
 | Oncologie |  |
DOI: 10.32604/oncologie.2021.014151
ARTICLE
Mir-1247 Affects the Proliferation, Invasion and Apoptosis of Osteosarcoma Cells through SOX9
1Department of Orthopaedics, Jinan Zhangqiu District Hospital of TCM, Jinan, 250200, China
2Department of Internal Medicine, Jinan Zhangqiu District Hospital of TCM, Jinan, 250200, China
3Chinese Medicine Practitioner, Jinan Zhangqiu District Hospital of TCM, Jinan, 250200, China
*Corresponding Author: Lu Cao. Email: changnianlei699939@163.com
Received: 03 September 2020; Accepted: 23 December 2020
ABSTRACT:
Objective: This study aimed to explore the miR-1247-mediated promotion of osteosarcoma (OS) cell proliferation by SOX9. Methods: We recruited 97 OS patients admitted to our hospital (the observation group, OG) and 82 healthy people undergoing physical examinations (the control group, CG) over the same period into this study. The expression of miR-1247 and SOX9 in human OS cells in vitro was tested to determine the effect of miR-1247 and SOX9 on OS and to analyze the relationship between miR-1247 and SOX9. Results: We detected lowly expressed miR-1247 and highly expressed SOX9 in the peripheral blood of OS patients (P < 0.05). Both miR-1247 and SOX9 showed good performances in diagnosing OS. OS cells (143B cells) in the miR-1247-mimics group showed markedly lower cell proliferation and invasion rates and higher apoptosis rates than cells in the miR-1247-inhibitor group and the miR-NC group (P < 0.05). 143B cells in the sh-SOX9 group showed higher proliferation and invasion rates and lower apoptosis rates than cells in the si-SOX9 group and the NC group (P < 0.05). SOX9 protein concentrations were lower in cells in the miR-1247-mimics group than in cells in the miR-1247-inhibitor group and the miR-NC group (P < 0.05), with higher SOX9 protein concentrations in the miR-1247-inhibitor group than in the miR-NC group (P < 0.05). Conclusion: MiR-1247 is lowly expressed in OS. It can promote the proliferation and invasion of OS cells and accelerate the progression of OS by mediating SOX9.
Keywords: miR-1247; SOX9; osteosarcoma
Osteosarcoma (OS) is a primary cancerous tumor in the bone, affecting 3.4 cases per million individuals worldwide every year [1]. OS is featured with a low five-year survival rate, a higher metastasis rate, a high degree of malignancy, and a poor prognosis [2]. Common in adolescents, OS often occurs in the distal femur, proximal tibia, and distal tibia [3]. A previous study suggests that the histological grading of OS is related to malignant mesenchymal cells [4]. OS mainly manifests as persistent local pain with a stepwise increase in the pain intensity, which seriously affects the normal life of patients [5]. Early OS is usually treated by surgical resection in the form of amputation, autograft, or allograft, aiming to remove tumors through complete resection [6]. Improvements in medical standards in recent years have provided various treatment regimens for OS, including surgery, neoadjuvant therapy, and adjuvant chemotherapy [7]. However, the prognosis of OS is not very favorable, due to the high recurrence rate and metastasis rate [8]. Currently, gene-targeted therapy is promising in clinical cancer treatment [9].
MicroRNAs (miRNAs) are small endogenous RNAs consisting of 21–24 nucleotides [10]. They have many functions in tumor progression, metabolism, and endocrine [11]. With multiple targets, miRNAs can regulate a variety of oncogenes or tumor suppressor genes [12]. As a member of the miRNA family, miR-1247 is highly related to the differentiation and phenotype of human articular chondrocytes [13]. A previous study has identified the tumor-suppressing role of miR-1247 in pancreatic cancer [14]. Another study indicates that matrix-induced down-regulation of miR-1247 stimulates the progression of prostate cancer [15]. But the specific mechanism of action underlying miR-1247 remains unclear. SOX9 is an important transcription factor of the SOX family, which is involved in DNA binding, transcriptional regulation, protein interaction, etc. [16]. It has been revealed that SOX9 plays key roles in many tumors. The study by Dreval [17] suggests that miR-1247 can regulate SOX9 in patients with acute kidney injury. Therefore, we speculate that miR-1247 could affect OS progression by regulating SOX9. Here, we investigated the roles of miR-1247 and SOX9 in OS, seeking to enlighten new ideas and direction for future diagnosis and treatment of OS.
1.1 Basic Information of Participants
A prospective analysis was performed on 97 OS patients treated in our hospital from April 2017 to April 2019 and 82 healthy people undergoing health examinations over the same period. We assigned patients with OS to the observation group (OG) and healthy people to the control group (CG). This study has obtained the ethical approval from the ethics committee of our hospital. All participants signed the written informed consent.
1.2 Exclusion and Inclusion Criteria
Inclusion criteria: Patients with previously untreated OS diagnosed by the results of laboratory test, imaging tests, and histopathologic examinations in our hospital; patients with complete medical data; healthy people with normal physical examination results; healthy people with no previous history of major diseases; participants who agreed to cooperate with this study.
Exclusion criteria: Patients undergoing tumor treatment prior to this study; patients with immune diseases, major organ dysfunction, low treatment compliance, or drug allergies; patients who were transferred to another hospital. The comparison between the two groups in basic data such as age, sec, and BMI showed no statistical differences (P > 0.05). We drew 4 ml of fasting venous blood from each participant and kept the sample still at room temperature for 30 min before centrifugation at 400×g for 10 min. The upper serum was collected for detection.
Human OS cells (143B cells) and human normal osteoblasts (hFOB1.19 cells) were offered by BeNa Culture Collection, an agent company of ATCC. We cultured cells in the DMEM medium supplemented by 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin mixture and incubated at 37°C with 5% CO2. Cells (80% confluent) were digested with trypsin for about 1 min and then cultured in new media in a split ratio of 1:3 (1 passage every 2 days).
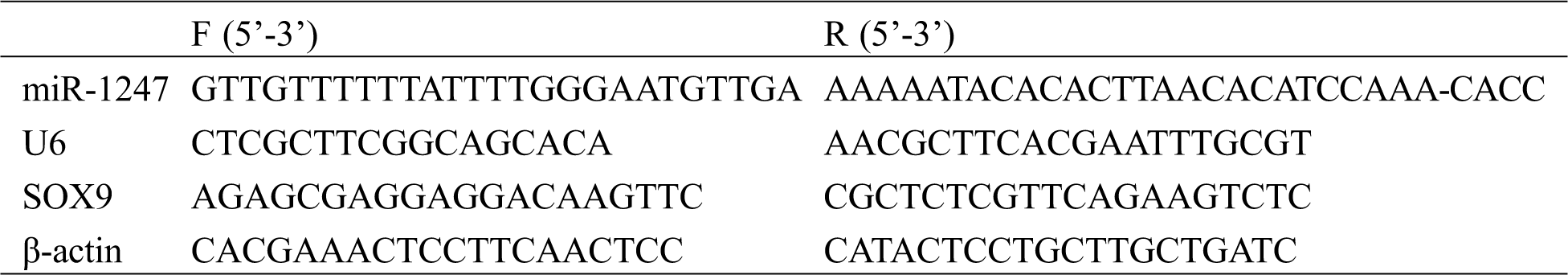
Total RNA was isolated from the blood, followed by reverse transcription and PCR amplification. PCR conditions: 94°C for 30 s, 94°C for 5 s, 60°C for 15 s, and 72°C for 10 s. The results were analyzed using 2–ΔΔct. Primer sequence as shown in Tab. 1.
143B cells were transfected with miR-1247-inhibitor, miR-1247-mimics, miR-NC, si-SOX9, sh-SOX9, and NC, respectively, following the instructions of the Lipofectamine™ 2000 kit.
Twenty μL of MTT solvent (5 g/L) was added to each well of the transwell plate and the supernatant was discarded 4 h later. Then we put 150 μL of dimethyl sulfoxide (DMSO) to each well and shook the plate to dissolve the crystals. Finally, the absorbance (A) value of each well was measured at 490 nm.
Transfected cells were seeded onto the 6-well plate (106 cells/well) and then cultured in the serum-free medium when reaching 75% confluency for a whole night. The density of the cell suspension was adjusted to 105 cells/mL. We added 10 μL of the cell suspension to the apical chamber and 600 μL of serum-containing medium to the basolateral chamber and cultured the transwell system overnight. We took out the transwell insert and detached cells remaining in the apical chamber. After PBS washing, cells in the basolateral chamber were mixed with methanol for 30 min for fixation and mixed with 0.1% crystal violet for 20 min for staining, followed by PBS washing. The number of invading cells was checked under a microscope.
After being mixed with 0.25% trypsin for digestion, transfected cells were washed twice with PBS. Next, 100 μL of binding buffer was added to make the cell suspension at a density of 1 × 106 cells/mL. Then, we added Annexin V-FITC and PI to the suspension and incubated at room temperature with no exposure to light. Finally, cell apoptosis was tested on the FC500MCL flow cytometer. The test was performed 3 times to determine the mean value.
We isolated protein lysate from cells with the protein extraction buffer, followed by incubation on ice for 30 min and centrifugation at 12000×g at 4°C for 10 min. After electrophoresis separation by 10% SDS-PAGE, proteins were transferred to the PVDF membrane, followed by staining and PBST washing for 5 min. Then we blocked the membrane with 5% skimmed milk for 1 h and incubated with the specific primary antibody (1:1000) at 4°C for a whole night. The membrane was washed to remove the primary antibody. Then we incubated the membrane with the horseradish peroxidase-labeled goat anti-rabbit secondary antibody (1:5000) at 37°C for 1 h, followed by 3 PBS washing, 5 min each time. The membrane was dried with filter paper and mixed with the ECL reagent for color development in the dark. The protein bands were scanned and analyzed the gray values on the Quantity One software.
Data was statistically analyzed on SPSS24.0 (IBM, Armonk, NY, USA). Data visualization was performed using Graphpad 8 software (La Jolla, CA, USA). The count data was denoted by the percentage (%) and compared between any two groups by the chi-square test. The measurement data was represented by the mean ± standard deviation and compared between any two groups by the t-test. The comparison between multiple groups was analyzed by the one-way ANOVA and the LSD post hoc test. The comparison between multiple time points was analyzed by the repeated measurement ANOVA and the Bonferroni post hoc test. The diagnostic value of genes was analyzed on the receiver operating characteristic (ROC) curve. P < 0.05 indicates a statistically significant difference.
3.1 Clinical Significance of miR-1247 in OS
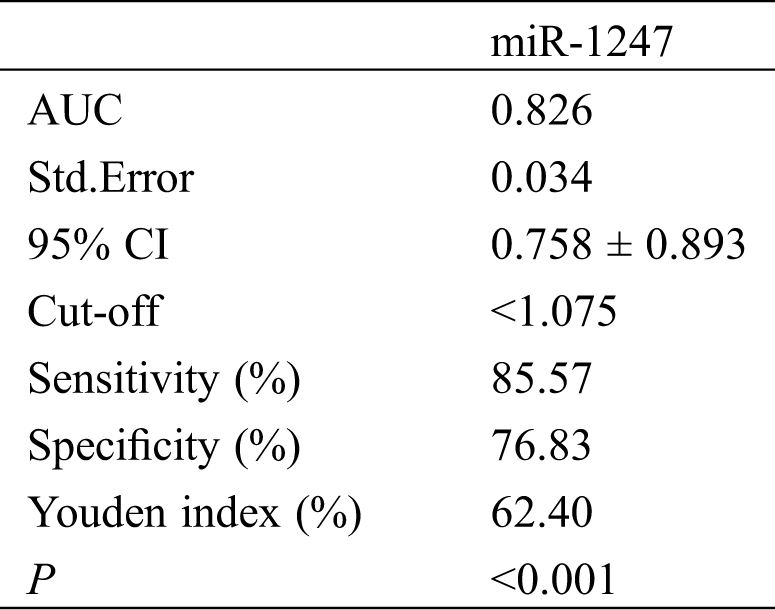
We detected low miR-1247 expression in the peripheral blood of OS patients (P < 0.05). The ROC curve demonstrated that the sensitivity of miR-1247 at the cut-off value of 1.075 for diagnosing OS was 85.57% and the specificity was 76.83%, as shown in Tab. 2 and Fig. 1.
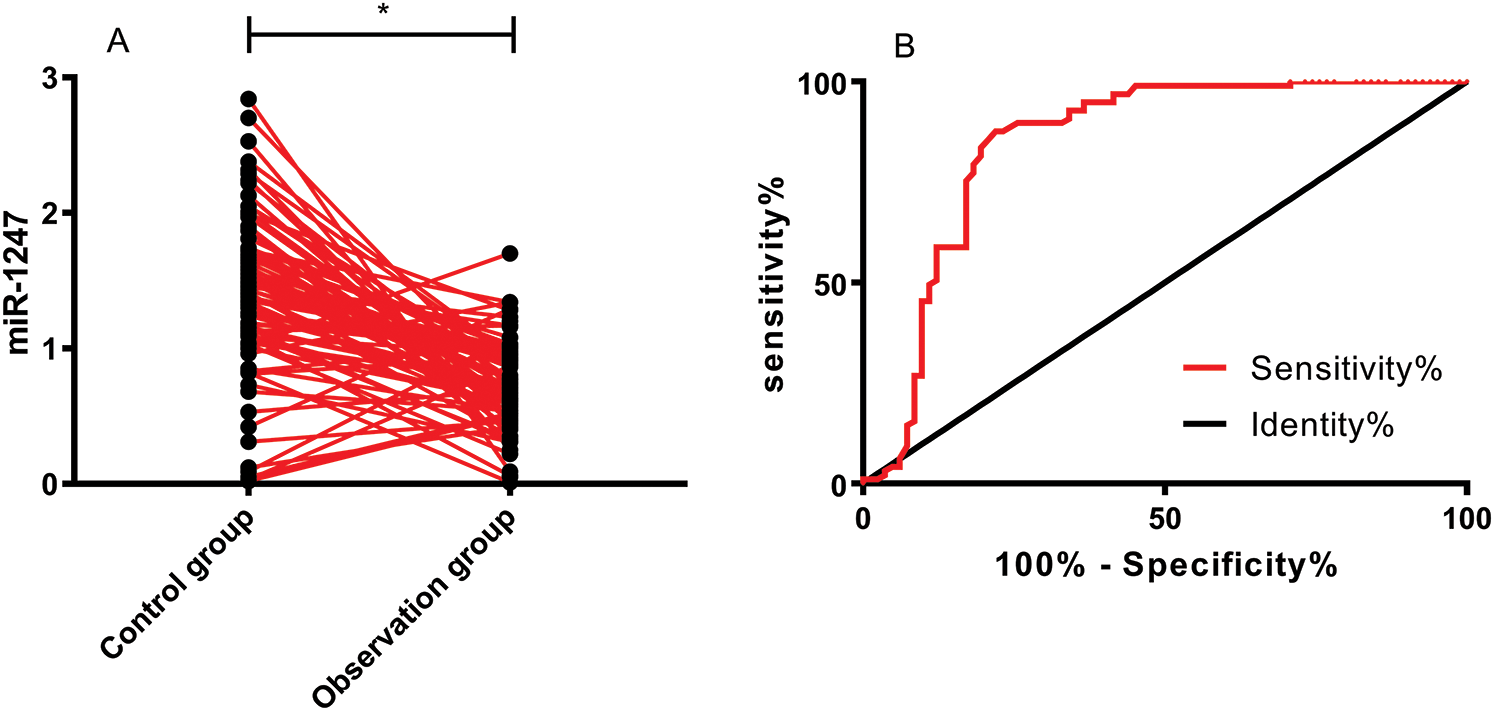
Figure 1: Clinical significance of miR-1247 in OS. (A) Expression of miR-1247 in OG and CG. *P < 0.05. (B) ROC curve demonstrating the performance of miR-1247 in diagnosing OS
Table 2: Diagnostic performance of miR-1247 for OS
3.2 Clinical Significance of SOX9 in OS
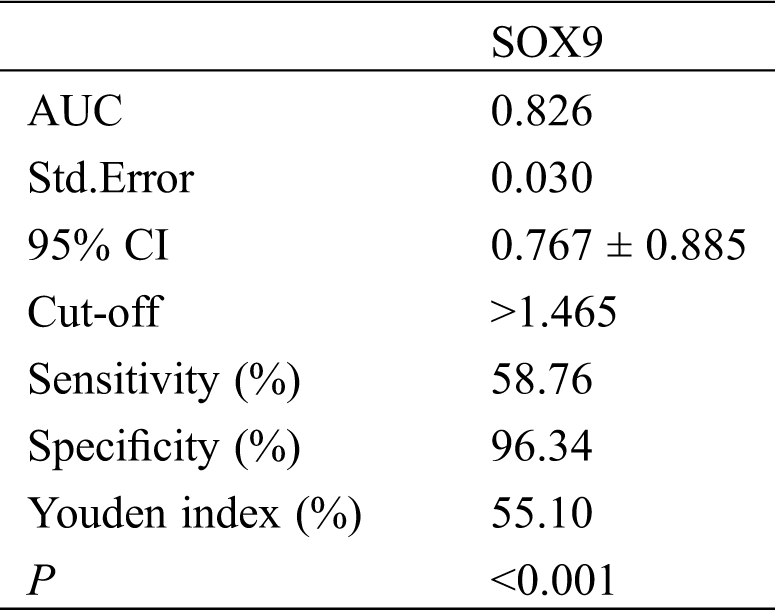
PCR results revealed high SOX9 mRNA expression in the peripheral blood of OS patients (P < 0.05). The ROC curve demonstrated that the sensitivity of SOX9 at the cut-off value of 1.465 for diagnosing OS was 58.76% and the specificity was 96.34%, as shown in Tab. 3 and Fig. 2.
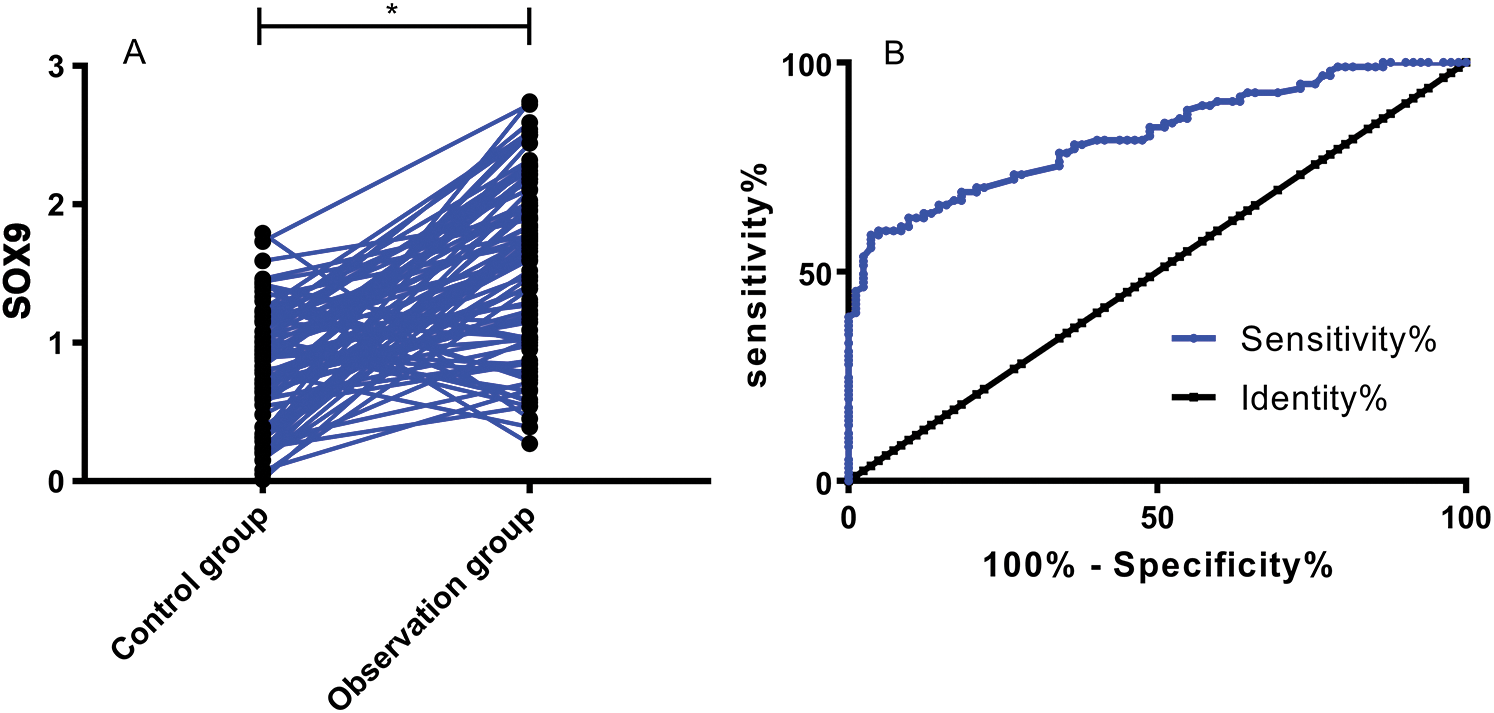
Figure 2: Clinical significance of SOX9 in OS. (A) Expression of SOX9 in OG and CG. *P < 0.05. (B) ROC curve demonstrating the performance of SOX9 in diagnosing OS
Table 3: Diagnostic performance of SOX9 for OS
We detected markedly lower expression of miR-1247 in 143B cells than in hFOB1.19 cells (P < 0.05). The miR-1247-mimics group showed markedly lower cell proliferation and invasion rates and higher apoptosis rates than the miR-1247-inhibitor group and the miR-NC group (P < 0.05), with the miR-1247-inhibitor group showing the highest cell proliferation and invasion rates and the lowest apoptosis rates (P < 0.05), as shown in Fig. 3.
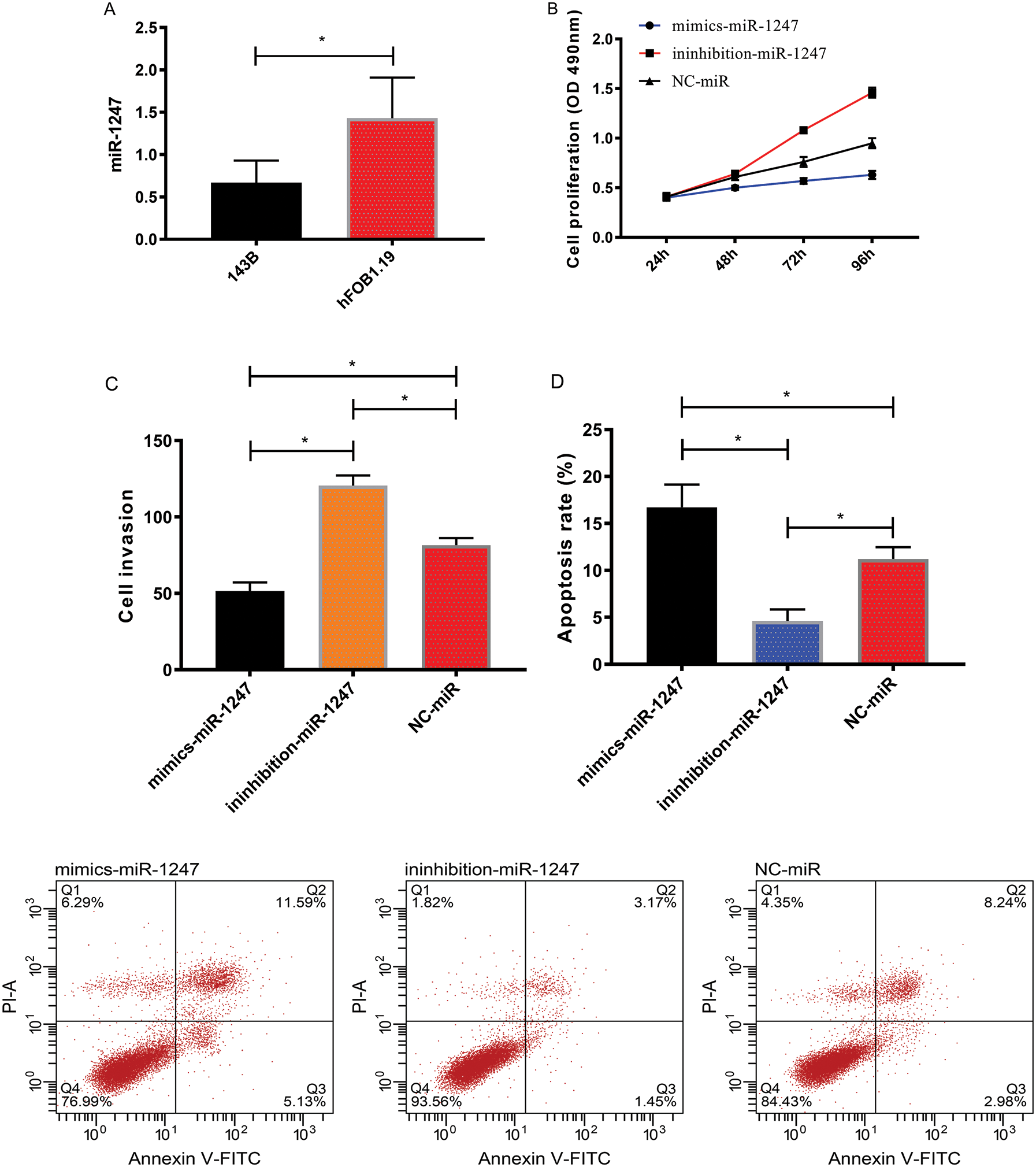
Figure 3: Effect of miR-1247 on OS. (A) MiR-1247 expression in 143B and hFOB1.19 cells. (B) Proliferation of 143B cells. (C) Invasion of 143B cells. (D) Apoptosis of 143B cells. *P < 0.05
We detected markedly higher expression of SOX9 in 143B cells than in hFOB1.19 cells (P < 0.05). The sh-SOX9 group showed higher cell proliferation and invasion rates and lower apoptosis rates than the si-SOX9 group and the NC group (P < 0.05), with the si-SOX9 group showing the lowest cell proliferation and invasion rates and the highest apoptosis rates (P < 0.05), as shown in Fig. 4.
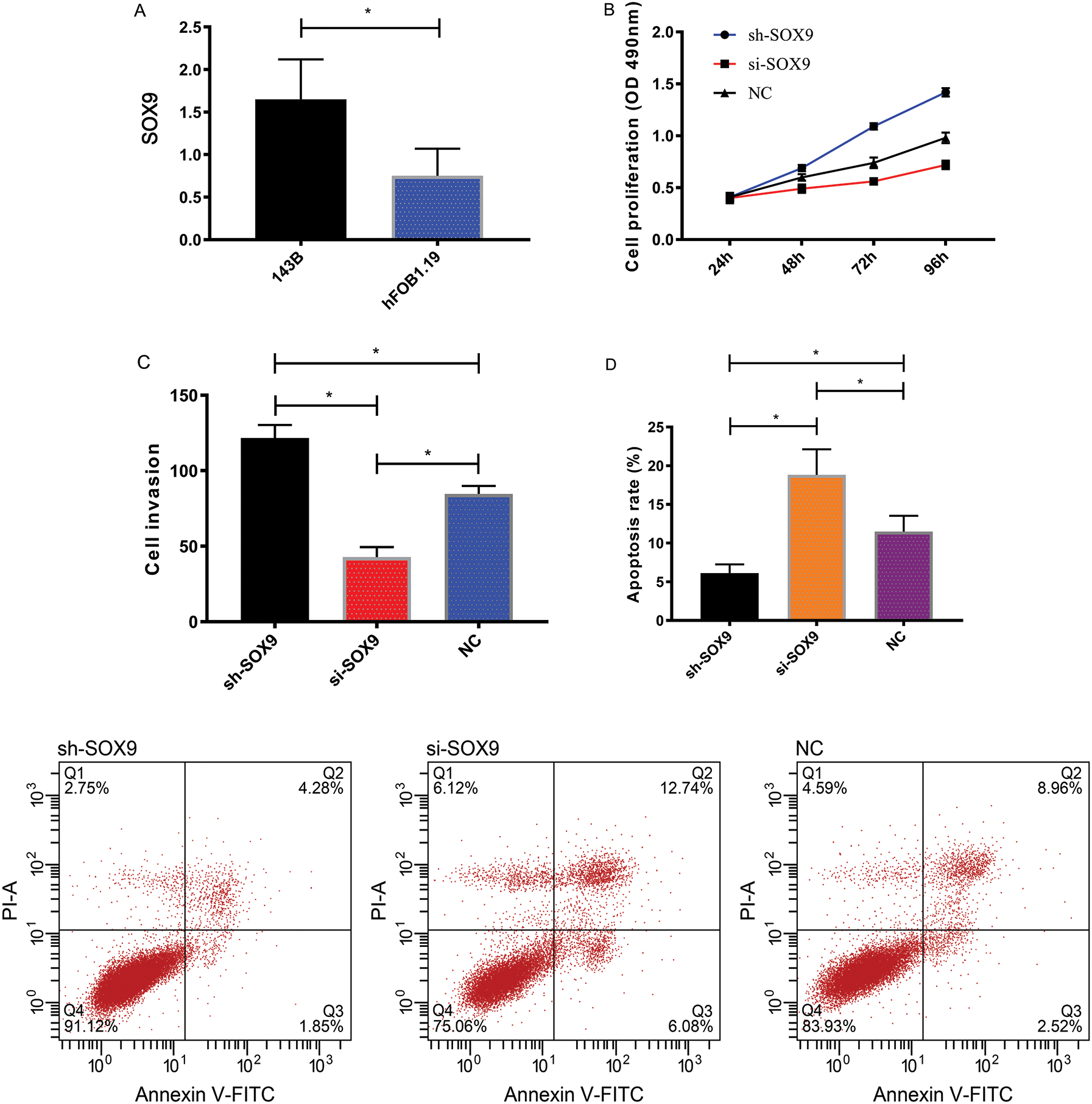
Figure 4: Effect of SOX9 on OS. (A) SOX9 expression in 143B and hFOB1.19 cells. (B) Proliferation of 143B cells. (C) Invasion of 143B cells. (D) Apoptosis of 143B cells. *P < 0.05
3.5 Relationship between miR-1247 and SOX9
SOX9 protein concentrations in 143B cells were the lowest in the miR-1247-mimics group, followed by the miR-NC group and the miR-1247-inhibitor group, and the differences between any two groups were statistically significant (all P < 0.05), as shown in Fig. 5.
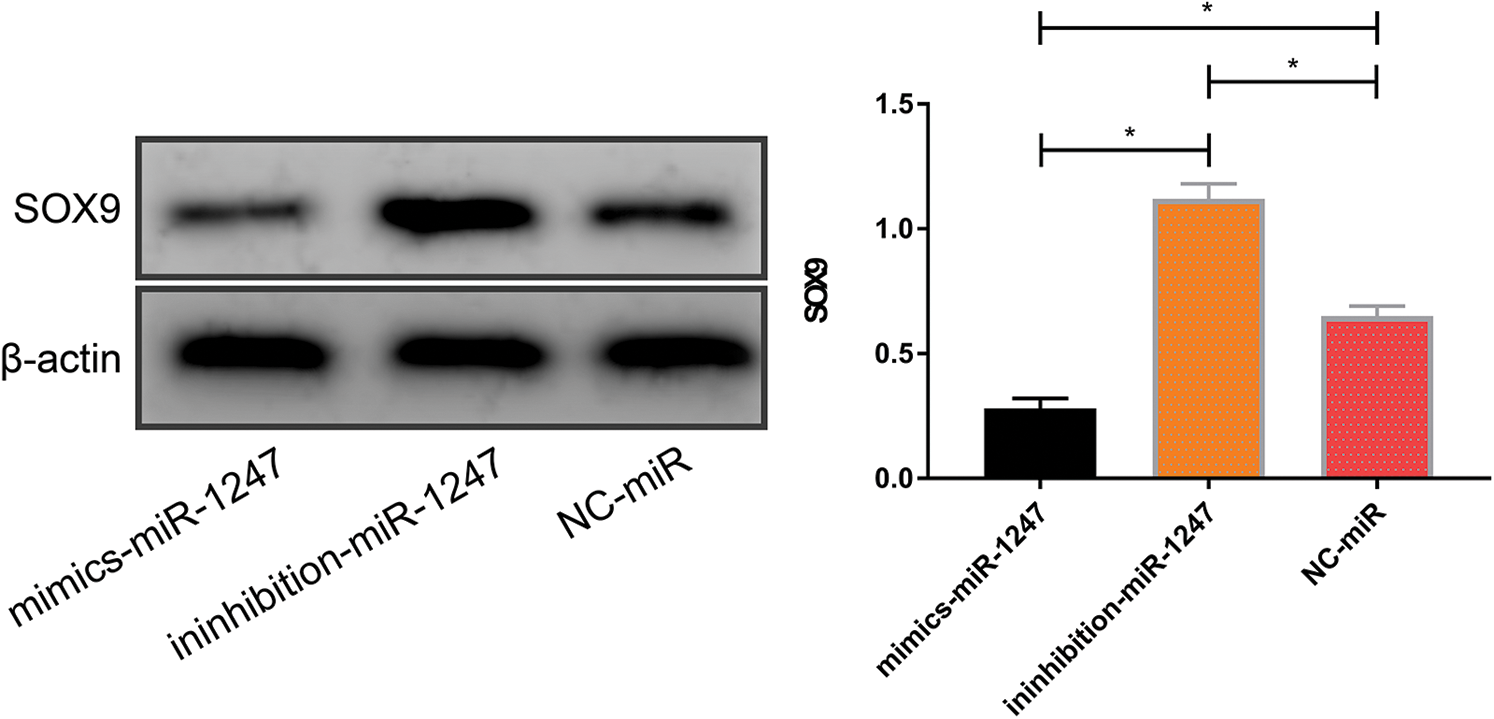
Figure 5: SOX9 protein concentrations in 143B cells after transfection with miR-1247-mimics, miR-1247-inhibitor, and miR-NC. *P < 0.05
Following the growing OS incidence year by year, the negative impacts of OS on patients are increasingly serious [18]. OS tends to affect adolescents or children [19]. The understanding of the pathological mechanism of OS in the clinic is effective for preventing the occurrence of OS. The molecular research on tumors is an existing hot topic in the clinic. Studies at home and abroad have revealed the involvement of miRNAs in tumors [20]. But the specific function of miR-1247 in OS needs to be further explored. Here we analyzed the effect of miR-1247 on OS cells, aiming to provide a reliable theoretical experimental basis for the OS diagnosis and treatment, which may become a breakthrough in OS researches.
To confirm the role of miR-1247 in OS, we tested the expression profiles of miR-1247 in OS patients and healthy participants. We detected lowly expressed miR-1247 in the peripheral blood of OS patients, which suggests that miR-1247 is linked to the development and progression of OS. A former study has revealed that miR-1247 expression is lowered in liver cancer [21], suggesting that miR-1247 expression is consistent in tumors and supports our results. We plotted the ROC curve to demonstrate the diagnostic performance of miR-1247 for OS. When the cut-off value hit 1.075, the sensitivity of miR-1247 for diagnosing OS was 85.57% and the specificity was 76.83%, indicating that miR-1247 is a promising diagnostic marker for OS. A past study has confirmed that miR-1247 is abnormally expressed in breast cancer [22]. We speculate that the confidence interval of miR-1247 in diagnosing various tumors can be calculated based on the big data analysis of clinical cases. MiR-1247 can also make up for the poor efficiency of traditional tumor markers in identifying tumor types [23]. Subsequently, we also detected highly expressed SOX9 in OS patients, which can initially suggest that SOX9 is related to the occurrence of OS. SOX9 can affect the cartilage osteogenesis to some extent [24]. OS occurrence is closely related to osteogenesis and bone destruction [25]. Such findings support our results from the side. The ROC curve of SOX9 revealed similar diagnostic performance to miR-1247 for OS. So, we speculate that the joint detection of miR-1247, SOX9, and conventional tumor markers in the future can remarkably enhance the early diagnosis rate of OS and improve the prognosis of patients.
The specific mechanism of miR-1247 and SOX9 in OS has not been figured out yet, so here we interfered with miR-1247 and SOX9 expression in OS cells to analyze the effects of the two genes on OS cells. In this study, 143B cells transfected with miR-1247-mimics/si-SOX9 showed markedly lower proliferation and invasion rates and markedly higher apoptosis rates than 143B cells transfected with miR-1247-inhibitor/sh-SOX9. Such results suggest that the up-regulation of miR-137 or down-regulation of SOX9 can effectively suppress OS progression, so they are promising therapeutic targets for OS in the future. The most effective treatment existing for OS is radical surgery, which is accompanied by a high probability of amputation [26], posing negative impacts on the growth of most OS patients (teens or children). Targeted therapy can achieve effective responses in cancer treatment without damaging the integrity of limbs, which is a great breakthrough for future clinical treatment of OS. This study merely revealed the potential of miR-1247 and SOX9 to work as therapeutic targets for OS, but did not illustrate the specific mechanism of action of miR-1247 and SOX9 in OS. As we mentioned above, we speculate that miR-1247 may regulate SOX9 to affect OS. Here we discovered that SOX9 expression was reduced in 143B cells transfected with miR-1247-mimics and enhanced in 143B cells transfected with miR-1247-inhibitor. Such results support our speculation that lowly expressed miR-1247 in OS promotes SOX9 expression to facilitate OS development and progression.
This study carried out a rudimentary analysis of the role of miR-1247 in OS and confirmed that miR-1247 regulates OS development by mediating SOX9. But there are still many deficiencies. For example, as mentioned above, the clinical value of miR-1247 and SOX9 needs to be verified by big data analysis of clinical cases and the ability of the two genes to function as therapeutic targets for OS needs to be tested by more in-depth experiments. We did not follow up on the prognosis of OS patients in this study, so we cannot illustrate the influence of miR-1247 and SOX9 on the prognosis of OS. Besides, we did not conduct nude mice tumorigenesis experiments, so we cannot explore the actual effects of miR-1247 and SOX9 on the tumorigenesis of OS. We will address such problems in future researches.
In summary, miR-1247 is lowly expressed in OS. It can promote OS cell proliferation and invasion and accelerate OS progression by mediating SOX9.
Funding Statement: The author(s) received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Lindsey, B. A., Markel, J. E., Kleinerman, E. S. (2017). Osteosarcoma overview. Rheumatology and Therapy, 4(1), 25–43. DOI 10.1007/s40744-016-0050-2. [Google Scholar] [CrossRef]
2. Misaghi, A., Goldin, A., Awad, M., Kulidjian, A. A. (2018). Osteosarcoma: A comprehensive review. SICOT-J, 4(1), 12. DOI 10.1051/sicotj/2017028. [Google Scholar] [CrossRef]
3. Harrison, D. J., Geller, D. S., Gill, J. D., Lewis, V. O., Gorlick, R. et al. (2017). Current and future therapeutic approaches for osteosarcoma. Expert Review of Anticancer Therapy, 18(1), 39–50. DOI 10.1080/14737140.2018.1413939. [Google Scholar] [CrossRef]
4. Rickel, K., Fang, F., Tao, J. (2017). Molecular genetics of osteosarcoma. Bone, 102, 69–79. DOI 10.1016/j.bone.2016.10.017. [Google Scholar] [CrossRef]
5. Sayles, L. C., Breese, M. R., Koehne, A. L., Leung, S. G., Lee, A. G. et al. (2019). Genome-informed targeted therapy for osteosarcoma. Cancer Discovery, 9(1), 46–63. DOI 10.1158/2159-8290.CD-17-1152. [Google Scholar] [CrossRef]
6. Whelan, J. S., Davis, L. E. (2018). Osteosarcoma, chondrosarcoma, and chordoma. Journal of Clinical Oncology, 36(2), 188–193. DOI 10.1200/JCO.2017.75.1743. [Google Scholar] [CrossRef]
7. Taran, S. J., Taran, R., Malipatil, N. B. (2017). Pediatric osteosarcoma: An updated review. Indian Journal of Medical and Paediatric Oncology: Official Journal of Indian Society of Medical & Paediatric Oncology, 38(1), 33–43.DOI 10.4103/0971-5851.203513. [Google Scholar] [CrossRef]
8. Chen, R., Wang, G., Zheng, Y., Hua, Y., Cai, Z. (2017). Long non-coding RNAs in osteosarcoma. Oncotarget, 8(12), 20462–20475. DOI 10.18632/oncotarget.14726. [Google Scholar] [CrossRef]
9. Dang, H. (2020). Diagnostic performance of serum miR-586 and miR-493 in patients with osteosarcoma. Oncologie, 22(1), 35–42. DOI 10.32604/oncologie.2020.012485. [Google Scholar] [CrossRef]
10. Chipman, L. B., Pasquinelli, A. E. (2019). miRNA targeting: Growing beyond the seed. Trends in Genetics, 35(3), 215–222. DOI 10.1016/j.tig.2018.12.005. [Google Scholar] [CrossRef]
11. Michlewski, G., Cáceres, J. F. (2019). Post-transcriptional control of miRNA biogenesis. RNA–A Publication of the RNA Society, 25(1), 1–16. DOI 10.1261/rna.068692.118. [Google Scholar] [CrossRef]
12. Vishnoi, A., Rani, S. (2017). MiRNA biogenesis and regulation of diseases: An overview. In: Rani, S. (ed.MicroRNA profiling. Methods in molecular biology. USA: Humana Press. [Google Scholar]
13. Fang, T., Lv, H., Lv, G., Li, T., Wang, C. et al. (2018). Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nature Communications, 9(1), 1–13. DOI 10.1038/s41467-017-02088-w. [Google Scholar] [CrossRef]
14. Yi, J. M., Kang, E. J., Kwon, H. M., Bae, J. H., Kang, K. et al. (2017). Epigenetically altered miR-1247 functions as a tumor suppressor in pancreatic cancer. Oncotarget, 8(16), 26600–26612. DOI 10.18632/oncotarget.15722. [Google Scholar] [CrossRef]
15. Taddei, M. L., Cavallini, L., Ramazzotti, M., Comito, G., Pietrovito, L. et al. (2019). Stromal-induced downregulation of miR-1247 promotes prostate cancer malignancy. Journal of Cellular Physiology, 234(6), 8274–8285. DOI 10.1002/jcp.27679. [Google Scholar] [CrossRef]
16. Wang, L., Zhang, Z., Yu, X., Li, Q., Wang, Q. et al. (2020). SOX9/miR-203a axis drives PI3K/AKT signaling to promote esophageal cancer progression. Cancer Letters, 468, 14–26. DOI 10.1016/j.canlet.2019.10.004. [Google Scholar] [CrossRef]
17. Dreval, K., de Conti, A.,Furuya, S., Beland, F. A., Rusyn, I. et al. (2017). miR-1247 blocks SOX9-mediated regeneration in alcohol-and fibrosis-associated acute kidney injury in mice. Toxicology, 384, 40–49. DOI 10.1016/j.tox.2017.03.004. [Google Scholar] [CrossRef]
18. Xiao, X., Wang, W., Li, Y., Yang, D., Li, X. et al. (2018). HSP90AA1-mediated autophagy promotes drug resistance in osteosarcoma. Journal of Experimental & Clinical Cancer Research, 37(1), 201. DOI 10.1186/s13046-018-0880-6. [Google Scholar] [CrossRef]
19. Yang, Y., Han, L., He, Z., Li, X., Yang, S. et al. (2018). Advances in limb salvage treatment of osteosarcoma. Journal of Bone Oncology, 10, 36–40. DOI 10.1016/j.jbo.2017.11.005. [Google Scholar] [CrossRef]
20. Cortini, M., Avnet, S., Baldini, N. (2017). Mesenchymal stroma: Role in osteosarcoma progression. Cancer Letters, 405, 90–99. DOI 10.1016/j.canlet.2017.07.024. [Google Scholar] [CrossRef]
21. Chu, Y., Fan, W., Guo, W., Zhang, Y. (2017). miR-1247-5p functions as a tumor suppressor in human hepatocellular carcinoma by targeting Wnt3. Oncology Reports, 38(1), 343–351. DOI 10.3892/or.2017.5702. [Google Scholar] [CrossRef]
22. Zhang, P., Fan, C., Du, J., Mo, X., Zhao, Q. (2018). Association of miR-1247-5p expression with clinicopathological parameters and prognosis in breast cancer. International Journal of Experimental Pathology, 99(4), 199–205. DOI 10.1111/iep.12287. [Google Scholar] [CrossRef]
23. Yu, L. L., Zhu, J., Liu, J. X., Jiang, F., Ni, W. K. et al. (2018). A comparison of traditional and novel methods for the separation of exosomes from human samples. BioMed Research International, 2018(1), 1–9. DOI 10.1155/2018/3634563. [Google Scholar] [CrossRef]
24. Lefebvre, V., Angelozzi, M., Haseeb, A. (2019). SOX9 in cartilage development and disease. Current Opinion in Cell Biology, 61, 39–47. DOI 10.1016/j.ceb.2019.07.008. [Google Scholar] [CrossRef]
25. Wang, W. T., Qi, Q., Zhao, P., Li, C. Y., Yin, X. Y. et al. (2018). miR-590-3p is a novel microRNA which suppresses osteosarcoma progression by targeting SOX9. Biomedicine & Pharmacotherapy, 107, 1763–1769. DOI 10.1016/j.biopha.2018.06.124. [Google Scholar] [CrossRef]
26. Kimura, Y., Tomihara, K., Tachinami, H., Imaue, S., Nakamori, K. et al. (2017). Conventional osteosarcoma of the mandible successfully treated with radical surgery and adjuvant chemotherapy after responding poorly to neoadjuvant chemotherapy: A case report. Journal of Medical Case Reports, 11(1), 1–6. DOI 10.1186/s13256-017-1386-0. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |