
 | Oncologie |  |
DOI: 10.32604/Oncologie.2021.014153
ARTICLE
Serum Long Non-Coding RNA CCAT2 is a Potential Diagnostic and Prognostic Marker for Gastric Cancer
1Department of Oncology, Jinan Sixth People’s Hospital, Jinan, China
2Department of Gynecology, Jinan Sixth People’s Hospital, Jinan, China
3Department of Traditional Chinese Medicine, West Hospital, Jinan, China
4Department of Urology, Jinan Sixth People’s Hospital, Jinan, China
5Department of General Surgery, Second Hospital, Shandong University, Jinan, China
*Corresponding Author: Guanghua Li. Email: qpu77zg@163.com
Received: 03 September 2020; Accepted: 30 January 2021
ABSTRACT:
Aim: This study aims to explore the role of serum long non-coding RNA (lncRNA) colon cancer-associated transcript 2 (CCAT2) as a diagnostic marker for gastric cancer (GC). Methods: We recruited 76 patients with GC admitted to our hospital (the observation group, OG) and 83 healthy volunteers undergoing physical examinations during the same period (the control group, CG). CCAT2 expression was tested and its correlation with clinicopathological characteristics of GC was analyzed. We also explored the value of CCAT2 in assessing the treatment efficacy, predicting the fatality, and evaluating the prognosis of patients. Results: We detected higher CCAT2 levels in OG than in CG and the levels in OG decreased after treatment (p < 0.05). CCAT2 was highly predictive of treatment outcome and fatality in HCC. CCAT2 levels varied notably among GC patients with different pathological stages, lymph node metastasis, tumor sizes, and differentiation degrees (p < 0.05). The prognosis of patients was better in the low CCAT2 group than that in the high CCAT2 group (p < 0.05). Conclusion: CCAT2 is highly expressed in GC and affects GC progression. CCAT2 is a potential new blood marker for GC considering its high efficiency in predicting treatment outcome and fatality of patients.
Keywords: RNA; colon cancer-associated transcript 2; gastric cancer; assessment; marker
Gastric cancer (GC) is a frequent malignancy in the digestive tract, which originates from gastric mucosa epithelium [1] and tops the ranking of cancer incidence [2]. GC generally occurs in the antrum and pylorus of the stomach, triggered by factors like poor living conditions, unhealthy diets, and Helicobacter pylori infections [3]. In 2012, there were approximately 950,000 new GC cases, including 720,000 deaths, seriously threatening patients’ lives and affecting family well-being [4]. GC incidence varies geographically in China, with a markedly higher incidence in the northwest and eastern coastal areas than in the southern areas [5]. GC is found more frequently in people over 50 years old, and in more males than females. In recent years, the increasing work pressure and changing dietary habits lead to a declined age of onset and increased incidence year by year [6]. GC can develop in any part of the stomach, with the vast majority cases starting in gastric gland cells. Due to non-specific symptoms of upper abdominal discomfort and belching in the early stage, GC is often misdiagnosed as chronic gastric diseases such as gastritis and gastric ulcer [7]. Therefore, most cases have already developed to the advanced stage at the time of correct diagnosis, resulting in a poor prognosis of patients [8]. At present, primary treatment methods for GC include surgery and chemoradiotherapy, which have progressed and improved the prognosis of patients because the diagnosis of CG is getting more accurate [9]. However, there is still a lack of early diagnostic markers for individualized treatment, prognostic assessment and prediction of postoperative recurrence, as well as survival improvement [10]. Therefore, the search for accurate biomarkers is still a hot and difficult issue in clinical research.
Long non-coding RNAs (lncRNAs) are a series of RNAs with over 200 nt in length involved in gene transcription, post-transcriptional and epigenetic regulation of genes in many cancers [11]. Colon cancer-associated transcript 2 (CCAT2) is an RNA transcript with 1,752 bp, which was first discovered and identified in 2013 at the highly conserved chromosomal region 8q24 [12]. CCAT2, as a lncRNA, has been discussed in many studies about tumors [13]. It is abnormally expressed in liver cancer, breast cancer, GC, etc. [14]. However, the application of CCAT2 in the development, efficacy assessment, and prognosis prediction of GC has been rarely studied. Here we explored the role of CCAT2 as a serum marker for GC, aiming to provide a reliable theoretical basis for future diagnosis and treatment.
We conducted a prospective analysis of 76 patients with GC admitted to our hospital (the observation group, OG) and 83 healthy volunteers undergoing physical examinations during the same period (the control group, CG). This study was approved by the medical ethics committee of Jinan Sixth People’s Hospital, Jinan, China. All participants signed the written informed consent.
2.1 Inclusion and Exclusion Criteria
Inclusion criteria for OG: Patients showing clinical symptoms of GC and diagnosed with GC by laboratory tests and biopsy of our hospital; patients receiving radical resection in our hospital after diagnosis; patients with complete medical data; patients who cooperate with our investigation; patients with no previous adjuvant treatment prior to the admission; all volunteers in CG were healthy people with normal physical examination results in our hospital, aged 30 to 70 years.
Exclusion criteria for OG: Patients with other tumors, cardiovascular and cerebrovascular diseases, chronic diseases, mental diseases, or autoimmune diseases; patients during lactation or pregnancy; patients with visual and speech disorders; patients with physical disabilities who had been bedridden for a long time and could not take care of themselves; patients who were transferred to other hospitals; patients who died during treatment.
2.2 qRT-PCR Quantifies Serum CCAT2 Levels
Patients from OG received chemotherapy in our hospital. Five mL fasting venous blood was collected from each patient before and after treatment and from each volunteer and kept at room temperature for 30 min, followed by centrifugation for 10 min at 4000 rpm to collect the upper serum. Total RNA extraction was performed using the EasyPure miRNA kit (TransGen Biotech Co., Ltd., Beijing, China, ER601-01). Its integrity, purity, and concentration were detected by ultraviolet spectrophotometer and agarose gel electrophoresis. The extracted total RNA was reversely transcribed using TransScript Green miRNA Two-Step qRT-PCR SuperMix (TransGen Biotech Co., Ltd., Beijing, China, AQ202-01) in strict accordance with the kit instructions. The acquired cDNA was collected for PCR amplification. The primer sequences are shown in Tab. 1. The qPCR amplification was performed in a 20-μLsystem (1 μL cDNA, 0.4 μL forward primer, 0.4 μL reverse primer, 10 μL 2×TransTaq® Tip Green qPCR SuperMix, 0.4 μL Passive Reference Dye (50×), 7.8 μL ddH2O) under conditions of pre-denaturation at 94°C for 30 s, followed by 40 cycles of denaturation at 94°C for 5 s, annealing and extension at 60°C for 30 s. Three replicate wells were set for each sample and the experiment was performed in triplicate. In this study, GAPDH worked as the internal control, and the data was analyzed using 2−ΔΔct.

Data were statistically analyzed on SPSS22.0 and visualized on Graphpad7. The count data were expressed as percentage and intra-group comparison was conducted using the chi-square test. The measurement data were expressed as mean ± standard deviation and inter-group comparison was conducted using the independent samples t-test, denoted by t. The comparison between multiple groups was analyzed by the one-way ANOVA. Through the ROC curve analysis, the cut-off value, that is, the critical value or the disease’s “threshold”, was obtained. Cut-off values are used as a bound to determine the critical value of a value for predicting the best state of a situation. Therefore, taking the cut-off value of 1.749 as the boundary, we determined the predictive effect of CCAT2 on prognosis of patients using ROC curves that demonstrated CCAT2 levels in dead and surviving patients, and grouped patients with different CCAT2 expression. In this way, we can understand the changes in the prognosis and survival of patients both above and below the critical value. The survival rate was calculated by the Kaplan-Meier estimator and compared by the Log-rank test. p < 0.050 indicates statistical differences.
3.1 General Data of Participants
As shown in Tab. 1, the difference in both groups was marked in concentrations of CEA and CA19-9 (p < 0.05), but was not notable in age, BMI, sex, smoking, drinking, exercise habits, place of residence, ethnicity, and family medical history (p > 0.05).
Table 1: General data in both groups [n (%)]
3.2 CCAT2 Expression in Both Groups and Changes of CCAT2 Expression in OG
As shown in Fig. 1, CCAT2 levels were markedly higher in OG than in CG (p < 0.05), and after treatment, CCAT2 levels remarkably decreased in OG (p < 0.05).
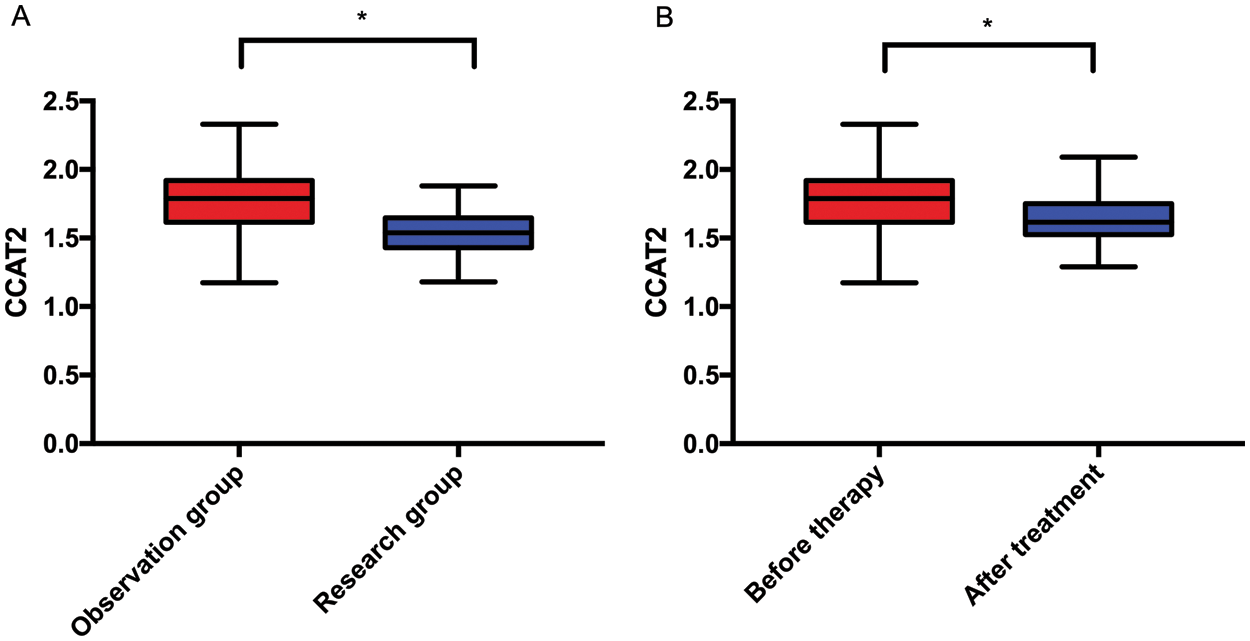
Figure 1: CCAT2 levels in OG and CG. A. CCAT2 levels in both groups. B. CCAT2 levels in OG before and after treatment. Note: *p < 0.05
3.3 CCAT2 in Assessing Treatment Efficacy in Patients with GC
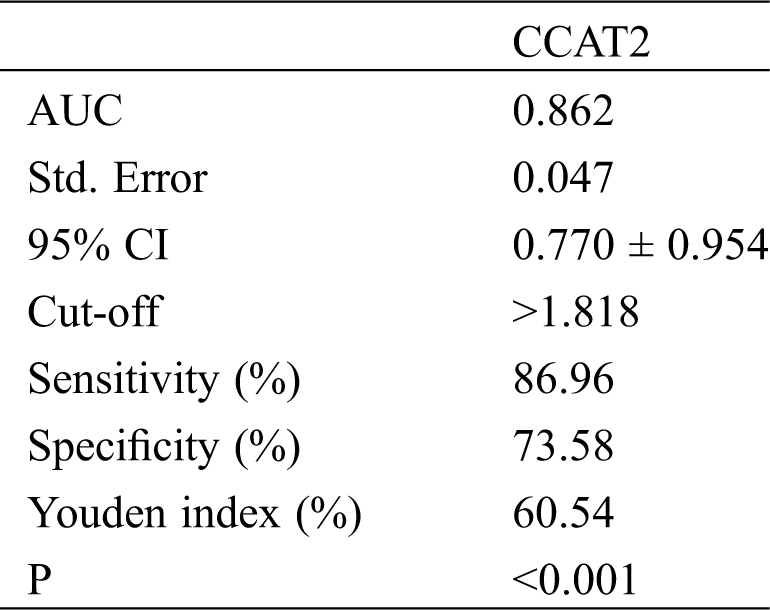
According to the treatment outcome, patients with complete or partial response were divided into Group A (53 cases) while those with stable or progressive disease were divided into Group B (23 cases). The diagnostic value of CCAT2 for GC was analyzed by the ROC curve. As shown in Fig. 2 and Tab. 2, the diagnostic sensitivity was 86.96% and the specificity was 73.58% when the cut-off value hit 1.818.
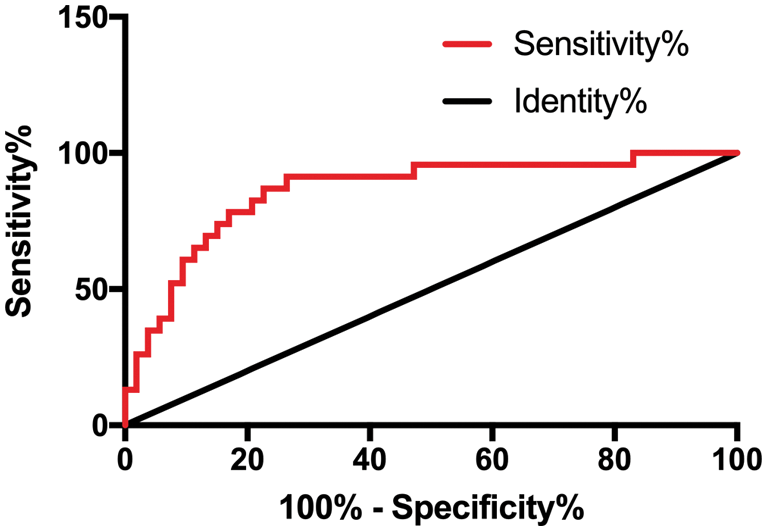
Figure 2: CCAT2 in assessing treatment efficacy in patients with GC
Table 2: CCAT2 in assessing treatment efficacy in patients with GC
3.4 Relationship between Serum CCAT2 Expression and Clinicopathological Characteristics of GC
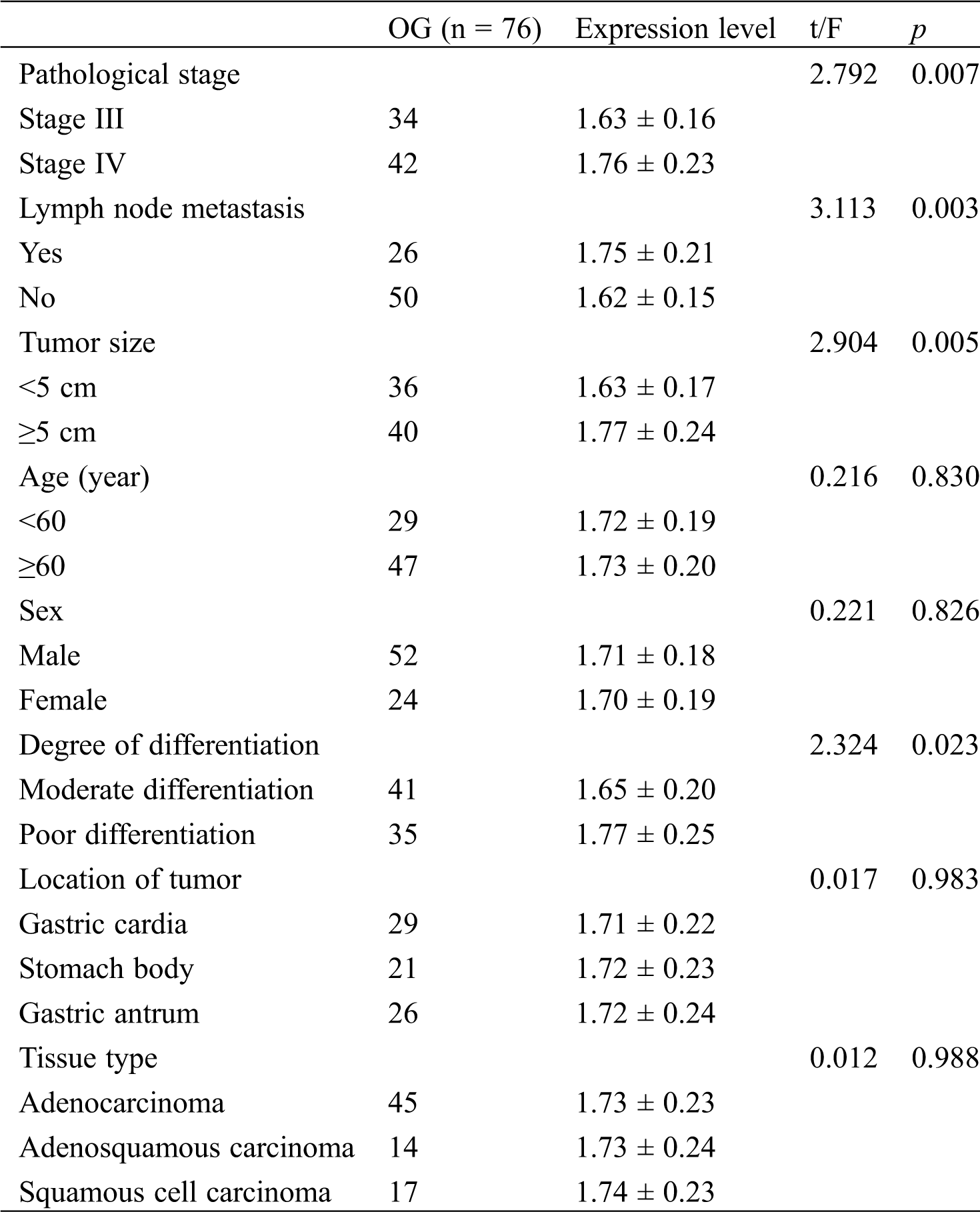
The relationship between serum CCAT2 expression and clinicopathological characteristics of GC was analyzed. As shown in Tab. 3, the difference in CCAT2 levels was not notable among patients with different age, sex, location of tumor, or tissue type (p > 0.05) but was marked among patients with different pathological stage, lymph node metastasis, tumor size, or degree of differentiation (p < 0.05).
Table 3: Relationship between serum CCAT2 expression and clinicopathological characteristics of GC
3.5 CCAT2 in Predicting Fatality of Patients with GC
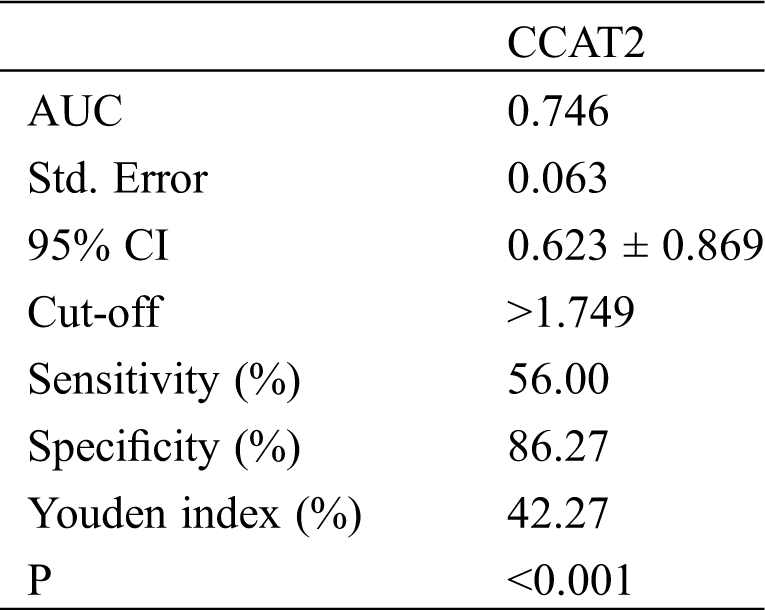
Patients were divided into the death group (25 cases) and the survival group (51 cases) after treatment. The predicting value of CCAT2 for GC fatality was analyzed by the ROC curve. As shown in Fig. 3 and Tab. 4, the predictive sensitivity was 56.00% and the specificity was 86.27% when the cut-off value hit 1.749.
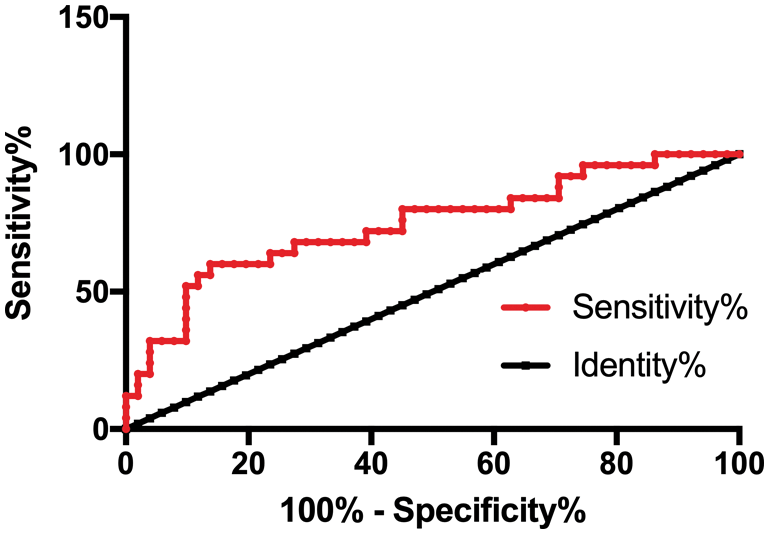
Figure 3: CCAT2 in predicting the fatality of patients with GC
Table 4: CCAT2 in predicting the fatality of patients with GC
3.6 Effect of CCAT2 on Prognosis of Patients with GC
All 76 patients in OG (100%) were successfully followed up for 3 years. Patients were divided into the high CCAT2 group (CCAT2 level > 1.749, n = 20) and the low CCAT2 group (CCAT2 level ≤ 1.749, n = 56) based on the cut-off value. As shown in Fig. 4, patient prognosis in the low CCAT2 group was superior to that in the high CCAT2 group (p < 0.001).
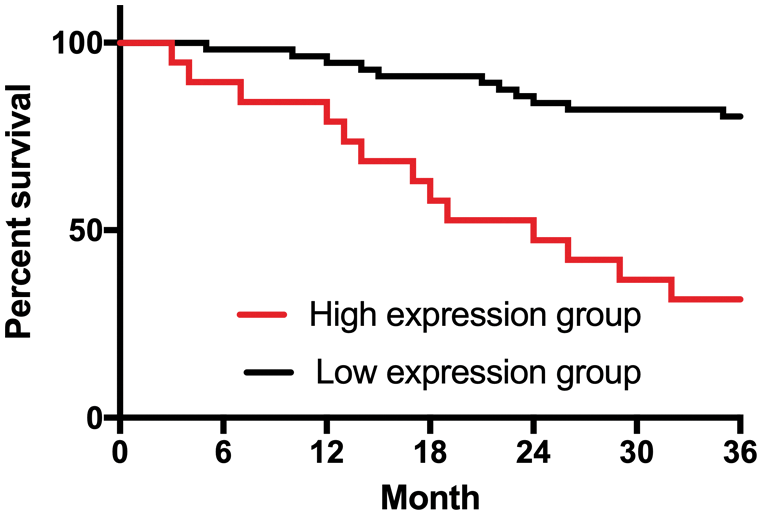
Figure 4: Effect of CCAT2 on the prognosis of patients with GC
GC is one of the most prevalent cancers worldwide, ranking fourth for cancer incidence and second for mortality [15]. The early symptoms of GC vary among patients, but it is generally diagnosed at the advanced stage with malignant hyperplasia and extensive metastasis [16]. Treatment for early GC has been markedly improved, but the long-term survival of patients with advanced GC remains low [17]. The lack of early diagnostic markers for GC is one of the main reasons for the low survival of patients [18]. Many lncRNAs, including CCAT2, are involved in the development and progression of GC [19]. A former study suggests that CCAT2 is a potent biomarker for the diagnosis and prognosis prediction of liver cancer [20]. It is also indicated that CCAT2 can affect the development of breast cancer [21]. However, the application of CCAT2 in the development, efficacy assessment, and prognosis prediction of GC has been rarely studied. Here we explored the role of CCAT2 in GC, aiming to provide a reliable theoretical basis for future diagnosis and treatment.
In this study, CCAT2 was highly expressed in patients with GC and lowly expressed in healthy individuals, and its levels in patients decreased after treatment, indicating the involvement of CCAT2 in the development and progression of GC. CCAT2 is highly expressed in many tumors [22]. Sarrafzadeh et al. [23] reveals that CCAT2 is expressed at higher levels in breast cancer. Zhao et al. [24] suggests that CCAT2 overexpression can promote tumorigenesis of non-small cell lung cancer. The study by Wu et al. [25] explores the role of lncRNA CCAT2 in the growth and apoptosis of cervical cancer cells and suggests that CCAT2 can stimulate the proliferation and survival of cervical cancer cells. The above-mentioned studies all support the results of this study. LncRNAs can regulate gene expression, modulate the development of tumors, and predict the prognosis of patients [26]. CCAT2 is an essential lncRNA located on chromosome 8q24. It is highly expressed in microsatellite stable (MSS) colorectal cancer tumor tissues and affects the expression of MYC, but its expression level in normal colon tissues is extremely low. CCAT2 is highly expressed in various tumors [27]. CCAT2 can suppress the proliferation of liver cancer cells and bladder cancer cells [28], as well as promote the migration, proliferation, and survival of cervical cancer cells, which enables it to work as a marker for the poor prognosis of various tumors [29]. Such studies confirm the important clinical significance and potential value of CCAT2 in various tumors. Here we found that CCAT2 was effective in predicting the occurrence of GC, which suggests the possibility of using CCAT2 as a screening index for GC to increase its early diagnosis rate. Compared with traditional imaging methods, the detection of CCAT2 is more convenient and intuitive, which does not rely on the clinician’s experience to analyze the imaging results. What’s more, the peripheral blood samples can be stored for a long time, facilitating clinical review at any time. Here we found that CCAT2 was relevant to the pathological stage, lymph node metastasis, tumor size, and degree of differentiation of patients with CG, and we detected higher CCAT2 levels in patients with more serious conditions (tumor stage IV, the presence of lymph nodes metastasis, tumor size ≥ 5 cm, or poor differentiation). From this, we speculate that CCAT2 may be involved in GC development. According to previous research, we find that CCAT2 mostly functions as a cancer-promoting gene in tumors. So we speculate that the CCAT2 levels in GC can stimulate the activation of GC cells, but we do not conduct basic experiments to confirm this due to limited conditions. We will explore further the effect of CCAT2 on GC cells to obtain more comprehensive experimental results. To further confirm the clinical significance of CCAT2 in GC, we conducted a follow-up and found that CCAT2 could predict the 3-year death of patients with GC with a sensitivity of 56.00% and a specificity of 86.27%. We divided patients into the high and low CCAT2 groups in the light of the cut-off value. The 3-year survival rate was markedly lower in the high CCAT2 group than in the low CCAT2 group, indicating that CCAT2 is also effective in assessing the prognosis of GC patients. We speculate that high CCAT2 expression indicates a high risk of death, so a high CCAT2 level may be used as a marker for poor prognosis. The study by Fu et al. [30] also reveals that CCAT2 serves as a marker for poor prognosis of liver cancer, which suggests the bright prospect of CCAT2 in GC and other tumors.
In summary, CCAT2 is highly expressed in GC and affects its progression. CCAT2 is a potential new blood marker for GC considering its high efficiency in predicting treatment outcome and fatality of patients.
Due to limited experimental conditions, there are some limitations. For example, we did not conduct basic experiments to verify the mechanism of action of CCAT2 in GC. Besides, we need to expand our sample size to obtain the most accurate cut-off value on the ROC curve. In addition, longer follow-up time is required to fully explore the impact of CCAT2 on the prognosis and prognosis of GC patients.
Funding Statement: The author(s) received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Wagner, A. D., Syn, N. L. X., Moehler, M., Grothe, W., Yong, W. P. et al. (2017). Chemotherapy for advanced gastric cancer. Cochrane Database of Systematic Reviews, 8(8), CD004064. DOI 10.1002/14651858.CD004064.pub4. [Google Scholar] [CrossRef]
2. Sitarz, R., Skierucha, M., Mielko, J., Offerhaus, G. J. A., Macjejewski, R. et al. (2018). Gastric cancer: Epidemiology, prevention, classification, and treatment. Cancer Management and Research, 10, 239–248. DOI 10.2147/CMAR.S149619. [Google Scholar] [CrossRef]
3. Chen, S., Li, T. W., Zhao, Q. F., Xiao, B. X., Guo, J. M. et al. (2017). Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clinica Chimica Acta, 466(2), 167–171. DOI 10.1016/j.cca.2017.01.025. [Google Scholar] [CrossRef]
4. Choi, I. J., Kook, M. C., Kim, Y. I., Cho, S. J., Lee, J. Y. et al. (2018). Helicobacter pylori therapy for the prevention of metachronous gastric cancer. New England Journal of Medicine, 378(12), 1085–1095. DOI 10.1056/NEJMoa1708423. [Google Scholar] [CrossRef]
5. Guilford, P. J., Holyoake, A. J. (2019). Markers for detection of gastric cancer. United States Patent US10179935. [Google Scholar]
6. Kim, S. T., Cristescu, R., Bass, A. J., Kim, K. M., Odegaard, J. et al. (2018). Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nature Medicine, 24(9), 1449–1458. DOI 10.1038/s41591-018-0101-z. [Google Scholar] [CrossRef]
7. Rugge, M., Genta, R. M., Mario, F. D., EI-Omar, E. M., EI-Serag, H. B. et al. (2017). Gastric cancer as preventable disease. Clinical Gastroenterology and Hepatology, 15(12), 1833–1843. DOI 10.1016/j.cgh.2017.05.023. [Google Scholar] [CrossRef]
8. Coburn, N., Cosby, R., Klein, L., Knight, G., Malthaner, R. et al. (2018). Staging and surgical approaches in gastric cancer: A systematic review. Cancer Treatment Reviews, 63(1), 104–115. DOI 10.1016/j.ctrv.2017.12.006. [Google Scholar] [CrossRef]
9. Moss, S. F. (2017). The clinical evidence linking Helicobacter pylori to gastric cancer. Cellular and Molecular Gastroenterology and Hepatology, 3(2), 183–191. DOI 10.1016/j.jcmgh.2016.12.001. [Google Scholar] [CrossRef]
10. Jun, J. K., Choi, K. S., Lee, H. Y., Suh, M., Park, B. et al. (2017). Effectiveness of the Korean National Cancer Screening Program in reducing gastric cancer mortality. Gastroenterology, 152(6), 1319–1328. DOI 10.1053/j.gastro.2017.01.029. [Google Scholar] [CrossRef]
11. Wang, Z. H., Yang, B., Zhang, M., Guo, W. W., Wu, Z. Y. et al. (2018). lncRNA epigenetic landscape analysis identifies EPIC1 as an oncogenic lncRNA that interacts with MYC and promotes cell-cycle progression in cancer. Cancer Cell, 33(4), 706–720. DOI 10.1016/j.ccell.2018.03.006. [Google Scholar] [CrossRef]
12. Lang, H. L., Hu, G. W., Zhang, B., Kuang, W., Chen, Y. et al. (2017). Glioma cells enhance angiogenesis and inhibit endothelial cell apoptosis through the release of exosomes that contain long non-coding RNA CCAT2. Oncology Reports, 38(2), 785–798. DOI 10.3892/or.2017.5742. [Google Scholar] [CrossRef]
13. Wu, Z. J., Li, Y., Wu, Y. Z., Wang, Y., Nian, W. Q. et al. (2017). Long non-coding RNA CCAT2 promotes the breast cancer growth and metastasis by regulating TGF-beta signaling pathway. European Review for Medical and Pharmacological Sciences, 21(4), 706–714. [Google Scholar]
14. Wu, S. W., Hao, Y. P., Qiu, J. H., Zhang, D. B., Yu, C. G. et al. (2017). High expression of long non-coding RNA CCAT2 indicates poor prognosis of gastric cancer and promotes cell proliferation and invasion. Minerva Medica, 108(4), 317–323. [Google Scholar]
15. Hao, N. B., He, Y. F., Li, X. Q., Wang, K., Wang, R. L. (2017). The role of miRNA and lncRNA in gastric cancer. Oncotarget, 8(46), 81572–81582. DOI 10.18632/oncotarget.19197. [Google Scholar] [CrossRef]
16. Nie, F. Q., Yu, X., Huang, M. D., Wang, Y. F., Xie, M. et al. (2017). Long noncoding RNA ZFAS1 promotes gastric cancer cells proliferation by epigenetically repressing KLF2 and NKD2 expression. Oncotarget, 8(24), 38227–38238. DOI 10.18632/oncotarget.9611. [Google Scholar] [CrossRef]
17. Sohn, B. H., Hwang, J. E., Jang, H. J., Lee, H. S., Oh, S. C. et al. (2017). Clinical significance of four molecular subtypes of gastric cancer identified by the cancer genome atlas project. Clinical Cancer Research, 23(15), 4441–4449. DOI 10.1158/1078-0432.CCR-16-2211. [Google Scholar] [CrossRef]
18. Pan, L., Liang, W., Fu, M., Huang, Z. H., Li, X. et al. (2017). Exosomes-mediated transfer of long noncoding RNA ZFAS1 promotes gastric cancer progression. Journal of Cancer Research and Clinical Oncology, 143(6), 991–1004. DOI 10.1007/s00432-017-2361-2. [Google Scholar] [CrossRef]
19. Wu, S. W., Hao, Y. P., Qiu, J. H., Zhang, D. B., Yu, C. G. et al. (2017). High expression of long non-coding RNA CCAT2 indicates poor prognosis of gastric cancer and promotes cell proliferation and invasion. Minerva Medica, 108(4), 317–323. [Google Scholar]
20. Zhou, N., Si, Z. Z., Li, T., Chen, G. S., Zhang, Z. Q. et al. (2016). Long non-coding RNA CCAT2 functions as an oncogene in hepatocellular carcinoma, regulating cellular proliferation, migration and apoptosis. Oncology Letters, 12(1), 132–138. DOI 10.3892/ol.2016.4580. [Google Scholar] [CrossRef]
21. Wu, Z. J., Li, Y., Wu, Y. Z., Wang, Y., Nian, W. Q. et al. (2017). Long non-coding RNA CCAT2 promotes the breast cancer growth and metastasis by regulating TGF-beta signaling pathway. European Review for Medical and Pharmacological Sciences, 21(4), 706–714. [Google Scholar]
22. Foßelteder, J., George, A. C., Martin, P. (2018). Long non-coding RNA CCAT2 as a therapeutic target in colorectal cancer. Expert Opinion on Therapeutic Targets, 22(12), 973–976. [Google Scholar]
23. Sarrafzadeh, S., Geranpayeh, L., Tasharrofi, B., Soudyab, M., Nikpayam, E. et al. (2017). Expression study and clinical correlations of MYC and CCAT2 in breast cancer patients. Iranian Biomedical Journal, 21(5), 303–311. DOI 10.18869/acadpub.ibj.21.5.303. [Google Scholar] [CrossRef]
24. Zhao, Z., Wang, J., Wang, S. F., Chang, H., Zhang, T. W. et al. (2017). LncRNA CCAT2 promotes tumorigenesis by over-expressed Pokemon in non-small cell lung cancer. Biomedicine & Pharmacotherapy, 87(2), 692–697. DOI 10.1016/j.biopha.2016.12.122. [Google Scholar] [CrossRef]
25. Wu, L., Jin, L., Zhang, W., Zhang, L. (2016). Roles of long non-coding RNA CCAT2 in cervical cancer cell growth and apoptosis. Medical Science Monitor: International Medical Journal of Experimental and Clinical Research, 22, 875. [Google Scholar]
26. Mattioli, K., Volders, P. J., Gerhardinger, C., Lee, J. C., Maass, P. G. et al. (2019). High-throughput functional analysis of lncRNA core promoters elucidates rules governing tissue specificity. Genome Research, 29(3), 344–355. DOI 10.1101/gr.242222.118. [Google Scholar] [CrossRef]
27. Xie, P. M., Cao, H. Y., Li, Y., Wang, J. H., Cui, Z. M. et al. (2018). Knockdown of lncRNA CCAT2 inhibits endometrial cancer cells growth and metastasis via sponging miR-216b. Cancer Biomarkers, 21(1), 123–133. DOI 10.3233/CBM-170388. [Google Scholar] [CrossRef]
28. Cao, X., Xu, J., Yue, D. (2018). LncRNA-SNHG16 predicts poor prognosis and promotes tumor proliferation through epigenetically silencing p21 in bladder cancer. Cancer Gene Therapy, 25(1), 10–17. DOI 10.1038/s41417-017-0006-x. [Google Scholar] [CrossRef]
29. Lin, J., Nong, L. L., Li, M. Q., Yang, F. C., Wang, S. H. et al. (2019). LINC00052 inhibits tumor growth, invasion and metastasis by repressing STAT3 in cervical carcinoma. European Review for Medical and Pharmacological Sciences, 23(11), 4673–4679. [Google Scholar]
30. Fu, C. B., Xu, X., Lu, W. J., Nie, L., Yin, T. et al. (2019). Increased expression of long non-coding RNA CCAT2 predicts poorer prognosis in patients with hepatocellular carcinoma. Medicine, 98(42), e17412. DOI 10.1097/MD.0000000000017412. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |