 Open Access
Open Access
ARTICLE
A Content-Based Medical Image Retrieval Method Using Relative Difference-Based Similarity Measure
1 Department of Computer Science, Faculty of Computing and Information Technology, King Abdulaziz University–Rabigh, Rabigh, 21589, Saudi Arabia
2 Department of Information Systems, Faculty of Computing and Information Technology, King Abdulaziz University, Jeddah, 21589, Saudi Arabia
3 Department of Information Technology, Faculty of Computing and Information Technology, King Abdulaziz University–Rabigh, Rabigh, 21589, Saudi Arabia
* Corresponding Author: Ali Ahmed. Email:
Intelligent Automation & Soft Computing 2023, 37(2), 2355-2370. https://doi.org/10.32604/iasc.2023.039847
Received 20 February 2023; Accepted 04 May 2023; Issue published 21 June 2023
Abstract
Content-based medical image retrieval (CBMIR) is a technique for retrieving medical images based on automatically derived image features. There are many applications of CBMIR, such as teaching, research, diagnosis and electronic patient records. Several methods are applied to enhance the retrieval performance of CBMIR systems. Developing new and effective similarity measure and features fusion methods are two of the most powerful and effective strategies for improving these systems. This study proposes the relative difference-based similarity measure (RDBSM) for CBMIR. The new measure was first used in the similarity calculation stage for the CBMIR using an unweighted fusion method of traditional color and texture features. Furthermore, the study also proposes a weighted fusion method for medical image features extracted using pre-trained convolutional neural networks (CNNs) models. Our proposed RDBSM has outperformed the standard well-known similarity and distance measures using two popular medical image datasets, Kvasir and PH2, in terms of recall and precision retrieval measures. The effectiveness and quality of our proposed similarity measure are also proved using a significant test and statistical confidence bound.Keywords
In the fields of data science and information retrieval, the concepts of similarity measures or metrics have been widely used. While the similarity measure describes how data samples or objects are closed or related to each other, the dissimilarity measure can tell us how distinct these objects or data are. In the content-based image retrieval (CBIR) in general, or specifically in the content-based medical image retrieval (CBMIR), similarity calculation is considered one of the important stages or phases of the retrieval process [1,2]. Any content-based medical image retrieval (CBMIR) method has the ability to represent medical images using their numerical attributes or features extracted from the basic image descriptors, such as color, texture and shape. This step of pre-processing or feature extraction was considered an offline process, because it was performed first and it was independent from the other components of retrieval systems. The second and most important stage, which is considered the core of any information retrieval system, is the retrieval process, which compromises two main stages or phases, the similarity calculation and the ranking process [3]. Feature extraction and representation are considered key to the success of any CBMIR system, because all the subsequent processes and operations, such as query analysis or reformulation, or any fusion method depends on these numerical values. The semantic gap between low level information, in terms of the main characteristics or image features captured by a machine and high level human perception, is still very large and a great deal of research has been conducted to reduce this gap [4].
Many solutions have been proposed over the past two decades to reduce this gap and enhance the retrieval performance of (CBMIR) systems. These solutions focus either on an offline process and features representation and fusion methods, or on contributing in the stage of image query similarity calculation. In addition, this gap could reduce by developing a new similarity measure or by applying some methods for improving the retrieval results, such as relevance feedback and query expansion [5–10]. Our recent contributions to this field may be found in [11,12]. The rest of this manuscript is organized as follows. In the next section, the research background and some examples of related works are provided. Our methodology and proposed similarity measure is explained in Section 3. The experimental results and research conclusion are presented in Sections 4 and 5, respectively.
The fusion process is one of the most frequently used methods for enhancing and improving the retrieval performance of many (CBMIR) systems. It has been applied in many (CBMIR) systems, and in several ways, but in general it can be divided into two categories: early fusion and late fusion [13,14]. Early fusion, which is also known as data fusion, feature fusion or the join features model, is considered to be a simple fusion process because it is done offline and is independent from any similarity measures, and it is done before any decision concerning the similarity process has been made. More of the benefits and advantages of this type of fusion can be found in [15,16]. In late fusion [17], features are not fused in advance, but some kinds of query evaluation and results merging are performed. Relevance feedback, query specification, and preprocessing and result post-processing, are the most common types of late fusion [18–20].
The authors applied early and late fusions in different ways: a fuse salient dominant color descriptor (SDCD) histogram, with a pyramidal histogram of visual words (PHOW) for background and foreground color problem solution, were used [21]. Wu et al. [22] combined visual and textual information to enhance the retrieval performance results. They merged (SIFT) descriptors, local binary patterns (LBP), Gabor texture and Tamura texture based on simple early fusion using multiple-kernel learning. Wei et al. [23] proposed a hybrid fusion approach for global features and local features, in their studies, four low orders Zernike moments (ZMs) are used to obtain the local features, while contour curvature and histograms of centrism distances (HCD) are used for local features. Similarly, Yu et al. [24] did the same work by combining both shape and texture features. They extracted shape features using exponent moment descriptors and a localized angular phase histogram was used for texture features extraction.
In many studies, the authors implemented (early) fusion by combining features vectors from multiple features domains, such as color, texture and shape, in unweighted or unequal form [25,26]. In contrast, this study applied unweighted fusion and treated each domain of features as un-equal, based on different weighted factors. The fusion process applied here is based on the weighted fusion process using variant weighted factors. Further, a fusion process can applied for statistical features extracted from color or texture information and it can also be applied for accurate features extracted using powerful and useful feature extraction tools, such as pre-trained convolution neural networks (CNNs) [27]. The deep learning method and convolutional neural networks (CNNs) have been widely used in medical feature extraction, as in [28–30], due to their powerful computation capabilities. They have also been successfully applied in other related healthcare applications, as in [31–33]. Recent model of fusion used rules of texture energy measures in the hybrid wavelet domain found in [34]. In addition to the fusion method, developing new similarity measures that give better results and outperform the well-known similarity coefficient groups is quite a challenge. Small additional mathematical factors or parameters, along with numbers of zero and nonzero features values could gain significant enhancement. This study utilized relative difference, one of the most well-known mathematical concepts, to develop a novel relative difference-based similarity (RDBSM) measure for (CBMIR), as well as for (CBIR) in general. The main novelty and contribution of the paper is summarized as follows:
a) Proposed a novel relative difference-based similarity measure (RDBSM) for content-based image retrieval (CBMIR). The proposed similarity measure gives a similarity value in a range of 0 to 1.
b) Develop an efficient retrieval method for (CBMIR) based on an unweighted fusion model of color and texture feature descriptors using the (RDBMS) similarity measure.
c) Enhance the retrieval performance of (CBMIR) methods using the (RDBSM) similarity measures and weighted fusion strategy for accurate features extracted using SqueezeNet and ResNet-18 pre-trained (CNNs).
3.1 Proposed Similarity Measure
This study proposes novel relative difference-based similarity measures (RDBSM) for content-based medical image retrieval (CBMIR). The new similarity measure is based on relative difference (RD) which will be computed first, and then used in our similarity formula. From quantitative science there are two ways to find and compute the change or difference into two quantities: these are absolute reference and relative difference. Absolute reference is equal to the new value minus the reference value. The size of the absolute change compared to the reference value is expressed as a fraction called the relative difference, as in Eq. (1) in the following:
One way of determining the relative difference (RD) between two numbers is to divide their absolute difference by their maximum absolute value. For two numbers, A and B, the relative difference (RD) is given by the following:
Another useful way is to divide the absolute difference by one of the number’s functional values, such as the absolute value of their arithmetical mean, as follows:
To avoid the division by zero in the following equation, when the two numbers have the same magnitude but the opposite sign, the equation was modified as follows:
In this study, the relative difference (RD) between two images features vectors is calculated first, and then this value is used to obtain the similarity value. For two images features vectors
where
For each pair of images (query image and single image from database) the above relative difference (RD) value will be used in computation of the similarity in our proposed similarity measure, as follows:
where
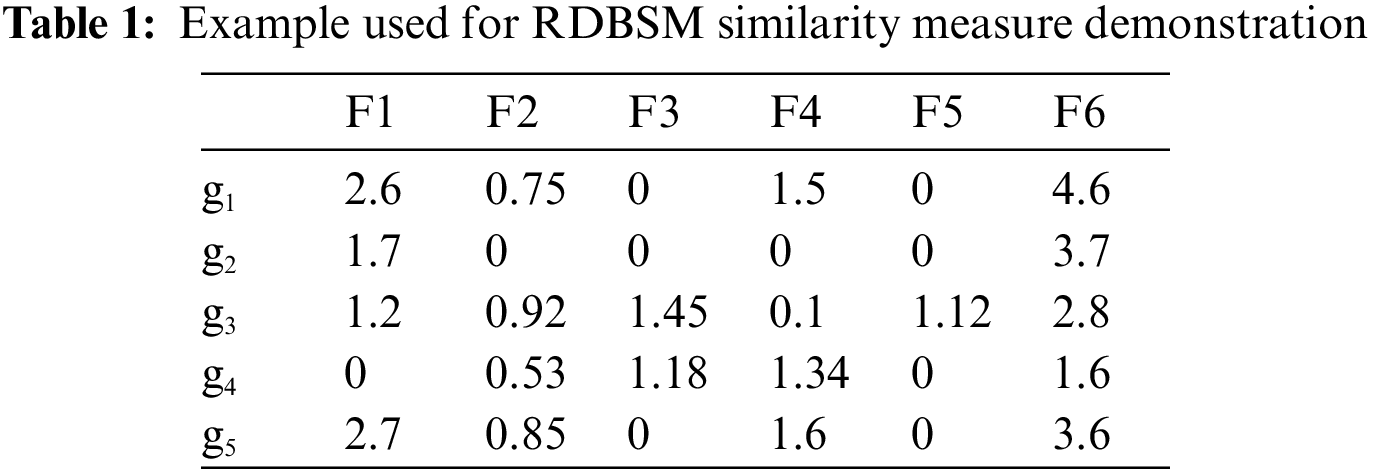
The relative difference (RD) between
For
Following the same steps, relative difference (RD) values between
3.2 Feature Extraction and Similarity Calculation
The general framework of content-based medical image retrieval (CBMIR) used in this study is shown in Fig. 1. This framework is divided into two stages or phases: the feature extraction (or offline) phase and the similarity and ranking (or online) phase. The stage of feature extraction is a major stage of any content-based image retrieval. For this purpose, features of medical images are extracted from the properties of color, shape or texture, and later the deep convolution artificial neural networks (CNNs) attracted the researchers to use them for feature extraction, due to their effectiveness in dealing with images and extracting accurate features that lead to a retrieval system with good results. In this first study, eighteen color features are extracted and combined with twelve texture features results in a total of thirty features used to represent each image in the image database. Color features were extracted using color moments functions in Eqs. (8) to (13) (six values from each channel of color images), while Eqs. (14) to (17) were used to extract texture features (four values from each channel of color images). The six statistical color moment functions used to extract color features, as described below, are well-known [35] and were used successfully in our previous studies [11,12]. For any image with a dimension of (M,N) in our dataset, let Vij the density value of pixel at ith and jth column. Then, numerical features values are given, as follows:
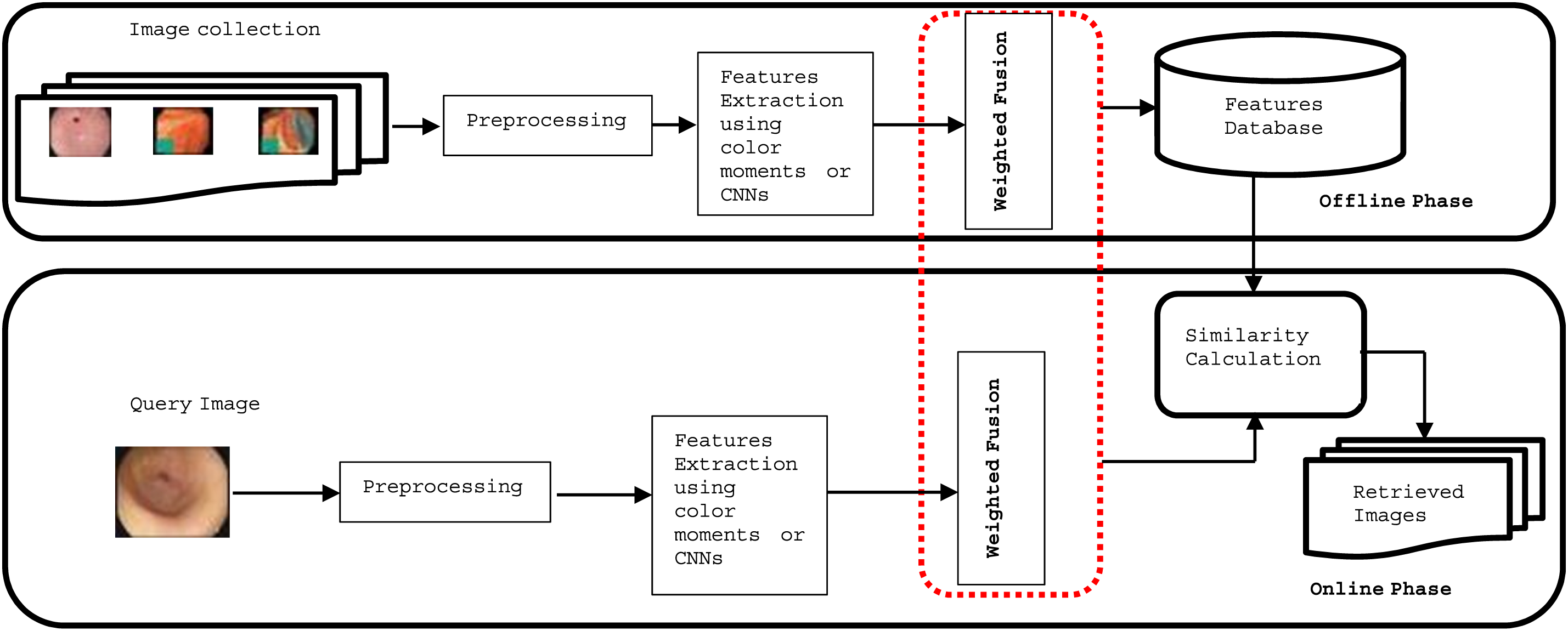
Figure 1: Main framework of proposed CBMIR
Similarly, for the texture features the Gray-level co-occurrence matrix (GLCM) method proposed earlier by [36] is used. Let P(i, j) = (P(i, j))/R is (i, j)th entry in normalized matrix for unique gray levels of the quantized medical image (where R is maximum gray value). Also, let Ng be the number of distinct gray levels of the quantized image, with μ and σ as the mean and standard deviations, respectively. Then texture features are extracted using the following equations:
All color medical images in our dataset are converted from RGB to HSV space model [37], then each of the color moment functions or texture functions used to extract a single value from each channel of H, S and V result in eighteen color features and twelve texture features.
3.3 Weighted Fusion Retrieval Method
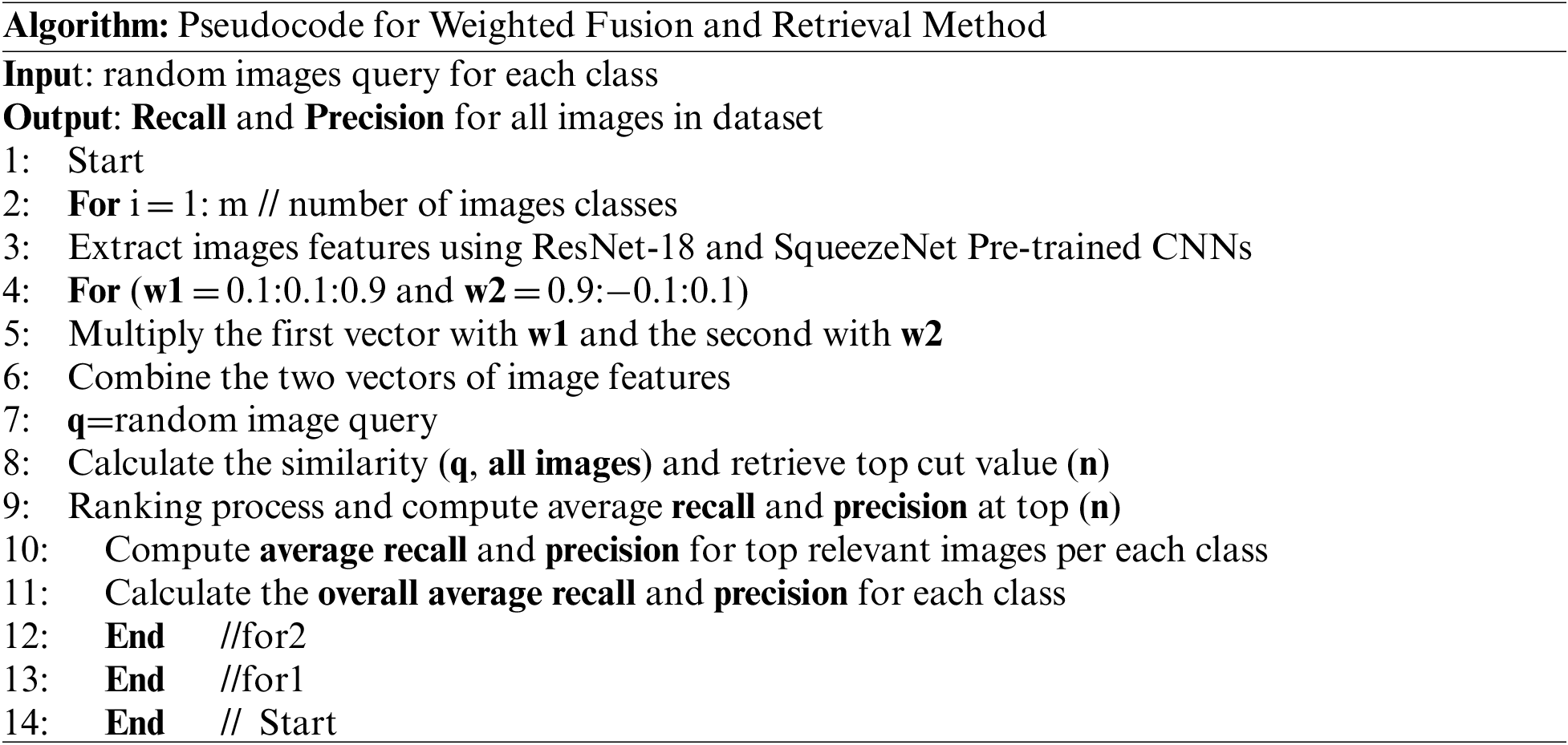
The feature fusion method for combining color and texture features in the previous section is known as the normal or unweighted fusion method. This type of fusion takes into account the equal importance and effect of both types of features vectors. In this section, we implemented weighted fusion methods for the two features vectors extracted, using two of the most well-known and effective pre-trained convolution neural networks (CNNs) models. Here, the medical images features extracted based on ResNet-18 [38] and SqueezeNet [39], pre-trained convolution neural networks (CNNs) models were fused using different fusion ratios ranging from 0.1 to 0.9 alternately for each of the two types of features vectors, as shown in the following algorithm. The similarity using an (RDBSM) similarity measure applies after each time of the fusion process and each retrieval result and performance were calculated, as in the proposed weighted fusion algorithm shown below.
This study uses two medical image datasets, Kvasir [40] and PH2 [41]. The first version of Kvasir, which has 4,000 images, was used in the first scenario of the color and texture features-based retrieval method, while the recent version of 8,000 images was used for the pre-trained convolution neural networks (CNNs) features-based retrieval method. Both versions of these medical images are divided into eight classes, showing anatomical landmarks and pathological findings or endoscopic procedures in the GI tract. The second dataset used in this study is PH2, which is used for melanoma detection and diagnosis. The 200 RGB dermoscopic images in this dataset, which were created through a collaborative study between a group of Portuguese universities and hospitals, are classified into three classes: 80 common nevi, 80 atypical nevi, and 40 melanomas. Figs. 2a and 2b show samples of images for these datasets. As required by pre-trained convolution neural networks (CNNs) models, a resizing process was performed. For ResNet-18, model images were resized to 224 × 224 × 3, while SqueezeNet restricted the input layer to 227 × 227 × 3 size.
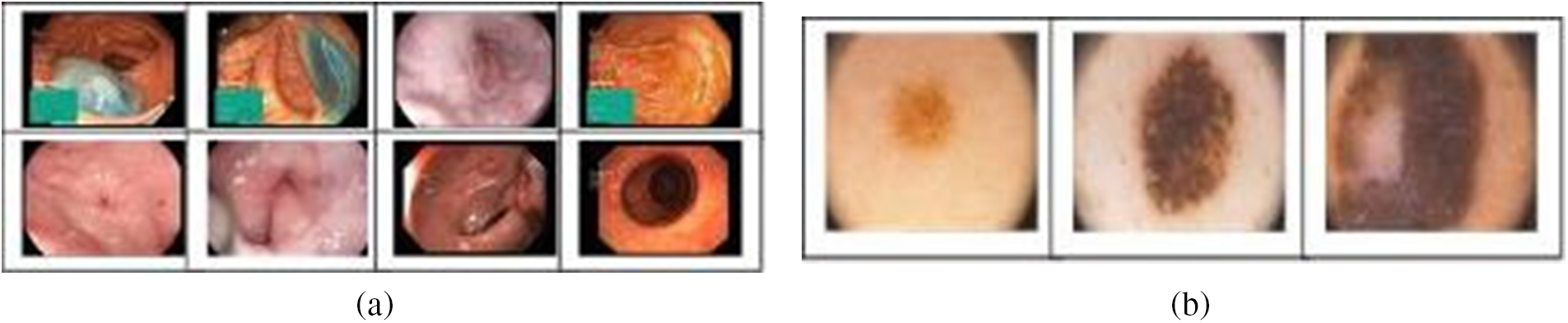
Figure 2: Image sample for (a) Kvasir dataset; (b) PH2 dataset
This study uses two common performance evaluation measures, recall and precision, which have been used and are still being used to evaluate many documents or content-based image retrieval methods. The general formula of precision and recall is given by the following equations:
The precision values shown in our results tables were computed in the top ten and top twenty retrieved images. However, recalls and precisions at different cut-off values are used for plotting some performance graphs.
3.6 Based Methods for Comparison
The result of the first experiment that uses our proposed similarity measure for unweighted features fusion color and texture descriptors is compared with six common standard similarity and distance measures used in the information retrieval area [42]. These measures are Jaccard, Cosine, Canberra, Chebychev, Euclidean and Pearson Correlation. For the second scenario which bases on weighted fusion method described in Section 3.3, our finding and result is compared with six based methods: CBGIR-GPD [43], MIRS [44], OCAM [45], SIFT-mLBP [46], VLAD [47] and RFRM [48]. The first study uses a modified version of ResNet-18 for generating binary hash codes for Kvasir, the second study is based on wavelet optimization and adaptive block truncation coding, and the third study uses opponent class adaptive margin loss method. The fifth study implements relevance feedback Bayesian network after SIFT-modified LBP descriptor for multi-modal medical images and the last study applies relevance feedback retrieval method based on voting process.
4 Experimental Results and Discussion
In this study, two experiments were conducted to test and evaluate the proposed similarity measures. In the first experiment, the color and texture features extracted by traditional statistical methods were used. As in most retrieval models, this experiment takes place in two stages, calculating the similarity values between the query image and all images in the database, followed by a ranking process and computing the precision of the retrieval method. Five images were selected randomly from each class to represent the queries. After the ranking stage, an average recall and precision of five images for each class was calculated, and finally an average recall and precision were calculated for all eight classes for the Kvasir dataset (and also for all three classes for the PH2 dataset). Average recall and precision at the top 10 and top 20 retrieved images were calculated and the results for two datasets are presented in Tables 2 and 3 for both image databases, respectively. From the first observation of Table 2 of the Kvasir dataset it is clear that the retrieval precision of (RDBSM) is superior to all the other six retrieval measures. For both average precision and average precision of eight classes, (RDBSM) has the best retrieval value. A similar conclusion can be drawn from Table 3 for the PH2 dataset, with best average precision.
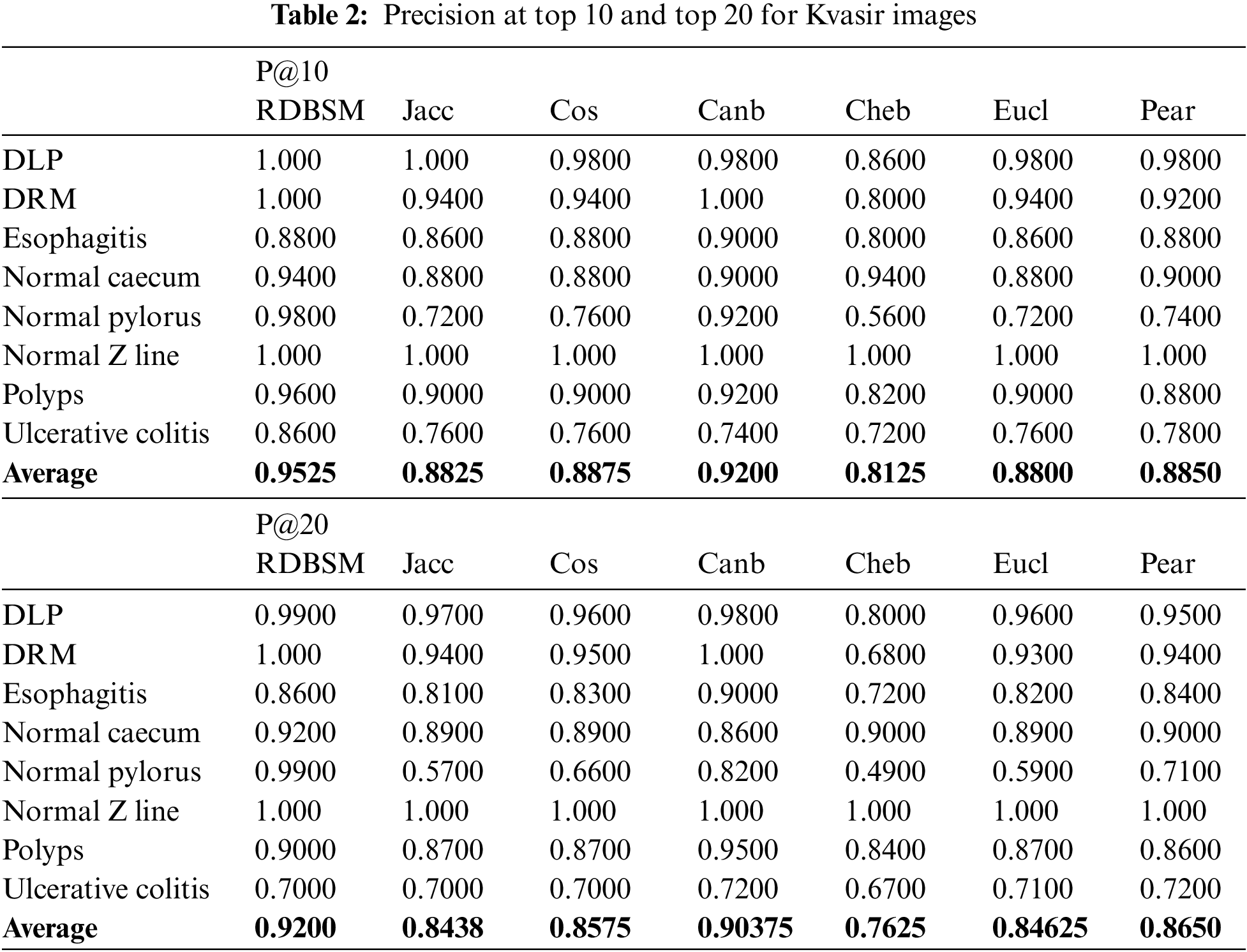
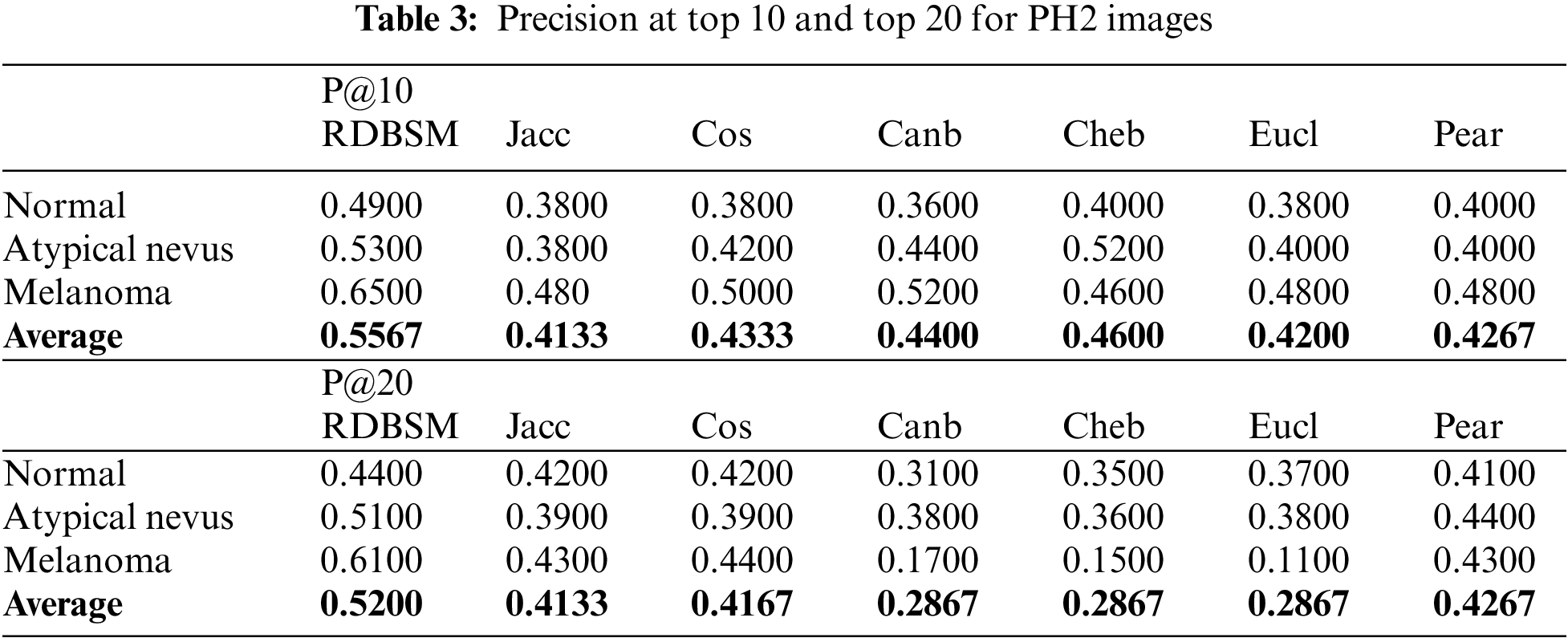
Moreover, Figs. 3a and 3b display the average values of recalls and precisions at various cut-off values. This gives more space to compare between all the seven similarity measures; obviously, the upper plotted lines indicate a good retrieval performance. Additionally, and because Tables 2 and 3 show the precision values for the top 10 and top 20 only, average precision values at different top retrieved images (from 100 to 5 images) for both datasets are shown in Figs. 4a and 4b. To prove the effectiveness of these proposed similarity measures, a Kendall W concordance test [49] and the statistical certainty of the mean, lower and upper bounds of confidence intervals are used. The Kendall W test is widely used for the ranking and comparison of retrieval models and coefficients. In this test, the medical images classes and their average precisions represent judges, while the different retrieval methods are considered objects. The input for this test are the average precision values, and the outputs of the test are the Kendall coefficient (W) and the associate level of ranking or significance. The results of this test for two datasets are shown in Table 4. The significance and ranking of the seven similarity measures show that our proposed RDBSM retrieval measure comes at the top of ranking at a best confidence of 45.4% and 89% for both datasets, respectively. Finally, additional statistical certainty in terms of the mean, lower and upper bounds of the confidence intervals of the seven similarity measures is used for more performance comparative purposes.
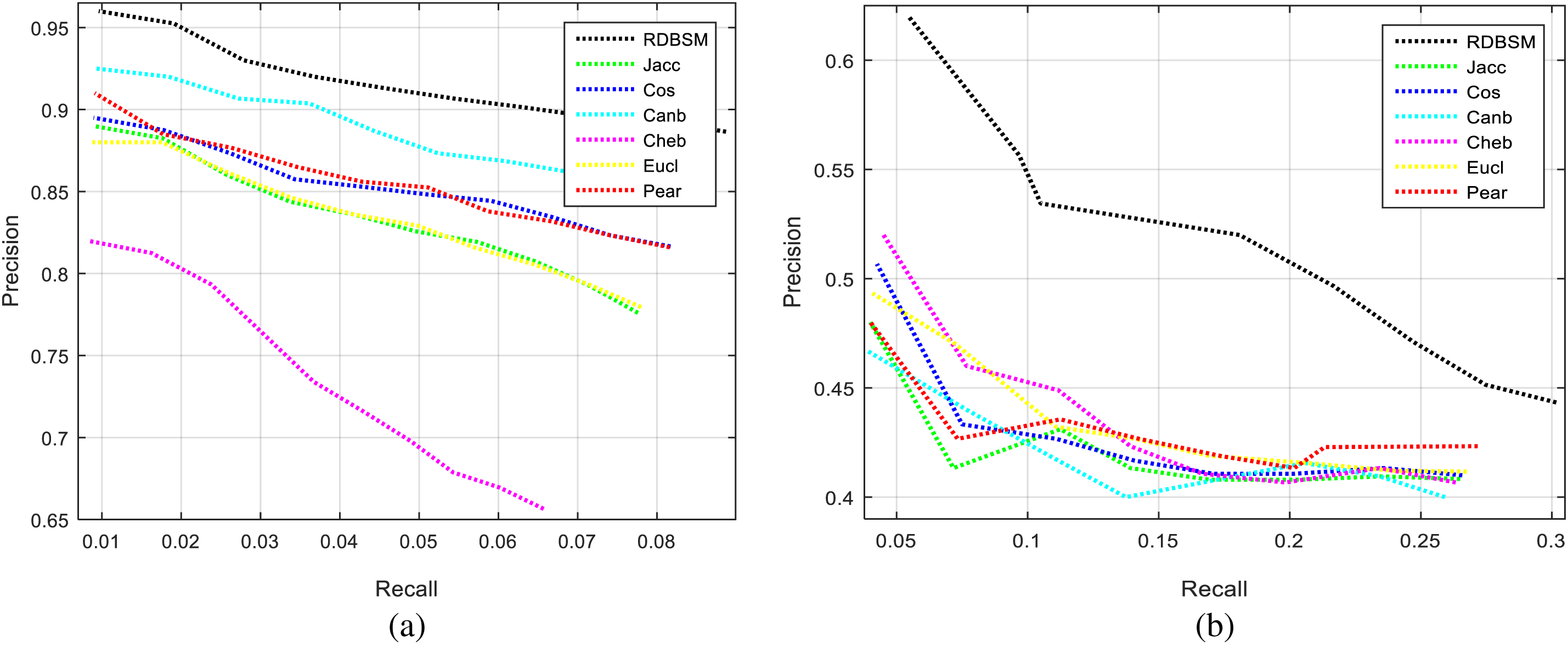
Figure 3: Average recall and precision for (a) Kvasir images; (b) PH2 images
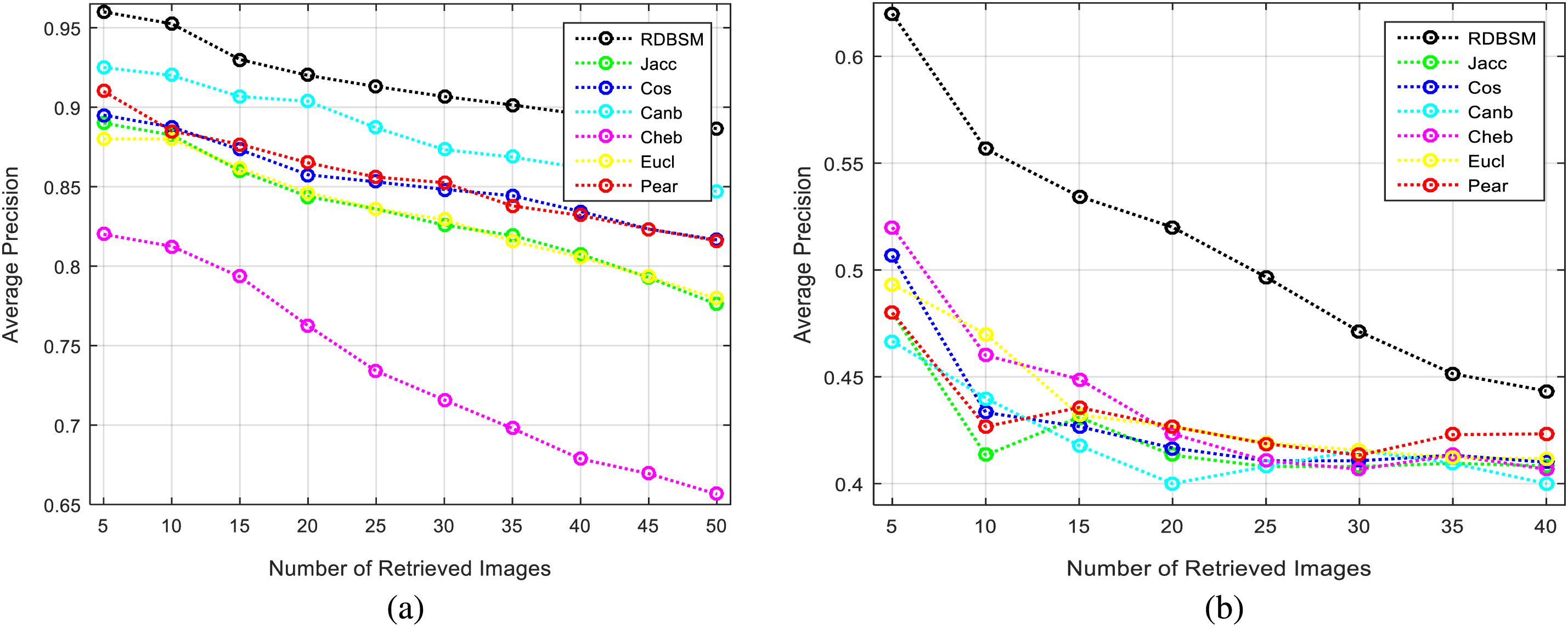
Figure 4: Average precision at different top images for (a) Kvasir images; (b) PH2 images
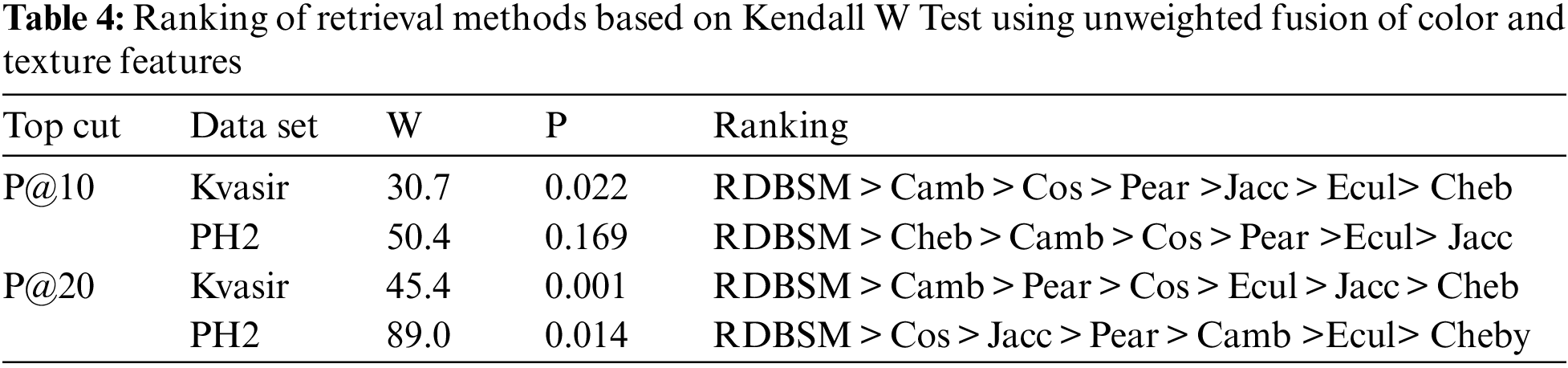
The results for this measure are shown in Figs. 5a and 5b for the two medical images datasets. Observation of the two figures, along with previous rankings of the Kendall W test, give good confidence of effectiveness and good performance of this similarity measure, and it could perform well for other types of object similarity. A second experiment was conducted to implement and use the (RDBSM), to investigate and develop an adaptive fusion method of two features vectors generated from well-known pre-trained convolution neural networks (CNNs) models. In contrast, by using the normal and unweighted fusion method as in the previous experiment, here, different values of weight ratios are used with two features vectors. After each fusion process, the average precision was calculated and the results of all the possibilities are shown in Table 5. Comparison of our results and finding with different retrieval approaches is shown in Table 6. And finally samples of top retrieved images for some classes of two datasets (for both scenarios) are shown in Figs. 6 and 7.
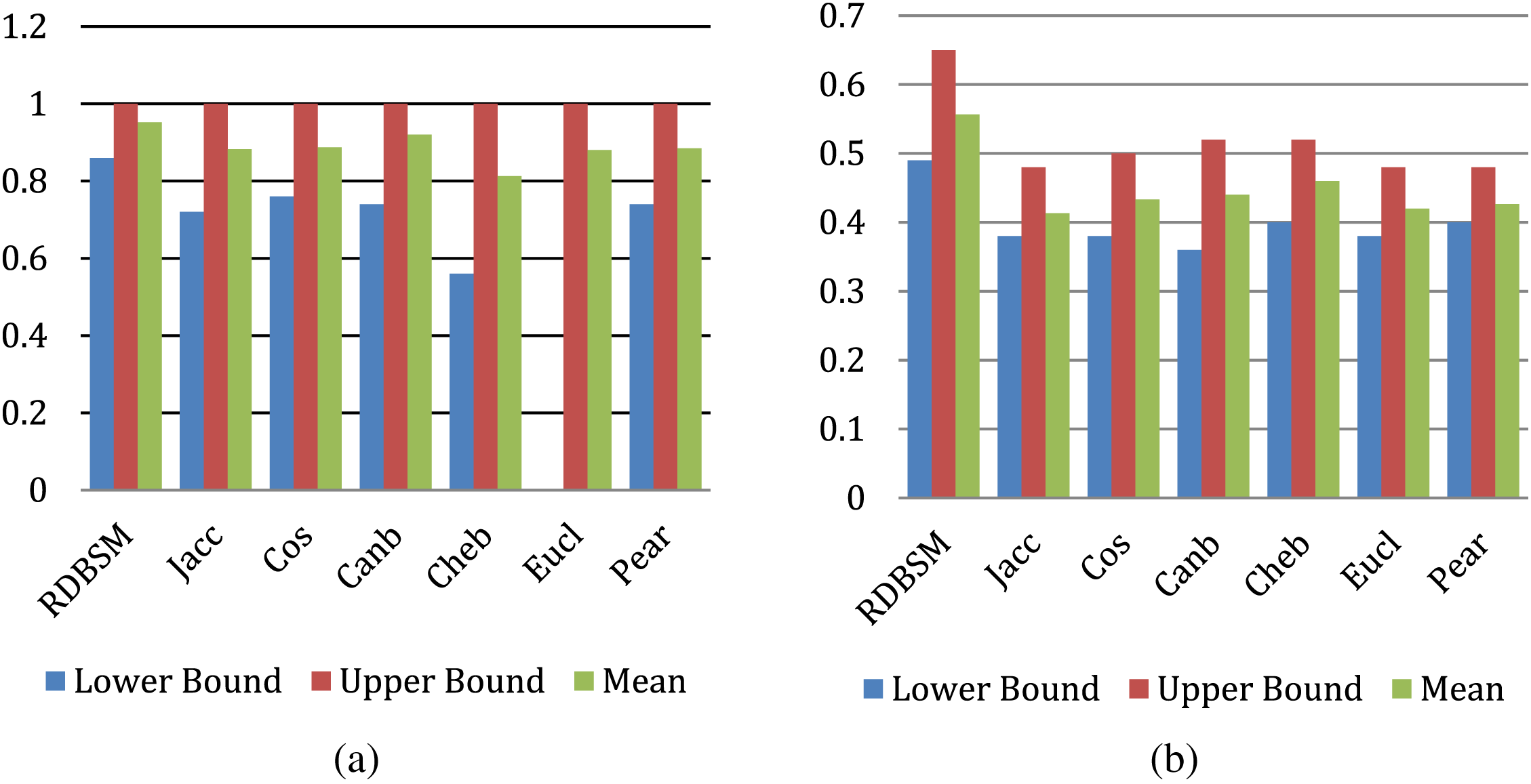
Figure 5: Precisions performance bounds for (a) Kvasir images; (b) PH2 images
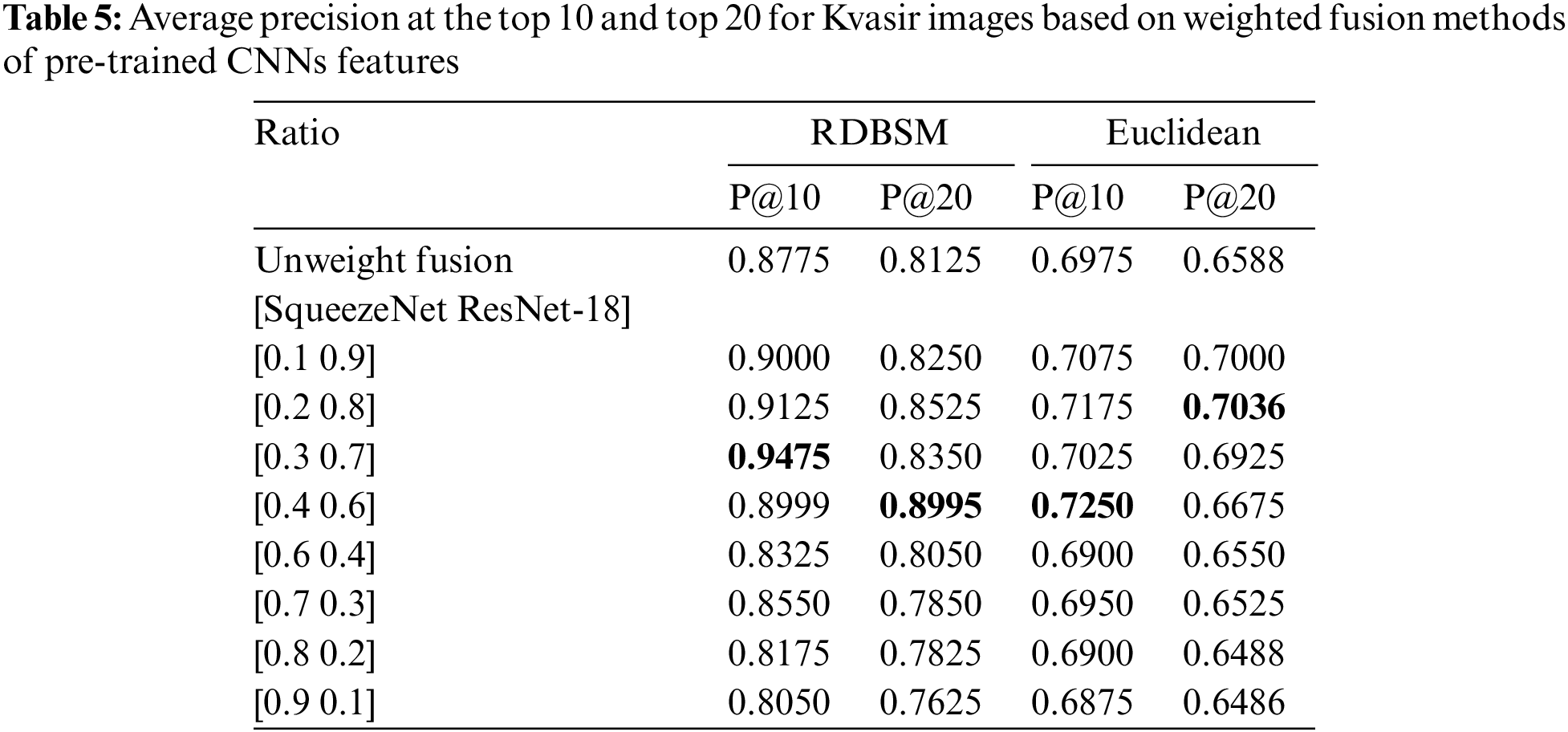
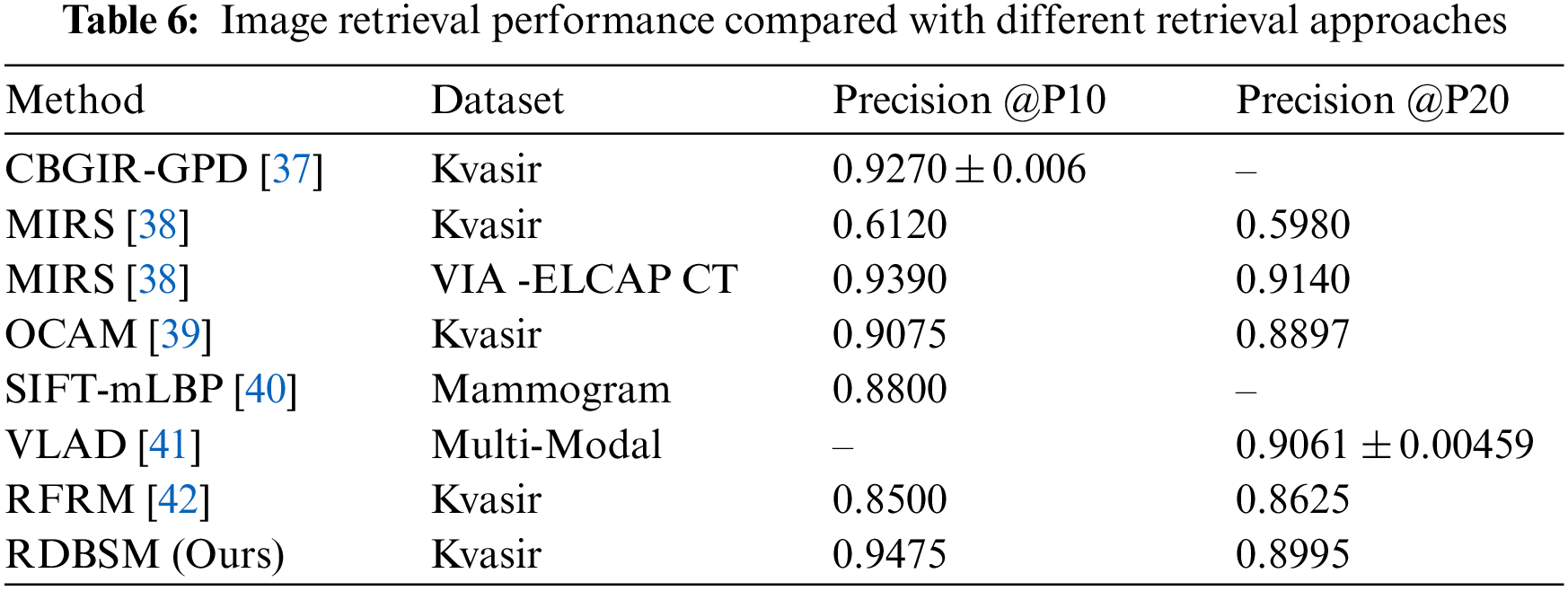
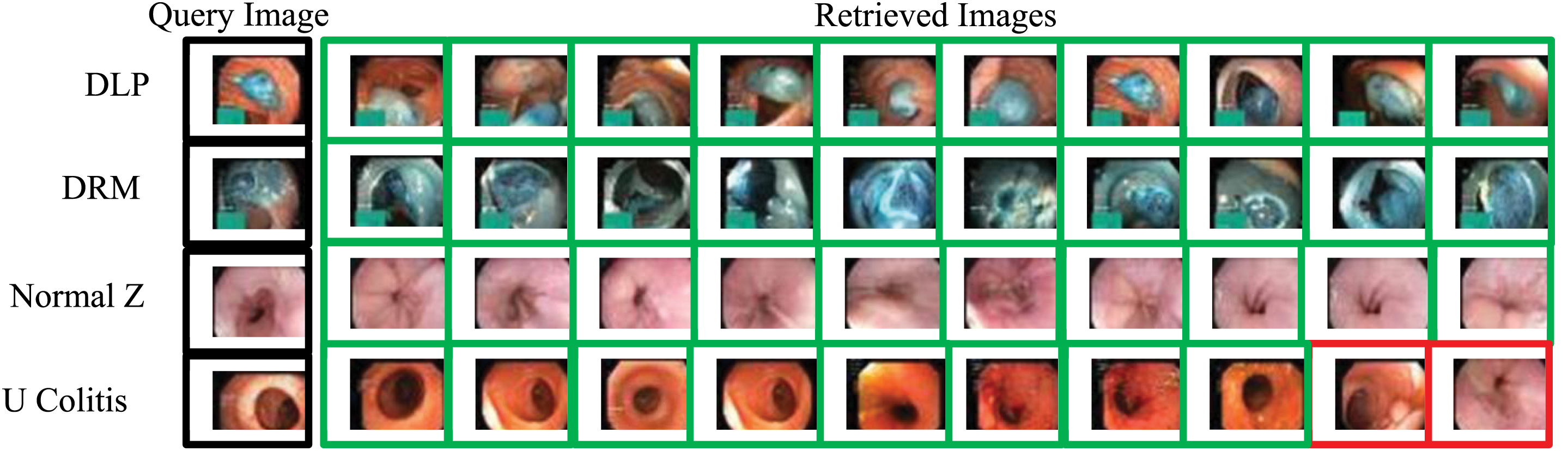
Figure 6: Samples image retrieved for some classes of Kvasir dataset using RDBSM and weighted fusion methods of pre-trained CNNs features (Red frames show false retrieved images)
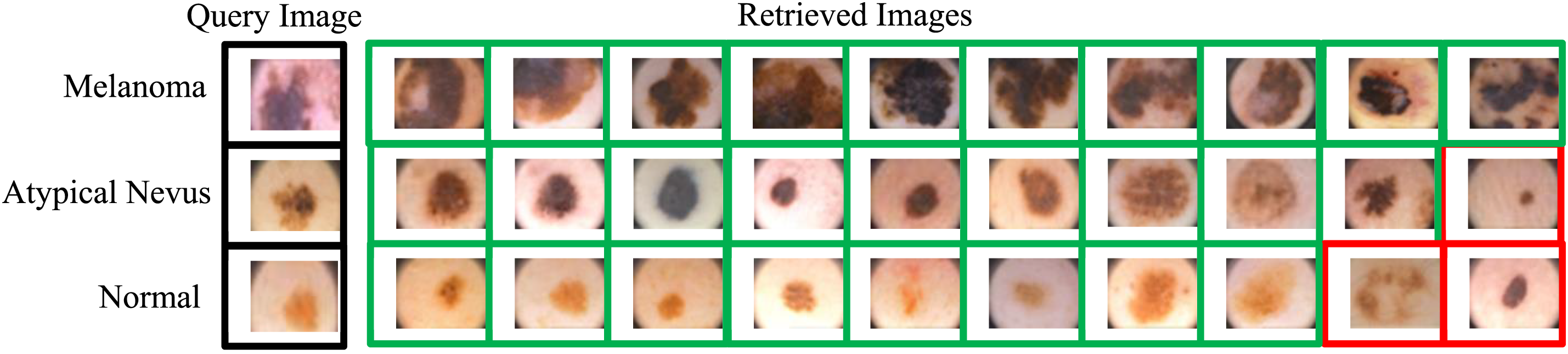
Figure 7: Samples image retrieved for all classes of PH2 dataset using RDBSM and unweighted fusion methods of color and texture features (Red frames show false retrieved images)
In this study, a new similarity measure based on the relative difference and relationship of match features count was proposed for content-based medical image retrieval (CBMIR). The proposed similarity measure was examined first by using similarity in a content-based medical image retrieval (CBMIR) system based on unweighted fusion of color and texture features. The proposed similarity measure has good retrieval performance compared with well-known standard similarity, distance and correlation coefficients. The second part of the contribution of this study is developing a weighted fusion method based on highly informative and accurate features extracted using SqueezeNet and ResNet-18 pre-trained convolution neural networks (CNNs) models. After implementing our proposed similarity measures for the two retrieval method scenario, it is clear that the average retrieval precision was improved to 95.25% and 55.67% at the top 10 images for Kvasir and PH2 images, respectively, using unweighted fusion for color and texture features. Also, for the weighted fusion method, our proposed similarity measure outperformed some based methods by achieving average precision of 94.75%. Additionally, further research could be conducted to develop simpler and high performance similarity or distance measures based on any other mathematical concepts. Finally, the use of multiple similarity measures could be utilized for use as a late fusion method.
Acknowledgement: This research was funded by the Deanship of Scientific Research (DSR) at King Abdulaziz University, Jeddah, Saudi Arabia, Under Grant No. (G: 146-830-1441).
Funding Statement: Authors received the Grant Fund described above for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. I. M. Hameed, S. H. Abdulhussain and B. M. Mahmmod, “Content-based image retrieval: A review of recent trends,” Cogent Engineering, vol. 8, pp. 1–37, 2021. [Google Scholar]
2. C. B. Akgül, D. L. Rubin, S. Napel, C. F. Beaulieu, H. Greenspan et al., “Content-based image retrieval in radiology: Current status and future directions,” Journal of Digital Imaging, vol. 24, pp. 208–222, 2011. [Google Scholar]
3. A. Latif, A. Rasheed, U. Sajid, J. Ahmed, N. Ali et al., “Content-based image retrieval and feature extraction: A comprehensive review,” Mathematical Problems in Engineering, vol. 2019, pp. 1–22, 2019. [Google Scholar]
4. Y. Liu, D. Zhang, G. Lu and W. -Y. Ma, “A survey of content-based image retrieval with high-level semantics,” Pattern Recognition, vol. 40, pp. 262–282, 2007. [Google Scholar]
5. R. C. Veltkamp and M. Tanase, “A survey of content-based image retrieval systems,” in Content-Based Image and Video Retrieval, Multimedia Systems and Applications Series, vol. 21. Boston, MA: Springer, pp. 47–101, 2002. [Google Scholar]
6. H. Müller, N. Michoux, D. Bandon and A. Geissbuhler, “A review of content-based image retrieval systems in medical applications—clinical benefits and future directions,” International Journal of Medical Informatics, vol. 73, pp. 1–23, 2004. [Google Scholar]
7. M. Oussalah, “Content based image retrieval: Review of state of art and future directions,” in The Proc. of Int. Conf. on First Workshops on Image Processing Theory, Tools and Applications, Sousse, Tunisia, pp. 1–10, 2008. [Google Scholar]
8. F. Rajam and S. Valli, “A survey on content based image retrieval,” Life Science Journal, vol. 10, pp. 2475–2487, 2013. [Google Scholar]
9. N. Bhowmik, R. González, V. Gouet-Brunet, H. Pedrini and G. Bloch, “Efficient fusion of multidimensional descriptors for image retrieval,” in The Proc. of Int. Conf. on IEEE Int. Conf. on Image Processing (ICIP), Paris, France, pp. 5766–5770, 2014. [Google Scholar]
10. D. Jiang and J. Kim, “Image retrieval method based on image feature fusion and discrete cosine transform,” Applied Sciences, vol. 11, no. 5701, pp. 1–28, 2021. [Google Scholar]
11. A. H. Osman, H. M. Aljahdali, S. M. Altarrazi and A. Ahmed, “SOM-LWL method for identification of COVID-19 on chest X-rays,” PLoS One, vol. 16, pp. 1–26, 2021. [Google Scholar]
12. A. Ahmed and S. J. Malebary, “Query expansion based on top-ranked images for content-based medical image retrieval,” IEEE Access, vol. 8, pp. 194541–194550, 2020. [Google Scholar]
13. L. T. Alemu and M. Pelillo, “Multi-feature fusion for image retrieval using constrained dominant sets,” Image and Vision Computing, vol. 94, pp. 1–35, 2020. [Google Scholar]
14. J. Li, B. Yang, W. Yang, C. Sun and J. Xu, “Subspace-based multi-view fusion for instance-level image retrieval,” The Visual Computer, vol. 37, pp. 619–633, 2021. [Google Scholar]
15. B. Bustos, S. Kreft and T. Skopal, “Adapting metric indexes for searching in multi-metric spaces,” Multimedia Tools and Applications, vol. 58, pp. 467–496, 2012. [Google Scholar]
16. P. Ciaccia and M. Patella, “Searching in metric spaces with user-defined and approximate distances,” ACM Transactions on Database Systems (TODS), vol. 27, pp. 398–437, 2002. [Google Scholar]
17. A. M. Mahmoud, H. Karamti and M. Hadjouni, “A hybrid late fusion-genetic algorithm approach for enhancing CBIR performance,” Multimedia Tools and Applications, vol. 79, pp. 20281–20298, 2020. [Google Scholar]
18. R. Baeza-Yates and B. Ribeiro-Neto, Modern Information Retrieval, vol. 463. New York: ACM press, 1999. [Google Scholar]
19. C. Carpineto and G. Romano, “A survey of automatic query expansion in information retrieval,” ACM Computing Surveys (CSUR), vol. 44, pp. 1–50, 2012. [Google Scholar]
20. S. Wang, X. Liu, L. Liu, S. Zhou and E. Zhu, “Late fusion multiple kernel clustering with proxy graph refinement,” IEEE Transactions on Neural Networks and Learning Systems, vol. 79, pp. 20281–20298, 2021. [Google Scholar]
21. L. Mansourian, M. T. Abdullah, L. N. Abdullah, A. Azman and M. R. Mustaffa, “An effective fusion model for image retrieval,” Multimedia Tools and Applications, vol. 77, pp. 16131–16154, 2018. [Google Scholar]
22. H. Wu and L. He, “Combining visual and textual features for medical image modality classification with ℓp− norm multiple Kernel learning,” Neurocomputing, vol. 147, pp. 387–394, 2015. [Google Scholar]
23. C. -H. Wei, Y. Li, W. -Y. Chau and C. -T. Li, “Trademark image retrieval using synthetic features for describing global shape and interior structure,” Pattern Recognition, vol. 42, pp. 386–394, 2009. [Google Scholar]
24. J. Yu, Z. Qin, T. Wan and X. Zhang, “Feature integration analysis of bag-of-features model for image retrieval,” Neurocomputing, vol. 120, pp. 355–364, 2013. [Google Scholar]
25. S. Unar, X. Wang and C. Zhang, “Visual and textual information fusion using Kernel method for content based image retrieval,” Information Fusion, vol. 44, pp. 176–187, 2018. [Google Scholar]
26. A. Ahmed and S. Mohamed, “Implementation of early and late fusion methods for content-based image retrieval,” International Journal of Advanced and Applied Sciences, vol. 8, no. 7, pp. 97–105, 2021. [Google Scholar]
27. A. Ahmed, A. O. Almagrabi and A. H. Osman, “Pre-trained convolution neural networks models for content-based medical image retrieval,” International Journal of Advanced and Applied Sciences, vol. 9, no. 12, pp. 11–24, 2022. [Google Scholar]
28. A. Ahmed, “Medical image classification using pre-trained convolutional neural networks and ssupport vector machine,” International Journal of Computer Science & Network Security, vol. 21, pp. 1–6, 2021. [Google Scholar]
29. A. Ahmed, “Pre-trained CNNs models for content based image retrieval,” International Journal of Advanced Computer Science and Applications (IJACSA), vol. 8, no. 7, pp. 200, 2021. [Google Scholar]
30. A. Ahmed, “Classification of gastrointestinal images based on transfer learning and denoising convolutional neural networks,” in The Proc. of Int. Conf. on Data Science and Applications: (ICDSA), Singapore, vol. 1, pp. 631–639, 2022. [Google Scholar]
31. P. Kumar, R. Kumar, G. P. Gupta, R. Tripathi, A. Jolfaei et al., “A Blockchain-orchestrated deep learning approach for secure data transmission in IoT-enabled healthcare system,” Journal of Parallel and Distributed Computing, vol. 172, pp. 69–83, 2023. [Google Scholar]
32. R. Kumar, P. Kumar, M. Aloqaily and A. Aljuhani, “Deep learning-based blockchain for secure zero touch networks,” IEEE Communications Magazine, vol. 61, no. 2, pp. 96–102, 2022. [Google Scholar]
33. P. Kumar, R. Kumar, S. Garg, K. Kaur, Y. Zhang et al., “A secure data dissemination scheme for IoT-based e-health systems using AI and blockchain,” in The IEEE Global Communications Conf. (GLOBECOM), Rio de Janeiro, Brazil, pp. 1397–1403, 2022. [Google Scholar]
34. C. R. Mohan and S. Kiran, “Improved procedure for multi-focus image quality enhancement using image fusion with rules of texture energy measures in the hybrid wavelet domain,” Applied Sciences, vol. 13, no. 4, pp. 2138–2162, 2023. [Google Scholar]
35. P. Maheshwary and N. Srivastava, “Prototype system for retrieval of remote sensing images based on color moment and gray level co-occurrence matrix,” International Journal of Computer Science Issues, IJCSI, vol. 3, pp. 20–23, 2009. [Google Scholar]
36. J. S. Weszka, C. R. Dyer and A. Rosenfeld, “A comparative study of texture measures for terrain classification,” IEEE Transactions on Systems, Man, and Cybernetics, vol. 6, no. 4, pp. 269–285, 1976. [Google Scholar]
37. U. Erkut, F. Bostancıoğlu, M. Erten, A. M. Özbayoğlu and E. Solak, “HSV color histogram based image retrieval with background elimination,” in The Proc. of Int. Conf. on 1st Int. Informatics and Software Engineering Conf. (UBMYK), Ankara, Turkey, pp. 1–5, 2019. [Google Scholar]
38. K. Zhang, W. Zuo, Y. Chen, D. Meng and L. Zhang, “Beyond a Gaussian denoiser: Residual learning of deep cnn for image denoising,” IEEE Transactions on Image Processing, vol. 26, pp. 3142–3155, 2017. [Google Scholar] [PubMed]
39. K. Gopalakrishnan, S. K. Khaitan, A. Choudhary and A. Agrawal, “Deep convolutional neural networks with transfer learning for computer vision-based data-driven pavement distress detection,” Construction and Building Materials, vol. 157, pp. 322–330, 2017. [Google Scholar]
40. K. Pogorelov, K. R. Randel, C. Griwodz, S. L. Eskeland, T. de Lange et al., “Kvasir: A multi-class image dataset for computer aided gastrointestinal disease detection,” in The Proc. of the 8th ACM on Multimedia Systems Conf., Taipei, Taiwan, pp. 164–169, 2017. [Google Scholar]
41. T. Mendoncÿa, P. Ferreira, J. Marques, A. Marcÿal and J. Rozeira, “A dermoscopic image database for research and benchmarking,” in The Presentation in Proc. of PH2 (IEEE EMBC), Osaka, Japan, pp. 5437–5440, 2013. [Google Scholar]
42. A. S. Shirkhorshidi, S. Aghabozorgi and T. Y. Wah, “A comparison study on similarity and dissimilarity measures in clustering continuous data,” PLoS One, vol. 10, no. 12, pp. 1–20, 2015. [Google Scholar]
43. H. Hu, W. Zheng, X. Zhang, X. Zhang, J. Liu et al., “Content based gastric image retrieval using convolutional neural networks,” International Journal of Imaging Systems and Technology, vol. 31, pp. 439–449, 2021. [Google Scholar]
44. H. Kasban and D. Salama, “A robust medical image retrieval system based on wavelet optimization and adaptive block truncation coding,” Multimedia Tools and Applications, vol. 78, pp. 35211–35236, 2019. [Google Scholar]
45. Ş. Öztürk, E. Celik and T. Cukur, “Content-based medical image retrieval with opponent class Adaptive margin loss,” arXiv Preprint arXiv:2211.15371, 2022. [Google Scholar]
46. B. Satish and K. Supreethi, “Content based medical image retrieval using relevance feedback Bayesian network,” in The Proc. of Int. Conf. on Electrical, Electronics, Communication, Computer, and Optimization Techniques (ICEECCOT), Mysuru, India, pp. 424–430, 2017. [Google Scholar]
47. E. Spyromitros-Xioufis, S. Papadopoulos, I. Y. Kompatsiaris, G. Tsoumakas and I. Vlahavas, “A comprehensive study over VLAD and product quantization in large-scale image retrieval,” IEEE Transactions on Multimedia, vol. 16, pp. 1713–1728, 2014. [Google Scholar]
48. A. Ahmed, “Implementing relevance feedback for content based medical image retrieval,” IEEE Access, vol. 8, pp. 79969–79976, 2020. [Google Scholar]
49. S. Sidney, “Nonparametric statistics for the behavioral sciences,” The Journal of Nervous and Mental Disease, vol. 125, pp. 497–498, 1957. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools