
 | Oncologie |  |
DOI: 10.32604/Oncologie.2021.014790
ARTICLE
The mtDNA Microsatellite Instability Indicated the Prognosis of Chinese Patients with Non-Hodgkin’s Lymphoma
1Hematology Department, The Fourth Hospital of Hebei Medical University, Shijiazhuang, China
*Corresponding Author: Lili Wu. Email: wzzyyxl@sina.com
Received: 29 October 2020; Accepted: 28 January 2021
Abstract: This study aimed to investigate the relationship and diagnostic potential between microsatellite instability (MSI) caused by a 9-bp GGGGGTAGA insertion/deletion in mitochondrial DNA (mtDNA) at position 8272 and clinical features in Chinese patients diagnosed with non-Hodgkin’s lymphoma. The 9-bp GGGGGTAGA insertion/deletion in the non-d-loop area of the mitochondrial DNA at the 8272 locus was detected by PCR in 139 non-Hodgkin’s lymphoma patient samples. GGGGGTAGA insertion/deletion was found in a total of 16 cases (11.5%), including nine T cell lymphoma and seven B cell lymphoma cases. The 5-year survival rate was 33.3% in patients with non-Hodgkin’s lymphoma and MSI, and was 26.7% in non-Hodgkin’s lymphoma patients lacking MSI. In B-cell lymphoma, the 5-year survival rate was 42.9% in patients harboring the 9-bp insertion/deletion, and was 36.8% in patients lacking MSI. In T-cell lymphoma, the 5-year survival rate was 55.6% in patients with the 9-bp insertion/deletion, and was 16.4% in patients lacking MSI (p < 0.05). The difference between 5-year survival rates was statistically significant only in T-cell lymphoma patients. In conclusion, the mtDNA GGGGGTAGA indel/variation at position 8272 can be used as an independent prognostic marker for T-cell lymphoma.
Keywords: Mitochondrial microsatellite instability; non-Hodgkin’s lymphoma; prognosis
Lymphoma is a hematological malignancy. Lymphoma is divided into two categories which include Hodgkin’s lymphoma and non-Hodgkin’s lymphoma (NHL) [1]. NHL includes numerous subtypes. NHL develops from abnormal lymphocytes, and there are two main types of lymphocyte: B-cell lymphocytes and T-cell lymphocytes. B-cell lymphoma develops from abnormal B-cell lymphocytes, while T-cell lymphoma develops from abnormal T-cell lymphocytes. The prognosis of each subtype is different with several criteria that must be met during evaluation. At present, the most commonly used method for the prognosis of NHL is the international prognostic index (IPI score). IPI score has limitations and still cannot meet the clinical needs. Therefore, it is important to identify new prognostic indicators to potentially develop individualized treatment of NHL.
A microsatellite is a highly polymorphic, short tandem repeat nucleotide sequence that exists in prokaryotic and eukaryotic genomes [2]. Microsatellite instability (MSI) occurs when the repetitive unit changes in length due to insertions or deletions [3,4]. Most changes in length are between 1–6 bp. MSI is a form of genetic instability that includes nuclear and mitochondrial MSI. Recent investigations have shown that mitochondrial MSI (mtMSl) plays an important role in the occurrence and development of a variety of cancers [5–7]. Currently, the relationship between mtMSI and lymphoma has not been reported.
The purpose of this study was to investigate the relationship between MSI and clinical features and prognosis of NHL caused by a 9-bp insertion/deletion (also called indel/variation). Our results suggest that MSI could be a prognostic indicator for T-cell lymphoma.
The study was approved by the Human Tissue Research Committee of Fourth Hospital, Hebei Medical University. All patients provided written informed consent for the collection of samples and subsequent analyses. The case samples were collected from 139 NHL patients diagnosed by immunohistochemistry methods in the hematology department at Fourth Hospital, Hebei Medical University, between January 2005 and January 2010. T cell lymphoma patients accounted for 69 cases (49.6%) which included those diagnosed with ALK T-cells lymphoma, peripheral T-cells lymphoma (non-specified type), and T-lymphoblastoid cell lymphoma. B-cell lymphoma cases accounted for 70 patients (50.4%), including 28 cases of inert lymphoma such as small lymphocytic lymphoma, follicular lymphoma and marginal lymphoma, and 42 cases of invasive lymphomas such as diffuse large B-cell and mantle cell lymphoma, and B-lymphoblastoid cell lymphoma. Peripheral venous blood samples were taken from 98 healthy adults (control subjects) who visited Fourth Hospital medical examination center at Hebei Medical University.
DNA was extracted using the kit from Genemed (San Antonio, TX, USA), and was stored at −80°C. PCR was performed according to the protocol supplied with the PCR Master Mix Kit (Promega Corporation, Fitchburg, WI, USA). Primers were designed based on database (http://www.mitomap.org): forward 5’-ACCACTTTCACCGCTACACG-3’; reverse 5’-TTTAGTTGGGGCATTTCACTG-3’. PCR conditions were 94°C for 5 min, followed by 35 cycles of 40 s at 94°C, 40 s at 58°C, and 40 s at 72°C, and final elongation at 72°C for 5 min. PCR products were purified prior to sequencing. The sequencing results were compared to the 15.4 kb wild type non-D-Loop region nucleotide sequence located at locus 577–16023. This sequence is described on the human mitochondrial database (http://www.mitomap.org). Cycle sequencing was performed with the Dye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystem, Foster City, CA), and the products were then separated on an ABIPRISM Genetic Analyzer 3100 (Applied Biosystem).
SPSS 19.0 software was used for statistics calculations. A probability of p < 0.05 was considered to indicate statistically significance. The survival curve was calculated using the Kaplan–Meier method. Multivariate survival analysis was performed using the Cox proportional hazards model. The differences between the various groups were analyzed by chi square test and Fisher’s exact probability method.
There were 139 patients with non-Hodgkin’s lymphoma in experimental group. Representative sequencing results of PCR products were shown in Fig. 1.
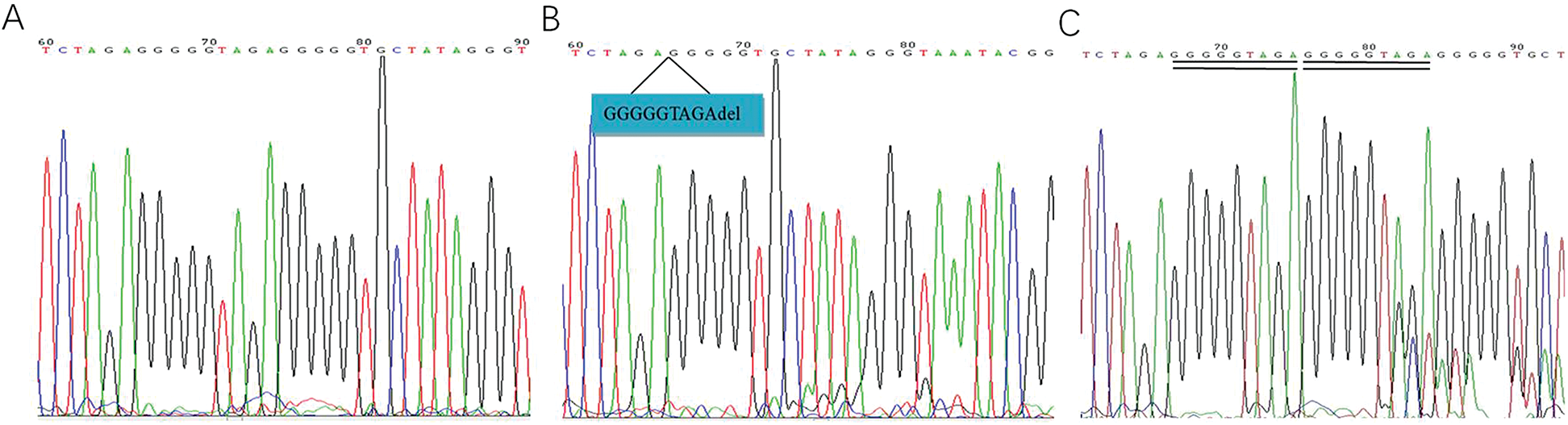
Figure 1: Representative sequencing results of PCR products. A. Normal control. B. mtDNA8272GGGGGTAGAdeletion. C. mtDNA8272GGGGGTAGA insert
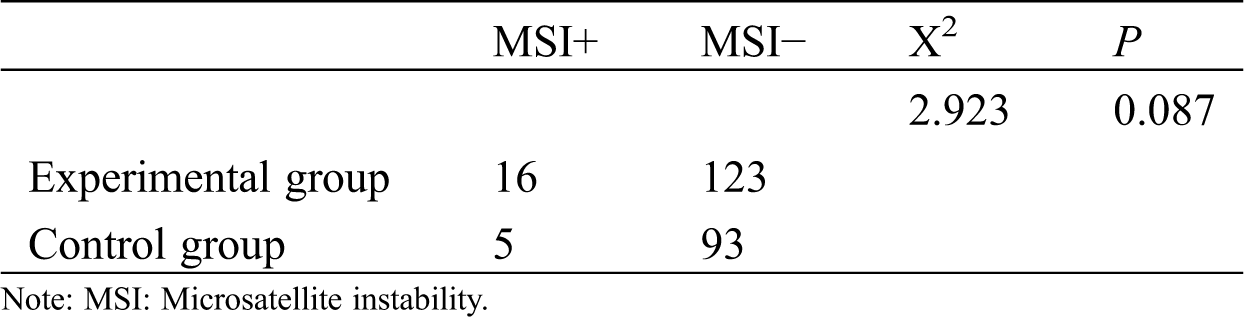
Among these patients, the GGGGGTAGA insertion/deletion occurred in 16 cases (11.5%). T cell lymphoma with GGGGGTAGA mitochondrial DNA (mtDNA) insertion/deletion at position 8272 accounted for nine cases, while B cell lymphoma with mtDNA GGGGGTAGA insertion/deletion at the same position accounted for seven cases. Among 98 control group, GGGGGTAGA deletion occurred in five cases (χ2 = 2.923, p = 0.087, Tab. 1).
Table 1: The presence of mtDNA8272 GGGGGTAGA in two groups
The incidence of mtDNA GGGGGTAGA insertion/deletion at the 8272 locus was 13.0% in T cell lymphoma and 10% in B cell lymphoma samples (χ2 = 0.316, p = 0.574). There was no correlation between the 9-bp insertion/deletion of mtDNA at position 8272 and the typing of non-Hodgkin’s lymphoma. The incidence of GGGGGTAGA mtDNA insertion/deletion was 7.9% in invasive B-cell lymphoma and 12.5% in inert B-cell lymphoma. Chi-square test showed that the GGGGGTAGA insertion/deletion rate of mtDNA was different (χ2 = 0.409), but the difference was not statistically significant (p = 0.403).
The 5-year survival rate was 33.3% among the 139 non-Hodgkin’s lymphoma patients. MSI was identified in 123 cases (88.5%). The 5-year survival rate in those who presented with MSI was 26.7%, and the median survival was 20 months (95% CI = 15.5–24.7, Fig. 2). There was no significant difference between the two groups (p = 0.795).
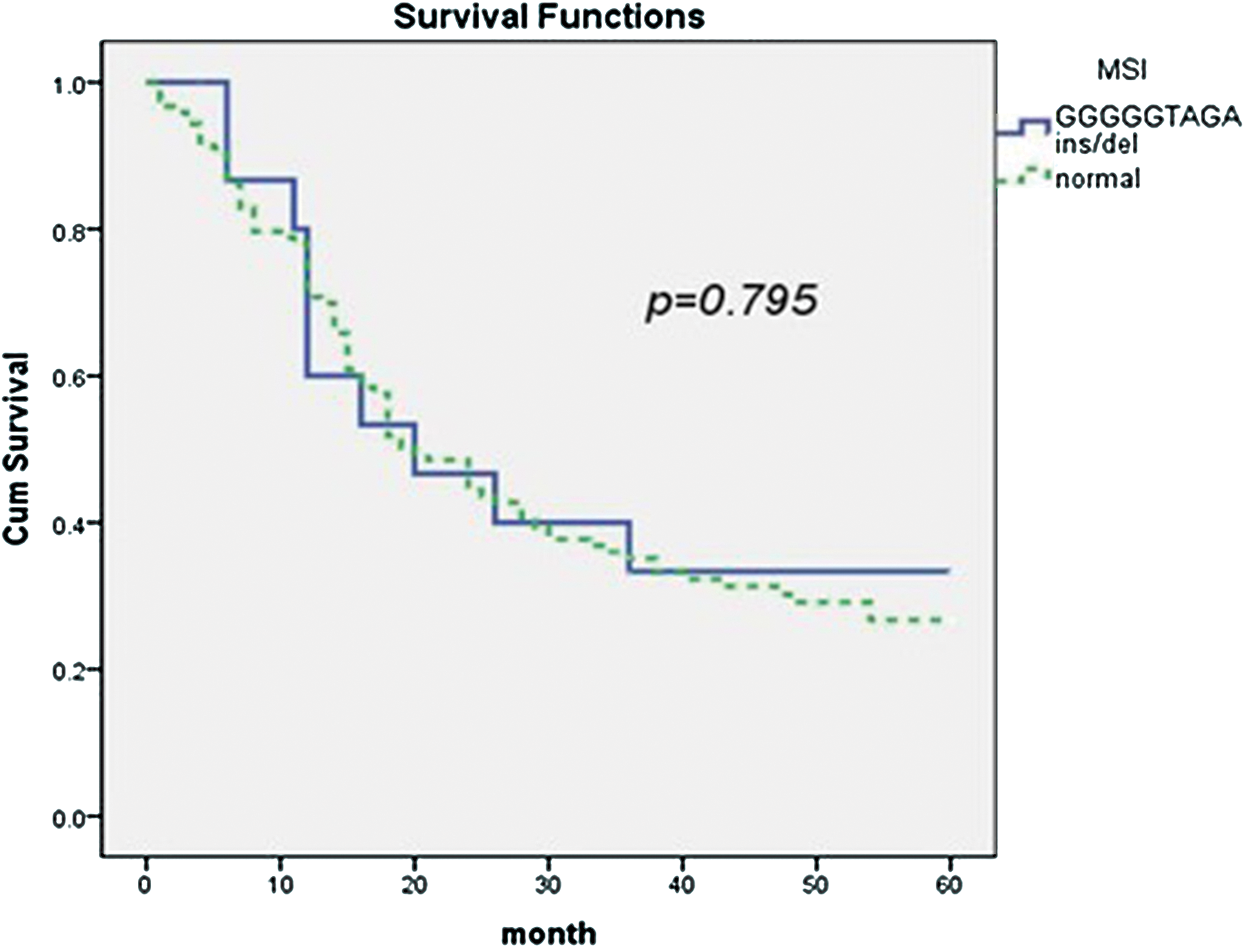
Figure 2: The association of mtDNA 8272GGGGGTAGA insertion/deletion and prognosis of non-Hodgkin’s lymphoma
Among the 70 cases of B-cell lymphoma, seven cases were positive for MSI. The 5-year survival rate was 42.9% (95% CI = 10.3–61.6), and the median survival was 36 months. In the 63 cases negative for MSI, the 5-year survival rate was 36.8%, and the median survival was 24 months (95% CI = 12.0–35.9). The difference was not statistically significant between two groups (p = 0.672, Fig. 3).
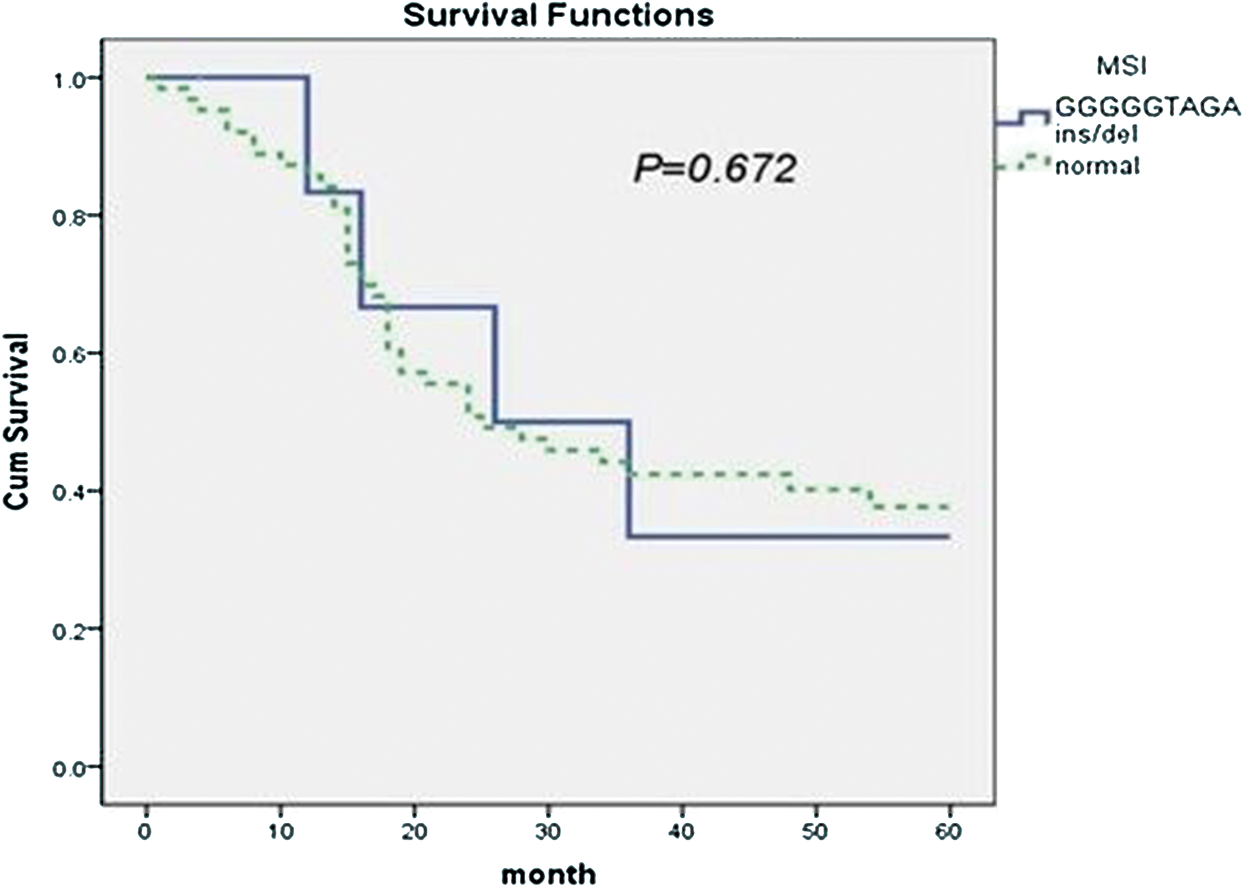
Figure 3: The association of mtDNA 8272GGGGGTAGA insertion/deletion and prognosis of B-cell lymphoma
The 9-bp insertion sequence was found at mtDNA position 8272 in nine of 69 patients with T-cell lymphoma. The 5-year survival rate was 55.6%, and the median survival was 18 months (95% CI = 10.9–25.0). In the 60 cases negative for MSI, the 5-year survival rate was 16.4%, and the median survival was 15 months (95% CI = 8.6–21.3). The difference was statistically significant (p = 0.046, Fig. 4).
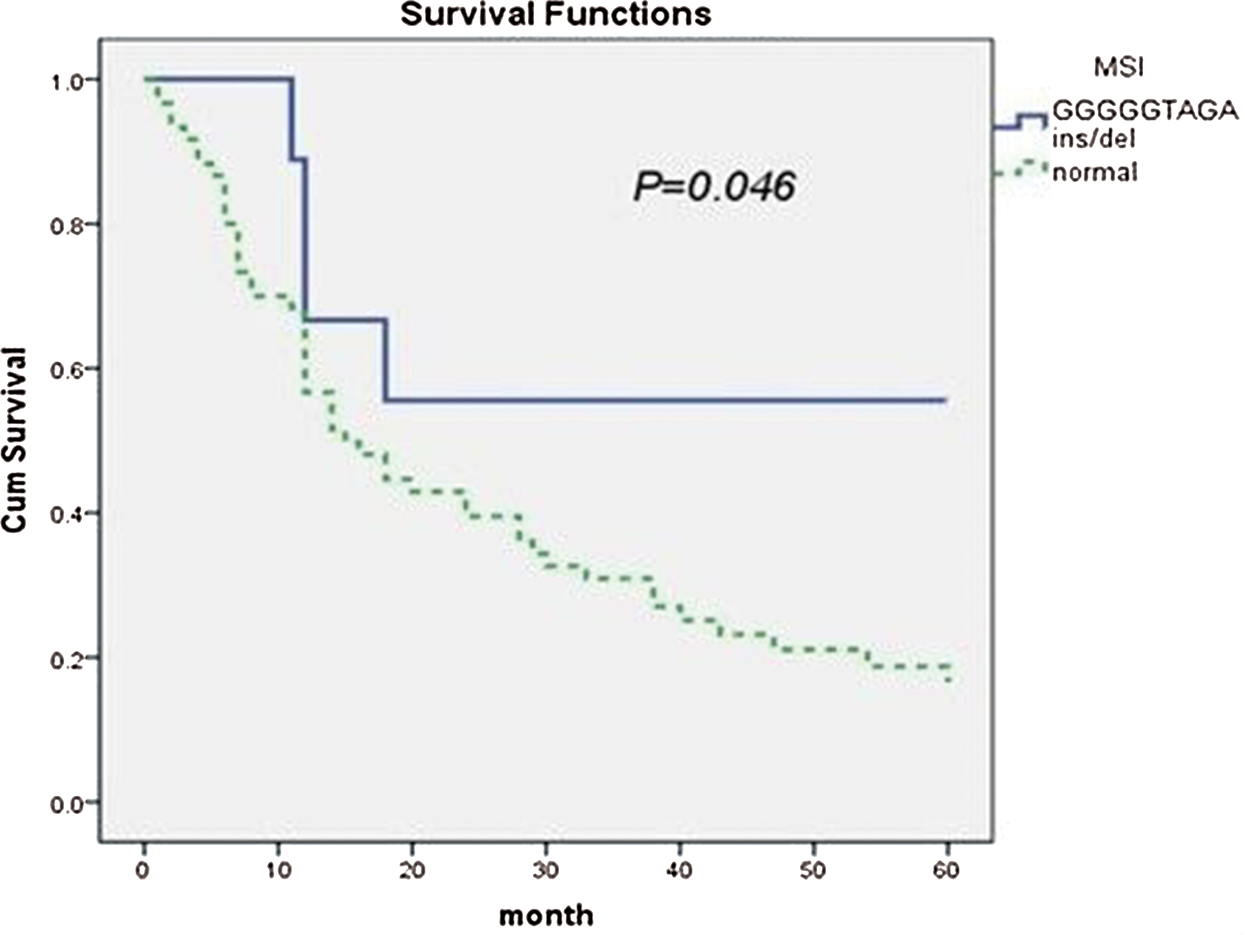
Figure 4: The association of mtDNA 8272GGGGGTAGA insertion/deletion and prognosis of T-cell lymphoma
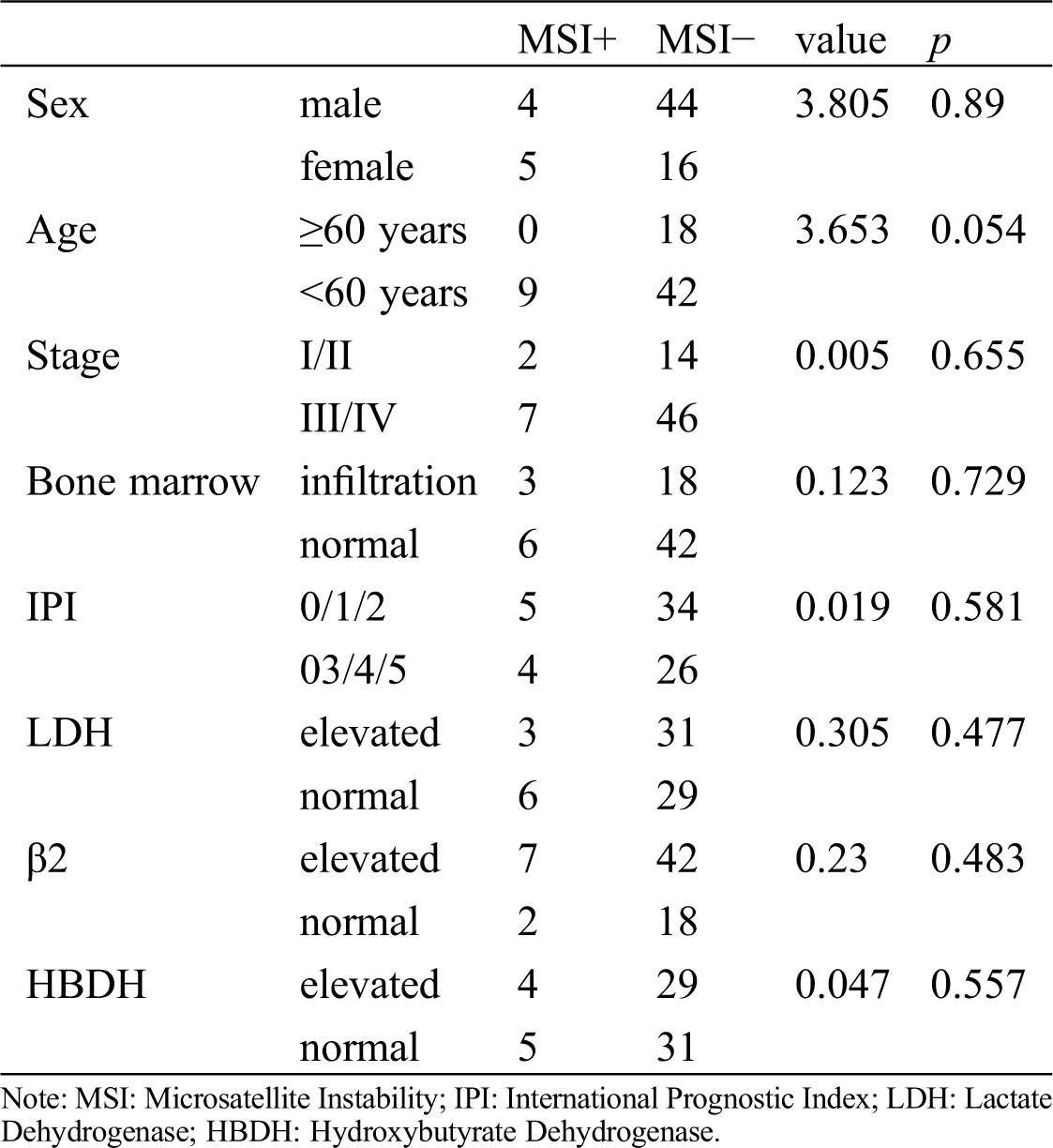
There was no correlation between clinical characteristics of T cell lymphoma such as IPI score, plasma level of lactate dehydrogenase (LDH), hydroxybutyrate dehydrogenase (HBDH), stage, typing, β2 microglobulin and bone marrow infiltration and GGGGGTAGA mtDNA indel at the 8272 locus (Tab. 2).
Table 2: Association of mtDNA8272GGGGGTAGA insertion/deletion with clinical characteristics of T-cell lymphoma patients
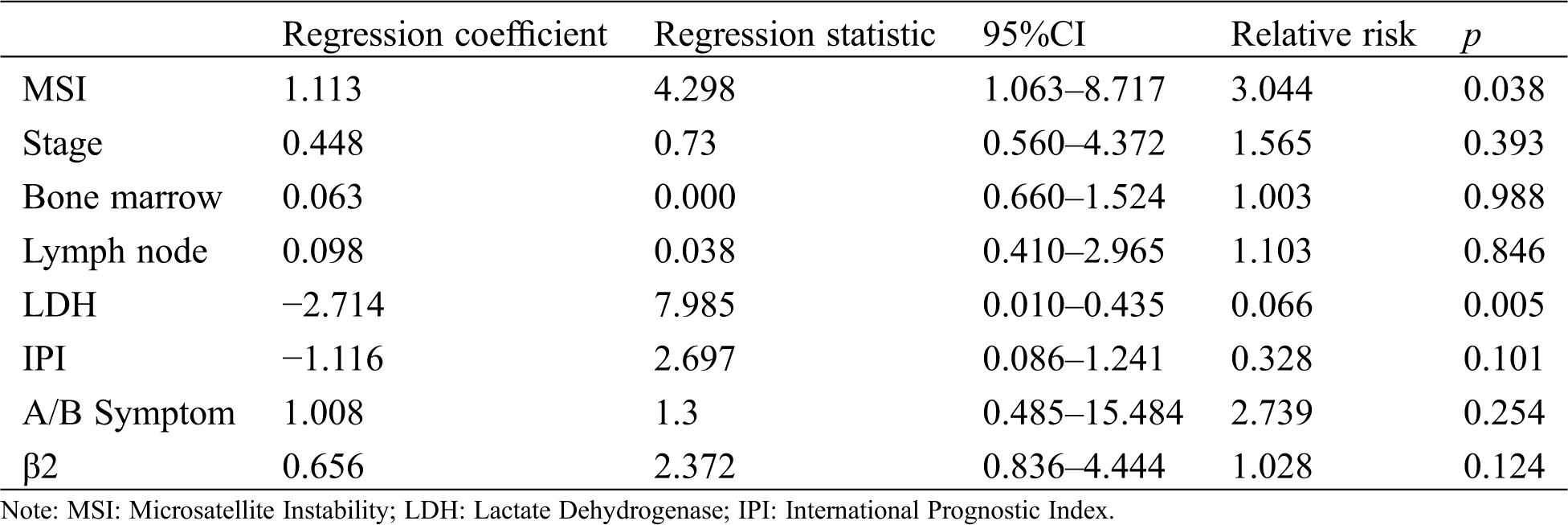
A multi-factor COX regression model analysis showed that the presence of the 9-bp insertion/deletion at mtDNA site 8272 and LDH value had significant impact on the survival rate over 5 years (p < 0.05, Tab. 3).
Table 3: Cox Multiple factor analysis of T-cell lymphoma patients
A microsatellite is a highly polymorphic nucleotide sequence with short, tandem repeats that is widely present in prokaryotic and eukaryotic genomes, and MSI was first detected in Lynch syndrome, or hereditary nonpolyposis colorectal cancer (HNPCC) [8] It is believed that the defect in the coding regions of mismatch repair (MMR) genes is an important molecular mechanism of MSI positive tumors [9]. To date, MSI is reported in many malignant tumors, such as gastric cancer, breast cancer, thyroid carcinoma, nasopharyngeal carcinoma and renal cell carcinoma [10–14].
mtMSI is a simple sequence repeat that occurs in the mtDNA. The prognosis of mtMSI and non-Hodgkin’s lymphoma is seldom reported [15]. In this study, we found no correlation between the 9-bp GGGGGTAGA mtDNA insertion/deletion and the 139 non-Hodgkin’s lymphoma patients who were tested for this sequence. The patients with the 9-bp GGGGGTAGA mtDNA insertion/deletion had better prognosis. Multi-variate analysis showed that the insertion/deletion of the 9-bp GGGGGTAGA mtDNA sequence was an independent prognostic factor of T-cell lymphoma.
The role of MSI in the development of lymphoma is uncertain, however, the function of the 9-bp GGGGGTAGA mtDNA insertion/deletion may be explained as below. The mitochondrial DNA is an important part of energy metabolism and oxidative phosphorylation. The 9-bp GGGGGTAGA sequence is located at position 8272 of mitochondrial DNA, which is upstream of the respiratory chain structure region. According to upstream and downstream primers, this region amplifies a 200-bp gene fragment specifically between cytochrome c oxidase II (COII) and mitochondrial TK2 thymidine kinase 2 (MTTK) [16]. The insertion or deletion of the 9-bp GGGGGTAGA mtDNA sequence may result in abnormal expression of upstream or downstream genes associated with structural components of the respiratory chain such as the ATP synthase 8 (MT-ATP8), the ATP synthase 6 (MT-ATP6), cytochrome c oxidase III (MT-CO3), and cytochrome b (MT-CYB) genes. Therefore, the insertion/deletion of the 9-bp GGGGGTAGA mtDNA affects the structure of the respiratory chain and reduces the level of oxidative phosphorylation and oxidative stress. Lymph nodes are glands with strong oxidative phosphorylation potential. It is speculated that the insertion/deletion of the 9-bp GGGGGTAGA mtDNA sequence can lead to the development of lymph node diseases.
In this study we found that MSI, IPI score, LDH, HBDH, stages, typing, β2 Globulin, and bone marrow infiltration could be independent prognostic factors of T-cell lymphoma. The clinical features associated with IPI score, stage, β2-microglobulin and bone marrow infiltration may affect the survival of patients with T-cell lymphoma, but we found no correlation between MSI and T-cell lymphoma, perhaps due to the heterogeneity of T-cell lymphoma. In the T-cell lymphoma cases included in this study, nine GGGGGTAGA insertion/deletion sequences were found. The GGGGGTAGA sequence was found to have been inserted in three, and deleted in six cases. No further correlative analysis was performed due to the low number of cases identified in this study.
This study showed no significant difference in the incidence and survival of patients with B-cell lymphoma and a 9-bp GGGGGTAGA mtDNA insertion/deletion, consistent with the results reported by Lee et al. [17]. However, the sample size in this study was low, and the number of sites from which patients were recruited was limited. Further studies are needed to verify our findings.
In conclusion, while there was no correlation between the GGGGGTAGA indel at the 8272 mtDNA locus and the outcome of non-Hodgkin’s and B-cell lymphomas. A correlation was found between the presentation of T-cell lymphoma and the GGGGGTAGA indel at mtDNA position 8272. LDH and the GGGGGTAGA indel at mtDNA position 8272 can be used as independent prognostic markers for T-cell lymphoma.
Funding Statement: The author(s) received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Jemal, A., Bray, F., Center, M. M., Ferlay, J., Ward, E. et al. (2011). Global cancer statistics. CA: A Cancer Journal for Clinicians, 61(2), 69–90. [Google Scholar]
2. Yu, C., Hong, H., Zhang, S., Zong, Y., Ma, J. et al. (2019). Identification of key genes and pathways involved in microsatellite instability in colorectal cancer. Molecular Medicine Reports, 19(3), 2065–2076. [Google Scholar]
3. Kang, J. U., Koo, S. H. (2011). Assessment of the beneficial loci and prognostic implications of microsatellite instability in gastric carcinoma. Molecular Medicine Reports, 4(6), 1175–1181. [Google Scholar]
4. Nenmer, A. O., Al Anazi, M. S., Bhat, R. S., Warsy, A. S., Babay, Z. A. et al. (2018). Association between preterm birth risk and polymorphism and expression of the DNA repair genes OGG1 and APE1 in Saudi women. Biocell, 42(1), 1–6. DOI 10.32604/biocell.2018.07005. [Google Scholar] [CrossRef]
5. Bianchi, M. S., Bianchi, N. O., Bailliet, G. (1995). Mitochondrial DNA mutations in normal and tumor tissues from breast cancer patients. Cytogenetic and Cell Genetics, 71(1), 99–103. DOI 10.1159/000134072. [Google Scholar] [CrossRef]
6. Park, S. Y., Shin, M. G., Kim, H. R., Oh, J. Y., Kim, S. H. et al. (2009). Alteration of mitochondrial DNA sequence and copy number in nasal polyp tissue. Mitochondrion, 9(5), 318–325. DOI 10.1016/j.mito.2009.04.006. [Google Scholar] [CrossRef]
7. Shin, M. G., Kajigaya, S., Tarnowka, M., McCoy, J. P., Levin, Jr, B. C. et al. (2004). Mitochondrial DNA sequence heterogeneity in circulating normal human CD34 cells and granulocytes. Blood, 103(12), 4466–4477. DOI 10.1182/blood-2003-11-3949. [Google Scholar] [CrossRef]
8. Locker, G. Y., Hamilton, S., Harris, J., Jessup, J. M., Kemeny, N. et al. (2006). ASCO, 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. Journal of Clinical Oncology, 24(33), 5313–5327. DOI 10.1200/JCO.2006.08.2644. [Google Scholar] [CrossRef]
9. Li, D., Zhou, X., Huang, L., Ji, X., Wang, D. (2019). XRCC1 Arg399Gln and Arg194Trp polymorphisms regulate XRCC1 expression and chemoresistance of non-small cell lung cancer cells. Biocell, 43(3), 139–144. DOI 10.32604/biocell.2019.06460. [Google Scholar] [CrossRef]
10. Leite, M., Corso, G., Sousa, S., Milanezi, F., Afonso, L. P. et al. (2011). MSI phenotype and MMR alterations in familial and sporadic gastric cancer. International Journal of Cancer, 128(7), 1606–1613. DOI 10.1002/ijc.25495. [Google Scholar] [CrossRef]
11. Basso, D., Navaglia, F., Fogar, P., Zambon, C. F., Greco, E. et al. (2007). DNA repair pathways and mitochondrial DNA mutations in gastrointestinal carcinogenesis. Clinica Chimica Acta, 381(1), 50–55. DOI 10.1016/j.cca.2007.02.020. [Google Scholar] [CrossRef]
12. Lacroix-Triki, M., Lambros, M. B., Geyer, F. C., Suarez, P. H., Reis-Filho, J. S. et al. (2010). Absence of microsatellite instability in mucinous carcinomas of the breast. International Journal of Clinical and Experimental Pathology, 4(1), 22–31. [Google Scholar]
13. Santos, J. C., Bastos, A. U., Cerutti, J. M., Ribeiro, M. L. (2013). Correlation of MLH1 and MGMT expression and promoter methylation with genomic instability in patients with thyroid carcinoma. BMC Cancer, 13(1), 79. DOI 10.1186/1471-2407-13-79. [Google Scholar] [CrossRef]
14. Bai, Y., Guo, Z., Xu, J., Zhang, J., Cui, L. et al. (2016). The 9-bp deletion at position 8272 in region V of mitochondrial DNA is associated with renal cell carcinoma outcome. Mitochondrial DNA, 27(3), 1973–1975. [Google Scholar]
15. Friedbeng, E. C. (2001). How nucleotide excision repair protects against cancer. Nature Reviews Cancer, 1(1), 22–33. DOI 10.1038/35094000. [Google Scholar] [CrossRef]
16. Wrischnik, L. A., Higuchi, R. G., Stoneking, M., Erlich, H. A., Arnheim, N. et al. (1987). Length mutations in human mitochondrial DNA: Direct sequencing of enzymatically amplified DNA. Nucleic Acids Research, 15(2), 529–542. DOI 10.1093/nar/15.2.529. [Google Scholar] [CrossRef]
17. Lee, H., Kim, J. J., Kim, J. H., Lee, J. H., Son, H. J. et al. (2006). Microsatellite instability in gastric B-cell lymphoma. Korean Journal of Gastroenterology, 47(3), 205–212. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |