 Open Access
Open Access
ARTICLE
Preliminary Study on the Treatment Efficiency of Pasteurized Lime Thermal Alkaline Hydrolysis for Excess Activated Sludge and Reduction of Tetracycline Resistance Genes
1
School of New Energy Materials and Chemistry, Leshan Normal University, Leshan, 614000, China
2
South Sichuan Pollution Control and Resource Recovery Research Center, Leshan Normal University, Leshan, 614000, China
* Corresponding Author: Maoxia Chen. Email:
Journal of Renewable Materials 2023, 11(10), 3711-3723. https://doi.org/10.32604/jrm.2023.027826
Received 16 November 2022; Accepted 03 January 2023; Issue published 10 August 2023
Abstract
Thermal alkaline hydrolysis is a common pretreatment method for the utilization of excess activated sludge (EAS). Owing to strict environment laws and need for better energy utilization, new methods were developed in this study to improve the efficiency of pretreatment method. Direct thermal hydrolysis (TH), pasteurized thermal hydrolysis (PTH), and alkaline pasteurized thermal hydrolysis (PTH + CaO and PTH + NaOH) methods were used to treat EAS. Each method was compared and analyzed in terms of dissolution in ammonium nitrogen (NH4 + -N) and soluble COD (SCOD) in EAS. Furthermore, the removal of tetracycline resistance genes (TRGs) and class 1 transposon gene intI1 from EAS was investigated. The NH4 + -N and SCOD concentrations in EAS treated by PTH were 1.24 and 2.58 times higher than those of TH. However, the removal efficiency of total TRGs and intI1 between the groups was comparable. The SCOD concentration of the PTH + NaOH group was 4.37 times higher than that of the PTH group, and the removal efficiency of total TRGs was increased by 9.52% compared with that by PTH. The NH4 + -N and SCOD concentrations of the PTH + CaO group could reach 85.04% and 92.14% of the PTH + NaOH group, but the removal efficiency of total TRGs by PTH + CaO was 19.78% lower than that by PTH + NaOH. Thus, to reduce the financial cost in actual operation, lime (CaO) can be used instead of a strong alkali (NaOH), and pasteurized steam at 70°C instead of conventional high-temperature heating to treat EAS. This study provides a reference for the development of alkaline hydrolysis under moderate temperatures along with the removal of TRGs in EAS.Keywords
Excess activated sludge (EAS) is effluent solid waste from biological sewage treatment systems; it is high in moisture content, organic matter, and perishables, and contains many parasitic eggs and pathogenic microorganisms. Its entry into the ecosystem poses an environmental risk. According to the latest sewage treatment statistics of the Ministry of Housing and Urban-Rural Development (Beijing, China), an average of 10,000 m3 of sewage treatment will produce 2.07 tons of dry EAS in China, and a total of 1.33 × 107 t of dry EAS was produced in a year [1]. Currently, commonly used EAS treatment methods include incineration, safe landfill, composting, or anaerobic digestion. Increasing research is devoted to the safe recycling of nutrients in EAS, including carbon, nitrogen, and phosphorus, compared to EAS as a solid waste to be treated [2,3]. Among the EAS treatment methods, composting and anaerobic digestion for methane production are effective ways to utilize the carbon present in EAS [4,5]. However, to improve the efficiency of composting and anaerobic digestion, pretreatment of EAS using thermal hydrolysis (TH) and alkali hydrolysis, which can promote the dissolution of nitrogen, organic matter, and other nutrients from sludge, are generally required [6,7]. Thermal hydrolysis can disintegrate the floc structure of sludge and break up sludge cells, to release and hydrolyze the organic matter [8]. Alkaline hydrolysis can also damage cells and dissolve nutrients, and compared to thermal hydrolysis, it is conducive to the recovery of soluble macromolecules such as carbohydrates and proteins [9]. The combination of thermal hydrolysis and alkaline hydrolysis considerably improves the sludge hydrolysis effect [10]. Conventional thermal alkaline hydrolysis requires a strong alkali and maintains the sludge at a high temperature (above 100°C), which leads to high resource and energy consumption [11]. Currently, there is a strong emphasis on carbon emission reduction and environmental protection worldwide, because of this the requirement for low-cost and high-efficiency EAS thermal alkaline digestion technology is increasing. To reduce the cost of thermal alkaline hydrolysis, regulating the operational temperature and using low-cost alkali sources are effective strategies. For example, Zheng et al. [12] found that when the thermal alkaline hydrolysis temperature of the EAS was 70°C, the total treatment cost was only 47.75% of that when the temperature was 130°C. Tang et al. [13] used Ca(OH)2 as the base source for alkaline thermal hydrolysis of EAS, which can produce high-quality liquid fertilizer, with high economic benefits.
Effluent and EAS from wastewater treatment facilities are also major sources of antibiotic resistance genes (ARGs) [14,15]. When the treatment objects are pharmaceutical wastewater and aquaculture wastewater, the detection of ARGs in sludge and effluent is particularly significant [16,17]. Thus, the discharge of effluent and EAS poses the risk of increased antibiotic resistance in the receiving environment. Studies have found that different types of ARGs were widely detected in WWTP effluent and EAS, with the majority (>99%) of ARGs present in EAS [18–20]. ARGs can still be detected in EAS after physical (thermal, microwave, and drying), chemical (alkaline, ozone, and hydrogen peroxide), and biological treatments (composting, anaerobic digestion, and aerobic digestion), and their release into the environment poses the risk of ARG propagation and proliferation [21,22].
In terms of the reduction of ARGs in EAS, tetracycline resistance genes (TRG) (tet), sulfonamide resistance genes (sul), β-lactam resistance genes (bla), macrolide resistance genes (erm), and quinolone resistance genes (qnr) are the most studied genes [23]. To analyze horizontal gene transfer of ARGs, mobile genetic elements (MGE), most typically integrator genes (intI), are also usually examined simultaneously [23]. TRGs are the most commonly found ARGs in wastewater and sludge from WWTPs [24], and more than 22 TRGs have been identified in wastewater and EAS, with resistance mechanisms mainly including ribosomal protection protein class (RPP), exocytosis pump class (EFF), and enzyme modification class [25]. Therefore, TRGs have been used as model ARGs for ARG attenuation studies in EAS [26,27].
Wu et al. [10] used three temperatures (room temperature, 70°C, and 120°C) and two pH (10 and 11) conditions for alkaline-thermal digestion of EAS and found that the best reduction of four TRGs was achieved at 70°C and pH 11. Hence, in this study, we used lime, which is used as a traditional conditioner for full-scale treatment and lab-scale scientific research for EAS [7,28], instead of strong alkali, and pasteurized steam heating to 70°C, instead of conventional high-temperature heating, for thermal alkaline hydrolysis of EAS to come up with a cost-effective solution. Moreover, the decrease in ARGs through this process was also investigated focusing on TRGs.
2.1 Excess Activated Sludge (EAS)
In this study, EAS (moisture content 97.8%, pH 6.7, ORP-289 mV) was obtained from a sludge thickening tank of a domestic wastewater treatment plant (WWTP) in Sichuan Province, China. This WWTP adopts the treatment process of “CASS + fiber carousel filter.” The EAS was filtered through a fine mesh sieve to remove large impurities. Ten liters of filtered EAS were placed in a 10-L plastic container; after sedimentation for 1 h, the supernatant was discharged, 10 of tap water was added, and the samples were stirred for 1 min. Thereafter, the operation of “sedimentation, draining supernatant, adding water, and stirring” was performed two times to achieve the purpose of sludge washing. Finally, the supernatant was discharged until the final volume of EAS was 3 L, when the mixed liquid suspended solid (MLSS) of EAS was 8756.24 ± 145.21 mg/L.
Five sets of experiments were run with 200 mL of the washed EAS placed in 500-mL conical flasks with a breathable plug for each set, as further described. (1) Control activated sludge (CAS), no modulation and treatment were done. (2) Direct TH, the conical flask was capped and placed in a constant temperature water bath shaker (SHY-2A; BILON, Shanghai, China) at 70°C and shaken at 160 rpm for 1 h [12]. (3) Pasteurized thermal hydrolysis group (PTH), the conical bottle was placed in an electromagnetic mixer (85-2A; Xinrui, Changzhou, Jiangsu, China) with a temperature probe, and the stirring speed was set to 160 rpm. The electric furnace heated the flask containing distilled water, and the steam was introduced through a hose to bring the sludge temperature to 70°C and maintain it. (4) Pasteurized thermal alkaline hydrolysis (PTH + NaOH), and NaOH solids (AR) were added to adjust the pH of the EAS to 11; other operations were the same as those of the experimental group (3) [10]. (5) Pasteurized lime thermal hydrolysis (PTH + CaO), to investigate the feasibility of using lime instead of a strong alkali in thermal alkaline hydrolysis, 3.2 g of lime (CaO, 83.4%) was added to the flask with the final addition ratio of 16 kg/t, and steam was passed to bring the sludge temperature to 70°C and maintain it for 1 h; other operations were the same as those of experimental group (3).
Each experimental group had three parallel repetitions. After the treatment, pH was measured using the electrode method and the mixture was centrifuged at 14,000 rpm for 2 min. The supernatant was used to detect chemical indexes, and the sludge was used to extract DNA for the detection and analysis of TRGs.
Determination of NH4+-N and SCOD concentrations was based on Nessler’s reagent colorimetry method (HJ 535-2009) and potassium dichromate method (HJ 828-2017), respectively, which were performed using a UV spectrophotometer (DR6000, HACH, Loveland, CO, USA).
Before DNA extraction from the sludge, the samples were pre-washed thrice using PBS (pH 8.0) and 1.7% polyvinylpyrrolidone K30 to remove impurities such as humic acid from the sludge. The total DNA in the sludge was extracted using the FastDNA SPIN Kit for Soil (MP Biomedicals, CA, USA) according to the instructions provided with the kit. The concentration and mass of the extracted DNA solution were determined using an ultra-micro spectrophotometer (MD2000D; Thermo Fisher Scientific, MA, USA). The value of OD260/OD280 between 1.8 and 2.0 indicated successful DNA extraction. The extracted DNA solution was stored at −20°C.
Five typical TRGs of EAS, namely, 2 RPP-TRGs (tetW, tetS), 2 EFF-TRGs (tetZ, tetC), and 1 enzyme-modified TRGs (tetX), as well as one typical MGE (class 1 transposon gene, intI1) were examined for all experimental setups. Additionally, total 16S rRNA genes were quantified to characterize the microbial population of the sludge. The primers for the above genes are listed in Table 1.
Fluorescence-based quantitative PCR (ABI 7500, ABI, US) was used for the quantitative detection of the above target genes. PCR was performed with reaction mixtures of final volume 20 μL, containing 10 μL qPCR mix (2× SYBR Green PCR Master Mix kit, ABI), 0.5 μL F-primer (10 μM, Shanghai Sangon), 0.5 μL R-primer (10 μM, Shanghai Sangon), 1 μL BSA (10 mg/mL, Shanghai Sangon), 3 μL DNA solution, and 5 μL ddH2O. Each assay was performed in triplicates.
The absolute abundance of the target gene was calculated based on the gene copies per dry weight solid and relative abundance was calculated as the ratio of the absolute abundance of the target gene to that of the 16S rRNA gene. A one-way ANOVA was used to compare the significant changes in the absolute and relative abundance of target genes among the experimental groups. Before the ANOVA analysis, the data were tested for normality (Kolmogorov–Smirnov test) and homogeneity of variance (two-sample variance test). A two-sample t-test was used to compare the significant differences between groups. Pearson’s correlation was used to calculate the correlation coefficients between the indicators. The analyses were conducted using OriginPro version 2021 software, and the significance level was set at 0.05. Principal component analysis (PCA) was performed using Canoco 5 software. Heatmaps were plotted using TBtools software [34].
3.1 Measurement of Physicochemical Components
As shown in Fig. 1A, the pH of control (CAS) was 6.6 ± 0.1. The pH of EAS after direct TH and PTH did not change significantly compared with that of CAS possibly because the humic acid in sludge possesses good buffering properties, and the acid–base and redox conditions are not easily altered by subtle changes in the environment [35]. The sludge after TH has a relatively neutral pH (7.0 ± 0.1), which facilitates subsequent bioavailability and land use of sludge [36,37]. The pH of the mixture after thermal alkaline hydrolysis treatment using NaOH (PTH + NaOH) was the highest, reaching 9.2 ± 0.2; the pH of the thermal alkaline hydrolysis treatment group using lime (PTH + CaO) was 8.6 ± 0.3. After PTH + NaOH treatment, the pH was significantly lower than the initial pH 11, indicating that the thermal alkaline hydrolysis system consumes a large amount of alkali.
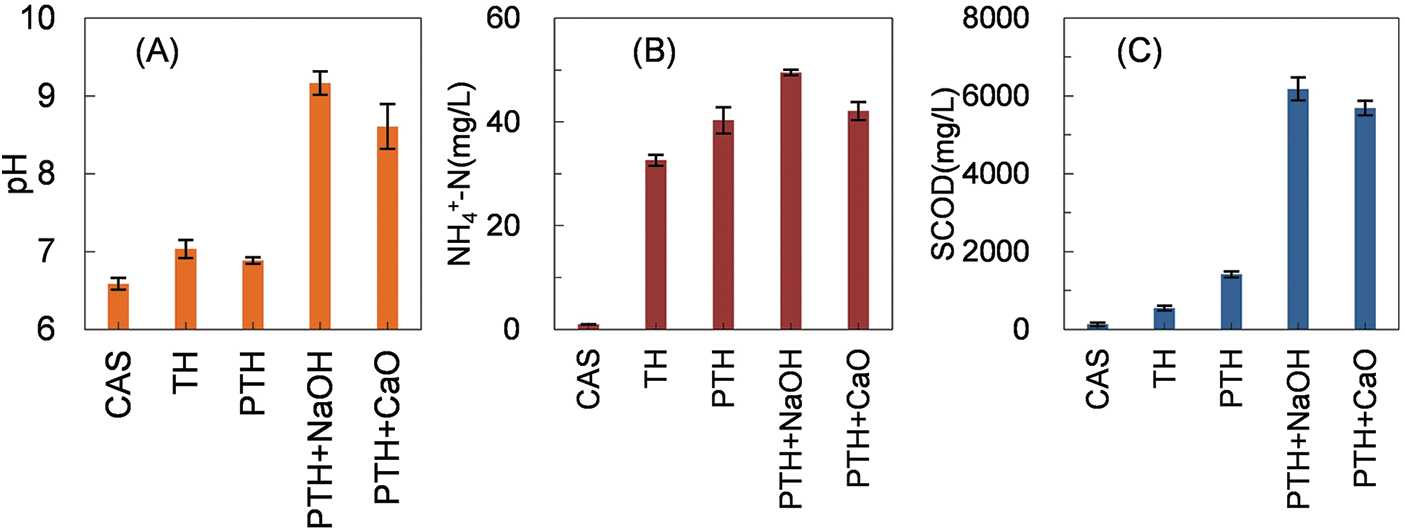
Figure 1: Changes in (A) pH, (B) NH4+-N, and (C) SCOD after different treatments of EAS
Compared with those in the control group, NH4+-N and SCOD leached from the sludge in all groups, as shown in Figs. 1B and 1C. The NH4+-N and SCOD concentrations were 32.6 ± 1.1 and 548.6 ± 61.7 mg/L, respectively, for TH. Under the same temperature conditions, when pasteurization was used instead of direct heating (PTH), the concentration of NH4+-N and SCOD increased to 40.3 ± 2.6 and 1414.0 ± 80.7 mg/L respectively, which were 1.24- and 2.58-times higher than those in the TH group. After the two thermal hydrolysis treatments, the NH4+-N and SCOD dissolution concentrations in the systems were remarkably lower than those in the study of Li et al. [38]. The reason may be that the thermal hydrolysis temperature in this experiment was only 70°C, which is considerably lower than the thermal hydrolysis temperature of 90°C–210°C used in Li et al.’s study. TH breaks down the sludge and releases a large amount of organic matter, mainly including proteins, polysaccharides, and lipids [39]. Among them, the hydrolysis of proteins increased the NH4+-N concentration in the solution [40]. Although the temperature was the same for TH and PTH, with the change in heating method from TH to PTH, more cell breakage occurred in EAS. This may be because in the PTH process, the water vapor passed into the system condenses and releases a large amount of heat, and the heat gained by the sludge contacting the steam instantaneously is higher than that in TH. Moreover, the steam passed into the system helps in agitation, which increases the heat and mass transfer efficiency. Therefore, pasteurization is also commonly used in water treatment and solid waste treatment [41,42]. However, PTH has the problem of water consumption, and increased volume of sludge after treatment due to water vapor influx.
With the addition of NaOH to the pasteurized thermal hydrolysis (PTH + NaOH), the NH4+-N and SCOD dissolved concentrations in the mixture were 49.5 ± 0.5 and 6175.4 ± 295.4 mg/L (Figs. 1B and 1C), which were 4.37- and 1.23-times higher than those in the PTH group, respectively. Wu et al. [10] performed thermal alkaline digestion of EAS at a medium temperature of 70°C and pH 11, and the dissolved SCOD was approximately 7400 mg/L, which is similar to the present experimental results of the PTH + NaOH and PTH + CaO groups. Dai et al. [43] carried out hot alkaline hydrolysis of sludge, and set the temperature to 135°C and pH to 12; the SCOD reached approximately 9000 mg/L after treatment. It is apparent that higher temperature and pH conditions are more favorable for SCOD dissolution. The dissolved NH4+-N and SCOD concentrations in the thermal alkaline digestion treatment using CaO (PTH + CaO) instead of NaOH as the base source were 42.1 ± 1.8 and 5690.0 ± 187.6 mg/L, respectively (Figs. 1B and 1C). Although the dissolved NH4+-N and SCOD concentrations in the PTH + CaO group were lower than those in the PTH + NaOH group, the NH4+-N and SCOD concentrations reached 85.04% and 92.14% in the PTH + NaOH group, respectively. PTH + CaO is more advantageous for the actual application in sludge treatment and resource utilization because it is relatively cheaper than PTH + NaOH and has sufficiently high nutrient dissolution performance.
3.2 Changes in the Absolute and Relative Abundance of TRGs and intI1
As is shown in Figs. 2A–2E, all target TRGs were detected in CAS with absolute abundances ranging from 5.38 × 104 to 7.08 × 108 copies/g of dry solid, and the range of absolute TRG abundance (Fig. 2F) was similar to that reported previously [44,45]. tetC had the highest absolute abundance (7.08 × 108 ± 3.34 × 105 copies/g of dry solid) among the five TRGs detected in CAS, followed by tetX (Fig. 2G). Moreover, tetC remained the major TRGs even after thermal and alkali-thermal treatments. In a previous survey on TRG contamination in soil samples, tetC presented the highest abundance among all TRGs [46]. Notably, all TRGs were still detected in the treated EAS with absolute abundances ranging from 5.38 × 104 to 5.12 × 108 copies/g of dry solid. It is evident that the discharge of treated EAS samples into the natural environment still poses the risk of TRG transmission and proliferation.
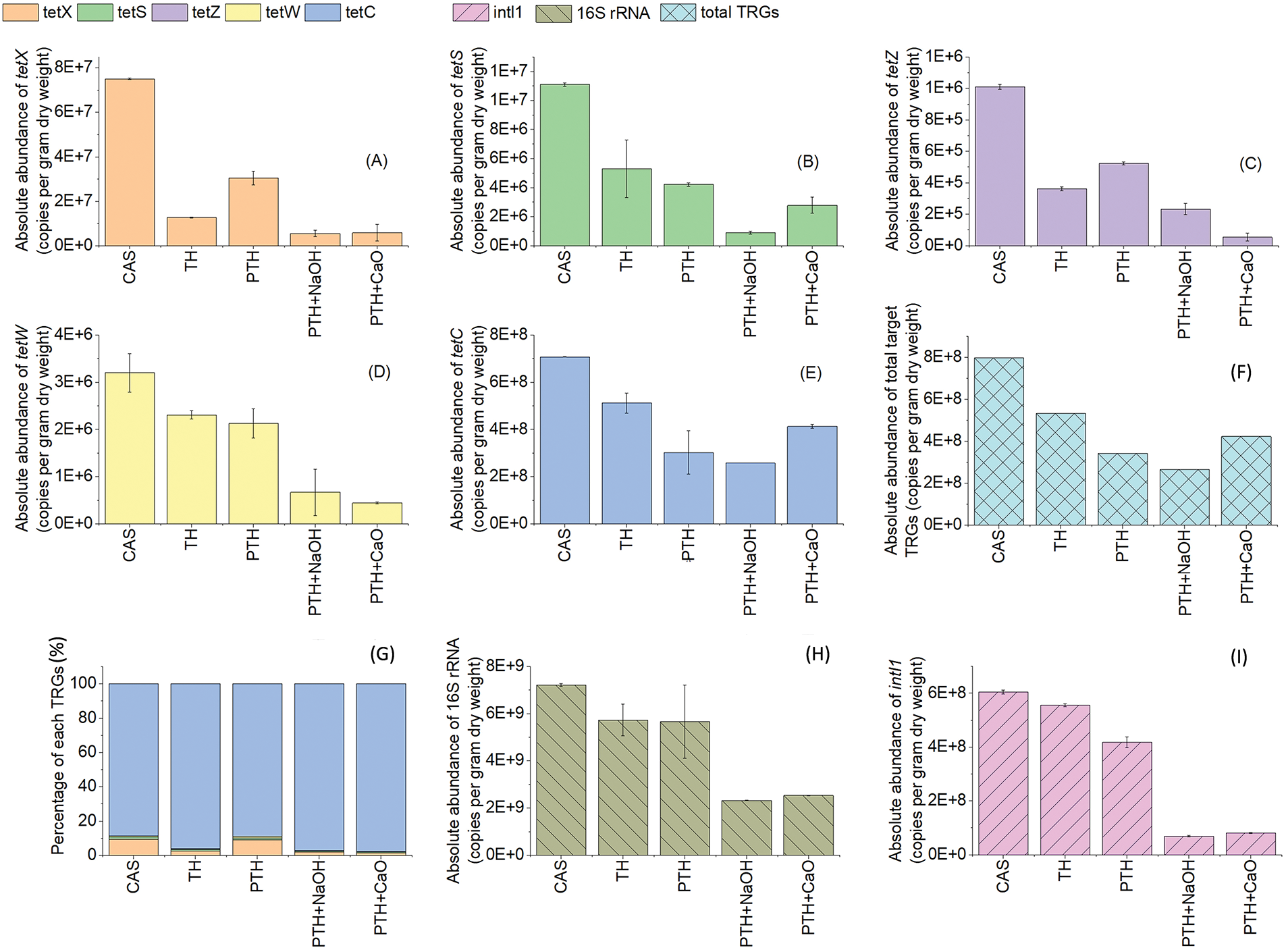
Figure 2: Changes in (A–F) the absolute abundance of TRGs, (G) the percentage of individual TRG after different treatments of EAS, and Changes in (H–I) the absolute abundance of 16S rRNA gene and intI1 gene
As is shown in Figs. 2A–2F, the two-sample t-test results showed that TH significantly reduced the absolute abundance of all TRGs and intI1 (P = 0.7697, P > t = 0.3008) with removal efficiencies of 27.64%–83.09% and 8.23%, respectively, compared to CAS. The removal efficiencies of TRGs and intI1 in the PTH group were 33.42%–62.23% and 30.90%, respectively. Compared with those by TH, the removal efficiencies of tetS, tetW, tetC and intI1 by PTH increased by 5.81%–29.50%. However, the removal efficiencies of PTH were 23.70% and 15.88% lower for tetX and tetZ vis-à-vis those of TH, respectively. Overall comparison using the two-sample t-test showed that the absolute abundance of TRGs and intI1 in the TH and PTH groups was not significantly different (P = 0.6924, P > t = 0.4613). This finding indicates that both kinds of thermal treatments remove TRGs and intI1, and the use of pasteurization instead of direct heating does not further enhance the removal of TRGs and intI1.
On the basis of PTH, alkali was added for alkali-thermal treatment. When NaOH was used as the base source, the removal rates of tetX, tetS, tetZ, tetW, tetC, and intI1 were 92.74%, 91.99%, 77.02%, 79.08%, 63.63%, and 88.62%, respectively. Compared with those by PTH, the removal efficiencies of TRGs and intI1 by PTH + NaOH were significantly (P < 0.05) increased by 6.48%–45.66% and 57.72%, respectively. With lime as the base source, the removal rates of tetX, tetS, tetZ, tetW, tetC, and intI1 were 92.30%, 75.02%, 94.70%, 86.15%, 41.57%, and 86.56%, respectively. In this study, the removal of most TRGs after alkali-thermal treatment was improved (except for tetC) in the PTH + CaO group, with an increase of 6.48%–57.72%, compared with that in the PTH group, which indicated that alkali-thermal treatment was better for the removal of TRGs than PTH.
A comparative analysis revealed that the removal efficiencies of absolute abundance of total TRGs, 16S rRNA, and intI1 were 66.84%, 67.91%, and 88.62% for PTH + NaOH, which were 19.78%, 3.02%, and 2.05% higher than those of PTH + CaO group, respectively. This finding indicates that NaOH is better for alkali-thermal treatment than lime, especially for TRG reduction.
Overall, thermal treatment and alkali-thermal treatment reduced the relative abundance of most TRGs (except tetC) and intI1, as shown in Fig. 3. After treatment, the relative abundance changes in TRGs and intI1 were clustered according to the treatment method, and the relative abundance changes in the target genes caused by alkali-thermal treatment were more comparable to those caused by thermal treatment. In addition, in terms of target gene types, the relative abundance changes in tetW (an RPP-TRG) were closer to the relative abundance changes in intI1, tetX (an enzyme modified TRG) clustered with tetZ (an EFF-TRG), and tetS (RPP-TRG) clustered with tetC (EFF-TEGs). This finding indicates that the relative abundance changes in TRGs during thermal treatment and alkali-thermal treatment were not related to the resistance mechanism of TRGs. A direct relationship between changes in ARGs and resistance mechanisms has not been reported previously [47,48].
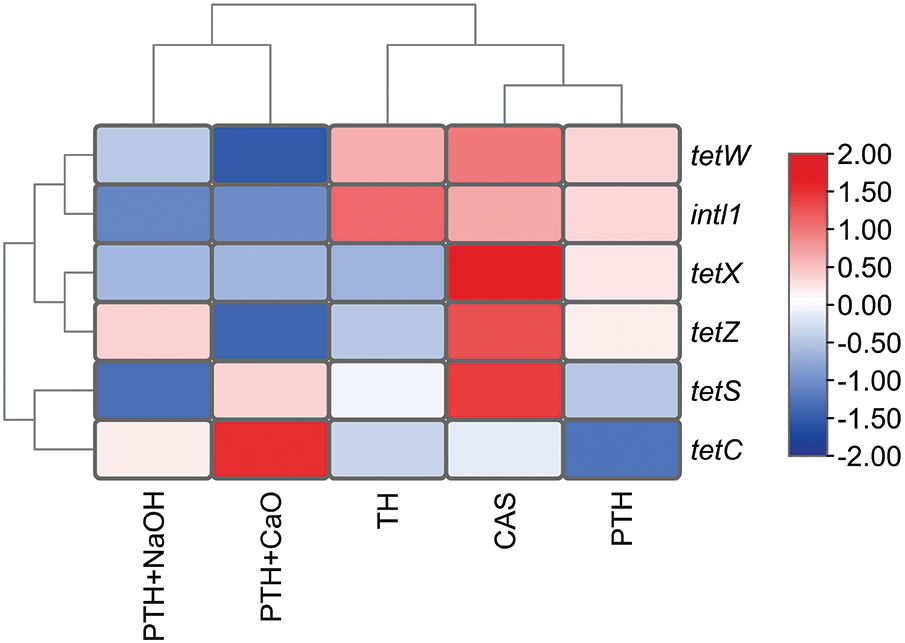
Figure 3: Heatmap of relative abundance of TRGs, intI1, and 16S rRNA genes after different treatments of EAS
In summary, TH at a medium temperature (70°C) can remove TRGs and intI1 from EAS, and the addition of alkali (NaOH or lime) can significantly increase the removal efficiency (except for tetC). Among them, alkali thermal treatment with NaOH was more effective in reducing the absolute and relative abundance of target genes compared to lime.
3.3 Correlation between Physicochemical Indexes and Target Genes
During the thermal and/or alkali treatment of EAS, pH showed a trend similar to that of SCOD and NH4+-N (Fig. 1). Moreover, it was significantly positively correlated with SCOD (R = 0.9831, P < 0.05), indicating that the addition of alkali to the TH group could promote the leaching of organic matter from the sludge. The results presented in Sections 3.1 and 3.2 revealed that pH showed opposite trends for all the target genes—pH was significantly negatively correlated with the absolute abundance of tetW (R = −0.9398, P < 0.05), intI1 (R = −0.9600, P < 0.05), and 16S rRNA (R = −0.9744, P < 0.05), indicating that alkali dosing can decrease bacterial abundance and reduce the potential of horizontal gene transfer. It has also been reported that the addition of alkali facilitates a reduction in TRGs during EAS treatment. For example, Huang et al. [49] reduced the target TRGs by 0.70–1.31 orders of magnitude at pH 10 compared with that under neutral conditions during anaerobic digestion of EAS. A study by Xiao et al. [50] showed that alkaline treatment was effective in destroying bacteria including antibiotic-resistance bacteria. This explains the improved reduction in TRGs from EAS after alkaline treatment in this study.
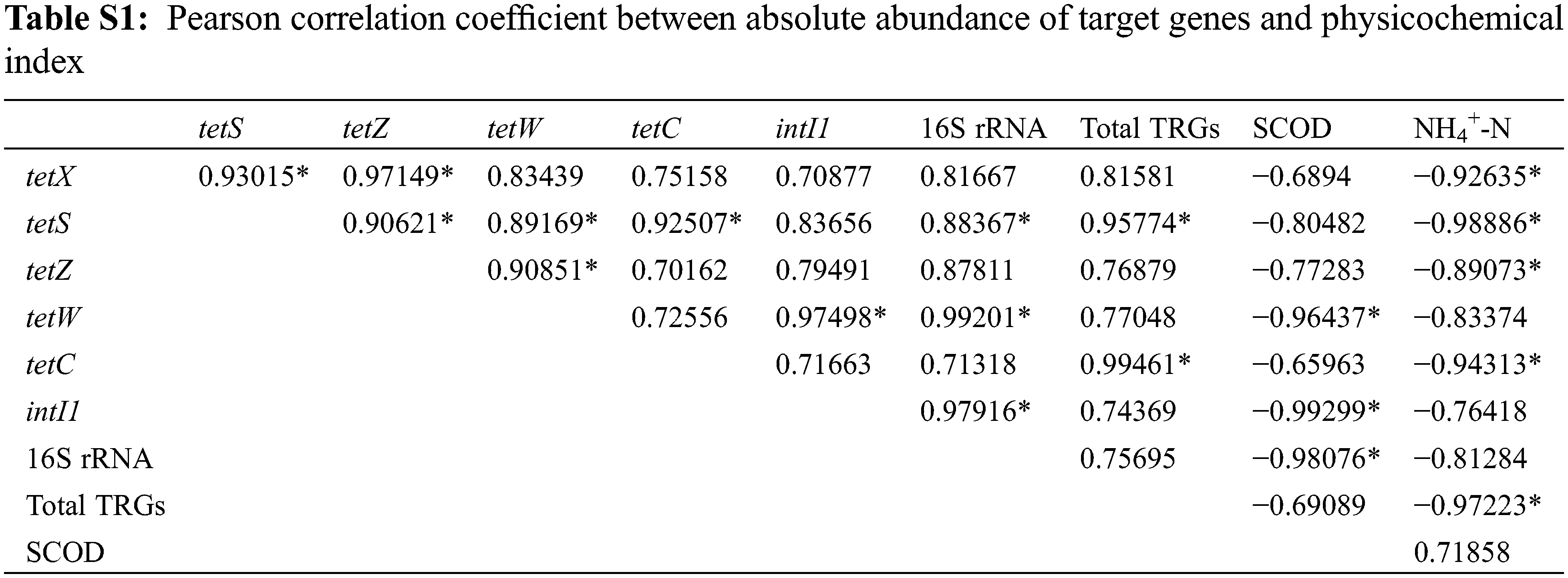
To investigate the relationship between target genes and physicochemical parameters, Pearson correlation coefficients between the absolute abundance of target genes and physicochemical index parameters were calculated (Table S1), and a heatmap was correspondingly drawn (Fig. 4A). Fig. 4A shows that NH4+-N was significantly negatively correlated with the absolute abundance of tetX (R = −0.9264, P < 0.05), tetS (R = −0.9889, P < 0.05), tetZ (R = −0.8907, P < 0.05), and tetC (R = −9431, P < 0.05); SCOD was significantly negatively correlated with the absolute abundance of tetW (R = −0.9644, P < 0.05) and intI1 (R = −0.9930, P < 0.05). SCOD was significantly negatively correlated with the absolute abundance of 16S rRNA (R = −0.9808), indicating cell lysis of microbes in EAS during the treatment. To visualize the correlation and clustering among target genes, a PCA analysis was performed on the absolute abundance of target genes and physicochemical indexes (Fig. 4B). As shown in Fig. 4B, target genes and physicochemical indices were significantly altered in EAS after thermal hydrolysis (in the upper right part of the PCA figure) and alkaline thermal hydrolysis (in the lower left part of the PCA figure) compared with that after CAS (in the lower right part of PCA figure). The target genes and physicochemical indexes of EAS were remarkably changed by alkali addition compared with those by thermal hydrolysis alone, which was consistent with the results in Sections 3.1 and 3.2. Moreover, the arrows of the target genes are well clustered, which indicated there was a good correlation between the target genes, but the overall trend of the target genes and the physicochemical indicators (NH4+-N and SCOD) was opposite.

Figure 4: Heatmap of (A) correlation coefficients between physicochemical indexes and target genes, and (B) principal component analysis (PCA) of physicochemical indexes and target genes (blue arrows)
Note: * in the heatmap (A) indicates P < 0.05; totlTRGs in B indicates the absolute abundance of total TRGs.
3.4 Economic and Technological Analyses
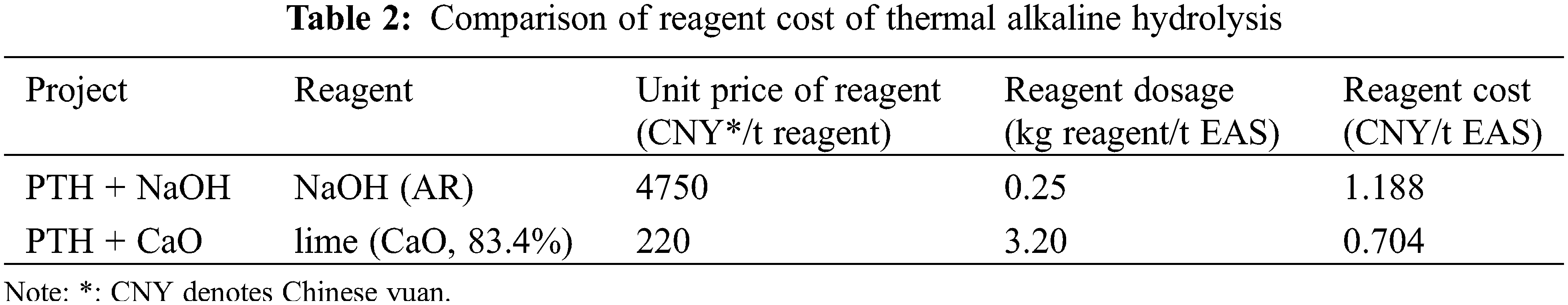
The difference in the cost between the two thermal alkaline hydrolysis processes of EAS was due to the diversity in the alkaline source (Table 2). The reagent cost when lime was used as the alkaline source was 0.704 CNY/t EAS, which was 0.484 CNY/t EAS lower than that when NaOH was used as the alkaline source. Gao et al. [51] used NaOH as an alkali source to conduct thermal alkali hydrolysis (pH12, 120°C) of EAS (moisture content 99%), and the total treatment cost was approximately 36 CNY/kg dry solid. Although lime is commonly used as a conditioner in EAS adjustment, researchers are still trying to find more economical or beneficial alternatives to solid waste resource utilization. For example, Gonzalez-Tolivia et al. [52] added basic oxygen furnace slag instead of lime as a stabilizer of EAS, resulting in a degree of nutrient dissolution similar to that achieved with the addition of lime. In the future study, we will focus on optimizing the lime dosage, steam inlet speed, and other factors to further explore cost-reduction strategies.
The alkali and/or thermal treatment of EAS releases nutrients from EAS and reduces the TRGs and intI in EAS. The released amount of nutrients was enhanced using pasteurization vis-à-vis direct heating. However, the use of pasteurization heating instead of direct heating did not promote the reduction in target genes. Adding alkali on the basis of thermal hydrolysis can promote the dissolution of nutrients and the reduction of target genes. Additionally, the efficacy of NaOH as an alkali source was higher than that of CaO. The leaching NH4+-N and SCOD concentrations in the PTH + CaO group were close to those in the PTH + NaOH group, with NH4+-N and SCOD concentrations of 85.04% and 92.14%, respectively, in the PTH + NaOH group, but the removal efficiency of the absolute abundance of total TRGs was 19.78% lower than that of the PTH + NaOH group. Considering the economic cost, relatively lower pH of the mixture after treatment, and the effective reduction of TRGs, the pasteurized lime thermal alkaline hydrolysis treatment (PTH + CaO) has more advantages than other experimental treatments in practical application. However, further research is needed to optimize the parameters in the process of alkaline hydrolysis. This study provides data reference for the development of economic and effective sludge treatment technology.
Acknowledgement: We thank Editage (www.editage.cn) for English language editing.
Funding Statement: This work was supported by the Key R&D Projects of the Sichuan Provincial Department of Science and Technology in 2022 (No. 2022YFS0457) and Innovation and Entrepreneurship Training Program for College Students (No. 202210649050).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Ministry of Housing and Urban Rural Development of the People’s Republic of China. Statistical Yearbook of Urban and Rural Construction in 2020. https://www.mohurd.gov.cn/gongkai/fdzdgknr/sjfb/tjxx/index.html. [Google Scholar]
2. Costanzo, D. N., Cesaro, A., Capua, D. F., Esposito, G. (2021). Exploiting the nutrient potential of anaerobically digested sewage sludge: A review. Energies, 14(23), 8149. https://doi.org/10.3390/en14238149 [Google Scholar] [CrossRef]
3. Yu, W., Duan, H., Wang, Z., Yang, J., Yuan, Z. et al. (2022). Transforming anaerobically digested sludge into high-quality biosolids with an integrated physiochemical approach. Resources, Conservation and Recycling, 184(1), 106416. https://doi.org/10.1016/j.resconrec.2022.106416 [Google Scholar] [CrossRef]
4. Cui, G., Fu, X., Bhat, A. S., Tian, W., Lei, X. et al. (2022). Temperature impacts fate of antibiotic resistance genes during vermicomposting of domestic excess activated sludge. Environmental Research, 207(10), 112654. https://doi.org/10.1016/j.envres.2021.112654 [Google Scholar] [PubMed] [CrossRef]
5. Wang, T., Xing, Z., Zeng, L., Peng, C., Shi, H. et al. (2022). Anaerobic codigestion of excess sludge with chicken manure with a focus on methane yield and digestate dewaterability. Bioresource Technology Reports, 19(2), 101127. https://doi.org/10.1016/j.biteb.2022.101127 [Google Scholar] [CrossRef]
6. Cai, C., Hu, C., Yang, W., Hua, Y., Li, L. et al. (2021). Sustainable disposal of excess sludge: Post-thermal hydrolysis for anaerobically digested sludge. Journal of Cleaner Production, 321(5), 128893. https://doi.org/10.1016/j.jclepro.2021.128893 [Google Scholar] [CrossRef]
7. Lu, Y., Meng, X., Wang, J., Dieketseng, Y. M., Xiao, Y. et al. (2022). Bioleaching rather than chemical conditioning using Fe[III]/CaO or polyacrylamide mitigates antibiotic resistance in sludge composting via pre-removing antibiotic resistance genes and limiting horizontal gene transfer. Waste Management, 137(24), 89–99. https://doi.org/10.1016/j.wasman.2021.10.029 [Google Scholar] [PubMed] [CrossRef]
8. Ngo, L. P., Udugama, A. I., Gernaey, V. K., Young, R. B., Baroutian, S. (2021). Mechanisms, status, and challenges of thermal hydrolysis and advanced thermal hydrolysis processes in sewage sludge treatment. Chemosphere, 281, 130890. https://doi.org/10.1016/j.chemosphere.2021.130890 [Google Scholar] [PubMed] [CrossRef]
9. Nguyen, K. V., Chaudhary, K. D., Dahal, H. R., Trinh, N., Kim, H. et al. (2021). Review on pretreatment techniques to improve anaerobic digestion of sewage sludge. Fuel, 285, 119105. https://doi.org/10.1016/j.fuel.2020.119105 [Google Scholar] [CrossRef]
10. Wu, X., Hu, Y., Liao, Z., Cheng, J., Chen, Y. (2019). Efficiency and the change of antibiotic resistance genes during thermal-alkaline hydrolysis and mesophilic two-phase anaerobic digestion of excess sludge. Acta Scientiae Circumstantiae, 39(7), 2088–2098. https://doi.org/10.13671/j.hjkxxb.2019.0124 [Google Scholar] [CrossRef]
11. Zhang, Z., Li, X., Liu, H., Zamyadi, A., Guo, W. et al. (2022). Advancements in detection and removal of antibiotic resistance genes in sludge digestion: A state-of-art review. Bioresource Technology, 344(1), 126197. https://doi.org/10.1016/j.biortech.2021.126197 [Google Scholar] [PubMed] [CrossRef]
12. Zheng, X. (2021). Research on enhancing thermal alkaline hydrolysis of sludge by external reflux/calcium ion and improving its dewatering performance (MA Thesis). Hefei University of Technology, Hefei, Anhui. https://doi.org/10.27101/d.cnki.ghfgu.2021.000723 [Google Scholar] [CrossRef]
13. Tang, Y., Xie, H., Sun, J., Li, X., Zhang, Y. et al. (2022). Alkaline thermal hydrolysis of sewage sludge to produce high-quality liquid fertilizer rich in nitrogen-containing plant-growth-promoting nutrients and biostimulants. Water Research, 211(12), 118036. https://doi.org/10.1016/j.watres.2021.118036 [Google Scholar] [PubMed] [CrossRef]
14. Zhang, J., Yang, M., Zhong, H., Liu, M., Sui, Q. et al. (2018). Deciphering the factors influencing the discrepant fate of antibiotic resistance genes in sludge and water phases during municipal wastewater treatment. Bioresource Technology, 265, 310–319. https://doi.org/10.1016/j.biortech.2018.06.021 [Google Scholar] [PubMed] [CrossRef]
15. Wang, R., Ji, M., Zhai, H., Guo, Y., Liu, Y. (2021). Occurrence of antibiotics and antibiotic-resistant genes in WWTP effluent-receiving water bodies and reclaimed wastewater treatment plants. Science of the Total Environment, 796(3), 148919. https://doi.org/10.1016/j.scitotenv.2021.148919 [Google Scholar] [PubMed] [CrossRef]
16. Obayiuwana, A., Ogunjobi, A., Ibekwe, A. (2021). Prevalence of antibiotic resistance genes in pharmaceutical wastewaters. Water, 13(13), 1731. https://doi.org/10.3390/w13131731 [Google Scholar] [CrossRef]
17. Chen, T., Zhang, S., Zhu, R., Zhao, M., Zhang, Y. et al. (2022). Distribution and driving factors of antibiotic resistance genes in treated wastewater from different types of livestock farms. The Science of the Total Environment, 849, 157837. https://doi.org/10.1016/j.scitotenv.2022.157837 [Google Scholar] [PubMed] [CrossRef]
18. Zhai, W., Yang, F., Mao, D., Luo, Y. (2016). Fate and removal of various antibiotic resistance genes in typical pharmaceutical wastewater treatment systems. Environmental Science & Pollution Research International, 23(12), 12030–12038. https://doi.org/10.1007/s11356-016-6350-9 [Google Scholar] [PubMed] [CrossRef]
19. Xu, Y., Hou, M., Li, Y., Huang, L., Ruan, J. et al. (2017). Distribution of tetracycline resistance genes and AmpC β-lactamase genes in representative non-urban sewage plants and correlations with treatment processes and heavy metals. Chemosphere, 170(2), 274–281. https://doi.org/10.1016/j.chemosphere.2016.12.027 [Google Scholar] [PubMed] [CrossRef]
20. Zhu, T., Su, Z., Lai, W., Zhang, Y., Liu, Y. (2021). Insights into the fate and removal of antibiotics and antibiotic resistance genes using biological wastewater treatment technology. Science of the Total Environment, 776, 145906. https://doi.org/10.1016/j.scitotenv.2021.145906 [Google Scholar] [CrossRef]
21. Wang, J., Chen, X. (2020). Removal of antibiotic resistance genes (ARGs) in various wastewater treatment processes: An overview. Critical Reviews in Environmental Science and Technology, 17(4), 1–60. https://doi.org/10.1080/10643389.2020.1835124 [Google Scholar] [CrossRef]
22. Goulas, A., Belhadi, D., Descamps, A., Andremont, A., Benoit, P. et al. (2020). How effective are strategies to control the dissemination of antibiotic resistance in the environment? A systematic review. Environmental Evidence, 9(1), 1–32. https://doi.org/10.1186/s13750-020-0187-x [Google Scholar] [CrossRef]
23. Ezeuko, S. A., Ojemaye, O. M., Okoh, O. O., Okoh, I. A. (2021). Technological advancement for eliminating antibiotic resistance genes from wastewater: A review of their mechanisms and progress. Journal of Environmental Chemical Engineering, 9(5), 106183. https://doi.org/10.1016/j.jece.2021.106183 [Google Scholar] [CrossRef]
24. Li, R., Jay, A. J., Stenstrom, K. M. (2019). Fate of antibiotic resistance genes and antibiotic-resistant bacteria in water resource recovery facilities. Water Environment Research, 91(1), 5–20. https://doi.org/10.1002/wer.1008 [Google Scholar] [PubMed] [CrossRef]
25. Li, J., Fu, X., Liu, Y., Zhou, W., Liu, M. (2014). New research advances in tetracycline drug efflux pump in bacteria. Heilongjiang Animal Science and Veterinary, 2014(23), 48–50. https://doi.org/10.13881/j.cnki.hljxmsy.2014.1407 [Google Scholar] [CrossRef]
26. Zhou, C., Wu, J., Liu, B., Ma, W., Yang, S. et al. (2022). (Micro) nanoplastics promote the risk of antibiotic resistance gene propagation in biological phosphorus removal system. Journal of Hazardous Materials, 431(5), 128547. https://doi.org/10.1016/j.jhazmat.2022.128547 [Google Scholar] [PubMed] [CrossRef]
27. Lu, W., Wang, M., Wu, J., Jiang, Q., Jin, J. et al. (2022). Spread of chloramphenicol and tetracycline resistance genes by plasmid mobilization in agricultural soil. Environmental Pollution, 260, 113998. https://doi.org/10.1016/j.envpol.2020.113998 [Google Scholar] [PubMed] [CrossRef]
28. Wen, Q., Ma, M., Hou, H., Yu, W., Gui, G. et al. (2022). Recirculation of reject water in deep-dewatering process to influent of wastewater treatment plant and dewaterability of sludge conditioned with Fe2+/H2O2, Fe2+/Ca(ClO)2, and Fe2+/Na2S2O8: From bench to pilot-scale study. Environmental Research, 203(124), 111825. https://doi.org/10.1016/j.envres.2021.111825 [Google Scholar] [PubMed] [CrossRef]
29. Aminov, R. I., Garrigues-Jeanjean, N., Mackie, R. I. (2001). Molecular ecology of tetracycline resistance: Development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins. Applied and Environmental Microbiology, 67(1), 22–32. https://doi.org/10.1128/AEM.67.1.22-32.2001 [Google Scholar] [PubMed] [CrossRef]
30. Aminov, R. I., Chee-Sanford, J. C., Garrigues, N., Teferedegne, B., Krapac, I. J. et al. (2002). Development, validation, and application of PCR primers for detection of tetracycline efflux genes of gram-negative bacteria. Applied and Environmental Microbiology, 68(4), 1786–1793. https://doi.org/10.1128/AEM.68.4.1786-1793.2002 [Google Scholar] [PubMed] [CrossRef]
31. Ng, L. K., Martin, I., Alfa, M., Mulvey, M. (2001). Multiplex PCR for the detection of tetracycline resistant genes. Molecular and Cellular Probes, 15(4), 209–215. https://doi.org/10.1006/mcpr.2001.0363 [Google Scholar] [PubMed] [CrossRef]
32. Barraud, O., Baclet, M. C., Denis, F., Ploy, M. C. (2010). Quantitative multiplex real-time PCR for detecting class 1, 2 and 3 integrons. The Journal of Antimicrobial Chemotherapy, 65(8), 1642–1645. https://doi.org/10.1093/jac/dkq167 [Google Scholar] [PubMed] [CrossRef]
33. Wang, X., Wang, W., Zhang, Y., Sun, Z., Zhang, J. et al. (2019). Effect of environmental factors on nitrogen removal and functional genes expression of strain Pseudomonas sp. China Environmental Science, 39(10), 4377–4386. https://doi.org/10.19674/j.cnki.issn1000-6923.2019.0511 [Google Scholar] [CrossRef]
34. Chen, C., Chen, H., Zhang, Y., Thomas, R. H., Frank, H. M. et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Molecular Plant, 13(8), 1194–1202. https://doi.org/10.1016/j.molp.2020.06.009 [Google Scholar] [PubMed] [CrossRef]
35. Sun, J. H., Zhang, S. T., Yao, Y. Y. (2015). On the buffering performance of activated sludge. Journal of Safety and Environment, 15(1), 199–202. https://doi.org/10.13637/j.issn.1009-6094.2015.01.041 [Google Scholar] [CrossRef]
36. Zhang, J., Sui, Q., Lu, T., Zhong, H., Shen, P. et al. (2019). Sludge bio-drying followed by land application could control the spread of antibiotic resistance genes. Environment International, 130, 104906. https://doi.org/10.1016/j.envint.2019.104906 [Google Scholar] [PubMed] [CrossRef]
37. Zhao, X., Wang, Z., Xu, T., Feng, Z., Liu, J. et al. (2021). The fate of antibiotic resistance genes and their influential factors during excess sludge composting in a full-scale plant. Bioresource Technology, 342(7717), 126049. https://doi.org/10.1016/j.biortech.2021.126049 [Google Scholar] [PubMed] [CrossRef]
38. Li, X., Xiao, X., Liu, Y., Fang, G., Wang, P. et al. (2022). Analysis of organic matter conversion behavior and kinetics during thermal hydrolysis of sludge and its anaerobic digestion performance. Journal of Environmental Management, 305(23), 114408. https://doi.org/10.1016/j.jenvman.2021.114408 [Google Scholar] [PubMed] [CrossRef]
39. Xu, Q., Duan, N., Lin, C., Zhai, Z. (2019). Effect of hydrothermal pretreatment of dehydrated sludge with different temperature. China Biogas, 37(1), 31–35. https://doi.org/10.3969/j.issn.1000-1166.2019.01.007 [Google Scholar] [CrossRef]
40. Yan, Y., Zhang, Y., Gao, J., Qin, L., Liu, F. et al. (2022). Intracellular and extracellular sources, transformation process and resource recovery value of proteins extracted from wastewater treatment sludge via alkaline thermal hydrolysis and enzymatic hydrolysis. The Science of the Total Environment, 852(17–22), 158512. https://doi.org/10.1016/j.scitotenv.2022.158512 [Google Scholar] [PubMed] [CrossRef]
41. Zaman, S., Yousuf, A., Begum, A., Bari, L. M., Rabbani, S. K. (2019). Evaluation of adaptive low cost solar water pasteurization device for providing safe potable water in rural households. Journal of Water and Health, 17(2), 274–286. https://doi.org/10.2166/wh.2019.268 [Google Scholar] [PubMed] [CrossRef]
42. Zhang, D., Santha, H., Pallansch, K., Novak, T. J., Wang, Z. W. (2020). Repurposing pre-pasteurization as an in situ thermal hydrolysis pretreatment process for enhancing anaerobic digestion of municipal sludge: A horizontal comparison between temperature-phased and standalone thermophilic or mesophilic anaerobic digestion. Environmental Science: Water Research & Technology, 6(12), 3316–3325. https://doi.org/10.1039/D0EW00633E [Google Scholar] [CrossRef]
43. Dai, Q., Zhang, W., Yu, P., Yi, H., Liu, J. et al. (2018). Effects of heat and heat-alkaline treatments on disintegration and dissolved organic matter in sludge. Environmental Science, 39(5), 2283–2288. https://doi.org/10.13227/j.hjkx.201709052 [Google Scholar] [PubMed] [CrossRef]
44. Mohammed, A., Tian, Z., Zhang, Y., Yang, M., Yin, W. et al. (2021). Hydrothermal pretreatment of oxytetracycline fermentation residue: Removal of oxytetracycline and increasing the potential for anaerobic digestion. Environmental Engineering Research, 26(4), 1–9. https://doi.org/10.4491/eer.2020.258 [Google Scholar] [CrossRef]
45. Huang, K., Chen, J., Guan, M., Xia, H., Lin, L. (2020). Effects of biochars on the fate of antibiotics and their resistance genes during vermicomposting of dewatered sludge. Journal of Hazardous Materials, 379(5), 122767. https://doi.org/10.1016/j.jhazmat.2020.122767 [Google Scholar] [PubMed] [CrossRef]
46. Zou, S., Li, Q., He, Z. M. (2012). A preliminary study on the tetracycline resistance genes in the livestock soil, South China. Acta Scientiarum Naturalium Universitatis Sunyatseni, 51(6), 87–91. [Google Scholar]
47. Zhang, J., Mao, F., Loh, K., Gin, K. Y., Dai, Y. et al. (2018). Evaluating the effects of activated carbon on methane generation and the fate of antibiotic resistant genes and class I integrons during anaerobic digestion of solid organic wastes. Bioresource Technology, 249, 729–736. https://doi.org/10.1016/j.biortech.2017.10.082 [Google Scholar] [PubMed] [CrossRef]
48. Chen, M., Chang, J., Jiang, T., Wei, S., Zou, J. et al. (2022). Lime addition significantly attenuates tetracycline resistance genes and class 1 integrons in dewatered sludge by affecting bacterial profiles. Journal of Environmental Chemical Engineering, 10(3), 107429. https://doi.org/10.1016/j.jece.2022.107429 [Google Scholar] [CrossRef]
49. Huang, H., Chen, Y., Zheng, X., Su, Y., Wan, R. et al. (2016). Distribution of tetracycline resistance genes in anaerobic treatment of waste sludge: The role of pH in regulating tetracycline resistant bacteria and horizontal gene transfer. Bioresource Technology, 218(5), 1284–1289. https://doi.org/10.1016/j.biortech.2016.07.097 [Google Scholar] [PubMed] [CrossRef]
50. Xiao, B., Liu, C., Liu, J., Guo, X. (2015). Evaluation of the microbial cell structure damages in alkaline pretreatment of waste activated sludge. Bioresource Technology, 196(8), 109–115. https://doi.org/10.1016/j.biortech.2015.07.056 [Google Scholar] [PubMed] [CrossRef]
51. Gao, J., Weng, W., Yan, Y., Wang, Y., Wang, Q. (2020). Comparison of protein extraction methods from excess activated sludge. Chemosphere, 249(4), 126107. https://doi.org/10.1016/j.chemosphere.2020.126107 [Google Scholar] [PubMed] [CrossRef]
52. González-Tolivia, E., Collad, E., Oulego, P., Díaz, M. (2022). BOF slag as a new alkalizing agent for the stabilization of sewage sludge. Waste Management, 153(2), 335–346. https://doi.org/10.1016/j.wasman.2022.09.009 [Google Scholar] [PubMed] [CrossRef]
Supplementary Materials
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF
 Downloads
Downloads
 Citation Tools
Citation Tools