 Open Access
Open Access
ARTICLE
Faster Region Based Convolutional Neural Network for Skin Lesion Segmentation
1 Department of Computer Science and Engineering, St. Joseph’s College of Engineering, Chennai, Tamil Nadu, India
2 Department of Computer Science and Engineering, B. S. Abdur Rahman Crescent Institute of Science and Technology, Chennai, Tamil Nadu, India
3 Department of Artificial Intelligence and Data Science, Saveetha Engineering College (Autonomous), Chennai, Tamil Nadu, India
4 Department of Electronics and Communication Engineering, St. Joseph’s College of Engineering, Chennai, Tamil Nadu, India
* Corresponding Author: G. Murugesan. Email:
Intelligent Automation & Soft Computing 2023, 36(2), 2099-2109. https://doi.org/10.32604/iasc.2023.032068
Received 05 May 2022; Accepted 12 July 2022; Issue published 05 January 2023
Abstract
The diagnostic interpretation of dermoscopic images is a complex task as it is very difficult to identify the skin lesions from the normal. Thus the accurate detection of potential abnormalities is required for patient monitoring and effective treatment. In this work, a Two-Tier Segmentation (TTS) system is designed, which combines the unsupervised and supervised techniques for skin lesion segmentation. It comprises preprocessing by the median filter, TTS by Colour K-Means Clustering (CKMC) for initial segmentation and Faster Region based Convolutional Neural Network (FR-CNN) for refined segmentation. The CKMC approach is evaluated using the different number of clusters (k = 3, 5, 7, and 9). An inception network with batch normalization is employed to segment melanoma regions effectively. Different loss functions such as Mean Absolute Error (MAE), Cross Entropy Loss (CEL), and Dice Loss (DL) are utilized for performance evaluation of the TTS system. The anchor box technique is employed to detect the melanoma region effectively. The TTS system is evaluated using 200 dermoscopic images from the PH2 database. The segmentation accuracies are analyzed in terms of Pixel Accuracy (PA) and Jaccard Index (JI). Results show that the TTS system has 90.19% PA with 0.8048 JI for skin lesion segmentation using DL in FR-CNN with seven clusters in CKMC than CEL and MAE.Keywords
Integrating image-based perception and reasoning requires highly skilled radiologists in the medical domain. As these analytical skills differ from each other and to reduce interpretive errors, Computer aided diagnosis systems have been developed recently. An ensemble approach is discussed in [1] for segmenting skin lesions. It uses DeeplabV3 and mask R Convolution Neural Network (CNN). Also, the performance of the system is compared with U-Net and SegNet. The attention-based network is described in [2] using DenseNet to segment skin lesions. It has segmentation and discriminator modules. The attention module extracts features from the skin lesion and suppresses other regions. The combination of adversarial feature loss and Jaccard distance loss is used while updating the weights in the network.
A multi-scale attention CNN is described in [3] for skin lesion segmentation. The attention module is the spatial and channel-based residual module that enhances the segmentation results. Also, a weighted cross entropy loss is used to improve the system further. A mutual bootstrapping model is discussed in [4] for segmenting skin lesions. It comprises coarse, mask-guided, and enhanced segmentation networks. The coarse network provides the lesion masks for the mask-guided network for the classification. All these networks mutually transferred the obtained information for effective segmentation of skin lesions. A feature learning framework is discussed in [5] with decision fusion for segmenting skin lesions. This framework is placed at the top of CNN, where the discriminative features are extracted based on the correlation between the lesions and their informative context. Also, decision fusion is performed to select the decisions from the classification layers. A dual attention module is described in [6] for segmenting skin lesions. The shape irregularity and boundary continuity are analyzed using pixel-wise and global average pooling respectively. It captures the multi-scale features for the segmentation along with the spatial information.
A dense deconvolution network is implemented in [7] for skin lesion segmentation. It uses residual learning with chained residual pooling to capture the rich textural information. Also, fusion takes place in the residual pooling that uses local and global contextual information. To refine the prediction results, hierarchical supervision is introduced. Active contour and YoloV4 network-based approach for segmenting skin lesions is described in [8]. At first, the artifacts are removed using morphological operations and then enhanced. After preprocessing, the YoloV4 object detector is employed to segment the regions with infected or non-infected regions, and then an active contour is used.
The conditional random fields by deep CNN for skin lesion segmentation are discussed in [9]. It utilizes different CNN models to obtain better performance along with the probabilistic inferences obtained from the conditional random fields. A fuzzy-based lesion segmentation rule is implemented to refine the lesion boundary. A saliency map-driven approach is discussed in [10] for segmenting skin lesions. The saliency score of each region in the dermoscopic images is provided based on the regional contrast, multi-level segmentation, and random forest regressor. A level set approach then refines the obtained initial mask. A fully convolutional network is implemented in [11] for skin lesion segmentation. The colour information from multiple colour spaces; RGB, HSV, and CIELAB are utilized and a smaller kernel deeper CNN is employed for the segmentation. Adversarial learning of dense residual networks is designed in [12] for skin lesion segmentation. A dense residual block is introduced in the encoder-decoder network to train the network effectively. The relationship between the adjacent pixels is also captured, and a multi-scale loss function is employed.
A colour based U-net model is discussed in [13] for skin lesion segmentation. The single input U-net model is modified to accept multi-inputs from different colour spaces. The obtained features from each encoder network are fused using a channel-wise attention module, and the decoder network generates the lesion maps. A semi-supervised system is discussed in [14] for skin lesion segmentation. It uses a mean teacher scheme for confidence-aware learning from dermoscopic images. The true class probability-based confidence level is employed for the segmentation. A fractal-based system is discussed in [15] for skin cancer diagnosis. It uses differential box-counting to obtain the fractal dimension, and parametric and non-parametric classifiers are employed for skin cancer detection.
In this paper, a skin lesion segmentation approach by Two-Tier Segmentation (TTS) is presented with the help of Colour K-Means Clustering (CKMC) and Faster Region based Convolutional Neural Network (FR-CNN). The rest of the paper is as follows: The design of the TTS approach is discussed in Section 2. The performances of the TTS approach with different loss functions and a different number of clusters (k = 3, 5, 7, and 9) are discussed in Section 3, and the conclusion is made in the last Section.
The proposed TTS system architecture is shown in Fig. 1. It consists of preprocessing and TTS stages. The former stage uses median filtering, and the later stage uses CKMC for initial segmentation and FR-CNN for refined segmentation.
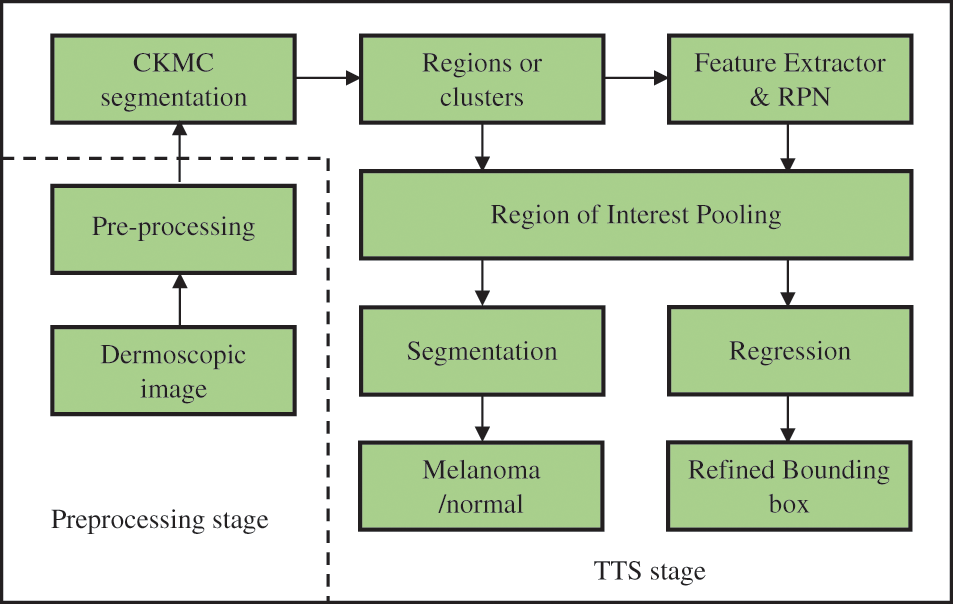
Figure 1: Proposed TTS system
A simple median filtering [16,17] is employed in this stage. In many pattern recognition approaches, it is often used to remove noises due to its simplicity and easy computation. It smoothes the images while keeping the important information in it. Fig. 2 shows the median filtered outputs for sample dermoscopic images.
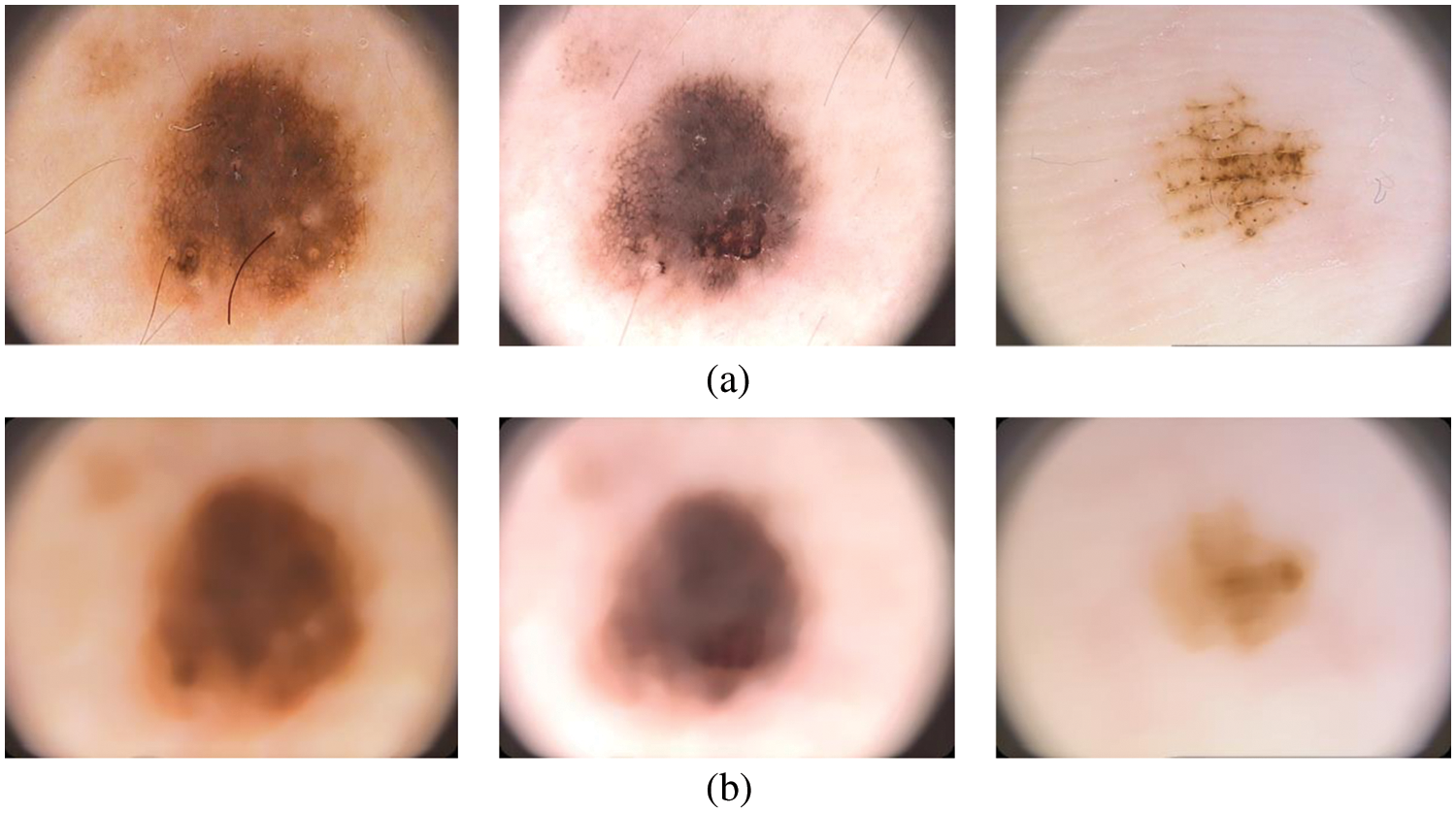
Figure 2: (a) Dermoscopic images (b) preprocessed image
It uses a square-shaped window and is placed over the images. Then finds the median value of pixels over the window and replace the center pixel with the median. The window is moved from left to right and top to bottom until the whole dermoscopic image is processed. The size of the square-shaped windows is 21 × 21.
2.2 Colour K-means Clustering in TTS System
This is the first tier system in TTS using the CKMC approach. The conventional KMC is performed on gray scale images, whereas CKMC is on colour images. The CKMC algorithm uses L*a*b* colour space as the visual differences can be easily quantified. The L*a*b* colour space where L (lightness of the colour), a (red minus green), and b (green minus blue) is defined by [17].
where
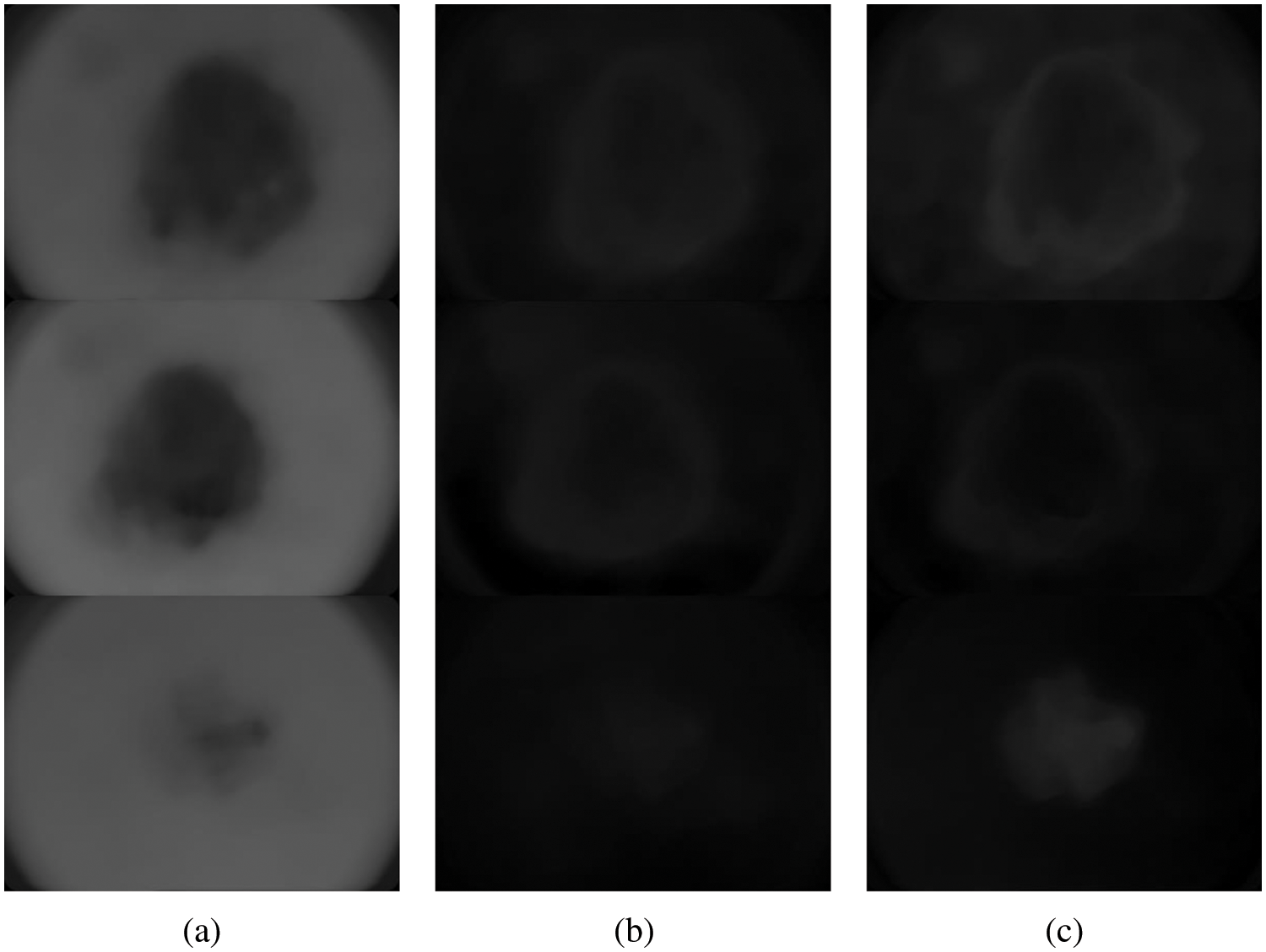
Figure 3: Different components of L*a*b* colour space (a) L (b) a* and (c) b*
The CKMC approach uses only the L and a* components as they only provide the colour information. The algorithm for the CKMC approach is as follows:
1. Choose the initial clusters based on the k value.
2. The CKMC has two steps;
3. a) Assignment: Compute the least squared Euclidean distances between the data points and the clusters. Assign the clustered index to the data points which have the least distance.
b) Update: The mean values in each new cluster are updated in this step.
The above steps are iterated until there are no changes in step 2a or it converges in step 2b. Different k values (3, 5, 7, and 9) are used in this study to segment the melanoma region initially. The CKMC algorithm converges only when there are no changes in step 2a or a finite number of iterations (number of iterations: 30). Figs. 4a and 4b show the results obtained by setting five and seven clusters in the CKMC segmentation approach.
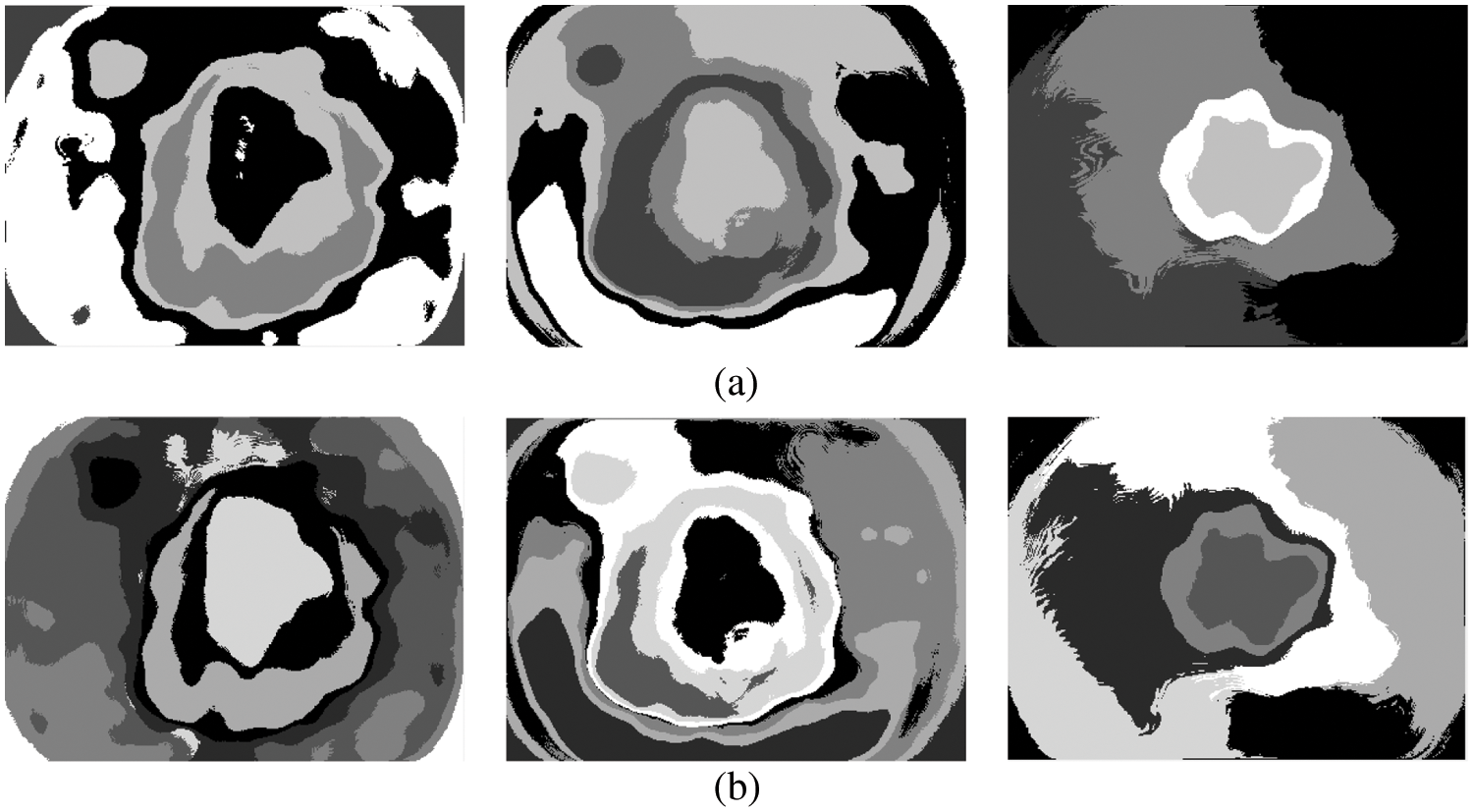
Figure 4: (a) 5 clusters or regions (b) 7 clusters
This is the second-tier system in TTS using the FR-CNN approach [18]. The inception architecture is used to extract deep features with batch normalization. Inception architecture is used to avoid overfitting and make the gradient updating very easy. In order to increase the training speed, the image size is reduced to 128 × 128 pixels before applying the FR-CNN. Fig. 5 shows the inception architecture where C is the convolution operation. The fully connected layer uses back propagation with three loss functions such as Mean Absolute Error (MAE), Cross Entropy Loss (CEL), and Dice Loss (DL). The loss functions definitions are as follows:
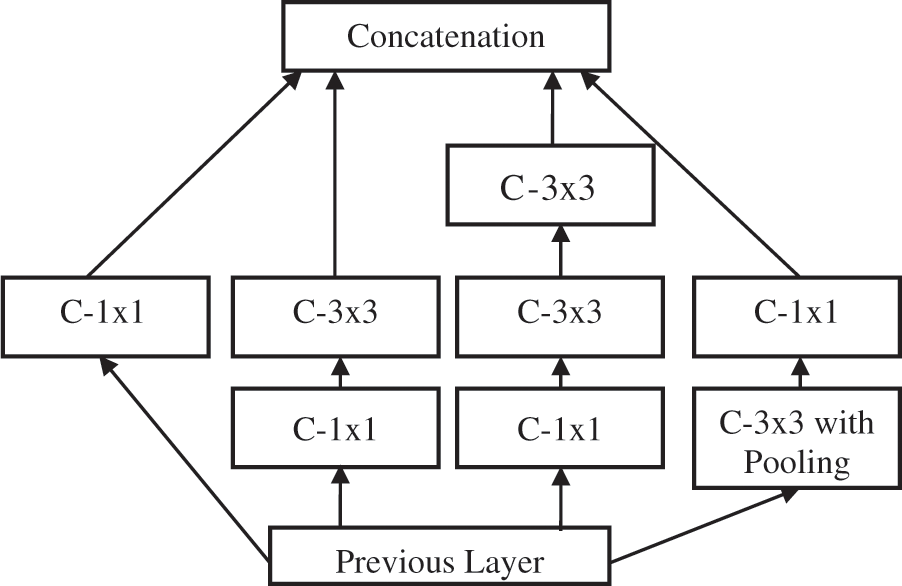
Figure 5: Inception architecture
The number of epochs used to train the FR-CNN approach is 30, with a learning rate of 0.001 and momentum of 0.8. Also, batch normalization is employed to stabilize the learning process with a mini-batch size of 8.
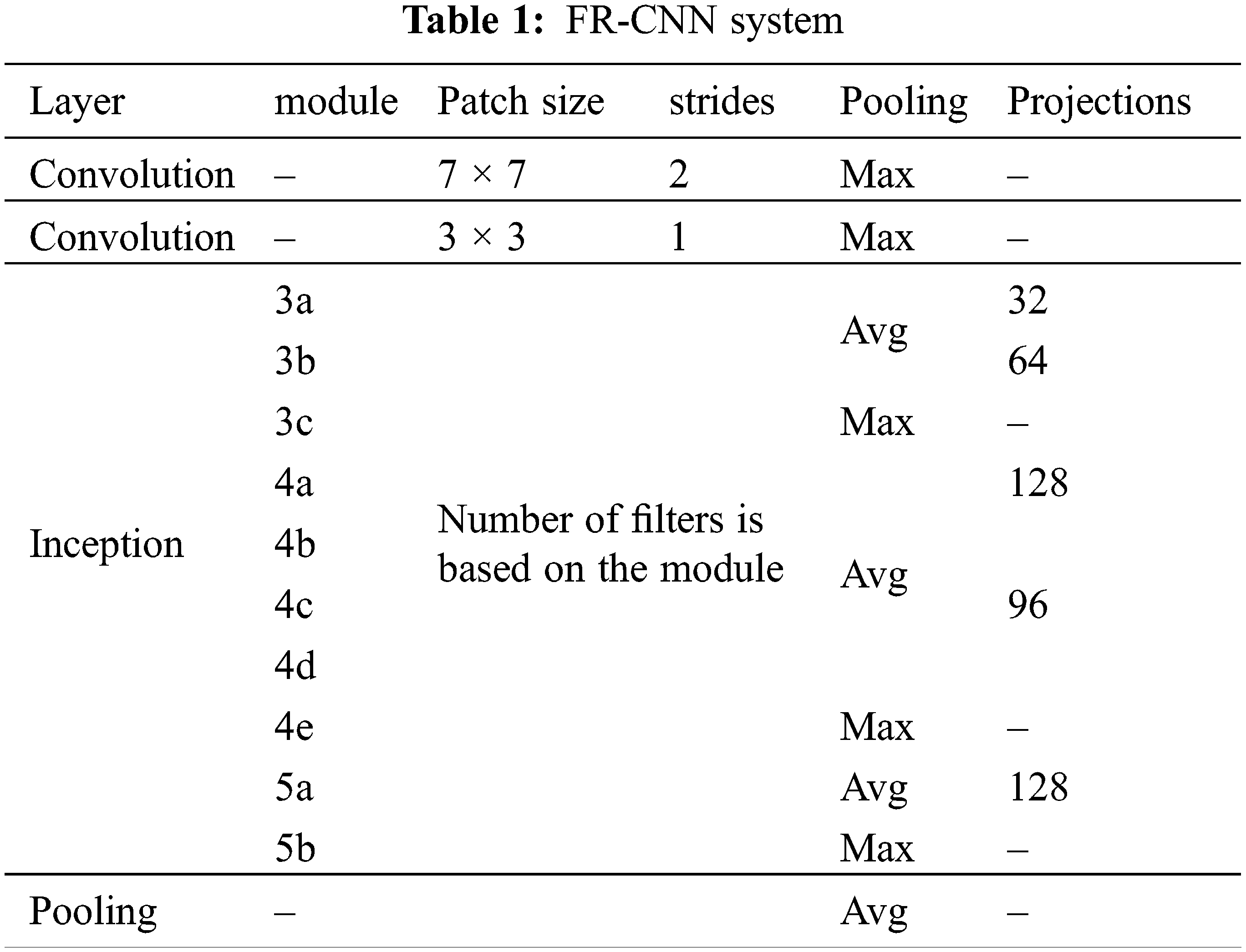
The inception architecture reduces the computational complexity by factorizing higher convolution operations (7 × 7) by more than one small convolution operation (two 3 × 3). The anchor box technique is used in RPN to detect objects of different shapes. It uses different scales and aspect ratios. Thousands of anchor boxes are created to predict the melanoma region. The intersection over union property (greater than 40%) is utilized to detect melanoma. Table 1 shows the important configuration details of FR-CNN architecture.
Though FR-CNN can be used directly for the segmentation, the proposed system operates in two stages. The first stage identifies the melanoma region by CKMC, and the FR-CNN fine-tunes the same region. As the FR-CNN uses only the pre-determined melanoma region, the proposed FR-CNN is faster than the system using the whole image. The distance or the number of pixels moved over the input by the subsequent convolution operation is termed stride. The max-pooling and average-pooling layers for the features f in the feature space F are defined below:
The dermoscopic images from the PH2 [19,20] database are utilized to analyze the TTS approach for melanoma detection. The original size of each dermoscopic image is 768 × 500 pixels. The PH2 has 200 images (120 abnormal (80 benign and 40 melanoma) and 80 normal). Fig. 6 shows a sample image in three categories in the PH2 database. A random split approach is employed with 70:30 ratios to obtain training and testing samples. Thus, 84 abnormal and 56 normal images are used for training, and the remaining 36 abnormal and 24 normal images are used to test the efficiency of the FR-CNN system.
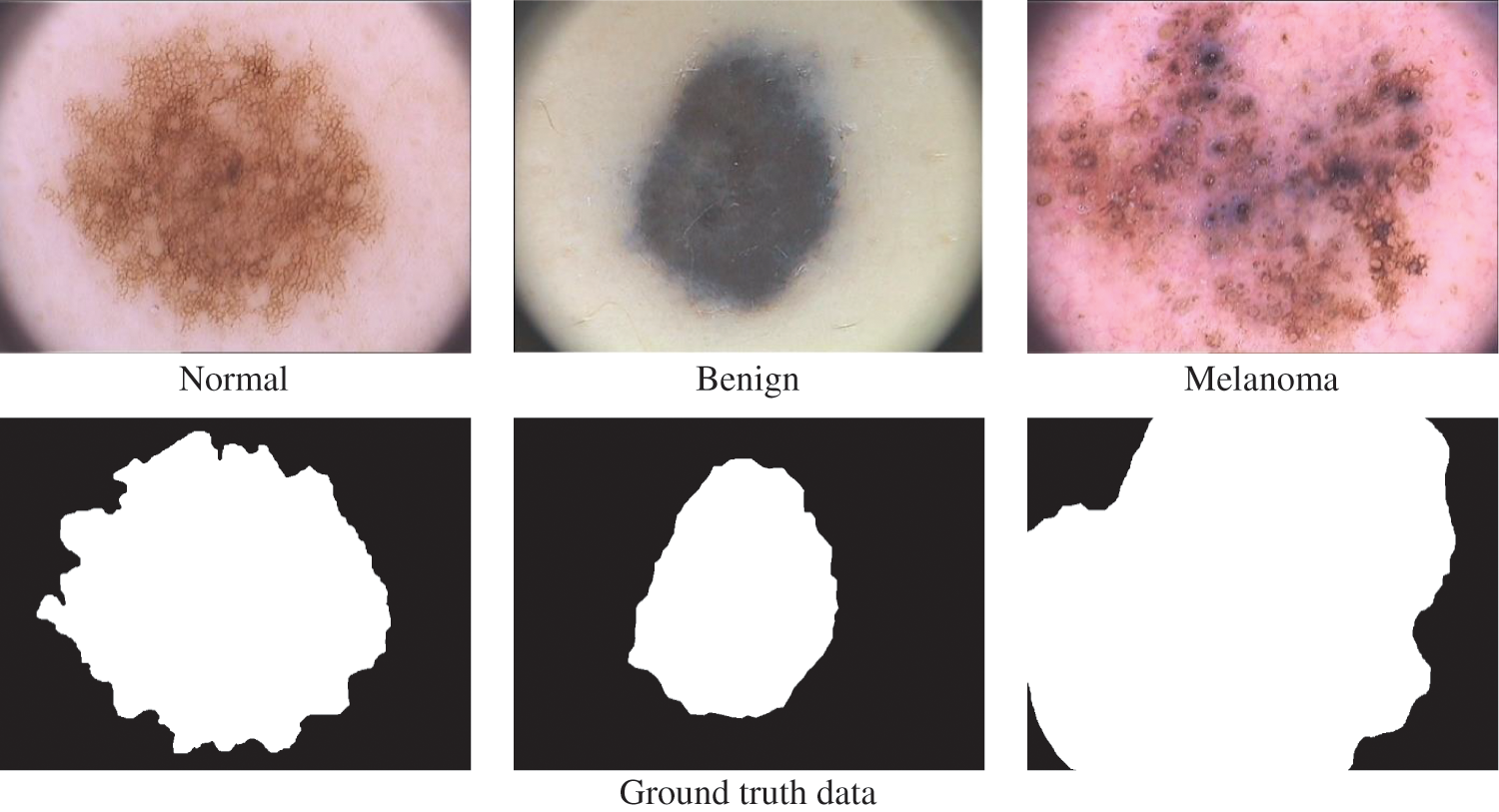
Figure 6: PH2 database images
The commonly used metrics of segmentation based approaches are Pixel Accuracy (PA) and Jaccard Index (JI) [21]. Their definitions are as follows:
PA: It is the ratio between the number of pixels correctly classified as lesion and the total number of pixels marked as lesion in the respective ground truth data.
JI: It shows the similarity and diversity between two sets A and B. It is defined as
Also, Precision (P) and Recall (R) matrices are used for the analysis. These two terms are defined in Eqs. (6) and (7).
The term ‘True Positive’ in Eq. (11) is the number of pixels correctly classified as melanoma compared to the ground truth image. Similarly, the term is ‘False Positive’ is the number of pixels corresponding to the normal region is misclassified as melanoma and ‘False Negative’ in Eq. (12) is the number of pixels corresponding to the melanoma region is misclassified as normal. Fig. 7 shows the final segmentation from the FR-CNN system using the TTS approach.
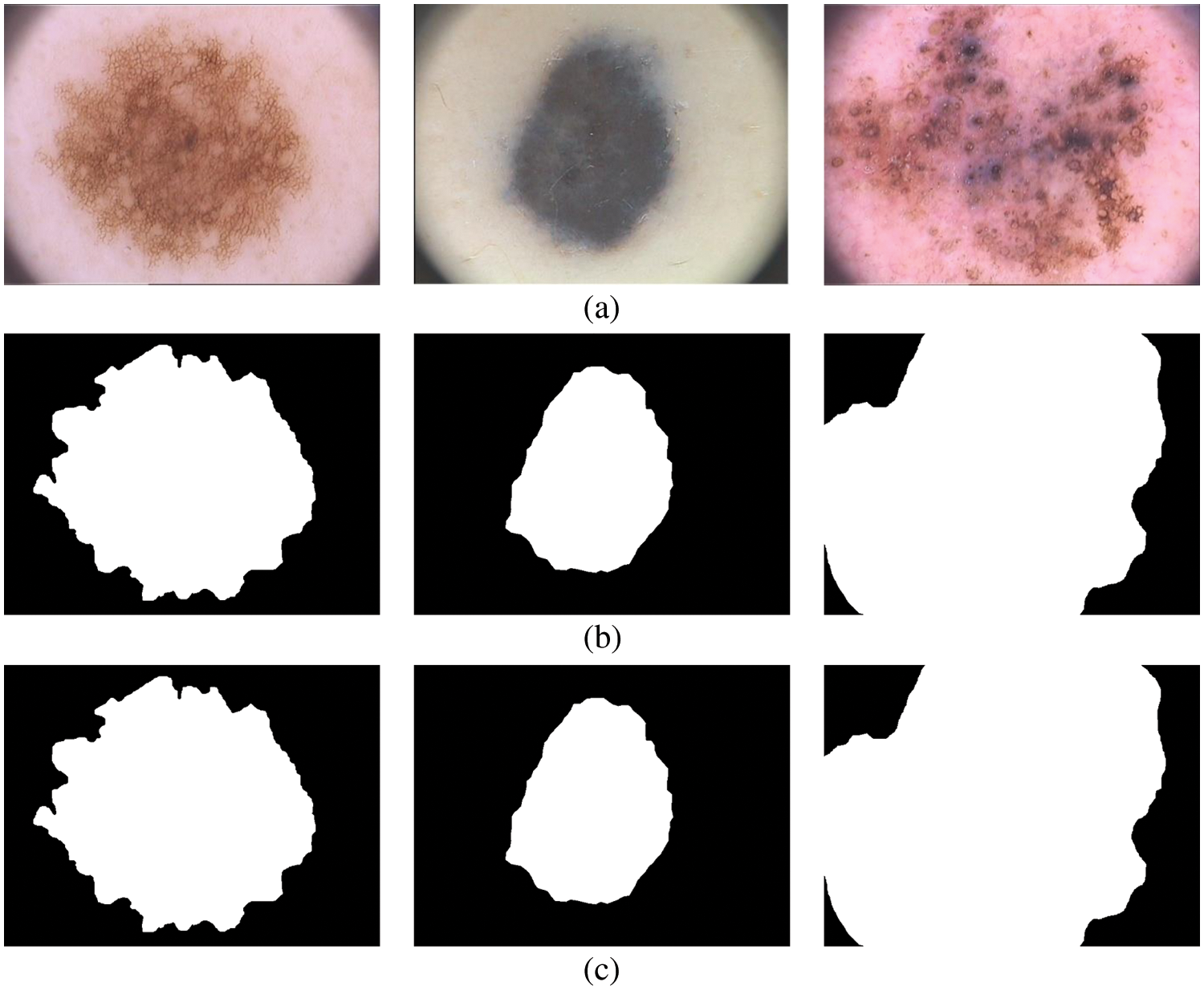
Figure 7: Segmented results (a) input image (b) 5 clusters (c) 7 clusters
Any deep learning network design requires an optimization function (Adam optimizer) for training the network. The model error is computed using a loss function. This work uses three loss functions to analyze the TTS approach; MAE, CEL, and DL. Table 2 shows the performances of the TTS approach.
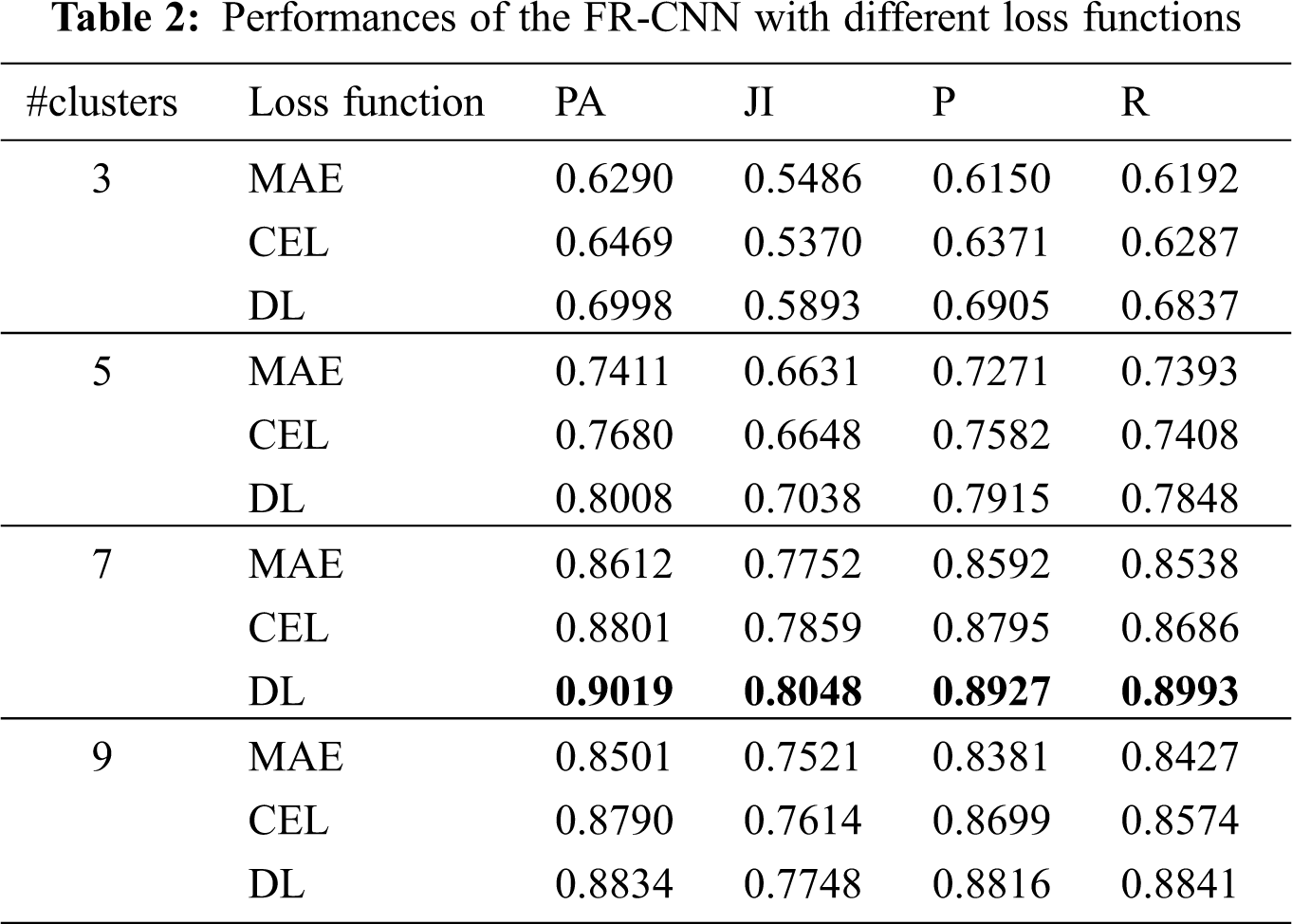
It can be seen from Table 2 that the DL function in the Adam optimizer provides better segmentation outputs than other loss functions in Adam optimizer. The system achieves 90.19% of PA with 0.8048 JI when the RPN uses the CKMC approach with seven clusters as inputs. The CEL in the Adam optimizer provides 88% PA with 0.7859 JI for the same inputs, whereas it is 86.12% (PA) and 0.7752 (JI) for MAE in Adam optimizer. Fig. 8 shows the performances of DL for skin lesion segmentation using the TTS system with 7 clusters, and Fig. 9 shows the comparison of loss functions with different k values in CKMC in terms of PA.
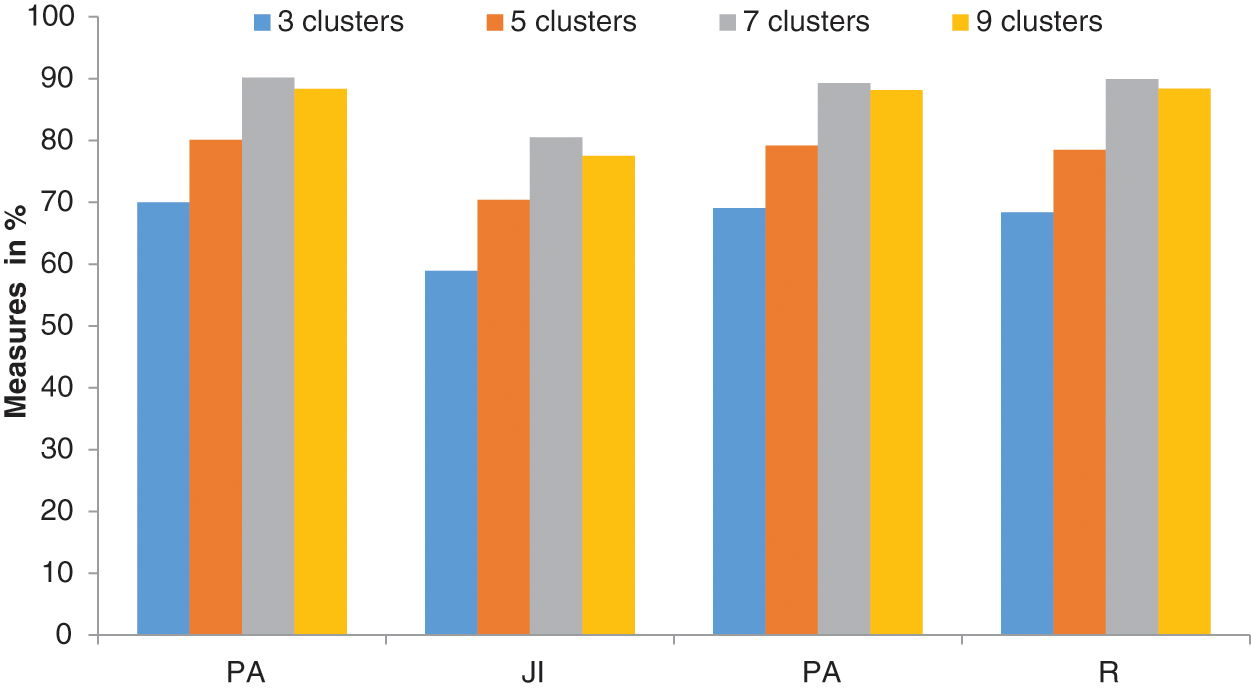
Figure 8: Performances of DL in the FR-CNN for skin lesion segmentation using the TTS system
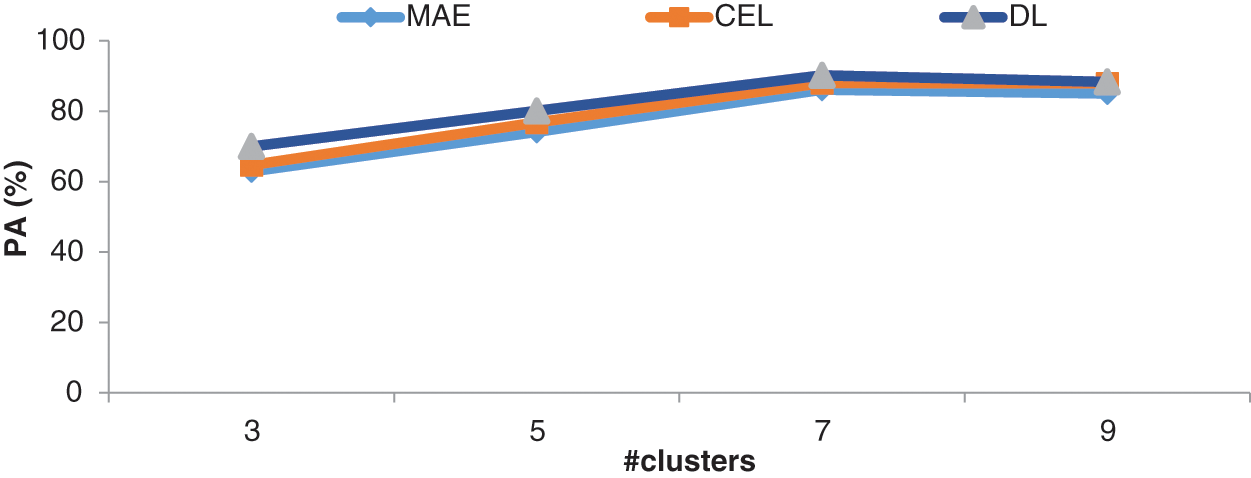
Figure 9: Comparison of loss functions with different k values in the CKMC in terms of PA
It can be seen from Figs. 8 and 9, DL has more promising performances than MAE and CEL, irrespective of the number of clusters used in CKMC. Also, CEL has more discriminating power than MAE.
The accurate segmentation of skin lesions is necessary to diagnose skin cancer. In this work, an efficient TTS approach is designed using unsupervised (CKMC) and supervised (FR-CNN) algorithms. CKMC obtains the initial segments of lesions, and the regions are fine-tuned and classified by the FR-CNN approach. Different k values in CKMC and different loss functions are analyzed for the performance of the TTS approach for skin lesion segmentation. Results show that among the different k values, k = 7 provides better performance in terms of PA, JI, P, and R with DL in FR-CNN. The system achieves 90.19% of PA with 0.8048 JI when the RPN uses seven clusters and DL. The MAE provides only 86.12% PA with 0.7752 JI for the same inputs, whereas it is 88% (PA) and 0.7859 (JI) for CEL. It is concluded that DL is more suitable for skin lesion segmentation.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. M. Goyal, A. Oakley, P. Bansal, D. Dancey and M. H. Yap, “Skin lesion segmentation in dermoscopic images with ensemble deep learning methods,” IEEE Access, vol. 8, pp. 4171–4181, 2020. [Google Scholar]
2. Z. Wei, H. Song, L. Chen, Q. Li and G. Han, “Attention based DenseUnet network with adversarial training for skin lesion segmentation,” IEEE Access, vol. 7, pp. 136616–136629, 2019. [Google Scholar]
3. Y. Jiang, S. Cao, S. Tao and H. Zhang, “Skin lesion segmentation based on multi-scale attention convolutional neural network,” IEEE Access, vol. 8, pp. 122811–122825, 2020. [Google Scholar]
4. Y. Xie, J. Zhang, Y. Xia and C. Shen, “A mutual bootstrapping model for automated skin lesion segmentation and classification,” IEEE Transactions on Medical Imaging, vol. 39, no. 7, pp. 2482–2493, 2020. [Google Scholar]
5. X. Wang, X. Jiang, H. Ding and J. Liu, “Bi-directional dermoscopic feature learning and multi-scale consistent decision fusion for skin lesion segmentation,” IEEE Transactions on Image Processing, vol. 29, pp. 3039–3051, 2020. [Google Scholar]
6. H. Wu, J. Pan, Z. Li, Z. Wen and J. Qin, “Automated skin lesion segmentation via an adaptive dual attention module,” IEEE Transactions on Medical Imaging, vol. 40, no. 1, pp. 357–370, 2021. [Google Scholar]
7. H. Li, X. He, F. Zhou, Z. Yu, D, Ni et al., “Dense deconvolutional network for skin lesion segmentation,” IEEE Journal of Biomedical and Health Informatics, vol. 23, no. 2, pp. 527–537, 2019. [Google Scholar]
8. S. Albahli, N. Nida, A. Irtaza, M. H. Yousaf and M. T. Mahmood, “Melanoma lesion detection and segmentation using YOLOv4-DarkNet and active contour,” IEEE Access, vol. 8, pp. 198403–198414, 2020. [Google Scholar]
9. Y. Qiu, J. Cai, X. Qin and J. Zhang, “Inferring skin lesion segmentation with fully connected CRFs based on multiple deep convolutional neural networks,” IEEE Access, vol. 8, pp. 144246–144258, 2020. [Google Scholar]
10. M. Jahanifar, N. Zamani Tajeddin, B. Mohammadzadeh Asl, A. Gooya, “Supervised saliency map driven segmentation of lesions in dermoscopic images,” IEEE Journal of Biomedical and Health Informatics, vol. 23, no. 2, pp. 509–518, 2019. [Google Scholar]
11. Y. Yuan and Y. C. Lo, “Improving dermoscopic image segmentation with enhanced convolutional-deconvolutional networks,” IEEE Journal of Biomedical and Health Informatics, vol. 23, no. 2, pp. 519–526, 2019. [Google Scholar]
12. W. Tu, X. Liu, W. Hu and Z. Pan, “Dense residual network with adversarial learning for skin lesion segmentation,” IEEE Access, vol. 7, pp. 77037–77051, 2019. [Google Scholar]
13. R. Ramadan and S. Aly, “CU-Net: A new improved multi-input color U-Net model for skin lesion semantic segmentation,” IEEE Access, vol. 10, pp. 15539–15564, 2022. [Google Scholar]
14. Z. Xie, E. Tu, H. Zheng, Y. Gu and J. Yang, “Semi-supervised skin lesion segmentation with learning model confidence,” in IEEE Int. Conf. on Acoustics, Speech and Signal Processing, Toronto, ON, Canada, pp. 1135–1139, 2021. [Google Scholar]
15. S. Jacob and J. D. Rosita, “Fractal model for skin cancer diagnosis using probabilistic classifiers,” International Journal of Advances in Signal and Image Sciences, vol. 7, no. 1, pp. 21–29, 2021. [Google Scholar]
16. V. V. Lakshmi and J. S. Jasmine, “A hybrid artificial intelligence model for skin cancer diagnosis,” Computer Systems Science and Engineering, vol. 37, no. 2, pp. 233–245, 2021. [Google Scholar]
17. R. Thamizhamuthu and D. Manjula, “Skin melanoma classification system using deep learning,” Computers, Materials & Continua, vol. 68, no. 1, pp. 1147–1160, 2021. [Google Scholar]
18. S. Anitha and T. R. Ganesh Babu, “An efficient method for the detection of oblique fissures from computed tomography images of lungs,” Journal of Medical Systems, vol. 43, no. 8, pp. 1–13, 2019. [Google Scholar]
19. PH2 Database. Available: https://www.fc.up.pt/addi/ph2%20database.html. [Google Scholar]
20. T. Mendonça, P. M. Ferreira, J. S. Marques, A. R. Marcal and J. Rozeira, “PH2-A dermoscopic image database for research and benchmarking,” in 35th Annual Int. Conf. on Engineering in Medicine and Biology Society, Osaka, Japan, pp. 5437–5440, 2013. [Google Scholar]
21. S. Justin and M. Pattnaik, “Skin lesion segmentation by pixel by pixel approach using deep learning,” International Journal of Advances in Signal and Image Sciences, vol. 6, no. 1, pp. 12–20, 2020. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools