
 | Oncologie |  |
DOI: 10.32604/Oncologie.2021.016900
ARTICLE
HBx Downregulates miR-422a Expression via Activation of FOXG1/Q1/E1 in HepG2 Cells
1Organ Transplantation Center, Sichuan Provincial People’s Hospital, University of Electronic Science and Technology of China, Chengdu, 610072, China
2Chinese Academy of Sciences Sichuan Translational Medicine Research Hospital, Chengdu, 610072, China
3Department of Hepatobiliary Surgery, Sichuan Provincial People’s Hospital, University of Electronic Science and Technology of China, Chengdu, 610072, China
*Corresponding Author: Gang Wu. Email: wugang@med.uestc.edu.cn
Received: 07 April 2021; Accepted: 21 May 2021
Abstract: microRNA-422a (miR-422a) is downregulated in many hematopoietic tumors and solid tumors including hepatocellular carcinoma. We previously demonstrated that hepatitis B virus X protein (HBx) downregulated expression of miR-422a in HCC cell line HepG2 in vitro. However, we explore the mechanisms underlying this action. Forkhead box proteins (FOX) G1/Q1/E1 are known to negatively regulate miR-422a expression, and this prompted us to determine whether HBx suppresses miR-422a expression via activation of FOXG1/Q1/E1. The relationship between FOXG1/Q1/E1 and miR-422a in HepG2 cells stably expressing HBx was assessed with qRT-PCR. The correlation between HBx and FOXG1/Q1/E1 was determined with qRT-PCR and western blot in vitro. The cell cycle and CCK-8 assays were used to elucidate the consequence of miR-422a transfection in HepG2-hbx cells. FOXG1/Q1/E1 activated by HBx was found to be responsible, at least in part, for the downregulation of miR-422a in HepG2 cells. miR-422a transfection hampered the growth of HepG2-hbx cells by arresting cells in G1 phase. Both FOXG1/Q1/E1 and miR-422a may be suitable molecular targets for treatment of HBV-infected HCC.
Keywords: HBx; hepatocellular carcinoma; FOXG1/Q1/E1; miR-422a
miRNAs play important roles in the regulation of gene expression post-transcriptionally and are extensively involved in tumorigenesis [1]. Deregulation of miRNAs has been observed in multiple human diseases including hepatocarcinogenesis. miRNAs can be tumor suppressors or tumor promoters according to their expression and functions in specific tumors [2]. miR-422a downregulation can be observed in many hematopoietic tumors and solid tumors including hepatocellular carcinoma [3,4], leading to the activation of such oncogenes as MAPK1, PIK3CA, CD73, IGF1/IGF1R, and FOXG1/Q1/E1 [4–9].
miRNA dysregulation in tumors can be attributed to genomic variations, epigenetic modification, and chaos in biogenesis process. In addition to known regulators, miRNAs are also regulated by protein-coding genes, especially those encoding transcription factors (TFs). p53 promotes the maturation of miR-16-1, miR-143, and miR-145, which are tumor suppressors, in response to DNA damage [10]. c-Myc has been found to transcriptionally upregulate the onco-cluster of miR-17-92, while such tumor suppressors as let-7, miR-34a, and miR-16 were suppressed [11]. miR-422a silences the expression of FOXG1/Q1/E1, which is its own upstream regulator [4].
Because it is an onco-protein encoded by hepatitis B virus (HBV), hepatitis B virus X protein (HBx) is extensively involved in the initiation and progression of hepatocarcinogenesis [12]. HBx deregulates gene expression of hepatocytes by activating cell signaling pathways in cytoplasm and by binding TFs in nucleus, thereby contributing to malignant transition in hepatocytes [13]. Our team, for the first time, reported that HBx downregulated miR-16 family in HepG2 cells via activation of c-Myc, expanding its regulation of gene expression from protein-coding to non-coding genes [14]. Other teams have reported that the HBx transcript itself can directly mediate miR-15a/miR-16-1 repression in hepatocytes [15]. In addition to binding protein, HBx can also bind the DLEU2 lncRNA to activate gene expression in hepatocytes [16].
In a previous work, based on the pleiotropic functions of HBx, we explored their effects on the miRNA expression of hepatocytes. As in another report [17], miR-422a was downregulated by HBx in HepG2 cells in a microarray [14]. However, the mechanisms underlying the repression of miR-422a by HBx were not clearly established. In this research, we further confirmed that miR-422a was repressed by HBx transfection in HepG2 cells using qRT-PCR. As expected, FOXG1, Q1, and E1 were activated by HBx in vitro. Accordingly, si-FOXG1/Q1/E1 transfection hampered the HBx-induced miR-422a repression. Inversely, miR-422a transfection downregulated the expression of FOXG1/Q1/E1 in HepG2-hbx cells. Finally, miR-422a mimic transfection hampered the proliferation of HepG2-hbx cells by arresting cells in the G1 phase. In brief, we observed a preliminary HBx-FOXG1/Q1/E1-miR-422a pathway in HepG2-hbx cells. miR-422a may be an original molecular targeted agent suitable for treating HBV+ HCC via silencing FOXG1/Q1/E1.
2.1 Cell Culture and Transfection
Untransfected HepG2 cells and HepG2 cells stably transfected with HBx expressing plasmid (HepG2-hbx) or empty vector (HepG2-vc) were constructed and cultured as described by our team [14]. The RNA nucleotides were transfected using Lipofectamine 3000 (Invitrogen). Except where otherwise specified, 50 nM of RNA nucleotides were transiently transfected in all experiments.
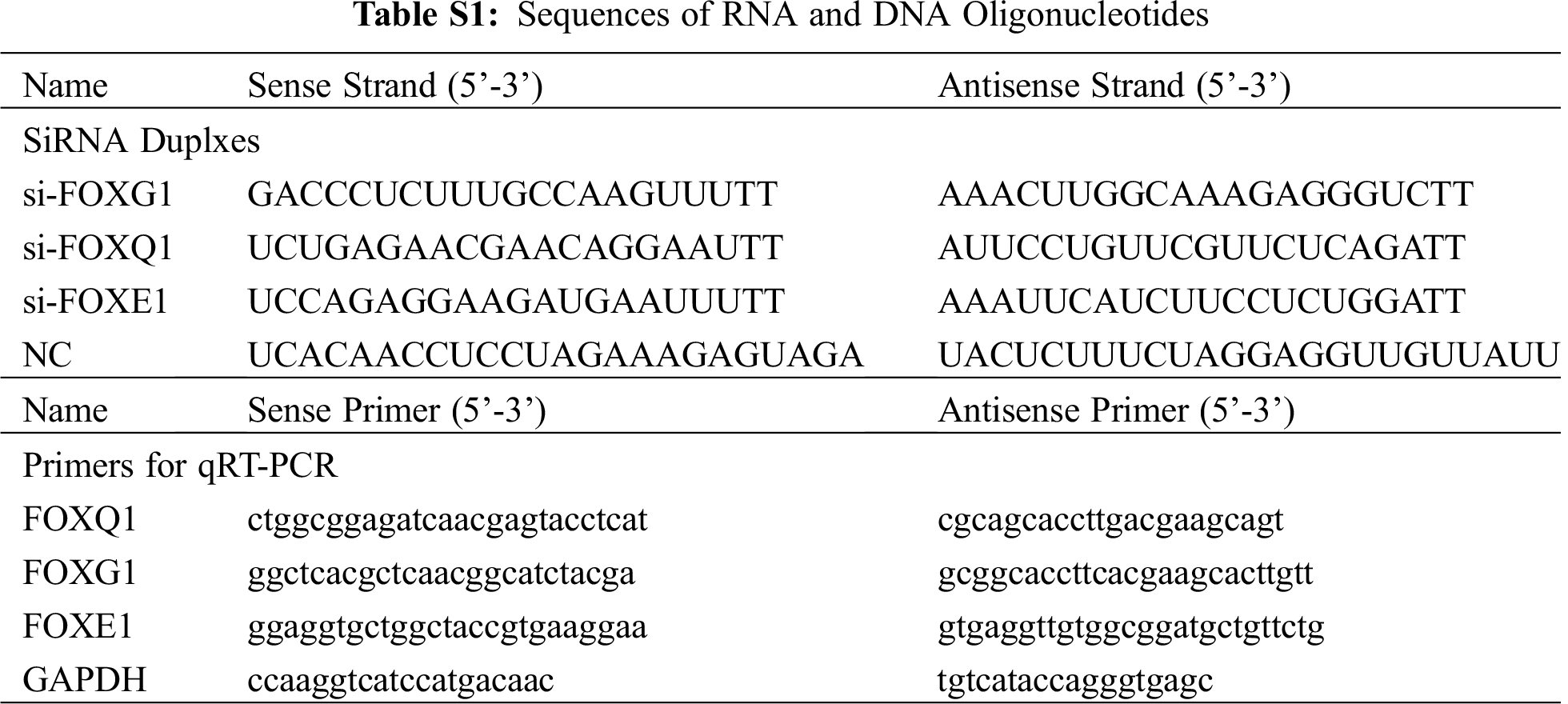
FOXG1/Q1/E1-specific siRNAs, negative control (NC), miR-422a mimic, and miRNA NC were purchased from Genepharma, Shanghai, China. The sequences of siRNAs and primers for PCR are provided in Tab. S1.
RNA was extracted using TRIzol reagent (Invitrogen). cDNA was synthesized with a PrimeScript™ RT Reagent Kit (Takara Biomedical Technology, Beijing, China). qRT-PCR for FOXG1/Q1/E1 and GAPDH were performed with TB Green® Premix Ex Taq™ II (Takara Biomedical Technology). The detection kits for miR-422a and U6 were provided by Genepharma; total cellular protein was extracted with RIPA buffer. The antibodies used in western blot were for FOXG1/Q1/E1 (ab18259, ab51340, ab236661; Abcam), HBx (sc-71239; Santa Cruz) and beta-actin (A1978; Sigma-Aldrich).
2.4 Cell Proliferation and Cell Cycle Assays
The growth of cells was detected with CCK-8 kit; the distribution of DNA content was determined using a commercial kit (KGA511, KeyGEN BioTECH, Nanjing, China).
All data analysis was performed with SPSS 19.0, and p < 0.05 was set as significance level. The qRT-PCR results were analyzed using a Student’s t-test.
3.1 miR-422a was Suppressed by HBx in HepG2 Cells
To confirm previous results that had been found using a microarray method [14], we evaluated the significant miR-422a downregulation in HepG2 cells with stable and transient HBx transfection using qRT-PCR (Figs. 1A–1B). The western-blot results confirmed the HBx protein expression in HepG2 cells (Fig. 1C).
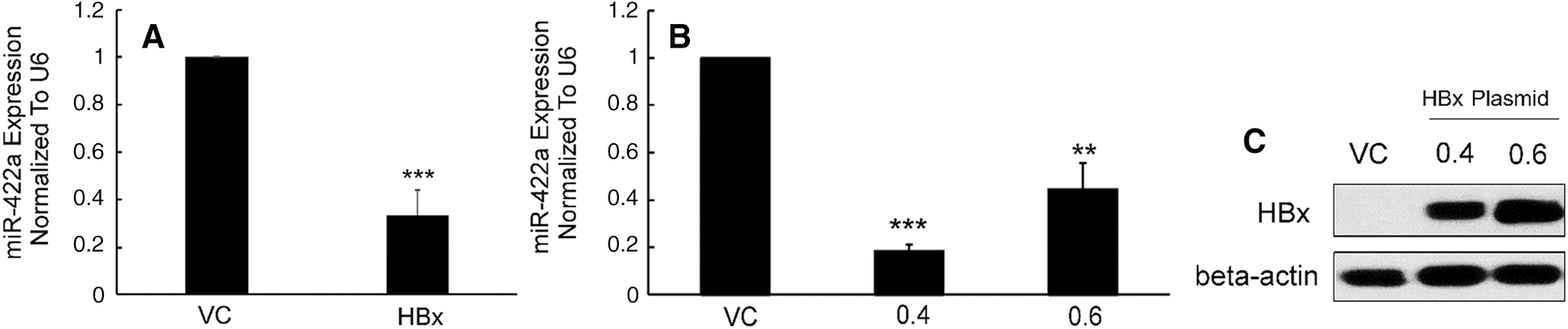
Figure 1: Stable and transient HBx transfection downregulated miR-422a expression in HepG2 cells. A) miR-422a expression was assessed in HepG2-hbx and HepG2-vc cells. B) HepG2 cells were transiently transfected with HBx plasmid (0.4 or 0.6 µg) or control. miR-422a expression was normalized to U6. C) Western-blot results confirmed the HBx protein expression in HepG2 cells after the transient transfection with HBx expressing plasmid
3.2 FOXG1/Q1/E1 was Upregulated in HepG2-hbx Cells
To clearly establish the mechanisms by which HBx downregulates miR-422a in HepG2 cells, we began by quoting a report that described a double-negative feedback loop between miR-422a and FOXG1/Q1/E1 in HCC [4]. We hypothesized that FOXG1, Q1, and E1 were activated by HBx and responsible for the miR-422a downregulation in HepG2 cells. As anticipated, qRT-PCR and western blot results confirmed the significant upregulation of FOXG1, Q1, and E1 in HepG2-hbx cells (Figs. 2A–2D). Moreover, the efficiency of si-FOXG1/Q1/E1 was also evaluated (Figs. 2A–2C). Collectively, our results, for the first time, showed that HBx could induce FOXG1/Q1/E1 activation in HepG2 cells in vitro.
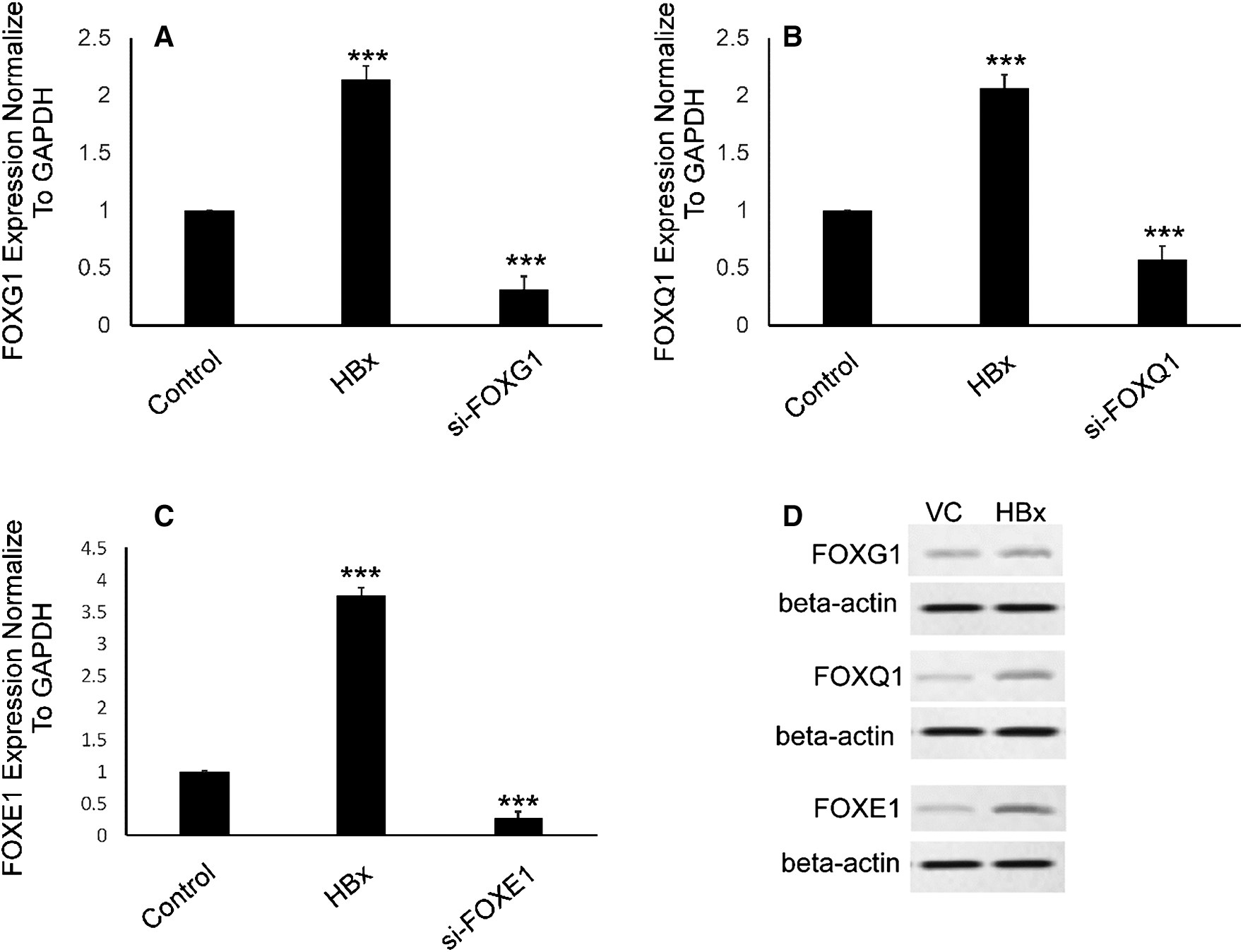
Figure 2: FOXG1, Q1, and E1 were upregulated in HepG2-hbx cells. A–C) qRT-PCR confirmed the elevation of FOXG1, Q1, and E1 mRNA in HepG2-hbx cells, while the FOXG1, Q1, and E1-specific siRNA efficiently silenced their expression. Expression of each was normalized to GAPDH. D) Western blot results validated the upregulation of FOXG1, Q1, and E1 protein in HepG2-hbx cells. Representative pictures are presented and beta-actin was used as reference gene
3.3 miR-422a and FOXG1/Q1/E1 Showed Antagonistic Effects against Each Other in HepG2-hbx Cells
In a previous work, we showed the existence of an HBx-Lin28B-let-7 pathway in HepG2 cells [18]. We hypothesized that there was also an HBx-FOXG1/Q1/E1-miR-422a pathway in HepG2 cells. We confirmed that loss-of-FOXG1/Q1/E1 function using specific siRNAs significantly reactivated the expression of miR-422a in HepG2-hbx cells (Fig. 3D). Correspondingly, we verified that ectopically expressed miR-422a mimics could suppress the protein expression of FOXG1/Q1/E1 in HepG2-hbx cells (Figs. 3A–3C, three duplicates). In conclusion, we suggest that there may be not only a feed-forward HBx-FOXG1/Q1/E1-miR-422a pathway but also negative feedback between miR-422a and FOXG1/Q1/E1 (Fig. 4).
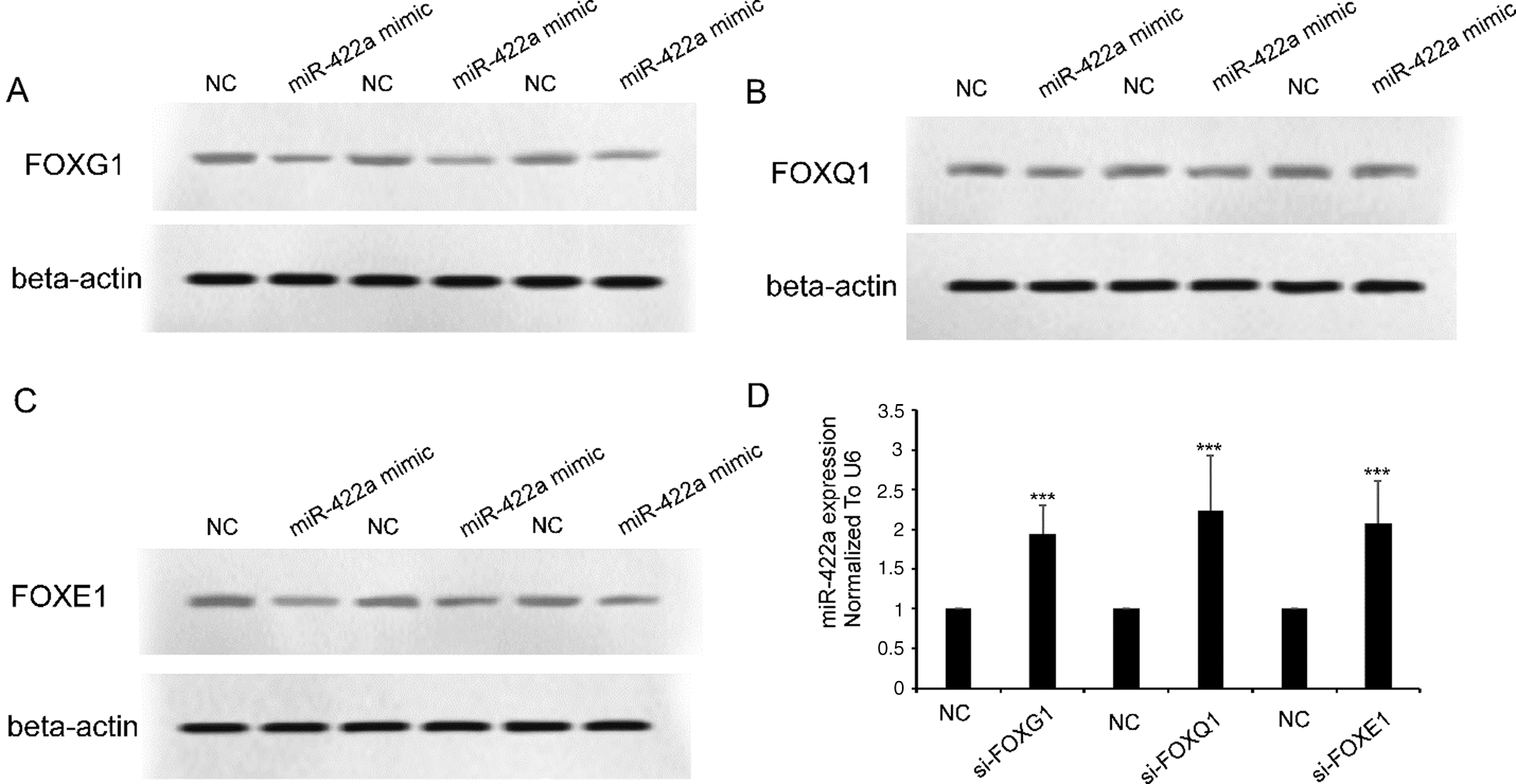
Figure 3: miR-422a and FOXG1/Q1/E1 antagonized against each other in HepG2-hbx cells. A–C) Western blot results showed that miR-422a transfection suppressed FOXG1/Q1/E1 expression in HepG2-hbx cells. D) miR-422a expression was rescued by FOXG1/Q1/E1-specific siRNA transfection in HepG2-hbx cells
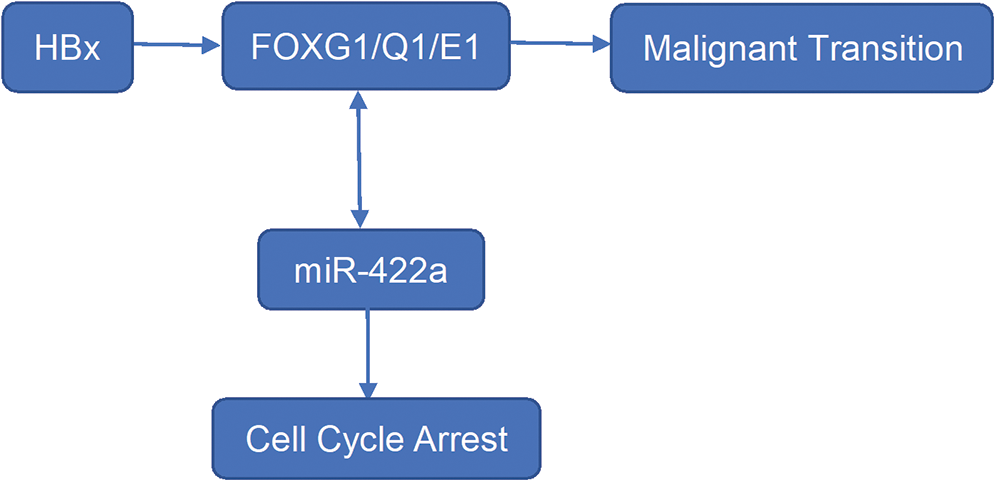
Figure 4: Preliminary HBx-FOXG1/Q1/E1-miR-422a pathway
3.4 miR-422a Hampered the Proliferation of HepG2-hbx Cells by Arresting Cells in G1 Phase
To clearly establish the significance of the FOXG1/Q1/E1-mediated miR-422a downregulation in HCC, we sought to determine the consequence of restoring miR-422a in HepG2-hbx cells in vitro. CCK-8 analysis showed that, compared with NC-transfected group, ectopically expressed miR-422a significantly hampered HepG2-hbx cells growth at 48 h and 72 h after transfection (Fig. 5A). Cell cycle assay confirmed that miR-422a arrested HepG2-hbx cells in the G1 phase (Fig. 5B).
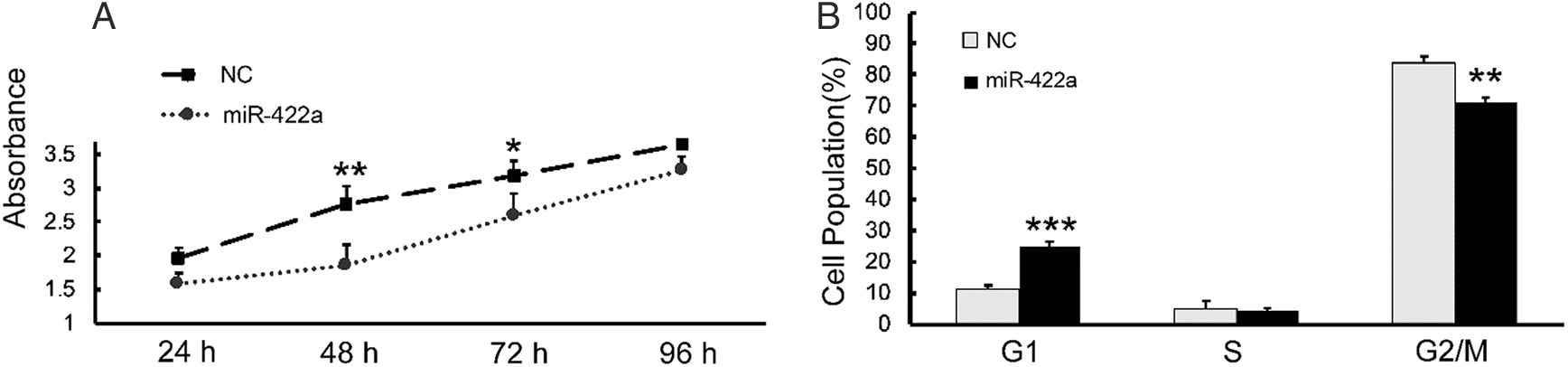
Figure 5: miR-422a hampered the proliferation of HepG2-hbx cells by arresting cells in G1 phase. A) The ectopic expression of miR-422a significantly hampered the proliferation of HepG2-hbx cells at 48 and 72 h. B) Cell cycle analysis indicated that the HepG2-hbx cells were arrested at G1 phase by miR-422a mimic
HBx participates in the initiation and progression of HCC by deregulating coding genes of host hepatocytes. Increasing evidence confirmed that HBx can also regulate the non-coding genes including miRNA, lncRNA, and circRNA. We previously confirmed that HBx induced extensive miRNA downregulation in HepG2 cells including miR-422a with a microarray method [14], which contributed to carcinogenesis by re-activating multiple oncogenes including MAPK1, PIK3CA, CD73, IGF1/IGF1R, and FOXG1/Q1/E1 [4–9]. FOXG1/Q1/E1 can also transcriptionally suppress miR-422a expression in HCC [4]. miR-422a is also the target of long non-coding RNAs, such as OIP5-AS1, NT5E, DUXAP8, LINC00313, D63785 LINC01133, and LINC00858 [3,19–25]. However, the mechanisms underlying miR-422a repression by HBx in HepG2 cells were not clearly established [14,17]. We confirmed the miR-422a downregulation in HepG2-hbx cells with qRT-PCR and found a tentative HBx-FOXG1/Q1/E1-miR-422a pathway in HepG2 cells in vitro.
FOX proteins are a superfamily of evolutionarily conserved transcriptional regulators, a loss or gain of FOX function can alter cell fate and promote tumorigenesis as well as cancer progression [26]. Several key members of the FOXA, FOXC, FOXM, FOXO, and FOXP subfamilies are heavily implicated in cancer, driving initiation, maintenance, progression, and drug resistance [27]. FOXM1, FOXC1, and FOXG1/Q1/E1 are involved in the initiation and progression of HCC [4,28–32]. For the first time, we found that HBx transfection activated FOXG1, Q1, and E1 expression in HepG2-hbx cells. FOXG1/Q1/E1 and miR-422a antagonize each other, striking a balance under normal conditions. Imbalance between FOXG1/Q1/E1/miR-422a can promote hepatocarcinogenesis [4]. Recently, we also identified a similar mechanism that HBx downregulated let-7 via activation c-Myc-Lin28B pathway [18]. Our results again show the significance of the HBx-TFs-miRNA mechanism in transformation of HCC cells in vitro.
Our results elucidated the critical roles played by FOXG1/Q1/E1 in HBx-induced miR-422a downregulation. Targeting miR-422a/FOXG1/Q1/E1 loop might be a prospective treatment for HBV+ HCC patients.
Acknowledgement: We would like to thank Prof. Jie Wang for revising this manuscript.
Data Availability: The data are available upon special request to the corresponding authors.
Author Contributions: Xiaofan Deng, Yamei Yang, and Xianfeng Gan performed the experiments and constructed the figures. Gang Wu designed the experiments and composed the manuscript.
Funding Statement: This work was supported by grants from the National Natural Science Foundation of China (81302161), the Key Program of Department of Science and Technology of Sichuan Province (2020YJ0450), and the Health and Family Planning Commission of Sichuan Province (150215).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Slack, F. J., Chinnaiyan, A. M. (2019). The role of non-coding RNAs in Oncology. Cell, 179, 1033–1055. [Google Scholar]
2. Wong, C. M., Tsang, F. H., Ng, I. O. (2018). Non-coding RNAs in hepatocellular carcinoma: molecular functions and pathological implications. Nature Reviews Gastroenterology & Hepatology, 15, 137–151. [Google Scholar]
3. Wei, F. F., Yang, L., Jiang, D. D., Pan, M., Tang, G. Y. et al. (2020). Long noncoding RNA DUXAP8 contributes to the progression of hepatocellular carcinoma via regulating miR-422a/PDK2 axis. Cancer Medicine, 9, 2480–2490. [Google Scholar]
4. Zhang, J., Yang, Y., Yang, T., Yuan, S. X., Wang, R. Y. et al. (2015). Double-negative feedback loop between microRNA-422a and forkhead box (FOX)G1/Q1/E1 regulates hepatocellular carcinoma tumor growth and metastasis. Hepatology, 61, 561–573. [Google Scholar]
5. Wei, W. T., Nian, X. X., Wang, S. Y., Jiao, H. L., Wang, Y. X. et al. (2017). miR-422a inhibits cell proliferation in colorectal cancer by targeting AKT1 and MAPK1. Cancer Cell International, 17, 91. [Google Scholar]
6. Liang, H. Q., Wang, R. J., Jin, Y., Li, J. W., Zhang, S. (2016). MiR-422a acts as a tumor suppressor in glioblastoma by targeting PIK3CA. American Journal of Cancer Research, 6, 1695–1707. [Google Scholar]
7. Wang, J. Y., Yang, H. O., Si, Y. R., Hu, D. Z., Yu, Y. et al. (2017). Iodine promotes tumorigenesis of thyroid cancer by suppressing Mir-422a and up-regulating MAPK1. Cellular Physiology and Biochemistry, 43, 1325–1336. [Google Scholar]
8. Wang, H. Y., Tang, C. Y., Na, M., Ma, W., Jiang, Z. F. et al. (2017). miR-422a inhibits glioma proliferation and invasion by targeting IGF1 and IGF1R. Oncology Research, 25, 187–194. [Google Scholar]
9. Bonnin, N., Armandy, E., Carras, J., Ferrandon, S., Battiston-Montagne, P. et al. (2016). MiR-422a promotes loco-regional recurrence by targeting NT5E/CD73 in head and neck squamous cell carcinoma. Oncotarget, 7, 44023–44038. [Google Scholar]
10. Hermeking, H. (2012). MicroRNAs in the p53 network: micromanagement of tumour suppression. Nature Reviews Cancer, 12, 613–626. [Google Scholar]
11. Buendia, M. A., Bourre, L., Cairo, S. (2012). Myc target miRs and liver cancer: small molecules to get Myc sick. Gastroenterology, 142, 214–218. [Google Scholar]
12. Chaturvedi, V. K., Singh, A., Dubey, S. K., Hetta, H. F., John, J. et al. (2019). Molecular mechanistic insight of hepatitis B virus mediated hepatocellular carcinoma. Microbial Pathogenesis, 128, 184–194. [Google Scholar]
13. Feitelson, M. A., Lee, J. (2007). Hepatitis B virus integration, fragile sites, and hepatocarcinogenesis. Cancer Letters, 252, 157–170. [Google Scholar]
14. Wu, G., Yu, F. Y., Xiao, Z. Y., Xu, K., Xu, J. Y. et al. (2011). Hepatitis B virus X protein downregulates expression of the miR-16 family in malignant hepatocytes in vitro. British Journal of Cancer, 105, 146–153. [Google Scholar]
15. Wang, Y. L., Jiang, L., Ji, X., Yang, B., Zhang, Y. et al. (2013). Hepatitis B viral RNA directly mediates down-regulation of the tumor suppressor microRNA miR-15a/miR-16-1 in hepatocytes. Journal of Biological Chemistry, 288, 18484–18493. [Google Scholar]
16. Salerno, D., Chiodo, L., Alfano, V., Floriot, O., Cottone, G. et al. (2020). Hepatitis B protein HBx binds the DLEU2 lncRNA to sustain cccDNA and host cancer-related gene transcription. Gut, 69, 2016–2024. [Google Scholar]
17. Liu, Y., Zhao, J. J., Wang, C. M., Li, M. Y., Han, P. et al. (2009). Altered expression profiles of microRNAs in a stable hepatitis B virus-expressing cell line. Chinese Medical Journal, 122, 10–14. [Google Scholar]
18. Wu, G., Huang, P. B., Ju, X. M., Li, Z. X., Wang, Y. Y. (2015). Lin28B over-expression mediates the repression of let-7 by hepatitis B virus X protein in hepatoma cells. International Journal of Clinical and Experimental Medicine, 8, 15108–15116. [Google Scholar]
19. Xie, R. J., Liu, L. F., Lu, X. Z., Hu, Y. (2020). LncRNA OIP5-AS1 facilitates gastric cancer cell growth by targeting the miR-422a/ANO1 axis. Acta Biochimica et Biophysica Sinica, 52, 430–438. [Google Scholar]
20. Dong, L. Y., Zheng, J. N., Gao, Y., Zhou, X. T., Song, W. Z. et al. (2020). The circular RNA NT5E promotes non-small cell lung cancer cell growth via sponging microRNA-134. Aging (Albany NY), 12, 3936–3949. [Google Scholar]
21. Zhu, S. P., Wang, J. Y., Wang, X. G., Zhao, J. P. (2017). Long intergenic non-protein coding RNA 00858 functions as a competing endogenous RNA for miR-422a to facilitate the cell growth in non-small cell lung cancer. Aging (Albany NY), 9, 475–486. [Google Scholar]
22. Wu, Y., Zhi, L. R., Zhao, Y., Yang, L. L., Cai, F. M. (2020). Knockdown of circular RNA UBAP2 inhibits the malignant behaviours of esophageal squamous cell carcinoma by microRNA-422a/Rab10 axis. Clinical and Experimental Pharmacology and Physiology, 47, 1283–1290. [Google Scholar]
23. Zhou, Z. X., Lin, Z. J., He, Y. Q., Pang, X., Wang, Y. et al. (2018). The long noncoding RNA D63785 regulates chemotherapy sensitivity in human gastric cancer by targeting miR-422a. Molecular Therapy-Nucleic Acids, 12, 405–419. [Google Scholar]
24. Zeng, H. F., Qiu, H. Y., Feng, F. B. (2018). Long noncoding RNA LINC01133 functions as an miR-422a sponge to aggravate the tumorigenesis of human Osteosarcoma. Oncology Research, 26, 335–343. [Google Scholar]
25. Yan, D. G., Liu, N., Chao, M., Tu, Y. Y., Liu, W. S. (2019). SP1-induced upregulation of long noncoding RNA LINC00313 contributes to papillary thyroid cancer progression via the miR-422a. European Review for Medical and Pharmacological Sciences, 23, 1134–1144. [Google Scholar]
26. Myatt, S. S., Lam, E. W. (2007). The emerging roles of forkhead box (Fox) proteins in cancer. Nature Reviews Cancer, 7, 847–859. [Google Scholar]
27. Lam, E. W., Brosens, J. J., Gomes, A. R., Koo, C. Y. (2013). Forkhead box proteins: tuning forks for transcriptional harmony. Nature Reviews Cancer, 13, 482–495. [Google Scholar]
28. Huang, W. J., Chen, Z. Q., Zhang, L., Tian, D. A., Wang, D. W. et al. (2015). Interleukin-8 induces expression of FOXC1 to promote transactivation of CXCR1 and CCL2 in hepatocellular carcinoma cell lines and formation of metastases in mice. Gastroenterology, 149, 1053–1067. [Google Scholar]
29. Calvisi, D. F., Pinna, F., Ladu, S., Pellegrino, R., Simile, M. M. et al. (2009). Forkhead box M1B is a determinant of rat susceptibility to hepatocarcinogenesis and sustains ERK activity in human HCC. Gut, 58, 679–687. [Google Scholar]
30. Huang, W. J., Chen, Z. Q., Shang, X., Tian, D. A., Wang, D. W. et al. (2015). Sox12, a direct target of FoxQ1, promotes hepatocellular carcinoma metastasis through up-regulating Twist1 and FGFBP1. Hepatology, 61, 1920–1933. [Google Scholar]
31. Li, Y., Lu, L. Q., Tu, J., Zhang, J., Xiong, T. et al. (2020). Reciprocal regulation between forkhead box M1/NF-kB and Methionine Adenosyltransferase 1A drives liver cancer. Hepatology, 72, 1682–1700. [Google Scholar]
32. Xia, L. M., Huang, W. J., Tian, D. A., Zhang, L., Qi, X. S. et al. (2014). Forkhead box Q1 promotes hepatocellular carcinoma metastasis by transactivating ZEB2 and VersicanV1 expression. Hepatology, 59, 958–973. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |