
 | Oncologie |  |
DOI: 10.32604/Oncologie.2021.016155
ARTICLE
Alteration of Ornithine Metabolic Pathway in Colon Cancer and Multivariate Data Modelling for Cancer Diagnosis
1The First Affiliated Hospital of Jinzhou Medical University, Jinzhou, 121000, China
2General Hospital of Benxi Iron and Steel, Liaoning Healthcare Group, Benxi, 117000, China
3Benxi Central Hospital, Benxi, 117000, China
4Key Laboratory of Liaoning Tumor Clinical Metabolomics, Jinzhou, 121000, China
5Dalian Institute of Chemical Physics, Chinese Academy of Sciences, Dalian, 116023, China
*Corresponding Author: Zhitu Zhu. Email: zhuzhitu0416@163.com
#These authors contributed equally to this work
Received: 11 February 2021; Accepted: 19 April 2021
Abstract: It is noteworthy that colon cancer is the fourth place in new cases and the fifth in mortalities according to global cancer statistics 2018. Tumorigenesis displays specific correlation with metabolic alterations. A variety of metabolites, including ornithine (Orn), are related to colon cancer according to sources of disease metabolic information retrieval in human metabolome database. The metabolic regulation of Orn pathway is a key link in the survival of cancer cells. In this study, the plasma Orn levels in colon cancer patients and healthy participants were measured by liquid chromatography tandem mass spectrometry, and the metabolic disturbances of Orn in colon cancer were identified. Based on exploring the pathway structure of Orn metabolism via MetaboAnalyst and Kyoto Encyclopedia of Genes and Genomes database, we found that the upstream and downstream metabolites included arginine (Arg) and N1, N12-diacetylspermine (DiAcSpm). We observed that the Arg-Orn-DiAcSpm metabolic pathway was up-regulated in colon cancer through pathway analysis. We used multivariate data modelling to build a regression diagnosis model based on the three metabolites for colon cancer, and the diagnosis capability of this model was analyzed via receiver operating characteristic curve. This study provides a theoretical basis for further feature description of tumor metabolic pathway, which may lead to discover new therapeutic targets and drugs against colon cancer. Multivariate data modelling is expected to be a novel technology for developing noninvasive screening tool of cancer.
Keywords: Metabolic pathway; colon cancer; ornithine; multivariate data modelling
According to the statistics, more than 1.8 million new colorectal cancer cases were diagnosed worldwide in 2018, and colorectal cancer is the third leading cause of cancer deaths in females and the fourth in males. Furthermore, the colon cancer is the fourth most common cancer in new cases [1].
Metabolic alterations are the key links in occurrence and development of tumors, which are driven by the genetically relevant mutations that promote oncogenesis [2]. Cell metabolic change is the idiosyncrasy of carcinoma, which is helpful for malignant transformation and tumor maintenance [3]. Metabonomics technique can be used for global profiling of metabolites in human body, and provides specific metabolic profiles to guide in-depth understanding of metabolic reaction endpoint information of diseases. Metabolite quantitative analysis technology has great potential in drug development and disease diagnosis [4]. Plasma is a collection area of metabolites in the human body. The changes of plasma metabolites in cancer patients may reflect the metabolic regulations in the process of cell carcinogenesis [5]. Recent studies show the significant differences in serum glycolysis, protein, and lipid metabolism between colorectal cancer patients and healthy participants [6]. By identifying the key metabolic pathways, the related mechanisms of tumorigenic process could be further studied.
Human Metabolome Database (HMDB) is a Web-based metabolomic database. It is the most complete and comprehensive database to collect human metabolites and metabolism data in the world. Through data retrieval of the biomedical information describing metabolites and diseases of human body in this database, we observed that the levels of various metabolites including ornithine (Orn) changed significantly in colorectal cancer.
Orn that is a non-proteinogenic amino acid is involved in many biosynthesis pathways in the human body. Some researches have shown that Orn metabolism is remarkably altered in many cancers. Orn plays a special role in tumor growth, invasion and metastasis [7,8]. Tan et al. [9] has found that Orn is down-regulated in colorectal cancer via gas chromatography time-of-flight mass spectrometry. Uchiyama et al. [10] has shown that Orn concentration is lower in serum of patients with colorectal cancer through capillary electrophoresis-time-of-flight mass spectrometry. However, the interesting question of why the metabolic perturbation of Orn appears in colorectal cancer has not yet been answered. In addition, the metabolic alteration of Orn pathway plays specific roles in survival of human esophageal chemotherapy-resistant cancer stem cells [11]. But there are limited data on the regulations of Orn metabolic pathway in colon cancer.
Liquid chromatography tandem mass spectrometry (LC-MS/MS) is a superior high-resolution biomolecular analysis technology, which has the advantages of fast scanning speed, strong selectivity and high sensitivity. It can realize rapid, micro and high-throughput analysis of metabolites. This technology has been widely applied to analyze the levels of metabolites in blood and urine samples of cancer patients and in tumor tissues [12–14].
Recent studies have demonstrated that many low-molecular metabolites may be markers for displaying biological mechanisms during the tumorigenic process and for diagnosing cancer diseases [15–17]. However, there is a lack of appropriate analytical methods to further evaluate the application potential of these molecular markers. Multivariate correlation analysis of metabolites would be helpful for clarifying their values in the pathogenesis, diagnosis and treatment of cancer. Accordingly, it is very desirable to develop an efficient mathematical model of multivariate correlation analysis for cancer diagnosis using low-molecular metabolites.
In this study, we used LC-MS/MS technology to verify the plasma Orn metabolic perturbation in patients with colon cancer. The metabolic pathway of Orn in human body was identified by MetaboAnalyst and Kyoto Encyclopedia of Genes and Genomes (KEGG) database, and the upstream and downstream relevant metabolites of Orn metabolism were analyzed to reveal the mechanisms of the alterations in Orn level of colon cancer. We developed a mathematical model based on the biomolecules in Orn metabolic pathway for diagnosing colon cancer through multivariate data modelling. Our study showed that the metabolic pathway of Orn participated in human colonic carcinogenesis and the combined metabolic profile of biomolecules in this pathway might be a novel diagnosis tool for early screening colon cancer.
2.1 Clinical Sample Collection
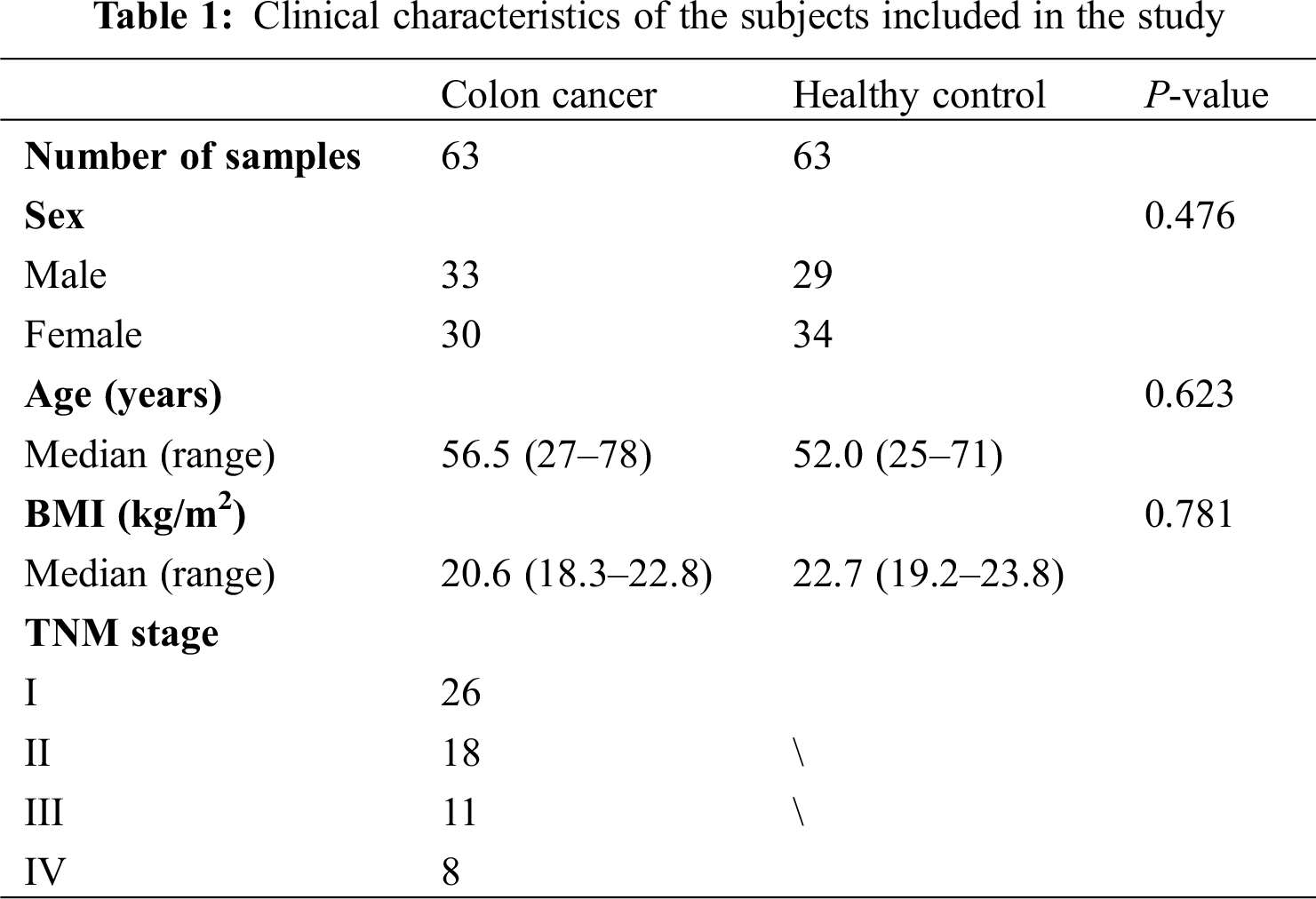
This study was approved by the Ethics Committee of the First Affiliated Hospital of Jinzhou Medical University and conducted in accordance with the principles of Helsinki Declaration. Each patient or healthy person involved in this study signed an informed consent. We collected blood and urine samples of 63 patients with colon cancer and 63 healthy participates from the First Affiliated Hospital of Jinzhou Medical University. There were no age, gender and body mass index (BMI) differences between the two groups. The inclusion criteria of all the subjects were as follows: blood samples and urine samples provided by all subjects were taken before surgery, chemotherapy, radiotherapy or other relevant treatments; the diagnosis of cancer patients was confirmed by histopathology, and cancer stage was classified according to the Tumor-Node-Metastasis (TNM) classification of American Joint Committee on Cancer (AJCC); tests of the liver and kidney functions of subjects showed normal findings; weight gain or loss of all subjects with normal BMI was less than 10%. All subjects had no hyperthyroidism, diabetes, hyperlipidemia and tuberculosis. The detailed baseline characteristics of subjects were shown in Tab. 1.
Specimens from fasting blood samples of cancer patients and healthy participants were collected in vacuum blood collection tubes and immediately stored at 4°C. Subsequently, the specimens were produced into dried blood spot (DBS) samples on filter papers and kept at –80°C until measurement. The urine samples of all the subjects were stored at –20°C until the sample preparation.
A disc with a diameter of 3 mm was punched out from each DBS paper at room temperature. Adding 100 μL working solution into each well of 96-well plates containing DBS discs, shaking for 20 min and centrifuging at 1500 × g for 2 min were used to remove interferences from samples. After dried in nitrogen airflow, the filtrate was collected using new 96-well plates containing 60 μL 1-butanol/acetyl chloride mixture and 100 μL mobile phase (80% acetonitrile aqueous solution) for LC-MS/MS detection.
LC-MS/MS analysis was used to detect the concentrations of Orn and arginine (Arg) in plasma, which was conducted with AB Sciex 4000 QTrap (AB Sciex, Framingham, MA, USA) and LC-20A (Shimadzu, Kyoto, Japan) as detectors.
Urine samples adjusted to room temperature were centrifuged for 5 min at 3000 rpm, and the supernatant was diluted with distilled deionized water in the ratio of 1:4. The collected samples were incubated with anti-diacetylspermine antibody in 96-well plates at room temperature for 1 h. After incubation, the wells were washed with wash solution. The incubation process was repeated with horseradish peroxidase (HRP)-conjugated anti-rabbit antibody, then wells were washed again. Subsequently, incubation with ortho-phenylenediamine was 10 min. The 1 mol/l sulfuric acid was added to stop the reaction, and the absorbance was measured at a wavelength of 490 nm. Correction of N1, N12-diacetylspermine (DiAcSpm) concentration with urine creatinine concentration (mg/dl) was used to be the actual measurement.
Concentration of urinary DiAcSpm was measured by the enzyme-linked immunosorbent assay (ELISA) in 7600-020 automated biochemical analyser (HITACHI, Tokyo, Japan).
Student’s t-test was performed to analyze the metabolic differences of Orn, Arg and DiAcSpm between colon cancer and healthy control groups. Metabolic profile discrimination of the three metabolites between the two groups was presented via partial least squared discriminant analysis (PLS-DA) of multivariate correlation analysis (SIMCA-P software v13.0, Umetrics, Umea, Sweden). The diagnosis model of colon cancer was established by logistic regression analysis, and its diagnostic ability was confirmed via the receiver operating characteristic (ROC) curve.
2.7 Metabolism Data Retrieval and Pathway Analysis
HMDB (http://hmdb.ca/) was performed for online retrieval of metabolic information related to colorectal cancer. The metabolic pathway of Orn was identified by database sources of MetaboAnalyst (http://www.metaboanalyst.ca/) and KEGG (http://www.genome.jp/kegg/).
3.1 Metabolic Information Related to Colon Cancer in HMDB
Data sets of human colorectal cancer metabolism in HMDB were downloaded and analyzed. All the differential expression metabolites in blood samples from adult patients with colorectal cancer were listed in Fig. 1, one of which was Orn.
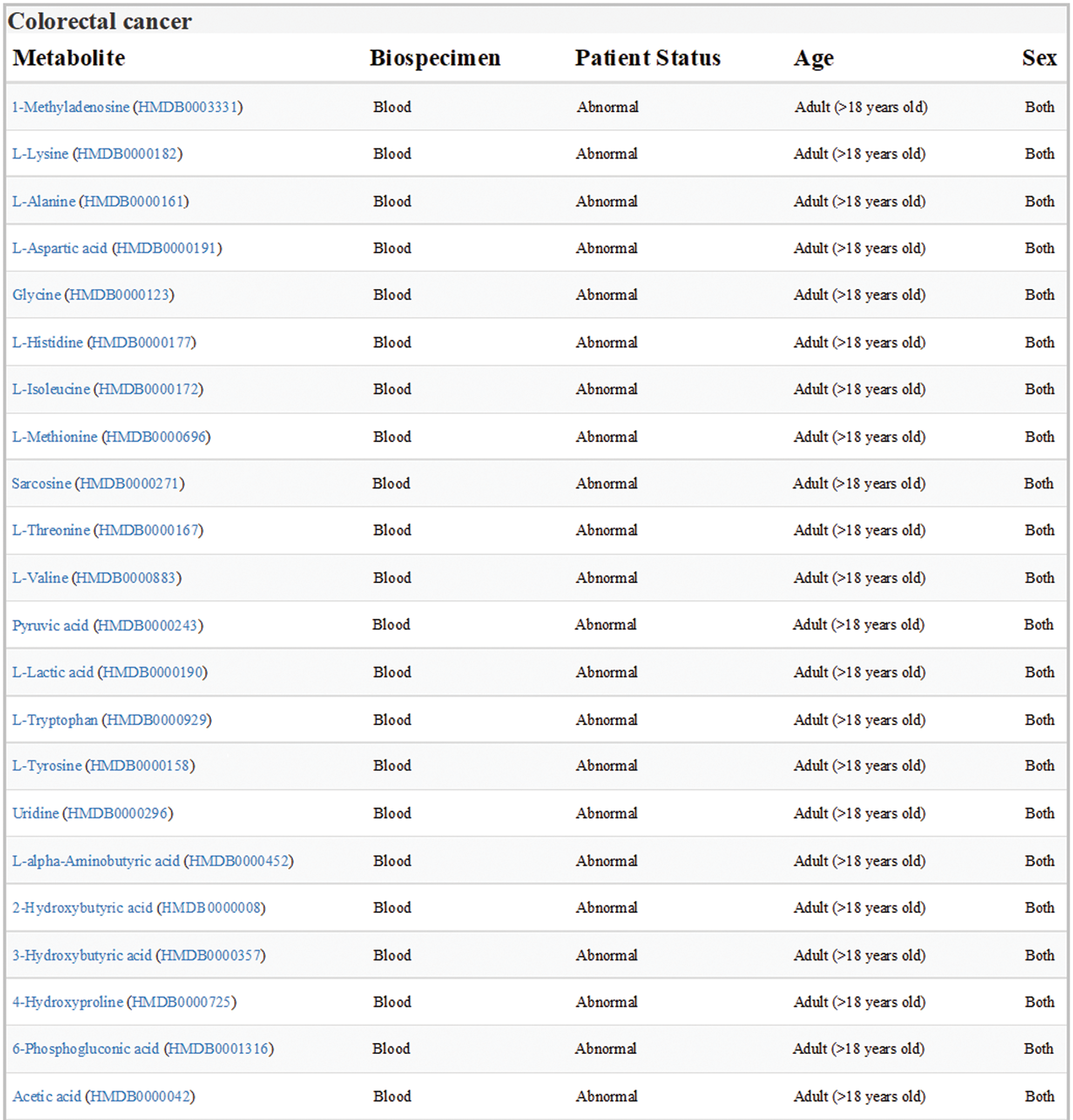
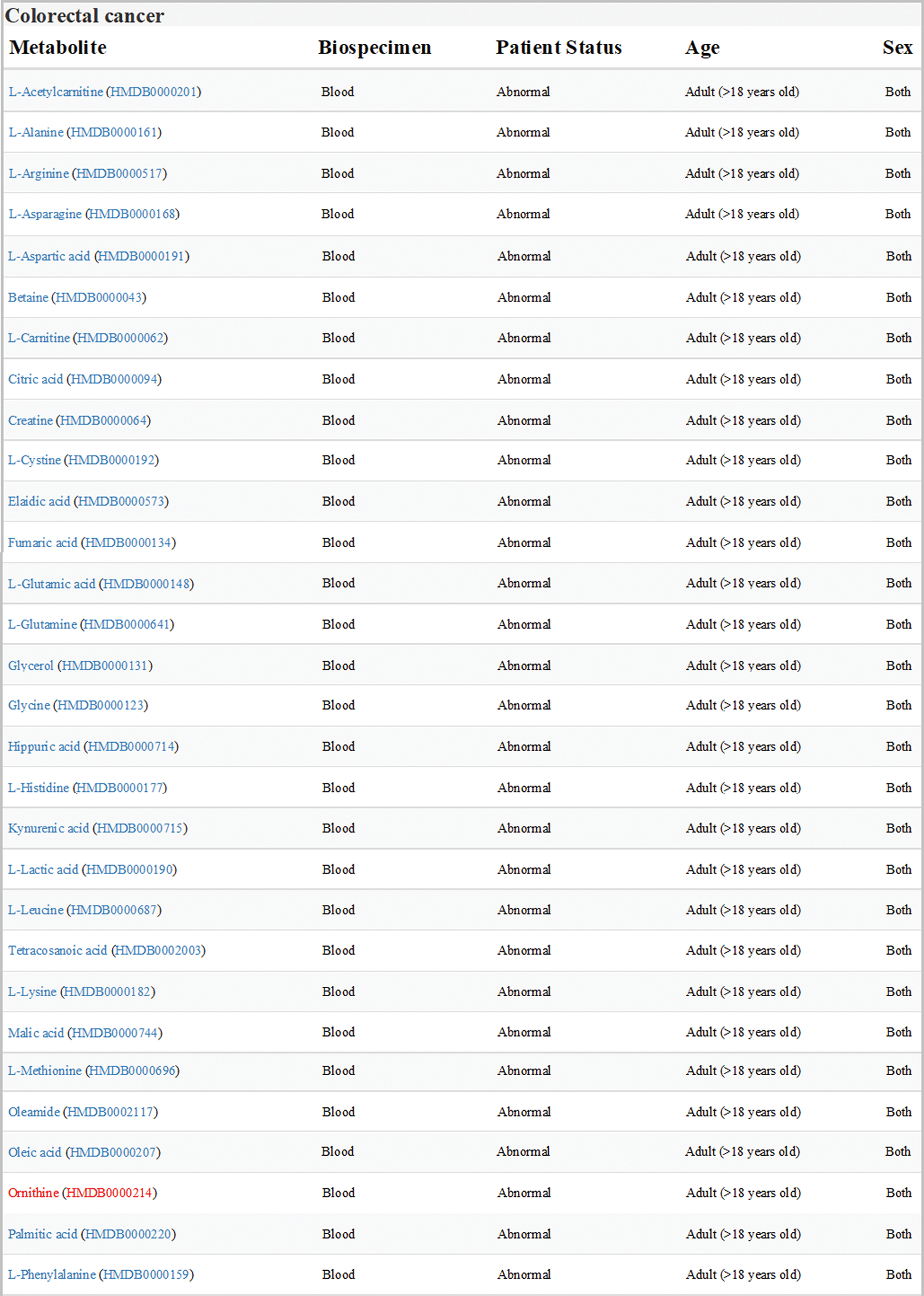
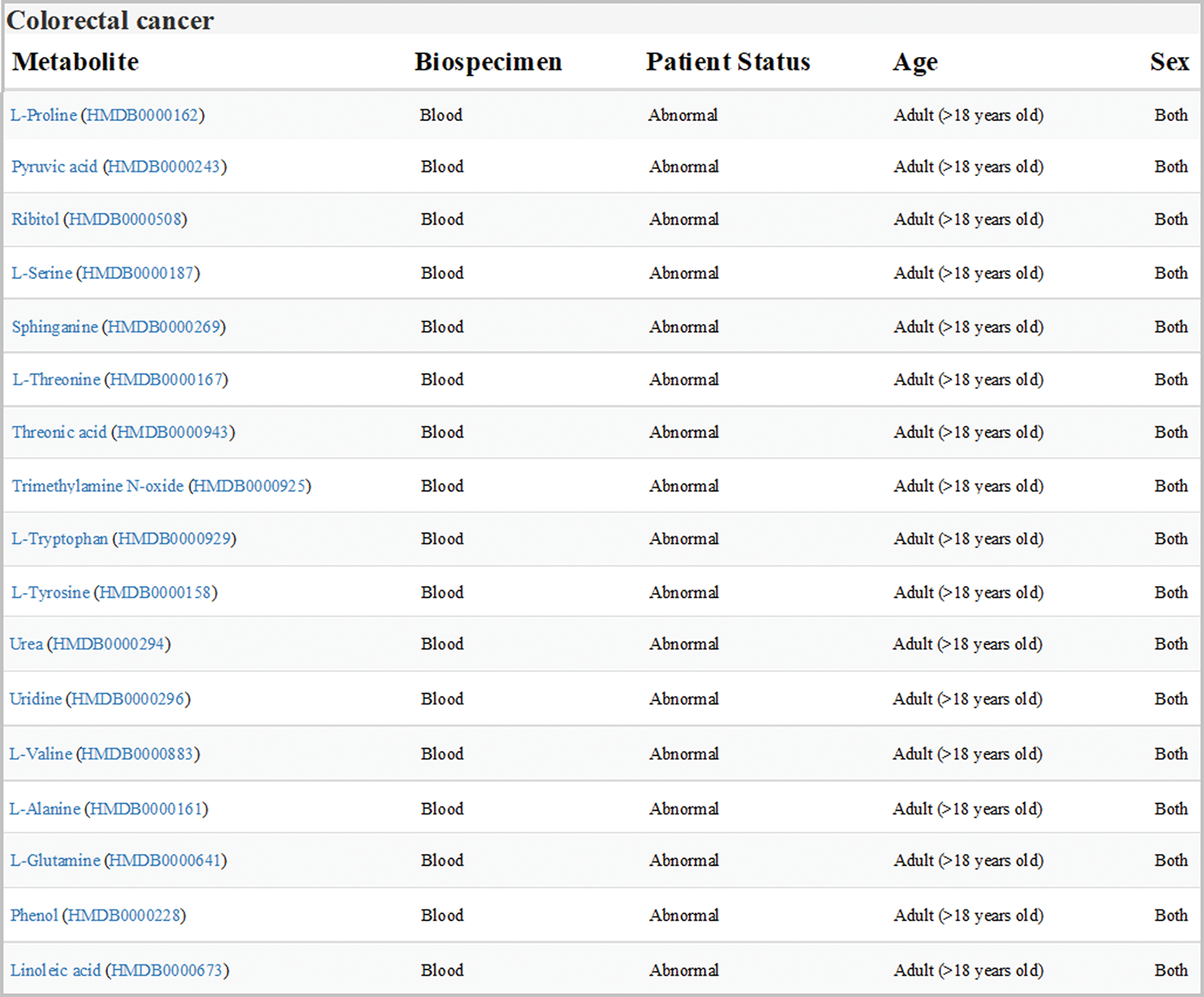
Figure 1: Expression of metabolites related to colon cancer in HMDB (version 4.0; hmdb.ca). Data sets showed all the differential expression metabolites in blood samples from adult patients with colorectal cancer. Orn was labeled in red
3.2 Orn Metabolic Alterations in Colon Cancer
LC-MS/MS analysis of blood samples showed that the metabolic level of Orn significantly decreased in colon cancer patients as compared to the healthy controls (Fig. 2A). The result of this study was similar to the previous reports.
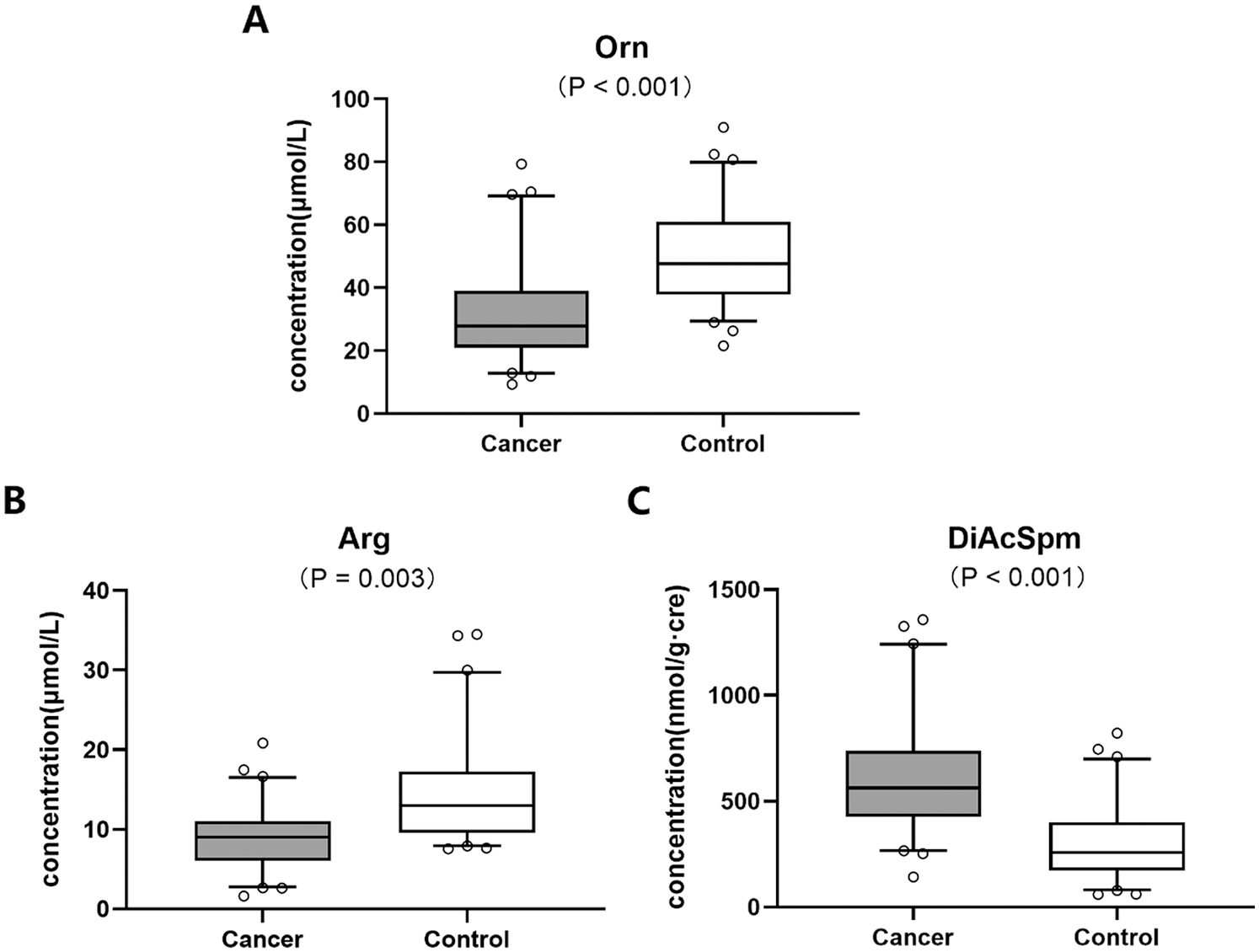
Figure 2: Metabolic levels of biomolecules including (A) Orn, (B) Arg and (C) DiAcSpm in Orn pathway between colon cancer and healthy control groups
We further analyzed the correlation between Orn and clinicopathological factors in colon cancer group. Tab. 2 presented that Orn was remarkably associated with tumor invasion depth and its level decreased in T3-4 stage cases compared with the T1-2 stage (29.46 ± 16.07 vs. 36.12 ± 20.09, P = 0.025). There was a negative correlation between Orn and lymphatic invasion status. Lymphatic invasion-positive group showed significantly lower level of Orn than the negative group (28.49 ± 12.63 vs. 35.54 ± 17.85, P = 0.018). Analysis of the association between Orn and TNM stage demonstrated that its level in Stage IV cases was lower than that in Stage I (27.36 ± 14.27 vs. 37.24 ± 20.61, P < 0.001).
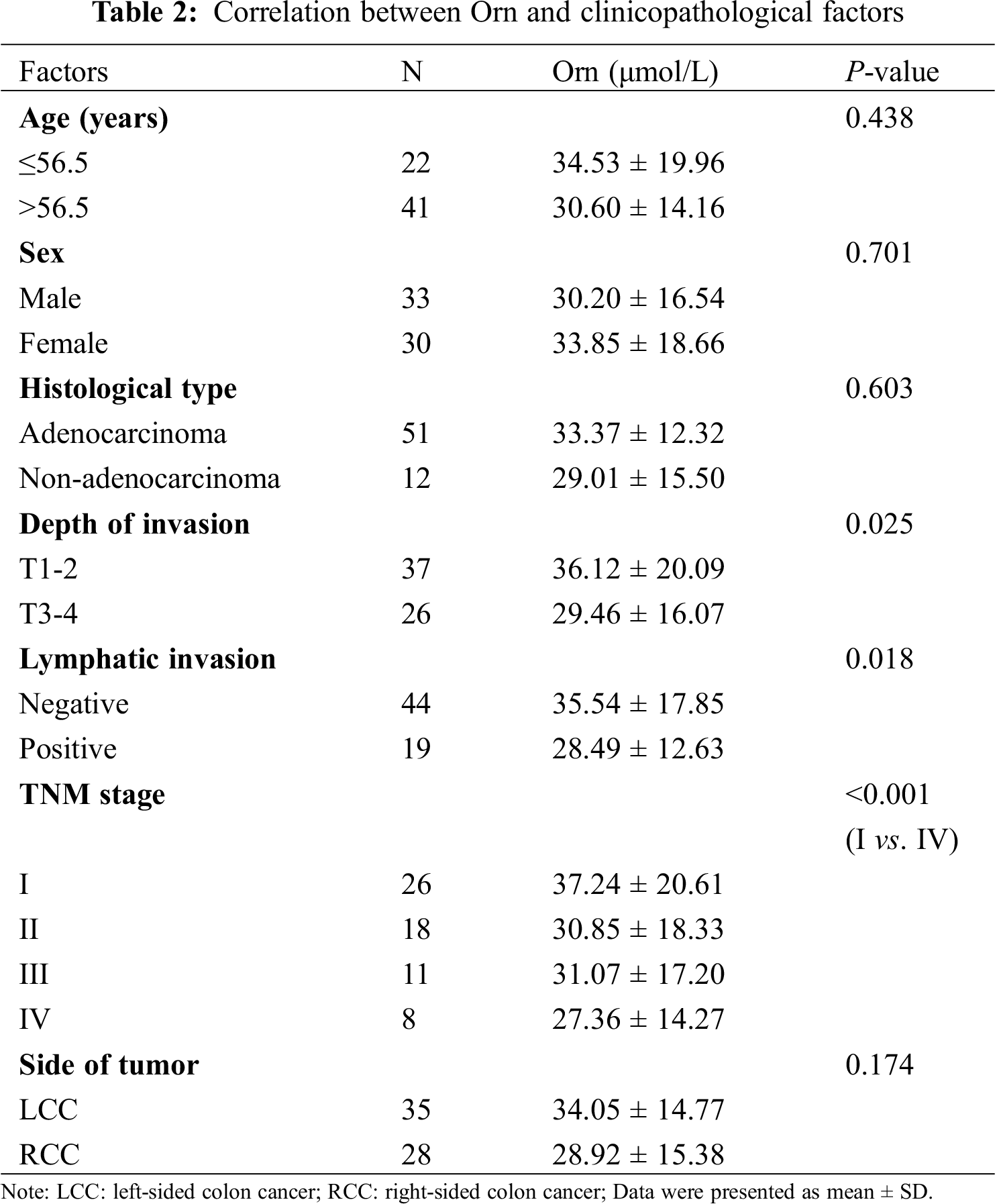
There have been evidences that left-sided colon cancer (LCC) represents differences in biology, pathology and epidemiology as compared to right-sided colon cancer (RCC) [18]. On the basis of this finding, our study also examined the metabolic levels of Orn in the tumor location of colon cancer, but no significant difference between RCC and LCC groups was evident (28.92 ± 15.38 vs. 34.05 ± 14.77, P = 0.174). Moreover, none of the other clinicopathological factors including age (P = 0.438), sex (P = 0.701), and histological type (P = 0.603) were significantly correlated to Orn.
3.3 Determination of Orn Metabolic Pathway
The functional enrichment analysis of Orn was performed by MetaboAnalyst. Fig. 3A presented the affected pathways of Orn based on all the previous studies data. Two of the most important Orn metabolic pathways were urea cycle and polyamine metabolism. A network of Orn metabolism was drawn according to the metabolic pathway information provided by KEGG database (Fig. 3B). The pathways inside the network, which involved urea cycle and polyamine metabolism, joined the metabolites. We observed that the upstream metabolites of Orn metabolic pathway contained Arg and the downstream metabolites included DiAcSpm.
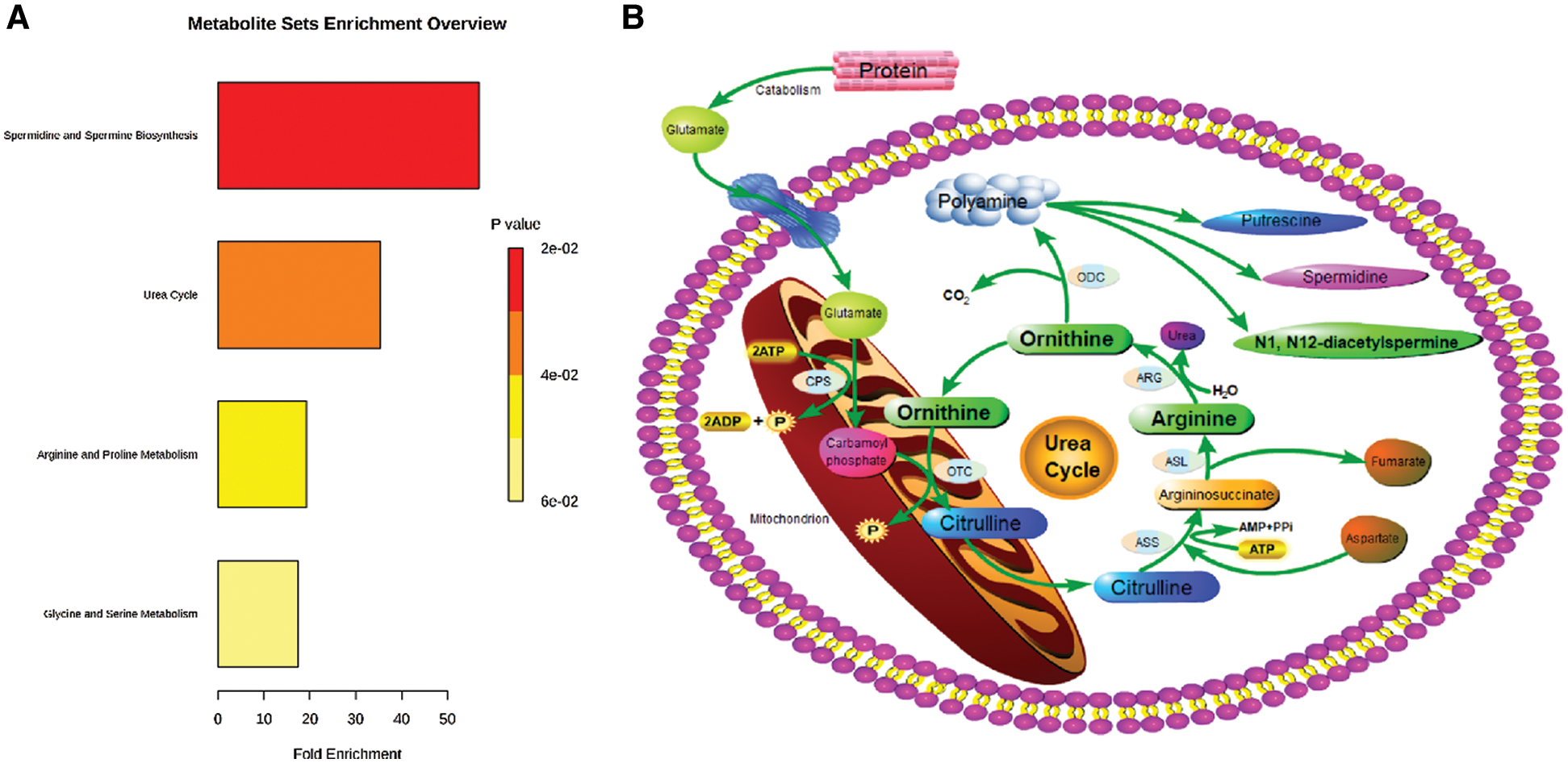
Figure 3: Affected metabolic pathways of Orn in human body. (A) Functional enrichment analysis of Orn in MetaboAnalyst (version 4.0; www.metaboanalyst.ca). (B) The network of Orn metabolic pathways. CPS: carbamoyl phosphate synthetase; OTC: ornithine transcarboxylase; ASS: arginine succinate synthetase; ASL: argininosuccinate lyase; ARG: arginase; ODC: ornithine decarboxylase
3.4 Analysis of Metabolites in Orn Metabolic Pathway
The results based on the analysis of Arg and DiAcSpm metabolisms exhibited that the concentration of plasma Arg decreased (Fig. 2B) while urine DiAcSpm increased (Fig. 2C) in colon cancer group compared with the controls.
We examined the correlation between the two metabolites and clinicopathological factors of the patients with colon cancer (Tab. 3). Lymphatic invasion-positive group showed significantly lower level of Arg than the negative group (7.71 ± 1.95 vs. 9.18 ± 2.97, P = 0.020). This study also proved that Arg level was lower in Stage IV than in Stage I cases (7.11 ± 1.78 vs. 9.32 ± 3.13, P = 0.006).
As shown in Tab. 3, DiAcSpm was positively associated with the lymphatic invasion status. DiAcSpm level in lymphatic invasion-positive group was significantly higher than that in the negative group (702.38 ± 269.47 vs. 557.00 ± 258.55, P = 0.007). Analysis of the association between DiAcSpm and TNM stage indicated that its level increased with the elevation of TNM stage giving values of 528.89 ± 256.43 (Stage I), 612.52 ± 270.38 (Stage II), 666.37 ± 303.57 (Stage III), and 730.61 ± 212.68 (Stage IV), respectively. There was a significant difference in the level of DiAcSpm between Stages I and IV cases (P < 0.001).
The other clinicopathological factors including age, sex, histological type, depth of invasion, and side of tumor did not significantly correlate with Arg and DiAcSpm.
3.5 Multivariate Analysis of Metabolites
The PLS-DA model was constructed based on the metabolic data of the three metabolites. Score scatter 3D plot presented a discrimination of the combined metabolic profile of Arg, Orn and DiAcSpm between colon cancer and control groups (Fig. 4A). The model displayed satisfactory modeling (R2Ycum = 0.837) and predictive (Q2cum = 0.751) abilities. Chance permutation test at 200 times (Fig. 4B) indicated that no over-fitting appeared in this model with y-axis intercept values of −0.0214 (R2) and −0.135 (Q2), respectively.
We used Arg, Orn and DiAcSpm to construct a regression diagnosis model for diagnosing colon cancer. The levels of these three metabolites showed significant differences between the two groups in logistic regression analysis giving values of P < 0.001 (Arg), P = 0.023 (Orn) and P < 0.001 (DiAcSpm), respectively. The regression equation was y = 0.044 × Orn + 0.408 × Arg − 0.009 × DiAcSpm − 2.013.
Efficiency of the diagnosis model based on logistic regression analysis was assessed via ROC curve. The area under the curve (AUC) was 0.880, and the model displayed sensitivity and specificity of 85.7% and 81.0%, respectively. The cut-off value was 0.667 (Fig. 4C).
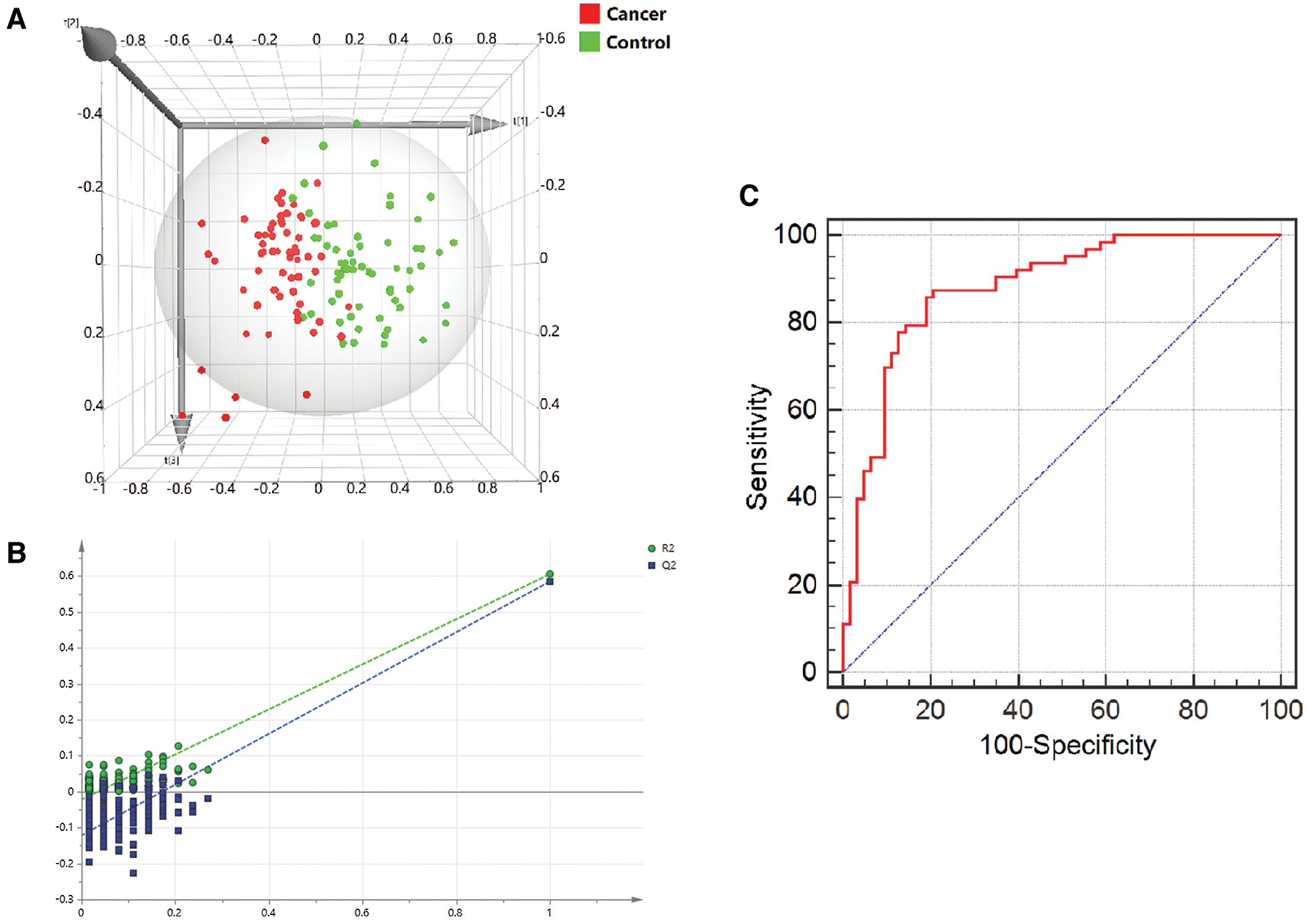
Figure 4: Multivariate correlation analysis of metabolites. (A) 3D score plot presented the discrepancy of a combined metabolic profile of Arg, Orn and DiAcSpm between the two groups. The x, y and z axes were the first 3 principal calculated components for the PLS-DA model. (B) A 200-time permutation test was used to validate the corresponding model. There was no over-fitting in the model. (C) ROC curve based on logistic regression analysis showed the efficiency of the diagnosis model
This study further verified that the concentration of Orn significantly decreased in plasma of patients with colon cancer compared with the healthy participants. Through the identification of correlation between Orn and clinicopathological factors in colon cancer group, we demonstrated that Orn was closely associated with the depth of tumor invasion, lymphatic invasion status and TNM stage. The results showed that the level of Orn in advanced colon cancer tended to be lower than that in early stage carcinoma, suggesting that Orn was negatively correlated with the tumor burden in colon cancer.
In order to further reveal the possible mechanisms of Orn metabolism changes in colon cancer, we used MetaboAnalyst and KEGG database to find the Orn metabolic pathway in human body, and observed the upstream and downstream metabolites including Arg and DiAcSpm. We analyzed the metabolic alterations of Arg and DiAcSpm in colon cancer. The results proved that Arg in plasma was down-regulated and DiAcSpm in urine was up-regulated in patients with colon cancer.
As the precursor of Orn synthesis, Arg is converted into Orn and urea through urea cycle. Arg can inhibit the growth of tumor cells by downregulating the expression of anti-apoptotic genes Survivin and Bcl-2 [19]. Arg depletion may selectively inhibit the motility and invasion of colon cancer cells [20]. In addition, Arg intake can improve the prognosis of colorectal cancer patients [21]. Arginine succinate synthetase 1 (ASS1) is the key enzyme in the progress of Arg biosynthesis. Recent studies have shown that the ability of tumor cells synthesizing Arg is reduced because of the ASS1 deficiency. The low concentration of Arg in cancer patients may contribute to the growth of tumors [22,23]. Therefore, these metabolic changes could lead to a decrease in the source of Orn synthesis in colon cancer patients.
Orn is converted into polyamines by ornithine decarboxylase (ODC), and polyamines mainly include putrescine, spermine and spermidine in human body. DiAcSpm with little individual variation is the derivative of spermine. DiAcSpm has strong correlation with the tumorigenesis, and its concentration is significantly higher in various tumors. DiAcSpm appears to accumulate in tissues at a particular stage during the development of cell carcinogenesis [24–26]. Moreover, activity of ODC which is a key enzyme in the biosynthesis of polyamines is elevated in colorectal cancer. Overexpression of ODC could cause enhanced proliferation to promote tumor formation [27]. Thus, it suggests that the ODC-DiAcSpm pathway is overexpressed in colon cancer patients and this change may enhance the Orn catabolism.
In this study, we further proved that the source of Orn synthesis was reduced and the catabolism was enhanced in patients with colon cancer. These metabolic regulations might explain the reason of a decrease in the level of Orn metabolism during the colonic carcinogenesis.
In the urea cycle, arginase catalyzes the synthesis of Orn from Arg. Ma et al observed that arginase was overexpressed in colon cancer, and this change was remarkably associated with Stages III–IV tumors [28]. In the polyamine metabolism, it has been demonstrated that ODC activity is elevated obviously in tumor tissue and the alteration is accompanied with elevation of TNM stage in colorectal cancer patients [29]. We examined the metabolic levels of Arg and DiAcSpm in different pathological stages of colon cancer patients. The results showed that compared with those in Stage I group, the Arg level decreased whereas DiAcSpm increased in Stage IV colon cancer. Thus, we could conclude that Arg-Orn-DiAcSpm pathway which might play important roles in the development and progression of carcinoma is up-regulated in colon cancer and this up-regulation is greater in advanced stage tumor.
The derived polyamine metabolites in Orn pathway play a certain role in the biosynthesis of cancer stem cells, and polyamines can promote the growth of cancer cells. The Orn metabolic pathway of cancer stem cells exposed to chemotherapy drugs changes significantly, which may produce a marked effect by inhibiting the activity of histone demethylase. This inhibition can regulate gene expression level and then affects cell survival to resist drug therapy [11]. Hence, the change of Orn metabolic pathway is a key link in the process of cancer cell proliferation.
Through multivariate data modelling, we investigated the characteristic of a combined metabolic profile of biomolecules in Orn pathway. The results of PLS-DA in multivariate statistical analysis presented significant differences in the metabolic profile of a combination of Arg, Orn and DiAcSpm between colon cancer and healthy control groups. We further used these three metabolites to construct a regression analysis model for diagnosing colon cancer. ROC curve analysis of the regression model showed its good diagnostic ability. As compared to a combination of carcinoembryonic antigen (CEA), carbohydrate antigen 19-9 (CA19-9) and alpha fetoprotein (AFP) for diagnosing colorectal cancer with sensitivity of 80.43%, 69.57% and 73.91%, respectively, our regression diagnostic model had preferable predictive capacity [30]. It suggests that the three metabolites’ quantitative analysis can successfully discriminate colon cancer patients and healthy participants. Arg, Orn and DiAcSpm may form an efficient diagnosis model of colon cancer.
In short, utilizing the metabolome database combined with clinical sample test, we verified the significant alterations in the level of Orn metabolism in patients with colon cancer. Through analysis of metabolic pathway, we observed the metabolic perturbations of Arg-Orn-DiAcSpm pathway and revealed the reason of the decrease in Orn level of colon cancer. The up-regulation of Orn metabolic pathway might be the key event in pathogenesis of colon cancer, which could provide theoretical support to discover new therapeutic targets and drugs against cancer. Based on the results from multivariate analysis, we found that the combined metabolic profile of Arg, Orn and DiAcSpm might be an excellent noninvasive screening tool of colon cancer. Multivariate data modelling is expected to offer new technical support for cancer diagnosis. The high standard deviation of the measurements in this study may be related to the limited sample size and individual differences. We will conduct prospective studies involving a large cohort of clinical samples from colon cancer patients and healthy controls to determine the validity of this diagnosis model. Further investigations should be completed to elucidate the oncological and biological significances of Orn metabolic pathway in cancer. It is necessary to research the effect of down-regulation of Orn metabolic pathway on cancer cells.
Funding Statement: This study was supported by the Education Science and Technology Innovation Foundation Key Project of Liaoning Provincial Department (XLYC1902026).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A. et al. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians, 68(6), 394–424. DOI 10.3322/caac.21492. [Google Scholar] [CrossRef]
2. Jones, R. G., Thompson, C. B. (2009). Tumor suppressors and cell metabolism: A recipe for cancer growth. Genes & Development, 23(5), 537–548. DOI 10.1101/gad.1756509. [Google Scholar] [CrossRef]
3. DeBerardinis, R. J., Thompson, C. B. (2012). Cellular metabolism and disease: What do metabolic outliers teach us? Cell, 148(6), 1132–1144. DOI 10.1016/j.cell.2012.02.032. [Google Scholar] [CrossRef]
4. Kosmides, A. K., Kamisoglu, K., Calvano, S. E., Corbett, S. A., Androulakis, I. P. (2013). Metabolomic fingerprinting: Challenges and opportunities. Critical Reviews in Biomedical Engineering, 41(3), 205–221. DOI 10.1615/CritRevBiomedEng.2013007736. [Google Scholar] [CrossRef]
5. Sreekumar, A., Poisson, L. M., Rajendiran, T. M., Khan, A. P., Cao, Q. et al. (2009). Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature, 457(7231), 910–914. DOI 10.1038/nature07762. [Google Scholar] [CrossRef]
6. Hashim, N., Ab-Rahim, S., Suddin, L. S., Saman, M., Mazlan, M. (2019). Global serum metabolomics profiling of colorectal cancer. Molecular and Clinical Oncology, 11(1), 3–14. [Google Scholar]
7. Gan, C., Huang, X., Wu, Y., Zhan, J., Zhang, X. et al. (2020). Untargeted metabolomics study and pro-apoptotic properties of B-norcholesteryl benzimidazole compounds in ovarian cancer SKOV3 cells. Journal of Steroid Biochemistry and Molecular Biology, 202(7), 105709. DOI 10.1016/j.jsbmb.2020.105709. [Google Scholar] [CrossRef]
8. Huynh, T. Y. L., Zareba, I., Baszanowska, W., Lewoniewska, S., Palka, J. (2020). Understanding the role of key amino acids in regulation of proline dehydrogenase/proline oxidase (prodh/pox)- dependent apoptosis/autophagy as an approach to targeted cancer therapy. Molecular and Cellular Biochemistry, 466(1–2), 35–44. DOI 10.1007/s11010-020-03685-y. [Google Scholar] [CrossRef]
9. Tan, B., Qiu, Y., Zou, X., Chen, T., Xie, G. et al. (2013). Metabonomics identifies serum metabolite markers of colorectal cancer. Journal of Proteome Research, 12(6), 3000–3009. DOI 10.1021/pr400337b. [Google Scholar] [CrossRef]
10. Uchiyama, K., Yagi, N., Mizushima, K., Higashimura, Y., Hirai, Y. et al. (2017). Serum metabolomics analysis for early detection of colorectal cancer. Journal of Gastroenterology, 52(6), 677–694. DOI 10.1007/s00535-016-1261-6. [Google Scholar] [CrossRef]
11. Koseki, J., Matsui, H., Konno, M., Nishida, N., Kawamoto, K. et al. (2016). A trans-omics mathematical analysis reveals novel functions of the ornithine metabolic pathway in cancer stem cells. Scientific Reports, 6(1), 20726. DOI 10.1038/srep20726. [Google Scholar] [CrossRef]
12. Gabriele, C., Cantiello, F., Nicastri, A., Crocerossa, F., Russo, G. I. et al. (2019). High-throughput detection of low abundance sialylated glycoproteins in human serum by TiO2 enrichment and targeted LC-MS/MS analysis: Application to a prostate cancer sample set. Analytical and Bioanalytical Chemistry, 411(3), 755–763. DOI 10.1007/s00216-018-1497-5. [Google Scholar] [CrossRef]
13. Beretov, J., Wasinger, V. C., Millar, E. K. A., Schwartz, P., Graham, P. H. et al. (2015). Proteomic analysis of urine to identify breast cancer biomarker candidates using a label-free LC-MS/MS approach. PLoS One, 10(11), e0141876. DOI 10.1371/journal.pone.0141876. [Google Scholar] [CrossRef]
14. Zhu, J., He, J., Liu, Y., Simeone, D. M., Lubman, D. M. (2012). Identification of glycoprotein markers for pancreatic cancer CD24+CD44+ stem-like cells using nano-LC-MS/MS and tissue microarray. Journal of Proteome Research, 11(4), 2272–2281. DOI 10.1021/pr201059g. [Google Scholar] [CrossRef]
15. Li, Y., Song, X., Zhao, X., Zou, L., Xu, G. (2014). Serum metabolic profiling study of lung cancer using ultra high performance liquid chromatography/quadrupole time-of flight mass spectrometry. Journal of Chromatography B-Analytical Technologies in the Biomedical and Life Sciences, 966, 147–153. DOI 10.1016/j.jchromb.2014.04.047. [Google Scholar] [CrossRef]
16. Fan, Y., Zhou, X., Xia, T. S., Chen, Z., Li, J. et al. (2016). Human plasma metabolomics for identifying differential metabolites and predicting molecular subtypes of breast cancer. Oncotarget, 7(9), 9925–9938. DOI 10.18632/oncotarget.7155. [Google Scholar] [CrossRef]
17. Gao, P., Zhou, C., Zhao, L., Zhang, G., Zhang, Y. (2016). Tissue amino acid profile could be used to differentiate advanced adenoma from colorectal cancer. Journal of Pharmaceutical and Biomedical Analysis, 118, 349–355. DOI 10.1016/j.jpba.2015.11.007. [Google Scholar] [CrossRef]
18. Brulé, S. Y., Jonker, D. J., Karapetis, C. S., O’Callaghan, C. J., Moore, M. J. et al. (2015). Location of colon cancer (right-sided versus left-sided) as a prognostic factor and a predictor of benefit from cetuximab in NCIC CO.17. European Journal of Cancer, 51(11), 1405–1414. DOI 10.1016/j.ejca.2015.03.015. [Google Scholar] [CrossRef]
19. Shu, X. L., Xu, H., Yu, T. T., Zhong, J. X., Lei, T. (2014). Regulation of apoptosis in human gastric cancer cell line SGC-7901 by L-arginine. Panminerva Medica, 56(3), 227–231. [Google Scholar]
20. Al-Koussa, H., Al-Haddad, M., Abi-Habib, R., El-Sibai, M. (2019). Human recombinant arginase I [HuArgI (Co)-PEG5000]-induced arginine depletion inhibits colorectal cancer cell migration and invasion. International Journal of Molecular Sciences, 20(23), 6018. DOI 10.3390/ijms20236018. [Google Scholar] [CrossRef]
21. Karimian, J., Hadi, A., Salehi-Sahlabadi, A., Kafeshani, M. (2019). The effect of arginine intake on colorectal cancer: A systematic review of literatures. Clinical Nutrition Research, 8(3), 209–218. DOI 10.7762/cnr.2019.8.3.209. [Google Scholar] [CrossRef]
22. Bowles, T. L., Kim, R., Galante, J., Parsons, C. M., Virudachalam, S. et al. (2008). Pancreatic cancer cell lines deficient in argininosuccinate synthetase are sensitive to arginine deprivation by arginine deiminase. International Journal of Cancer, 123(8), 1950–1955. DOI 10.1002/ijc.23723. [Google Scholar] [CrossRef]
23. Sahu, D., Gupta, S., Hau, A. M., Nakashima, K., Leivo, M. Z. et al. (2017). Argininosuccinate synthetase 1 loss in invasive bladder cancer regulates survival through general control nonderepressible 2 kinase-mediated eukaryotic initiation factor 2α activity and is targetable by pegylated arginine deiminase. American Journal of Pathology, 187(1), 200–213. DOI 10.1016/j.ajpath.2016.09.004. [Google Scholar] [CrossRef]
24. Enjoji, M., Nakamuta, M., Arimura, E., Morizono, S., Kuniyoshi, M. et al. (2004). Clinical significance of urinary N, -diacetylspermine levels in patients with hepatocellular carcinoma. International Journal of Biological Markers, 19(4), 322–327. DOI 10.1177/172460080401900411. [Google Scholar] [CrossRef]
25. Yamaguchi, K., Nakamura, M., Shirahane, K., Konomi, H., Torara, N. et al. (2005). Urine diacetylspermine as a novel tumour maker for pancreatobiliary carcinomas. Digestive and Liver Disease, 37(3), 190–194. DOI 10.1016/j.dld.2004.10.006. [Google Scholar] [CrossRef]
26. Takahashi, Y., Sakaguchi, K., Horio, H., Hiramatsu, K., Moriya, S. et al. (2015). Urinary N1, N12-diacetylspermine is a non-invasive marker for the diagnosis and prognosis of non-small-cell lung cancer. British Journal of Cancer, 113(10), 1493–1501. DOI 10.1038/bjc.2015.349. [Google Scholar] [CrossRef]
27. Zell, J. A., Ziogas, A., Ignatenko, N., Honda, J., Qu, N. et al. (2009). Associations of a polymorphism in the ornithine decarboxylase gene with colorectal cancer survival. Clinical Cancer Research, 15(19), 6208–6216. DOI 10.1158/1078-0432.CCR-09-0592. [Google Scholar] [CrossRef]
28. Ma, Z., Lian, J., Yang, M., Wuyang, J., Zhao, C. et al. (2019). Overexpression of Arginase-1 is an indicator of poor prognosis in patients with colorectal cancer. Pathology Research and Practice, 215(6), 152383. DOI 10.1016/j.prp.2019.03.012. [Google Scholar] [CrossRef]
29. Hoshino, Y., Terashima, S., Teranishi, Y., Terashima, M., Kogure, M. et al. (2007). Ornithine decarboxylase activity as a prognostic marker for colorectal cancer. Fukushima Journal of Medical Science, 53(1), 1–9. DOI 10.5387/fms.53.1. [Google Scholar] [CrossRef]
30. Wang, Y. R., Yan, J. X., Wang, L. N. (2014). The diagnostic value of serum carcino-embryonic antigen, alpha fetoprotein and carbohydrate antigen 19-9 for colorectal cancer. Journal of Cancer Research and Therapeutics, 10, 307–309. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |