studies of circRNAs in cancer autophagy are still in their infancy, and more researches are needed to verify their functions and mechanisms (Fig. 2c).
2.4 Regulating the Transcription and Translation
CircRNAs can regulate gene expression transcription or post-transcription level [34]. The circRNAs part in the nucleus acts as a transcription or splicing regulator, interferes with gene expression, and participates in other splicing and transcription processes [35]. CircRNAs in human cells can regulate the transcription of the parental gene in a cis-acting manner [5]. For example, the circRNAs produced by the ANKRD52 gene can form a complex with RNA polymerase II (pol II), and the complex binds to the promoter region of the ANKRD52 to enhance transcription [36]. CircRNAs used to be considered non-coding RNAs, but later studies have found that circRNAs can participate in translation when they have internal ribosomal entry sites (IRES) or open reading frames (ORFs) [37,38]. CircRNAs can encode regulatory peptides, and there may be hidden proteomes encoded by circRNAs [8]. CircLgr4 encodes circLgr4-peptide for CRC targeted therapy [39]. The spanning junction open reading frame of circ-FBXW7 is driven by the internal ribosome entry site encoding FBXW7-185aa, and the expression of circ-FBXW7 is positively correlated with overall survival in glioblastoma patients [40] (Fig. 2d).
3 The Role of Circulating circRNA in Cancers
Compared with other methods, obtaining peripheral blood from patients is relatively simple and non-invasive. The expression of circulating circRNAs can be detected to diagnose cancers or predict the progress of cancers. Many studies have verified that the expression of circRNAs in plasma and serum is related to the development of cancers, and it is believed that circulating circRNAs can be used as biomarkers for cancer diagnosis and prognosis.
3.1 Respiratory System Cancers
3.1.1 The Role of Circulating circRNA in Lung Cancer
Lung cancer is the leading cause of cancer death worldwide, most of which is non-small cell lung cancer (NSCLC). NSCLC includes lung adenocarcinoma (LUAD), squamous cell carcinoma, and large cell carcinoma. LUAD accounts for approximately 50% of all types of lung cancer, and the 5-years survival rate is less than 20%. Therefore, it is necessary to find biomarkers for early diagnosis of LUAD [41–44]. In the current study, hsa_circ_0005962 in LUAD plasma and cells was up-regulated, while hsa_circ_0086414 was down-regulated. Based on the study results and the receiver operating characteristic (ROC), the authors believed that two circRNA molecules can improve the diagnostic accuracy of LUAD [45]. Hsa_circ_0013958 was further confirmed to be up-regulated in all LAUD plasma. Therefore, hsa_circ_0013958 can be used as a potential non-invasive biomarker for the early detection of LAUD [46]. Regarding the study of serum circRNA, the expression level of circMAN1A2 in the serum of lung cancer patients was higher than healthy controls. Considering that serum circMAN1A2 can be used as a serum biomarker for lung cancer [47]. Lu et al. [48] found that plasma hsa_circ_0001715 expression was an independent prognostic factor for overall survival (OS) of LUAD. Other studies have found that the effect of cisplatin combined with gemcitabine on NSCLC chemotherapy could be determined by detecting the expression of serum circPVT1 [49]. The above studies have shown that the expression of circulating circRNA in lung cancer can reflect the progress of cancers, but these studies are more common in LUAD, while other types of lung cancer are less.
3.1.2 The Role of Circulating circRNA in Nasopharyngeal Carcinoma
Nasopharyngeal carcinoma (NPC) is a malignant cancer that occurs in nasopharyngeal epithelial tissue, and most patients with NPC are already in middle-stage and late-stage at the time of diagnosis [50,51]. In order to diagnose NPC earlier, people have explored whether circRNAs in the blood could be used as non-invasive biomarker. CircMAN1A2 can be up-regulated in the serum of patients with NPC. The area under the receiver operating characteristic curve (AUC) of circMAN1A2 in the serum of patients is 0.911, indicating that it can be used as an effective diagnostic biomarker for NPC [47]. Compared with the healthy control group, circRNA_0000285 in serum samples of NPC patients increased significantly. Univariate and multivariate analysis indicated that circRNA_0000285 may be an independent prognostic factor for the prognosis of NPC patients, so it may be a new biomarker for NPC and participate in the radiosensitivity [52]. We believe that circulating circRNA can provide new ideas for the diagnosis of NPC.
3.2.1 The Role of Circulating circRNA in Gastric Cancer
GC is a major malignant cancer with high morbidity and mortality. Although this situation has been alleviated in recent years with the development of medical technology, the prognosis is still poor [53,54]. To date ,the five-years survival rate of advanced GC is very poor, so early diagnosis is important [55]. At present, gastroscopy is widely used for early detection of GC, but there would be omissions. The specificity and sensitivity of carcinoembryonic antigen (CEA), carbohydrate antigen199 (CA199), carbohydrateantigen724 (CA724) and other serum biomarkers which commonly used in clinical are not high enough, so new biomarkers need to be explored for use alone or in combination for early detection [56]. Some studies have found that the expression of plasma circRNA is different between GC patients and healthy people. For example, hsa_circ_0000520 is down-regulated in GC tissues, and the detection in plasma is also down-regulated. Besides, its expression in plasma is related to CEA [57]. There are some studies to prove the value of plasma circRNA in diagnosis. Hsa_circ_0021087 and hsa_circ_0005051 were down-regulated in GC tissues and plasma. There was a significant difference in the expression of plasma hsa_circ_0021087 between GC patients before and after the operation. So it can be used as a non-invasive biomarker for GC diagnosis [58]. Lu et al. [59] found that the expression of plasma hsa_circ_0006848 has a good diagnostic value, and the plasma level of hsa_circ_0006848 in postoperative patients is significantly higher than that in preoperative patients. Hsa_circ_0006848 in plasma may be a promising diagnostic biomarker for early gastric cancer (EGC) [59]. Besides, some people believe that plasma circRNA has higher diagnostic accuracy than tissues, and the combined circRNA has good diagnostic efficacy for GC [60]. In summary, circulating circRNA could be used as a diagnostic and prognostic biomarker for GC.
3.2.2 The Role of Circulating circRNA in Hepatocellular Carcinoma
Hepatocellular carcinoma (HCC) is one of the most common malignant tumor in the world, with a high mortality rate [61]. Alpha-fetoprotein (AFP) is the most commonly used biomarker for HCC, but its specificity and sensitivity are not perfect [62]. Many studies have shown that HCC can be diagnosed by detecting the expression level of circRNA in plasma [63]. For example, hsa_circ_0027089 is up-regulated in the plasma of HCC patients compared with healthy people, and it can be used in combination with serum AFP as a biomarker of hepatitis B-related HCC [64]. Other studies have found that the plasma level of hsa_circ_0003998 in patients with HCC is significantly higher than that in patients with hepatitis B and healthy controls, and the plasma hsa_circ_0003998 level in postoperative patients is significantly lower than that in preoperative patients. Consequently, it can be used as a new potential biomarker for the diagnosis and prognosis of HCC [65]. There are similar studies in serum, for example, circRNA_101237 was up-regulated in the serum of HCC patients compared with the healthy controls. Additionally, univariate and multivariate analysis showed that serum circRNA_101237 level is an independent predictor of survival prognosis in HCC patients [66]. Other studies have pointed out that plasma circRNA can diagnose HCC. The experimental results showed that the level of plasma hsa_circ_0001445 can be used as an indicator for determining HCC [67]. High expression of serum circRNA_101237 is related to poor prognosis of liver cancers patients. Univariate and multivariate analysis can determine that serum circRNA_101237 level is an independent predictor of survival prognosis in HCC patients [68]. These studies could find that the expression of plasma or serum circRNA is different in healthy controls and HCC patients, which can be used in combination with serum AFP to diagnose and assess the stage of HCC.
3.2.3 The Role of Circulating circRNA in Colorectal Cancer
Colorectal cancer (CRC) is one of the most common malignant gastrointestinal cancers, and some patients will find liver metastases during examination [69]. Although the diagnosis and treatment methods have been mastered, the mortality of colorectal cancers is still high [70]. Early diagnosis may improve the survival ability of CRC patients to a certain extent. The diagnostic efficiency of CEA and CA199, which are commonly used at present, needs to be improved, so it is necessary to explore new diagnostic methods to treat as soon as possible [71]. For example, circVAPA was up-regulated in the plasma of CRC patients by qRT-PCR, and circVAPA levels were related to the adverse clinical-pathological characteristics of CRC. The AUC is 0.724, indicating that the plasma level of circVAPA can be used as a promising biomarker for CRC detection [72]. The expression of hsa_circ_0007534 in the plasma of CRC patients was significantly increased compared with the healthy control group. The increased expression of hsa_circ_0007534 in plasma is associated with the clinical classification, metastatic phenotype, and poor differentiation of CRC patients. The high expression of hsa_circ_0007534 is positively correlated with the poor prognosis of CRC patients [73]. Plasma circ-STIL1 levels are related to tumor growth and progression. Plasma circCCDC66 and circ-ABCC1 levels are reduced in CRC precursor lesions. Considering circ-CCDC66 and circ-STIL can be used to diagnose early CRC [74]. In these studies, circRNAs were highly expressed in the plasma of CRC patients. The expression of plasma circRNAs in CRC could reflect the progress of the tumor, which can be used in combination with CA199 and CEA to diagnose CRC.
3.2.4 The Role of Circulating circRNA in Esophageal Cancer
Esophageal cancer is the eighth most common cancers in the world and is widely considered to be a genetic disease with high incidence and high mortality in Asia [75]. Esophageal squamous cell carcinoma (ESCC) is one of the main subtypes and originates from esophageal epithelial cells [76,77]. In recent years, there have been many studies on circulating circRNA in ESCC. Recent studies have found that ESCC tissues can secrete circRNA into plasma. Patients with high plasma circ-SLC7A5 were associated with high tumor-node-metastasis (TNM) stage, and overall survival was often shorter than patients with high levels [77]. Hu et al. [78] suggested that plasma CIRCGSK3β expression could detect ESCC. The up-regulated expression of plasma circGSK3β was positively correlated with the late clinical stage and poor prognosis of ESCC patients [78]. Huang et al. [79] experimentally verified that the expression of plasma hsa_circ_0004771 was decreased in patients with ESCC after surgery. This finding suggests that the increase in plasma hsa_circ_0004771 may be related to the tumor [79]. Thus, we consider that plasma circRNAs as non-invasive biomarkers for ESCC and may provide diagnostic and prognostic value for it.
3.2.5 The Role of Circulating circRNA in Pancreatic Cancer
Pancreatic cancer is a malignant cancer of the digestive system, with insipid onset and rapid development, leading to delay and difficulty in early diagnosis and poor prognosis. 90% of pancreatic cancers are pancreatic ductal adenocarcinomas (PDAC) [80,81]. Currently, CA199 is a commonly used serum biomarker for PDAC [82]. In recent years, there have been some studies on the combined diagnosis of circulating circRNA and CA199 for pancreatic cancer. For example, the expression of circ-LDLRAD3 in serum samples of pancreatic cancer is higher than that of healthy volunteers, and circ-LDLRAD3 is associated with pancreatic cancers metastasis. Besides, the serum level of circ-LDLRAD3 is closely related to the level of blood CA199, and the combination of circ-6909D3 and CA199 can improve the diagnostic value [83]. By detecting the expression of circulating circRNA in PDAC patients, Shao et al. [84] found that their overexpression can increase the resistance of normal PANC-1 and PACA-2 cells to gemcitabine. It can be seen that circulating circRNA has diagnostic value for PDAC, and we hope that more studies will support this idea in the future.
3.3 Reproductive and Urinary System Cancers
3.3.1 The Role of Circulating circRNA in Breast Cancer
Breast cancer is a common cancer among women worldwide [85]. Although the treatment of breast cancer has made significant progress, its mortality rate is still high [86]. Early screening of breast cancer is very helpful for treatment and prognosis, and current studies have shown that the expression level of plasma circRNA can reflect the progression of breast cancer. For example, cell-free RNA was used from the plasma samples of four breast cancers patients, and amplified qRT-PCR was used to detect circCNOT2. All samples showed detectable variable levels of circCNOT2, indicating that circRNAs in plasma [87]. CircRNAs can be used as biomarkers for breast cancer, and joint diagnosis can improve accuracy. For instance, the expressions of hsa_circ_0069094, hsa_circ_0079876, hsa_circ_0017650, and hsa_circ_0017536 in the plasma of patients with breast cancer were significantly higher than those of healthy controls. When combining hsa_circ_0069094 and hsa_circ_0017650, the AUC becomes 0.8469, and when combining all four circRNAs, the AUC is 0.8397 [86]. Furthermore, the diagnostic accuracy of hsa_circ_0001785 plasma was higher than CEA and cancer antigen-153 (CA153). The expression level of plasma hsa_circ_0001785 was closely related to histological grade, TNM stage and histological level, and its expression in plasma was significantly lower after surgery than before surgery [88]. The above researches have shown that plasma circRNA has great potential in the diagnosis of breast cancer.
3.3.2 The Role of Circulating circRNA in Ovarian Cancer
Ovarian cancer (OC) is one of the most common gynecological cancer in the world [89]. Although the surgical level has improved, the five-years survival rate is still not satisfactory [90]. CircRNAs have a long half-life in body fluids, so people are actively exploring circRNAs as biomarkers for OC diagnosis. A study found that 178 differentially expressed circRNAs were detected in the serum of OC patients, of which 175 were up-regulated and 3 were down-regulated and these circulating circRNAs may be related to the development of OC [91]. Circulating circRNAs may be valuable diagnostic biomarkers for early OC and have a potential role in disease progression. For example, circMAN1A2 is down-regulated in the ovary and can be used as a biomarker for ovarian cancer [47]. CircSETDB1 can be used as a new type of non-invasive biomarker to detect the progress of serous ovarian cancers (SOC) and predict the response of high-grade SOC to chemotherapy and relapse [92]. Hu et al. [90] found that circBNC2 is down-regulated in the plasma of epithelial ovarian cancers (EOC) patients, and they believed that circBNC2 may become a new biomarker for EOC. Thus, detecting the expression of circulating circRNA can predict the therapeutic effect of OC. We believe that circRNAs can be used as a biomarker for diagnosis and treatment of OC in the future.
3.3.3 The Role of Circulating circRNA in Prostate Cancer
Prostate cancer (PCa) is one of the leading cancers that cause death among men worldwide [93]. Currently, serum prostate-specific antigen (PSA) is still the standard biomarker for the diagnosis and treatment of PCa. However, PSA testing often leads to overdiagnosis and overtreatment due to its poor specificity [94]. It is proved that the expression of circZMIZ1 in the plasma of patients with PCa is higher than that in matched benign prostatic hyperplasia (BPH) patients. Thus, CircZMIZ1 can be considered as valuable biomarkers in PCa plasma [95]. Kong et al. [96] found that circFOXO3 is up-regulated in PCa tissues and serum samples, experiments have verified that circFOXO3 can be used as a promising biomarker for PCa. The combined diagnosis of circulating circRNA and PSA can be considered to make up for their shortcomings.
3.3.4 The Role of Circulating circRNA in Bladder Cancer
Bladder cancer is a common cancer of the male urinary system with high mortality [97]. Bladder cancer is classified as muscle-invasive bladder cancers (MIBC) and non-muscle invasive bladder cancer (NMBIC) [98]. CircRNAs can be detected in the serum of bladder cancer patients and is relatively stable. The levels of circFARSA, circSHKBP1, and circBANP in the serum of bladder cancer patients are significantly higher than those of healthy controls. Serum circFARSA and circBANP levels can be used as prognostic factors in patients with bladder cancer recurrence [99]. Compared with healthy controls, hsa_circ_000285 in serum is significantly reduced. Moreover, in patients with cisplatin-resistant bladder cancers, the expression of hsa_circ_000285 is lower than that of patients who are sensitive to cisplatin [100]. Therefore, circulating circRNA can be used as a biomarker for the diagnosis of bladder cancers, and some can also reflect drug resistance, which is very helpful for treatment.
3.4.1 The Role of Circulating circRNA in Leukemia
Leukemia is a malignant tumor of the blood system, which can be divided into acute leukemia and chronic leukemia. In recent years, it has been found that circRNA is involved in leukemia progression, so there are some studies exploring plasma or serum circRNA as biomarker. For instance, Circ-RPL15 in Chronic lymphocytic leukemia (CLL) plasma samples were significantly up-regulated, and ROC analysis showed that circ-RPL15 may be a potential biomarker for screening CLL [101]. The expression of circ_100053 in the serum was significantly higher than that of the healthy control group. The high expression of circ_100053 indicates a poor prognosis and imatinib resistance in patients with chronic myeloid leukemia (CML) [102]. We hope that more studies will explore the role of circulating circRNA in acute leukemia. From the above experimental studies, we believe that circulating circRNA can reflect the prognosis and resistance of CLL, which provides a new target for the future treatment of it.
3.4.2 The Role of Circulating circRNA in Multiple Myeloma
Multiple myeloma (MM) is a hematologic malignancy associated with plasma cells [103,104]. It is the second common hematologic malignancy with high recurrence rate and poor prognosis [105]. To search for potential diagnostic and therapeutic prognostic targets, Yu et al. [106] found that circ-MyBL2 was significantly reduced in MM tissue and serum samples compared to normal samples, and that its low level was associated with higher clinical staging, then speculated that serum circ-MyBL2 had good accuracy in the diagnosis of MM. We believe that circulating circRNAs, as diagnostic biomarkers of MM, have great potential in diagnosis and monitoring of prognosis.
3.4.3 The Role of Circulating circRNA in Lymphoma
Lymphoma include Hodgkin lymphoma and non-Hodgkin lymphoma. Diffused large B-cell lymphoma (DLBCL) is the most common non-Hodgkin lymphoma in adults, accounting for about 30% of all cases [107,108]. It was found that plasma circAPC levels were significantly decreased in DLBCL patients compared with healthy controls, suggesting that circAPC is a new proliferation inhibitor, and restoration of its expression may be an effective treatment for DLBCL [109]. Circulating circRNA can be used as a biomarker of lymphoma, and it has clinical significance in treatment. However, there are few studies related to lymphoma at present, we believe that there can be more researches on treatment and prognosis in the future.
The above studies have confirmed the role of circulating circRNA in cancers of different systems. The main functions, sources and expressions have been summarized in Tab. 1.
Based on previous studies on cancers biomarkers, circulating circRNAs were found as biomarkers for human cancers [110]. Current researches demonstrate that circRNA is rich in content, high in stability, and stable in peripheral blood [111]. Evidence has been that circRNAs expression in plasma or serum is associated with cancer progression, and a variety of circRNAs can be detected in blood samples collected from cancer patients [37]. Compared with other invasive tests, the method of detecting circRNAs by collecting patient blood is less harmful and more convenient, so circulating circRNAs can be used as noninvasive biomarkers for cancers.
Circulating circRNAs, mainly used to diagnose cancers, serves as biomarkers for monitoring cancer progression. Based on evidence drawn from the above studies, the expression level of circulating circRNAs also aids diagnosing cancers. Some authors believe that the combination of circulating circRNA and existing markers can improve the accuracy of detection. For example, Huang et al. believe that hsa_circ_0000745 plays an important role in GC. The combination of its plasma expression level and CEA level would provide a promising diagnostic marker for this malignant tumor [112]. In addition, circulating circRNAs can be utilized as dynamic indicators to monitor cancer progression. Circulating circRNAs are often expressed differently in cancer patients than in healthy controls, especially as the cancer progresses. CircRNAs can be conducive to treating cancers owe to its vital part in tumor genesis, proliferation, metastasis, invasion, stem cell regulation, and radiation resistance [113]. Statistical analysis prove that circulating circRNAs are related to the prognosis of cancers. The researches mentioned above have shown a correlation between circulating circRNAs and cancer prognosis, and their expression level can be detected to predict the prognosis of patients.
Circulating circRNA, with its biological functions, can be employed as biomarkers for diagnosis, treatment and prognosis of cancers. In addition, the detection of circulating circRNA levels in patients can be performed easily and cost-effectively with a non-invasive method. However, the researches of circulating circRNAs in cancers are far from enough since there lacks studies on the role of circulating circRNA in cancer treatment. If new progress and advances are made in treatment, the survival rate of patients should be better improved. There are many researches in the cancer of digestive system, but much fewer in other systems. Therefore, more experiments are still needed for verification. Through these existing studies, circulating circRNAs could be expected to serve as biomarkers for futuristic cancer diagnosis and treatment.
Funding Statement: This work was financially supported by the National Natural Science Foundation of China (81873978), the Key Project of Social Development in Jiangsu Province (BE2019691), the Sixth Talent Peaks Project of Jiangsu Province (2018-WSW-068), the Chinese Post-Doctoral Science Foundation (2018M642298), the Post-Doctoral Science Foundation of Jiangsu Province (2021K012A) and the Foundation of Nantong Science and Technology Bureau (MS2019021, MS22018010).
Conflicts of Interest:The authors declare that they have no conflict of interest to report regarding the present study.
1. Zaidi, S. A., Shahzad, F., Batool, S. (2020). Progress in cancer biomarkers monitoring strategies using graphene modified support materials. Talanta, 210(1), 120669. DOI 10.1016/j.talanta.2019.120669. [Google Scholar] [CrossRef]
2. Li, J., Sun, D., Pu, W., Wang, J., Peng, Y. (2020). Circular RNAs in cancer: Biogenesis, function, and clinical significance. Trends Cancer, 6(4), 319–336. DOI 10.1016/j.trecan.2020.01.012. [Google Scholar] [CrossRef]
3. Li, Y., Zeng, X., He, J., Gui, Y., Zhao, S. et al. (2018). Circular RNA as a biomarker for cancer: A systematic meta-analysis. Oncology Letters, 16(3), 4078–4084. DOI 10.3892/ol.2018.9125. [Google Scholar] [CrossRef]
4. Zhu, L., Li, N., Sun, L., Zheng, D., Shao, G. J. G. (2020). Non-coding RNAs: The key detectors and regulators in cardiovascular disease. Genomics, 113(1), 1233–1246. DOI 10.1016/j.ygeno.2020.10.024. [Google Scholar] [CrossRef]
5. Qu, S., Yang, X., Li, X., Wang, J., Gao, Y. et al. (2015). Circular RNA: A new star of noncoding RNAs. Cancer Letters, 365(2), 141–148. DOI 10.1016/j.canlet.2015.06.003. [Google Scholar] [CrossRef]
6. Long, F., Lin, Z., Li, L., Ma, M., Lu, Z. et al. (2021). Comprehensive landscape and future perspectives of circular RNAs in colorectal cancer. Molecular Cancer, 20(1), 26. DOI 10.1186/s12943-021-01318-6. [Google Scholar] [CrossRef]
7. Hao, S., Cong, L., Qu, R., Liu, R., Zhang, G. et al. (2019). Emerging roles of circular RNAs in colorectal cancer. OncoTargets and Therapy, 12, 4765–4777. DOI 10.2147/OTT. [Google Scholar] [CrossRef]
8. Lei, M., Zheng, G., Ning, Q., Zheng, J., Dong, D. (2020). Translation and functional roles of circular RNAs in human cancer. Molecular Cancer, 19(1), 30. DOI 10.1186/s12943-020-1135-7. [Google Scholar] [CrossRef]
9. Xu, Y., Yu, J., Huang, Z., Fu, B., Tao, Y. et al. (2020). Circular RNA hsa_circ_0000326 acts as a miR-338-3p sponge to facilitate lung adenocarcinoma progression. Journal of Experimental & Clinical Cancer Research, 39(1), 57. DOI 10.1186/s13046-020-01556-4. [Google Scholar] [CrossRef]
10. Li, J., Yang, J., Zhou, P., Le, Y., Zhou, C. et al. (2015). Circular RNAs in cancer: Novel insights into origins, properties, functions and implications. American Journal of Cancer Research, 5(2), 472–480. [Google Scholar]
11. Liu, X., Abraham, J., Cheng, Y., Wang, Z., Wang, Z. et al. (2018). Synthetic circular RNA functions as a miR-21 sponge to suppress gastric carcinoma cell proliferation. Molecular Therapy-Nucleic Acids, 13, 312–321. DOI 10.1016/j.omtn.2018.09.010. [Google Scholar] [CrossRef]
12. Zhang, M., Wang, T., Xiao, G., Xie, Y. (2020). Large-scale profiling of RBP-circRNA interactions from public CLIP-seq datasets. Genes, 11(1), 54. DOI 10.3390/genes11010054. [Google Scholar] [CrossRef]
13. Jiang, Y., Zhang, Y., Chu, F., Xu, L., Wu, H. (2020). Circ_0032821 acts as an oncogene in cell proliferation, metastasis and autophagy in human gastric cancer cells in vitro and in vivo through activating MEK1/ERK1/2 signaling pathway. Cancer Cell International, 20(1), 74. DOI 10.1186/s12935-020-1151-0. [Google Scholar] [CrossRef]
14. Lu, M., Wu, Y., Zeng, B., Sun, J., Li, Y. et al. (2020). CircEHMT1 inhibits metastatic potential of breast cancer cells by modulating miR-1233-3p/KLF4/MMP2 axis. Biochemical and Biophysical Research Communications, 526(2), 306–313. DOI 10.1016/j.bbrc.2020.03.084. [Google Scholar] [CrossRef]
15. Tran, A. M., Chalbatani, G. M., Berland, L., Cruz de Los Santos, M., Raj, P. et al. (2020). A new world of biomarkers and therapeutics for female reproductive system and breast cancers: Circular RNAs. Frontiers in Cell and Developmental Biology, 8, 50. DOI 10.3389/fcell.2020.00050. [Google Scholar] [CrossRef]
16. Hansen, T. B., Kjems, J., Damgaard, C. K. (2013). Circular RNA and miR-7 in cancer. Cancer Research, 73(18), 5609–5612. DOI 10.1158/0008-5472.CAN-13-1568. [Google Scholar] [CrossRef]
17. Chi, G., Xu, D., Zhang, B., Yang, F. (2019). Matrine induces apoptosis and autophagy of glioma cell line U251 by regulation of circRNA-104075/BCL-9. Chemico-Biological Interactions, 308(Suppl. 5), 198–205. DOI 10.1016/j.cbi.2019.05.030. [Google Scholar] [CrossRef]
18. Zhao, Q., Li, W., Pan, W., Wang, Z. (2021). CircRNA 010567 plays a significant role in myocardial infarction via the regulation of the miRNA-141/DAPK1 axis. Journal of Thoracic Disease, 13(4), 2447–2459. DOI 10.21037/jtd-21-212. [Google Scholar] [CrossRef]
19. Memczak, S., Papavasileiou, P., Peters, O., Rajewsky, N. (2015). Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS One, 10(10), e0141214. DOI 10.1371/journal.pone.0141214. [Google Scholar] [CrossRef]
20. Jeck, W. R., Sorrentino, J. A., Wang, K., Slevin, M. K., Burd, C. E. et al. (2013). Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA, 19(2), 141–157. DOI 10.1261/rna.035667.112. [Google Scholar] [CrossRef]
21. Feng, Y., Wang, Q., Shi, C., Liu, C., Zhang, Z. (2019). Does circular RNA exert significant effects in? Ovarian Cancer, 29(2), 161–170. DOI 10.1615/CritRevEukaryotGeneExpr.2019025941. [Google Scholar] [CrossRef]
22. Li, R., Jiang, J., Shi, H., Qian, H., Zhang, X. et al. (2020). CircRNA: A rising star in gastric cancer. Cellular and Molecular Life Sciences, 77(9), 1661–1680. DOI 10.1007/s00018-019-03345-5. [Google Scholar] [CrossRef]
23. Xie, R., Zhang, Y., Zhang, J., Li, J., Zhou, X. (2020). The role of circular RNAs in immune-related diseases. Frontiers in Immunology, 11, 545. DOI 10.3389/fimmu.2020.00545. [Google Scholar] [CrossRef]
24. Wang, W., Li, Y., Li, X., Liu, B., Han, S. et al. (2020). Circular RNA circ-FOXP1 induced by SOX9 promotes hepatocellular carcinoma progression via sponging miR-875-3p and miR-421. Biomedicine & Pharmacotherapy, 121(6), 109517. DOI 10.1016/j.biopha.2019.109517. [Google Scholar] [CrossRef]
25. Mo, W., Jiang, J., Zhang, L., Lu, Q., Li, J. et al. (2020). Circular RNA hsa_circ_0000467 promotes the development of gastric cancer by competitively binding to microRNA miR-326-3p. Biomed Research International, 2020(2), 4030826. DOI 10.1155/2020/4030826. [Google Scholar] [CrossRef]
26. Chen, L., Kong, R., Wu, C., Wang, S., Liu, Z. et al. (2020). Circ-MALAT1 functions as both an mRNA translation brake and a microrna sponge to promote self-renewal of hepatocellular cancer stem cells. Advanced Science, 7(4), 1900949. DOI 10.1002/advs.201900949. [Google Scholar] [CrossRef]
27. Deng, G., Mou, T., He, J., Chen, D., Lv, D. et al. (2020). Circular RNA circRHOBTB3 acts as a sponge for miR-654-3p inhibiting gastric cancer growth. Journal of Experimental & Clinical Cancer Research, 39(1), 1. DOI 10.1186/s13046-019-1487-2. [Google Scholar] [CrossRef]
28. Wang, Z., Lei, X., Wu, F. X. (2019). Identifying cancer-specific circRNA-RBP binding sites based on deep learning. Molecules, 24(22), 4035. DOI 10.3390/molecules24224035. [Google Scholar] [CrossRef]
29. Huang, A., Zheng, H., Wu, Z., Chen, M., Huang, Y. (2020). Circular RNA-protein interactions: Functions, mechanisms, and identification. Theranostics, 10(8), 3503–3517. DOI 10.7150/thno.42174. [Google Scholar] [CrossRef]
30. Dong, W., Dai, Z., Liu, F., Guo, X., Ge, C. et al. (2019). The RNA-binding protein RBM3 promotes cell proliferation in hepatocellular carcinoma by regulating circular RNA SCD-circRNA 2 production. EBioMedicine, 45, 155–167. DOI 10.1016/j.ebiom.2019.06.030. [Google Scholar] [CrossRef]
31. Vera-Ramirez, L., Vodnala, S. K., Nini, R., Hunter, K. W., Green, J. E. (2018). Autophagy promotes the survival of dormant breast cancer cells and metastatic tumour recurrence. Nature Communications, 9(1), 1994. DOI 10.1038/s41467-018-04070-6. [Google Scholar] [CrossRef]
32. Li, Y. J., Lei, Y. H., Yao, N., Wang, C. R., Hu, N. et al. (2017). Autophagy and multidrug resistance in cancer. Chinese Journal of Cancer, 36(1), 52. DOI 10.1186/s40880-017-0219-2. [Google Scholar] [CrossRef]
33. Gan, X., Zhu, H., Jiang, X., Obiegbusi, S. C., Yong, M. et al. (2020). CircMUC16 promotes autophagy of epithelial ovarian cancer via interaction with ATG13 and miR-199a. Molecular Cancer, 19(1), 45. DOI 10.1186/s12943-020-01163-z. [Google Scholar] [CrossRef]
34. Zhou, S. Y., Chen, W., Yang, S. J., Xu, Z. H., Hu, J. H. et al. (2019). The emerging role of circular RNAs in breast cancer. Bioscience Reports, 39(6), 7. DOI 10.1042/BSR20190621. [Google Scholar] [CrossRef]
35. Ruan, Y., Li, Z., Shen, Y., Li, T., Zhang, H. et al. (2020). Functions of circular RNAs and their potential applications in gastric cancer. Expert Review of Gastroenterology & Hepatology, 14(2), 85–92. DOI 10.1080/17474124.2020.1715211. [Google Scholar] [CrossRef]
36. Zhang, Y., Zhang, X. O., Chen, T., Xiang, J. F., Yin, Q. F. et al. (2013). Circular intronic long noncoding RNAs. Molecular Cell, 51(6), 792–806. DOI 10.1016/j.molcel.2013.08.017. [Google Scholar] [CrossRef]
37. Naeli, P., Pourhanifeh, M., Karimzadeh, M., Shabaninejad, Z., Movahedpour, A. et al. (2020). Circular RNAs and gastrointestinal cancers: Epigenetic regulators with a prognostic and therapeutic role. Critical Reviews in Oncology Hematology, 145(2), 102854. DOI 10.1016/j.critrevonc.2019.102854. [Google Scholar] [CrossRef]
38. Wan, B., Liu, B., Lv, C. (2020). Progress of research into circular RNAs in urinary neoplasms. PeerJ, 8(40), e8666. DOI 10.7717/peerj.8666. [Google Scholar] [CrossRef]
39. Zhi, X., Zhang, J., Cheng, Z., Bian, L., Qin, J. (2019). circLgr4 drives colorectal tumorigenesis and invasion through Lgr4-targeting peptide. International Journal of Cancer. DOI 10.1002/ijc.32549. [Google Scholar] [CrossRef]
40. Yang, Y., Gao, X., Zhang, M., Yan, S., Sun, C. et al. (2018). Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. Journal of the National Cancer Institute, 110(3), 304–315. DOI 10.1093/jnci/djx166. [Google Scholar] [CrossRef]
41. Wang, J., Zhao, X., Wang, Y., Ren, F., Sun, D. et al. (2020). circRNA-002178 act as a ceRNA to promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death & Disease, 11(1), 32. DOI 10.1038/s41419-020-2230-9. [Google Scholar] [CrossRef]
42. Hang, D., Zhou, J., Qin, N., Zhou, W., Ma, H. et al. (2018). A novel plasma circular RNA circFARSA is a potential biomarker for non-small cell lung cancer. Cancer Medicine, 7(6), 2783–2791. DOI 10.1002/cam4.1514. [Google Scholar] [CrossRef]
43. Tan, S., Sun, D., Pu, W., Gou, Q., Guo, C. et al. (2018). Circular RNA F-circEA-2a derived from EML4-ALK fusion gene promotes cell migration and invasion in non-small cell lung cancer. Molecular Cancer, 17(1), 138. DOI 10.1186/s12943-018-0887-9. [Google Scholar] [CrossRef]
44. Di, X., Jin, X., Li, R., Zhao, M., Wang, K. (2019). CircRNAs and lung cancer: Biomarkers and master regulators. Life Sciences, 220(Suppl. 5), 177–185. DOI 10.1016/j.lfs.2019.01.055. [Google Scholar] [CrossRef]
45. Liu, X. X., Yang, Y. E., Liu, X., Zhang, M. Y., Li, R. et al. (2019). A two-circular RNA signature as a noninvasive diagnostic biomarker for lung adenocarcinoma. Journal of Translational Medicine, 17(1), 50. DOI 10.1186/s12967-019-1800-z. [Google Scholar] [CrossRef]
46. Zhu, X., Wang, X., Wei, S., Chen, Y., Chen, Y. et al. (2017). hsa_circ_0013958: A circular RNA and potential novel biomarker for lung adenocarcinoma. FEBS Journal, 284(14), 2170–2182. DOI 10.1111/febs.14132. [Google Scholar] [CrossRef]
47. Fan, C. M., Wang, J. P., Tang, Y. Y., Zhao, J., He, S. Y. et al. (2019). circMAN1A2 could serve as a novel serum biomarker for malignant tumors. Cancer Science, 110(7), 2180–2188. DOI 10.1111/cas.14034. [Google Scholar] [CrossRef]
48. Lu, G., Cui, J., Qian, Q., Hou, Z., Xie, H. et al. (2020). Overexpression of hsa_circ_0001715 is a potential diagnostic and prognostic biomarker in. Lung Adenocarcinoma, 13, 10775–10783. DOI 10.2147/ott.S274932. [Google Scholar] [CrossRef]
49. Lu, H., Xie, X., Chen, Q., Cai, S., Liu, S. et al. (2021). Clinical significance of circPVT1 in patients with non-small cell lung cancer who received cisplatin combined with gemcitabine chemotherapy. Tumori Journal, 107(3), 204–208. DOI 10.1177/0300891620941940. [Google Scholar] [CrossRef]
50. Li, H., You, J., Xue, H., Tan, X., Chao, C. (2020). CircCTDP1 promotes nasopharyngeal carcinoma progression via a microRNA‐320b/HOXA10/TGFβ2 pathway. International Journal of Molecular Medicine, 45(3), 836–846. DOI 10.3892/ijmm.2020.4467. [Google Scholar] [CrossRef]
51. Zhong, Q., Huang, J., Wei, J., Wu, R. (2019). Circular RNA CDR1as sponges miR-7-5p to enhance E2F3 stability and promote the growth of nasopharyngeal carcinoma. Cancer Cell International, 19(1), 252. DOI 10.1186/s12935-019-0959-y. [Google Scholar] [CrossRef]
52. Shuai, M., Hong, J., Huang, D., Zhang, X., Tian, Y. (2018). Upregulation of circRNA_0000285 serves as a prognostic biomarker for nasopharyngeal carcinoma and is involved in radiosensitivity. Oncology Letters, 16(5), 6495–6501. DOI 10.3892/ol.2018.9471. [Google Scholar] [CrossRef]
53. Xia, T., Pan, Z., Zhang, J. (2020). CircSMC3 regulates gastric cancer tumorigenesis by targeting miR-4720-3p/TJP1 axis. Cancer Medicine, 9(12), 4299–4309. DOI 10.1002/cam4.3057. [Google Scholar] [CrossRef]
54. Lu, J., Wang, Y. H., Yoon, C., Huang, X. Y., Xu, Y. et al. (2020). Circular RNA circ-RanGAP1 regulates VEGFA expression by targeting miR-877-3p to facilitate gastric cancer invasion and metastasis. Cancer Letters, 471, 38–48. DOI 10.1016/j.canlet.2019.11.038. [Google Scholar] [CrossRef]
55. Liu, J., Li, Z., Teng, W., Ye, X. (2020). Identification of downregulated circRNAs from tissue and plasma of patients with gastric cancer and construction of a circRNA-miRNA-mRNA network. Journal of Cellular Biochemistry, 121(11), 4590–4600. DOI 10.1002/jcb.29673. [Google Scholar] [CrossRef]
56. Huang, C., Liu, Z., Xiao, L., Xia, Y., Huang, J. et al. (2019). Clinical significance of serum CA125, CA19-9, CA72-4, and fibrinogen-to-lymphocyte ratio in gastric cancer with peritoneal dissemination. Frontiers in Oncology, 9, 1159. DOI 10.3389/fonc.2019.01159. [Google Scholar] [CrossRef]
57. Sun, H., Tang, W., Rong, D., Jin, H., Fu, K. et al. (2018). Hsa_circ_0000520, a potential new circular RNA biomarker, is involved in gastric carcinoma. Cancer Biomarkers, 21(2), 299–306. DOI 10.3233/CBM-170379. [Google Scholar] [CrossRef]
58. Han, L., Zhang, X., Wang, A., Ji, Y., Cao, X. et al. (2020). A dual-circular RNA signature as a non-invasive diagnostic biomarker for gastric cancer. Frontiers in Oncology, 10, 184. DOI 10.3389/fonc.2020.00184. [Google Scholar] [CrossRef]
59. Lu, J., Zhang, P. Y., Xie, J. W., Wang, J. B., Lin, J. X. et al. (2019). Circular RNA hsa_circ_0006848 related to ribosomal protein L6 acts as a novel biomarker for early gastric cancer. Disease Markers, 2019(10), 3863458–3863413. DOI 10.1155/2019/3863458. [Google Scholar] [CrossRef]
60. Li, J., Li, H., Lv, X., Yang, Z., Gao, M. et al. (2019). Diagnostic performance of circular RNAs in human cancers: A systematic review and meta-analysis. Molecular Genetics & Genomic Medicine, 7(7), e00749. DOI 10.1002/mgg3.749. [Google Scholar] [CrossRef]
61. Ding, Y., Fang, A., Yan, J., Duan, J., Wang, N. et al. (2019). Selective downregulation of distinct circRNAs in the tissues and plasma of patients with primary hepatic carcinoma. Oncology Letters, 18(5), 5255–5268. DOI 10.3892/ol.2019.10908. [Google Scholar] [CrossRef]
62. Wei, W., Liu, M., Ning, S., Wei, J., Zhong, J. et al. (2020). Diagnostic value of plasma HSP90alpha levels for detection of hepatocellular carcinoma. BMC Cancer, 20(1), 6. DOI 10.1186/s12885-019-6489-0. [Google Scholar] [CrossRef]
63. Weng, Q., Chen, M., Li, M., Zheng, Y. F., Shao, G. et al. (2019). Global microarray profiling identified hsa_circ_0064428 as a potential immune-associated prognosis biomarker for hepatocellular carcinoma. Journal of Medical Genetics, 56(1), 32–38. DOI 10.1136/jmedgenet-2018-105440. [Google Scholar] [CrossRef]
64. Zhu, K., Zhan, H., Peng, Y., Yang, L., Gao, Q. et al. (2019). Plasma hsa_circ_0027089 is a diagnostic biomarker for hepatitis B virus-related hepatocellular carcinoma. Carcinogenesis, 41(3), 296–302. DOI 10.1093/carcin/bgz154. [Google Scholar] [CrossRef]
65. Qiao, G. L., Chen, L., Jiang, W. H., Yang, C., Yang, C. M. et al. (2019). Hsa_circ_0003998 may be used as a new biomarker for the diagnosis and prognosis of hepatocellular carcinoma. OncoTargets and Therapy, 12, 5849–5860. DOI 10.2147/OTT. [Google Scholar] [CrossRef]
66. Zhou, S., Wei, J., Wang, Y., Liu, X. (2020). Cisplatin resistance-associated circRNA_101237 serves as a prognostic biomarker in hepatocellular carcinoma. Experimental and Therapeutic Medicine, 19(4), 2733–2740. DOI 10.3892/etm.2020.8526. [Google Scholar] [CrossRef]
67. Zhang, X., Zhou, H., Jing, W., Luo, P., Qiu, S. et al. (2018). The circular RNA hsa_circ_0001445 regulates the proliferation and migration of hepatocellular carcinoma and may serve as a diagnostic biomarker. Disease Markers, 2018(1), 3073467–3073469. DOI 10.1155/2018/3073467. [Google Scholar] [CrossRef]
68. Zhou, S., Wei, J., Wang, Y., Liu, X. (2020). Cisplatin resistance-associated circRNA_101237 serves as a prognostic biomarker in hepatocellular carcinoma. Experimental and Therapeutic Medicine, 19(4), 2733–2740. DOI 10.3892/etm.2020.8526. [Google Scholar] [CrossRef]
69. Wang, X., Ren, Y., Ma, S., Wang, S. (2020). Circular RNA 0060745, a novel circRNA, promotes colorectal cancer cell proliferation and metastasis through miR-4736 sponging. OncoTargets and Therapy, 13, 1941–1951. DOI 10.2147/OTT. [Google Scholar] [CrossRef]
70. Slaby, O. (2016). Non-coding RNAs as biomarkers for colorectal cancer screening and early detection. Advances in Experimental Medicine and Biology, 937, 153–170. DOI 10.1007/978-3-319-42059-2. [Google Scholar] [CrossRef]
71. Zhang, J., Wang, Y., Zhao, T., Li, Y., Tian, L. et al. (2020). Evaluation of serum MUC5AC in combination with CA19-9 for the diagnosis of pancreatic cancer. World Journal of Surgical Oncology, 18(1), 31. DOI 10.1186/s12957-020-1809-z. [Google Scholar] [CrossRef]
72. Li, X. N., Wang, Z. J., Ye, C. X., Zhao, B. C., Huang, X. X. et al. (2019). Circular RNA circVAPA is up-regulated and exerts oncogenic properties by sponging miR-101 in colorectal cancer. Biomedicine & Pharmacotherapy, 112(6), 108611. DOI 10.1016/j.biopha.2019.108611. [Google Scholar] [CrossRef]
73. Zhang, W., Yang, S., Liu, Y., Wang, Y., Lin, T. et al. (2018). Hsa_circ_0007534 as a blood-based marker for the diagnosis of colorectal cancer and its prognostic value. International Journal of Clinical and Experimental Pathology, 11(3), 1399–1406. [Google Scholar]
74. Lin, J., Cai, D., Li, W., Yu, T., Mao, H. et al. (2019). Plasma circular RNA panel acts as a novel diagnostic biomarker for colorectal cancer. Clinical Biochemistry, 74(2), 60–68. DOI 10.1016/j.clinbiochem.2019.10.012. [Google Scholar] [CrossRef]
75. Xu, Z., Tie, X., Li, N., Yi, Z., Shen, F. et al. (2020). Circular RNA hsa_circ_0000654 promotes esophageal squamous cell carcinoma progression by regulating the miR-149-5p/IL-6/STAT3 pathway. IUBMB Life, 72(3), 426–439. DOI 10.1002/iub.2202. [Google Scholar] [CrossRef]
76. Zhang, Z., Li, X., Xiong, F., Ren, Z., Han, Y. (2020). Hsa_circ_0012563 promotes migration and invasion of esophageal squamous cell carcinoma by regulating XRCC1/EMT pathway. Journal of Clinical Laboratory Analysis, 34(8), e23308. DOI 10.1002/jcla.23308. [Google Scholar] [CrossRef]
77. Wang, Q., Liu, H., Liu, Z., Yang, L., Zhou, J. et al. (2020). Circ-SLC7A5, a potential prognostic circulating biomarker for detection of ESCC. Cancer Genetics, 240(1), 33–39. DOI 10.1016/j.cancergen.2019.11.001. [Google Scholar] [CrossRef]
78. Hu, X., Wu, D., He, X., Zhao, H., He, Z. et al. (2019). circGSK3beta promotes metastasis in esophageal squamous cell carcinoma by augmenting beta-catenin signaling. Molecular Cancer, 18(1), 160. DOI 10.1186/s12943-019-1095-y. [Google Scholar] [CrossRef]
79. Huang, E., Fu, J., Yu, Q., Xie, P., Yang, Z. et al. (2020). CircRNA hsa_circ_0004771 promotes esophageal squamous cell cancer progression via miR-339-5p/CDC25A axis. Epigenomics, 12(7), 587–603. DOI 10.2217/epi-2019-0404. [Google Scholar] [CrossRef]
80. Hao, L., Rong, W., Bai, L., Cui, H., Zhang, S. et al. (2019). Upregulated circular RNA circ_0007534 indicates an unfavorable prognosis in pancreatic ductal adenocarcinoma and regulates cell proliferation, apoptosis, and invasion by sponging miR-625 and miR-892b. Journal of Cellular Biochemistry, 120(3), 3780–3789. DOI 10.1002/jcb.27658. [Google Scholar] [CrossRef]
81. Reni, M., Peretti, U., Zanon, S., Macchini, M., Balzano, G. et al. (2020). Time to CA19-9 nadir: a clue for defining optimal treatment duration in patients with resectable pancreatic ductal adenocarcinoma. Cancer Chemother Pharmacol, 85(4), 641–650. DOI 10.1007/s00280-020-04047-7. [Google Scholar] [CrossRef]
82. Wang, A., Sun, B., Wang, M., Shi, H., Huang, Z. et al. (2020). Predictive value of CONUT score combined with serum CA199 levels in postoperative survival of patients with pancreatic ductal adenocarcinoma: A retrospective study. PeerJ, 8(13), e8811. DOI 10.7717/peerj.8811. [Google Scholar] [CrossRef]
83. Yang, F., Liu, D. Y., Guo, J. T., Ge, N., Zhu, P. et al. (2017). Circular RNA circ-LDLRAD3 as a biomarker in diagnosis of pancreatic cancer. World Journal of Gastroenterology, 23(47), 8345–8354. DOI 10.3748/wjg.v23.i47.8345. [Google Scholar] [CrossRef]
84. Shao, F., Huang, M., Meng, F., Huang, Q. (2018). Circular RNA signature predicts gemcitabine resistance of pancreatic ductal adenocarcinoma. Frontiers in Pharmacology, 9, 584. DOI 10.3389/fphar.2018.00584. [Google Scholar] [CrossRef]
85. Cao, L., Wang, M., Dong, Y., Xu, B., Chen, J. et al. (2020). Circular RNA circRNF20 promotes breast cancer tumorigenesis and warburg effect through miR-487a/HIF-1alpha/HK2. Cell Death & Disease, 11(2), 145. DOI 10.1038/s41419-020-2336-0. [Google Scholar] [CrossRef]
86. Li, Z., Chen, Z., Hu, G., Zhang, Y., Feng, Y. et al. (2020). Profiling and integrated analysis of differentially expressed circRNAs as novel biomarkers for breast cancer. Journal of Cellular Physiology, 235(11), 7945–7959. DOI 10.1002/jcp.29449. [Google Scholar] [CrossRef]
87. Smid, M., Wilting, S., Uhr, K., Rodríguez-González, F., de Weerd, V. et al. (2019). The circular RNome of primary breast cancer. Genome Research, 29(3), 356–366. DOI 10.1101/gr.238121.118. [Google Scholar] [CrossRef]
88. Yin, W. B., Yan, M. G., Fang, X., Guo, J. J., Xiong, W. et al. (2018). Circulating circular RNA hsa_circ_0001785 acts as a diagnostic biomarker for breast cancer detection. Clinica Chimica Acta, 487(11), 363–368. DOI 10.1016/j.cca.2017.10.011. [Google Scholar] [CrossRef]
89. Xu, F., Ni, M., Li, J., Cheng, J., Zhao, H. et al. (2020). Circ0004390 promotes cell proliferation through sponging miR-198 in ovarian cancer. Biochemical and Biophysical Research Communications, 526(1), 14–20. DOI 10.1016/j.bbrc.2020.03.024. [Google Scholar] [CrossRef]
90. Hu, Y., Zhu, Y., Zhang, W., Lang, J., Ning, L. (2019). Utility of plasma circBNC2 as a diagnostic biomarker in epithelial ovarian cancer. OncoTargets and Therapy, 12, 9715–9723. DOI 10.2147/OTT. [Google Scholar] [CrossRef]
91. Wang, J., Wu, A., Yang, B., Zhu, X., Teng, Y. et al. (2020). Profiling and bioinformatics analyses reveal differential circular RNA expression in ovarian cancer. Gene, 724, 144150. DOI 10.1016/j.gene.2019.144150. [Google Scholar] [CrossRef]
92. Wang, W., Wang, J., Zhang, X., Liu, G. (2019). Serum circSETDB1 is a promising biomarker for predicting response to platinum-taxane-combined chemotherapy and relapse in high-grade serous ovarian cancer. OncoTargets and Therapy, 12, 7451–7457. DOI 10.2147/OTT. [Google Scholar] [CrossRef]
93. Greene, J., Baird, A. M., Casey, O., Brady, L., Blackshields, G. et al. (2019). Circular RNAs are differentially expressed in prostate cancer and are potentially associated with resistance to enzalutamide. Scientific Reports, 9(1), 10739. DOI 10.1038/s41598-019-47189-2. [Google Scholar] [CrossRef]
94. Xia, Q., Ding, T., Zhang, G., Li, Z., Zeng, L. et al. (2018). Circular RNa expression profiling identifies prostate cancer- specific circrnas in prostate cancer. Cellular Physiology and Biochemistry, 50(5), 1903–1915. DOI 10.1159/000494870. [Google Scholar] [CrossRef]
95. Jiang, H., Lv, D., Song, X., Wang, C., Yu, Y. et al. (2020). Upregulated circZMIZ1 promotes the proliferation of prostate cancer cells and is a valuable marker in plasma. Neoplasma, 67(1), 68–77. DOI 10.4149/neo_2019_190213N116. [Google Scholar] [CrossRef]
96. Kong, Z., Wan, X., Lu, Y., Zhang, Y., Huang, Y. et al. (2020). Circular RNA circFOXO3 promotes prostate cancer progression through sponging miR-29a-3p. Journal of Cellular and Molecular Medicine, 24(1), 799–813. DOI 10.1111/jcmm.14791. [Google Scholar] [CrossRef]
97. Yang, C., Li, Q., Chen, X., Zhang, Z., Mou, Z. et al. (2020). Circular RNA circRGNEF promotes bladder cancer progression via miR-548/KIF2C axis regulation. Sedentary Life and Nutrition, 12(8), 6865–6879. DOI 10.18632/aging.103047. [Google Scholar] [CrossRef]
98. Shen, C., Wu, Z., Wang, Y., Gao, S., Da, L. et al. (2020). Downregulated hsa_circ_0077837 and hsa_circ_0004826, facilitate bladder cancer progression and predict poor prognosis for bladder cancer patients. Cancer Medicine, 9(11), 3885–3903. DOI 10.1002/cam4.3006. [Google Scholar] [CrossRef]
99. Pan, J., Xie, X., Li, H., Li, Z., Ren, C. et al. (2019). Detection of serum long non-coding RNA UCA1 and circular RNAs for the diagnosis of bladder cancer and prediction of recurrence. International Journal of Clinical and Experimental Pathology, 12(8), 2951–2958. [Google Scholar]
100. Chi, B., Zhao, D., Liu, L., Yin, X., Wang, F. et al. (2019). Downregulation of hsa_circ_0000285 serves as a prognostic biomarker for bladder cancer and is involved in cisplatin resistance. Neoplasma, 66(2), 197–202. DOI 10.4149/neo_2018_180318N185. [Google Scholar] [CrossRef]
101. Wu, Z., Sun, H., Liu, W., Zhu, H., Fu, J. et al. (2020). Circ-RPL15: A plasma circular RNA as novel oncogenic driver to promote progression of chronic lymphocytic leukemia. Leukemia, 34(3), 919–923. DOI 10.1038/s41375-019-0594-6. [Google Scholar] [CrossRef]
102. Lei, P., Chen, J. J., Liao, C. S., Liu., G. H., Zhou, M. (2019). High circ_100053 predicts a poor outcome for chronic myeloid leukemia and is involved in imatinib resistance. Oncology Research. DOI 10.3727/096504018x15412701483326. [Google Scholar] [CrossRef]
103. Xu, H., Yin, Q., Shen, X., Ju, S. (2020). Long non-coding RNA CCAT2 as a potential serum biomarker for diagnosis and prognosis of multiple myeloma. Annals of Hematology, 99(9), 2159–2171. DOI 10.1007/s00277-020-04161-9. [Google Scholar] [CrossRef]
104. Zhang, Y., Pisano, M., Li, N., Tan, G., Sun, F. et al. (2021). Exosomal circRNA as a novel potential therapeutic target for multiple myeloma-related peripheral neuropathy. Cellular Signalling, 78(5), 109872. DOI 10.1016/j.cellsig.2020.109872. [Google Scholar] [CrossRef]
105. Chen, F., Wang, X., Fu, S., Wang, S., Fu, Y. et al. (2020). Circular RNA circ-CDYL sponges miR-1180 to elevate yes-associated protein in multiple myeloma. Experimental Biology and Medicine, 245(11), 925–932. DOI 10.1177/1535370220918191. [Google Scholar] [CrossRef]
106. Yu, S., Ai, L., Wei, W., Pan, J. (2021). circRNA circ-MYBL2 is a novel tumor suppressor and potential biomarker in multiple myeloma. Human Cell, 34(1), 219–228. DOI 10.1007/s13577-020-00441-8. [Google Scholar] [CrossRef]
107. Chen, X., Xie, X., Zhou, W. (2020). CircCFL1/MiR-107 axis targeting HMGB1 promotes the malignant progression of diffuse large B-cell lymphoma tumors. Cancer Management and Research, 12, 9351–9362. DOI 10.2147/CMAR.S263222. [Google Scholar] [CrossRef]
108. Shi, Y., Ding, D., Qu, R., Tang, Y., Hao, S. (2020). Non-coding RNAs in diffuse large B-cell lymphoma. OncoTargets and Therapy, 13, 12097–12112. DOI 10.2147/OTT.S281810. [Google Scholar] [CrossRef]
109. Hu, Y., Zhao, Y., Shi, C., Ren, P., Wei, B. et al. (2019). Acircular RNA from APC inhibits the proliferation of diffuse large B-cell lymphoma by inactivating Wnt/β-catenin signaling via interacting with TET1 and miR-888. Sedentary Life and Nutrition, 11(19), 8068–8084. DOI 10.18632/aging.102122. [Google Scholar] [CrossRef]
110. Wu, Z., Sun, H., Li, J., Jin, H. (2019). Circular RNAs in leukemia. Aging, 11(134757–4771. DOI 10.18632/aging.102091. [Google Scholar] [CrossRef]
111. Han, Y., Xia, S., Zhang, Y., Zheng, J., Li, W. (2017). Circular RNAs: A novel type of biomarker and genetic tools in cancer. Oncotarget, 8(38), 64551–64563. DOI 10.18632/oncotarget.18350. [Google Scholar] [CrossRef]
112. Huang, M., He, Y., Liang, L., Huang, Q., Zhu, Z. (2017). Circular RNA hsa_circ_0000745 may serve as a diagnostic marker for gastric cancer. World Journal of Gastroenterology, 23(34), 6330–6338. DOI 10.3748/wjg.v23.i34.6330. [Google Scholar] [CrossRef]
113. Chen, B., Huang, S. (2018). Circular RNA: An emerging non-coding RNA as a regulator and biomarker in cancer. Cancer Letters, 418, 41–50. DOI 10.1016/j.canlet.2018.01.011. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |