 Journal of Renewable Materials
Journal of Renewable Materials
 Journal of Renewable Materials Journal of Renewable Materials |  |
DOI: 10.32604/jrm.2021.015288
ARTICLE
Differentiation of Fungal Destructive Behaviour of Wood by the White-Rot Fungus Fomes fomentarius by MALDI-TOF Mass Spectrometry
1Department of Wood Sciences and Engineering, Technical Faculty of No. 1, Mazandaran Branch, Technical and Vocational University (TVU), Sari, Iran
2Department of Industrial Chemistry, ENSTIB-LERMAB, University of Lorraine, Epinal, France
3Department of Wood Biology, University of Hamburg, Hamburg, Germany
4Department of Mycology and Plant Pathology, College of Agronomic Sciences, Agricultural and Natural Resources University, Sari, Iran
5Department of Wood Science and Technology, Biotechnical Faculty, University of Ljubljana, Ljubljana, Slovenia
*Corresponding Authors: Ehsan Bari. Email: bari_lenzites@yahoo.com; Antonio Pizzi. Email: antonio.pizzi@univ-lorraine.fr
Received: 06 December 2020; Accepted: 09 December 2020
Abstract: There are many methods to identify and recognize the molecular and behavioural differences between organisms. One of the methods for the detection and identification of unknown organisms as well as intermolecular and intramolecular structural differences is MALDI-TOF mass spectrometry. Therefore, differentiation of Fomes fomentarius decay capabilities on the chemical properties of the wood cell wall of the tree species Quercus castaneifolia, Juglans regia, and Carpinus betulus were used to determine and characterize the destructive behaviour of F. fomentarius decay by MALDI-TOF mass spectrometry. The results showed that the fungus had more significant destructive behaviour on J. regia than the other species. For this evidence, completely removal of xylan hemicellulose fragment +Na+ at peak 1227 Da and severe digestion of fragment of glucomannan hemicellulose at peak 1477–1480 Da that it seems that signs of soft-rot patterns were obtained from the decayed sample of J. regia, while these were incremental and unchanged for C. betulus and Q. castaneifolia, respectively. However, C. betulus had different peaks of atomic mass than J. regia and Q. castaneifolia wood, respectively. These results showed that this technique could be useful for separating and identifying unknown compounds of the wood cell wall attacked by fungi relying on their biological behaviour.
Keywords: White-rot; Fomes fomentarius; MALDI-TOF; fungal destructive behaviours
There are a variety of methods of analysis to apply to fungi biological behaviours. For wood materials, brown-, white- and soft-rot fungi, are responsible for their degradation [1–3]. These groups have different strategies to attack and decompose the wood cell wall [4]. White-rot fungi can degrade the wood cell wall either by selective patterns preferring the lignin or by simultaneous white-rot such as Fomes fomentarius degrading all three cell wall components [3]. In this regards, different hosts (tree species) may show differences in rot types. White-rot fungi, mostly basidiomycetes, prefer hardwoods whereas most brown-rot species prefer softwoods.
Several methods were used for the detection of fungal destructive behaviours including gravimetric analysis (for example, Wälchli [5]), loss of dynamic modulus of elasticity [6–9], microscopy, e.g., light, polarized and electron microscopies [4,10–12], NMR and FTIR spectroscopy [13–19], microcalorimetry [9,20–23], ergosterol assay [9,21], analysis of flourescein diacetate (FDA) [9], chemical analysis [8,24,25], electronic nose [26], electron paramagnetic resonance and synchrotron X-ray fluorescence [27], immunodiagnostics [28,29], bending vibration technique [30]. Although these techniques provide structural information, their molecular selectivity is quite limited. One approach that helps to overcome this lack of molecular specificity is mass spectrometry, specifically matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry (MALDI-TOF-MS) [31].
Research on biofuels from agricultural and forest waste or lignocellulosic biomass has recently received considerable attention [32]. However, cellulose, hemicellulose, and lignin present complex molecular arrangements. This means that these materials are difficult to be enzymatically digested, thus limiting their bioconversion to biofuel, differently from corn starch and simple sugars [33,34]. Glucose (C6H12O6), mannose (C6H12O6), xylose (C5H10O5), glucuronic acid (C6H10O7), and methyl glucuronic acid (C7H12O7) are monomer units forming complex polymeric structures in the cell walls of wood. MALDI-TOF-MS often ionizes different carbohydrates by non-specific fragmentation. This renders spectral interpretation rather complex. This complexity manifests itself due to the similarity between various sugars and monolignols. Thus, charged ions with equivalent mass to charge ratios are often observed in MALDI-ToF-MS analysis [32].
For instance, digestion of cell wall material with lichenase leads to the release of mixed-linkage glucan-derived oligosaccharides that can be analyzed with MALDI-TOF/MS [35,36]. Furthermore, with slightly altered experimental set-ups, glycosyltransferase and glycoside hydrolase activities can be assayed [32]. To isolate particular polysaccharides, however, further fractionation is required. The structure of isolated polysaccharides can be examined, for example, by determining their monosaccharide compositions to identify and quantify the component monosaccharides [37]. The carbon atoms involved in linking the different monosaccharides (linkage composition) is usually determined by methylation analysis [38]. Additionally, highly specific enzymes can be used to cleave polysaccharides into oligosaccharides, the structures of which can be further analyzed by matrix-assisted laser-desorption ionization time-of-flight mass spectrometry (MALDI-TOF) [39–41]. Analysis of oligosaccharides by MALDI-TOF MS is attractive because it is a rapid, sensitive technique that does not require derivatization of the oligosaccharides for detection. In MALDI-TOF-MS, biomolecules and even whole cells are embedded in a crystal of matrix molecules. Organism-specific signal patterns (“fingerprints”) in the mass range 2000–20000 Da were obtained, e.g., for the identification of bacteria and fungi [42,43]. With the additional screening of natural products in the range below 2000 m/z, different isolates of microorganisms have been detected [44–47].
It is clear that the microorganisms such as fungi have variety behaviour against their host and their response usually were seen by changing their secondary metabolisms such as production of unrespectable enzymes which causes for example switching from selective to simultaneous white rot [15,25,48,49] or white to soft rot [4]. However, these behaviour changing were seen only by residue effects in the affected wood cell walls with methods imaging [11,50–53] or spectral [18,54]. Thus, unfortunately. The applied methods could not show the accurate detail information of fungal destructive behaviour. The objective of the present study, therefore, was to detect the differentiate the white-rot fungus F. fomentarius decay behaviour on the chemical properties of natural decayed wood cell walls of Juglans regia, Carpinus betulus, and Quercus castaneifolia to determine and characterize the destructive behaviours of F. fomentarius decay using MALDI-TOF-MS.
2.1 Wood Samples Collection and Preparation
The relatively soft and colorless white solid natural decayed wood samples (approx. 10 × 5 × 3 cm) from walnut (Juglans regia L.), hornbeam (Carpinus betulus L.), and oak (Quercus castaneifolia C.A. Mey.) (about 60–120-year-old trees) at breast height were collected from the stem part below fruiting bodies of the Fomes fomentarius (L.) Fr. and were immediately transferred into an ice chamber at 4°C to the laboratory. About half of the decayed wood samples were cut and then oven dried at 103°C for 24 h. thereafter, the wood samples were milled by a hammer mill and wood powder was passed through a 60-mesh screen. The average of old year of the fungi on the trees was between 2 and 3 as these annual rings were seen on the fungal fruit bodies.
2.2 Isolation and Identification of the Causal Fungus
Fruiting bodies of Fomes fomentarius (L.) Fr. were first collected from Juglans regia, Carpinus betulus, and Quercus castaneifolia and identified by macro-and microscopic analysis according to Stancheva et al. [55] as well as by rDNA-ITS sequencing (Schmidt et al. [56], Bari [57]). Isolation and confirmation of the fungus causing the decay was carried out as described by Bari et al. [49].
2.3 Matrix Assisted Laser Desorption Ionization Time-of-Flight (MALDI-TOF) Analysis
The natural decayed and undecayed samples for matrix assisted laser desorption ionization time-of-flight (MALDI-TOF) analysis were prepared by first dissolving 7.5 mg of the wood powder in 1 mL of acetone/water. Then 3 µL of this solution was added to 3 µL of a 2.5-dihydroxy benzoic acid (DHB) matrix. The locations dedicated to the samples on the analysis plaque were first covered with 1.5 µL of a NaCl solution 0.1 M in 2:1 v/v methanol/water, as an analysis enhancer, and pre-dried. Then 1.5 µL of the sample solution was placed on its dedicated location and the plaque dried again. The reference substance used for the equipment calibration, up to 2000 Da, was red phosphorus. MALDI-TOF spectra were obtained using an Axima-Performance mass spectrometer from Shimadzu Biotech (Kratos Analytical Shimadzu Europe Ltd., Manchester, UK) using a linear polarity-positive tuning mode with precision is of ±0.1 D. The measurements were carried out, making 200 profiles per sample with two shots accumulated per profile.
3.1 MALDI-TOF Analysis of Juglans regia Wood
The results of matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) analysis of sound and decayed wood samples of Juglans regia wood samples by the white-rot fungus Fomes fomentarius in the ranges of m/z 10–1500 and 1200–1500 Da, respectively, are shown in Figs. 1A, 1B, 2A and 2B). According to the Figures, a great change took place during decay by the fungus, which resulted in the removal of the peak at 103.6 m/z Da. A considerable reduction was seen in peaks 303.4 and 360.2 m/z Da. Another change was related to the intensity of the peak at 38.6 Da, which increased after fungal decomposition (Figs. 1A and 1B). Detailed information of fungal enzymes influences on the wood cell walls components were obtained in ranges of m/z 1200–1500 Da (Figs. 2A and 2B). During degradation, some changes were obtained in decayed samples compared to the sound samples. Conversely, a considerable reduction of the intensity of the peak at 1374.2 m/z Da occurred in the decayed sample. Besides, the peak at 1480.2 m/z Da disappeared.
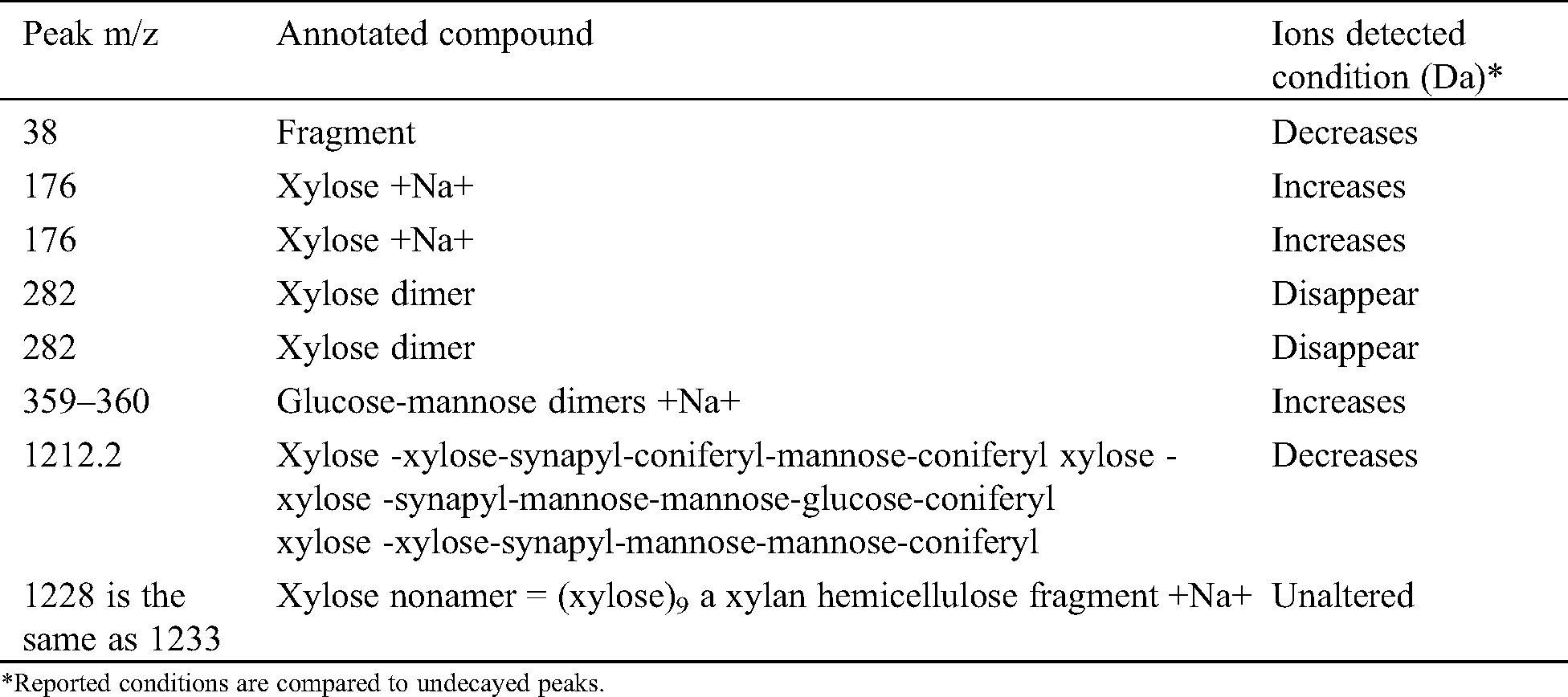
From Tab. 1 on J. regia the following may be deducted. The fungus attacks mainly the hemicelluloses and specifically xylans as shown by the disappearance of the peak at 1274 Da and the decrease of the xylose dimer at 303 Da. It seems that also the glucomannan hemicelluloses are affected as the glucose-mannose dimer decreases after the fungal attack. Furthermore, the reduction on the fungal attack of the peaks at 536 Da (slightly) and 698 Da (decreases a lot) and 1036 Da indicates that also the so-called lignin-hemicelluloses complexes are attacked by the fungus, both the lignin-xylan links and the lignin-glucomannan links. It is most likely that the fungus cleaves the linkages between the hemicelluloses and lignin to get and feed on the hemicelluloses fragments.
Table 1: MALDI TOF MS peaks of sound wood and wood decayed of Juglans regia by Fomes fomentarius
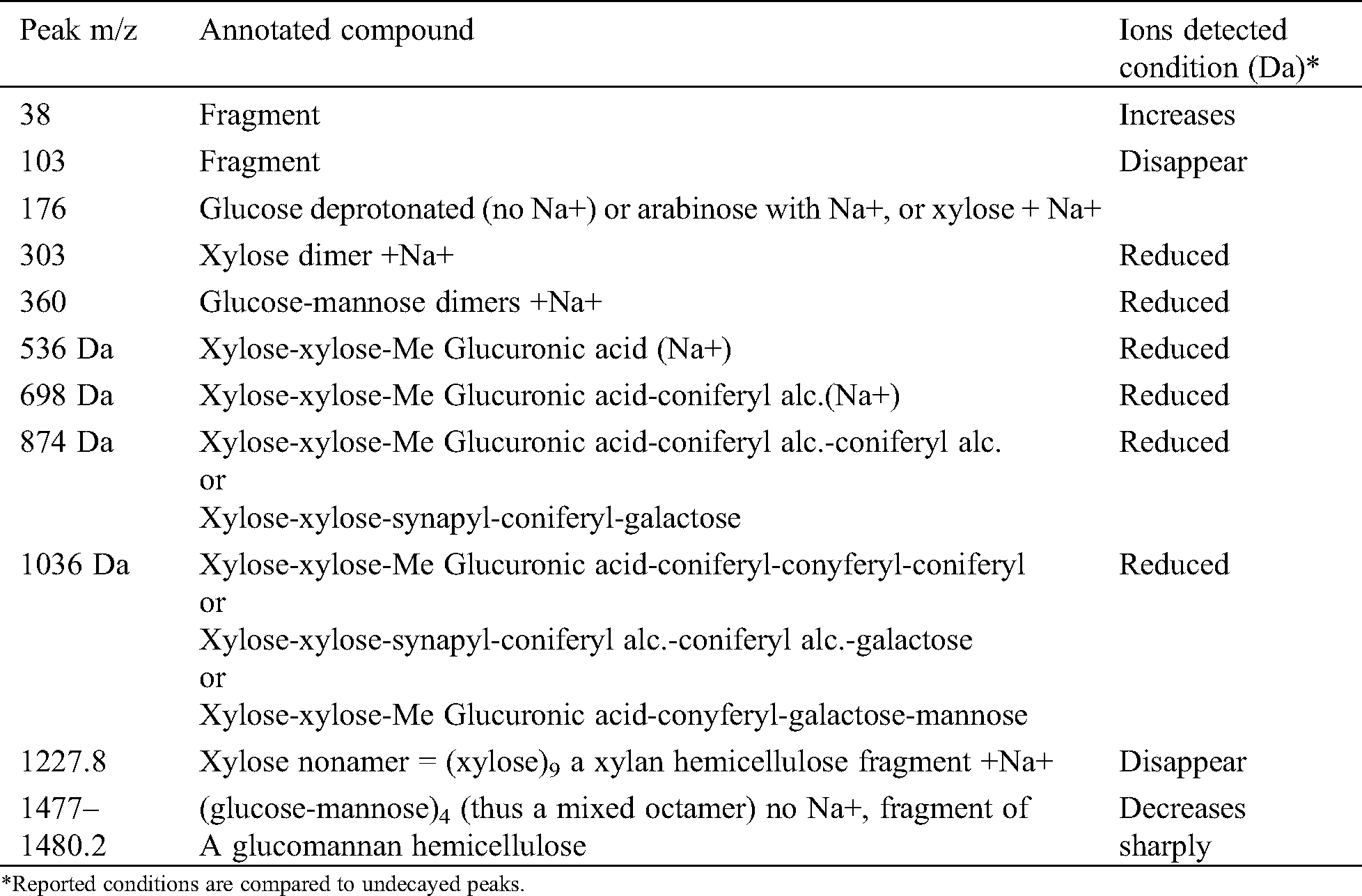
From Figs. 1 and 2 there is also an alternative explanation that appears likely. The series of peaks at 199, 360, 536, 698, 873, 1036, 1211, 1374 Da, are alternatively separated by 162 Da and 175 Da. This means a series of oligomers as follows: Glucose deprotonated+Na+(199 Da), glucose dimer no Na+(360 Da), (glucose)2-glucuronic acid (536 Da), (glucose)3-glucuronic acid (698 Da), (glucose)3-(glucuronic acid)2 (873 Da), (glucose)4-(glucuronic acid)2 (1036 Da); (glucose)4-(glucuronic acid)3 (1211 Da), (glucose)5-(glucuronic)2 (1374 Da). First of all what is indicated here is that glucose could well be a mix of glucose and mannose, thus indicating a series of oligomers of glucomannans derived by cleavage of this type of hemicellulose. As glucuronic acid is often a degradation product by oxidation of glucose such abundance of glucuronic acid indicates how the fungus attacks both glucomannan hemicelluloses as well as possibly cellulose itself by first oxidizing a carbohydrate unit to finally cleave the carbohydrate polymer be it a glucomannan or cellulose. Such an indication is equally valid as what indicated in Tab. 1, the two explanations and interpretations being quite probably both correct and the two manners of attack of the fungus quite possibly occurring simultaneously.
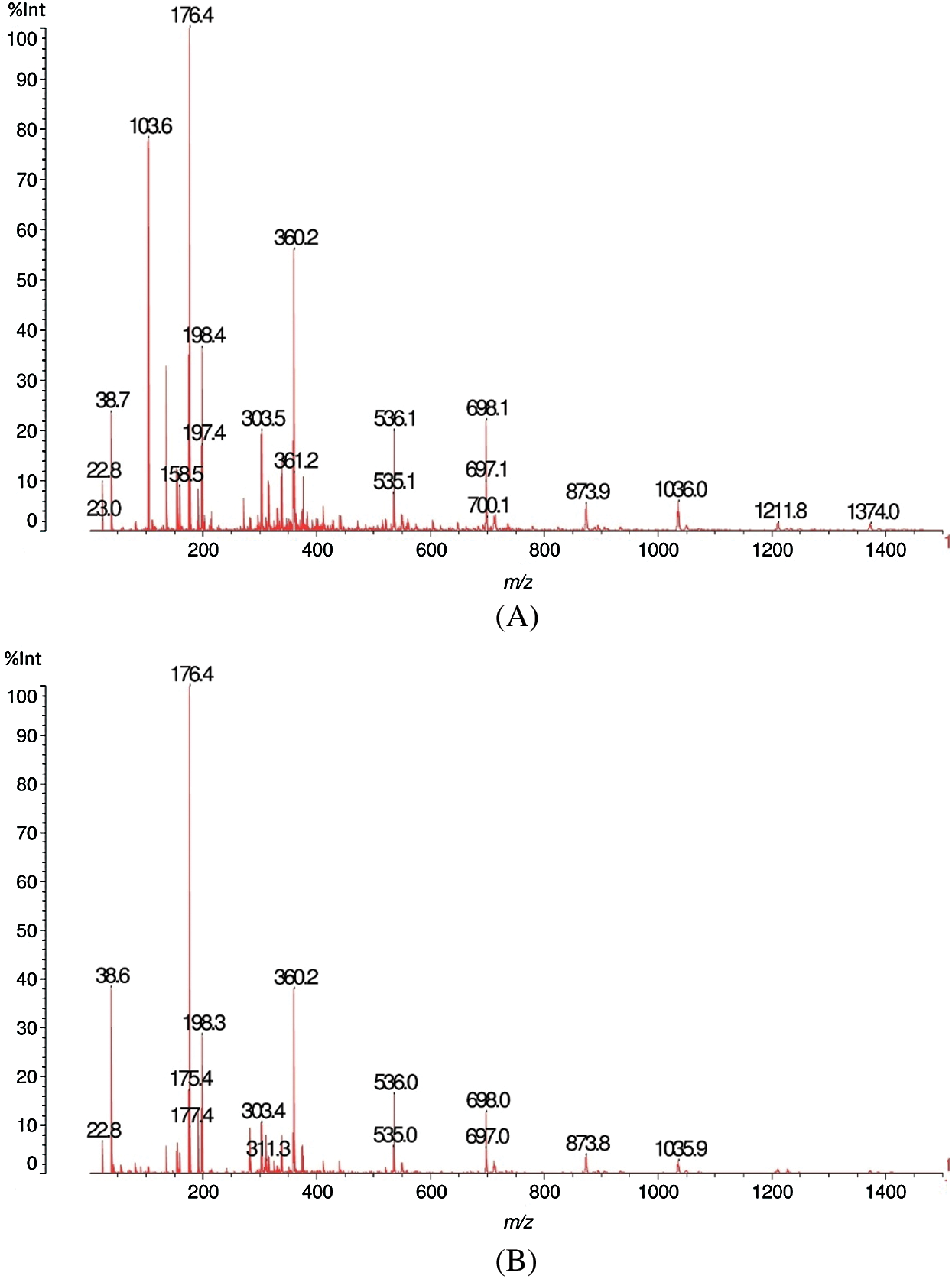
Figure 1: MALDI-TOF analysis results for sound (A) and decayed (B) of Juglans regia wood by Fomes fomentarius, m/z 10–1500 Da
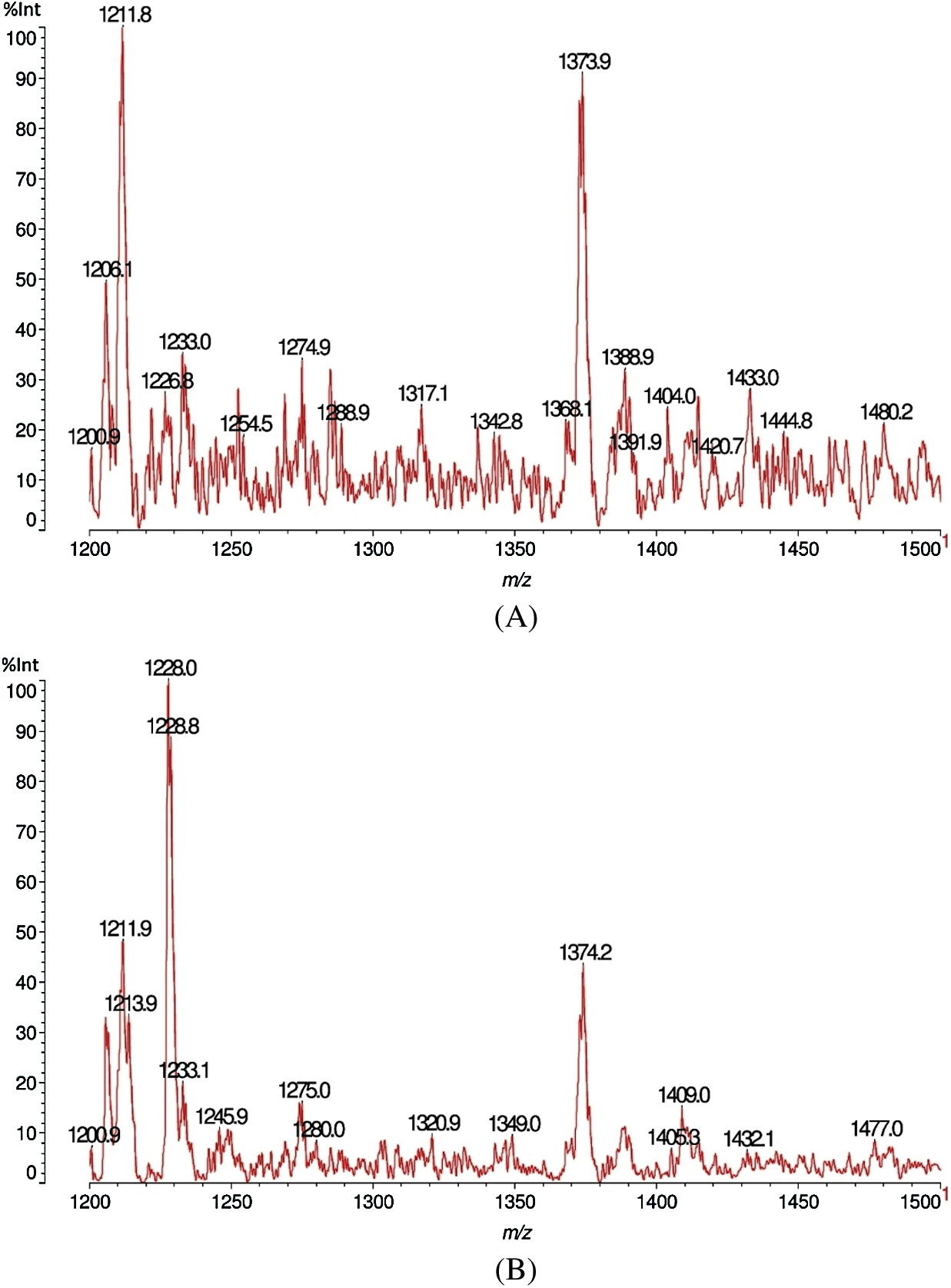
Figure 2: MALDI-TOF analysis results for sound (A) and decayed (B) of Juglans regia wood by Fomes fomentarius, m/z 1200–1500 Da
It is of particular interest, in this case, in which fragments obtained by the fungus attack, are relatively more varied, to determine the type of fragment formulas resulting and from which sites in the wood constituents these derive. Thus the peak at 536 da is a fragment of a xylan composed by two glucoses and of a glucuronic acid side chain (D-Xylp-1-4-β-D-Xylp-1-2-α-D-Me-GlupA), while a clear understanding of the site of attack of the fungus is deduced by the peak at 698 Da, this being either a xylose-xylose-MeGlucuronic acid-conyferyl alcohol fragment (D-Xylp-1-4-β-D-Xylp-1-2-α-D-Me-GlupA-conyferil alcohol) from a lignin-xylan carbohydrate complex where the benzyl ester linkage between the Methyl Glucuronic acid side chain of a xylan is still linked to a lignin unit of coniferyl alcohol, thus a fragment of structure produced as showed in Fig. 3.
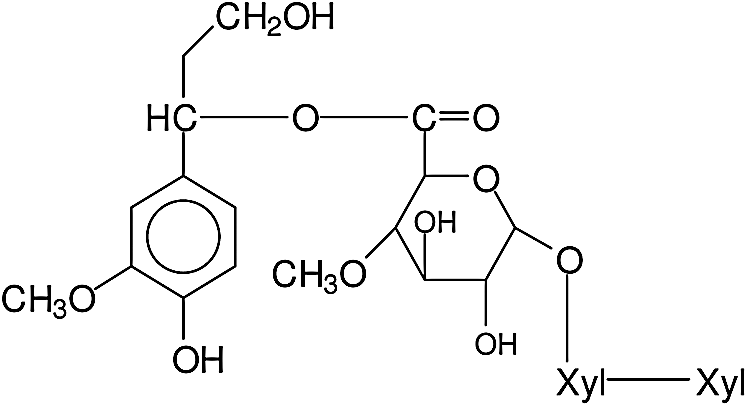
Figure 3: Structure of xylose-xylose-MeGlucuronic acid-conyferyl alcohol fragment
A similar pattern from a xylan-lignin complex is repeated for the 874 Da peak, this being either a fragment of xylose-xylose-MeGlucuronic acid-conyferyl alcohol-conyferyl alcohol, thus a lignin of two units of conyferyl alcohol linked, namely D-Xylp-1-4-β-D-Xylp-1-2-α-D-Me-GlupA-conyferil alcohol-conyferyl alcohol, of structure as showed in Fig. 4. Or a xylose-xylose-MeGlucuronic acid-coniferyl-galactose (D-Xylp-1-4-β-D-Xylp-1-2-α-D-Me-GlupA-conyferil alcohol-α-D-Galp) fragment in which the lignin unit from conyferyl alcohol is linked both to the xylan dimer fragment through the MethylGlucuronic acid and with the galactose side chain of a galactoglucomannan through a phenyl glycosidic linkage. The structure of which can be represented as showed in Fig. 5.
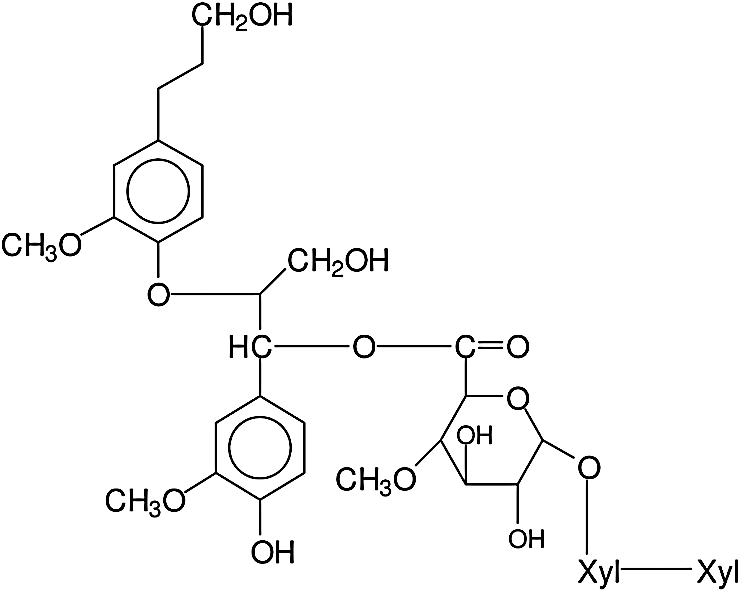
Figure 4: Structure of fragment of xylose-xylose-MeGlucuronic acid-conyferyl alcohol-conyferyl alcohol
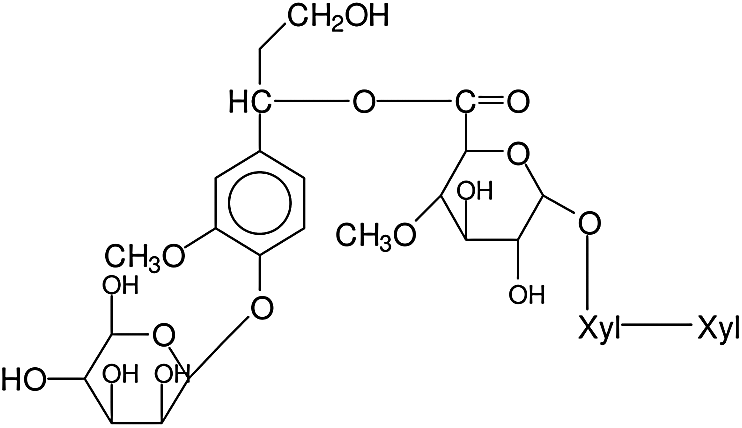
Figure 5: Structure of xylose-xylose-MeGlucuronic acid-coniferyl-galactose fragment
The peak at 1036 Da is either a fragment of a xylose-xylose-MeGlucuronic acid-coniferyl alc.-coniferyl alc.-coniferyl alc sequence, namely D-Xylp-1-4-β-D-Xylp-1-2-α-D-Me-GlupA-coniferil alc.-coniferyl alc.-coniferyl alc, thus again a fragment of a xylan linked by a benzyl ester linkage to a conyferyl alcohol lignin trimer. Or alternatively a xylose-xylose-Me Glucuronic acid-coniferyl-conyferyl-galactose fragment (D-Xylp-1-4-β-D-Xylp-1-2-α-D-Me-GlupA-coniferil alc.-coniferyl alc.-α-D-Galp) again showing an additional cut link between the galactose side chain and its glucomannan skeletal chain. Alternatively, even more telling, a xylose-xylose- Me Glucuronic acid-coniferyl alc.-galactose-mannose (D-Xylp-1-4-β-D-Xylp-1-2-α-D-Me-GlupA-conyferil alcohol-D-Galp-1-6-α-D-Manp) in which to the side chain of the galactose is still linked a mannose from the galactoglucomannan of origin before the fungal attack. The other peaks are oligomer fragments originating from the cleavage of xylan and galactoglucomannan hemicelluloses. Thus the peak at 1227.8 Da is a xylose nonamer = (xylose)9, thus a a xylan hemicellulose fragment +Na+, that can be represented as (-4-β-D-Xylp-1-)9. The peak at 1477–1480 Da is (glucose-mannose)4 (thus a mixed octamer) (no Na+), fragment of a glucomannan hemicellulose, representable in succinct formula as (-4-β-D-Glup-1-4-β-D-Manp-1-)4.
All the above indicates that the fungus cleaves the two main types of hemicelluloses, namely xylans and glucomannans but that it finds more difficult somehow to cleave the benzyl ester and phenyl glycosidic linkages of the so-called lignin-carbohydrates complex. If one compares this result for J. regia in Tab. 1, with the results of the other wood species in Tabs. 2 and 3 this pattern does not seem to appear, in Tabs. 2 and 3 mainly showing oligomer fragments of the main chains of the hemicelluloses cleaved. Thus the structures involved are for a xylan dimer fragment and for longer oligomer fragments as showed in Fig. 6.
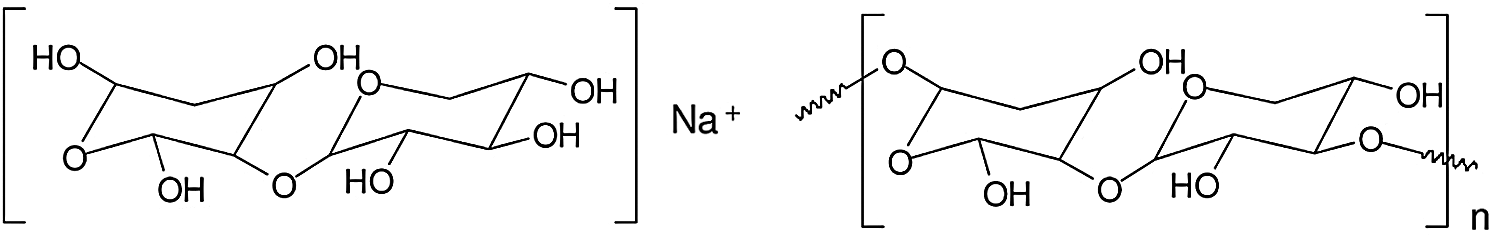
Figure 6: Structures involved are for a xylan dimer fragment and for longer oligomer fragments
3.2 MALDI-TOF Analysis of Carpinus betulus Wood
Figs. 7A, 7B, 8A and 8B shows the results of the MALDI-TOF analysis of sound and decayed Carpinus betulus wood samples by F. fomentarius. According to the Figs. 7A and 7B, the total conditions of the cell wall chemistry in the sound and decayed wood samples occurred in the 10–1500 Da range. After degradation, huge reductions of the 22.8, 42.7, and 38.7 m/z Da peaks occurred, while the intensity of the peaks at 176.4 and 198.5 m/z Da increased in the decayed sample. A sizable reduction was seen in the 282.3 m/z Da peak due to fungal activity. Figs. 8A and 8B showed the emergence of a sharp peak in the 1205–1207 m/z Da range. However, noticeable reductions took place for wood cell wall composition in relationship to peaks at 1409.1 and 1441.1 m/z Da after attack.
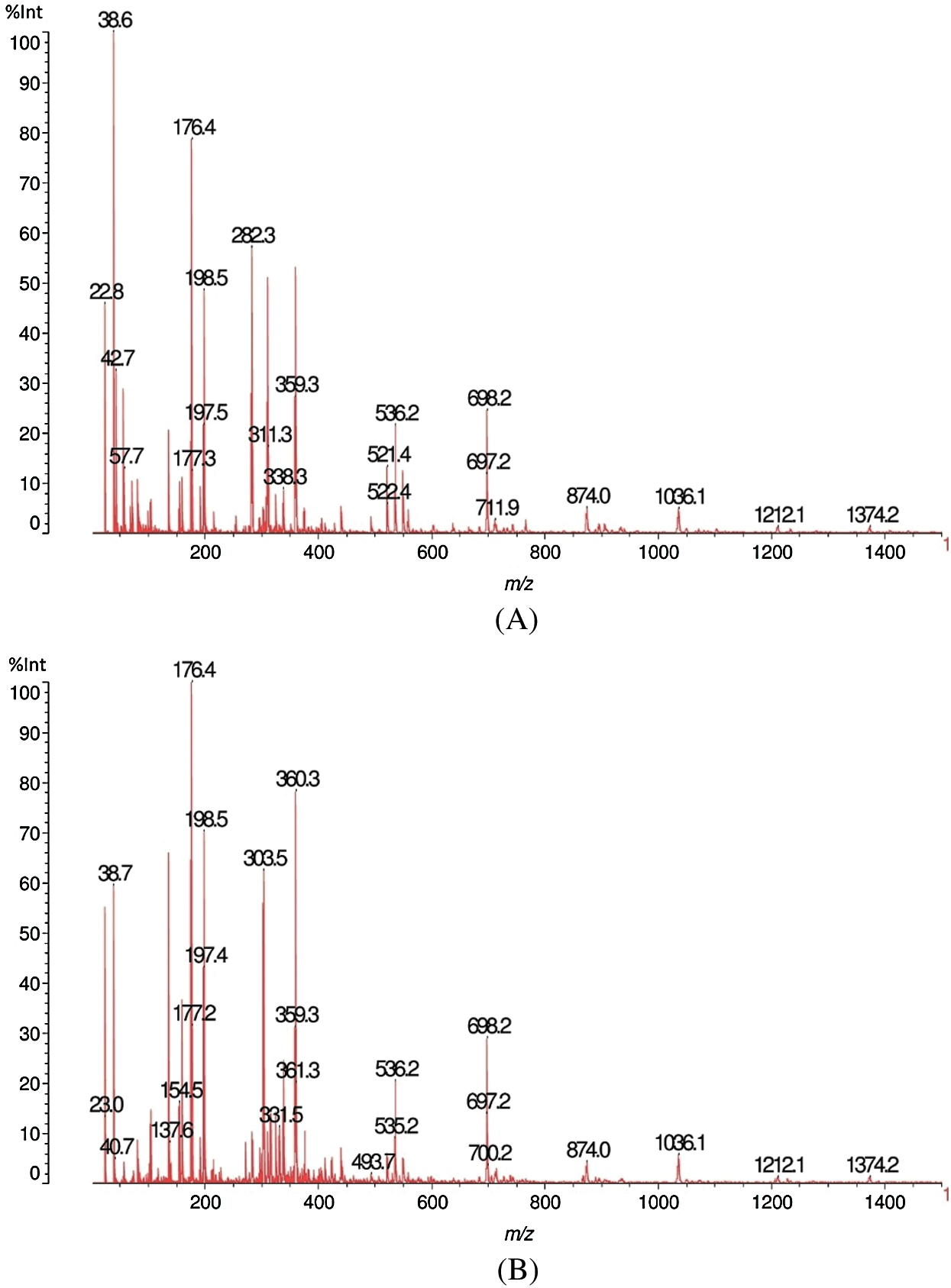
Figure 7: MALDI-TOF analysis results for sound (A) and decayed (B) of Carpinus betulus wood by Fomes fomentarius, m/z 10–1500 Da
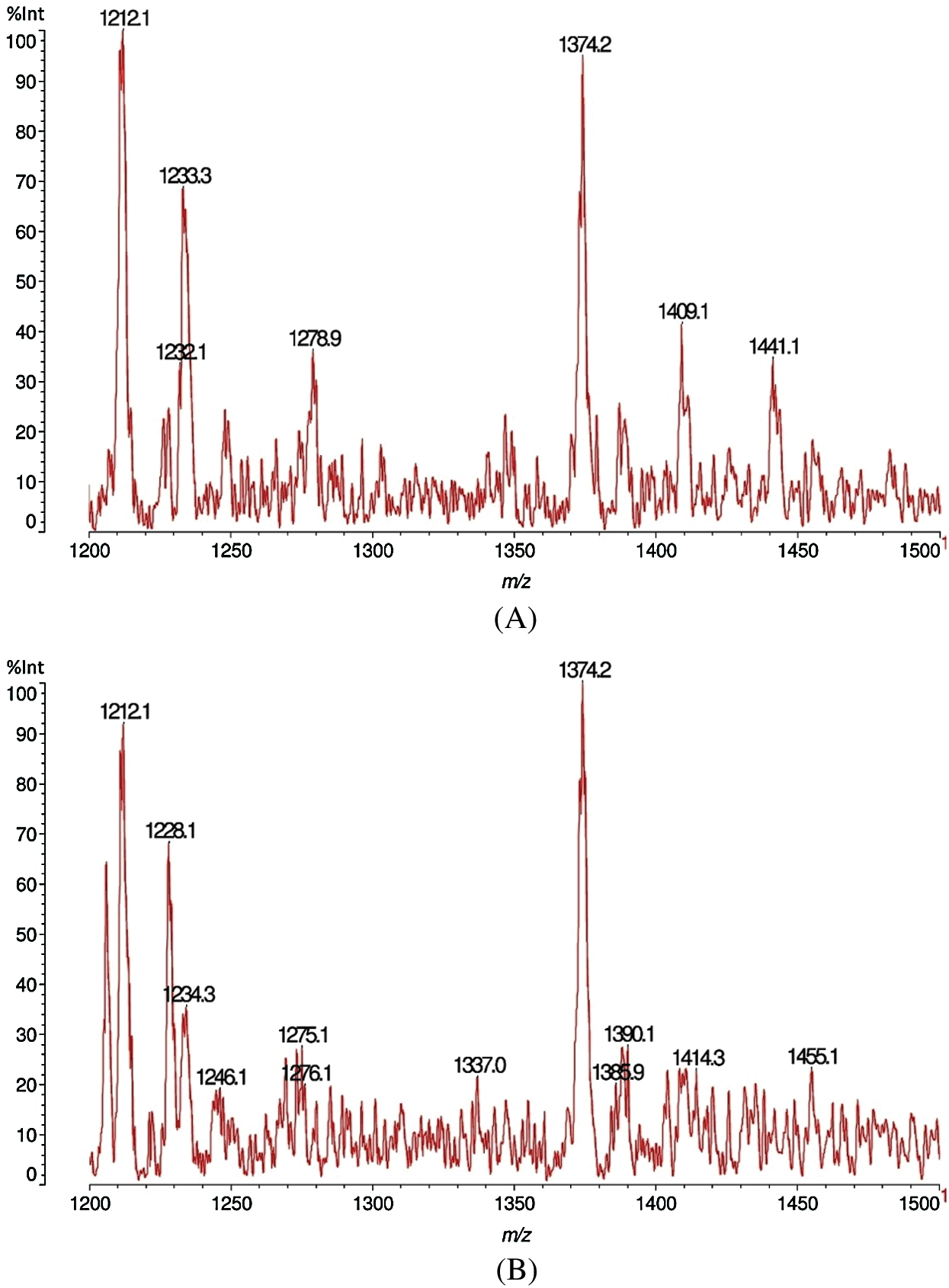
Figure 8: MALDI-TOF analysis results for sound (A) and decayed (B) of Carpinus betulus wood by Fomes fomentarius, m/z 1200–1500 Da
Tab. 2 explains the details of the components obtained from the C. betulus wood sample. The activation of the fungus on the C. betulus was somewhat similar to both the other two wood species since the fungus caused reduction of the intensity at 282.3 Da in both C. betulus and O. castaneifolia which is related to glucomannan hemicelluloses and affected as the glucose-mannose dimer decreases after attack. However, increasing in peak of 303.6 Da was also the same between C. betulus and J. regia that seems due to the action of fungal enzymes to cleave the xylose dimer. It is the nature of the fungus that attacks the xylose dimer and changes it to xylose monomer at 1205 Da by hydrolyses enzymes (endo-1,4-ß-xylanase and ß-xylosidase) causing an increase of this peak hence demonstrating the cleaving of xylans.
Table 2: MALDI TOF MS peaks of sound wood and wood decayed of Carpinus betulus by Fomes fomentarius

As for J. regia, the same series of alternating main peak differences of 162 Da and 175 Da alternating, with the same peak values, hence with the same oligomers being formed, is an alternative and equally valid explanation. It adds the further dimension of understanding of how the fungus cleaves the carbohydrate polymers, namely by degrading glucose by initial oxidation to glucuronic acid, this applicable to both glucomannan hemicelluloses as well as to cellulose itself.
3.3 MALDI-TOF Analysis Quercus castaneifolia Wood
The destructive behaviour of F. fomentarius on the Quercus castaneifolia wood sample analyzed by MALDI-TOF is shown in Figs. 9A, 9B, 10A and 10B. The graphs demonstrate that a severe reduction of intensity occurred for the peaks at 38.7 and 136.7 m/z Da. Equally, a very marked decrease was seen for the peak at 176 m/z Da in the decayed sample. Meanwhile, a new peak appeared in the range of 282.4 peaks of 282.3 m/z Da for the attacked wood sample (Figs. 9A and 9B). Moreover, by comparing the sound and degraded samples in the ranges of m/z 1200–1500 Da, it can be noticed that a large decrease occurred in the peaks at 1212.2 as well as 1374.3 m/z Da. In addition, a large peak appeared at 1228.3 m/z Da (Figs. 10A and 10B) after the fungal attack.
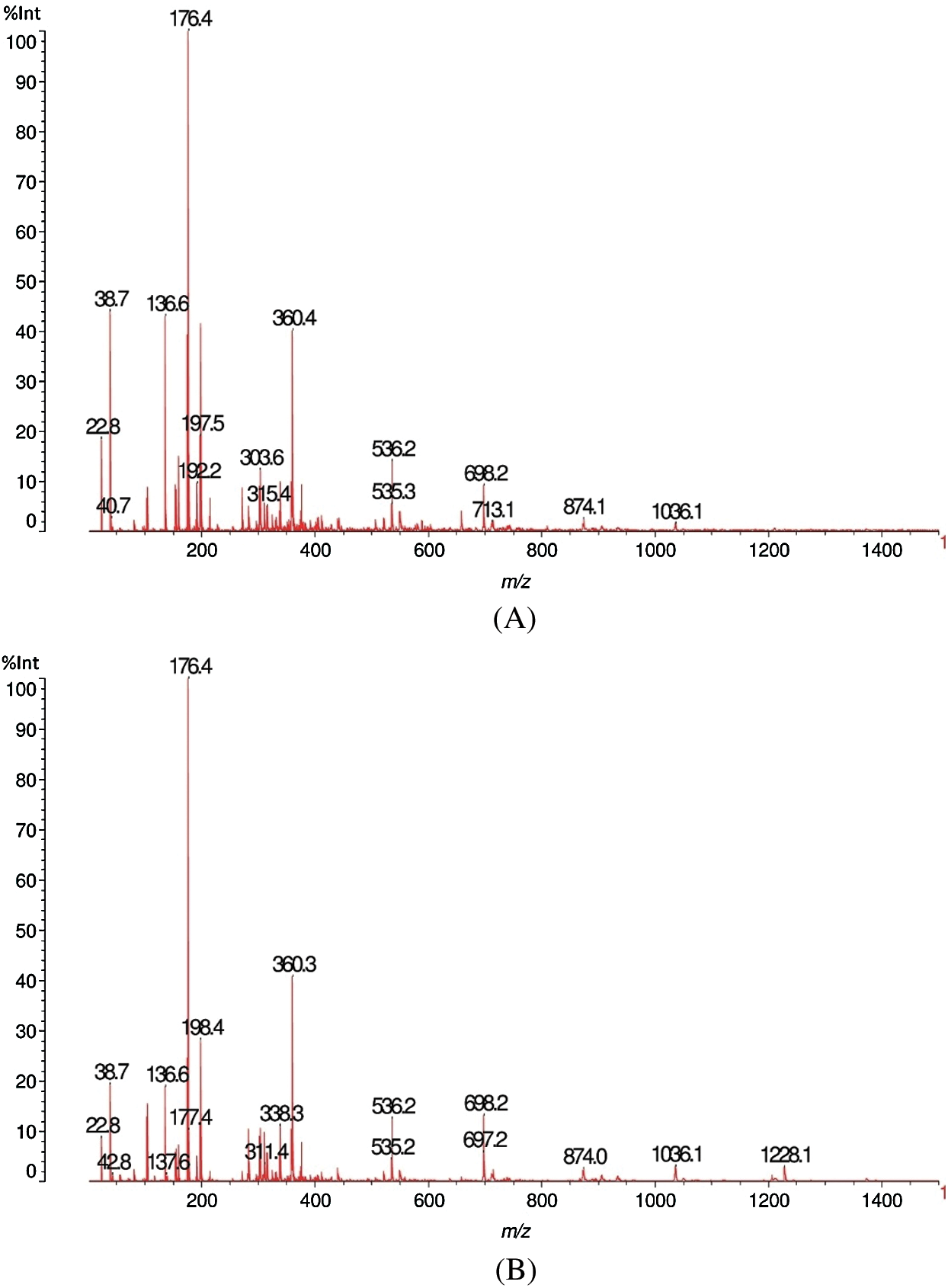
Figure 9: MALDI-TOF analysis results for sound (A) and decayed (B) of Quercus castaneifolia wood by Fomes fomentarius, m/z 10–1500 Da
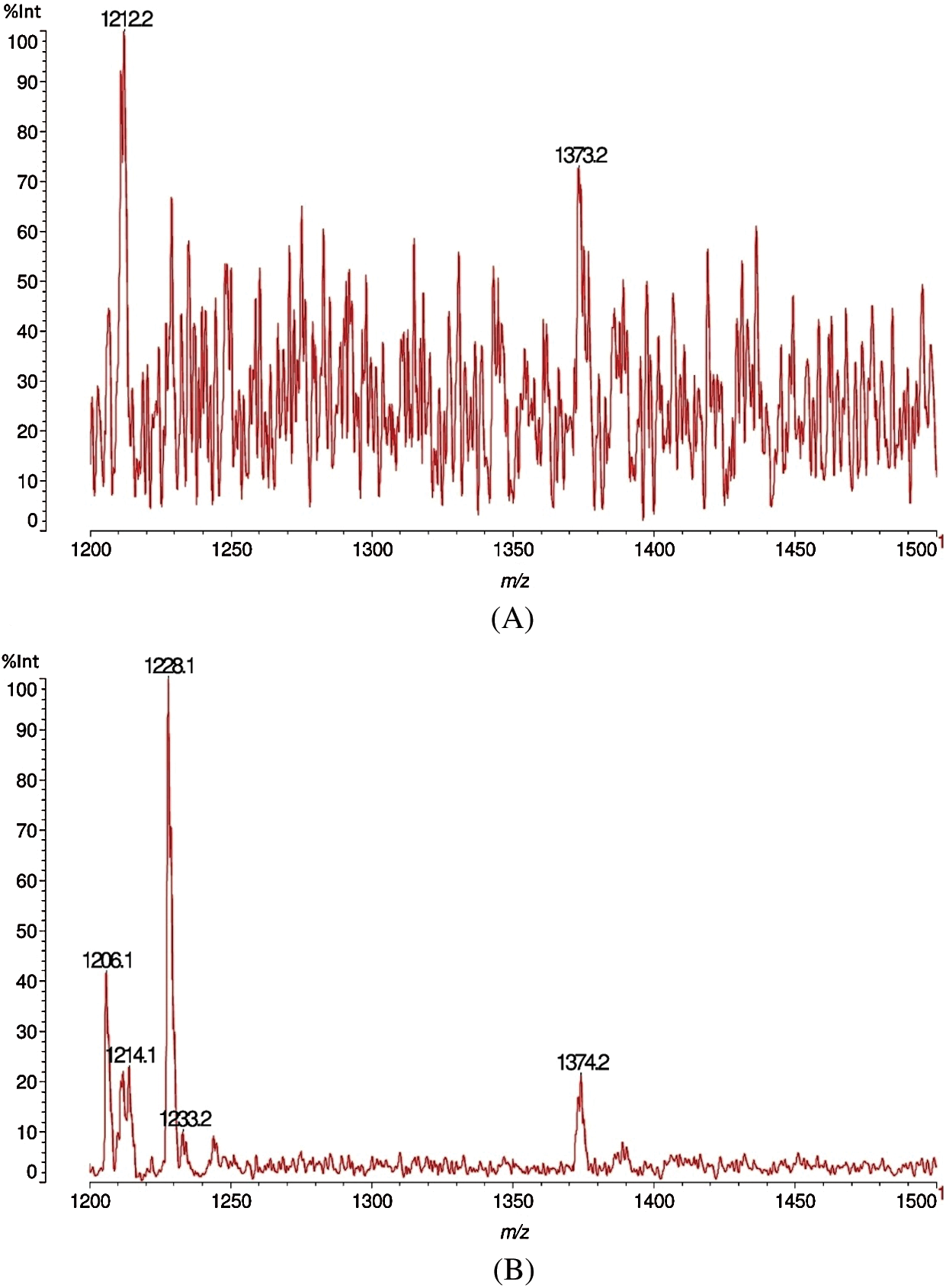
Figure 10: MALDI-TOF analysis results for sound (A) and decayed (B) of Quercus castaneifolia wood by Fomes fomentarius, m/z 1200–1500 Da
From Tab. 3 the fungal attack is the same as for J. regia, with the only difference that the xylose monomer increases. This is logical because as the xylans oligomers are depolymerized by the enzymes of the fungus the supply of xylose monomer does increase before serving as feed to the fungus. However, there are some differences from J. regia: The relative proportion of the glucose-mannose dimer at 359–360 Da does increase by the fungus attack. This is relative to the peaks that have decreased in intensity. This could just be one of the fungal biological strategies to justify a slower or more difficult decay. Alternatively, the fungus may not attack at all the glucomannans of this timber species. It is able to anyhow cleave them from the lignin-carbohydrates complex. This indicates that as for J. regia the fungus is able to cleave both the benzyl ester and benzyl-ether linkages between xylans and lignin, as well as the phenyl glycosidic linkage between glucomannans and lignin.
Table 3: MALDI TOF MS peaks of sound wood and wood decayed of Quercus castaneifolia by Fomes fomentarius
The fungal destructive behaviours of the white-rot fungus Fomes fomentarius was determined by its natural infection activation on the three wood species Juglans regia, Carpinus betulus, and Quercus castaneifolia by MALDI-TOF mass spectrometry. These tree species exhibited different durability, from the lowest (C. betulus) to the highest (Q. castaneifolia). From the peaks observed in the spectra, the results indicate that the fungus has a slightly different behaviour on the three wood species. The fungus caused severe degradation on C. betulus and Q. castaneifolia woods by attacking the lignin and carbohydrate in the cell walls and it seems that it created simultaneous white rot. However, the fungus, in spite of removing both lignin and cellulose, appears to degrade and consume the hemicellulose slightly more preferentially than in other wood species due to changes in mannose and arabinose in decayed wood sample of J. regia. The fungus moreover appears to cleave also the linkages between lignin and xylan and glucomannans in the so-called lignin-carbohydrate complex. Overall, F. fomentarius is a white-rot fungus capable of attacking most tree species by consumption of the cellulose, hemicelluloses, and lignin at a similar ratio [3,4]. Campbell [58] demonstrated that F. fomentarius is able to degrade much more cellulose in beech wood while the attack on pentosans is slightly more severe and the pentosans in the cellulose suffer a proportionally more significant depletion than the pentosans not associated with the cellulose. One of the signs in the simultaneous white rot observed is the severe degradation of carbohydrates in the decayed wood samples [3,59]. Generally, white-rot fungi apply several enzymatic systems such as five endo-1,4-ß-glucanases attacking the cellulose chain at random, thereby splitting the 1,4-ß-glucosidic linkages. Such an enzyme, as its exo-counterpart, is equally active on cleaving the 1,4-p-glucosidic linkages in glucomannan hemicelluloses. Equally, by exo-1,4-ß-glucanase releasing either cellobiose or glucose from the non-reducing end of the cellulose chain, and 1,4-ß-glucosidases hydrolyzing cellobiose and water-soluble cellodextrins to glucose [2]. Nevertheless, the investigated fungus caused considerable degradation in the hemicelluloses this being a sign of the soft-rot decay behaviour. The significant difference from white rot fungi is that soft-rot fungi do not attack the middle lamella and does not involve ligninases. The only enzymes used are cellulases and hemicellulases, and soft-rot is therefore characterized by an extensive loss in the carbohydrate polymers and consequently a significant reduction in the strength of the decayed wood. The rate by which soft-rot fungi decay wood appear to be strongly dependent on the content and spatial distribution of lignin, guaiacyl components of softwood lignin being apparently the biggest obstacle [52,59]. There is not any doubt to the capability as well as to the rot type (simultaneous white-rot) of degradation induced by F. fomentarius. However, what is important in the work presented is its decomposition capacities of wood cell wall chemistry as shown by the figures and individual tables. The secretion of fungal enzymes depends on accessing the fungal hyphae to the wood cell walls and then the ratio and type of their chemistry. Thus, it is logical that the intensity of the degradation capacity of the test fungus would be different in the three wood species tested while the age of the fruit bodies of fungus were almost similar as determine by number of their annual rings.
Fomes fomentarius decayed the three analyzed wood species in the natural condition as obtained by the MALDI-TOF MS analysis. Differentiation of fungal destructive behaviours demonstrated that the fungus presented a slightly different pattern of degradation in Carpinus betulus with the signs of the soft-rot attack systems being due to a more considerable degradation of carbohydrates especially hemicellulose (mannose, glucose and arabinose). Moreover, the experience in identifying compounds, without however being able to distinguish between isomers of identical molecular weight, has been proven for a number of wood derived polymers and compounds, such as polymeric carbohydrates and polyphenolic compounds [60,61]. Furthermore, the results of applied experiment, shows that MALDI-TOF MS is an analytical technique that can be used to help in identifying oligomers and oligomer residues of the main wood constituents of the wood cell wall attacked by microorganisms relying on their biological behaviour. However, future studies should investigate the behaviour of further white-rot fungi, also of those producing the selective type of white-rot.
Acknowledgement: We are grateful to Eng. Asghar Sistani, Department of Wood and Paper Science and Technology, University of Tehran, for preparing the wood samples.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Pournou, A. (2020). Biodeterioration of wooden cultural heritage organisms and decay mechanisms in aquatic and terrestrial ecosystems. Cham, Switzerland: Springer Nature. [Google Scholar]
2. Eriksson, K. E. L., Blanchette, R. A., Ander, P. (1990). Microbial and enzymatic degradation of wood and wood components. New York, USA: Springer-Verlag. [Google Scholar]
3. Schmidt, O. (2006). Wood and tree fungi. Biology, damage, protection and use. Berlin, Germany: Springer. [Google Scholar]
4. Schwarze, F. W. M. R., Engels, J., Mattheck, C. (2004). Fungal strategies of wood decay in trees. 2nd edition. Berlin, Germany: Springer. [Google Scholar]
5. Wälchli, O. (1970). Influence of the content of organic matter of soil on the degradation of wood by soft rot fungi. International Research Group in Wood Preservation, IRG/WP 27. [Google Scholar]
6. Machek, L., Edlund, M. L., Sierra-Alvarez, R., Militz, H. (1998). Acoustic technique for assessing decay in preservative treated wood. International Research Group in Wood Preservation, IRG/WP 98-20138. [Google Scholar]
7. Machek, L., Militz, H., Seierra-Alvarez, R. (1998). A dynamic approach to assess the modulus of elasticity in wood decay testing. International Research Group in Wood Preservation, IRG/WP 98-20139. [Google Scholar]
8. Mohebby, B., Militz, H. (2002). Soft rot decay in acetylated wood. International Research Group in Wood Preservation, IRG/WP 02-40231. [Google Scholar]
9. Mohebby, B. (2003). Biological attack of acetylated wood (Ph.D. Dissertation). Göttingen University, Germany. [Google Scholar]
10. Schwarze, F. W. M. R. (2007). Wood decay under the microscope. Fungal Biological Reviews, 21(4), 133–170. DOI 10.1016/j.fbr.2007.09.001. [Google Scholar] [CrossRef]
11. Bari, E., Schmidt, O., Oladi, R. A. (2015). Histological investigation of oriental beech wood decayed by Pleurotus ostreatus and Trametes versicolor. Forest Pathology, 45(5), 349–357. DOI 10.1111/efp.12174. [Google Scholar] [CrossRef]
12. Bari, E., Daniel, G., Yilgor, N., Kim, J. S., Tajick-Ghanbary, M. A. et al. (2020). Comparison of the decay behavior of two white-rot fungi in relation to wood type and exposure conditions. Microorganisms, 8, 1931. [Google Scholar]
13. Pandey, K. K., Pitman, A. J. (2003). FTIR studies of the changes in wood following decay by brown-rot and white-rot fungi. International Biodeteriorration and Biodegradation, 52(3), 151–160. DOI 10.1016/S0964-8305(03)00052-0. [Google Scholar] [CrossRef]
14. Mohebby, B. (2005). Attenuated total reflection infrared spectroscopy of white-rot decayed beech wood. International Biodeterioration and Biodegradation, 55(4), 247–251. DOI 10.1016/j.ibiod.2005.01.003. [Google Scholar] [CrossRef]
15. Bari, E., Taghiyari, H. R., Mohebby, B., Clausen, C. A., Schmidt, O. et al. (2015). Mechanical properties and chemical composition of beech wood exposed for 30 and 120 days to white-rot fungi. Holzforschung, 69(5), 587–593. DOI 10.1515/hf-2014-0057. [Google Scholar] [CrossRef]
16. Karim, M., Ghodskhah Daryaei, M., Torkaman, J., Oladi, R., Tajick Ghanbary, M. A. et al. (2016). In vivo investigation of chemical alteration in Oak wood decayed by Pleurotus ostreatus. International Biodeterioration and Biodegradation, 108, 127–132. DOI 10.1016/j.ibiod.2015.12.012. [Google Scholar] [CrossRef]
17. Karim, M., Ghodskhah Daryaei, M., Torkaman, J., Oladi, R., Tajick Ghanbary, M. A. et al. (2017). Natural decomposition of hornbeam wood decayed by the white rot fungus Trametes versicolor. Journal of Anais da Academia Brasileira de Ciências, 89(4), 2647–2655. DOI 10.1590/0001-3765201720160714. [Google Scholar] [CrossRef]
18. Bari, E., Mohebby, B., Naji, H. R., Oladi, R., Yilgor, N. et al. (2018). Monitoring the cell wall characteristics of degraded beech wood by white-rot fungi: Anatomical, chemical, and photochemical study. Maderas Ciencia y Tecnologia, 20, 1, 35–56. [Google Scholar]
19. Bari, E., Jamali, A., Nazarnezhad, N., Nicholas, D. D., Humar, M. (2019). An innovative method for the chemical modification of Carpinus betulus wood: A methodology and approach study. Holzforschung, 73(9), 839–846. DOI 10.1515/hf-2018-0242. [Google Scholar] [CrossRef]
20. Bjurman, J., Wadsö, L. (2000). Microcalorimetric measurements of metabolic activity of six decay fungi on spruce wood as a function of temperature. Mycologia, 92(1), 23–28. DOI 10.1080/00275514.2000.12061126. [Google Scholar] [CrossRef]
21. Mohebby, B., Mai, C., Militz, H. (2003). Soft rot decay in acetylated wood: Microcalorimetry and ergosterol assay in decayed wood. Proceedings, European Conference on Wood Modification, Ghent, Belgium, 197–202. [Google Scholar]
22. Yui, T., Uto, T. (2020). Docking and molecular dynamics study of the carbohydrate binding module from Trichoderma reesei Cel7A on the surfaces of the cellulose IIII crystal. Journal of Renewable Materials, 8(8), 863–878. DOI 10.32604/jrm.2020.010830. [Google Scholar] [CrossRef]
23. Bopenga Bopenga, C. S. A., Meyo Degboevi, H., Candelier, K., Edou Engonga, P., Dumarçay, S. et al. (2020). Characterization of extracts from the bark of the Gabon hazel tree (Coula edulis baill) for antioxidant, antifungal and anti-termite products. Journal of Renewable Materials, 9(1), 17–33. DOI 10.32604/jrm.2021.013366. [Google Scholar] [CrossRef]
24. Wang, W., Xu, Q., Wang, X., Guo, J., Cao, W. et al. (2020). Strength degradation of wood members based on the correlation of natural and accelerated decay experiments. Journal of Renewable Materials, 8(5), 565–577. DOI 10.32604/jrm.2020.09020. [Google Scholar] [CrossRef]
25. Bari, E., Nazarnezhad, N., Kazemi, S. M., Tajick Ghanbary, M. A., Mohebby, B. et al. (2015). Comparison of degradation capabilities of the white-rot fungi Pleurotus ostreatus and Trametes versicolor. International Biodeterioration and Biodegradation, 104, 231–237. DOI 10.1016/j.ibiod.2015.03.033. [Google Scholar] [CrossRef]
26. Nilsson, K. 1998). (. Detection of wood decay using electronic nose (Ph.D. Thesis). Swedish University of Agricultural Sciences, Uppsala, Sweden, 50. [Google Scholar]
27. Illman, B. L., Bajt, S. (1997). Nondestructive elemental analysis of wood biodeterioration using electron paramagnetic resonance and synchrotron X-ray fluorescence. International Biodeterioration and Biodegradation, 39(2–3), 235–243. DOI 10.1016/S0964-8305(97)00020-6. [Google Scholar] [CrossRef]
28. Clausen, C. (1996). Immunological detection of wood decay fungi, an overview of techniques developed from 1986 to the present. International Biodeterioration and Biodegradation, 39(2–3), 133–143. DOI 10.1016/S0964-8305(97)00017-6. [Google Scholar] [CrossRef]
29. Clausen, C., Green, F. (1998). Immunodiagnostic wood decay test. Techline: Decay process and bioprocessing, USDA Forest Products Laboratory, II-3. http://www.fpl.fs.fed.us. [Google Scholar]
30. Ouis, D. (2000). Detection of decay in logs through measuring the damping of bending vibrations by means of a room acoustical technique. Wood Science and Technology, 34(3), 221–236. DOI 10.1007/s002260000044. [Google Scholar] [CrossRef]
31. Karas, M., Hillenkamp, F. (1988). Laser desorption ionization of proteins with molecular masses exceeding 10,000 Daltons. Analytical Chemistry, 60(20), 2299–2301. DOI 10.1021/ac00171a028. [Google Scholar] [CrossRef]
32. Lunsford, K. A., Peter, G. F., Yost, R. A. (2011). Direct matrix-assisted laser desorption/ionization mass spectrometric imaging of cellulose and hemicellulose in Populus tissue. Analytical Chemistry, 83(17), 6722–6730. DOI 10.1021/ac2013527. [Google Scholar] [CrossRef]
33. Ragauskas, A. J., Williams, C. K., Davison, B. H., Britovsek, G., Cairney, J. et al. (2006). The path forward for biofuels and biomaterials. Science, 311(5760), 484–489. DOI 10.1126/science.1114736. [Google Scholar] [CrossRef]
34. McCann, M., Carpita, N. (2008). Designing the deconstruction of plant cell walls. Current Opinion in Plant Biology, 11(3), 314–320. DOI 10.1016/j.pbi.2008.04.001. [Google Scholar] [CrossRef]
35. McDonnell, L. A., Heeren, R. M. A. (2007). Imaging mass spectrometry. Mass Spectrometry Reviews, 26(4), 606–643. DOI 10.1002/mas.20124. [Google Scholar] [CrossRef]
36. Cha, S. W., Song, Z. H., Nikolau, B. J., Yeung, E. S. (2009). Direct profiling and imaging of epicuticular waxes on Arabidopsis thaliana by laser desorption/ionization mass spectrometry using silver colloid as a matrix. Analytical Chemistry, 81(8), 2991–3000. DOI 10.1021/ac802615r. [Google Scholar] [CrossRef]
37. Harris, P. J., Blakeney, A. B., Henry, R. J., Stone, B. A. (1988). Gas chromatographic determination of the monosaccharide composition of plant cell wall preparations. Journal of the Association of Official Analytical Chemists, 71, 272–275. [Google Scholar]
38. Harris, P. J., Blakeney, A. B., Henry, R. J., Stone, B. A. (1984). An improved procedure for the methylation analysis of oligosaccharides and polysaccharides. Carbohydrate Research, 127(1), 59–73. DOI 10.1016/0008-6215(84)85106-X. [Google Scholar] [CrossRef]
39. Lerouxel, O., Choo, T. S., Seveno, M., Usadel, B., Faye, L. et al. (2002). Rapid structural phenotyping of plant cell wall mutants by enzymatic oligosaccharide fingerprinting. Plant Physiology, 130(4), 1754–1763. DOI 10.1104/pp.011965. [Google Scholar] [CrossRef]
40. Hsieh, Y. S. Y., Harris, P. J. (2009). Xyloglucans of monocotyledons have diverse structures. Molecular Plant, 2(5), 943–965. DOI 10.1093/mp/ssp061. [Google Scholar] [CrossRef]
41. Günl, M., Kraemer, F., Pauly, M. (2011). Oligosaccharide mass profiling (OLIMP) of cell wall polysaccharides by MALDI-TOF/MS. Methods in Molecular Biology, 715, 43–54. [Google Scholar]
42. Schmidt, O., Kallow, W. (2005). Differentiation of indoor wood decay fungi with MALDI-TOF mass spectrometry. Holzforschung, 59(3), 74–377. DOI 10.1515/HF.2005.062. [Google Scholar] [CrossRef]
43. Pristaš, P., Kvasnová, S., Gáperová, S., Gašparcová, T. Gáper, J. et al. (2017). Application of MALDI-TOF mass spectrometry for in vitro identification of wood decay polypores. Forest Pathology, 47(5), e12352. DOI 10.1111/efp.12352. [Google Scholar] [CrossRef]
44. Erhard, M., von Döhren, H.,Jungblut, P. (1997). Rapid typing and elucidation of new secondary metabolites of intact cyanobacteria using MALDI-TOF mass spectrometry. Natural Biotechnology, 15(9), 906–909. DOI 10.1038/nbt0997-906. [Google Scholar] [CrossRef]
45. Erhard, M., Kallow, W., Dieckmann, R., Neuhof, T., von Doöhren, H. et al. (2001). Schnelles Naturstoffscreening, Strukturanalyse und Stammidentifizierung. BIOforum, 10, 734–735. [Google Scholar]
46. Fastner, J., Erhard, M., von Döhren, H. (2001). Determination of oligopeptide diversity within a natural population of Microcystis spp. (cyanobacteria) by typing single colonies by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Applied Environmental Microbiology, 67(11), 5069–5076. DOI 10.1128/AEM.67.11.5069-5076.2001. [Google Scholar] [CrossRef]
47. Walker, J., Fox, A. J., Edwards-Jones, V., Gordon, D. B. (2002). Intact cell mass spectrometry (ICMS) used to type methicillin-resistant Staphylococcus aureus: Media effects and interlaboratory reproducibility. Journal of Microbiology Methods., 48(2–3), 117–126. DOI 10.1016/S0167-7012(01)00316-5. [Google Scholar] [CrossRef]
48. Bari, E., Karim, M., Oladi, R., Tajick Ghanbary, M. A., Ghodskhah Daryaei, M. et al. (2017). Comparison between decay patterns of the white-rot fungus Pleurotus ostreatus in chestnut-leaved oak (Quercus castaneifolia) shows predominantly simultaneous attack both in vivo and in vitro. Forest Pathology, 47(4), e12338. DOI 10.1111/efp.12338. [Google Scholar] [CrossRef]
49. Bari, E., Ghodskhah Daryaei, M., Karim, M., Bahmani, M., Schmidt, O. et al. (2019). Decay of Carpinus betulus wood by Trametes versicolor—An anatomical and chemical study. International Biodeterioration and Biodegradation, 137, 68–77. DOI 10.1016/j.ibiod.2018.11.011. [Google Scholar] [CrossRef]
50. Daniel, G. (2003). Microview of wood under degradation by bacteria and fungi. In: Goodell, B., Nicholas, D. D., Schultz, T. P. (eds.Wood deterioration and preservation: Advances in our changing world; Advances in chemistry series. Washington, DC, USA: American Chemical Society, pp. 34–72. [Google Scholar]
51. Daniel, G. (2014). Fungal and bacterial biodegradation: White rots, brown rots, soft rots & bacteria. In: Schulz, T. P., Goodell, B., Nicholas, D. D. (eds.Deterioration and protection of sustainable biomaterials; Advances in chemistry series. Washington, DC, USA: American Chemical Society, pp. 23–58. [Google Scholar]
52. Daniel, G. (2016). Fungal degradation of wood cell walls. In: Kim, Y. S., Funada, R., Singh, A. P. (eds.Secondary xylem biology: Origins, function, and applications. Amsterdam, The Netherlands: Elsevier, pp. 131–167. [Google Scholar]
53. Bari, E., Oladi, R., Schmidt, O., Clausen, C. A., Ohno, K. et al. (2015). Influence of xylem ray integrity and degree of polymerization on bending strength of beech wood decayed by Pleurotus ostreatus and Trametes versicolor. International Biodeterioration and Biodegradation, 104, 299–306. DOI 10.1016/j.ibiod.2015.06.019. [Google Scholar] [CrossRef]
54. Pandey, K. K., Pitman, A. J. (2003). FTIR studies of the changes in wood chemistry following decay by brown-rot and white-rot fungi. International Biodeterioration and Biodegradation, 52(3), 151–160. DOI 10.1016/S0964-8305(03)00052-0. [Google Scholar] [CrossRef]
55. Stancheva, Y., Bencheva, S., Pavlidis, T., Ilieva, M. (2009). Atlas of wood decaying fungi. Sofia, Bulgaria: Pensoft Publishers (Sofia-Moscowpp. 350. [Google Scholar]
56. Schmidt, O., Gaiser, O., Dujesiefken, D. (2012). Molecular identification of decay fungi in the wood of urban trees. European Journal of Forest Research, 131(3), 885–891. DOI 10.1007/s10342-011-0562-9. [Google Scholar] [CrossRef]
57. Bari, E. (2014). Potential of biological degradation of Oriental beech wood by the white rot fungus Pleurotus ostreatus and the effects on mechanical and chemical properties and its comparison with standard the white-rot fungus Trametes versicolor (Master’s Thesis). Sari Agricultural Sciences and Natural Resources University, Sari, Iran, pp. 76. [Google Scholar]
58. Campbell, W. G. (1932). The chemistry of the white rots of wood. Biochemical Journal, 26(6), 1829–1838. DOI 10.1042/bj0261829. [Google Scholar] [CrossRef]
59. Kubicek, C. P. (2013). Fungi and lignocellulosic biomass. Chichester, UK: John Wiley & Sons, Inc., 305. [Google Scholar]
60. Pasch, H., Pizzi, A., Rode, K. (2001). MALDI-TOF mass spectrometry of polyflavonoid tannins. Polymer, 42(18), 7531–7539. DOI 10.1016/S0032-3861(01)00216-6. [Google Scholar] [CrossRef]
61. Pizzi, A., Pasch, H., Rode, K., Giovando, S. (2009). Polymer structure of commercial hydrolyzable tannins by matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry. Journal of Applied Polymer Science, 113(6), 3847–3859. DOI 10.1002/app.30377. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License,, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |