 Open Access
Open Access
ARTICLE
A Knowledge-Enhanced Disease Diagnosis Method Based on Prompt Learning and BERT Integration
School of Computer and Information Engineering, Shanghai Polytechnic University, Shanghai, 201209, China
* Corresponding Author: Hengyang Wu. Email:
Journal on Artificial Intelligence 2025, 7, 17-37. https://doi.org/10.32604/jai.2025.059607
Received 12 October 2024; Accepted 26 December 2024; Issue published 19 March 2025
Abstract
This paper proposes a knowledge-enhanced disease diagnosis method based on a prompt learning framework. Addressing challenges such as the complexity of medical terminology, the difficulty of constructing medical knowledge graphs, and the scarcity of medical data, the method retrieves structured knowledge from clinical cases via external knowledge graphs. The method retrieves structured knowledge from external knowledge graphs related to clinical cases, encodes it, and injects it into the prompt templates to enhance the language model’s understanding and reasoning capabilities for the task. We conducted experiments on three public datasets: CHIP-CTC, IMCS-V2-NER, and KUAKE-QTR. The results indicate that the proposed method significantly outperforms existing models across multiple evaluation metrics. Additionally, ablation studies confirmed the critical role of the knowledge injection module, as the removal of this module resulted in a significant drop in F1 score. The experimental results demonstrate that the proposed method not only effectively improves the accuracy of disease diagnosis but also enhances the interpretability of the predictions, providing more reliable support and evidence for clinical diagnosis.Keywords
Disease diagnosis is the process of identifying and confirming a patient’s illness by analyzing symptoms, medical history, physical examinations, and various medical test data to determine the underlying causes and guide treatment. Accurate diagnosis is not only the foundation for formulating treatment plans but also critical for preventing disease progression and transmission, improving the efficiency of medical resource utilization, and reducing healthcare costs.
Initially, people primarily relied on doctors’ experience and knowledge for diagnosis, using methods such as patient interviews, physical examinations, and laboratory tests. However, this simple and mechanical approach clearly could not produce optimal diagnostic results. To address issues of subjectivity, time consumption, and risks of misdiagnosis or missed diagnosis, knowledge engineering methods were later adopted [1]. These methods utilize rule-based matching techniques to determine text categories. However, in recent years, with the rapid development of technologies such as big data and cloud computing, the internet has experienced an explosive growth in information. The vast amount of textual data has posed significant challenges to traditional classification methods.
With the accumulation of large-scale clinical data, statistical analysis has emerged as a crucial tool for disease diagnosis. For instance, Lv et al. [2] and colleagues developed classification models for diagnosing sleep quality and diseases using methods such as multinomial logistic regression and discriminant analysis. These models are suitable for use during the early stages of diagnosis to categorize diseases. However, as data complexity and medical data volume increase exponentially, statistical methods face limitations in handling multidimensional and nonlinear relationships. Machine learning-based disease prediction models have reframed disease prediction as a classification problem [3–5]. However, these methods also exhibit constraints when dealing with complex multidimensional medical data, requiring extensive labeled datasets and often lacking generalizability across various disease contexts [6].
In addition, with the rapid advancement of artificial intelligence and natural language processing technologies, the use of deep learning models for text classification to assist in disease diagnosis has become a significant area of research. Many diagnostic tasks involve extracting and classifying disease-related information from unstructured text. Supported by NLP techniques, text classification models can efficiently process and analyze large volumes of clinical text data. Among them, the BERT model, as a powerful natural language processing tool, has demonstrated outstanding performance in many text classification tasks. Onan [6] proposed an innovative hierarchical graph-based text classification framework, which captures the complex relationships between nodes through a dynamic fusion mechanism of contextual node embeddings and the BERT model. Although these methods have achieved good results in classification accuracy and stability, they still require improvements in structured reasoning and integration of medical knowledge [7].
To address this issue, this paper proposes an integrated approach that combines pre-trained language models with knowledge graphs for extracting and processing structured knowledge from clinical texts, which is then applied to disease prediction tasks. While the focus of this study appears to be on text classification, the process plays a significant supporting role in diagnostic tasks. By classifying patient symptoms and medical history, the model identifies associations between specific symptoms and diseases, providing accurate diagnostic recommendations. Experimental results indicate that the proposed method achieves superior performance on publicly available disease diagnosis datasets. Furthermore, the experiments demonstrate that prompt-based knowledge injection effectively guides language models to grasp relevant medical knowledge, significantly enhancing reasoning performance. This approach transforms raw text into high-quality semantic vectors and maps these representations to specific terms through designed functions. This method not only enhances semantic expression but also provides rich contextual and structured knowledge support for disease prediction, improving the model’s performance and reliability in practical applications. The implementation steps and experimental results are elaborated in the subsequent sections.
The continuous advancement of information technology has driven the development of intelligent clinical decision support systems. The widespread application of machine learning models has significantly improved the effectiveness of these systems, particularly in the field of disease prediction. The evolution of these models has gone through several stages: from the early expert rule-based models, to models based on statistical analysis and case-based reasoning, and finally to the current advanced models utilizing machine learning and deep learning techniques [8]. At the same time, breakthroughs in the field of natural language processing (NLP) have introduced innovative tools and techniques, opening up new perspectives for disease diagnosis and providing unprecedented possibilities.
In these early disease diagnosis methods, rule-based research primarily relied on the analysis of medical literature and case data. Expert rule-based disease diagnosis methods involved the collection of expert diagnostic experiences to form disease diagnosis pathways, thus creating expert systems. A typical example of such an expert system is the MYCIN expert system [9], developed by Shortliffe in 1976, which became a foundational model for many subsequent expert systems in the medical field. However, many techniques are unable to explain the processes involved in disease monitoring and data inference. To address this, Aamir et al. [10] proposed a fuzzy rule-based diabetes classification system which combines fuzzy logic with the cosine amplitude method. They developed two fuzzy classifiers, and the proposed model demonstrated high predictive accuracy. Sanz et al. [11] further developed a new approach based on the Fuzzy Rule-Based Classification System (FRBCS) by integrating it with Interval-Valued Fuzzy Sets (IV-FRBCS). They validated the applicability and effectiveness of this method in medical diagnostic classification problems, demonstrating it potential in improving diagnostic accuracy in comple medical scenarios. With the accumulation of large-scale clinical data, statistical analysis has become an important method for disease diagnosis. Researchers use statistical analysis to uncover potential correlations between patient characteristics and medical indicators, thereby providing new perspectives and approaches for disease prediction and diagnosis. As a result, statistically based disease diagnosis methods have become increasingly significant in medical research and practice. Yadav et al. [12] discussed statistical modeling and prediction techniques for various infectious diseases, addressing the issue of single-variable data related to infectious diseases. They proposed fitting time series models and making predictions based on the best-fit model, offering a more accurate approach to forecasting the spread and development of infectious diseases.
Recently, neural network-based disease diagnosis methods have emerged and gradually become a hot topic of research. Researchers are focused on developing new algorithms and models that integrate multi-source information, such as clinical data, medical imaging, and biomarkers, to enhance the accuracy and reliability of diagnoses. These advancements aim to improve diagnostic capabilities by leveraging the power of deep learning in processing complex medical data. Wang and Li employed machine learning methods, combined with large-scale clinical databases, to develop a statistical-based disease prediction model. This model successfully achieved diagnostic predictions for multiple diseases, offering new insights into personalized disease diagnosis. Their approach represents a significant advancement in tailoring diagnoses to individual patient data. On the other hand, to address the challenge of assigning specific stages in the diagnosis of clinical diseases with long courses and staging characteristics, Ma et al. [13] proposed a gout staging diagnosis method based on deep reinforcement learning. They first used a candidate binary classification model library to accurately diagnose gout, and then refined the binary classification results by applying predefined medical rules for gout diagnosis. Additionally, some scholars have integrated diagnostic rules into deep neural networks for bladder cancer staging, significantly mitigating the drawback of cancer staging methods based on deep convolutional neural networks that tend to overlook clinicians’ domain knowledge and expertise. Additionally, to address the impact of imbalanced medical record samples on the training and predictive performance of disease prediction models, the academic community has proposed various solutions for training models on small and imbalanced datasets [14,15]. These methods aim to improve model robustness and accuracy when faced with limited or skewed data distributions, which is a common challenge in medical data analysis.
However, despite the numerous advantages brought by neural network-based disease diagnosis methods, there are still some shortcomings. First, the aforementioned methods often require large amounts of labeled data for training, and annotating medical data typically demands significant time and effort from expert physicians, making it costly and time-consuming. Second, due to the complexity and diversity of medical data, existing machine learning models may struggle to adapt well to various diseases and clinical scenarios, leading to insufficient generalization capability of the models. Through further reading and research, it was found that many scholars have proposed different improvement methods to address this challenge. For example, Luo et al. [16] first proposed a new multi-modal heterogeneous graph model to represent medical data, which helps address the label allocation challenges within the same cluster, enabling more precise targeting of desired medical information. Similarly, Xie et al. [17] proposed a knowledge-based dynamic prompt learning (KBDPT) method that integrates medical knowledge with pre-trained language models, enhancing diagnostic models’ expressiveness and generalizability. At the same time, Ge et al. [18]. proposed a domain knowledge-enhanced multi-label classification (DKEC) method for electronic health records, addressing the issue of previous work neglecting the incorporation of domain knowledge from medical guidelines. They introduced a label attention mechanism and a simple yet effective group training method based on label similarity. This method greatly improves applicability to minority (tail) class label distributions. Moreover, cross-modal knowledge enhancement approaches have gained attention. For instance, the DIE-CDK [19] method combines visual and textual information, leveraging cross-modal knowledge to enhance fine-grained classification, particularly for tasks involving complex features. Inspired by this approach, this study integrates medical knowledge graphs and multimodal information to improve model accuracy and adaptability in complex diagnostic tasks. Compared to existing methods, the proposed prompt-learning-based knowledge enhancement framework not only leverages cross-modal knowledge but also uses designed prompt templates to better understand structured information in medical knowledge graphs. This approach expands the model’s medical knowledge coverage, improves comprehension of complex disease characteristics, and significantly enhances model interpretability and robustness, demonstrating superior adaptability and accuracy in diagnostic tasks.
2.2 Application of the BERT Model in Disease Diagnosis
BERT (Bidirectional Encoder Representations from Transformers), proposed by Google in 2018, is an advanced pre-trained natural language processing (NLP) model. This model has achieved state-of-the-art results in various NLP tasks, including but not limited to text classification, named entity recognition (NER), and question-answering systems (QA). Recently, the research community has begun exploring the potential of the BERT model in the medical field. By integrating multi-source information such as clinical texts and medical literature, the BERT model can absorb and learn rich medical knowledge, thereby providing more precise auxiliary information in the disease diagnosis process. The bidirectional contextual modeling capability of the BERT model gives it a significant advantage in understanding the complex contexts of medical texts, offering comprehensive informational support for disease diagnosis.
In the research on medical named entity recognition algorithms, Tian et al. [7] first proposed a method based on pre-trained language models. This method utilizes the BERT model to generate sentence-level feature vector representations of short text data and combines it with recurrent neural networks (RNN) and transfer learning models to classify medical short texts. Xu et al. [20] developed a medical text classification model (CMNN) that combines BERT, convolutional neural networks (CNN), and bidirectional long short-term memory networks (BiLSTM) to address the efficiency and accuracy challenges in medical text classification. This model showed improvements in evaluation metrics such as accuracy, precision, recall, and F1 score compared to traditional deep learning models.
As advanced NLP technologies gradually penetrate the medical field, the BioBERT model has emerged. This model is specifically optimized for the biomedical domain, demonstrating significant advantages in the understanding and processing of medical texts. Sharaf et al. [21] provided a detailed analysis and overview of a systematic BioBERT fine-tuning method aimed at meeting the unique needs of the healthcare field. This approach includes annotating data for medical entity recognition and classification tasks, as well as applying specialized preprocessing techniques to handle the complexity of biomedical texts.
Recently, pre-trained language models have achieved significant success in many question-answering tasks [22,23]. However, while these models encompass a wide range of knowledge, they perform poorly in structured reasoning. Additionally, considering the previously mentioned issues, such as the low efficiency of medical data annotation, these limitations pose further challenges. This paper aims to enhance structured reasoning using knowledge graphs and leverage the BERT model’s outstanding performance in text classification tasks. The primary focus of this research is on how to effectively utilize language models and knowledge graphs for reasoning, thereby achieving the goal of significantly improving disease prediction accuracy.
Given clinical information and electronic medical records of a patient, where
This paper utilizes prompt learning to accomplish the task, converting it from a classification problem into a language modeling problem. The original classification problem is formulated as fitting
The overall method structure of this paper is shown in Fig. 1. The diagram presents an innovative method combining knowledge graphs with the BERT model to enhance semantic understanding in medical text analysis tasks. The approach primarily consists of the following components: a knowledge graph, an entity and relationship recognition layer, a context understanding and semantic enhancement layer, a knowledge integration and application layer, a sequence modeling network, and a masked language model (MLM) training task. This structure effectively improves semantic comprehension and precise identification of entity relationships in text.

Figure 1: Method structure diagram
Firstly, the knowledge graph module serves as a rich source of background knowledge, storing information such as disease names, symptoms, and treatments. By providing structured relational information between entities, the knowledge graph supports the model in understanding medical terms and their associations within the text. Based on this, the entity and relationship recognition layer identifies key entities (e.g., diseases and symptoms) in the text and determines the relationships between them. This layer extracts core medical information from the text, laying a foundation for subsequent semantic understanding. Next, the context understanding and semantic enhancement layer integrates structured information from the knowledge graph to enhance the semantic representation of entities and relationships. This enables the model to gain a more comprehensive understanding of the medical entities’ meanings. By leveraging the contextual information from the knowledge graph, this layer optimizes the representation of entities, facilitating more precise analysis and prediction in downstream tasks. The knowledge integration and application layer applies the information from the knowledge graph to specific medical analysis tasks, such as disease classification and symptom recognition, broadening the model’s scope of application. The sequence modeling network, comprising input, hidden, and output layers, processes the fused information from the knowledge graph’s entity vectors and BERT embeddings. This network structure captures the complex sequential and contextual dependencies among medical entities, improving the accuracy of entity representation and enhancing text sequence modeling, thereby delivering higher precision in medical text analysis. Finally, the model undergoes training using the masked language model (MLM) task. By randomly masking words in the text and predicting them, this task further improves the model’s semantic understanding within its context. The MLM task helps the model adapt better to complex medical text environments, enhancing its ability to recognize the contextual relationships of medical entities and their associations accurately.
In conclusion, this method achieves knowledge-driven semantic enhancement by deeply integrating knowledge graphs with the BERT model, providing more accurate and comprehensive semantic understanding in medical text analysis tasks. This approach has significant practical value for applications in medical text processing and prediction.
Clinical texts contain many conceptual and structured forms of knowledge, such as symptoms, diagnoses, and treatment plans. Knowledge retrieval is capable of identifying and extracting key medical concepts and relationships. For example, “type 2 diabetes” is a common chronic disease, characterized primarily by persistently elevated blood glucose levels. The diagnostic criteria include fasting blood glucose and HbA1c levels. Common symptoms include excessive thirst, hunger, frequent urination, weight loss, and fatigue. Through knowledge retrieval, it can extract structured knowledge such as causes, related complications, risk factors, and more. This type of knowledge plays an important auxiliary role in disease prediction. A knowledge graph is utilized to structure various types of medical information, providing comprehensive support for disease diagnosis. The knowledge graph employed in this study includes entities such as “disease,” “symptom,” “diagnostic method,” and “treatment.” The relationship types span categories like “causes,” “used for treatment,” and “exhibits symptoms.” For instance, the entity “hyperglycemia” is connected to “type 2 diabetes” through a “causes” relationship, while “type 2 diabetes” is also associated with “kidney disease” through a “complication” relationship. By leveraging this graph structure, the model enhances logical reasoning during the diagnostic process by utilizing these relationship pathways, leading to more accurate disease predictions. The structured advantages of knowledge graphs make them highly relevant and applicable in medical tasks, particularly in complex and diverse disease scenarios. They effectively address the limitations of models solely relying on data training. Therefore, this paper utilizes the following methods to achieve knowledge retrieval from the text:
Given the clinical text x of a patient, we process x by performing tasks such as tokenization and part-of-speech tagging, resulting in a set of vocabulary
Let the knowledge graph be
Next, for each pair of matched entities, we search for all possible reasoning paths P in the knowledge graph. Relationships in the knowledge graph can be represented in the form of triples
For each pair of matched entities
The following section presents a simplified example of a knowledge inference path related to “type 2 diabetes” as shown in Fig. 2. This visualization highlights the logical connections and relationships encoded in the knowledge graph, aiding the diagnostic reasoning process.

Figure 2: Example of a knowledge inference path for “Type 2 Diabetes”
Next, we will represent the structured knowledge along these reasoning paths.
The reasoning paths from the collection are concatenated into a single text sequence. This concatenated text is then represented as a vector
(1) Reasoning Path Representation
Each reasoning path
After converting the path into text, the pre-trained BERT model M is used to transform the text
(2) Text Vectorization Model Selection (BERT)
The input text
where:
The function fBERT transforms the text sequence
The BERT model outputs the vector representation of the text
Here,
(3) Output Vector Extraction
In the text vectorization model selection mentioned earlier, we used the BERT model to convert the input text sequence
The design of prompt templates plays a critical role in this method. In this study, templates incorporating soft tokens and masked tokens (MASK) were designed to enhance the model’s understanding of medical information. For instance, for the input “Type 2 diabetes is a chronic disease,” a corresponding template might be “Type 2 diabetes is a [MASK] chronic [SOFT] disease,” where [MASK] and [SOFT] represent key positions requiring the model’s reasoning. In this template, soft tokens help the model capture the diverse characteristics of different diseases, reducing noise during the template generation process and enhancing the model’s generalization ability. Masked tokens focus the model’s attention on critical reasoning content, minimizing distractions from irrelevant information and improving diagnostic accuracy. For example, when the input text is “The patient exhibits symptoms of hyperglycemia, suspected to have Type 2 diabetes,” the template can be constructed as “The patient exhibits [MASK] symptoms, suspected to have [SOFT] disease.” This template enables the model to extract the association between “hyperglycemia” and “Type 2 diabetes,” emphasizing specific symptoms while maintaining a comprehensive consideration of disease categories. The following sections detail the steps for constructing prompt templates and configuring specific tokens to ensure the model input conforms to BERT’s optimal format, enhancing its performance in diagnostic tasks.
(1) Construction of Prompt Template
To effectively utilize the BERT model for text vectorization, we first need to preprocess the original input text x to construct a proper prompt template
For example:
Original input text x: “Type 2 diabetes is a chronic metabolic disease.”
Prompt template
Generated preprocessed text
(2) Expression of Template Conversion
Here,
Original input text
Converted preprocessed text
(3) Vector Representation and Processing
Using the pre-trained model M, the preprocessed text
For example, for the original input text x: “Type 2 diabetes is a chronic metabolic disease.”, the generated vector set can be explained as follows:
Other vectors follow similarly, with each vector representing the respective words or tokens in the preprocessed text.
The text
Given the modified input
Feed
Extract the representation
Use a verbalizer function
The specific steps are as follows:
(1) Calculate the similarity between
(2) Based on the computed similarities, select the word with the highest probability as the prediction result.
(3) Vector Processing
The formula for replacing the original
where:
Finally, after replacing the original
To better understand how the construction of prompt templates affects model performance, this study compares different template designs in subsequent experiments. For instance, besides the designed template “Type 2 diabetes is a [MASK] chronic [SOFT] disease,” we also tested a simple template without soft tokens and masked markers: “The patient exhibits symptoms, suspected to have a disease.” In this comparative template, the model directly receives complete sentences without specific markers guiding its focus on particular features and diagnostic relationships.
In this paper, experiments were conducted on the CHIP-CTC, IMCS-V2-NER, and KUAKE-QTR datasets. CHIP-CTC, one of the experimental datasets, originates from a bench-marking task released at the CHIP2019 conference. As shown in Table 1, the number of samples in the CHIP-CTC, IMCS-V2-NER, and KUAKE-QTR datasets includes the training, validation, and test samples. All text data is sourced from real clinical trials, including 22,962 entries in the training set, 7682 entries in the validation set, and 10,000 entries in the test set. The dataset is available at https://github.com/zonghui0228/chip2019task3 (accessed on 26 December 2024). Although CHIP-CTC is primarily a short text classification dataset, its data includes medical information about symptoms, medical history, and other relevant details. These attributes allow us to test the model’s performance in medical text classification tasks, showcasing its ability to provide preliminary feature extraction capabilities for disease diagnosis.

The IMCS-V2-NER dataset, used as another experimental dataset, comes from the Named Entity Recognition task in the IMCS2 dataset developed by the School of Data Science at Fudan University. It includes 2472 entries in the training set, 833 entries in the validation set, and 811 entries in the test set. The dataset is available at https://github.com/lemuria-wchen/imcs21 (accessed on 26 December 2024). This dataset primarily serves to validate the model’s capability in medical entity and term recognition, which is crucial for extracting diseases, symptoms, and treatment plans in diagnostic tasks.
The KUAKE-QTR dataset, used as another experimental dataset, includes 24, 174 entries in the training set, 2913 entries in the validation set, and 5465 entries in the test set.
The dataset is available at https:/tianchi.aliyun.com/dataset/95414 (accessed on 26 December 2024). While this dataset mainly evaluates the alignment between query terms and landing page titles, its inclusion of medical terminology and entity matches also aids in testing the model’s ability to infer relationships between symptoms and diseases. Furthermore, although these datasets are not explicitly designed for disease diagnosis, they play a vital role in verifying the proposed model’s adaptability to multiple tasks and its performance in broader medical text processing scenarios. These datasets help validate the model’s effectiveness in extracting medical text information, laying the groundwork for extending its application to more complex real-world diagnostic tasks, such as clinical case reports and electronic health records.
When evaluating our proposed method, we established a comprehensive set of baseline models to ensure rigorous and fair comparison. These baselines were selected to represent robust benchmarks in the field, including both traditional machine learning algorithms and advanced deep learning techniques.
SVM [24]: Support Vector Machines (SVMs) classify text categories using hyperplanes, excelling at handling linear and nonlinear problems. SVMs are well-suited for tasks like sentiment analysis and spam detection.
CNN [25]: Convolutional Neural Networks (CNNs) extract local features from text through convolution layers, enabling quick identification of critical information. They are ideal for content understanding and classification tasks.
RNN [26]: Recurrent Neural Networks (RNNs) are particularly effective in text classification tasks due to their ability to process sequential data and capture long-term dependencies.
Attention [27]: The Attention mechanism assigns higher weights to important words or phrases, improving focus on critical information and optimizing classification performance.
BiLSTM [28]: Bidirectional Long Short-Term Memory Networks (BiLSTMs) combine forward and backward LSTM layers, offering advantages in capturing semantic dependencies within text.
BiRNN [28]: Bidirectional Recurrent Neural Networks (BiRNNs) capture information in both forward and backward text sequences, enhancing the understanding of long-term dependencies.
BERT [29]: BERT employs bidirectional encoding and pre-training to capture semantic and contextual features of text. It performs exceptionally well across various text classification tasks.
BioBERT [30]: BioBERT is a pre-trained model specifically designed for biomedical text. Trained on medical literature, it excels in understanding medical terminology and performing biomedical tasks. Compared to general-purpose BERT, BioBERT is better suited for processing medical and biological contexts.
These baseline models provide a reliable reference for evaluating our approach, enabling a clearer demonstration of its advantages in addressing specific text classification tasks.
This section presents our experimental results and provides an analysis of these outcomes. The evaluation metrics employed include precision, recall, and F1 score, which are commonly used in text classification experiments. Precision measures the proportion of true positives (TP) among all samples predicted as positive by the model, reflecting the accuracy of positive predictions. It is calculated using the formula:
To ensure the reliability of the experiments, we repeated each experiment three times and used the average values as the final results. The experimental results are shown in Table 2 and Figs. 3–6.

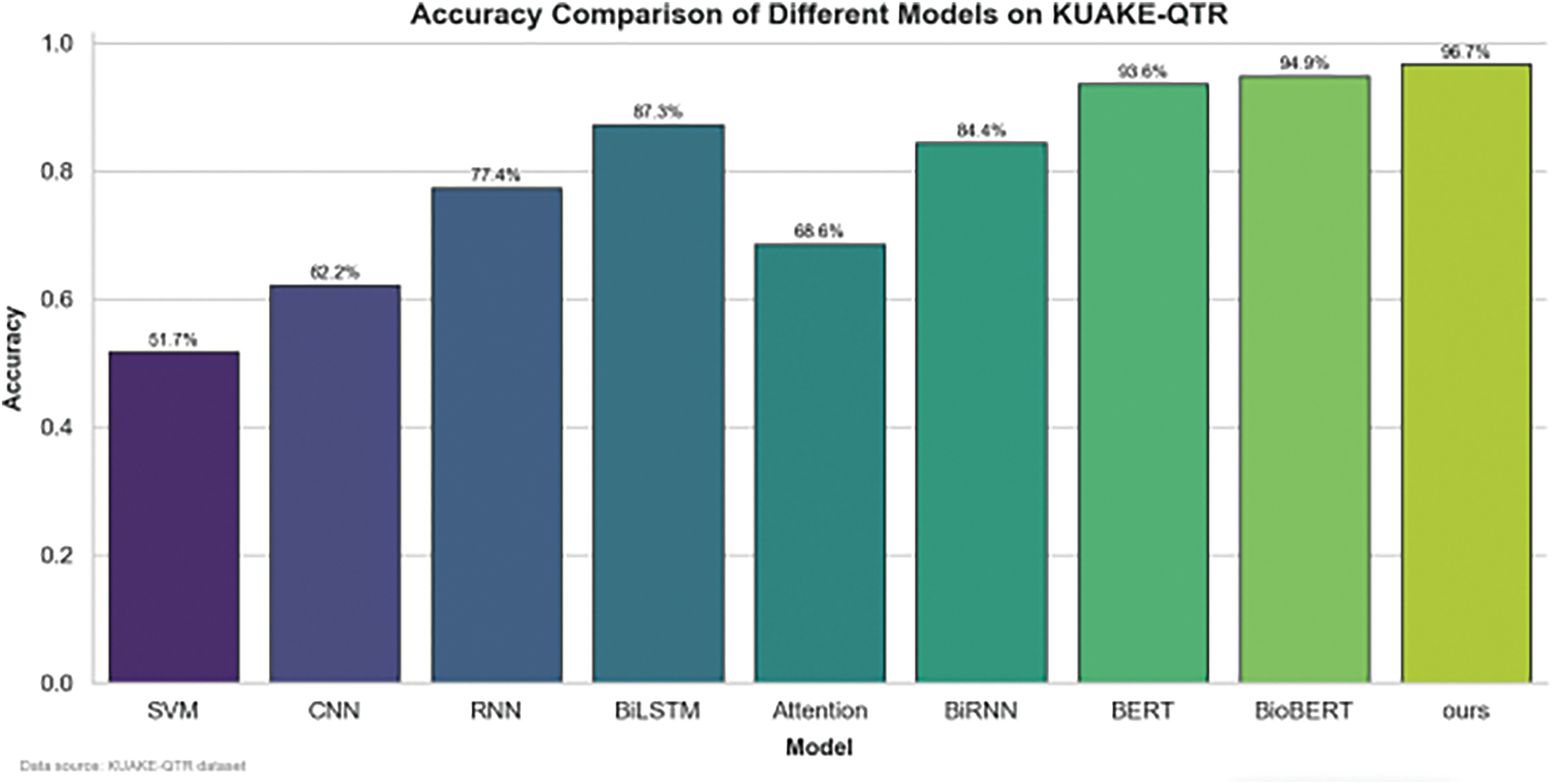
Figure 3: Performance of different benchmark models and the propoesd model on the KUAKE-QTR dataset

Figure 4: Performance of different benchmark models and the propoesd model on the CHIP-CTC dataset

Figure 5: Performance of different benchmark models and the proposed model on the IMCS-V2-NER dataset

Figure 6: Trend chart of various model F1 values on different datasets
In this study, we evaluated the performance of various models, including classical and deep learning approaches, across multiple datasets: KUAKE-QTR, IMCS-V2-NER, and CHIP-CTC. The primary metrics used for evaluation were accuracy and F1 score, providing a comprehensive insight into the models’ classification and generalization abilities.
6.1.2 Accuracy Comparison across Models
The bar charts show the accuracy comparison of models such as SVM, CNN, RNN, BLSTM, Attention, BiRNN, BERT, BioBERT, and a custom model (“ours”) across three different datasets. The custom model consistently outperformed the other models on all datasets, achieving the highest accuracy in each case: KUAKE-QTR: The “ours” model achieved an accuracy of 93.6%, surpassing models like BERT (91.1%) and BioBERT (92.1%). IMCS-V2-NER: The “ours” model reached 94.7% accuracy, again outperforming BERT (92.1%) and BioBERT (92.9%). CHIP-CTC: The custom model recorded an accuracy of 94.5%, followed closely by BERT at 92.1% and BioBERT at 93.2%.
These results illustrate that advanced models (BERT, DistBERT) and our custom model generally exhibit superior accuracy compared to traditional models like SVM and CNN, especially in complex language understanding tasks.
6.1.3 F1 Score Trend across Datasets
The line chart visualizes the F1 score trends for different models across the three datasets, reflecting consistency and robustness in performance. The “ours” model achieves the highest F1 score on all datasets, indicating its effectiveness in handling diverse data characteristics. Notably, the performance gap between classical and deep learning models widens significantly, emphasizing the importance of sophisticated architectures for improved natural language processing.
The experimental results demonstrate that our method achieved the best performance across all three datasets, proving its feasibility and effectiveness. Furthermore, these results highlight the significant impact of incorporating medical knowledge into the model through the prompt-learning framework in enhancing its performance.
This section aims to analyze the effectiveness of each module in our proposed method. To achieve this, we first remove the knowledge representation component from our method. As seen in the first row of Table 3, removing the knowledge representation results in a significant drop of 0.2 in the F1 score. This is because knowledge representation is a core component of our method, which injects structured knowledge obtained from the knowledge graph into the language model, thereby enhancing the model’s understanding of medical domain knowledge and improving performance on medical diagnostic tasks.

Subsequently, we modified the prompt template, changing it to: “The characteristics of Type 2 Diabetes include [MASK] nature, and it is also manifested as [SOFT] disease.” From rows 2 to 4 of Table 3, it can be observed that the prompt template used in our method achieves the best performance, while other prompt templates do not perform as well. We believe this may be due to the presence of excessive or irrelevant words in the prompt templates, which could introduce noise into the language model’s reasoning process.
This study conducts systematic ablation tests to evaluate the impact of knowledge enhancement modules, diverse prompt template designs, and label verbalization strategies on disease diagnosis performance. It compares the efficacy of basic symptom templates, explicit symptom-disease relation templates, and complex context templates in contextual encoding, while assessing label mapping approaches including direct term matching, expanded vocabulary, and synonym tolerance. Findings highlight that knowledge integration strengthens reasoning capabilities, whereas excessive contextual complexity introduces noise.
In this study, we designed various templates and verbalizers to enhance the model’s performance in medical text classification tasks. Basic Symptom Template: Focuses on symptom information for inference, suitable for simple symptom classification tasks. Symptom-Disease-Relation Template: Introduces symptom-disease associations, improving the model’s diagnostic accuracy. Complex Context Template: Handles more intricate contextual relationships, such as patient medical history or symptom combinations, enhancing the model’s performance in multidimensional scenarios. On Verbalizers, Direct Mapping Verbalizer: Simplifies reasoning by directly mapping symptoms to specific diseases. Extended Vocabulary Verbalizer: Expands the vocabulary to improve recognition of different expressions and synonyms. Synonym Tolerant Verbalizer: Strengthens the model’s understanding of synonyms and semantically similar terms, improving robustness across diverse expressions.
The combination of these templates and verbalizers allows the model to flexibly handle different layers of information in medical text, achieving favorable results in disease diagnosis tasks.
6.3 Model Interpretability Study
This section aims to analyze the interpretability of the model. Unlike general text classification tasks, in disease diagnosis tasks, users are additionally concerned with the interpretability of the results. An unexplainable prediction result is unacceptable to users. Therefore, to verify the model’s explainability across diverse clinical cases, we introduced a multi-case validation approach to ensure consistency and transparency in its reasoning processes. The cases include diabetes, hypertension, hyperlipidemia, and coronary heart disease, covering a variety of symptoms, medical histories, and complex comorbidity scenarios. This ensures the comprehensiveness and applicability of the explainability analysis. For example, a case involving a 55-year-old male patient with diabetes was selected to visualize the reasoning process through illustrative diagrams.
Using the clinical case in Fig. 7 as an example, we can observe the knowledge retrieved during the knowledge retrieval phase, including: “Type 2 diabetes is a common chronic disease,” “Its main characteristic is persistently elevated blood sugar levels,” and “Frequent drinking, eating, and urination are common symptoms.” From this knowledge, we can identify which key pieces of information from the clinical case are related to the final prediction. In other words, we can clearly understand which knowledge elements led the model to make the current prediction.

Figure 7: Disease diagnosis flowchart based on clinical cases
Through analyzing multiple clinical cases, we observed that the model consistently traced its reasoning paths across different conditions, ensuring transparency and reliability in the diagnostic process. This multi-case validation approach not only improves the model’s explainability in complex clinical scenarios but also provides clinicians with clearer reasoning references, thereby enhancing the model’s credibility.
In addition, to facilitate readers’ understanding, this paper not only explains the evaluation metrics but also demonstrates the advantages of the proposed method through relevant examples. Figs. 8–10 show the specific example.

Figure 8: Disambiguation of medical terms
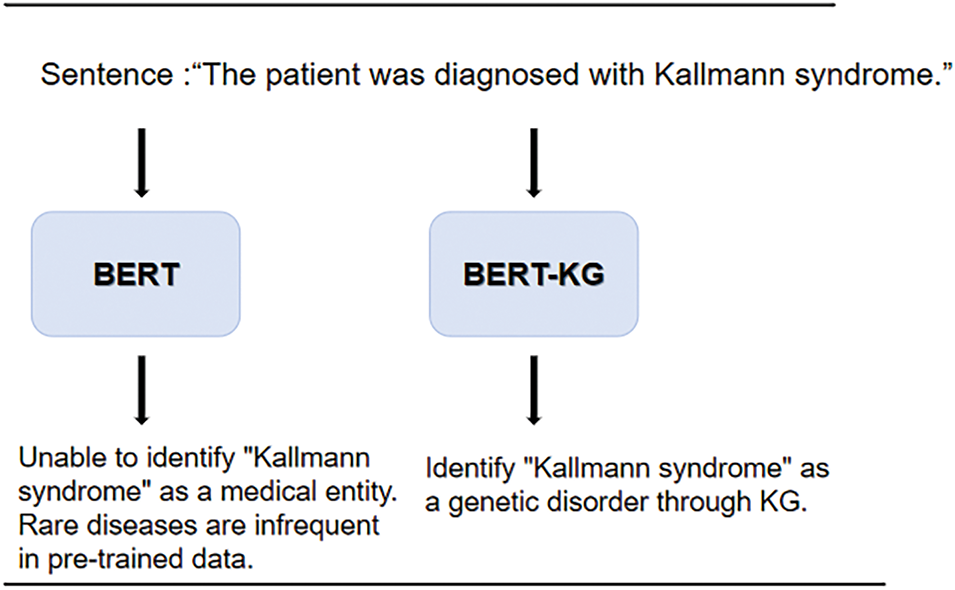
Figure 9: Long-tail medical entity recognition

Figure 10: Medical text classification
The specific examples presented show that the model proposed in this paper addresses the issues of insufficient domain knowledge and difficulties in long-tail entity recognition in existing models. The predicted disease conditions are closer to actual clinical outcomes. These examples further illustrate the innovation and superiority of our approach, allowing readers to intuitively understand the improvements and advantages over traditional methods.
In this paper, we propose a knowledge-enhanced disease diagnosis method based on a prompt learning framework. This method leverages an external knowledge graph to retrieve relevant knowledge from clinical cases and then encodes this structured knowledge into prompt templates. By incorporating this encoded knowledge, the language model’s understanding of the task is improved, resulting in more accurate disease diagnosis outcomes. Experimental results demonstrate that the proposed method effectively enhances the performance of language models in disease diagnosis tasks. Additionally, the model exhibits strong interpretability, providing users with supporting evidence related to the diagnostic results.
In the future, when integrating prompt learning with knowledge graphs for medical diagnosis, several practical challenges need to be addressed. Firstly, data privacy is a critical issue. Since medical data involves sensitive patient information, the construction of knowledge graphs must strictly comply with data protection laws to ensure patient data is not misused. Secondly, the maintenance and updating of medical knowledge graphs pose cost and technical challenges. Clinical medical knowledge evolves rapidly, and continuously incorporating the latest information into knowledge graphs to ensure diagnostic accuracy is a persistent difficulty. These challenges highlight the need for in-depth research on data privacy protections and knowledge update technologies when advancing the practical application of these methods, ensuring the safety and practicality of diagnostic tools.
We will explore additional methods for knowledge injection. Furthermore, we will investigate more advanced knowledge editing techniques to integrate medical knowledge into the reasoning process of language models.
Acknowledgement: We extend our gratitude to all individuals and institutions that supported this work.
Funding Statement: This research was supported by the National Natural Science Foundation of China (Grant No. 62162014).
Author Contributions: Zheng Zhang proposed the research problem and designed the research plan, implemented and optimized the model, processed and analyzed medical text data, optimized prompt templates and wrote the initial draft of the manuscript and made multiple rounds of revisions. Hengyang Wu, the corresponding author, provided the overall research framework and academic support, oversaw the research approach, manuscript content, and scientific rigor, ensured the accuracy of the results and the academic value of the publication, and supervised the revisions and final approval of the manuscript. Na Wang provided key guidance on research design and data processing methods, optimized the experimental design to ensure the accuracy and validity of the data, and assisted in model optimization and result analysis, and contributed important technical reviews of the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Due to the nature of this research, participants of this study did not agree for their data to be shared publicly, so supporting data is not available.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Qu C, Bi J. Application of association rules based on Apriori algorithm in disease diagnosis. China Comput Commun. 2015;16(16):8–9, 11 (In Chinese). [Google Scholar]
2. Lv X, Chen Q, Zhao Z, Yao D. Research on sleep quality evaluation and disease diagnosis based on statistical classification model. Value Eng. 2018;37(22):273–5 (In Chinese). [Google Scholar]
3. Chattopadhyay S, Davis RM, Menezes DD, Singh G, Acharya RU, Tamura T. Application of Bayesian classifier for the diagnosis of dental pain. J Med Syst. 2012;36(3):1425–39. doi:10.1007/s10916-010-9604-y. [Google Scholar] [CrossRef]
4. Battineni G, Sagaro GG, Chinatalapudi N, Amenta F. Applications of machine learning predictive models in the chronic disease diagnosis. J Pers Med. 2020;10(2):21. doi:10.3390/jpm10020021. [Google Scholar] [PubMed] [CrossRef]
5. Lin D, Vasilakos AV, Tang Y, Yao Y. Neural networks for computer-aided diagnosis in medicine: a review. Neurocomputing. 2016;216(6):700–8. doi:10.1016/j.neucom.2016.08.039. [Google Scholar] [CrossRef]
6. Onan A. Hierarchical graph-based text classification framework with contextual node embedding and BERT-based dynamic fusion. J King Saud Univ Comput Inf Sci. 2023;35(7):101610. doi:10.1016/j.jksuci.2023.101610. [Google Scholar] [CrossRef]
7. Tian H, Xu C. Research on medical short text classification algorithm based on BERT model. J Yili Norm Univ (Nat Sci Ed). 2021;15(4):50–7. [Google Scholar]
8. Hu M, Chen X, Sun Y, Shen X, Wang X, Yu T, et al. A disease prediction model based on dynamic sampling and transfer learning. Chin J Comput. 2019;42(10):2339–54. [Google Scholar]
9. Shortliffe E. Computer-based medical consultations: MYCIN. Vol. 2. New York, NY, USA: Elsevier; 2012. [Google Scholar]
10. Aamir KM, Sarfraz L, Ramzan M, Bilal M, Shafi J, Attique M. A fuzzy rule-based system for classification of diabetes. Sensors. 2021;21(23):8095. doi:10.3390/s21238095. [Google Scholar] [PubMed] [CrossRef]
11. Sanz JA, Galar M, Jurio A, Brugos A, Pagola M, Bustince H. Medical diagnosis of cardiovascular diseases using an interval-valued fuzzy rule-based classification system. Appl Soft Comput. 2014;20(4):103–11. doi:10.1016/j.asoc.2013.11.009. [Google Scholar] [CrossRef]
12. Yadav SK, Akhter Y. Statistical modeling for the prediction of infectious disease dissemination with special reference to COVID-19 spread. Front Public Health. 2021;9:645405. doi:10.3389/fpubh.2021.645405. [Google Scholar] [PubMed] [CrossRef]
13. Ma C, Pan C, Ye Z, Ren H, Huang H, Qu J. Gout staging diagnosis method based on deep reinforcement learning. Processes. 2023;11(8):2450. doi:10.3390/pr11082450. [Google Scholar] [CrossRef]
14. He H, Garcia EA. Learning from imbalanced data. IEEE Trans Knowl Data Eng. 2009;21(9):1263–84. doi:10.1109/TKDE.2008.239. [Google Scholar] [CrossRef]
15. Branco P, Torgo L, Ribeiro RP. A survey of predictive modeling on imbalanced domains. ACM Comput Surv. 2017;49(2):1–50. doi:10.1145/2907070. [Google Scholar] [CrossRef]
16. Luo F, Zhang Y, Wang X. IMAS++: an intelligent medical analysis system enhanced with deep graph neural networks. In: Proceedings of the 30th ACM International Conference on Information & Knowledge Management; 2021. p. 4754–8. doi:10.1145/3459637.3481966. [Google Scholar] [CrossRef]
17. Xie J, Li X, Yuan Y, Guan Y, Jiang J, Guo X, et al. Knowledge-based dynamic prompt learning for multi-label disease diagnosis. Knowl Based Syst. 2024;286:111395. doi:10.1016/j.knosys.2024.111395. [Google Scholar] [CrossRef]
18. Ge X, Williams RD, Stankovic JA, Alemzadeh H. DKEC: domain knowledge enhanced multi-label classification for electronic health records. arXiv:2310.07059. 2023. [Google Scholar]
19. Guo Y, Yu H, Ma L, Luo X, Xie S. DIE-CDK: a discriminative information enhancement method with cross-modal domain knowledge for fine-grained ship detection. IEEE Trans Circuits Syst Video Technol. 2024;34(11):10646–61. doi:10.1109/TCSVT.2024.3407057. [Google Scholar] [CrossRef]
20. Xu L, Li D, Zhang H, Tang D, Lei H. Medical text classification based on neural network. Comput Eng Sci. 2023;45(6):1116–22. [Google Scholar]
21. Sharaf S, Anoop VS. An analysis on large language models in healthcare: a case study of BioBERT. arXiv:2310.07282. 2023. [Google Scholar]
22. Raffel C, Shazeer N, Roberts A, Lee K, Narang S, Matena M, et al. Exploring the limits of transfer learning with a unified text-to-text transformer. J Mach Learn Res. 2020;21(140):1–67. [Google Scholar]
23. Liao W, Zeng B, Yin X, Wei P. An improved aspect-category sentiment analysis model for text sentiment analysis based on RoBERTa. Appl Intell. 2021;51(6):3522–33. doi:10.1007/s10489-020-01964-1. [Google Scholar] [CrossRef]
24. Cortes C, Vapnik V. Support-vector networks. Mach Learn. 1995;(20):273–97. [Google Scholar]
25. LeCun Y, Bottou L, Bengio Y, Haffner P. Gradient-based learning applied to document recognition. Proc IEEE. 1998;86(11):2278–324. doi:10.1109/5.726791. [Google Scholar] [CrossRef]
26. Elman J. Finding structure in time. Cogn Sci. 1990;14(2):179–211. doi:10.1207/s15516709cog1402_1. [Google Scholar] [CrossRef]
27. Bahdanau D. Neural machine translation by jointly learning to align and translate. arXiv:1409.0473. 2014. [Google Scholar]
28. Schuster M, Paliwal KK. Bidirectional recurrent neural networks. IEEE Trans Signal Process. 1997;45(11):2673–81. doi:10.1109/78.650093. [Google Scholar] [CrossRef]
29. Devlin J, Chang MW, Lee K, Toutanova K, Hulburd E, Liu D, et al. BERT: pre-training of deep bidirectional transformers for language understanding. arXiv:1810.04805. 2018. [Google Scholar]
30. Lee J, Yoon W, Kim S, Kim D, Kim S, So CH, et al. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. 2020;36(4):1234–40. doi:10.1093/bioinformatics/btz682. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools