
 | Oncologie |  |
DOI: 10.32604/Oncologie.2021.012489
ARTICLE
Comprehensive Network Analysis of the Molecular Regulation Mechanism for Breast Cancer Metastasis
1Emergency Internal Medicine, Jiading District Central Hospital Affiliated Shanghai University of Medicine & Health Sciences, Shanghai, 201800, China
2Oncology Department, Jiading Hospital of Traditional Chinese Medicine, Shanghai, 201800, China
3Oncology Department, Jiading District Central Hospital Affiliated Shanghai University of Medicine & Health Sciences, Shanghai, 201800, China
*Corresponding Author: Lizhen Liu. Email: hsg785101666@163.com
Received: 01 June 2020; Accepted: 20 August 2020
Abstract: Breast cancer is one of malignant severe diseases that cause cancer death in women. Although research about the pathogenesis and studies about treatment mechanisms in breast cancer have become clear focuses, we have no clear conclusion yet. Therefore, this research is based on a modular approach to explore key factors and molecular mechanisms that affect breast cancer metastasis. First of all, it is necessary to download breast cancer-related data on the GEO database, and we analyzed the difference between primary tumors and metastatic lesions to obtain differential gene expression profiles. On this basis, a series of bioinformatics analyses were performed to comprehensively, and they were presented to identify critical regulators in breast cancer metastasis. We have obtained a total of five co-expression modules, among which HECW1, FBN1, and other genes have effective regulation in dysfunction modules, and thus they would be recognized as driving genes for breast cancer metastasis. Module genes were significantly enriched in biological function, for instance, leukocyte-cell adhesion and negative regulation in the immune system process. At the same time, it substantially regulates signaling pathways, for example, fatty acid degradation, synthesis, and degradation of ketone bodies, and amino acid metabolism. Finally, we identified ncRNA pivots (including FENDRR, miR-19a-3p, and miR-26b-5p) and TF pivot (including NFKB1 and SP1) to regulate dysfunction modules significantly. Our study identified the coexpression network of genes involved in breast cancer metastasis. These results may be helpful to reveal the gene modules and regulatory factors of breast cancer. Importantly, we identified a long non-coding FENDRR that inhibits breast cancer metastasis through a fatty acid degradation signaling pathway, providing new directions and targets for subsequent studies.
Keywords: Breast cancer metastasis; enrichment analysis; co-expression analysis; regulatory factors
Breast cancer (BC) is one of the most common female malignancies in the world [1]. In the past few years, significant progress has been made in the treatment of breast cancer, but metastatic breast cancer (MBC) remains incurable, and as a result, mortality in breast cancer patients remains high [2,3]. Insomnia in the context of breast cancer is both an independent symptom and includes other symptom groups such as depression, anxiety, fatigue and pain [4]. Similarly, breast cancer patients have physical discomfort and pain that further affect disease progression, quality of life and prognosis survival [5]. After the diagnosis of breast cancer, the occurrence of complications of the nervous system is widespread, and neuropathic pain (NP) has always affected the quality of life of patients [6]. Also, in patients undergoing breast surgery, metastatic patients are more likely to experience infectious, respiratory, thromboembolic, cardiac, and hemorrhagic complications than non-metastatic patients [7]. Studies have shown that family history and therapeutic chest radiography are high-risk factors for breast cancer development [8]. In genetics, more than 70 single nucleotide polymorphisms (SNPs) are correlated with the development and progression of breast cancer [9]. Among them, the genetic polymorphisms of osteoprotegerin (OPG) and RANK-ligand (RANKL) are correlated with free survival of bone metastasis (BM) and BC risk, respectively [10]. At the same time, PI3KR1-gene polymorphism is associated with metastasis in breast cancer patients, and polymorphism can also be used for overall predictive scoring to detect early CNS (Central Nervous System) metastasis [11]. At present, many studies have explored the mechanism of breast cancer metastasis and made some progress.
IL-19 can directly promote proliferation and migration, which can provide a microenvironment for tumor development, and believe that IL-19 is a prognostic marker of breast cancer, and it may become a potential therapeutic target [12]. Also, adenovirus infection with CXCL12 (Protein Chemokines SDF) vector up-regulated CXCL12 expression significantly inhibited cell growth and reduced breast cancer cell migration, which is a potential prognostic marker for breast cancer patients [13]. Up-regulation of miR-96/miR-182 can reduce the level of Paladin protein, thereby inhibiting the migration and invasion of breast cancer cells [14]. On the other hand, at low concentrations, ROS can promote cancer cell survival by activating growth factors and MAP kinase (MAPK). At high frequencies, ROS produce oxidative stress that stimulates cell apoptosis [15]. In the treatment of breast cancer, Stages I and II breast cancers are usually treated with breast-conserving surgery and radiation therapy. Stage III breast cancer often requires induction of rejuvenation. The prognosis of patients with metastatic (Stage IV) breast cancer is reduced, so the treatment plan must balance the length of life and reduce the treatment of pain to set [16].
To explore the mechanism of breast cancer cell metastasis, we conducted a systematic modular analysis to determine the dysfunction modules and core molecules between them to exploit the most critical genes for breast cancer metastasis further. Collectively, our work details the long non-coding FENDRR inhibition of breast cancer metastasis through fatty acid degradation signaling pathways and identifies potential therapeutic targets and related biological processes. This provided a rich resource for future treatment and indicated a new direction for subsequent research.
We first collected a set of gene interpretation profiles for primary breast cancer and metastatic breast cancer from Gene Expression Omnibus (GEO) database [17], numbered GSE33116. The data set included 149 primary breast cancers and 23 metastatic breast cancers. We then screened the ncRNA mRNA regulatory pairs by setting the score ≥ 0.5 using the RAID v2.0 database [18]. In parallel, we obtained transcriptional regulators from the general database of transcriptional studies (TRUST v2 database) [19].
2.2 Differentially Expressed Gene
The background correction and normalization for GSE33116 were performed using lumi R package. Then the difference of metastatic breast cancers and primary breast cancers was analyzed using limma R package [20–22]. We obtained differentially expressed genes (DEGs) by setting p-value < 0.05.
The weighted gene co-expression network analysis (WGCNA) [23] was used to explore the synergistic expression behavior of DEGs. Based on the magnitude of the regulatory power of the gene in each dysfunctional module, we explored the essential genes that lead to dysfunctional modules, which are thought to be critical genes responsible for breast cancer cell proliferation and metastasis.
The functions and signal transduction involved in module genes were performed using enrichment analysis. Therefore, we performed a GO function and a KEGG pathway for the breast cancer metastasis-related module genes through the Clusterprofiler R package [24]. Besides, the BinGO and ClueGO functions of Cytoscape were used to analyze the integrated module network.
2.5 Transcription Factors and ncRNAs that Regulate Dysfunctional Modules
We have predicted regulatory roles for non-coding genes (ncRNA) and transcription factors (TF) in a dysfunctional module. Pivot regulator is calculated based on hypergeometric test and defined as a regulator with a significant regulatory function in the module with p-value < 0.01.
2.6 Quantitative Real-Time PCR
Total RNA of whole blood was extracted using Trizol. The RNA was then reverse transcribed into cDNA by using a reverse transcription kit. The PCR reaction was performed through SYBR qPCR Kit (Invitrogen, Carlsbad, CA, USA). The internal reference genes were beta-actin and U6.
3.1 Identifying Metastasis-Related Disorders in Breast Cancer
To observe molecular changes during breast cancer metastasis, we performed differential analysis for GSE33116. The DEGs between primary breast cancer and metastatic breast cancer was identified, leading to critical factors that may lead to breast cancer metastasis. The results showed that we had a total of 5,620 differential genes (Tab. S1), and we thought that these differential genes were correlated with breast cancer metastasis.
3.2 A Functional Module for Identifying Breast Cancer Metastasis
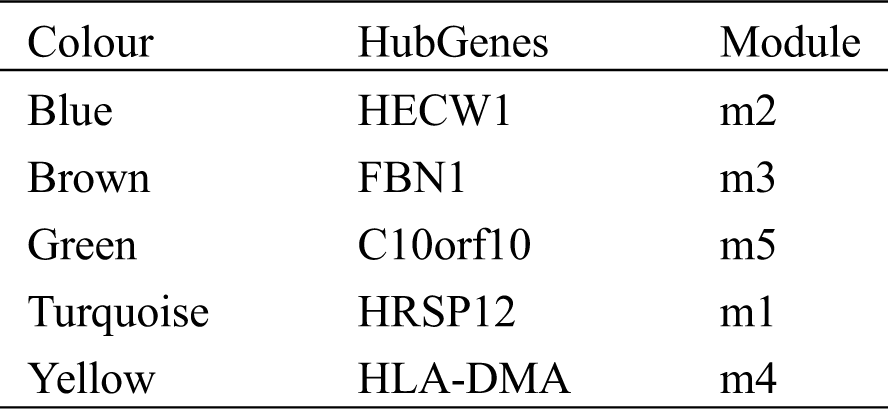
Initially, we constructed an expression profile matrix based on 5,620 DEGs for breast cancer in patient samples. Then, according to WGCNA, genes which exhibited significant group coexpression were observed in disease samples. We identified five coexpression modules with functional disorder (Figs. 1A and 1B). Based on the functional disorder module to identify the critical genes of each module, we obtained core genes based on HECW1, FBN1 (Tab. 1). Further correlating the module with phenotypic data, ME turquoise can be seen, while ME brown is associated with primary breast cancer, while ME blue is associated with metastatic breast cancer (Fig. 1C).
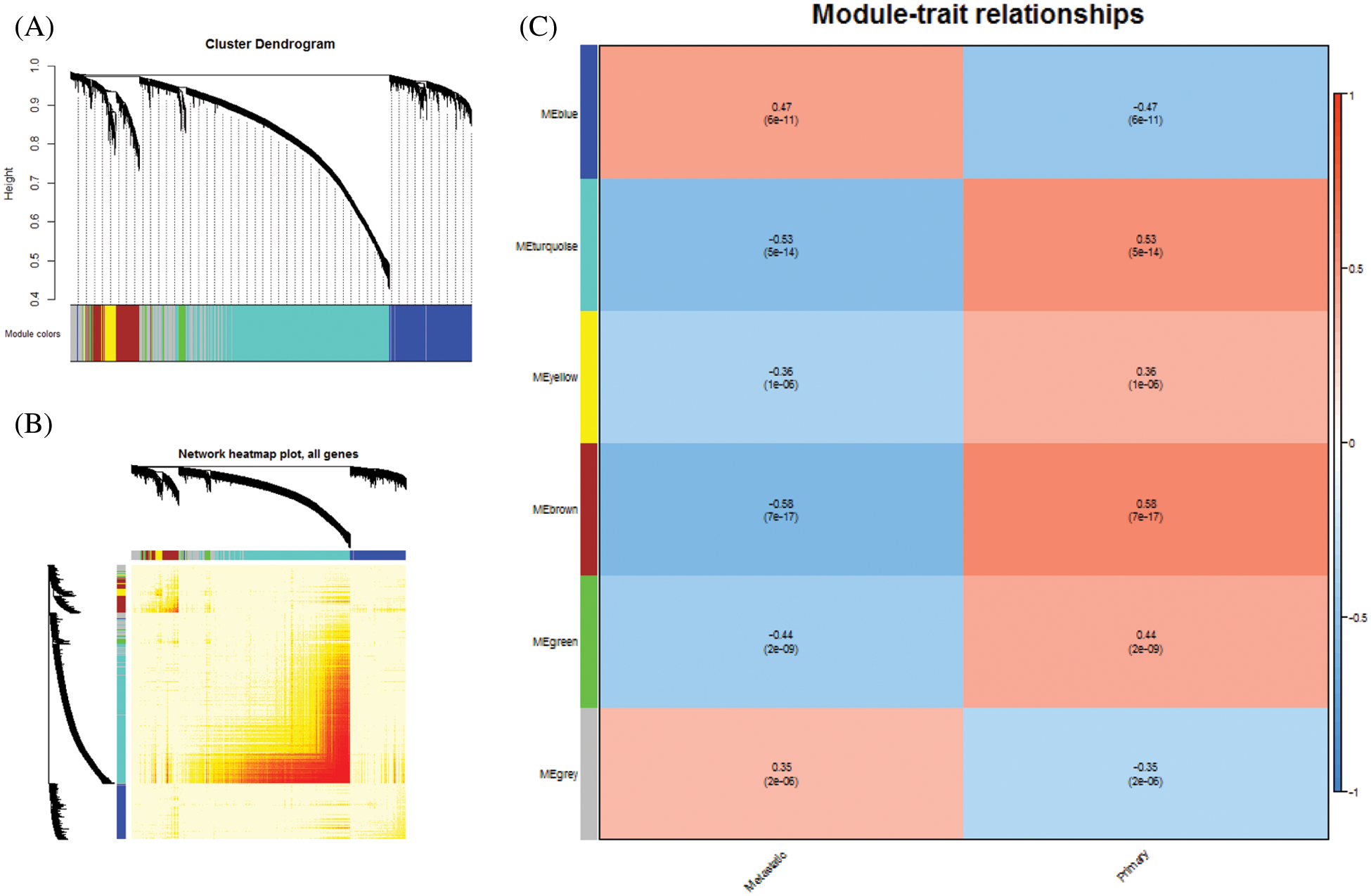
Figure 1: Coexpression analysis of differentially expressed genes. A. Module clustering tree of differentially expressed genes. Five colors represented five co-expression modules. B. Clustering Heat Map of Five Modules. C. Modular association of breast cancer phenotypes. The corresponding correlation coefficient maps the color of each cell. Number from −1 to 1, color from blue to white, and then to red
3.3 Module Genes Involved in Functions and Pathways
By performing GO function and KEGG pathway enrichment analysis on five modules, we obtained 28,448 biological processes, 3,454 cells, 5,961 molecular functions, and 45 KEGG pathways (Tab. S2, Figs. 2A and 2B). At the same time, we found that the purpose is mainly focused on the regulation of immune system processes, tumor necrosis factor response and bone marrow cell differentiation. Equally important, the enrichment results of the KEGG pathway reflect that breast cancer differential genes are mainly involved in fatty acid degradation, ketone body synthesis and degradation, and amino acid metabolism. For the functions and pathways that regulate the most genes in the dysfunctional module, it can be considered to play the most critical role in modules. We integrated the five modular networks and analyzed the biological functions and signaling pathways using Cytoscape software (Figs. 3A and 3B).
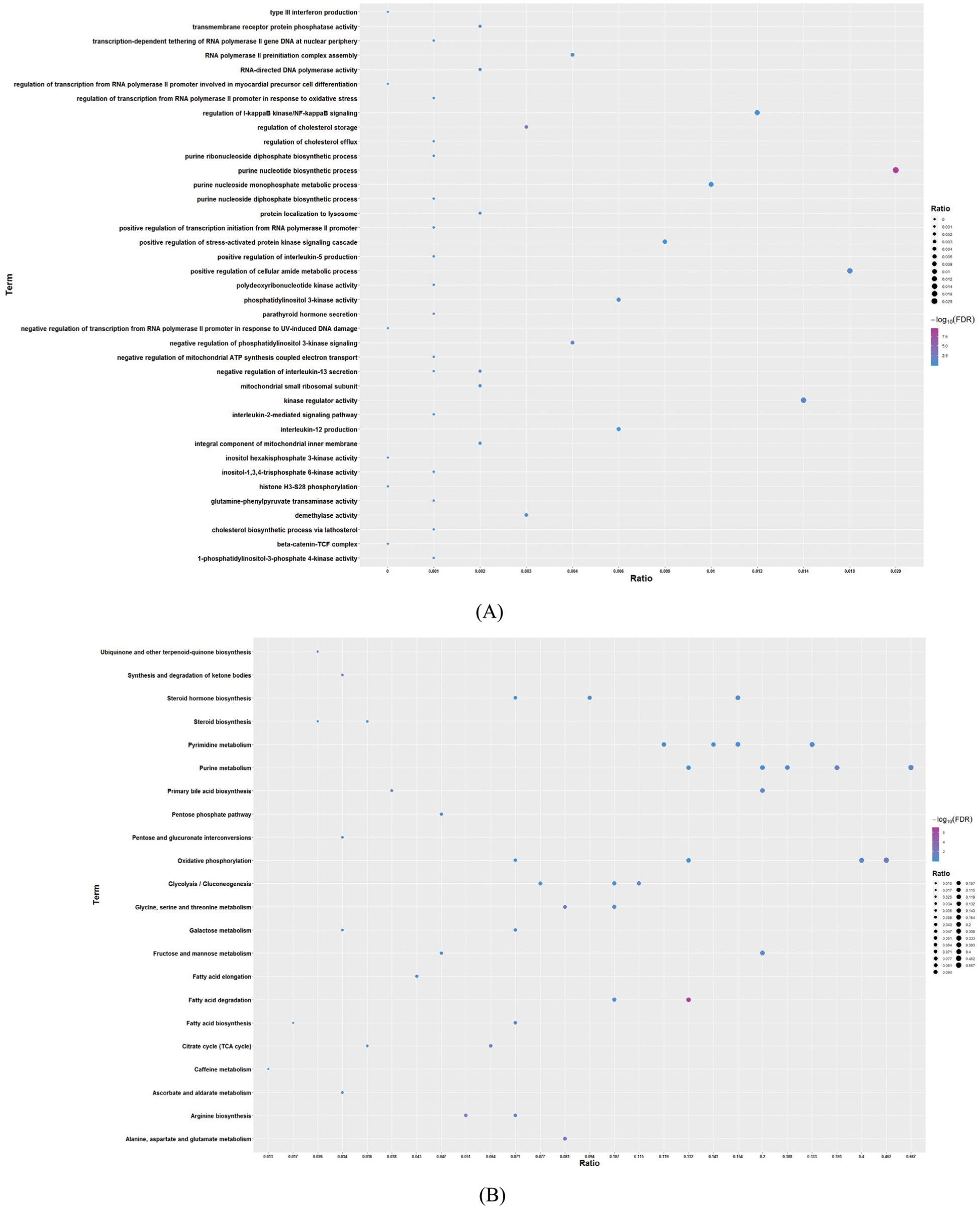
Figure 2: Enrichment analysis of GO and KEGG pathway for modular genes (excerpts). A. GO results of enrichment analysis. The larger the circle, the greater the number of genes. B. KEGG pathway results of enrichment analysis. The larger the circle, the greater the number of genes
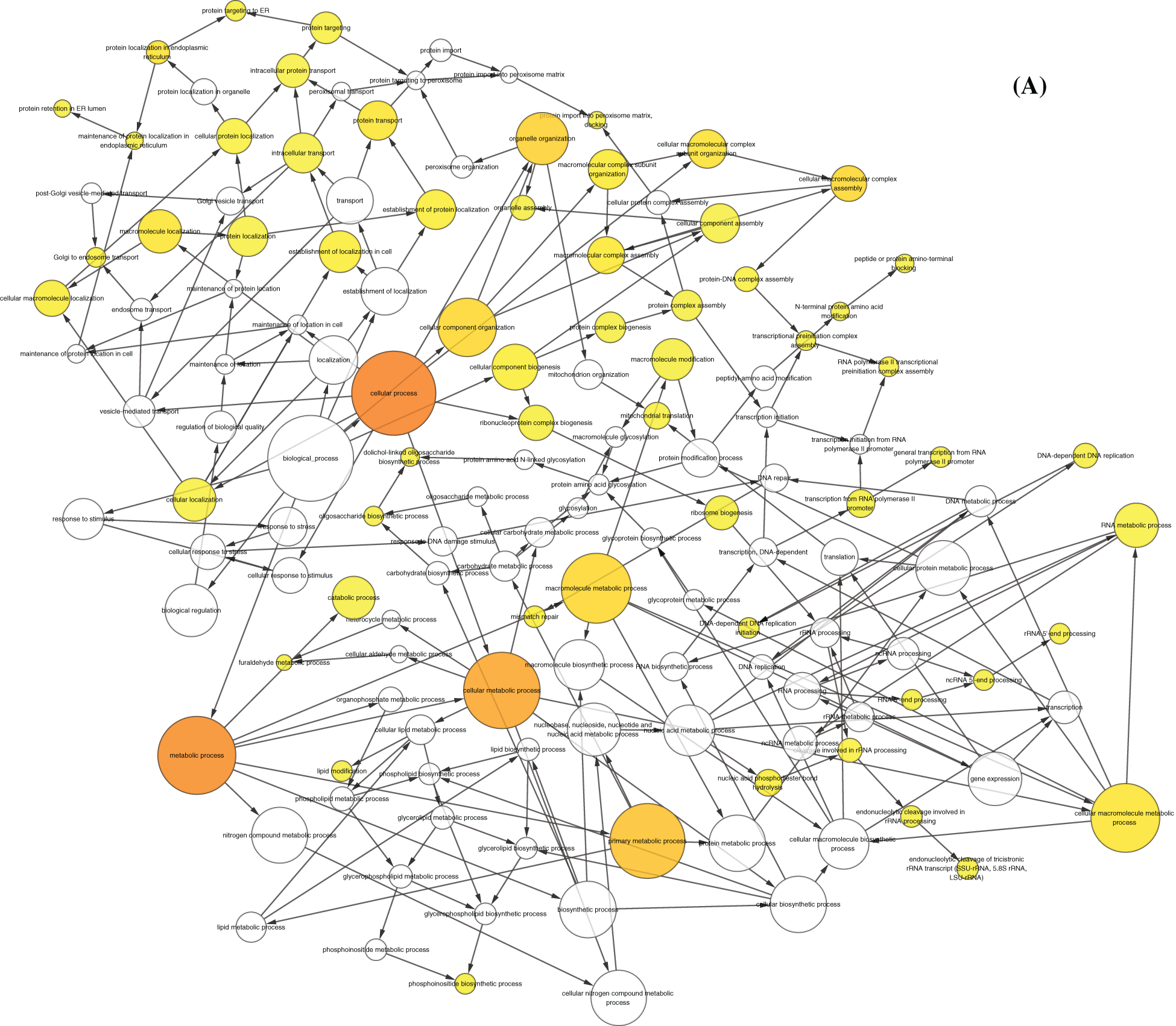

Figure 3: Function and KEGG pathway analysis of modular network. A. Network diagram of module participation function. The larger and darker the nodes, the more modules involved in the biological function. B. Network diagram of module participation signal transduction. The larger and darker the nodes, the more modules involved in the signal transduction signal pathway
3.4 TF and ncRNA that Drive Breast Cancer Progression
Based on the regulatory relationship between TF, ncRNA and module genes, a pivot analysis for module genes was performed to explore key regulators in breast cancer metastasis. The predicted results (Tabs. S3 and S4) showed that there were 962 ncRNAs involved 1261 ncRNA-module regulatory pairs, 92 transcription factors affected 102 TF and target genes pairs. Cytoscape software was used to demonstrate these interaction relationship networks (Figs. 4A and 4B). According to the statistical analysis, FENDER, miR-19a-3p, miR-26b-5p (ncRNA) and NFKB1, SP1 (TF) were identified as key regulators. They may regulate breast cancer cell proliferation and migration by mediating dysregulated expression of module genes. In addition, through qPCR method, we found the expression of key genes was consistent with the analysis results (Fig. 5).
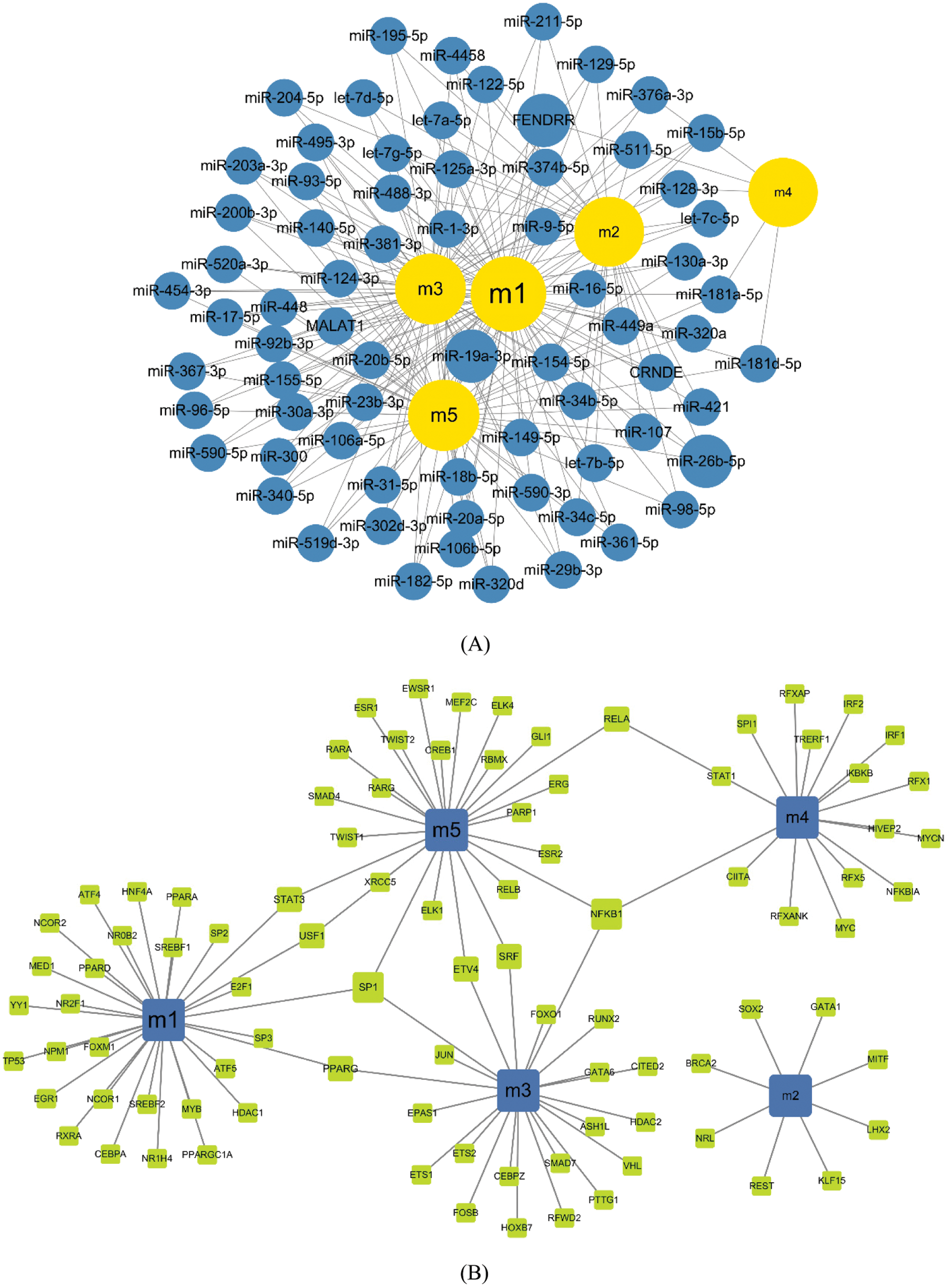
Figure 4: Modulatory effects of regulatory factors on the module. A. Modular regulation by ncRNA. Yellow nodes represent modules, and blue nodes represent ncRNA. The larger the nodes, the more modules they control. B. Modular regulation by TF. Blue nodes represent modules, and green nodes represent TF. The larger the nodes, the more modules they represent
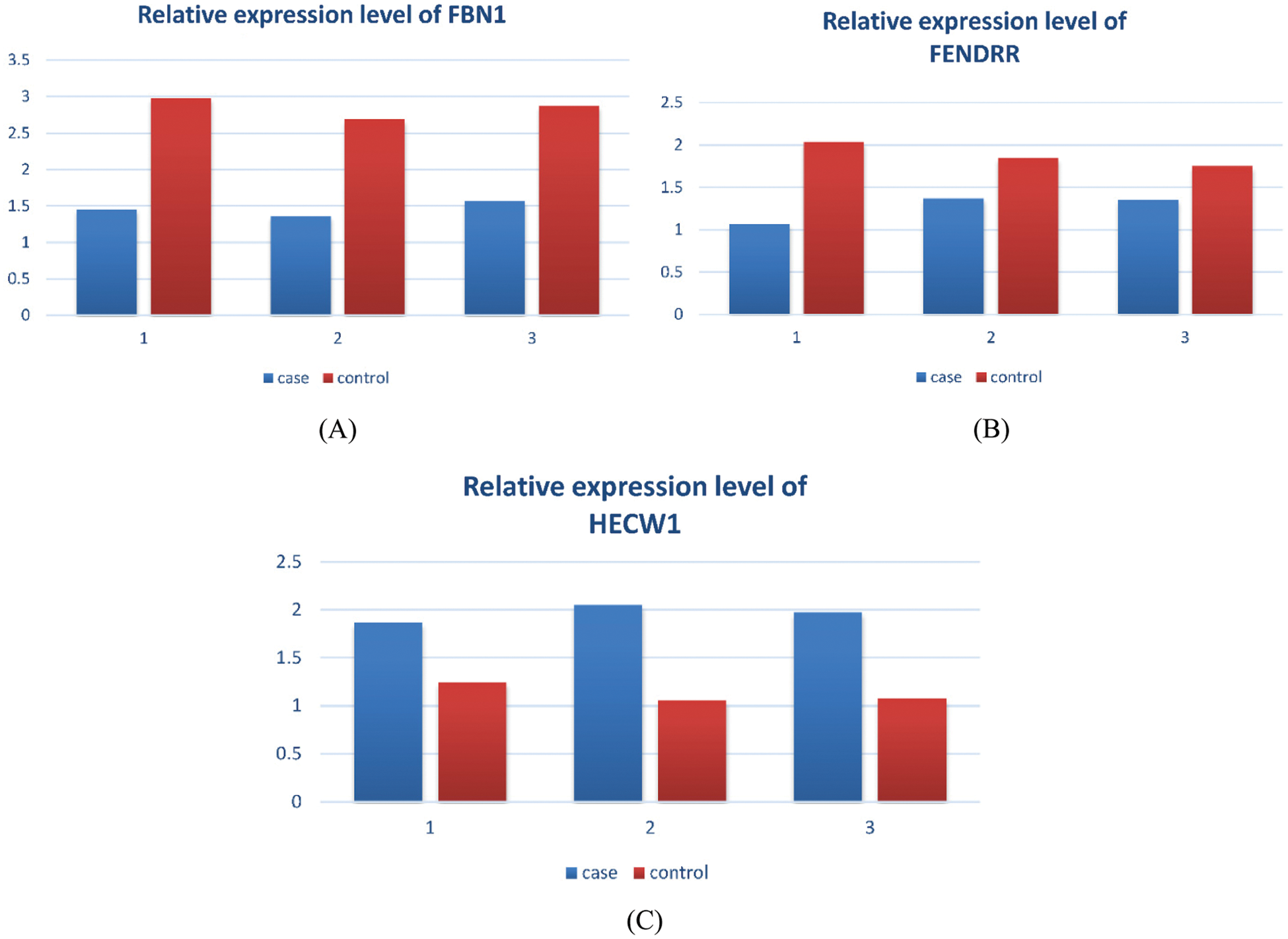
Figure 5: Expression levels of key genes. A. Relative expression level of FBN1. B. Relative expression level of FENDRR. C. Relative expression level of HECW1
Breast cancer, in women, it is commonly seen. Moreover, metastasis is the underlying cause of death in most breast cancer patients [25,26]. Despite significant advances in cancer research, breast cancer remains a significant health problem and a top priority for biomedical research [27]. More importantly, metastatic breast cancer is incurable regardless of age. Current treatments can only reduce symptoms and provide patients with the best quality of life for as long as possible [28]. In this study, we collected primary breast cancer and genes for metastasis to breast cancer based on the NCBI Gene Expression Omnibus database.
Differential gene expression profile data further analyzed the molecular mechanism of fine metastasis of breast cancer. Module genes were significantly involved in biological processes, for example, leukocyte-cell adhesion, regulation of cell-cell adhesion, negative regulation of the immune system process, response to tumor necrosis factor, and differentiation of bone marrow cells. At the same time, the module genes were also enriched in the signaling pathway, for example, fatty acid degradation, synthesis, and degradation of ketone bodies, and metabolism of alanine, aspartic acid, and glutamic acid. Studies have shown that fatty acid synthesis and oxidized proteins have significant functions in the proliferation, migration, and invasion of breast cancer cells. Thus, the imbalance of fatty acid metabolism is considered to be a component of the malignant transformation of breast cancer [29]. More importantly, most cancer cell types require fatty acids (FA), which are strictly related to the critical signaling molecules that stimulate BC cell proliferation [30]. Besides, studies have shown that ketone body formation and ketone inhibitors can be designed as novel therapies that are effective in treating tumor recurrence and metastatic disease in patients with advanced cancer [31].
On the other hand, critical metabolic pathways, for instance, like taurine, hypotaurine metabolism, alanine, aspartate, and glutamate pathways, are used for early diagnosis of breast cancer [32]. Besides, the module’s genes are also involved in the signal transduction. For example, it is transforming growth factor beta, apoptosis, cell cycle, NF-κB, Wnt, Hippo. The signal transductions are involved in the proliferation and migration of breast cancer cells. Among them, miR-153 inhibits the migration, invasion and epithelial-mesenchymal transition of breast cancer by regulating the transforming growth factor beta (TGF-β) signal transduction [33]. Overexpression of miR-99a-5p and decreased expression of CDC25A can inhibit proliferation and invasion of breast cancer cells, promote apoptosis, and significantly activate cell cycle progression [34]. Also, through NF-κB and Wnt/β-catenin signal transduction, Antrodia camphorata reverses EMT and inhibits invasion and metastasis of triple-negative breast cancer cells [35]. Down-regulation of long non-coding RNA inhibits invasion and migration of breast cancer cells via FAT4-dependent Hippo signal transduction [36].
On the other hand, we had extracted five hub genes in five modules, for instance, HECW1, and FBN1. Among them, HECW1 can regulate the expression of ErbB4 protein in T47D, and ErbB4 has a necessary effect on breast cancer metastasis by regulating the proliferation, survival, and differentiation of mammary epithelial cells [37]. At the same time, recent studies have shown that FBN1 is correlated with breast cancer or other tissue types of cancer [38]. Also, the expression level of C10orf10 is decreased in breast cancer tissues, and the low expression of C10orf10 may be an important prognostic factor for poor survival time of breast cancer patients [39]. Also, studies have found that some cytosines in the promoter region may have a particular significance for the expression of the p14.5 gene during cell proliferation and cancer development [40]. On the other hand, tumor cell expression of HLA-DM is correlated with Th1 profiles and predicts survival in breast cancer patients [41]. These genes, as critical genes in the dysfunction module, have a driving role in affecting breast cancer metastasis.
Also, we predicted that 962 ncRNAs participate in breast cancer metastasis mechanisms by regulating module genes. According to statistical analysis, we identified FENDRR, miR-19a-3p, and miR-26b-5p had significant effect on the dysfunctional modules and that they were the genes of the most regulated modules. Among them, FENDRR has been shown to inhibit breast cancer cell proliferation, promote apoptosis, and correlate with good prognosis of breast cancer [42]. By down-regulating the expression of the Fra-1 proto-oncogene, MicroRNA-19a-3p induces macrophage polarization to inhibit breast cancer progression and metastasis [43]. Meanwhile, miR-19a-3p (miR-19a) was identified as a mediator of cell proliferation inhibition by CAP in MCF-7 breast cancer cells [44]. Progesterone receptor A promotes invasion and metastasis of luminal breast cancer by inhibiting estrogen regulation of key microRNAs (miR-26b-5p, miR-92a-3p) [45]. Down-regulation of the TRPS1 protein of the miR-26b-5p transcriptional target is correlated with radiation exposure. After TRPS1 knockdown, the dysregulated genes are associated with DNA repair, mitosis, angiogenesis, and EMT pathways [46]. At the same time, other ncRNAs we identified may regulate breast cancer module genes, may also be involved in the primary process of breast cancer metastasis, which need to be verified by further molecular experiments.
Finally, we analyzed important transcription factors regulated module genes. Of these, NFKB1, SP1 significantly regulated three modules; however, these regulators are essential in metastatic breast cancer. In metastatic breast cancer, it was demonstrated that targeting NFATc2 and NFKB1/RELA interactions can appropriately regulate Ets1 gene interpretation. This is since Ets1-mediated metastatic breast cancer is a potential therapeutic target [47]. At the same time, NF-κB contributes to the definition of MMP1 in breast cancer spheres, leading to activation and disintegration of paracrine PAR1 in the lymphatic endothelial barrier in vitro [48]. Also, they provide a theoretical basis for the effective treatment of metastatic breast cancer in regulating the migration and invasion of breast cancer cell lines [49].
On the other hand, miR-3178 is a target in Sp1 in a variety of cancer cell models, and overexpression of miR-3178 can inhibit migration and invasion of cells in highly metastatic prostate cancer, lung cancer and breast cancer [50]. More importantly, oncogenic HBXIP accelerates the growth of breast cancer by co-activating Sp1 to control the transcriptional regulation of ZEB1 [51]. At the same time, for the provision of metastatic breast cancer dysfunction module, transcription factors may be involved in the primary process of asthma, but this needs to be confirmed by experiments. Finally, through the integrated landscape of the breast cancer staging mechanism, we obtained FENDRR that modulates the most dysfunctional module, which significantly regulates the highest signal transduction and degrades fatty acids. Furthermore, by modulating the fatty acid degradation pathway, we can conclude that FENDRR inhibits the proliferation and migration of breast cancer cells.
Funding Statement: The author(s) received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Ferlay, J., Shin, H. R., Bray, F., Forman, D., Mathers, C. et al. (2010). Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. International Journal of Gynecological Cancer, 127(12), 2893–2917. DOI 10.1002/ijc.25516. [Google Scholar] [CrossRef]
2. D’Hondt, R., Spoormans, I., Neyens, N., Mortier, N., Van Aelst, F. (2014). Survival of patients with metastatic breast cancer: A single-centre experience. Acta Clinica Belgica, 69(3), 194–199. [Google Scholar]
3. Maddams, J. B. D., Gavin, A., Steward, J., Elliott, J., Utley, M. et al. (2008). Cancer prevalence in the United Kingdom: Estimates for 2008. British Journal of Cancer, 101(3), 541–547. DOI 10.1038/sj.bjc.6605148. [Google Scholar] [CrossRef]
4. Maddams, J., Utley, M., Moller, H. (2015). Projections of cancer prevalence in the United Kingdom, 2010–2040. British Journal of Cancer, 107(7), 1195–1202. DOI 10.1038/bjc.2012.366. [Google Scholar] [CrossRef]
5. Luengo-Fernandez, R., Leal, J., Gray, A., Sullivan, R. (2013). Economic burden of cancer across the European Union: A population-based cost analysis. Lancet Oncology, 14(12), 1165–1174. [Google Scholar]
6. Fontes, F., Pereira, S., Castro-Lopes, J. M., Lunet, N. (2016). A prospective study on the neurological complications of breast cancer and its treatment: Updated analysis three years after cancer diagnosis. Breast, 29, 31–38. [Google Scholar]
7. Thompson, A., Brennan, K., Cox, A., Gee, J., Harcourt, D. et al. (2008). Evaluation of the current knowledge limitations in breast cancer research: A gap analysis. Breast Cancer Research, 10(2), R26. DOI 10.1186/bcr1983. [Google Scholar] [CrossRef]
8. Yalcin, B. (2013). Staging, risk assessment and screening of breast cancer. Experimental Oncology, 35(4), 238–245. [Google Scholar]
9. Melchor, L., Benitez, J. (2013). The complex genetic landscape of familial breast cancer. Human Genetics, 132(8), 845–863. DOI 10.1007/s00439-013-1299-y. [Google Scholar] [CrossRef]
10. Michailidou, K., Hall, P., Gonzalez-Neira, A., Ghoussaini, M., Dennis, J. et al. (2013). Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nature Genetics, 45(4), 353–361. DOI 10.1038/ng.2563. [Google Scholar] [CrossRef]
11. Sakoda, L. C., Jorgenson, E., Witte, J. S. (2013). Turning of COGS moves forward findings for hormonally mediated cancers. Nature Genetics, 45(4), 345–348. DOI 10.1038/ng.2587. [Google Scholar] [CrossRef]
12. Antoniou, A. C., Beesley, J., McGuffog, L., Sinilnikova, O. M., Healey, S. et al. (2010). Common breast cancer susceptibility alleles and the risk of breast cancer for BRCA1 and BRCA2 mutation carriers: Implications for risk prediction. Cancer Research, 70(23), 9742–9754. DOI 10.1158/0008-5472.CAN-10-1907. [Google Scholar] [CrossRef]
13. Ingham, S., Warwick, J., Byers, H., Lalloo, F., Newman, W. et al. (2013). Is multiple SNP testing in BRCA2 and BRCA1 female carriers ready for use in clinical practice? Results from a large Genetic Centre in the UK. Clinical Genetics, 84(1), 37–42. DOI 10.1111/cge.12035. [Google Scholar] [CrossRef]
14. Audeh, M. W., Carmichael, J., Penson, R. T., Friedlander, M., Powell, B. et al. (2010). Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: A proof-of-concept trial. Lancet, 376(9737), 245–251. DOI 10.1016/S0140-6736(10)60893-8. [Google Scholar] [CrossRef]
15. Turnbull, C., Seal, S., Renwick, A., Warren-Perry, M., Hughes, D. et al. (2012). Gene–gene interactions in breast cancer susceptibility. Human Molecular Genetics, 21(4), 958–962. DOI 10.1093/hmg/ddr525. [Google Scholar] [CrossRef]
16. Muller, H. M., Widschwendter, A., Fiegl, H., Ivarsson, L., Goebel, G. et al. (2003). DNA methylation in serum of breast cancer patients: An independent prognostic marker. Cancer Research, 63, 7641–7645. [Google Scholar]
17. Chlebowski, R. T., Anderson, G. L., Gass, M., Lane, D. S., Aragaki, A. K. et al. (2010). WHI Investigators. Estrogen plus progestin and breast cancer incidence and mortality in postmenopausal women. JAMA, 304(15), 1684–1692. DOI 10.1001/jama.2010.1500. [Google Scholar] [CrossRef]
18. Anderson, G. L., Chlebowski, R. T., Aragaki, A. K., Kuller, L. H., Manson, J. E. et al. (2012). Conjugated equine oestrogen and breast cancer incidence and mortality in postmenopausal women with hysterectomy: Extended follow-up of the Women’s Health Initiative randomised placebo-controlled trial. Lancet Oncology, 13(5), 476–486. DOI 10.1016/S1470-2045(12)70075-X. [Google Scholar] [CrossRef]
19. Wiseman, M. (2008). The second world cancer research fund/american institute for cancer research expert report. Food, nutrition, physical activity, and the prevention of cancer: A global perspective. Proceedings of the Nutrition Society, 15(3), 253–256. DOI 10.1017/S002966510800712X. [Google Scholar] [CrossRef]
20. Parkin, D. M., Boyd, L., Walker, L. C. (2011). The fraction of cancer attributable to lifestyle and environmental factors in the UK in 2010. British Journal of Cancer, 105(S2), S77–S81. DOI 10.1038/bjc.2011.489. [Google Scholar] [CrossRef]
21. Li, C. I., Chlebowski, R. T., Freiberg, M., Johnson, K. C., Kuller, L. et al. (2010). Alcohol consumption and risk of postmenopausal breast cancer by subtype: The women’s health initiative observational study. JCNI Journal of the National Cancer Institute, 18(18), 1422–1431. DOI 10.1093/jnci/djq316. [Google Scholar] [CrossRef]
22. Hansen, J., Stevens, R. G. (2012). Case–control study of shift-work and breast cancer risk in Danish nurses: Impact of shift systems. European Journal of Cancer, 48(11), 1722–1729. DOI 10.1016/j.ejca.2011.07.005. [Google Scholar] [CrossRef]
23. Anderson, A. S., Mackison, D., Boath, C., Steele, R. (2013). Promoting changes in diet and physical activity in breast and colorectal cancer screening settings: An unexplored opportunity for endorsing healthy behaviors. Cancer Prevention Research, 6(3), 165–172. DOI 10.1158/1940-6207.CAPR-12-0385. [Google Scholar] [CrossRef]
24. Huang, Z., Hankinson, S. E., Colditz, G. A., Stampfer, M. J., Hunter, D. J. et al. (1997). Dual effects of weight and weight gain on breast cancer risk. JAMA, 278(17), 1407–1411. DOI 10.1001/jama.1997.03550170037029. [Google Scholar] [CrossRef]
25. Bernstein, B. E., Birney, E., Dunham, I., Green, E. D., Gunter, C. et al. (2012). ENCODE project consortium. An integrated encyclopedia of DNA elements in the human genome. Nature, 489(7414), 57–74. DOI 10.1038/nature11247. [Google Scholar] [CrossRef]
26. Brennan, K., Garcia-Closas, M., Orr, N., Fletcher, O., Jones, M. et al. (2012). Intragenic ATM methylation in peripheral blood DNA as a biomarker of breast cancer risk. Cancer Research, 72(9), 2304–2313. DOI 10.1158/0008-5472.CAN-11-3157. [Google Scholar] [CrossRef]
27. Azad, N., Zahnow, C. A., Rudin, C. M., Baylin, S. B. (2013). The future of epigenetic therapy in solid tumours—lessons from the past. Nature Reviews Clinical Oncology, 10(5), 256–266. DOI 10.1038/nrclinonc.2013.42. [Google Scholar] [CrossRef]
28. Tsai, H. C., Li, H., Van Neste, L., Cai, Y., Robert, C. et al. (2012). Transient low doses of DNA-demethylating agents exert durable antitumor effects on hematological and epithelial tumor cells. Cancer Cell, 21(3), 430–446. DOI 10.1016/j.ccr.2011.12.029. [Google Scholar] [CrossRef]
29. Foster, C., Watson, M., Eeles, R., Eccles, D., Ashley, S. et al. (2007). Predictive genetic testing for BRCA1/2 in a UK clinical cohort: Three-year follow-up. British Journal of Cancer, 96(5), 718–724. DOI 10.1038/sj.bjc.6603610. [Google Scholar] [CrossRef]
30. Hilgart, J. S., Coles, B., Iredale, R. (2012). Cancer genetic risk assessment for individuals at risk of familial breast cancer. Cochrane Database of Systematic Reviews, CD003721(7), 7. DOI 10.1002/14651858.CD003721.pub3. [Google Scholar] [CrossRef]
31. Albada, A., Werrett, J., Van Dulmen, S., Bensing, J. M., Chapman, C. et al. (2011). Breast cancer genetic counselling referrals: How comparable are the findings between the UK and the Netherlands? Journal of Community Genetics, 2(4), 233–247. DOI 10.1007/s12687-011-0061-1. [Google Scholar] [CrossRef]
32. Wakefield, C. E., Meiser, B., Homewood, J., Peate, M., Taylor, A. et al. (2008). A randomized controlled trial of a decision aid for women considering genetic testing for breast and ovarian cancer risk. Breast Cancer Research and Treatment, 107(2), 289–301. DOI 10.1007/s10549-007-9539-2. [Google Scholar] [CrossRef]
33. Lindor, N. M., Goldgar, D. E., Tavtigian, S. V., Plon, S. E., Couch, F. J. (2013). BRCA1/2 Sequence variants of uncertain significance: A primer for providers to assist in discussions and in medical management. Oncologist, 15(5), 518–524. DOI 10.1634/theoncologist.2012-0452. [Google Scholar] [CrossRef]
34. Hallowell, N., Baylock, B., Heiniger, L., Butow, P. N., Patel, D. et al. (2012). Looking different, feeling different: Women’s reactions to risk-reducing breast and ovarian surgery. Familial Cancer, 18(2), 215–224. DOI 10.1007/s10689-011-9504-4. [Google Scholar] [CrossRef]
35. Watts, K. J., Meiser, B., Mitchell, G., Kirk, J., Saunders, C. et al. (2012). How should we discuss genetic testing with women newly diagnosed with breast cancer? Design and implementation of a randomized controlled trial of two models of delivering education about treatment-focused genetic testing to younger women newly diagnosed with breast cancer. BMC Cancer, 12(1), 320. DOI 10.1186/1471-2407-12-320. [Google Scholar] [CrossRef]
36. Chivers Seymour, K., Addington-Hall, J., Lucassen, A. M., Foster, C. L. (2010). What facilitates or impedes family communication following genetic testing for cancer risk? A systematic review and meta-synthesis of primary qualitative research. Journal of Genetic Counseling, 19(4), 330–342. DOI 10.1007/s10897-010-9296-y. [Google Scholar] [CrossRef]
37. Mireskandari, S., Sherman, K. A., Meiser, B., Taylor, A. J., Gleeson, M. et al. (2007). Psychological adjustment among partners of women at high risk of developing breast/ovarian cancer. Genetics in Medicine, 9(5), 311–320. DOI 10.1097/GIM.0b013e3180534293. [Google Scholar] [CrossRef]
38. Amir, E., Freedman, O. C., Seruga, B., Evans, D. G. (2010). Assessing women at high risk of breast cancer: A review of risk assessment models. JCNI Journal of the National Cancer Institute, 10(10), 680–691. DOI 10.1093/jnci/djq088. [Google Scholar] [CrossRef]
39. Dite, G. S., Mahmoodi, M., Bickerstaffe, A., Hammet, F., Macinnis, R. J. et al. (2013). Using SNP genotypes to improve the discrimination of a simple breast cancer risk prediction model. Breast Cancer Research and Treatment, 139(3), 887–896. DOI 10.1007/s10549-013-2610-2. [Google Scholar] [CrossRef]
40. Eriksson, L., Hall, P., Czene, K., Dos Santos, S. I., McCormack, V. et al. (2012). Mammographic density and molecular subtypes of breast cancer. British Journal of Cancer, 107(1), 18–23. DOI 10.1038/bjc.2012.234. [Google Scholar] [CrossRef]
41. Swerdlow, A. J., Cooke, R., Bates, A., Cunningham, D., Falk, S. J. et al. (2012). Breast cancer risk after supradiaphragmatic radiotherapy for Hodgkin’s lymphoma in England and Wales: A national cohort study. Journal of Clinical Oncology, 22(22), 2745–2752. DOI 10.1200/JCO.2011.38.8835. [Google Scholar] [CrossRef]
42. Aupperlee, M. D., Leipprandt, J. R., Bennett, J. M., Schwartz, R. C., Haslam, S. Z. (2013). Amphiregulin mediates progesterone-induced mammary ductal development during puberty. Breast Cancer Research, 15(3), R44. DOI 10.1186/bcr3431. [Google Scholar] [CrossRef]
43. Denkert, C., Bucher, E., Hilvo, M., Salek, R., Oresic, M. et al. (2012). Metabolomics of human breast cancer: New approaches for tumor typing and biomarker discovery. Genome Medicine, 4(4), 37. DOI 10.1186/gm336. [Google Scholar] [CrossRef]
44. Santen, R. J., Boyd, N. F., Chlebowski, R. T., Cummings, S., Cuzick, J. et al. (2007). Critical assessment of new risk factors for breast cancer: Considerations for development of an improved risk prediction model. Endocrine-Related Cancer, 14, 169–187. [Google Scholar]
45. Cuzick, J., Sestak, I., Bonanni, B., Costantino, J. P., Cummings, S. et al. (2015). Selective oestrogen receptor modulators in prevention of breast cancer: An updated meta-analysis of individual participant data. Lancet, 381, 1827–1834. [Google Scholar]
46. LaCroix, A. Z., Powles, T., Osborne, C. K., Wolter, K., Thompson, J. R. et al. (2010). Breast cancer incidence in the randomized PEARL trial of lasofoxifene in postmenopausal osteoporotic women. JCNI Journal of the National Cancer Institute, 102(22), 1706–1715. DOI 10.1093/jnci/djq415. [Google Scholar] [CrossRef]
47. Goss, P. E., Ingle, J. N., Ales-Martinez, J. E., Cheung, A. M., Chlebowski, R. T. et al. (2011). Exemestane for breast-cancer prevention in postmenopausal women. New England Journal of Medicine, 364(25), 2381–2391. DOI 10.1056/NEJMoa1103507. [Google Scholar] [CrossRef]
48. Decensi, A., Gandini, S., Serrano, D., Cazzaniga, M., Pizzamiglio, M. et al. (2007). Randomized dose-ranging trial of tamoxifen at low doses in hormone replacement therapy users. Journal of Clinical Oncology, 25(27), 4201–4209. DOI 10.1200/JCO.2006.09.4318. [Google Scholar] [CrossRef]
49. Rosner, B., Glynn, R. J., Tamimi, R. M., Chen, W. Y., Colditz, G. A. et al. (2013). Breast cancer risk prediction with heterogeneous risk profiles according to breast cancer tumor markers. American Journal of Epidemiology, 178(2), 296–308. DOI 10.1093/aje/kws457. [Google Scholar] [CrossRef]
50. Uray, I. P., Brown, P. H. (2011). Chemoprevention of hormone receptor-negative breast cancer: New approaches needed. Recent Results in Cancer Research, 188, 147–162. [Google Scholar]
51. Li, H., Wang, Z., Jiang, M., Fang, R. P., Shi, H. et al. (2018). The oncoprotein HBXIP promotes human breast cancer growth through down-regulating p53 via miR-18b/MDM2 and pAKT/MDM2 pathways. Acta Pharmacologica Sinica, 39(11), 1787–1796. DOI 10.1038/s41401-018-0034-6. [Google Scholar] [CrossRef]
Table S1: Differentially expressed genes between breast cancer samples and normal control samples.
Table S2: Biological functions and signaling pathways involving modular genes.
Table S3: NcRNA pivot that regulates modular genes.
Table S4: TF pivot that regulates modular genes.
Note: Original figures 2–3 are provided in supporting materials.
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |