 Open Access
Open Access
ARTICLE
Pancreas Segmentation Optimization Based on Coarse-to-Fine Scheme
1 School of Computer Science and Telecommunication, Jiangsu University, Zhenjiang, 212013, China
2 School of Computer Science, Jiangsu University of Science and Technology, Zhenjiang, 212013, China
* Corresponding Author: Zhe Liu. Email:
(This article belongs to the Special Issue: Cognitive Granular Computing Methods for Big Data Analysis)
Intelligent Automation & Soft Computing 2023, 37(3), 2583-2594. https://doi.org/10.32604/iasc.2023.037205
Received 27 October 2022; Accepted 06 February 2023; Issue published 11 September 2023
Abstract
As the pancreas only occupies a small region in the whole abdominal computed tomography (CT) scans and has high variability in shape, location and size, deep neural networks in automatic pancreas segmentation task can be easily confused by the complex and variable background. To alleviate these issues, this paper proposes a novel pancreas segmentation optimization based on the coarse-to-fine structure, in which the coarse stage is responsible for increasing the proportion of the target region in the input image through the minimum bounding box, and the fine is for improving the accuracy of pancreas segmentation by enhancing the data diversity and by introducing a new segmentation model, and reducing the running time by adding a total weights constraint. This optimization is evaluated on the public pancreas segmentation dataset and achieves 87.87% average Dice-Sørensen coefficient (DSC) accuracy, which is 0.94% higher than 86.93%, result of the state-of-the-art pancreas segmentation methods. Moreover, this method has strong generalization that it can be easily applied to other coarse-to-fine or one step organ segmentation tasks.Keywords
According to national cancer registry center (cancer hospital chinese academy of medical sciences) data released in 2022 [1], pancreatic cancer is a malignant tumor with a 5-year survival rate of 5 percent. Fortunately, early diagnosis and treatment can improve the prognosis of pancreatic cancer, then preventing its further deterioration. Accurate pancreas segmentation can help us reduce the impact of the other organs on the detection of tumor areas earlier and make it easier to treat it in advance to improve survival [2]. In addition, the segmented pancreas can be used for volumetric measurements, 3D reconstruction, and visualization, which can greatly benefit the pancreatic diseases diagnosis [3]. Meanwhile, compared to the segmentation of abdominal liver, spleen and kidney, the segmentation accuracy of pancreas is the lowest at present because of the following three reasons: (1) the small size of pancreas in abdominal image, (2) the blurred boundary between pancreas and its surrounding tissues, and (3) the high variabilities in shape, size and location between inter and intra slices, as shown in Fig. 1 [4]. Hence, the research of pancreas segmentation is extraordinary essential, and it is of great significance to improve the accuracy of pancreas segmentation.
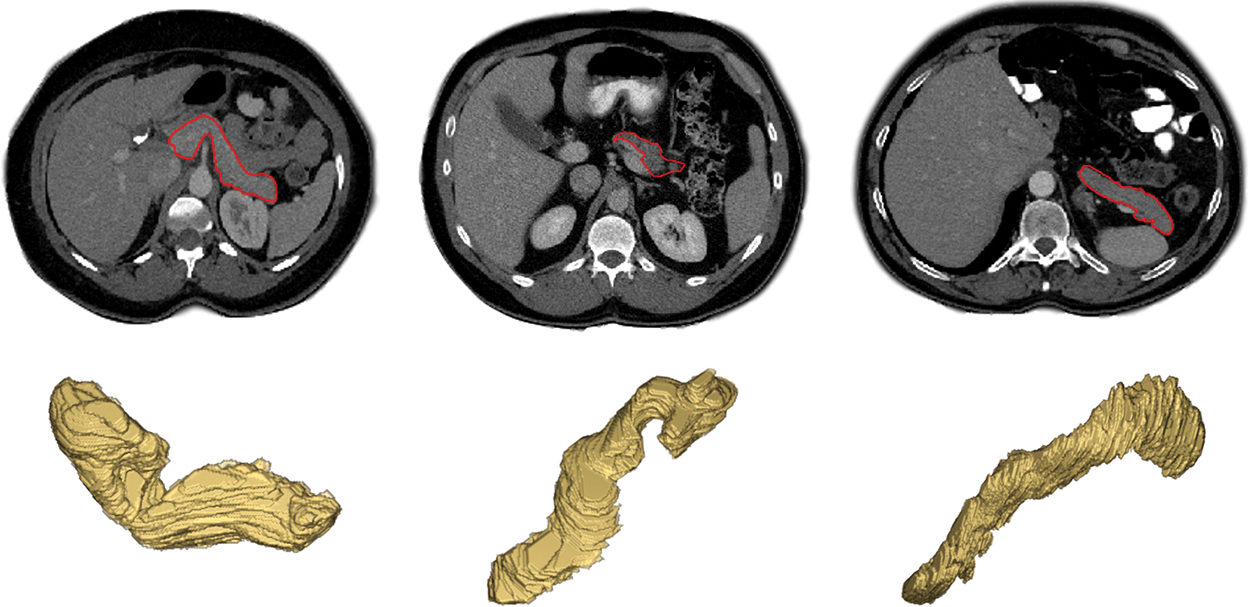
Figure 1: Examples of variations in appearance, shape, size and location of the pancreas as seen on contrast enhanced CT after removal of the image background by masking the patient’s body
Due to the non-invasive, high density resolution of CT scans, it is widely used in the diagnosis and detection of abdominal diseases. According to that, the pancreas segmentation based on CT images has attracted considerable attention in recent years, and a large number of segmentation approaches, which are mainly divided into two categories: tradition machine learning and deep learning based approaches, have been proposed for pancreas and tumor segmentation. In early machine learning, the image registration and atlas methods were proposed to achieve the pancreas segmentation based on the location obtained by locating other organs or vessels around the pancreas first [5–7], but the last segmentation accuracy is basically more than 60%, which is not satisfactory and the complexity of these algorithms is relatively high. While Farag et al. [8] proposed the super-pixel algorithm to obtain the general pancreas region and realize the final segmentation through random forest classification algorithm, which is a novel bottom-up segmentation strategy, and achieved a high accuracy of DSC 68.2% at that time. In another type of method [9], the seed point in an interactive manner was given and the segmentation was achieved by graph cut or region growth, which is a new pancreas segmentation method, but its segmentation accuracy is measured by Jaccard index (0.705 ± 0.054).
With the rapid development of machine learning, deep learning technique has shown its outstanding advantages in many fields including medical image processing, compared to traditional machine learning methods. Consequently, some deep learning methods based on the convolutional neural networks (CNNs) were proposed. It has been proved that using super-pixel algorithm to form candidate regions as an input to the CNN could achieve better pancreas segmentation results with the average DSC of 80% ± 4% [10]. Subsequently, Zhou et al. [11] proposed using the a novel CNN structure named full convolutional network (FCN) to do coarse-to-fine pancreas segmentation, of which, the network in the two stages were both FCN. The first stage is to achieve the initial segmentation, which means obtaining a smaller input region and the second is to do re-segmentation with the same FCN to get the final result. While Zheng et al. [12] also put forward a two-stage network framework (location stage and fine stage) to segment pancreas, which achieved more than 80% accuracy of DSC. From the other aspect, CNN combined with recurrent neural network (RNN) was also put forward to do pancreas segmentation [13], which combined CNN with RNN to obtain the context of the CT image and then completed the segmentation with the current good precision. Different from the methods above which are all 2-Dimensional, the 3-Dimensional segmentation method with more significant effect has been presented [14–16], which used 3D FCN or 3D U-Net to obtain the initial pancreas segmentation probability map to locate the pancreas, and then conducted the fine segmentation through the graph cut or 3D U-net. No matter 2D, 3D or traditional algorithms, most of the methods above are all coarse-to-fine two stage, and it is well-known that the coarse-to-fine method can always achieve the highest pancreas segmentation accuracy, in the field of pancreas segmentation, whether it is traditional or the state-of-the-art method used.
Despite the exciting success of deep learning technique in automatic pancreas segmentation, there are still two-fold drawbacks that might be addressed for further improvement. On one hand, the highest and state-of-art accuracy of pancreas segmentation is still not high enough to be used clinically. This may be because the domain knowledge of pancreas segmentation has not been fully explored, such as the region saliency information of the pancreas. On the other hand, CNN models are typically computationally expensive and require a large amount of memory and computation power. Then making it impossible to deploy it on natural devices in clinical applications.
To tackle these problems, this work proposes a novel learning optimization based on coarse-to-fine scheme for automatic pancreas segmentation. Our coarse-to-fine segmentation method can be divided into two key steps: coarse stage and fine stage. The coarse segmentation stage is responsible for locating the approximate position of the pancreas and increasing the proportion of the target region in the input image. While the fine is for improving the accuracy of pancreas segmentation by enhancing the data diversity and by introducing a new segmentation model, and reducing the running time by adding a total weights constraint. In the coarse stage, the ground-truth image and its one-to-one correspondence with the input image are fully utilized, and the background region is reduced through the minimum bounding-box of the ground-truth to increase the proportion of the target region in the input image. In the fine stage, we will study from the following three aspects for improving the fine stage segmentation accuracy when given a bounding box:
i) The preprocessing algorithms are used to expand the original data to four times, which can greatly enhance the data diversity.
ii) A new model is proposed to improve the semantic gap problem between the encoder and decoder part of the traditional U-net.
iii) A total weights constraint is come up with to strengthen the loss function, which cannot only simplify the network structure but also alleviate the over-fitting problem.
The remainder of this paper is organized as follows. Section 2 outlines our method including data preprocessing, Aug-net and new loss-function, and comprehensive experiments are presented in Section 3. Section 4 presents and discusses the experimental results. Finally, Section 5 summarizes the conclusions of this work.
The brief process of the typical coarse-to-fine framework [17] is shown in Fig. 2, where a, c and e indicate the original input CT slices, coarse stage outputs and fine stage outputs, respectively. The b and d are the deep neural networks which can be replaced with other segmentation networks.
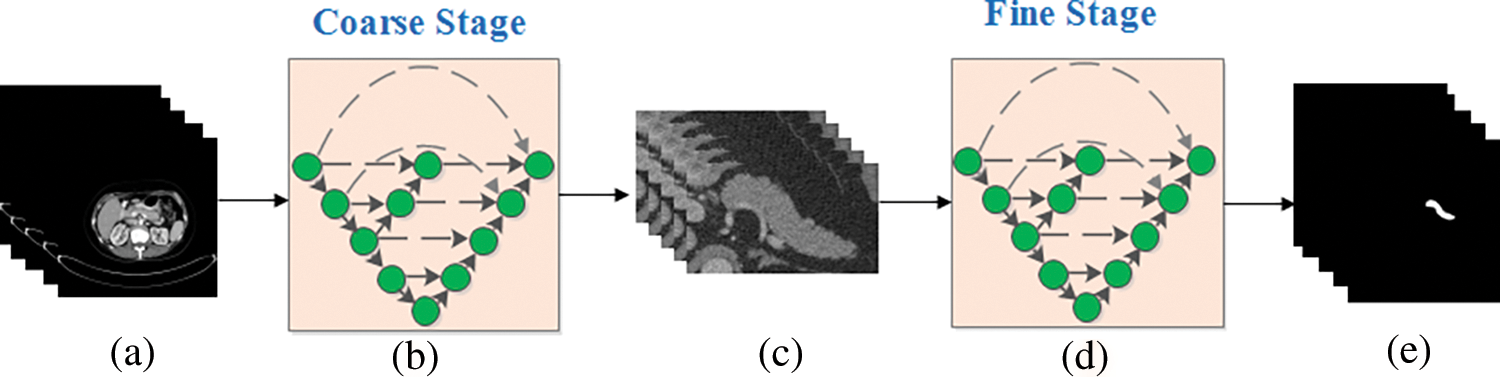
Figure 2: A typical example of the coarse-to-fine algorithm
Generally, the coarse-to-fine scheme can be divided into two stages: coarse and fine, in which, the coarse is to reduce the background region without pancreas and the fine is to achieve more accurate segmentation results with the new images obtained by the coarse stage as input. It can be seen from the figure that the same training strategy is applied in both two stages in traditional coarse-to-fine approach, which may have a negative impact on the final segmentation accuracy. Consequently, three methods focused on improving the fine stage segmentation accuracy are proposed in this paper where both of these methods discuss how to make full use of the fine stage based on the bounding box given, which is much different from the tradition.
The following comes with the introduction of some parameters and their settings that may appear in the fine segmentation stage. Firstly, the original input of the whole network, CT slice, is a 3D volume
2.1 Data Preprocessing with Three Algorithms
For improving the contrast of the specified region in and around pancreas, and lowering the influence of background, effective preprocessing of the data has carried out, which can be very conducive to speeding up the network convergence and improving the segmentation accuracy when taking use of the pre-processed data as inputs instead of the original data. In the following, three optimal algorithms presented are used to improve the proposed segmentation method during the training process which is shown in Fig. 3. Namely, from three aspects that diversifying data preprocessing pattern, improving network and constraining loss function. What illustrated in Fig. 3 is that when given bounding boxes, the designed model in the training phase is fitted to the data set by multiple iterations while the segmentation result in testing is obtained by the previously trained model.
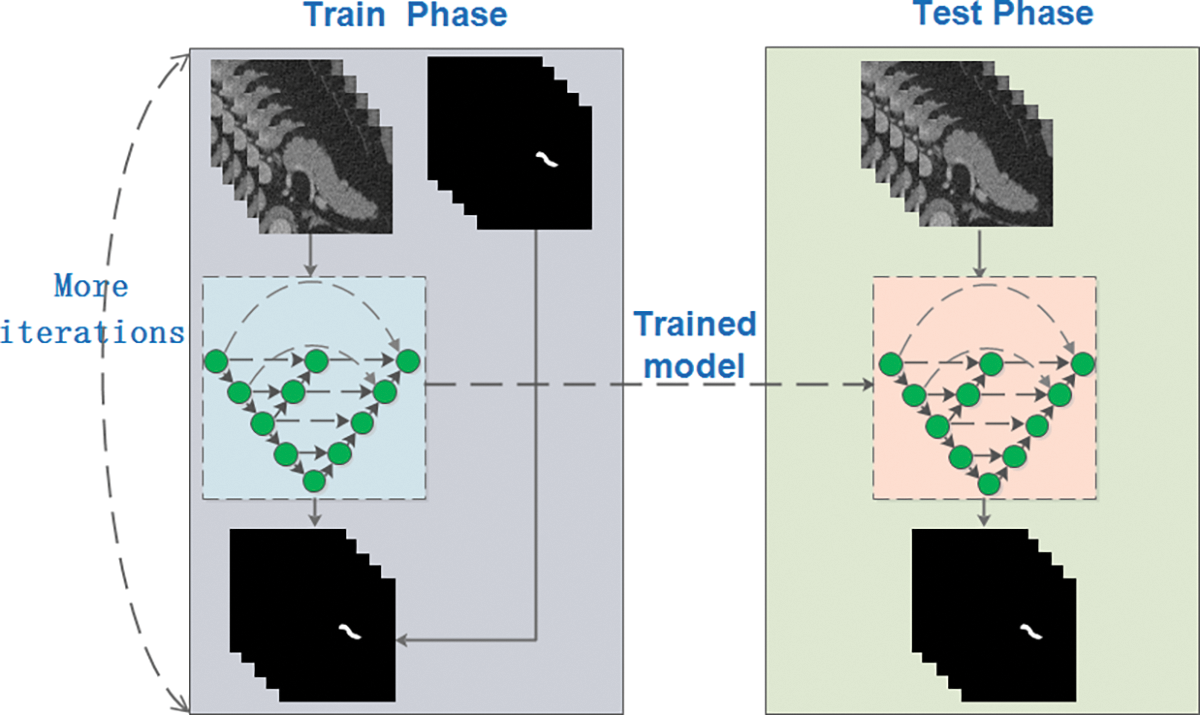
Figure 3: Flow chart of the proposed segmentation method
The details of the data preprocessing can be divided into the next three steps:
Step 1: Cropping the pancreas slices with bounding boxes generated by ground-truth labels because pancreas region only accounts for a small proportion in the raw abdominal CT 2D image (less than 5% [4]), and of which, the around margins are filled with the numbers sampled from {0, 1,..., 60}. This data enhancement method, which is conducive to regularizing the network and preventing over-fitting, is shown in Fig. 4 where a is the ground-truth label of the original data size and b which is taken as the input of the proposed network in the fine stage indicates a CT image cropped by adding the green bounding box in a to the raw 512 * 512 CT image correspondingly.
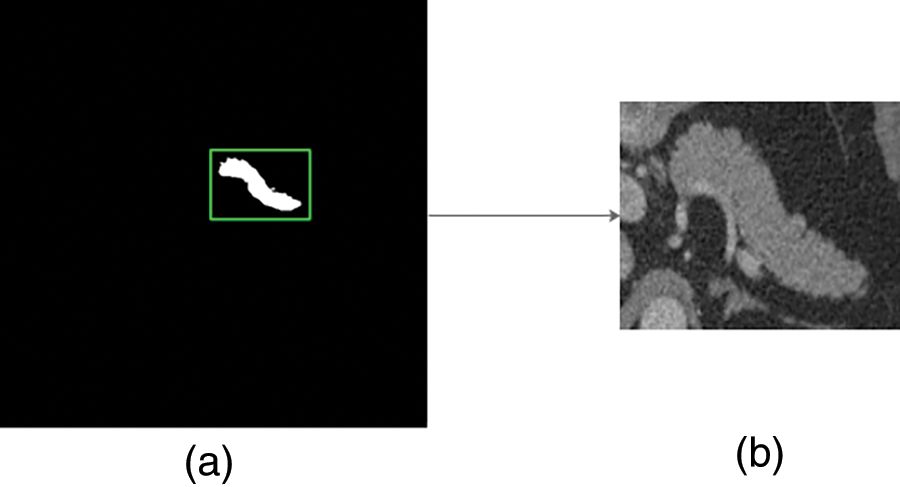
Figure 4: Process of pancreas cropping
Step 2: Referring to the common intensity distribution of pancreas, scaling the image pixel values which is first transformed to HU value [18] limited in [−55, 210] to the range [0, 1], because the raw CT value has a wide range of values(common [−1024, 1023]), which can increase the contrast of the pancreas-related region and speed up the gradient descent convergency after normalization to promote the performance. The comparisons of the images before and after transformation are shown in Fig. 5, where the left are the original CT images and the right are corresponding transformed CT images.
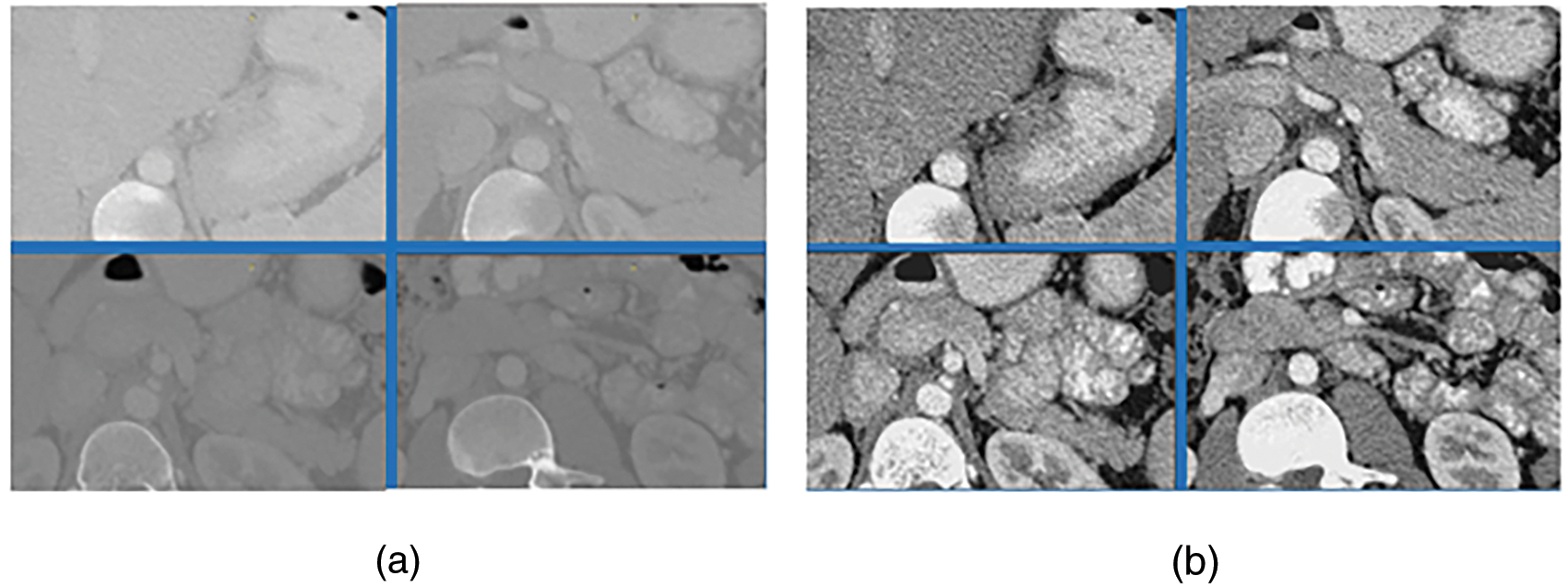
Figure 5: HU contrast pictures
Step 3: For increasing the diversity of the original dataset, applying the elastic deformation and small amplitude rotation to augment the data. Namely, deforming each small 8 * 8 region of the image and rotating it in randomly degrees in [−15, 15], meanwhile, conducting the same operation on the corresponding ground-truth label image, which can be seen in Fig. 6. The original CT image and the corresponding ground-truth label in Fig. 6 are placed in the first column, while the next three columns are the results generated based on elastic deformation, small amplitude rotation and both of above two operations respectively. In this manner, the training data numbers can be expanded to four times of the original through these behaviors.

Figure 6: A set of expanded data
After the effective data preprocessing, it is of great importance to establish a powerful network that can learn these data adequately. U-net, a classical medical image segmentation network with an encoding-decoding structure, extracts feature information of different scales in the encoding part and uses the long connection to supplement the decoder part to better restore the position and edge information of the target region. The long connection can fight the vanishing gradient problem and recover the pyramid level features learned. However, how to reduce the same pyramid level “semantic gap” is just as important to the final segmentation results. In this work, a short connection formed through a decoder combined with the encoding part is added in the proposed Aug-unet to overcome this problem, of which, the structure is shown in Fig. 7 where both short and long connections that can enhance the feature transferring and solve the “semantic gap” are used for different layers.
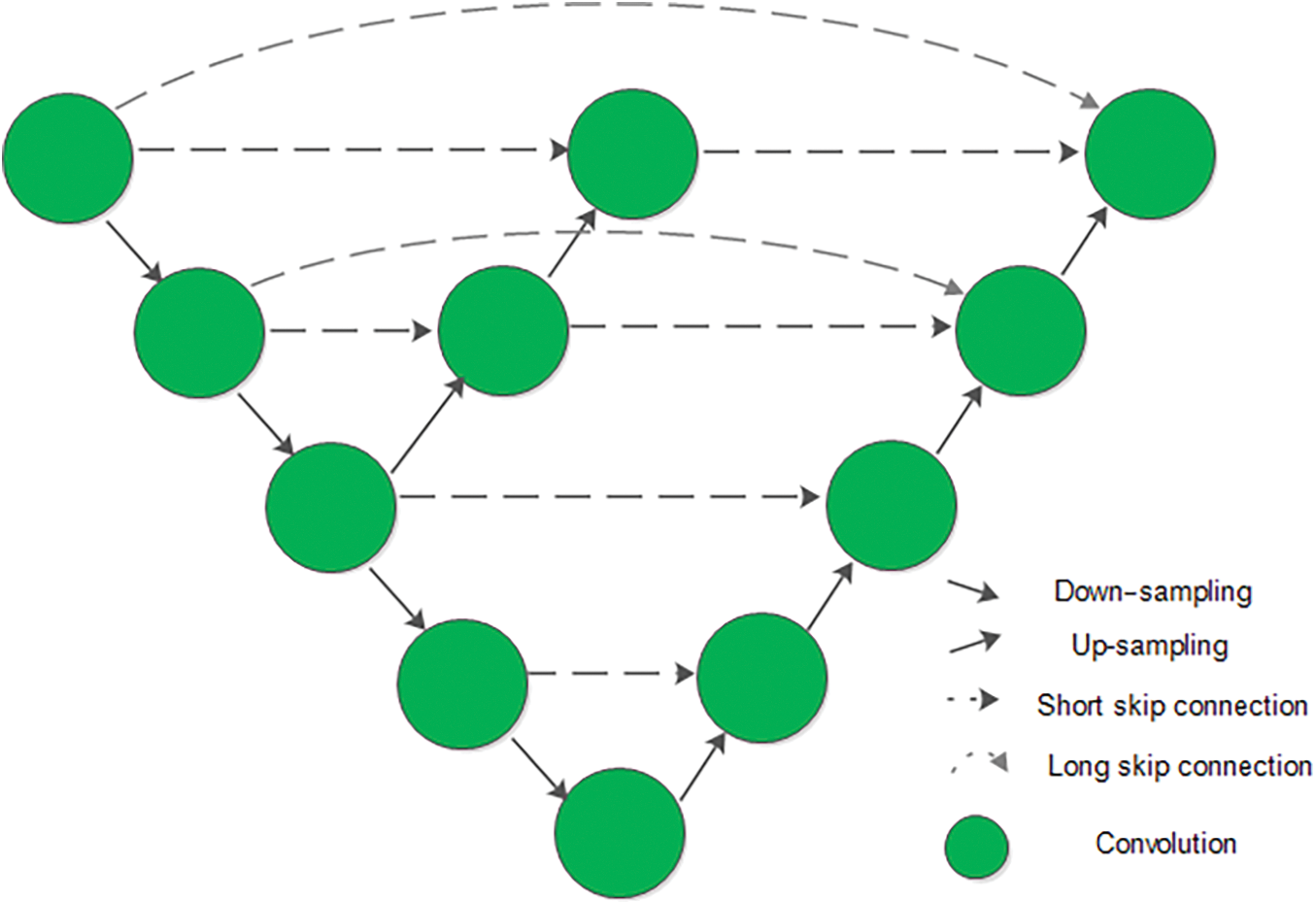
Figure 7: Structure of proposed Aug-unet
The intermediate decoding part is set to generate the different decoding information from the same scale feature maps, which is going to be added to the final decoding part. Combination of the two different connection structures, which can enhancement the traditional U-net, forms a novel network called Aug-unet in this paper.
2.3 Strengthened Loss Function
In the training process, the ultimate goal is to reduce the loss to improve the accuracy. The most common used loss function of segmentation task is formula (1). According to that in deep networks, the number of parameters far exceeding the amount of data leads to over-fitting, which makes the model perform better in training but worse in testing. Based on this, the original loss function is modified with regularization, which is shown as formula (2).
where
The national institutes of health (NIH) clinical center performed 82 abdominal contrast enhanced 3D CT scans from 53 male and 27 female subjects, which is public available (https://wiki.cancerimagingarchive.net/display/Public/Pancreas-CT) and the most widely used dataset for pancreas segmentation. So this public NIH pancreas dataset is also used in this work to fully validate our proposed method. The size of each CT scan is
The cross-validation strategy is applied to the whole training and test process, which divides the whole dataset into four folds including 21,21,20 and 20 volumes, respectively. Where three subsets are used to training process and the remaining subset is used to the testing.
The final effects of experiments are measured by computing the average DSC and standard deviation over all 82 cases. The DSC measures how similar image segmentation algorithm’s results with it’s corresponding ground-truth segmentation. It is easy to represent geometrically:
where
The software environment where to train the model is TensorFlow which is written in keras as the backend, and the hardware device is the GEFORCE RTX 2080 Ti GPU.
In the experiments, the input volume is generated through the ground-truth (oracle) bounding box of each training case. During the training process, the learning rate is set to
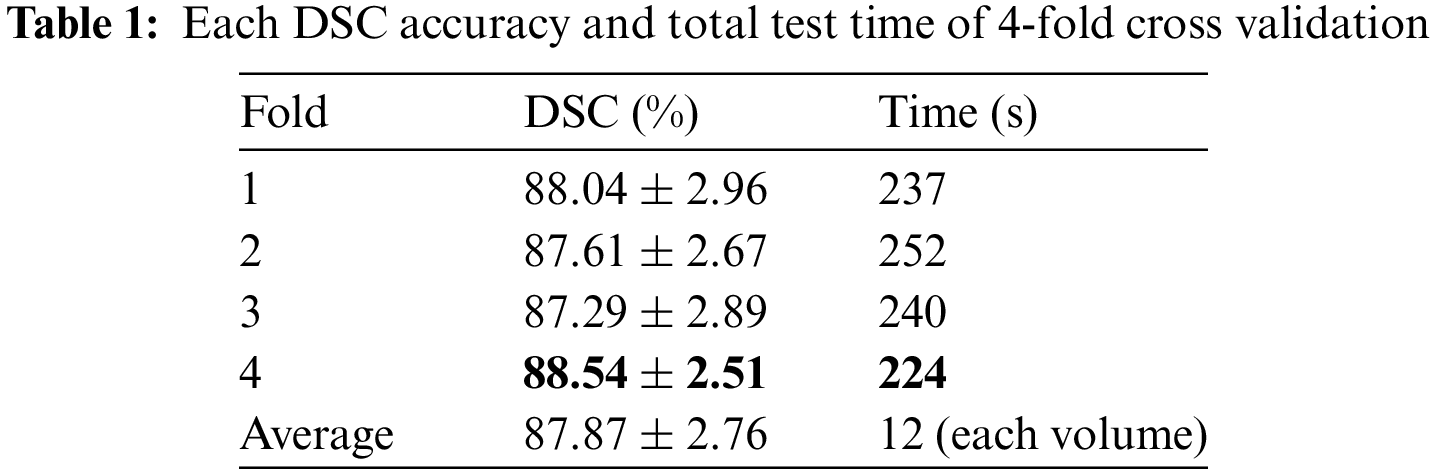
The final DSC accuracy obtained by 4-fold cross-validation verification is shown in Table 1 where it can be seen that the results of different test processes are similar, which can prove the effectiveness of the proposed method and avoid the probability problem that the experimental results may be accidental. In the evaluation indicators, apart from the DSC accuracy and standard deviation, the total time consumption of each test process is also counted to compute the average time of each patient case (seen from the last row of Table 1) which can be used to estimate the performance of other experiments and projects based on this algorithm.
To further demonstrate the performance of the proposed method, final result of the proposed method is compared with the eight state-of-the-art methods which are all training and testing the networks based on the NIH datasets with identical initialization. The comparison results are shown in Table 2. As can be seen from the Table 2, our proposed segmentation algorithm in the accuracy of DSC has shown its superiority over others, among which, increasing the DSC accuracy from 86.93% to 87.87% and decreasing the standard deviation from 4.92% to 2.76%, which proves the effectiveness and robustness of this method. In particular, the worst result of the presented method increases from a reported worst case of 61.82% to 74.16%.
Over time, the accuracy of the pancreas segmentation using the coarse-to-fine scheme is increasing, which can be viewed in Fig. 8, where x and y axis represent time and DSC (%) accuracy, respectively. It is intuitively observed that the segmentation accuracy is increasing steadily along the time axis and the proposed procedure has the highest accuracy by comparison.

Figure 8: Comparison of experimental results in different years
Finally, the visualized comparison between the predicted segmentation probability maps and the ground-truth masks is shown in Fig. 9, where the first two rows are the 3D reconstruction of the ground-truth labels (left) and the predict results (right). In the third row, red lines depict ground-truth labels and the blue lines indicate the predicted segmentation masks. It can be observed that the segmentation results of the proposed method are very close to the standard ground truth labels.

Figure 9: Comparison of a set of segmentation results
Motivated by that coarse-to-fine segmentation framework plays an important role in pancreas segmentation, a novel method based on coarse-to-fine scheme is proposed to achieve the pancreas segmentation results. The coarse stage has completed the target region location through the information of the ground-truth and improved the proportion of target region in the input image of the fine stage. The fine stage has improved the accuracy of pancreas segmentation by enhancing the data diversity and by adjusting the U-net, and decreased the time cost of the whole pancreas segmentation by adding a total weight constraint. Comprehensive experimental results on the NIH dataset compared with the state-of-art methods which are shown in Table 2 have demonstrated the effectiveness of this approach. Not only increasing the DSC accuracy by 0.94% from 86.93% but also reducing the standard deviation and the running time, which means that this procedure is more stable and robust. In addition, the proposed method can also be used for the segmentation tasks based on coarse-to-fine or one step of other organs such as the spleen, liver and kidney.
Our promotion at present is not enough for clinical application, and further research is needed to improve the accuracy in the next few years. We will use co-saliency detection techniques to unearth significant information from multi-stages and adjacent CT slices for further improving the pancreas segmentation performance.
Acknowledgement: The authors thank classmates in the laboratory for their continued support and the supervisor for his guidance and constant support rendered during this work.
Funding Statement: This work was supported by the National Natural Science Foundation of China [61772242, 61976106, 61572239]; the China Postdoctoral Science Foundation [2017M611737], the Six Talent Peaks Project in Jiangsu Province [DZXX-122], the Jiangsu Province Emergency Management Science and Technology Project [YJGL-TG-2020-8], the Key Research and Development Plan of Zhenjiang City [SH2020011], and Postgraduate Innovation Fund of Jiangsu Province [KYCX18_2257].
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. C. F. Xia, X. S. Dong, H. Li, M. M. Cao, D. A. Q. Sun et al., “Cancer statistics in China and United States, 2022: Profiles, trends and determinants,” Chinese Medical Journal, vol. 135, no. 5, pp. 584–590, 2022. [Google Scholar] [PubMed]
2. Y. Zhang, J. Wu, Y. Liu, Y. Chen, W. Chen et al., “A deep learning framework for pancreas segmentation with multi-atlas registration and 3D level-set,” Medical Image Analysis, vol. 68, no. 10, pp. 101884–101895, 2021. [Google Scholar] [PubMed]
3. D. W. Zhang, J. J. Zhang, Q. Zhang, J. G. Han, S. Zhang et al., “Automatic pancreas segmentation based on lightweight DCNN modules and spatial prior propagation,” Pattern Recognition, vol. 114, no. 1, pp. 31–45, 2021. [Google Scholar]
4. X. Yao, Y. Song and Z. Liu, “Advances on pancreas segmentation: A review,” Multimedia Tools and Applications, vol. 79, no. 9, pp. 6799–6821, 2019. [Google Scholar]
5. M. Hammon, A. Cavallaro, M. Erdt, P. Dankerl, M. Kirschner et al., “Model-based pancreas segmentation in portal venous phase contrast-enhanced CT images,” Journal of Digital Imaging, vol. 26, no. 6, pp. 1082–1090, 2013. [Google Scholar] [PubMed]
6. M. Erdt, M. Kirschner, K. Drechsler, S. Wesarg, M. Hammon et al., “Automatic pancreas segmentation in contrast enhanced CT data using learned spatial anatomy and texture descriptors,” in IEEE Int. Symp. on Biomedical Imaging: From Nano to Macro, Chicago, USA, pp. 2076–2082, 2011. [Google Scholar]
7. A. Shimizu, T. Kimoto, H. Kobatake, S. Nawano and K. Shinozaki, “Automated pancreas segmentation from three-dimensional contrast-enhanced computed tomography,” International Journal of Computer Assisted Radiology and Surgery, vol. 5, no. 1, pp. 85–98, 2010. [Google Scholar] [PubMed]
8. A. Farag, L. Lu, E. Turkbey, J. M. Liu and R. M. Summers, “A bottom-up approach for automatic pancreas segmentation in abdominal CT scans,” Abdominal Imaging: Computational and Clinical Applications, vol. 8676, no. 1, pp. 103–113, 2014. [Google Scholar]
9. H. Takizawa, T. Suzuki, H. Kudo and T. Okada, “Interactive segmentation of pancreases in abdominal computed tomography images and its evaluation based on segmentation accuracy and interaction costs,” Biomed Research International, vol. 2017, no. 1, pp. 239–244, 2017. [Google Scholar]
10. H. R. Roth, L. Lu, A. Farag, H. C. Shin, J. M. Liu et al., “Deeporgan: Multi-level deep convolutional networks for automated pancreas segmentation,” Medical Image Computing and Computer-Assisted Intervention, vol. 9349, no. 1, pp. 556–564, 2015. [Google Scholar]
11. Y. Zhou, L. Xie, W. Shen, Y. Wang, E. K. Fishman et al., “A fixed-point model for pancreas segmentation in abdominal CT scans,” in Int. Conf. on Medical Image Computing and Computer-Assisted Intervention, Quebec City, Quebec, Canada, pp. 693–701, 2017. [Google Scholar]
12. H. Zheng, L. J. Qian, Y. L. Qin, Y. Gu and J. Yang, “Improving the slice interaction of 2.5D CNN for automatic pancreas segmentation,” Medical Physics, vol. 47, no. 11, pp. 5543–5554, 2020. [Google Scholar] [PubMed]
13. J. Xue, K. L. He, D. Nie, E. Adeli, Z. S. Shi et al., “Cascaded multitask 3D fully convolutional networks for pancreas segmentation,” IEEE Transactions on Cybernetics, vol. 51, no. 4, pp. 2153–2165, 2021. [Google Scholar] [PubMed]
14. Z. Zhu, Y. Xia, W. Shen, E. K. Fishman and A. L. Yuille, “A 3D coarse-to-fine framework for volumetric medical image segmentation,” in Int. Conf. on 3D Vision, Verona, Italy, pp. 682–690, 2018. [Google Scholar]
15. H. R. Roth, H. Oda, X. Zhou, N. Shimizu, Y. Yang et al., “An application of cascaded 3D fully convolutional networks for medical image segmentation,” Computerized Medical Imaging and Graphics, vol. 66, no. 1, pp. 90–99, 2018. [Google Scholar] [PubMed]
16. M. Oda, N. Shimizu, H. R. Roth, K. i. Karasawa, T. Kitasaka et al., “3D FCN feature driven regression forest-based pancreas localization and segmentation,” in Computer Vision and Pattern Recognition, Quebec, Canada, pp. 222–230, 2017. [Google Scholar]
17. Y. Liu and S. Liu, “U-net for pancreas segmentation in abdominal CT scans,” in Int. Symp. on Biomedical Imaging, Washington, USA, pp. 203–222, 2018. [Google Scholar]
18. M. Li, F. Lian and S. Guo, “Multi-scale selection and multi-channel fusion model for pancreas segmentation using adversarial deep convolutional nets,” Journal of Digital Imaging, vol. 35, no. 1, pp. 47–55, 2022. [Google Scholar] [PubMed]
19. X. Glorot and Y. Bengio, “Understanding the difficulty of training deep feedforward neural networks,” Journal of Machine Learning Research, vol. 9, no. 1, pp. 249–256, 2010. [Google Scholar]
20. A. Farag, L. Lu, H. R. Roth, J. Liu, E. Turkbey et al., “Deep learning and convolutional neural networks for medical image computing,” in Advances in Computer Vision and Pattern Recognition, 1st ed., Cham, Switzerland: Springer, pp. 279–302, 2017. [Google Scholar]
21. Q. Yu, L. Xie, Y. Wang, Y. Zhou, E. K. Fishman et al., “Recurrent saliency transformation network: Incorporating multi-stage visual cues for small organ segmentation,” in Conf. on Computer Vision and Pattern Recognition, Salt Lake City, United States, pp. 8280–8289, 2018. [Google Scholar]
22. Y. Z. Man, Y. S. B. Huang, J. Y. Feng, X. Li and F. Wu, “Deep Q learning driven CT pancreas segmentation with geometry-aware U-net,” IEEE Transactions on Medical Imaging, vol. 38, no. 8, pp. 1971–1980, 2019. [Google Scholar] [PubMed]
23. J. W. Long, X. L. Song, Y. An, T. Li and J. Z. Zhu, “Parallel multi-scale network with attention mechanism for pancreas segmentation,” IEEJ Transactions on Electrical and Electronic Engineering, vol. 17, no. 1, pp. 110–119, 2022. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF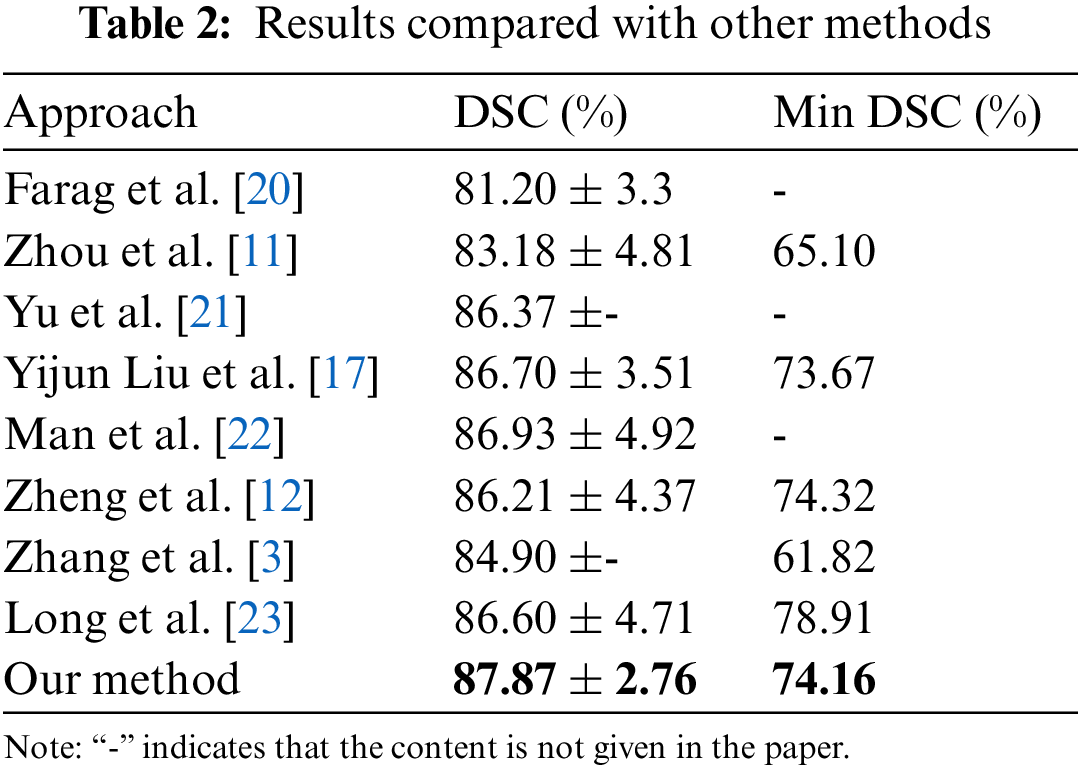
 Downloads
Downloads
 Citation Tools
Citation Tools