 Open Access
Open Access
ARTICLE
Early Detection Glaucoma and Stargardt’s Disease Using Deep Learning Techniques
Department of Electronics and Communication Engineering, Sri Shakthi Institute of Engineering and Technology, Coimabatore, 641062, Tamilnadu, India
* Corresponding Author: Somasundaram Devaraj. Email:
Intelligent Automation & Soft Computing 2023, 36(2), 1283-1299. https://doi.org/10.32604/iasc.2023.033200
Received 10 June 2022; Accepted 02 August 2022; Issue published 05 January 2023
Abstract
Retinal fundus images are used to discover many diseases. Several Machine learning algorithms are designed to identify the Glaucoma disease. But the accuracy and time consumption performance were not improved. To address this problem Max Pool Convolution Neural Kuan Filtered Tobit Regressive Segmentation based Radial Basis Image Classifier (MPCNKFTRS-RBIC) Model is used for detecting the Glaucoma and Stargardt’s disease by early period using higher accuracy and minimal time. In MPCNKFTRS-RBIC Model, the retinal fundus image is considered as an input which is preprocessed in hidden layer 1 using weighted adaptive Kuan filter. Then, preprocessed retinal fundus is given for hidden layer 2 for extracting the features like color, intensity, texture with higher accuracy. After extracting these features, the Tobit Regressive Segmentation process is performed by hidden layer 3 for partitioning preprocessed image within more segments by analyzing the pixel with the extracted features of the fundus image. Then, the segmented image was given to output layer. The radial basis function analyzes the testing image region of a particular class as well as training image region with higher accuracy and minimum time consumption. Simulation is performed with retinal fundus image dataset with various performance metrics namely peak signal-to-noise ratio, accuracy and time, error rate concerning several retina fundus image and image size.Keywords
In early recognition, image is the significant function for identifying of diverse health associated with patient. The eye disorder affects the optic nerve and eventually leads to fractional or complete vision loss. Hence, there is a strong need for early screening of eye disorders to avoid lasting vision misfortune. Several techniques are designed to accurate identification of eye disorders using fundus images.
Random Implication Image Classifier Technique (RIICT) was introduced by [1] it provides a better accuracy result, but the time consumption of disease detection was not reduced. Deep Convolutional Neural Networks (DCNN) classifier were developed by [2] However, the classifier failed to examine the performance of the disease detection with lesser time consumption. An integrated CNN and recurrent neural network (RNN) were developed in [3] but the filtering technique was not applied for improve Peak Signal to Noise Ratio (PSNR). Supervised learning methods were introduced in [4] for automatic detection using fundus images. However, the segmentation process was not performed to improve the detection accuracy. Deep Learning schemes was developed in [5] but the models were not learning the significant features to reduce the time consumption. Machine learning methods combined with metaheuristic approach were proposed by [6] to retinal image. However, the designed approach unsuccessful for providing optimality detection as well as classification by using high-resolution retinal images. A new generative adversarial network was developed in [7] it distinctively differentiates different kinds of fundus diseases. A wavelet-based glaucoma detection algorithm was designed in [8] for real-time screening systems. But the accurate detection was not achieved.
A contour transformation was developed in [9] it failed to reduce the time consumption of glaucoma classification. A new MFCN (multi task fully convolution network) was developed in [10] for removing features to glaucoma identification but model was unsuccessful for achieving image segmentation to improve the accuracy of glaucoma diagnosis further. Dolph-Chebyshev matched filter were designed by [11] on fundus images. Designed method increases the accuracy and minimizes the False Positive Fraction and time was not minimized. Glowworm Swarm Optimization was developed by [12] to automatically detect the retinal fundus images. But algorithm was unsuccessful for performing accurate finding of optic disc for automated glaucoma identification.
The Flexible Analytic Wavelet Transforms (FAWT) based new method was developed in [13] to classify glaucoma. But the method failed to perform the image preprocessing for accurate classification of glaucoma. A wavelet-based glaucoma detection method was introduced in [14] for real-time screening systems. But the method was not efficient for processing a large number of high-resolution retinal images. Recurrent Fully Convolutional Network (RFC-Net) was developed by [15] to optic disk with a minimum cross-entropy loss function. But the designed method failed to determine methods of optic disk tested in a larger database. An automatic two-stage glaucoma screening system was developed in [16] to classify glaucomatous images. But the time complexity analysis was not performed.
An Artificially Intelligent glaucoma expert system was introduced in [17] But the system was not feasible for accurate detection with a minimum error rate. Different classification and segmentation algorithms were developed in [18] However, the designed algorithms failed for precise evaluation of retinal to increase accuracy of glaucoma detection. A deep learning architecture was presented in [19] with higher accuracy. But the image quality enhancement process was not achieved for reducing time consumption. A region growing and locally adaptive Thresholding method was presented in [20] to optic disc. However, it was not efficient for enhancing accuracy and reducing computational time.
To solve existing issues, a novel technique called the MPCNKFTRS-RBIC model is developed and the key contributions summarized are given below:
• To improve Glaucoma and Stargardt’s Disease detection accuracy, an MPCNKFTRS-RBIC model is developed to significantly enhance the classification performance.
• A weighted adaptive Kuan filtering was utilized for extracting unnecessary noise as well as improves image feature. It aids to reduce mean square error as well as improve PSNR.
• To reduce disease detection time, the MPCNKFTRS-RBIC model performs feature extraction and segmentation. Max pooling operations are carried out for minimizing input image as well as remove color, texture, and intensity features. Next, Tobit regression was utilized for analyzing pixels and attains the segmented regions based on the Ruzicka similarity.
• Lastly, accuracy was increased with utilizing radial basis function for analyzing testing and training image region.
• Experimental analyses are carried out with various methods and image databases to determine the performance improvement of the MPCNKFTRS-RBIC model than the standard and other related approaches.
The article is summarized as follows. Section 2 provides methodology with an architecture diagram. Experimental settings are provided in Section 3, which gives details about the implementation. Section 4 illustrates results and discussion that includes a description of evaluative measures and performance analysis. The conclusion is presented in Section 5.
Glaucoma damages optic nerve and consequently blinds the vision. The fundus is used for collecting retinal images, as well as images are used to glaucoma. But early detection of glaucoma becomes a demanding issue. Glaucoma with various machine learning methods is most popular. However, it is not efficient to solve issues for accurate detection using lesser time as well as error rate. MPCNKFTRS-RBIC is utilized to disease identification of glaucoma.
Fig. 1 illustrates a basic architecture of the proposed MPCNKFTRS-RBIC model to identify the disease at earlier stages based on four different steps. The numerous training fundus images
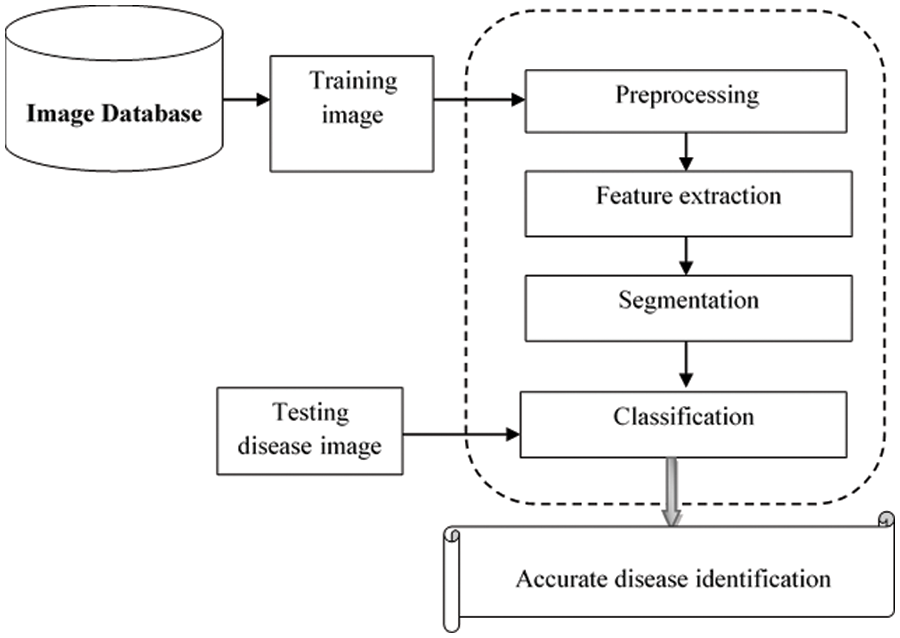
Figure 1: Architecture of the proposed MPCNKFTRS-RBIC model
Fig. 2 describes structure of Deep Convolutional Neural Network framework. Structure comprises several layers and neurons. In Connectedness neural network, the nodes were fully associated with another layer form entire network. The numerous training fundus images
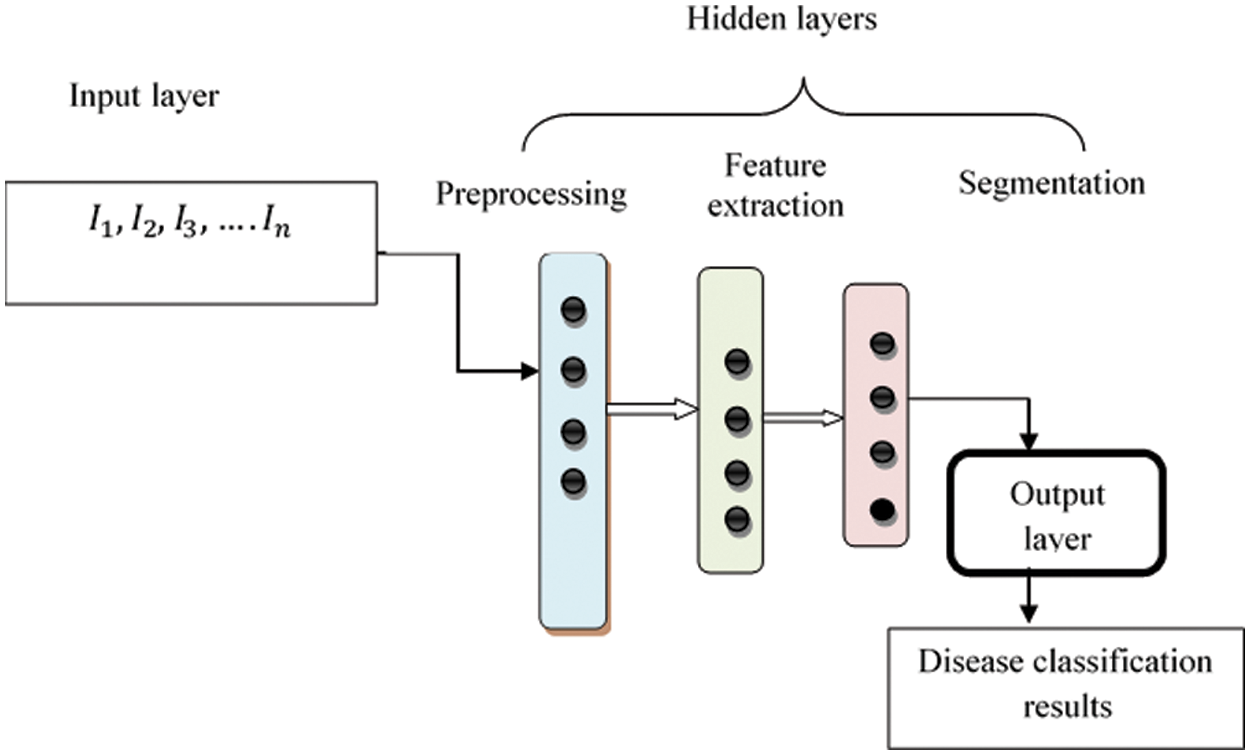
Figure 2: Structure of max pool convolution neural network
where ‘
2.1 Weighted Adaptive Kuan Filter
The proposed MPCNKFTRS-RBIC model starts to perform the image preprocessing. The preprocessing step involves image resize, noise removal and enhances the quality, resulting in improved disease identification accuracy. Consider retinal fundus is ‘
Fig. 3 illustrates the 3 × 3 filtering window, where nine pixels were arranged as well as central coordinates of a pixel in the window are represented as
where F symbolizes the filtering output, m indicates the local mean, w represents weighting function that allocate better weight to pixel is nearer to central pixel ‘
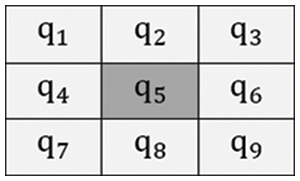
Figure 3: Filtering window
2.2 Max Pooling Operation-Based Feature Extraction
Behind preprocessing of input retinal fundus image, the feature extraction process was carried out within second hidden layer. If convolution layers carry out the feature maps, the dimensionality of the image size needs to reduce for extracting meaningful features. This is addressed by applying the Max Pooling operation. Max Pooling removes highest value of pixels in the given region. Therefore, output of max-pooling layer comprises important features of feature map.
Fig. 4, illustrates the operation of Max pooling operations by 4*4 channels with 2*2 kernels. In 4*4 channel, 2*2 with kernel focuses channel has four-pixel values

Figure 4: Operation of max pooling
where
where,
where β indicates the skewness. After extracting the color feature, texture features are extracted for finding the spatial illustration of color or intensities in each patch of an image. The texture is evaluated depending on the correlation between the pixels.
where M shows the correlation,
where
2.3 Ruzicka Indexed Tobit Regressive Segmentation
Tobit Regressive Segmentation process is performed within hidden layer 3 for partitioning preprocessed image within more segments through analyzing the pixel with the extracted features of the retinal fundus image. Tobit Regression is a statistical method that helps to determine the linear relationships between pixels in the image. Fig. 5 portrays the process of image segmentation using the Ruzicka indexed Tobit Regression algorithm. The regression function takes the input preprocessed image with extracted features. The regression functions analyze the pixels of the input images Ruzicka similarity index.
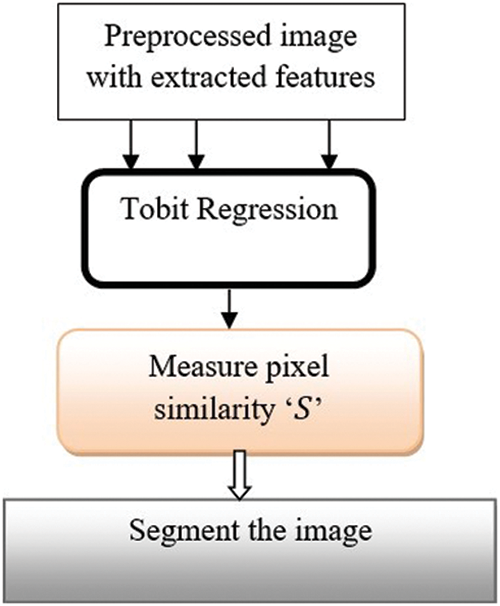
Figure 5: Regressive segmentation
The Ruzicka index was applied for identifying linear relationships among pixels. Ruzicka similarity is used for measuring the relationships between pixels.
where ‘
where ‘
Finally, the classification was performed at output layer. Radial basis function is applied for testing and training image by output layer for detecting disease using better accuracy.
where,
Algorithm 1: MPCNKFTRS-RBIC Model

Algorithm 1 provides step-by-step process of disease using better accuracy. Numerous retinal fundus images were given at input layer. Next, input is sent to first hidden layer, where preprocessing is performed using Kuan Filter to remove the noisy pixels. After that, the Max pooling operations are carried out for reducing input image. Further, Tobit regression is applied to analyze the image pixels and segment the images into different regions on similarity measure. Lastly, classification is done by output layer using radial basis function among testing and training image using better accuracy. Depend on analysis, accurate disease is carried out with minimum time consumption.
Simulation of MPCNKFTRS-RBIC and two methods such as Random Implication Image Classifier Technique (RIICT) [1], Deep convolutional neural networks (DCNN) [2] is implemented using Matrix Laboratory (MATLAB) simulator with the help of two datasets namely acrima database, https://figshare.com/articles/CNNs_for_Automatic_Glaucoma_Assessment_using_Fundus_Images_An_Extensive_Validation/7613135, retina image bank database: https://imagebank.asrs.org/file/28619/stargardt-disease.
In ACRIMA database 705 retinal fundus images are collected for disease detection Here, 396 retinal fundus images are categorized as glaucomatous or abnormal and 309 images are categorized as normal images. In the retina image bank database includes 352 retinal fundus images for identifying Stargardt’s diseases. The experiments are conducted with 200 retinal fundus images from acrima database and retina image bank database for ten iterations.
MPCNKFTRS-RBIC and two methods, such as RIICT [1] DCNN [2] are explained using different performance parameters namely Peak Signal to Noise Ratio, accuracy, Error rate, and time.
Peak Signal to noise ratio: It is calculated depending on the mean square error. It is calculated by difference among preprocessed image and original fundus image. It is expressed by,
where, ‘
Disease detection accuracy
where,
Error rate: It is calculated by proportion of amount of retinal fundus images incorrectly detected by normal or abnormal from the entire amount of input images. It is computed as,
where,
Disease detection time: It was measured by number of times consumed with algorithm for identifying the disease or normal image. The Disease detection time is measured as follows,
where,
Tabs. 1a and 1b shows the simulation results of peak signal to noise ratio using different sizes of images consumed over two databases such as acrima and retina image bank database. The observed results using two datasets indicate that the MPCNKFTRS-RBIC technique outperforms than other technique to achieve better peak signal-to-noise ratio compared with existing classification techniques RIICT [1] DCNN [2]. The ACRIMA database is employed for conducting experiment. Ten outcomes are obtained with different sizes of images. MPCNKFTRS-RBIC were compared with existing methods. MPCNKFTRS-RBIC performs a better peak signal to noise ratio as well as minimal mean square error. The average of ten outcomes indicates that the peak noise ratio of proposed method is considerably enhanced as 11% compared with RIICT [1] and 30% compared with DCNN [2]. Retina image bank database is applied to conduct the experiment with various image sizes, as shown in Tab. 1b. From the outcomes, MPCNKFTRS-RBIC outperforms well with other two existing techniques. Ten outcomes shows peak signal to noise ratio of proposed method is significantly improved as 11% compared with RIICT [1] and 24% compared with DCNN [2].
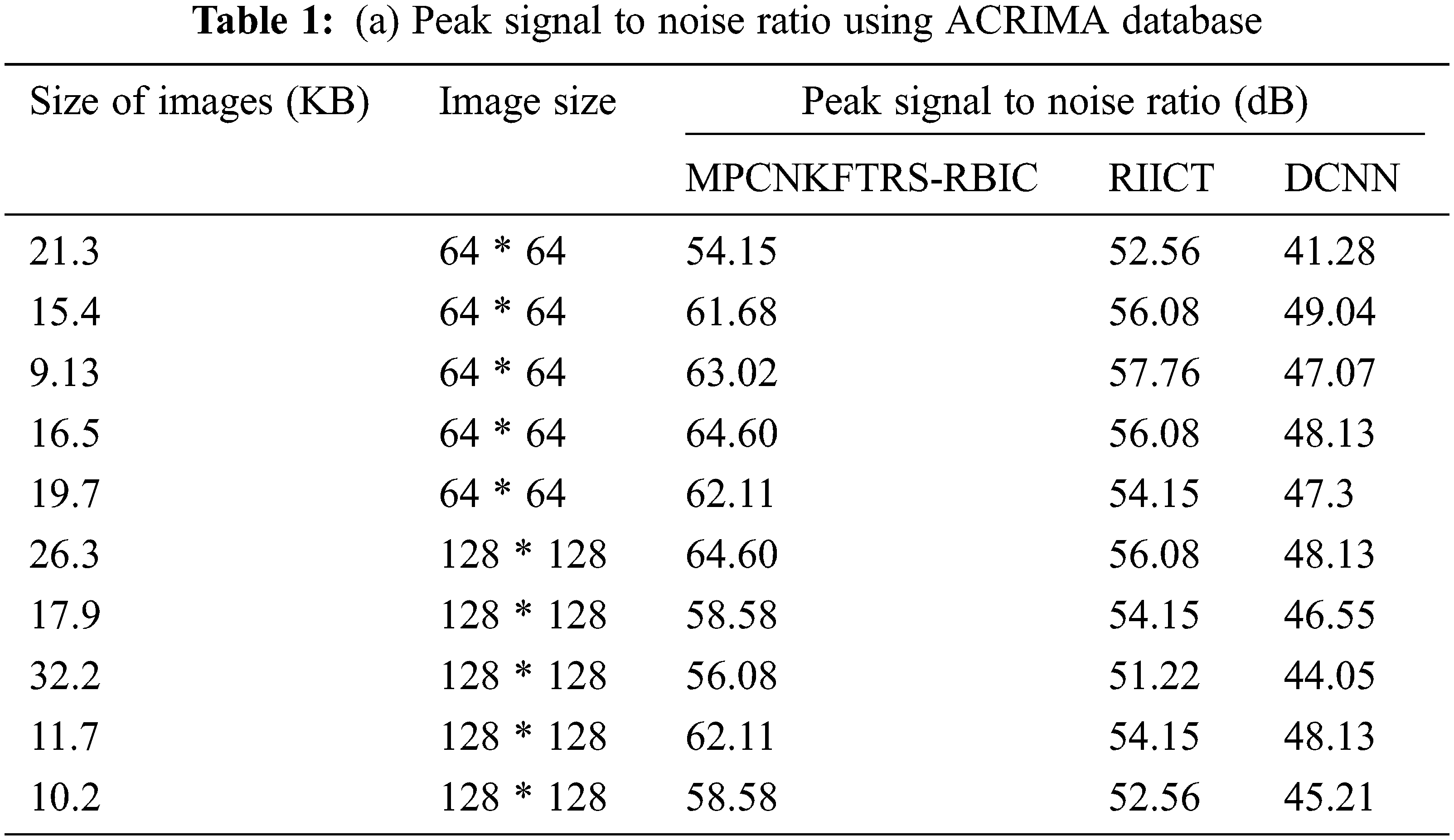
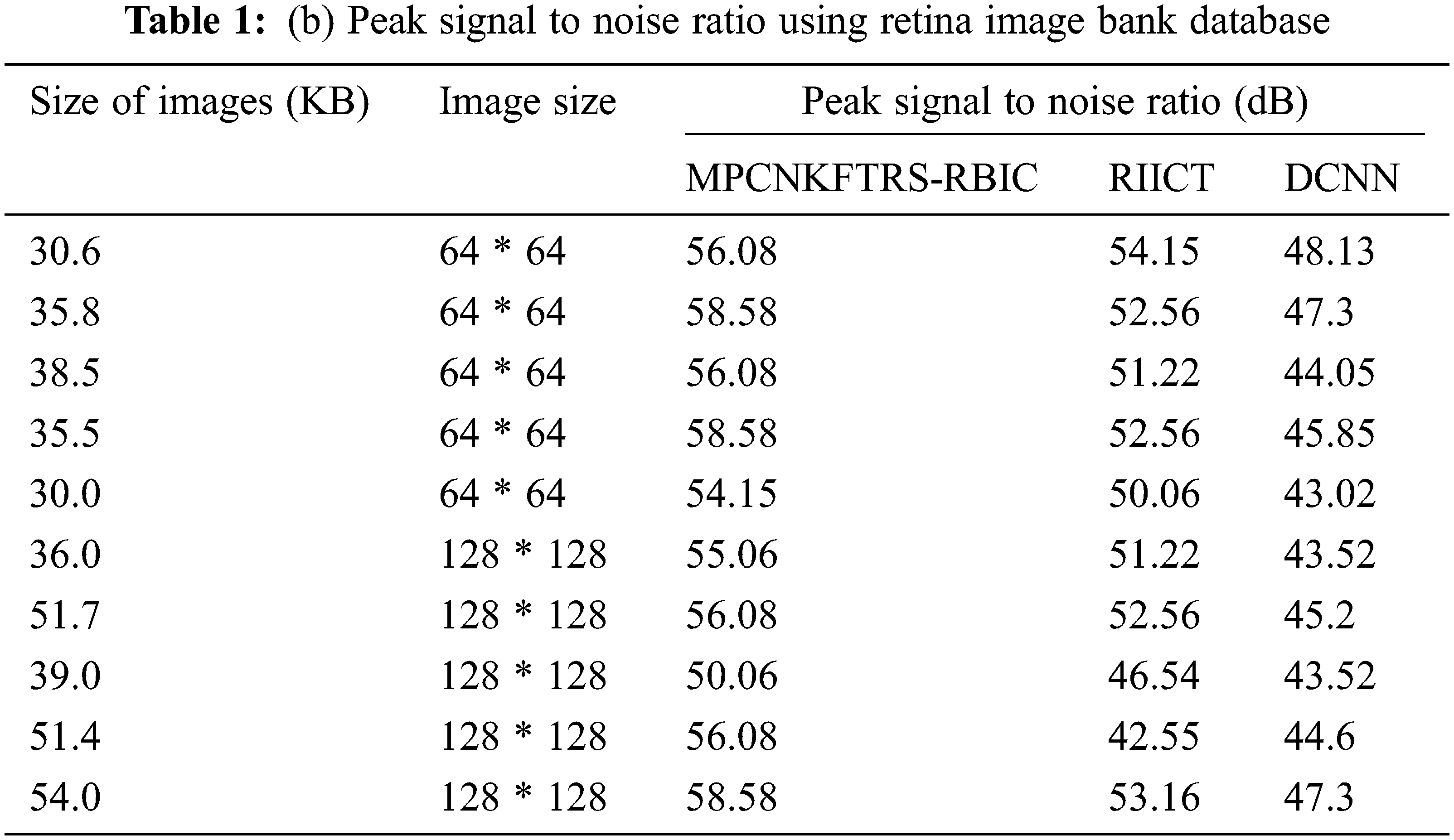
Figs. 6a and 6b reveals the comparatives analysis of peak signal to noise ratio vs. different sizes of images with various classification techniques. Figs. 6a and 6b demonstrates various sizes of retinal fundus image are taken as input for calculating peak signal to noise ratio. MPCNKFTRS-RBIC enhances performance outcomes compared with existing methods. It is performed with utilizing weighted adaptive Kuan filtering for finding noisy pixels and other pixels in the image. Noisy pixels are extracted over input image resulting in improved quality as well as lesser mean square error.
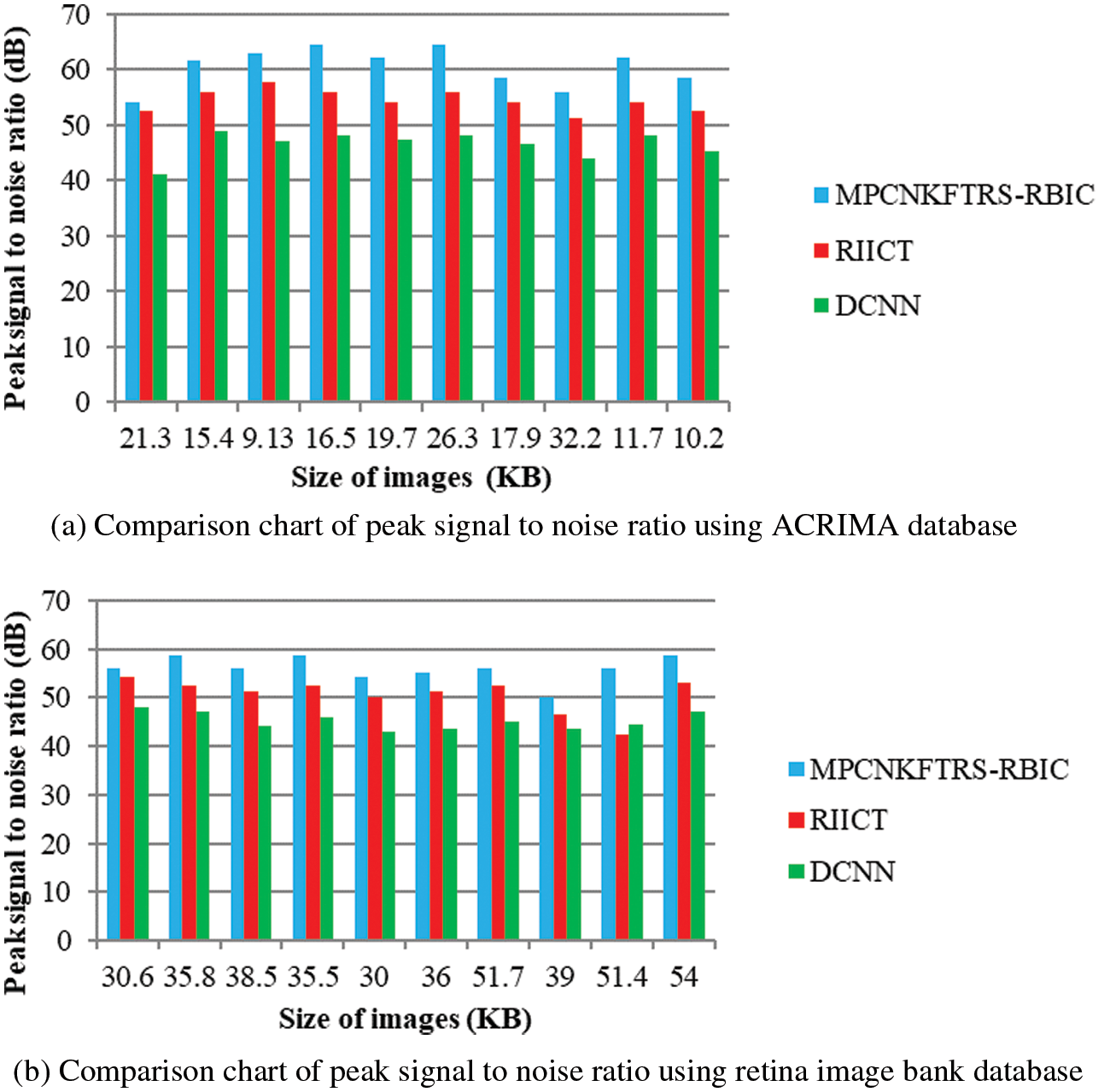
Figure 6: (a) Comparison chart of peak signal to noise ratio using ACRIMA database (b) Comparison chart of peak signal to noise ratio using retina image bank database
Tabs. 2a and 2b shows the comparative analysis of the disease detection accuracy of three classification techniques, namely MPCNKFTRS-RBIC technique, RIICT [1] DCNN [2] using two databases. MPCNKFTRS-RBIC enhances higher accuracy than the conventional methods. Let us consider that 200 retinal fundus images are taken from the two databases. By applying the acrima database, ten various outcomes are obtained. MPCNKFTRS-RBIC is compared with existing methods. MPCNKFTRS-RBIC method increases disease detection accuracy as 6% and 13% compared with RIICT [1] and DCNN [2], respectively. The proposed method is applied to various retinal image databases and accuracy is measured. The obtained outcomes represent disease detection accuracy of MPCNKFTRS-RBIC is enhanced as 6% and 14% than the conventional methods.
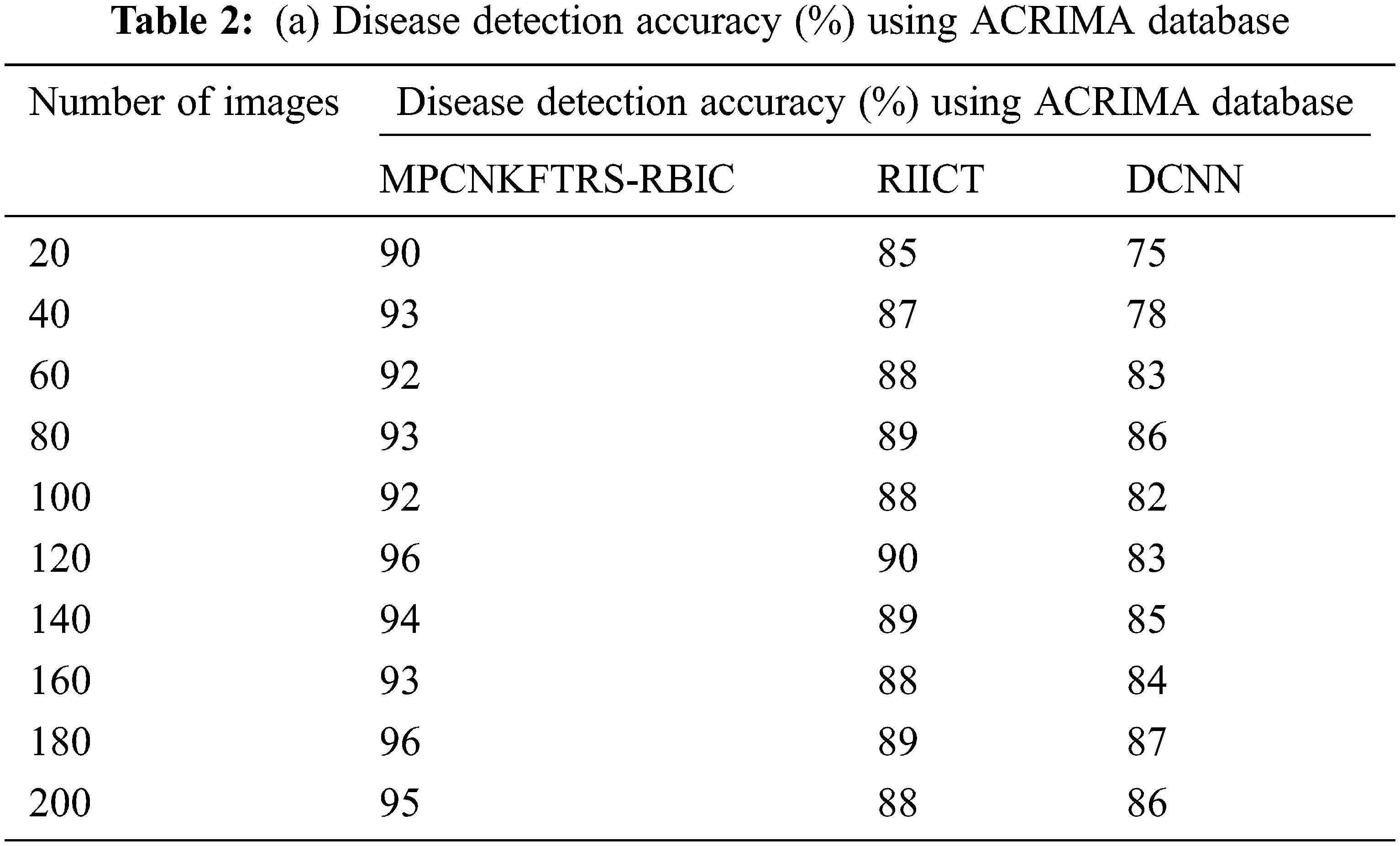
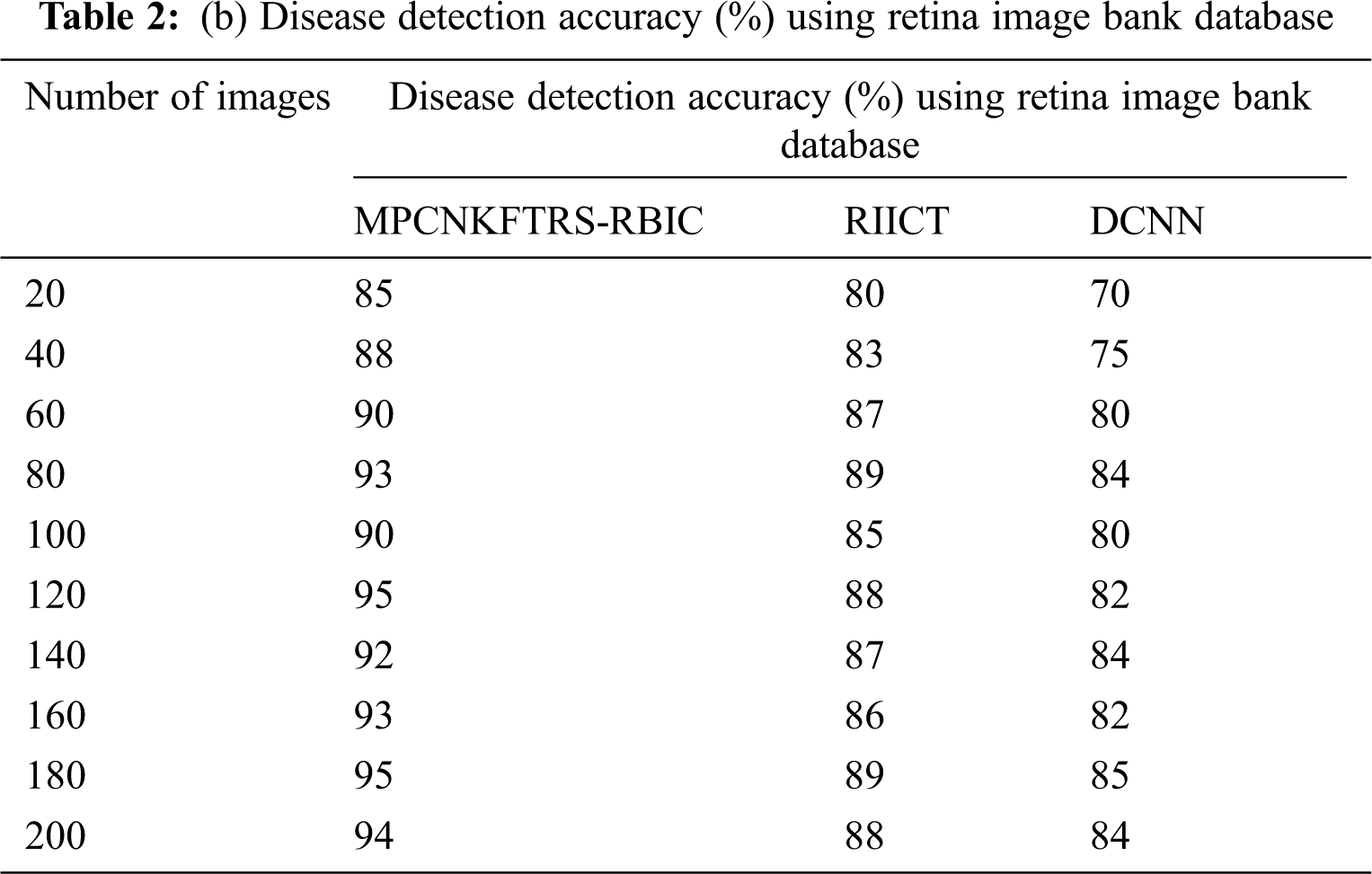
Figs. 7a and 7b depicts the performance comparison of disease detection accuracy using two different databases. The graphical chart shows that the MPCNKFTRS-RBIC technique provides a higher disease detection accuracy. Significant reason for this improvement is to apply the Tobit regression for image Segmentation by analyzing the pixel with the extracted features of the retinal fundus image using the Ruzhicka similarity measure. Then, the segmented image is given with output layer. The radial basis kernel function is used for estimating testing disease image region as well as the training image region and classifies the images as normal or abnormal with higher accuracy.
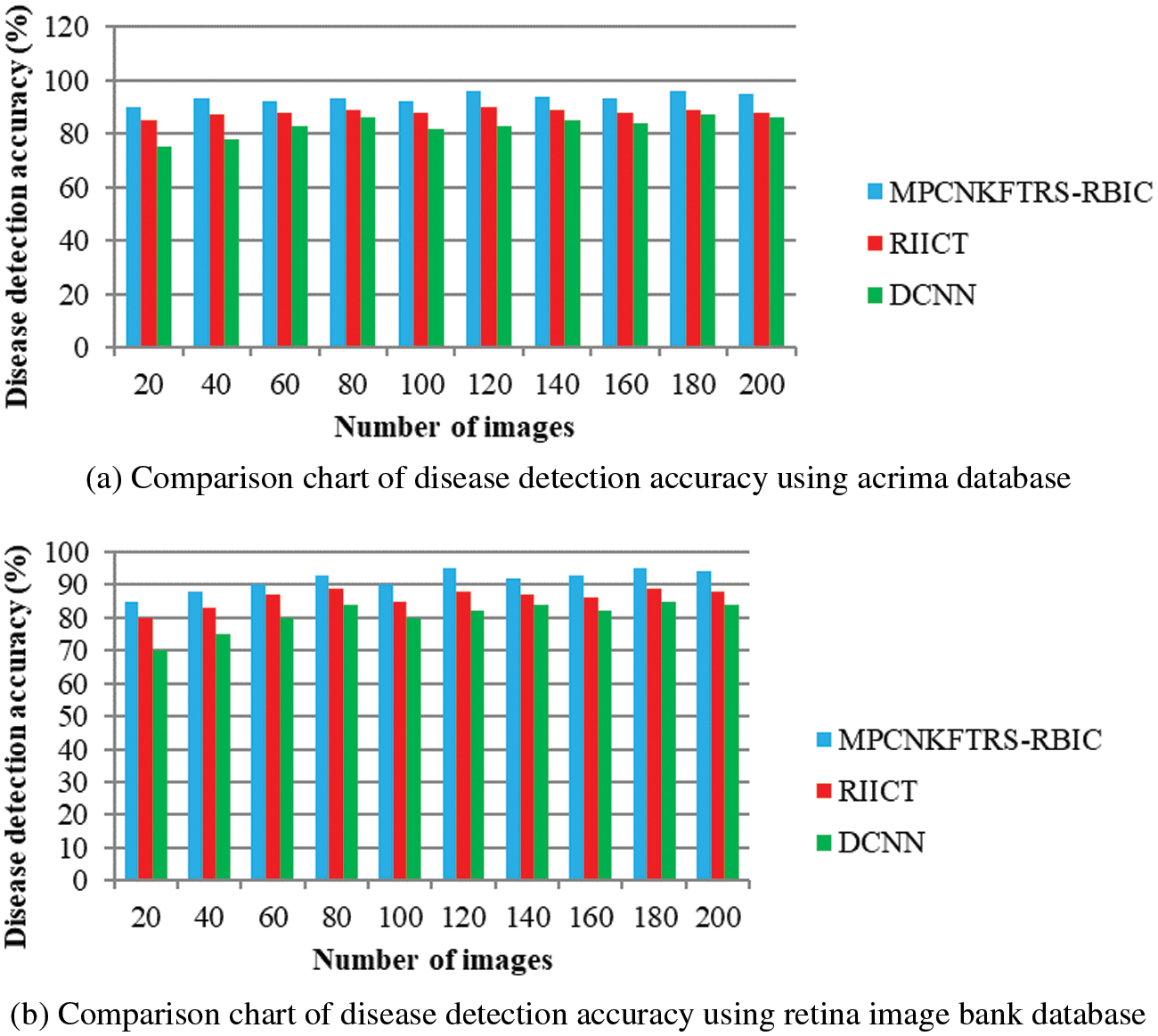
Figure 7: (a) Comparison chart of disease detection accuracy using acrima database (b) Comparison chart of disease detection accuracy using retina image bank database
Tabs. 3a and 3b shows the comparison results of error rate using three various methods proposed, MPCNKFTRS-RBIC and RIICT [1] and DCNN [2]. By comparing proposed MPCNKFTRS-RBIC technique with other methods, it is inferred that the error rate considerably displays better performance. The experiment is conducted with the ACRIMA database with 200 retinal fundus images. Compared with two existing methods, our proposed method reduces error rate as 45% compared with RIICT [1] and 65% compared with DCNN [2]. Similarly, experiment is conducted with the retina image bank database to calculate the error rate. Among the three methods, the error rate of proposed method is reduced as 40% and 57% compared with existing techniques.
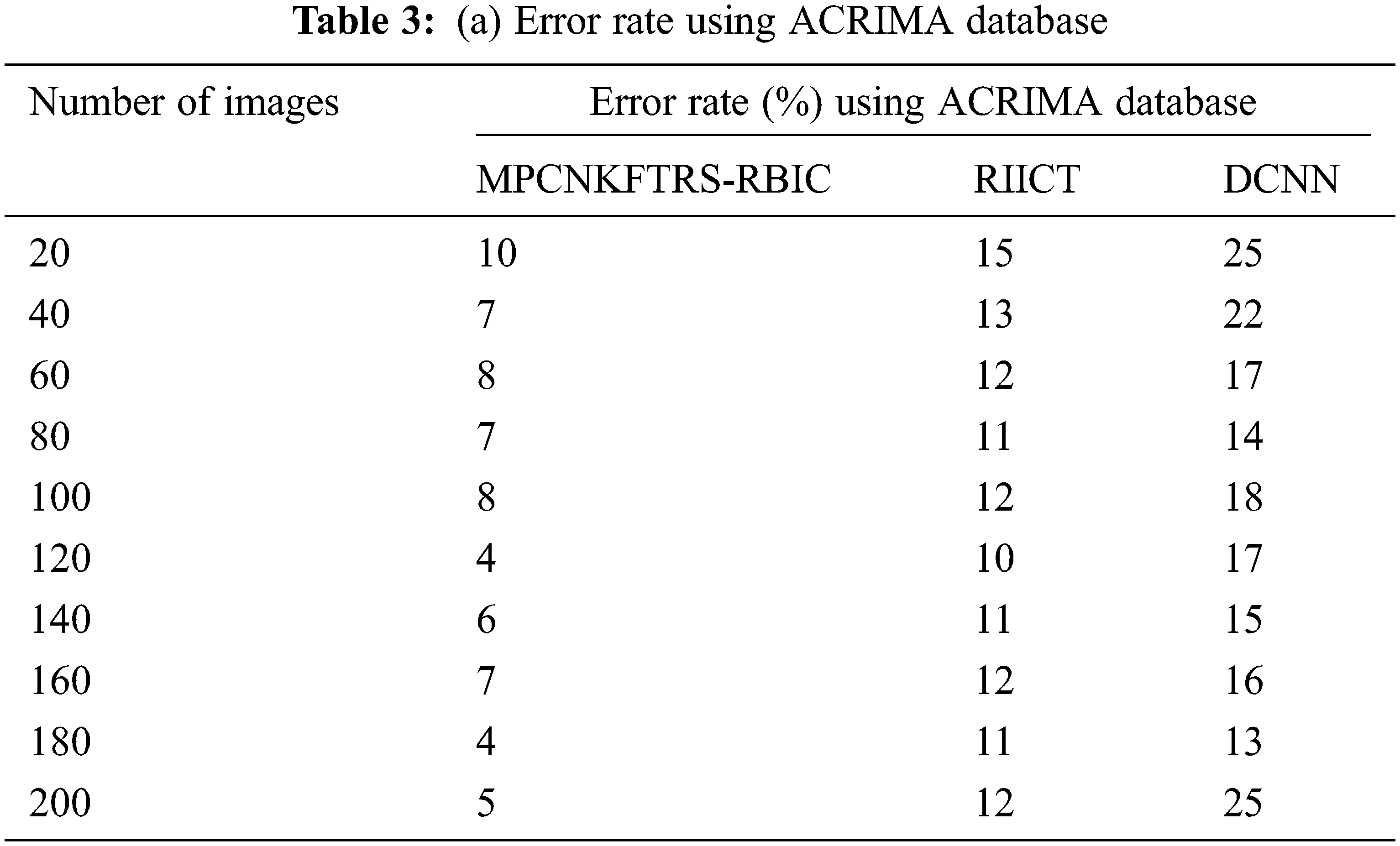
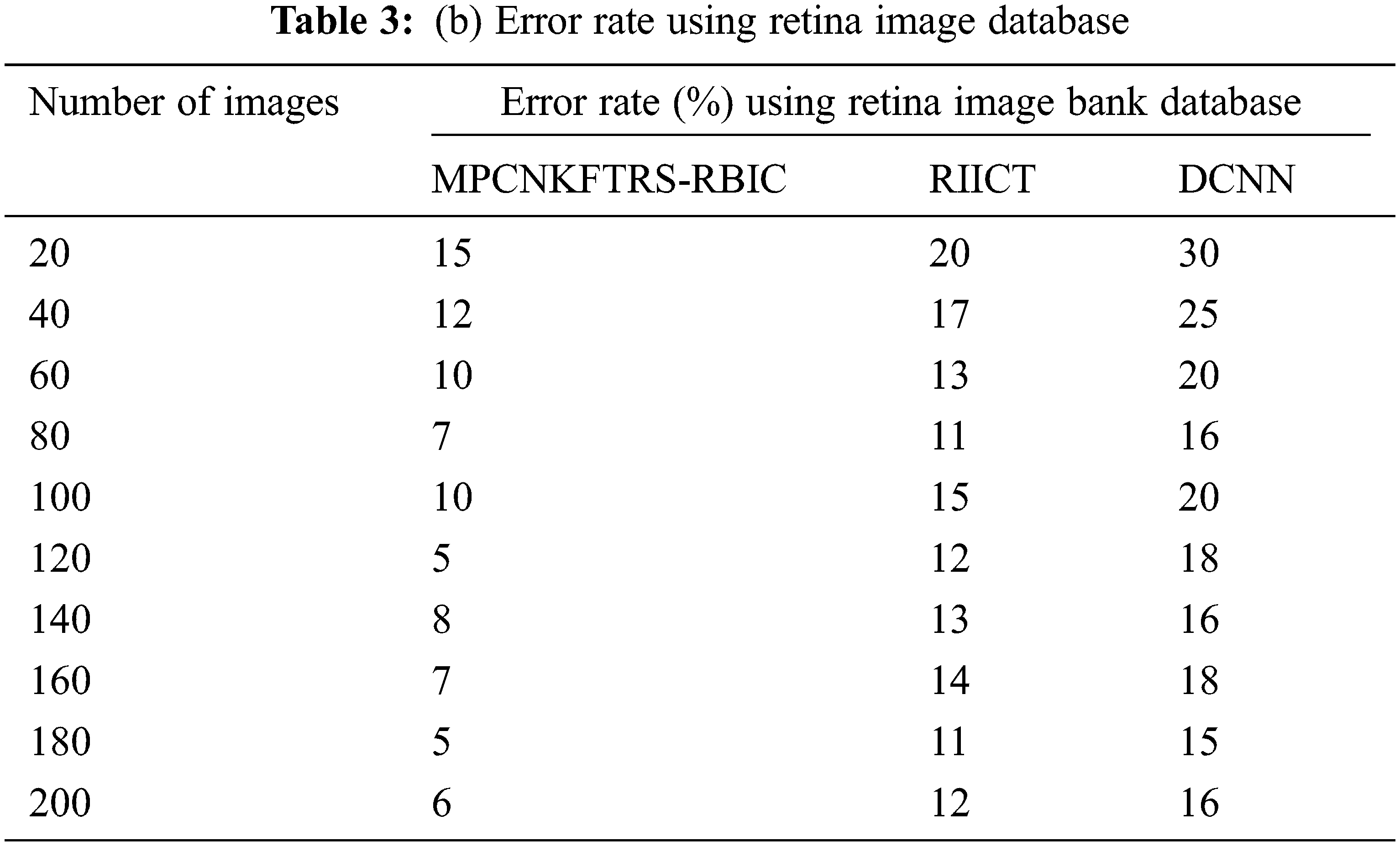
Figs. 8a and 8b portrays the error rate using 200 eye retinal fundus images using two databases. From graph, various error rate outcomes are obtained to three techniques with various input images. MPCNKFTRS-RBIC reduces the error rate with existing techniques. This is because the deep learning technique performs image preprocessing to enhance quality of disease detection. Besides, image segmentation process helps to partition the image within various segments. The infected area is accurately detected. Finally, the radial basis function matches the disease region with the training image, thus increasing the disease detection accuracy and minimizing the error rate.
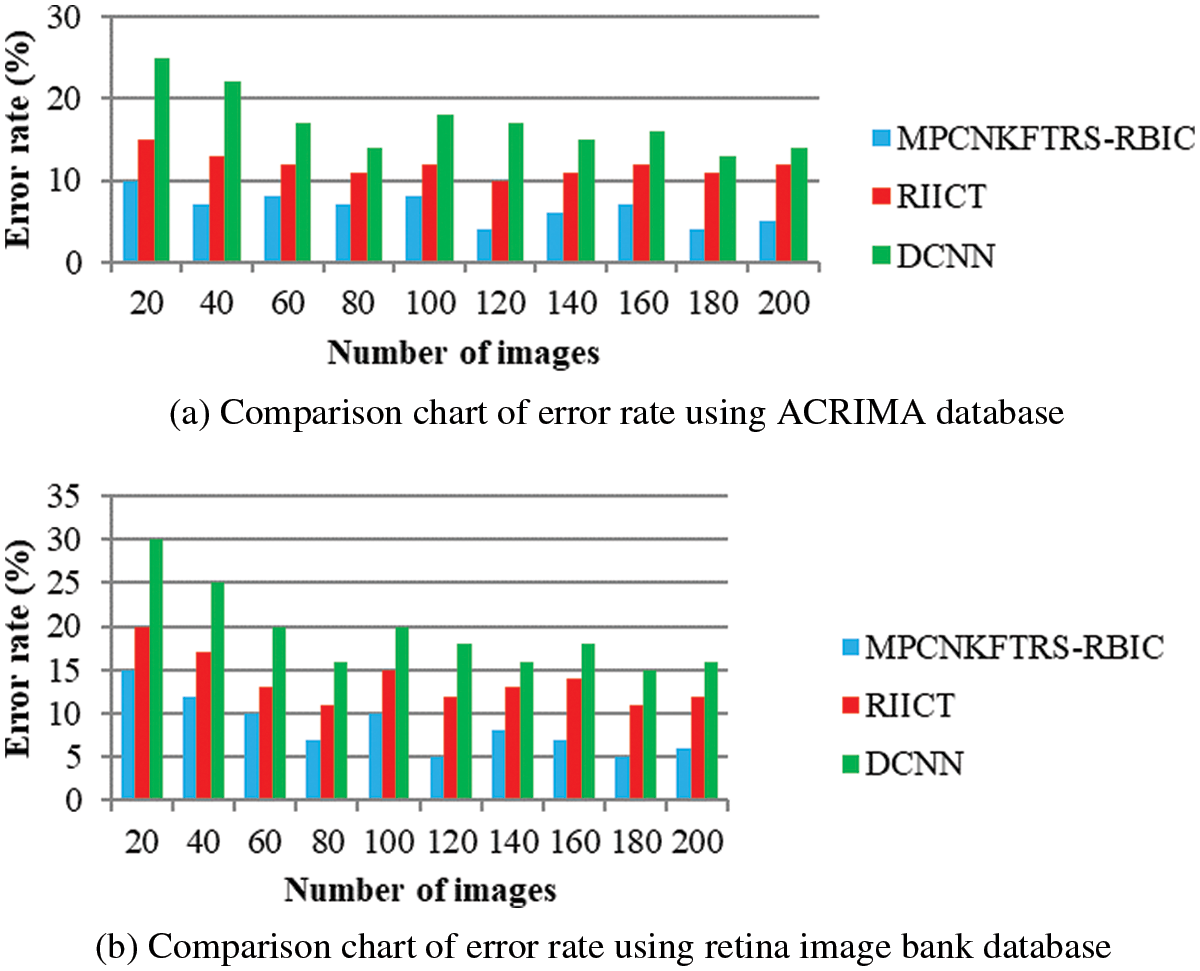
Figure 8: (a) Comparison chart of error rate using ACRIMA database (b) Comparison chart of error rate using retina image bank database
Tabs. 4a and 4b explains simulation of disease detection time with retinal fundus images with three different classification techniques, namely MPCNKFTRS-RBIC technique and RIICT [1] and DCNN [2]. The numbers of retinal fundus images taken from 20 to 200 are gathered from the ACRIMA database and retina image bank database. Ten various performance outcomes were accomplished by different counts of input images. Disease detection time of MPCNKFTRS-RBIC is significantly reduced than the other two conventional techniques. By applying the ACRIMA database, the disease detection time of MPCNKFTRS-RBIC model is reduced as 17% and 31% compared with [1,2]. Disease detection time of the MPCNKFTRS-RBIC technique using the retina image bank database is considerably reduced by 16% and 28% than the conventional classification techniques.
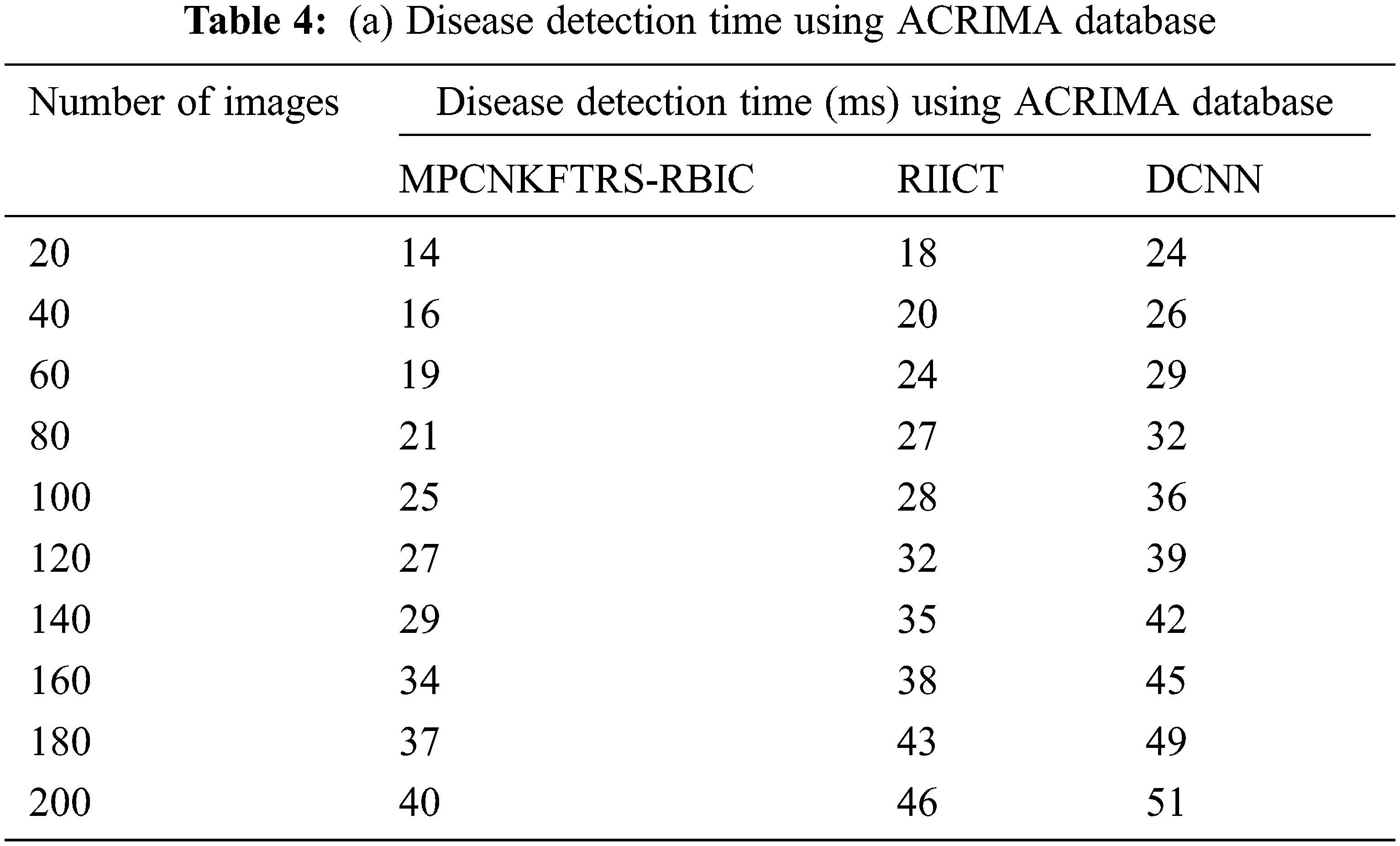
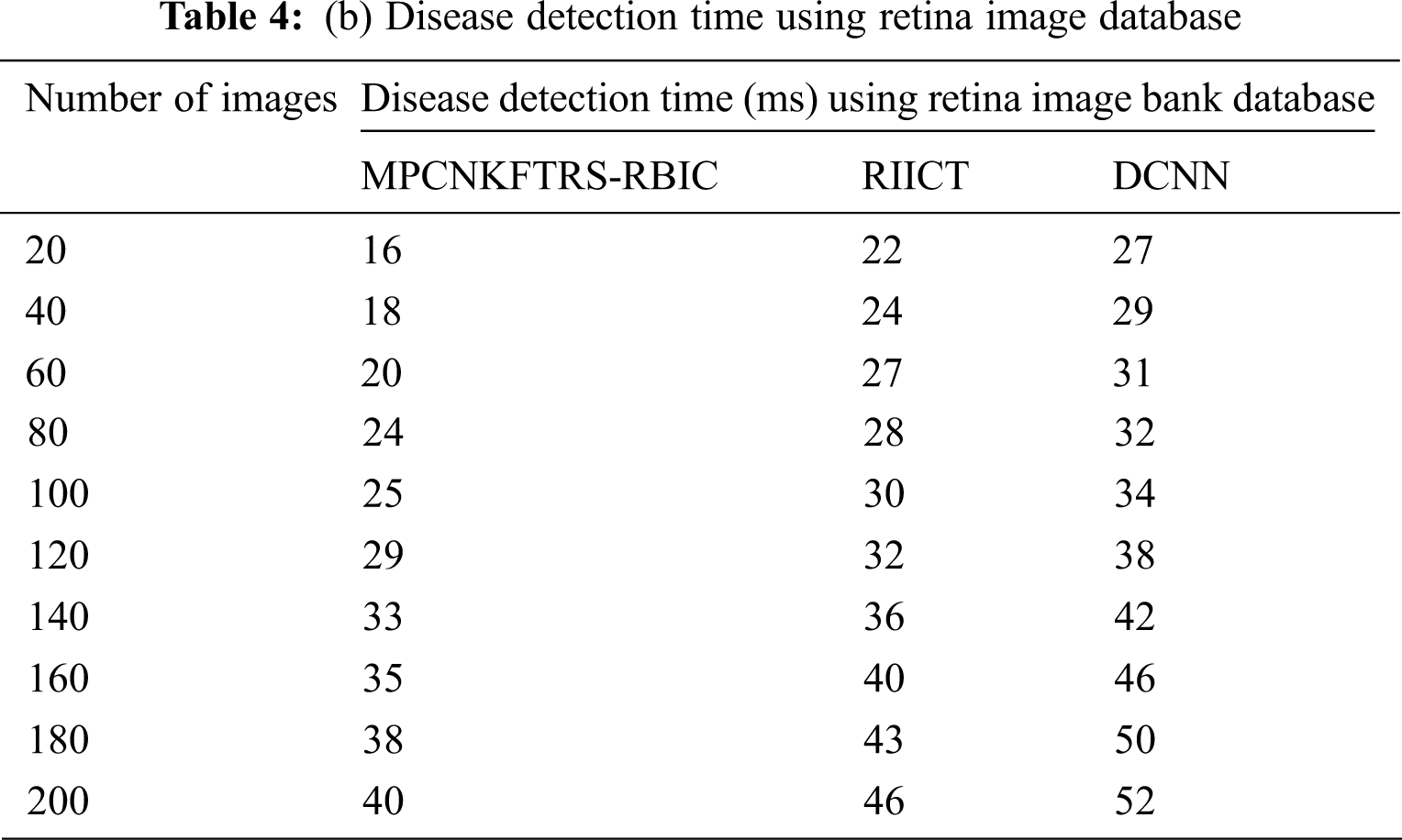
Figs. 9a and 9b illustrates the comparison results of disease detection time using two different databases. The disease detection time of three classification techniques gets increased while increasing the number of images. But comparatively MPCNKFTRS-RBIC technique reduces the disease detection time using two image databases. The reason behinds this improvement is to perform the feature extraction and segmentation. The Max pooling is performed for minimizing dimensionality of dataset and remove significant features. The Tobit regression is applied to analyze the pixels, and segment the images into different regions. Lastly, classification is achieved using radial basis to detect disease with minimum time consumption.
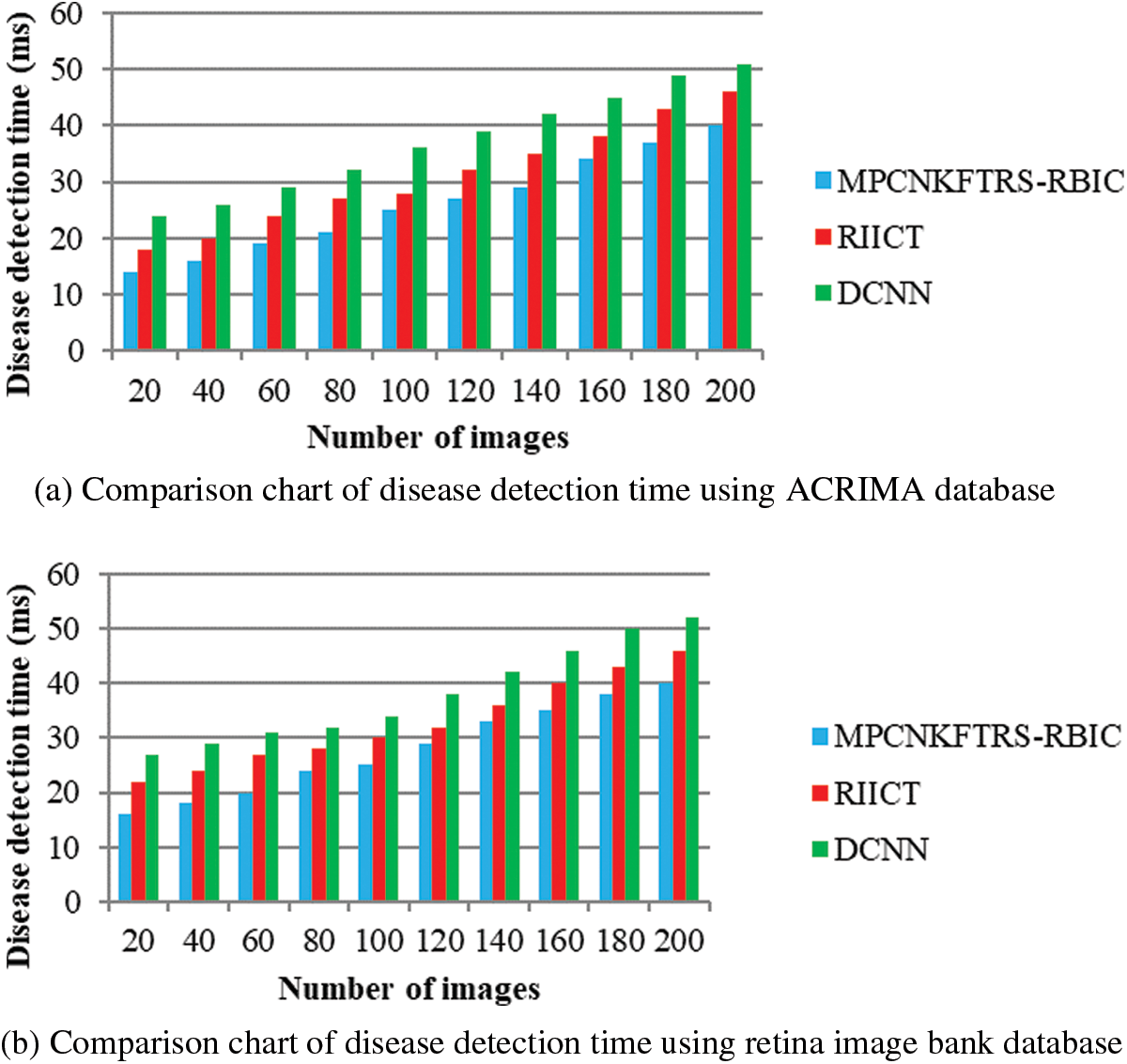
Figure 9: (a) Comparison chart of disease detection time using ACRIMA database (b) Comparison chart of disease detection time using retina image bank database
An efficient technique called the MPCNKFTRS-RBIC model is developed for improving the disease identification at an earlier stage using retinal fundus images with minimum time consumption. The MPCNKFTRS-RBIC technique performs four various processes to achieve the above contribution. The input retinal fundus images are preprocessed by applying the weighted adaptive Kuan filter for enhancing peak signal to noise ratio. After that, significant features are removed over preprocessed image. With significant features, segmentation is performed for partitioning images into different regions. Lastly, classification is done using radial basis for identifying normal or abnormal images by better accuracy. Simulation is performed using two image databases, as well as different metrics. This proposed model needs to be enhanced and trained with low resolution and highly degraded images. MPCNKFTRS-RBIC considerably increases accuracy as well as reduces the feature extraction time and error rate compared to other baseline approaches.
Acknowledgement: Authors liked to thank SSIET institute for their research support and anonymous reviewers for their insights about this article.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Y. Wang and S. Shan, “Accurate disease detection quantification of iris based retinal images using random implication image classifier technique,” Microprocessors and Microsystems, vol. 80, pp. 1–11, 2021. [Google Scholar]
2. S. Gheisari, S. Shariflou, J. Phu, P. J. Kennedy, A. Agar et al., “A combined convolutional and recurrent neural network for enhanced glaucoma detection,” Scientific Reports, vol. 11, no. 1, pp. 1–11, 2021. [Google Scholar]
3. N. T. Umpon, I. Poonkasem, S. Auephanwiriyakul and D. Patikulsila,“Hard exudate detection in retinal fundus images using supervised learning,” Neural Computing and Applications, vol. 32, no. 17, pp. 13079–13096, 2020. [Google Scholar]
4. J. Joo, Y. Shin, H. D. Kim, K. HwanJung, K. Hyung et al., “Development and validation of deep learning models for screening multiple abnormal findings in retinal fundus images,” Ophthalmology, vol. 127, no. 1, pp. 85–94, 2020. [Google Scholar]
5. D. Devarajan, S. M. Ramesh and B. Gomathy, “A metaheuristic segmentation framework for detection of retinal disorders from fundus images using a hybrid ant colony optimization,” Soft Computing, vol. 24, no. 17, pp. 13347–13356, 2020. [Google Scholar]
6. X. Hai, H. Lei, X. Zeng, Y. He, G. Chen et al., “AMD-GAN: Attention encoder and multi-branch structure based generative adversarial networks for fundus disease detection from scanning laser ophthalmoscopy images,” Neural Networks, vol. 132, no. 3, pp. 477–490, 2020. [Google Scholar]
7. L. A. Hamid, “Glaucoma detection from retinal images using statistical and textural wavelet features,” Journal of Digital Imaging, vol. 33, no. 1, pp. 151–158, 2020. [Google Scholar]
8. Z. Xie, T. Ling, Y. Yang, R. Shu and B. J. Liu, “Optic disc and cup image segmentation utilizing contour-based transformation and sequence labeling networks,” Journal of Medical Systems, vol. 44, no. 5, pp. 1–13, 2020. [Google Scholar]
9. U. Raghavendra, H. Fujita, S. V. Bhandary, A. Gudigar, J. H. Tan et al., “Deep convolution neural network for accurate diagnosis of glaucoma using digital fundus images,” Information Sciences, vol. 441, pp. 41–49, 2018. [Google Scholar]
10. D. A. Dharmawana, B. Poh Nga and S. Rahardja, “A new optic disc segmentation method using a modified dolph-chebyshev matched filter,” Biomedical Signal Processing and Control, vol. 59, pp. 1–10, 2020. [Google Scholar]
11. J. Pruthi, K. Khanna and S. Arora, “Optic cup segmentation from retinal fundus images using glowworm swarm optimization for glaucoma detection,” Biomedical Signal Processing and Control, vol. 60, pp. 1–12, 2020. [Google Scholar]
12. D. Parashar and D. K. Agrawal, “Automated classification of glaucoma stages using flexible analytic wavelet transform from retinal fundus images,” IEEE Sensors Journal, vol. 20, no. 21, pp. 12885–12894, 2020. [Google Scholar]
13. S. Wan, Y. Liang and Y. Zhang, “Deep convolutional neural networks for diabetic retinopathy detection by image classification,” Computers & Electrical Engineering, vol. 72, pp. 274–282, 2018. [Google Scholar]
14. S. Sreng, N. Maneerat, K. Hamamoto and K. Y. Win, “Deep learning for optic disc segmentation and glaucoma diagnosis on retinal images,” Applied Sciences, vol. 10, no. 14, pp. 1–19, 2020. [Google Scholar]
15. M. Juneja, S. Singh, N. Agarwal, S. Bali, S. Gupta et al., “Automated detection of glaucoma using deep learning convolution network,” Multimedia Tools and Applications, vol. 79, no. 21, pp. 15531–15553, 2020. [Google Scholar]
16. T. Pratap and P. Kokil, “Computer-aided diagnosis of cataract using deep transfer learning,” Biomedical Signal Processing and Control, vol. 53, no. 5, pp. 101533, 2019. [Google Scholar]
17. P. Mangipudi, H. M. Pandey and A. Choudhary, “Improved optic disc and cup segmentation in glaucomatic images using deep learning architecture,” Multimedia Tools and Applications, vol. 1, no. 3, pp. 1–21, 2021. [Google Scholar]
18. K. Wang, X. Zhang, S. Huang, F. Chen, X. Zhang et al., “Learning to recognize thoracic disease in chest x-rays with knowledge-guided deep zoom neural networks,” IEEE Access, vol. 8, pp. 159790–159805, 2020. [Google Scholar]
19. D. Somasundaram, S. Gnanasaravanan and N. Madian, “Automatic segmentation of nuclei from pap smear cell images: A step toward cervical cancer screening,” International Journal of Imaging Systems and Technology, vol. 30, no. 4, pp. 1209–1219, 2020. [Google Scholar]
20. S. Lu, “Accurate and efficient optic disc detection and segmentation by a circular transformation,” IEEE Transactions on Medical Imaging, vol. 30, no. 12, pp. 2126–2133, 2011. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools