 Open Access
Open Access
ARTICLE
Enhanced Diagnostic Precision: Deep Learning for Tumors Lesion Classification in Dermatology
1 Engineering Technical College, Al-Ayen University, Thi-Qar, 64001, Iraq
2 Information Technology College, University of Babylon, Hilla, 51001, Iraq
3 Information and Communication Technology Research Group, Scientific Research Center, Al-Ayen University, Thi-Qar, 64001, Iraq
4 College of Computer Sciences and Information Technology, University of Kerbala, Karbala, 56001, Iraq
5 Artificial Intelligence Engineering Department, College of Engineering, Al-Ayen University, Thi-Qar, 64001, Iraq
6 Computer Science, Bayan University, Erbil, 44001, Iraq
* Corresponding Authors: Rafid Sagban. Email: ; Saadaldeen Rashid Ahmed. Email:
Intelligent Automation & Soft Computing 2024, 39(6), 1035-1051. https://doi.org/10.32604/iasc.2024.058416
Received 11 September 2024; Accepted 10 December 2024; Issue published 30 December 2024
Abstract
Skin cancer is a highly frequent kind of cancer. Early identification of a phenomenon significantly improves outcomes and mitigates the risk of fatalities. Melanoma, basal, and squamous cell carcinomas are well-recognized cutaneous malignancies. Malignant We can differentiate Melanoma from non-pigmented carcinomas like basal and squamous cell carcinoma. The research on developing automated skin cancer detection systems has primarily focused on pigmented malignant type melanoma. The limited availability of datasets with a wide range of lesion categories has hindered in-depth exploration of non-pigmented malignant skin lesions. The present study investigates the feasibility of automated methods for detecting pigmented skin lesions with potential malignancy. To diagnose skin lesions, medical professionals employ a two-step approach. Before detecting malignant types with other deep learning (DL) models, a preliminary step involves using a DL model to identify the skin lesions as either pigmented or non-pigmented. The performance assessments accurately assessed four distinct DL models: Long short-term memory (LSTM), Visual Geometry Group (VGG19), Residual Blocks (ResNet50), and AlexNet. The LSTM model exhibited higher classification accuracy compared to the other models used. The accuracy of LSTM for pigmented and non-pigmented, pigmented tumours and benign classes, and melanomas and pigmented nevus classes was 0.9491, 0.9531, and 0.949, respectively. Automated computerized skin cancer detection promises to enhance diagnostic efficiency and precision significantly.Keywords
The rising incidence of skin cancer, particularly melanoma, underscores the urgent need for enhanced diagnostic techniques. While current practices like skin biopsies remain the gold standard for diagnosis, they are time-consuming and invasive. Automated deep learning (DL) systems promise to make diagnoses more accurate, especially when it comes to telling the difference between skin lesions that are coloured and those that are not. This study examines how well DL models like LSTM, VGG19, ResNet50, and AlexNet can sort different skin lesions, help find them earlier, and give dermatologists better diagnostic tools.
One of the most deadly skin cancers is melanoma. Globally, cutaneous malignancies account for 75% of mortality due to melanoma. Melanoma can inhibit the formation of melanin due to its impact on the cells responsible for melanin synthesis [1]. Given these circumstances, the early identification of melanoma assumes significant importance in the battle against cancer-related death, as shown in Fig. 1.
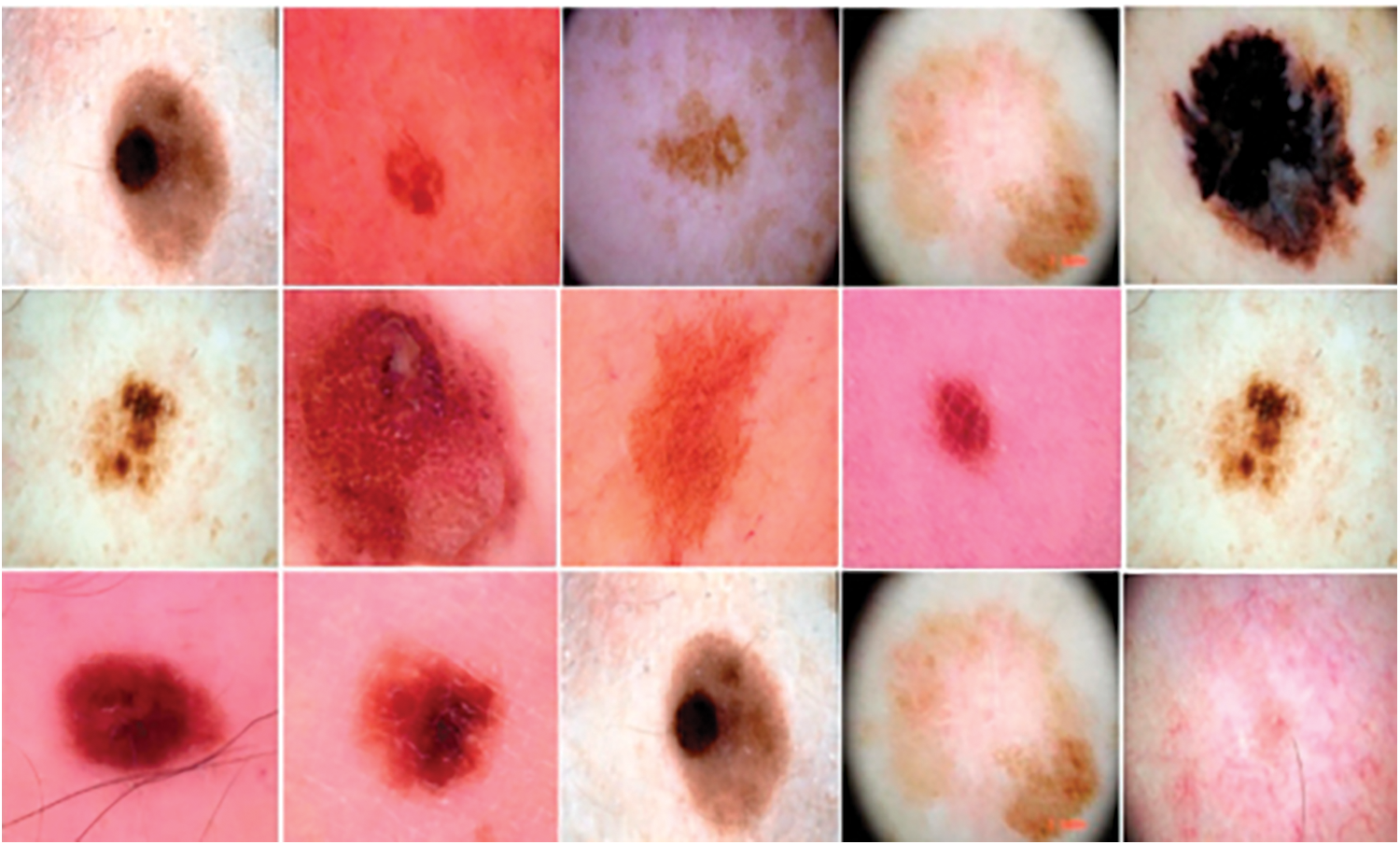
Figure 1: Some samples from the dataset
We can broadly classify skin lesions into two primary categories: pigmented and non-pigmented. Both melanoma and pigmented nevi are instances of pigmented lesions. Similarly, non-pigmented lesions encompass vascular issues, basal cell cancer, dermatofibroma, and actinic keratosis, the initial manifestation of squamous cell carcinoma, commonly referred to as Bowen’s disease. We can classify pigmented and non-pigmented lesions as benign or malignant. Melanoma, basal cell carcinoma, and actinic keratosis are all examples of skin conditions that are malignant, which means they can cause cancer.
Scanning is a commonly used non-invasive imaging method that studies the morphology of skin lesions. Fig. 1, the provided dermoscopy images, shows the melanoma skin lesion. Dermatoscopy is a useful way to find melanoma early on because pigmented lesions can sometimes show signs of not being pigmented. The people who discussed dermoscopy agreed to use a two-step diagnostic method to group skin lesion pigmentation seen with dermoscopy [2,3]. The implementation of this measure aimed to standardise dermoscopic nomenclature. After explaining the difference between pigmented and non-pigmented lesions, the next step in this process is to explain the difference between benign and malignant lesions.
Unfortunately, this instrument’s complex and time-consuming framework occasionally compromises its efficacy, as does its limited ability to consistently distinguish between inter- and intra-features. Furthermore, we can employ computational methods to expand the use of dermoscopy [4]. An arduous implementation of this idea involves using computer methods to categorise melanomas using artificial segmentation, considering factors such as size, colour, form, and texture (Fig. 2).
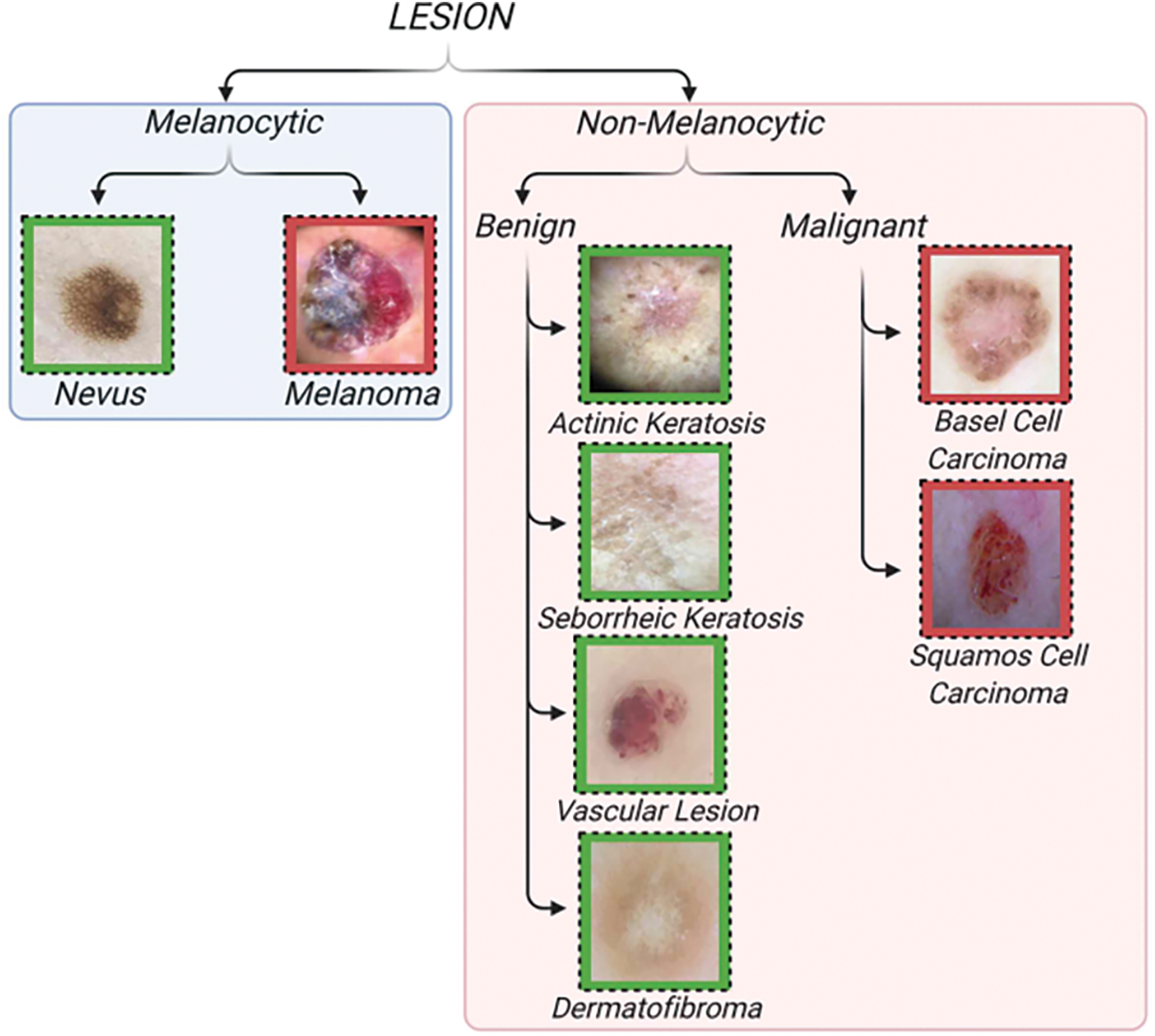
Figure 2: Classification of skin lesions [3]
The primary outcomes derived from this research are:
• Commodifying non-pigmented skin lesions augments dermatologists’ two-step dermoscopic procedure [3].
• In contrast to previous research that used clinical imaging to categorize malignant, non-pigmented skin lesions, the present study utilizes dermoscopy pictures [5]. The presence of photographic variability can influence the clinical imaging process.
• We used a collection of 7285 dermoscopy pictures to more accurately classify pigmented skin lesions that are not pigmented. This collection is substantially larger than the 167 [6] or 107 [7] photos used in earlier study projects.
• To broaden the classification of non-pigmented skin lesions, we have included vascular lesions in addition to the dermatofibroma categories. In the study above, we classified these two common forms of benign, non-pigmented skin lesions in clinical practice using only fourteen out of six hundred fifty photographs [7]. However, binary non-pigmented skin lesions classification has overlooked these groups in studies [6] and [8].
The following section outlines the organizational structure of the paper: Preceding this, one may encounter pertinent scholarly material about machine-learning approaches for classifying skin lesions. The next part explains the research techniques employed in this study and the database used for data collection and analysis. Finally, we evaluated and discussed the efficacy of the DL architecture.
Melanoma identification in dermoscopic images employs two primary strategies: deep learning (DL) and classical methods. Initial efforts relying on manually designed features exhibited limited effectiveness. For instance, Reference [9] introduced the California STEM Learning Network (CSLNet), a specialised deep convolutional neural network (CNN) trained on the ISIC-19 dataset using end-to-end learning that achieves superior performance across various metrics without relying on handcrafted features. Concurrently, Reference [10] proposed an algorithm to optimise wavelet networks by enhancing feature selection to improve melanoma detection effectiveness. However, both approaches struggled to distinguish melanoma subtypes and differentiate visual similarities between melanoma and non-melanoma images [1].
To address these challenges [11], we combined neural networks with genetic algorithms, emphasising segmentation methodologies. Border detection techniques extracted clinical features like border irregularity and asymmetry. However, this method faced issues resolving indistinct boundaries in unclear lesions, complicating melanoma diagnosis [12].
Han et al. [13] compiled over 20,000 macroscopic images spanning 12 disease classes and utilised a fine-tuned ResNet-152 model [14] to freeze early layers for feature extraction from manually cropped images. Similarly, Ma et al. [15] demonstrated using pre-trained ConvNets for classifying non-dermoscopic skin images through feature extraction. In addition, Reference [16] used a multi-stage, multi-scale method and a softmax classifier to sort melanoma lesions by pixel.
The utility of CNNs in melanoma detection lies in their ability to effectively perform identification, classification, and segmentation tasks. For example, Reference [17] developed a hybrid model integrating a CNN with sparse coding and a support vector machine (SVM) algorithm. A non-supervised CNN model based on AlexNet was also proposed by [7], aiming to extract features specific to melanoma.
A deep learning method for separating melanoma lesions first described by [4] used a fully convolutional network (FCN) with 19 layers. The convolution operator improved the accuracy, but FCNs had problems like being less flexible because they had fewer parameters, were less efficient than Maxpool operators, and took longer to train. We validated this approach using ISIC 2016 and PH datasets.
Expanding into more medical imaging, Reference [18] created HGANet, a CNN for automatically finding problems in the digestive tract using Kvasir datasets. This demonstrates the effectiveness of Deep Learning in medical image analysis. The study [19] reviewed smartphone applications for skin disease diagnosis using Convolutional Neural Networks (CNNs). It employs digital photographs and dermoscopy images as datasets. CNNs enhance classification accuracy without user intervention. Results show that while many apps assist in skin cancer detection, few offer direct computational analysis with varying performance metrics.
Deep learning systems (DLS) [20] identify skin disorders based on clinical cases (skin pictures and medical history). The DLS recognises 26 skin disorders—80% of primary-care skin problems. We created and tested the DLS using de-identified cases from 17 teledermatology clinics that were spread out over time.
The SPL analysis method is based on a deep convolutional neural network and works with wide-field photos [21]. It was tested on a dermatology dataset containing 38,283 images which included 133 patient images used in the ISBI 2016 dataset, which has 900 dermoscopy images [22].
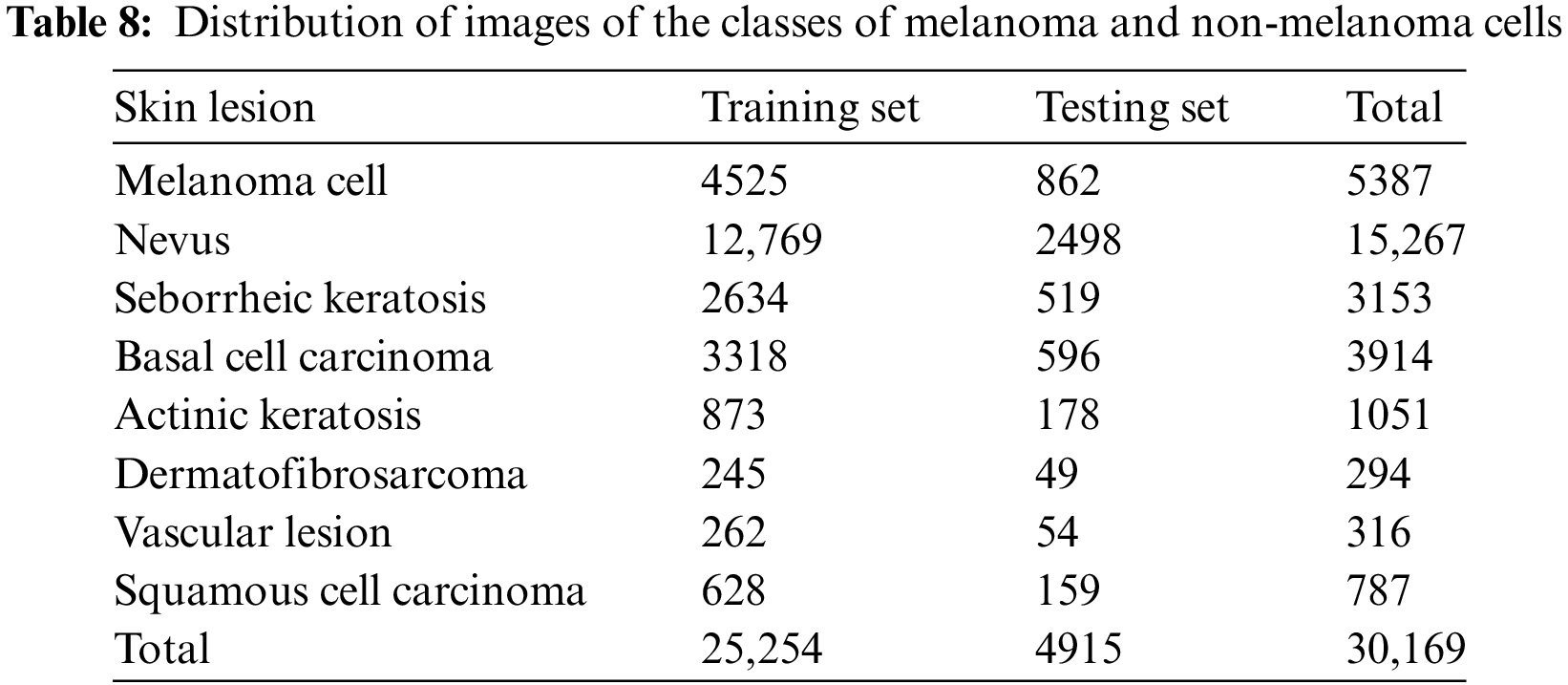
The PH2 dataset, which has 200 images split into three groups, to make sure that their methods worked.Different pre-trained state-of-the-art architectures (DenseNet 201, ResNet 152, Inception v3, InceptionResNet v2) were used by applied on applied 10,135 dermoscopy skin images in total (HAM10000: 10015, PH2: 120). This study utilises the 2019 ISIC Challenge dataset (Fig. 3) [8], encompassing 30,169 dermoscopy images across seven categories, facilitating DL frameworks for distinguishing pigmented and non-pigmented tumour lesions (Table 1).
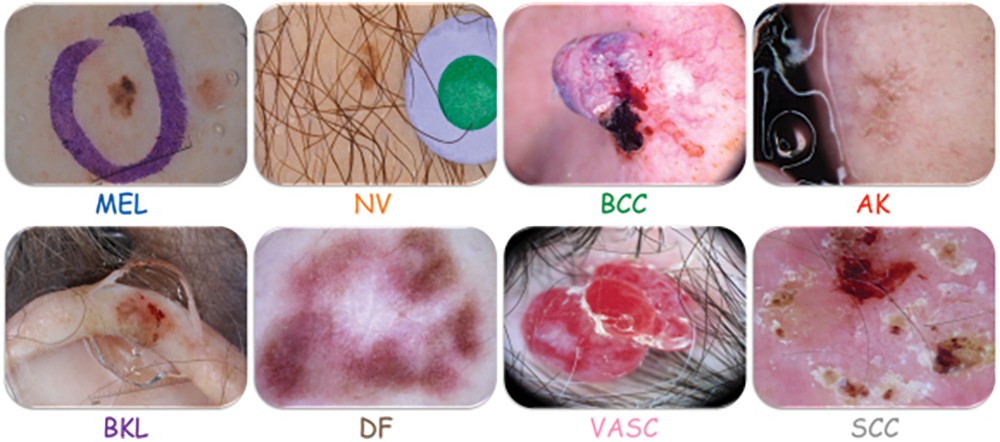
Figure 3: Skin lesions of ISIC 2019
The main goal of developing and testing four different deep learning (DL) models, namely Long Short-Term Memory (LSTM), VGG19, ResNet50, and AlexNet, is to distinguish between cancerous skin lesions and those that are not. The process involves three main phases: data preprocessing and augmentation, model development, and model evaluation.
3.1 Data Preprocessing and Augmentation
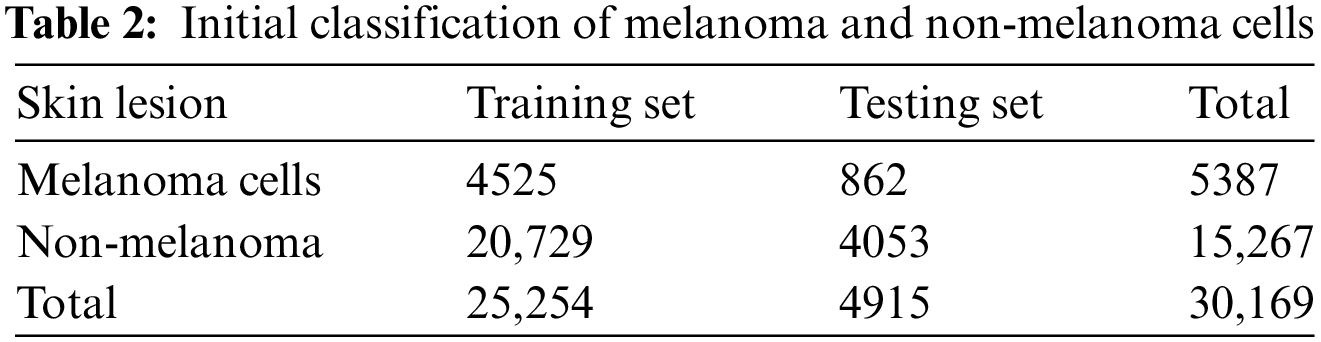
This work uses a collection of pictures of melanocytic and non-melanocytic skin lesions. The pictures came from the 2019 ISIC challenge collection, which is open to the public and has 25,331 pictures of different skin diseases. Table 2 illustrates the initial uneven distribution of the dataset’s pictures among the various groups.
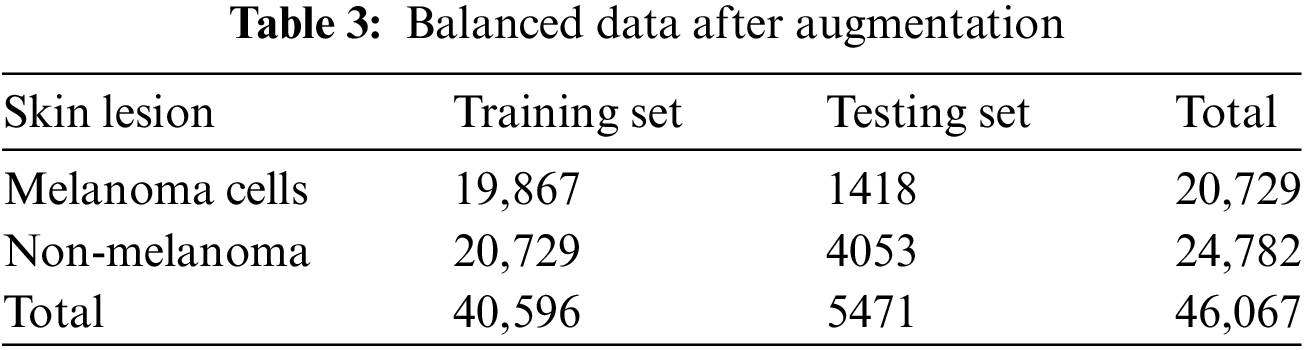
Different augmentation approaches [33–35] used data augmentation methods like image rotation, flipping, zooming, and scaling to fix the imbalance because they were fast on the computer. This process increased the dataset size and ensured that each category contained a more even distribution of images, as shown in Table 3. The augmentation process improved the model’s ability to generalise across different lesion types by exposing it to a wider range of variations in the training set.
Data preprocessing for this study involves resizing all images to a uniform dimension of 224 × 224 pixels to ensure consistency across the dataset and normalising pixel values to a range of [0, 1] by dividing by 255, which improves model convergence during training. We apply data augmentation techniques to enhance the diversity of the dataset. These include random rotations within a range of −30 to +30 degrees to simulate different orientations, horizontal and vertical flipping to create mirror images, zooming from 0.8 to 1.2 to simulate different camera distances, and shearing transformations to introduce perspective changes. Additionally, brightness adjustments between 0.8 and 1.2 account for lighting variations. At the same time, colour jittering modifies hues and saturations to simulate different lighting conditions. We also employ random cropping, which shifts the image width and height by 10% to focus on different parts of the lesions. We implement these techniques using Keras’ ImageDataGenerator, forming an augmentation pipeline that applies these transformations in real-time during model training through batch generation and the flow method.
We developed four DL models to classify skin lesions—LSTM, VGG19, ResNet50, and AlexNet. We trained each model to identify both pigmented and non-pigmented skin lesions. The pigmented category includes melanoma and pigmented nevus. In this group are basal cell carcinoma (BCC), actinic keratosis (AK), benign keratosis (BKL), dermatofibroma (DF), and vascular lesions (VASC). However, they are not coloured.
We employed the Keras deep learning framework for model implementation. The need to balance accuracy and computational efficiency drove the choice of architecture. We used already trained models (VGG19, ResNet50, and AlexNet) to make transfer learning easier. The ISIC dataset was used to fine-tune the models. We incorporated LSTM to capture temporal dependencies in lesion features, enhancing the model’s ability to classify images with subtle variations. The Machine Learning Models Training & Testing Framework for the 2019 ISIC Challenge Dataset is depicted in Fig. 4.
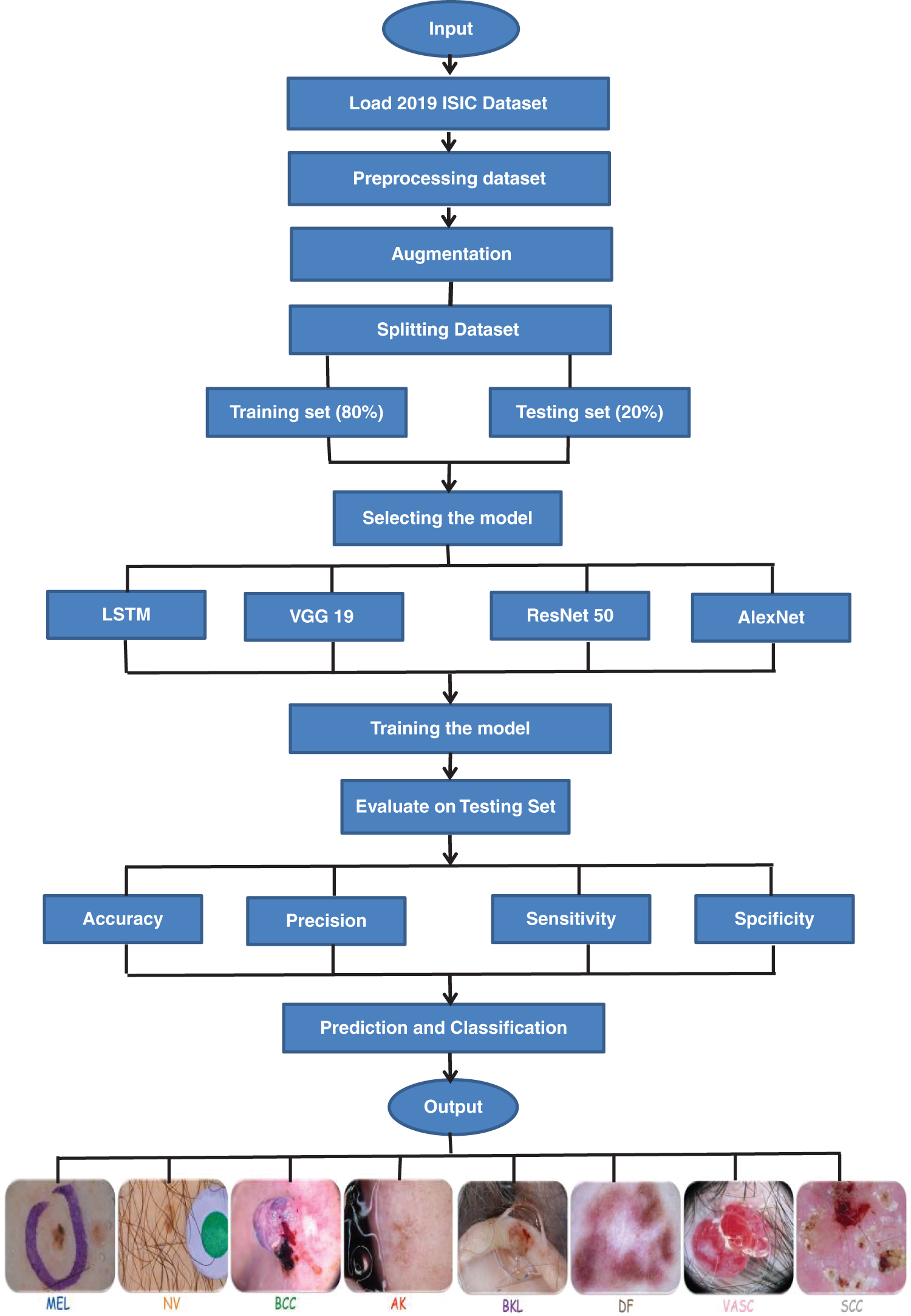
Figure 4: The framework of this study
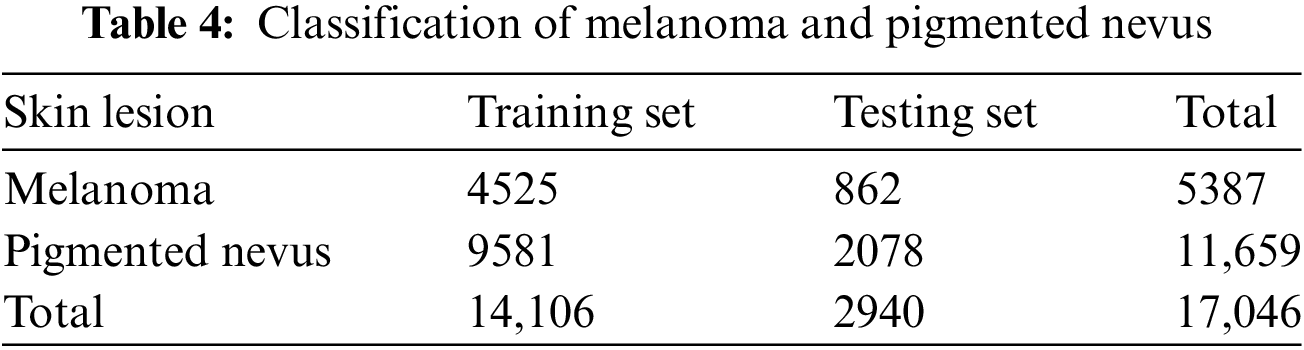
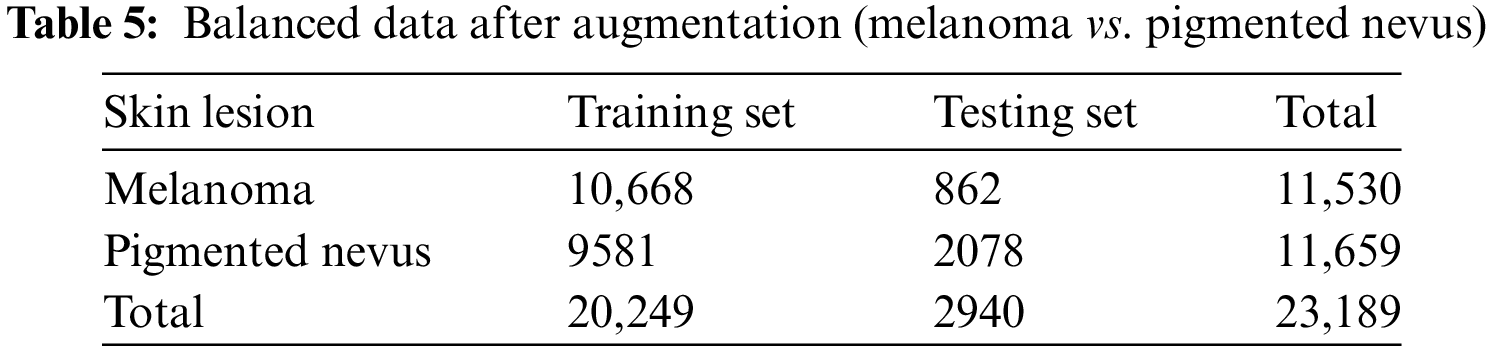
Additional augmentation techniques were applied to further balance the dataset between melanoma. They pigmented nevus, as shown in Tables 4 and 5.
DL models are used to classify benign and non-pigmented malignant tissues accurately. Non-pigmented benign tumours include benign keratosis, dermato-fibroma, and vascular lesions. Conversely, basal cell carcinoma and actinic keratosis (commonly known as Bowen’s disease) are two instances of non-pigmented malignant tumours. Table 6 displays the number of photographs allocated to each category.
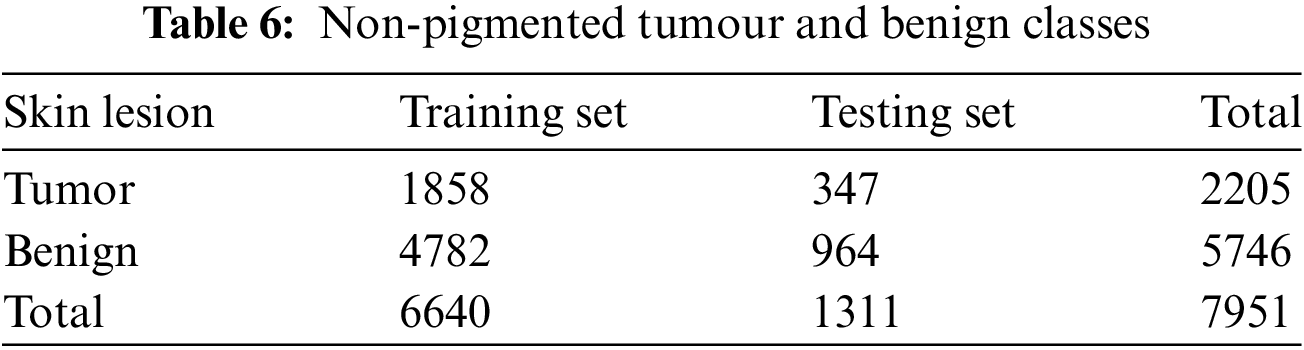
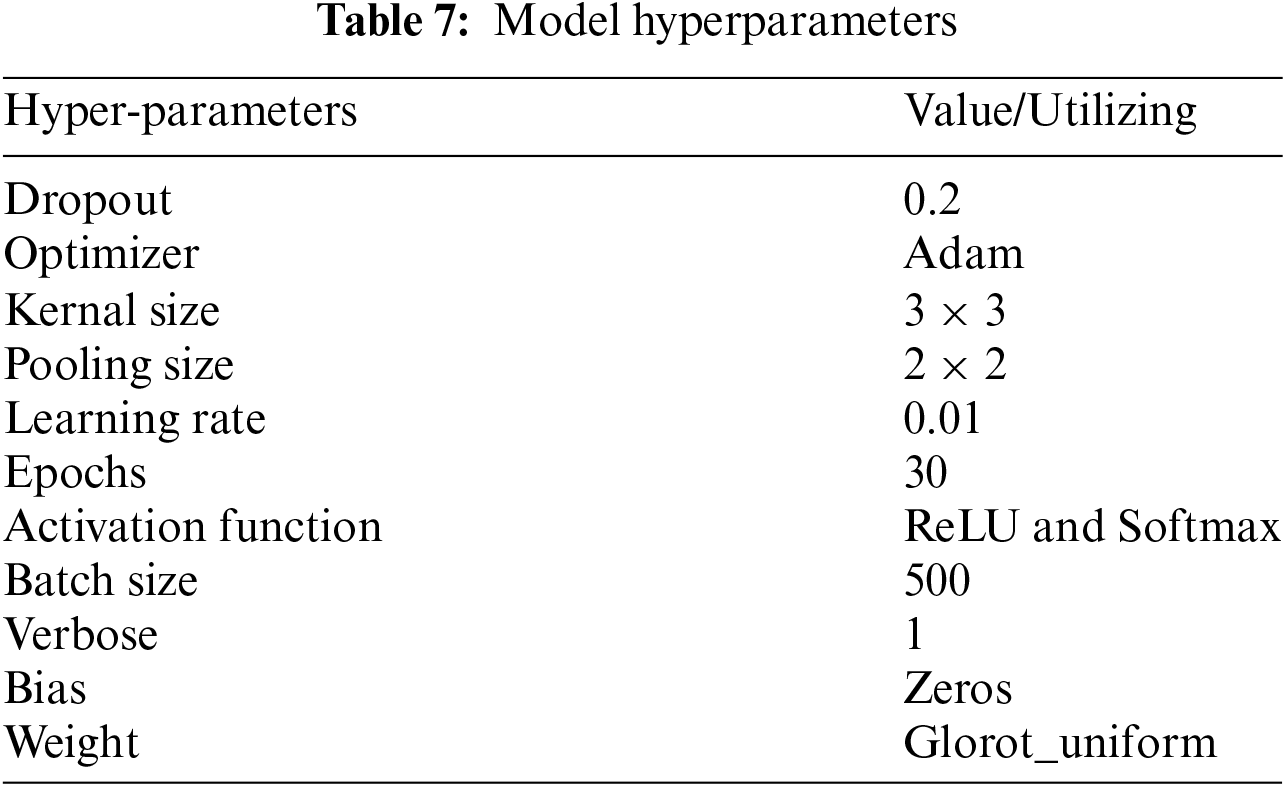
3.3 Hyperparameters and Training Configuration
The hyperparameters utilized in the training process are summarized in Table 7. These values were selected based on prior research and experimental tuning. The models were trained using the Adam optimizer, with a learning rate of 0.01 and a batch size of 500. The activation functions used were ReLU for hidden layers and Softmax for the output layer, allowing multiclass classification. The models were trained for 30 epochs, with early stopping to prevent overfitting.
We evaluated each model’s performance using accuracy, precision, recall, F1 score, and Area Under the Curve (AUC). We calculated these metrics for melanoma and non-pigmented lesion classification, ensuring consistency across evaluations.
We evaluate performance by using the freely available 2019 ISIC challenge dataset. The dataset consists of 25,331 images, divided into two sections. The first portion comprised 80%, or 20,265 images, of the training dataset, while the second comprised 20%, or 5066 images, for testing purposes. We evaluated each classifier on the original image test set to determine the model’s efficacy. The images illustrate many types of skin cancer, including melanoma, basal cell carcinoma, benign keratosis, dermatofibroma, actinic keratosis, and pigmented nevus. Table 8 comprehensively summarises the number of photos in each category.
While it is desirable to have high classification performance across all classes, accurately predicting all skin cancers, particularly those with a high mortality rate, such as melanoma, takes precedence over incorrectly predicting a benign lesion.
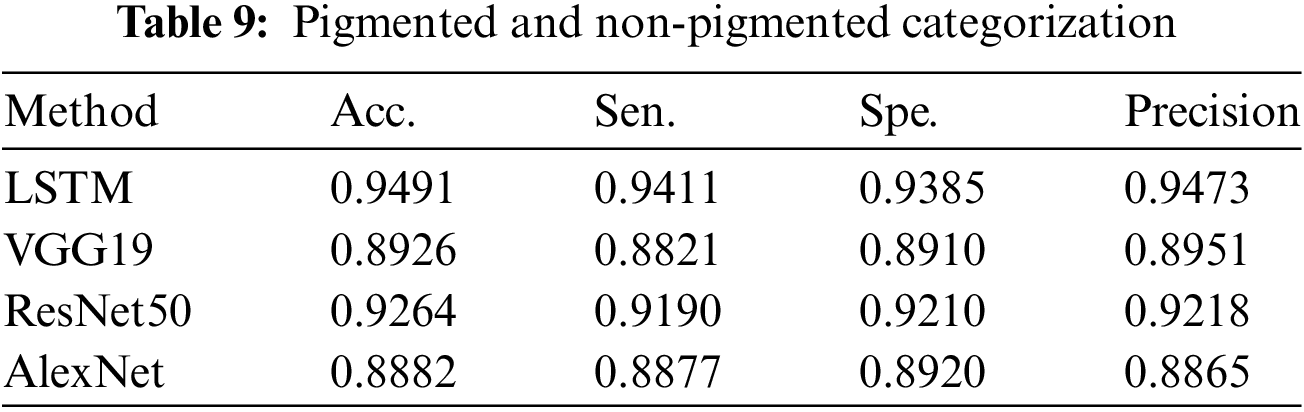
We assess the effectiveness of the DL-based classification strategy using proposed classification algorithms. We evaluate the efficacy of the DL models in categorizing pigmented and non-pigmented skin lesions. We perform this examination at the beginning of the operation. Later on, the approach achieved a high success rate in the classification, as shown in Table 9.
The next phase involves examining deep-learning models to classify skin lesions as melanoma or pigmented nevus. Finally, we assess the ability of the third DL model to classify non-pigmented malignant and benign skin disorders. This evaluation confirms the effectiveness of the recommended models.
4.3 Experiment Results: Pigmented vs. Non-Pigmented Classes
Table 9 summarises the results of categorizing pigmented cancers and benign classes. This table displays the count of classes and test photos used to evaluate the performance of the classification. Table 9 also displays the overall results of the categorization process.
4.4 Experimental Results: Pigmented Tumor vs. Benign Classes
Table 10 summarises the classification findings of pigmented tumours and benign classes by displaying the number of photos used for training and testing assessment results. This table also presents the overall classification results.
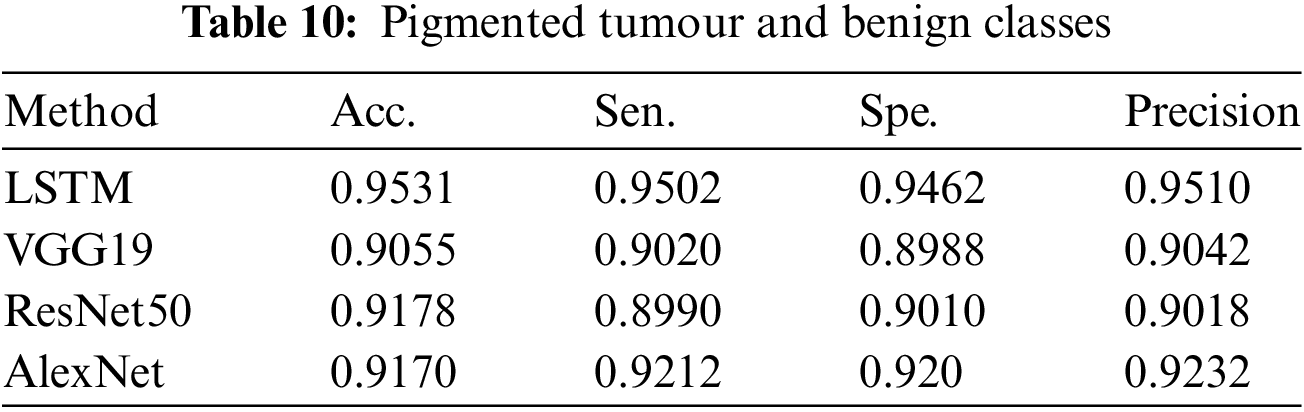
4.5 Experiment Results: Melanoma vs. Pigmented Nevus Classes
A summary of the findings from the categorization of pigmented nevus and melanoma can be seen in Table 11, which also contains the findings from the performance assessment.
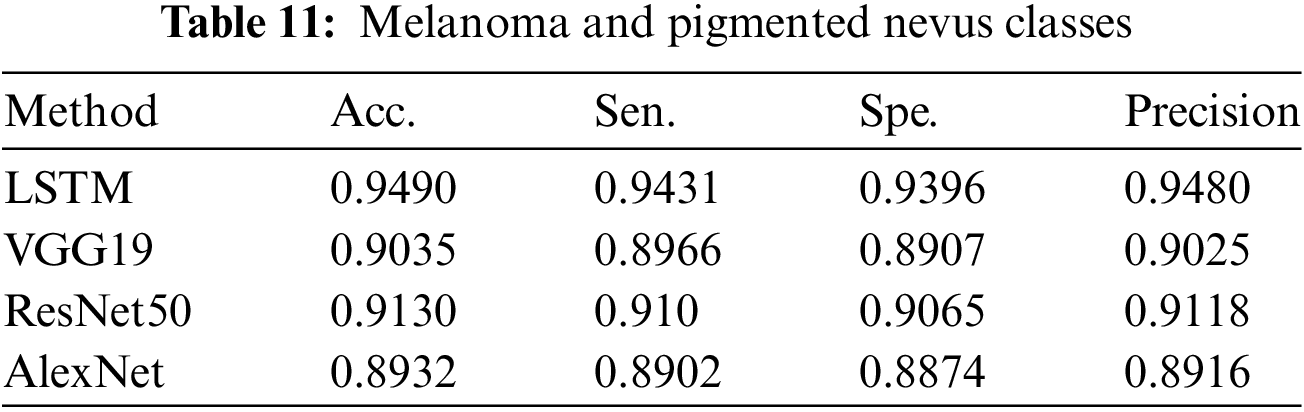
Three experiments were conducted. 1. Classification of pigmented and non-pigmented classes. 2. Classification of pigmented tumours and benign classes. 3. Classification of melanoma and pigmented nevus classes.
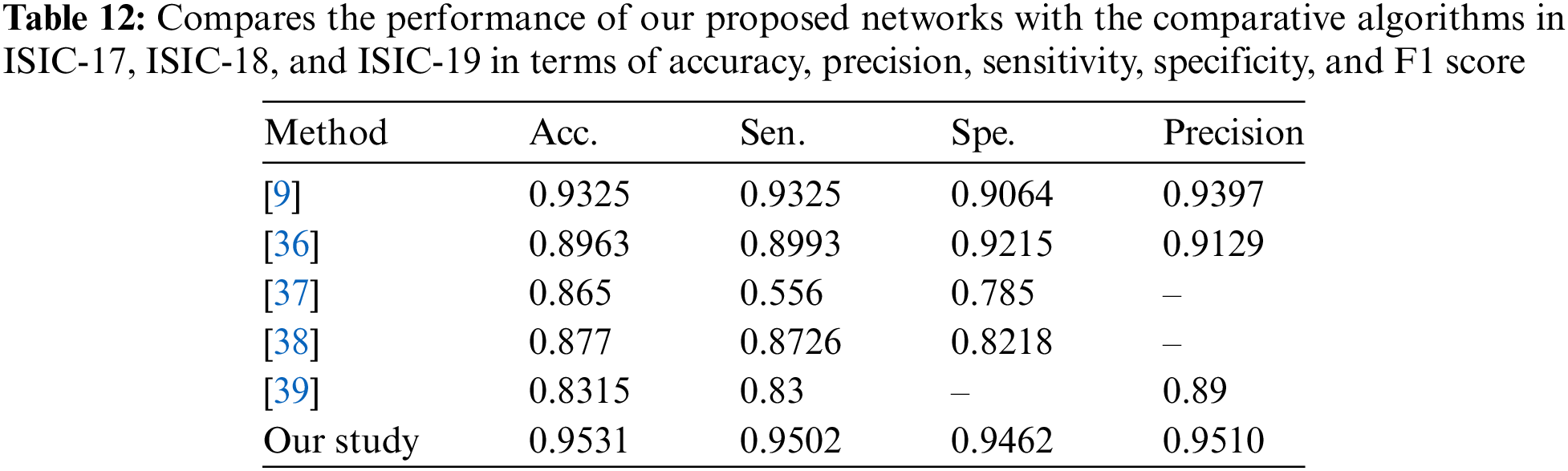
The information in Tables 9–11 shows that the complementing LSTM model did better than the other three models in the three classification tests we did on the ISIC 2019 dataset in terms of accuracy, sensitivity, and specificity. Table 12 also shows the comparison between our results and some previous related studies, which displays the superiority of our proposed networks over the rest of the networks.
The graphs illustrate the training results of the proposed models for distinguishing melanoma and pigmented cells, non-melanoma cells, tumour cells, and benign cells. Figs. 5–7 display these graphs.
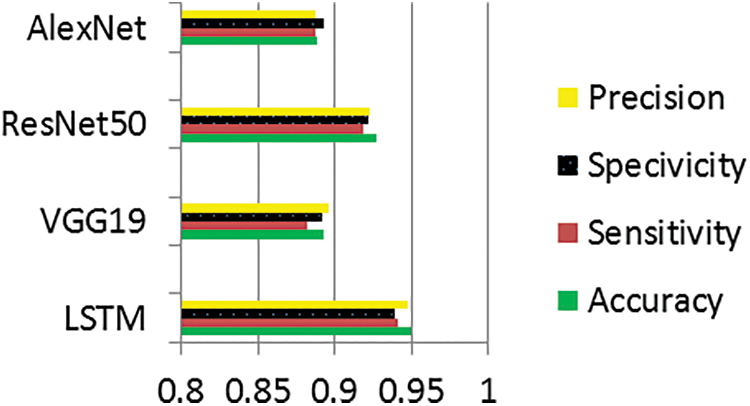
Figure 5: Performance evaluation of pigmented and non-pigmented classes
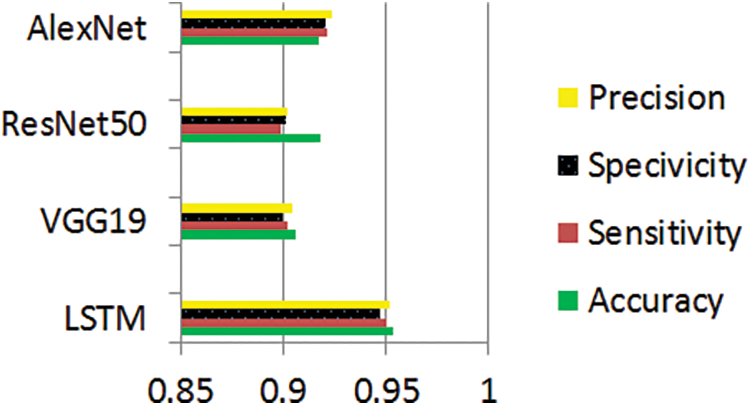
Figure 6: Performance evaluation of pigmented tumor and benign classes
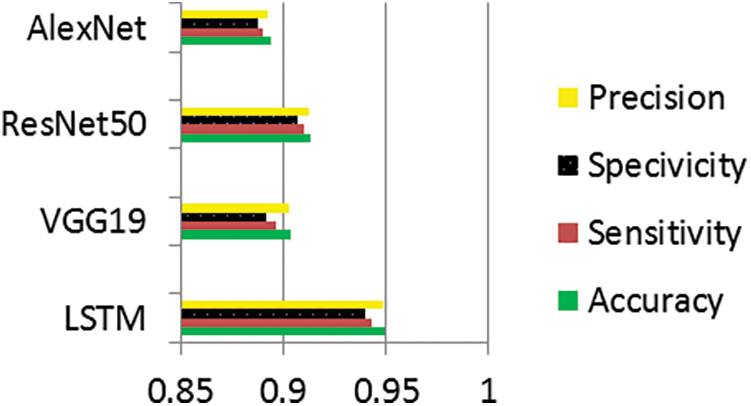
Figure 7: Performance evaluation of melanoma and pigmented nevus classes
Metadata, which includes sociodemographic information about patients, is needed to prove that imbalance or under-representation biases exist [40]. The most direct route to resolving this issue is, where feasible, to augment the dataset with photos and information from patients belonging to under-represented groups. Another option is to do further validation, such as prospective studies, to ensure the model’s resilience.
An intriguing conversation about the makeup of the training dataset emerged during [41]. The training dataset’s limitations in the clinical presentation spectrum, variability in picture collection settings, and limited clinical information raised concerns about the generality of automated diagnosis. These issues pertained to the fact that the training dataset had constraints. A common and often inherent issue with healthcare data is the under-representation of clinical or demographic categories, which may limit the model’s potential for generalisation.
Clinically, close-up photographs can effectively aid in the artificial categorisation of skin lesions. The first step that a doctor takes in determining whether or not to continue with dermoscopy is to perform a macroscopic evaluation of a lesion. It is possible that clinical photographs of the lesion can offer extra information that is not evident by dermoscopy. These additional data include the pearly look and shining surface of seborrhoeic keratosis, as well as the “stuck-on” aspect of the condition [42]. Smartphone apps could potentially use these datasets of clinical photographs to train their algorithms. However, even the most advanced of these algorithms still have a significant distance to go.
Shimizu et al. [6] introduced VGG19, a 19-layer convolutional neural network, as an approach for picture classification. The structure consists of three fully interconnected layers and sixteen convolutional layers. We dedicate the first three convolutional layers to classification and use the remaining sixteen layers for feature extraction.
He et al. [43] introduced ResNet, an enhanced CNN iteration that uses residual blocks. We designed this architecture to tackle the issue of gradient degradation in very deep networks. In these networks, the accuracy first reaches a saturation point. However, it then quickly deteriorates owing to a drop in gradient values.
AlexNet, an efficient, simple CNN Alex Krizhevsky et al. suggested the idea at the 2012 ImageNet Large Scale Visual Recognition Challenge (ILSVRC-2012) [44]. The architecture consists of convolutional, pooling, ReLU, and connected layers. Three layers make up AlexNet’s five convolutional layers: a pooling layer, an initial layer, an intermediate layer, and a final layer.
We have introduced a DL-based classification technique for skin lesions. It employs four forms of DL. We can classify skin lesions using the provided models as pigmented or non-pigmented. This classification enables the identification of types such as basal cell carcinoma and squamous cell carcinoma. The evaluation data indicates that the LSTM DL model achieves accuracy and sensitivity values. Furthermore, it demonstrates that the melanoma and pigmented nevus classes exhibit classification abilities compared to models.
Based on these results, using the other three models, it seems harder to tell the difference between skin lesions in the Melanoma and non-category than between Melanoma and pigmented nevus. This difficulty arises because both groups encompass a range of skin abnormalities. Additionally, each of these four models uses several training photographs. Moreover, we have determined that identifying a non-pigmented tumour skin lesion is more difficult than identifying a pigmented skin lesion. Moreover, our findings indicate that the LSTM model had greater effectiveness in this aspect than the other models used.
Limitations of the Method Used: The study’s models may not generalize well due to a limited and potentially biased dataset that doesn’t represent diverse skin types.
Shortcomings of This Method: The computational complexity and need for extensive labelled data can hinder real-time clinical application and widespread implementation.
Prospects for the Future: Future efforts should focus on diversifying datasets, integrating multimodal data for improved accuracy, and developing lightweight models for mobile use to enhance accessibility in dermatological care.
Acknowledgement: The authors express gratitude to the medical staff and nurses at Nasiriyah General Hospital and Turkish Hospital in Nasiriyah City for their essential support and insight into the research. I appreciate AL-Ayen University’s assistance with this research.
Funding Statement: The authors received no specific funding for this study.
Author Contributions: Conceptualization, Rafid Sagban; Methodology, Rafid Sagban; Software, Rafid Sagban; Validation, Saadaldeen Rashid Ahmed; Formal analysis and writing—original draft preparation, Haydar Abdulameer Marhoon; Writing—original draft preparation, Rafid Sagban. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The data can be accessed through the Kaggle website as it is open source from the following link: https://www.kaggle.com/datasets/andrewmvd/isic-2019 (accessed on 09 December 2024).
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. N. C. Codella et al., “Skin lesion analysis toward melanoma detection: A challenge at the 2017 International Symposium on Biomedical Imaging (ISBIhosted by the International Skin Imaging Collaboration (ISIC),” in 2018 IEEE 15th Int. Symp. Biomed. Imaging (ISBI 2018), Apr. 2018, pp. 168–172. doi: 10.1109/ISBI.2018.8363547. [Google Scholar] [CrossRef]
2. M. E. Celebi, N. Codella, and A. Halpern, “Dermoscopy image analysis: Overview and future directions,” IEEE J. Biomed. Health Inform., vol. 23, no. 2, pp. 474–478, Mar. 2019. doi: 10.1109/JBHI.2019.2895803. [Google Scholar] [PubMed] [CrossRef]
3. A. Yilmaz, G. Gencoglan, R. Varol, A. A. Demircali, M. Keshavarz and H. Uvet, “MobileSkin: Classification of skin lesion images acquired using mobile phone-attached hand-held dermoscopes,” J. Clin. Med., vol. 11, no. 17, 2022, Art. no. 5102. doi: 10.3390/jcm11175102. [Google Scholar] [PubMed] [CrossRef]
4. Y. Yuan, M. Chao, and Y. -C. Lo, “Automatic skin lesion segmentation using deep fully convolutional networks with jaccard distance,” IEEE Trans. Med. Imaging, vol. 36, no. 9, pp. 1876–1886, Sep. 2017. doi: 10.1109/TMI.2017.2695227. [Google Scholar] [PubMed] [CrossRef]
5. J. Dinnes et al., “Dermoscopy, with and without visual inspection, for diagnosing melanoma in adults,” Cochrane Database Syst. Rev., vol. 12, no. 12, 2018, Art. no. CD011902. doi: 10.1002/14651858.CD011902.pub2. [Google Scholar] [PubMed] [CrossRef]
6. K. Shimizu, H. Iyatomi, M. E. Celebi, K. -A. Norton, and M. Tanaka, “Four-class classification of skin lesions with task decomposition strategy,” IEEE Trans. Biomed. Eng., vol. 62, no. 1, pp. 274–283, Jan. 2015. doi: 10.1109/TBME.2014.2348323. [Google Scholar] [PubMed] [CrossRef]
7. S. Abdelouahed, Y. Filali, and A. Aarab, “An improved segmentation approach for skin lesion classification,” Stat. Optimiz. Inform. Comput., vol. 7, no. 2, pp. 456–467, May 2019. doi: 10.19139/soic.v7i2.533. [Google Scholar] [CrossRef]
8. C. Barata, M. E. Celebi, and J. S. Marques, “Explainable skin lesion diagnosis using taxonomies,” Pattern Recognit., vol. 110, Feb. 2021, Art. no. 107413. doi: 10.1016/j.patcog.2020.107413. [Google Scholar] [CrossRef]
9. I. Iqbal, M. Younus, K. Walayat, M. U. Kakar, and J. Ma, “Automated multiclass classification of skin lesions through deep convolutional neural network with dermoscopic images,” Comput. Med. Imaging Graph., vol. 88, 2021, Art. no. 101843. doi: 10.1016/j.compmedimag.2020.101843. [Google Scholar] [PubMed] [CrossRef]
10. Y. Peng, N. Wang, Y. Wang, and M. Wang, “Segmentation of dermoscopy image using adversarial networks,” Multimed. Tools Appl., vol. 78, no. 8, pp. 10965–10981, Sep. 2018. doi: 10.1007/s11042-018-6523-2. [Google Scholar] [CrossRef]
11. E. Pérez and S. Ventura, “An ensemble-based convolutional neural network model powered by a genetic algorithm for melanoma diagnosis,” Neural Comput. Appl., vol. 34, no. 13, pp. 10429–10448, Nov. 2021. doi: 10.1007/s00521-021-06655-7. [Google Scholar] [CrossRef]
12. A. Ameri, “A deep learning approach to skin cancer detection in dermoscopy images,” J. Biomed. Phys. Eng., vol. 10, no. 6, pp. 801–806, Dec. 2020. doi: 10.31661/jbpe.v0i0.2004-1107. [Google Scholar] [PubMed] [CrossRef]
13. S. S. Han, M. S. Kim, W. Lim, G. H. Park, I. Park and S. E. Chang, “Classification of the clinical images for benign and malignant cutaneous tumors using a deep learning algorithm,” J. Investig. Dermatol., vol. 138, no. 7, pp. 1529–1538, Jul. 2018. doi: 10.1016/j.jid.2018.01.028. [Google Scholar] [PubMed] [CrossRef]
14. M. Shafiq and Z. Gu, “Deep residual learning for image recognition: A survey,” Appl. Sci., vol. 12, no. 18, Sep. 2022, Art. no. 8972. doi: 10.3390/app12188972. [Google Scholar] [CrossRef]
15. Z. Ma and J. M. R. S. Tavares, “Effective features to classify skin lesions in dermoscopic images,” Expert. Syst. Appl., vol. 84, pp. 92–101, Oct. 2017. doi: 10.1016/j.eswa.2017.05.003. [Google Scholar] [CrossRef]
16. A. A. Adegun and S. Viriri, “Deep learning-based system for automatic melanoma detection,” IEEE Access, vol. 8, pp. 7160–7172, 2020. doi: 10.1109/ACCESS.2019.2962812. [Google Scholar] [CrossRef]
17. J. A. A. Salido and C. Ruiz Jr, “Using deep learning for melanoma detection in dermoscopy images,” Int. J. Mach. Learn. Comput., vol. 8, no. 1, pp. 61–68, Feb. 2018. doi: 10.18178/ijmlc.2018.8.1.664. [Google Scholar] [CrossRef]
18. I. Iqbal, K. Walayat, M. U. Kakar, and J. Ma, “Automated identification of human gastrointestinal tract abnormalities based on deep convolutional neural network with endoscopic images,” Intell. Syst. Appl., vol. 16, Nov. 1, 2022, Art. no. 200149. doi: 10.1016/j.iswa.2022.200149. [Google Scholar] [CrossRef]
19. E. Göçeri, “Impact of deep learning and smartphone technologies in dermatology: Automated diagnosis,” in 2020 Tenth Int. Conf. Image Process. Theory Tools Appl. (IPTA), Nov. 2020, pp. 1–6. doi: 10.1109/IPTA50016.2020.9286651. [Google Scholar] [CrossRef]
20. Y. Liu et al., “A deep learning system for differential diagnosis of skin diseases,” Nat. Med., vol. 26, no. 6, pp. 900–908, Jun. 2020. doi: 10.1038/s41591-020-0842-3. [Google Scholar] [PubMed] [CrossRef]
21. L. R. Soenksen et al., “Using deep learning for dermatologist-level detection of suspicious pigmented skin lesions from wide-field images,” Sci. Transl. Med., vol. 13, no. 581, Apr. 2021, Art. no. eabb3652. doi: 10.1126/scitranslmed.abb3652. [Google Scholar] [PubMed] [CrossRef]
22. A. Rezvantalab, H. Safigholi, and S. Karimijeshni, “Dermatologist-level dermoscopy skin cancer classification using different deep learning convolutional neural networks algorithms,” 2018, doi: 10.48550/arXiv.1810.10348. [Google Scholar] [CrossRef]
23. M. M. Musthafa, T. R. Mahesh, V. Vinoth Kumar, and S. Guluwadi, “Enhanced skin cancer diagnosis using optimized CNN architecture and checkpoints for automated dermatological lesion classification,” BMC Med. Imaging, vol. 24, no. 1, 2024, Art. no. 201. doi: 10.1186/s12880-024-01356-8. [Google Scholar] [PubMed] [CrossRef]
24. S. G. Malik, S. S. Jamil, A. Aziz, S. Ullah, I. Ullah and M. Abohashrh, “High-precision skin disease diagnosis through deep learning on dermoscopic images,” Bioengineering, vol. 11, no. 9, 2024, Art. no. 867. doi: 10.3390/bioengineering11090867. [Google Scholar] [PubMed] [CrossRef]
25. H. Guan et al., “Dermoscopy-based radiomics help distinguish basal cell carcinoma and actinic keratosis: A large-scale real-world study based on a 207-combination machine learning computational framework,” J. Cancer, vol. 15, no. 11, 2024, Art. no. 3350. doi: 10.7150/jca.94759. [Google Scholar] [PubMed] [CrossRef]
26. I. Matas, C. Serrano, F. Silva, A. Serrano, T. Toledo-Pastrana and B. Acha, “AI-driven skin cancer diagnosis: Grad-CAM and expert annotations for enhanced interpretability,” 2024, arXiv:2407.00104. [Google Scholar]
27. M. M. Saraiva et al., “Deep learning and high-resolution anoscopy: Development of an interoperable algorithm for the detection and differentiation of anal squamous cell carcinoma precursors—A multicentric study,” Cancers, vol. 16, no. 10, 2024, Art. no. 1909. doi: 10.3390/cancers16101909. [Google Scholar] [PubMed] [CrossRef]
28. G. H. Dagnaw, M. El Mouhtadi, and M. Mustapha, “Skin cancer classification using vision transformers and explainable artificial intelligence,” J. Med. Artif. Intell., 2024. doi: 10.21037/jmai. [Google Scholar] [CrossRef]
29. M. M. Hossain, M. M. Hossain, M. B. Arefin, F. Akhtar, and J. Blake, “Combining state-of-the-art pre-trained deep learning models: A noble approach for skin cancer detection using max voting ensemble,” Diagnostics, vol. 14, no. 1, 2023, Art. no. 89. doi: 10.3390/diagnostics14010089. [Google Scholar] [PubMed] [CrossRef]
30. H. Y. Huang, H. T. Nguyen, T. L. Lin, P. Saenprasarn, P. H. Liu and H. C. Wang, “Identification of skin lesions by snapshot hyperspectral imaging,” Cancers, vol. 16, no. 1, 2024, Art. no. 217. doi: 10.3390/cancers16010217. [Google Scholar] [PubMed] [CrossRef]
31. W. Yuan, Z. Du, and S. Han, “Semi-supervised skin cancer diagnosis based on self-feedback threshold focal learning,” Discover Oncol., vol. 15, no. 1, 2024, Art. no. 180. doi: 10.1007/s12672-024-01043-8. [Google Scholar] [PubMed] [CrossRef]
32. C. E. A. Tai, E. Janes, C. Czarnecki, and A. Wong, “Double-condensing attention condenser: Leveraging attention in deep learning to detect skin cancer from skin lesion images,” 2023, arXiv:2311.11656. [Google Scholar]
33. A. Mumuni and F. Mumuni, “Data augmentation: A comprehensive survey of modern approaches,” Array, vol. 16, 2022, Art. no. 100258. doi: 10.1016/j.array.2022.100258. [Google Scholar] [CrossRef]
34. K. Maharana, S. Mondal, and B. Nemade, “A review: Data pre-processing and data augmentation techniques,” Glob. Transit. Proc., vol. 3, no. 1, pp. 91–99, 2022. [Google Scholar]
35. K. Alomar, H. I. Aysel, and X. Cai, “Data augmentation in classification and segmentation: A survey and new strategies,” J. Imaging, vol. 9, no. 2, 2023, Art. no. 46. [Google Scholar]
36. M. Michalska-Ciekańska, “Multiclass skin lesions classification based on deep neural networks,” Informatyka, Automatyka, Pomiary w Gospodarce i Ochronie Środowiska, vol. 12, no. 2, pp. 40–54, 2022. doi: 10.1016/j.compmedimag.2018.05.004. [Google Scholar] [CrossRef]
37. B. Harangi, “Skin lesion detection based on an ensemble of deep convolutional neural networks,” 2017. doi: 10.1016/j.jbi.2018.08.006. [Google Scholar] [PubMed] [CrossRef]
38. A. Mahbod, G. Schaefer, I. Ellinger, R. Ecker, A. Pitiot and C. Wang, “Fusing fine-tuned deep features for skin lesion classification,” Comput. Med. Imaging Graph, vol. 71, pp. 19–29, Jan. 2019. doi: 10.1016/j.compmedimag.2018.10.007. [Google Scholar] [PubMed] [CrossRef]
39. S. S. Chaturvedi, K. Gupta, and P. S. Prasad, “Skin lesion analyzer: An efficient seven-way multiclass skin cancer classification using MobileNet,” in Advanced Machine Learning Technologies and Applications. Singapore: Springer. 2021, pp. 165–176. doi: 10.1007/978-981-15-3383-9_15. [Google Scholar] [CrossRef]
40. Z. C. Navarrete-Dechent, S. W. Dusza, K. Liopyris, A. A. Marghoob, A. C. Halpern and M. A. Marchetti, “Automated dermatological diagnosis: Hype or reality?” J. Investig. Dermatol., vol. 138, no. 10, Oct. 2018, Art. no. 2277. doi: 10.1016/j.jid.2018.04.040. [Google Scholar] [PubMed] [CrossRef]
41. S. S. Han et al., “Deep neural networks show an equivalent and often superior performance to dermatologists in onychomycosis diagnosis: Automatic construction of onychomycosis datasets by region-based convolutional deep neural network,” PLoS One, vol. 13, no. 1, Jan. 2018, Art. no. e0191493. doi: 10.1371/journal.pone.0191493. [Google Scholar] [PubMed] [CrossRef]
42. J. Yap, W. Yolland, and P. Tschandl, “Multimodal skin lesion classification using deep learning,” Exp. Dermatol., vol. 27, no. 11, pp. 1261–1267, Nov. 2018. doi: 10.1111/exd.13777. [Google Scholar] [PubMed] [CrossRef]
43. K. He, X. Zhang, S. Ren, and J. Sun, “Deep residual learning for image recognition,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit., 2016, pp. 770–778. doi: 10.1109/CVPR.2016.90. [Google Scholar] [CrossRef]
44. A. Krizhevsky, I. Sutskever, and G. E. Hinton, “ImageNet classification with deep convolutional neural networks,” Advanc Neural Informat Process Systems, vol. 25, 2012. [Google Scholar]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF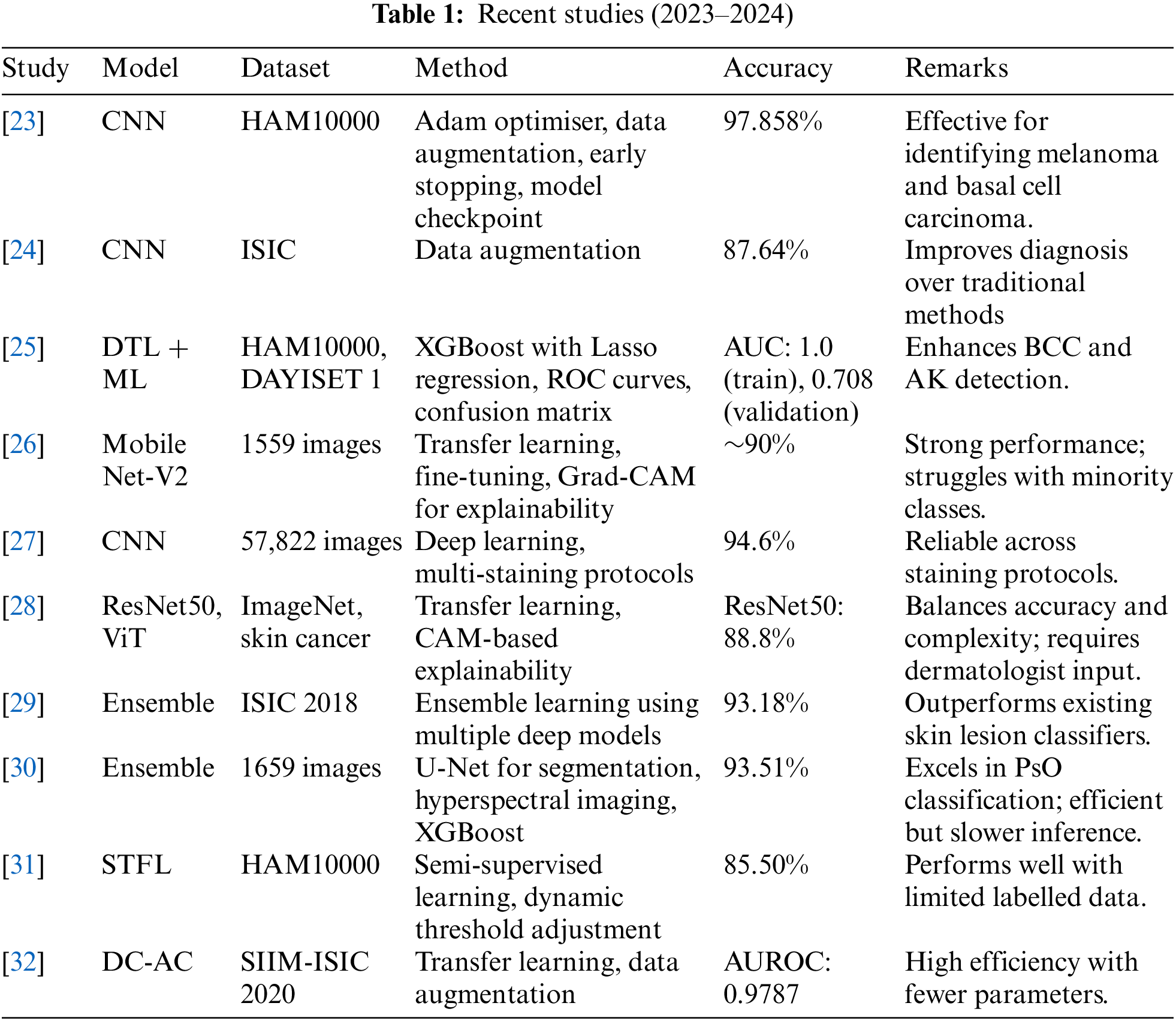
 Downloads
Downloads
 Citation Tools
Citation Tools