DOI:10.32604/iasc.2023.026341

| Intelligent Automation & Soft Computing DOI:10.32604/iasc.2023.026341 |  |
| Article |
Generative Deep Belief Model for Improved Medical Image Segmentation
Department of Computer Science, King Khalid University, Saudi Arabia
*Corresponding Author: Prasanalakshmi Balaji. Email: prengaraj@kku.edu.sa
Received: 22 December 2021; Accepted: 15 February 2022
Abstract: Medical image assessment is based on segmentation at its fundamental stage. Deep neural networks have been more popular for segmentation work in recent years. However, the quality of labels has an impact on the training performance of these algorithms, particularly in the medical image domain, where both the interpretation cost and inter-observer variation are considerable. For this reason, a novel optimized deep learning approach is proposed for medical image segmentation. Optimization plays an important role in terms of resources used, accuracy, and the time taken. The noise in the raw medical image are processed using Quasi-Continuous Wavelet Transform (QCWT). Then, feature extraction and selection are done after the pre-processing of the image. The features are optimally selected by the Golden Eagle Optimization (GEO) method. Specifically, the processed image is segmented accurately using the proposed Generative Heap Belief Network (GHBN) technique. The execution of this research is done on MATLAB software. According to the results of the experiments, the proposed framework is superior to current techniques in terms of segmentation performance with a valid accuracy of 99%, which is comparable to the other methods.
Keywords: Deep learning; optimization; segmentation; medical images; tumors; classification
Segmenting medical images is a critical part of image assessment. Numerous image analysis programs rely on automated image segmentation as a foundational feature [1]. The challenge of image segmentation in medical image analysis is thus critical. Anatomical structural studies, tumor, and lesion identification, and treatment planning before radiation therapy all need accurate segmentation in medical image processing [2]. Nevertheless, image segmentation is a time-consuming, specialized operation with low repeatability. As a result, image segmentation’s benefits and drawbacks are critical in image-guided surgery [3]. In medical image segmentation, traditional machine learning algorithms have shown some success in improving classification accuracy and resilience [4].
Tumor research is now the most significant area of study for academics. The brain is located at the center of the neurological system. As a result, when there is a problem with brain cells, it may lead to major health difficulties, and early detection is critical [5]. Tumor classification and Tumor-affected area identification are regarded as the most important activities in cancer classification. Different therapeutic imaging applications need an automated capacity to examine the organ’s anatomy and abnormalities [6]. Deep learning theory is well-suited to medical image segmentation due to of its generalizability and feature extraction capabilities [7].
Using Convolutional Neural Networks (CNNs) [8], Artificial Neural Network [9] for image classification, object recognition, and segmentation has been a huge success in recent years [10]. One reason for their growing popularity is their ability to extract low, mid, and high-level characteristics without involving manual efforts [11]. Lower layer filters extracts the local informtion, while more complicated objects are recognized by higher-layer filters, which build on previous layers’ work [12]. Deep learning has challenges when it comes to medical image segmentation. Medical image attributes cannot be used to build a deep learning network structure, and the deep learning model’s generalizability is limited [13]. In addition, it is challenging to collect enough training data due to the high annotation costs [14].
In this research, the main aim is to identify an accurate diagnosis of tumors from the human body using the medical image segmentation approach. At first in the preprocessing phase, the Quasi-Continuous Wavelet Transform(QCWT) method removes the noise. The different features are defined for extracting the features from the images. Then, Golden Eagle Optimization (GEO) is proposed for feature selection. Furthermore, the novel Generative Heap Belief Network (GHBN) is proposed for the final image segmentation. Finally, the various performance metric measures are used to validate the developed.
The organization of this article is as follows: Section 2 provides the recent literature related to this research.The problem definition is explained in Section 3. In Section 4, contributes to the the proposed model of medical image segmentation. Section 5 provides the consequences, discussion, and comparison of results obtained. Finally, the paper is concluded in the Section 6.
Various research works are held in previous studies for medical image segmentation. Some images are analyzed for this current research function, which is summarized as follows: Recently, different meta-heuristic methods have been developed for overcoming the image segmentation issues like Ant Colony Optimization (ACO) [15], Whale Optimization Algorithm (WOA) [16], Particle Swarm Optimization (PSO) [17], Genetic Algorithm (GA) [18], Firefly Optimization (FFO) [19], Cuckoo Search Algorithm (CSA) [20], etc. All the methods presented in the literature have certain restriction for segmentation of images. In computer vision applications and medical imaging systems, the performance of segmentation is estimated using Jaccard and Dice score index. For this reason, Eelbode et al. [21] investigated the theoretical function of metric loss and optimal weighting system for the optimized Jaccard and Dice score index using weighted cross-entropy method. The consequences of these approaches are validated with the six medical images. However, the variation of intensity tends to create false results.
Moreover, Calisto et al. [22] developed a self-adaptive based 2D to 3D ensemble method in fully convolutional networks method which is referred to as AdaEn-Net for the segmentation of 3D medical images. The multi-objective evolutionary algorithm is used to improve the accuracy of segmentation as well as reduce the parameter quantity. Indeed, only limited parameters are considered for the validation of image segmentation.
The image segmentation in medical imaging is improved by an Intelligent Fuzzy Level Set Method (IFLSM) and Quantum Particle Swarm Optimization developed by Radha et al. [23]. The execution of this work is done for the image segmentation of brain tissues. The contributed works have achieved the finest stability than the other methods. Yet, the computational efficiency and time consumption is very poor.
Consequently, multi-objective adaptive convolutional neural network (AdaResU-Net) is proposed by Calisto et al. [24] for the automatic segmentation of medical images. The model size and accuracy of the developed model is optimized by the multi-objective evolutionary algorithm (MEA). The observation shows that the proposed method has achieved superior performance than others. However, certain parameters are only considered for the system validation.
Recently, Mohamed et al. [25] developed a new hybrid thresholding method for the image segmentation of chest X-ray pictures from the COVID-19 patients. In this work, the Kapur’s entropy function is improved by the combination of Whale optimization with the Slime Mold algorithm (SMA). The performance of the hybrid model is estimated with the different X-ray images with the threshold rate of 30. However, the point of threshold finding is complex. The conventional research progression has certain drawbacks. Thus, new approaches are seriously required by proposing a novel algorithm for medical image segmentation using Magnetic Resonance Imaging (MRI) and Computer Tomography (CT) images.
In biomedical image segmentation, the problem is exacerbated by the following factors:
• As a result, crowd-computing in the biomedical image segmentation domain is almost impossible, since only professionals can contribute accurate annotations.
• High-throughput biomedical investigations generate numerous images, and annotating those images at the pixel level necessitates a large workforce.
• A general training dataset and transfer learning techniques can’t be used to solve all kinds of segmentation tasks due to the dramatic differences in biomedical images, e.g., different imaging modalities and specimens.
• Deep learning models need a particular set of training knowledge to obtain automated segmentation performances. Many topic experts may be required hundreds of hours to annotate on an authentic biomedical picture segmentation project due to these factors.
Anew deep learning model optimization technique that iteratively offers the most interesting annotation data to enhance the model’s segmentation performance is developed in this study. Because of these problems, designing a framework that can precisely incorporate a deep learning approach into an optimization procedure may be difficult. For example, a deep learning model must have a solid generalization capacity, and it must perform well during the whole training set development.
Additionally, segmentation output accuracy must be very high and precise. To overcome these challenges, this research designs a new image segmentation framework with different major components. Initially, the QCWT method has been reducing the noises in the dataset images during the pre-processing stage. Then, the various features are extracted from the feature extraction steps. Furthermore, the exact features are selected via Golden Eagle Optimisation (GEO) method. Also, accurate segmentation can take place using the proposed GBHN approach. The proposed methodology of image segmentation in the medical system is illustrated in Fig. 1.
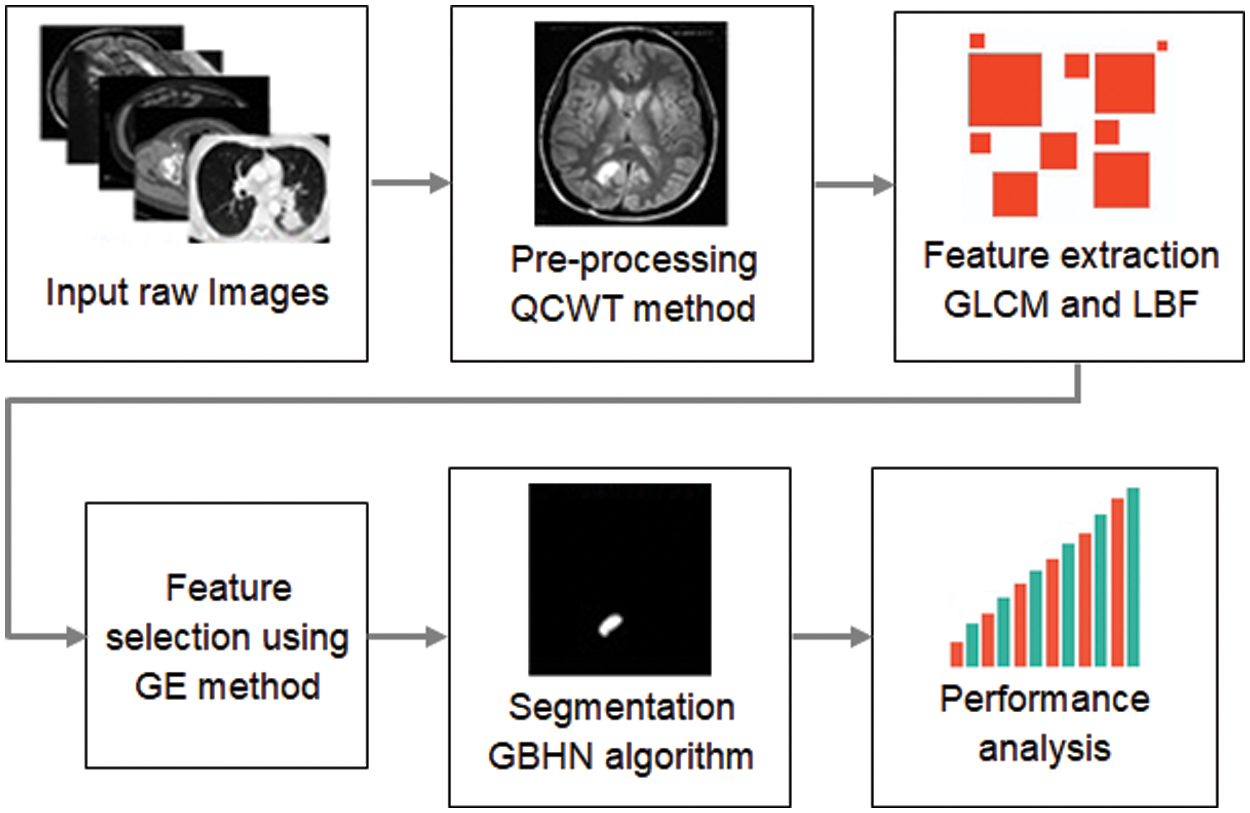
Figure 1: Proposed methodology of image segmentation in medical system
4.1 Pre-processing –QCWT Method
An image’s contaminants or distracting elements are removed during pre-processing. Noise reduction is one of the most difficult issues in medical image processing. There have been a number of ways to noise reduction explored in the past. The proposed QCWT is overcome the discrete wavelet transform function in pre-processing. This method of pre-processing is easy to design due to its simple basic technique. A dyadic sampling of scales and a non-redundancy frequency both are maintained by QCWT and in carrying uniform sampling time. In this transform, they contribute better in time-frequency localization. The low pass and high pass filters are applied to the data at every level and it generates two sequences for the upcoming level. The generated sequences have an equal length, and passed as the input sequence and it gives redundant information. There is no down sampling and hence there is no loss of informationoccurs in this technique. The function of QCWT is illustrated in Fig. 2.
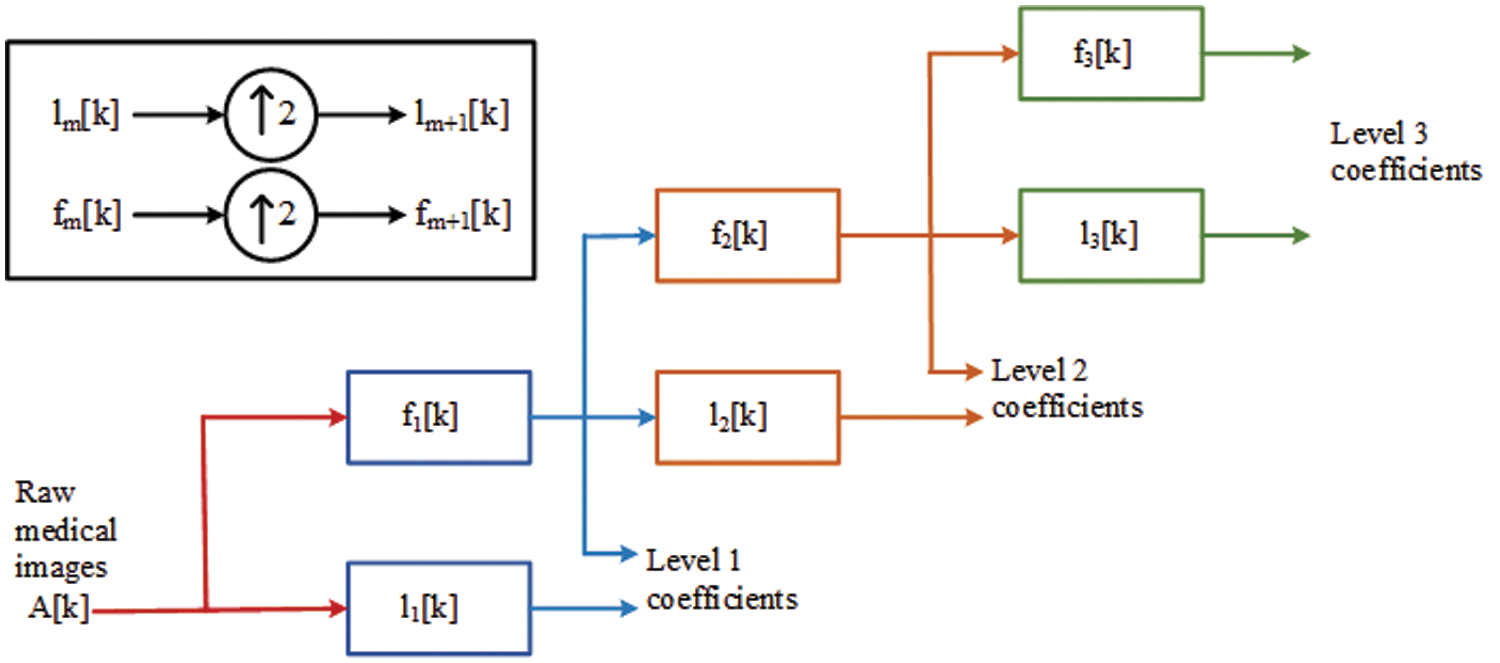
Figure 2: Quasi continuous wavelet transform (QCWT) in image pre-processing stage
A common conversion equation of QCWT based on coefficients
where,
where the wavelet coefficients are denoted by
From the Eq. (4), up-sampling the low pass filter
where,
After pre-processing, the segmented image is subjected to a feature extraction. To reduce the initial data set, the feature extraction step extracts the most significant characteristics. A person’s ability to achieve accurate categorization is greatly influenced by their ability to choose the best attributes. In this suggested research, a feature extraction approach based on Gray Level Co-occurrence Matrix (GLCM) and Local Binary Features (LBF) is considered. GLCM is a method to extract the pixel-based statistical and textural features from images. Using a GLCM, the Haralick texture features are computed which is a typical way to describe image texture. Haralick texture features are calculated from a Gray Level Co-occurrence Matrix, (GLCM), a matrix that counts the co-occurrence of neighboring gray levels in the image. The GLCM is a square matrix that has the dimension of the number of gray levels N in the region of interest (ROI).Since it is easy to implement and produces a collection of texture descriptors that can be interpreted.
4.3 Feature Selection using Golden Eagle Algorithm
Initialize the features from the extracted features from each cycle, which is expressed as
where
where
where
New positions of features that are more fit are stored in memory, and old positions are discarded if the new positions are superior. Unless otherwise specified, the memory is left untouched, but the feature value is relocated to a different location inside the system. This iteration picks a random feature point from the population, calculates strike vector, cruise vector, and ultimately the step vector and new position for the following iteration to circle about its most-visited place. Any of the termination requirements must be met for this loop to continue.
4.4 GBHN for Image Segmentation
The affected portion in the input medical images is segmented by the proposed GBHN method. This method is developed based on the combination of deep belief network along with the heap optimization. Deep belief network is a kind of deep learning algorithm and heap optimization is one of the recent natural inspired algorithms which are inspired from the corporative organization. The GBHN method has three different layers such as hidden, input as well as output layer. Each layers are separated as two section one is visible and another one is hidden. The visible layer enclosed with the input and output layer also, the hidden layer is enclosed with the hidden side of the network. Moreover, the learning parameters in the deep functions are optimized by the heap optimization. The visible node
The hidden and visible network energy part is denoted as
The weight rate of the segmentation node
where, the part of gradient
where, the number of iteration is represented as
where
where,
where,
where the learning rate is denoted as
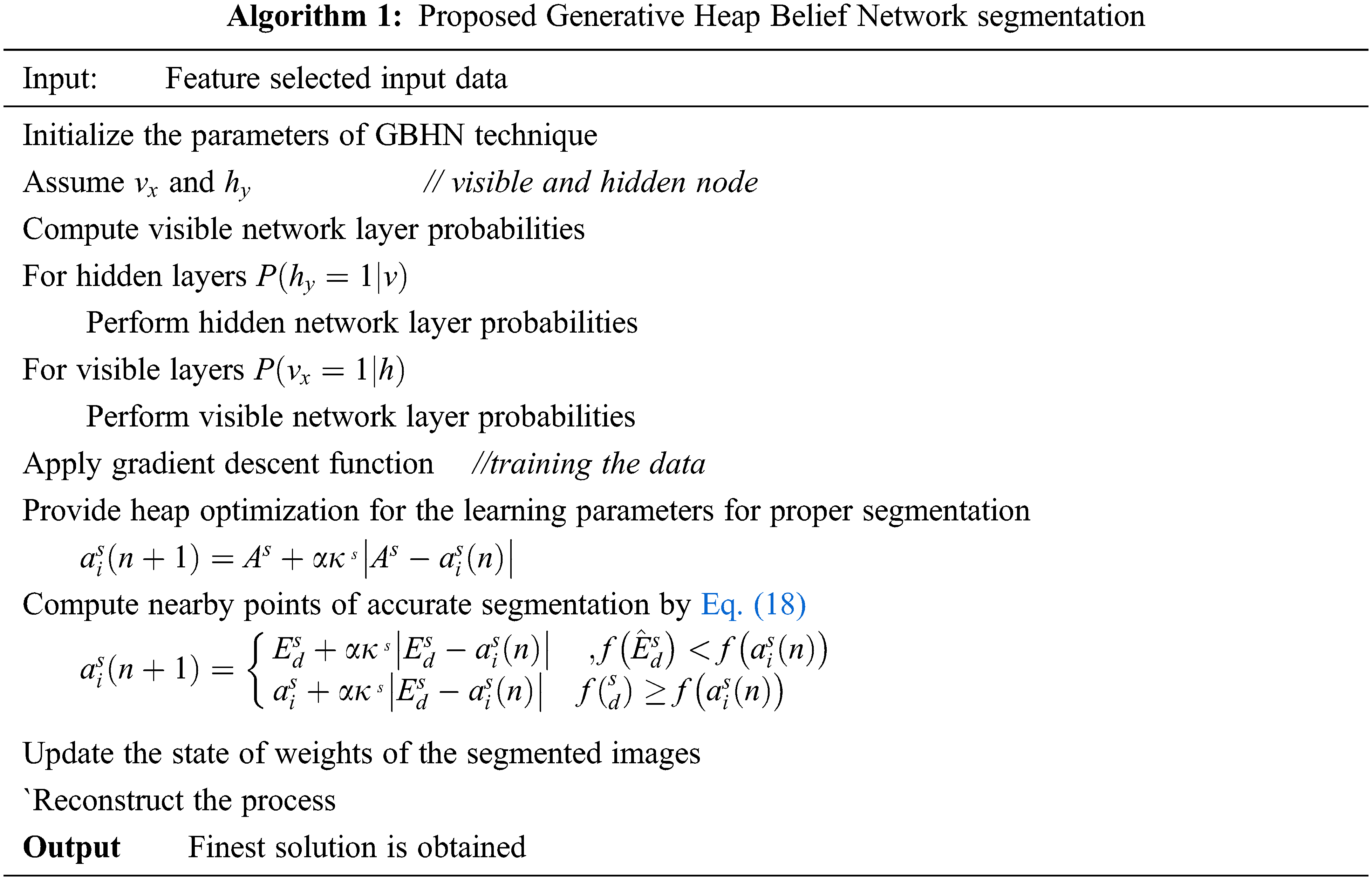
Then the consequences show the effective segmentation for medical images. The procedure for the proposed GBHN segmentation method is detailed in algorithm 1.
The flow chart of proposed medical image segmentation is illustrated in Fig. 3. The process of each phase in segmentation of medical images for the affected portion identification is explored till the final solution is achieved.
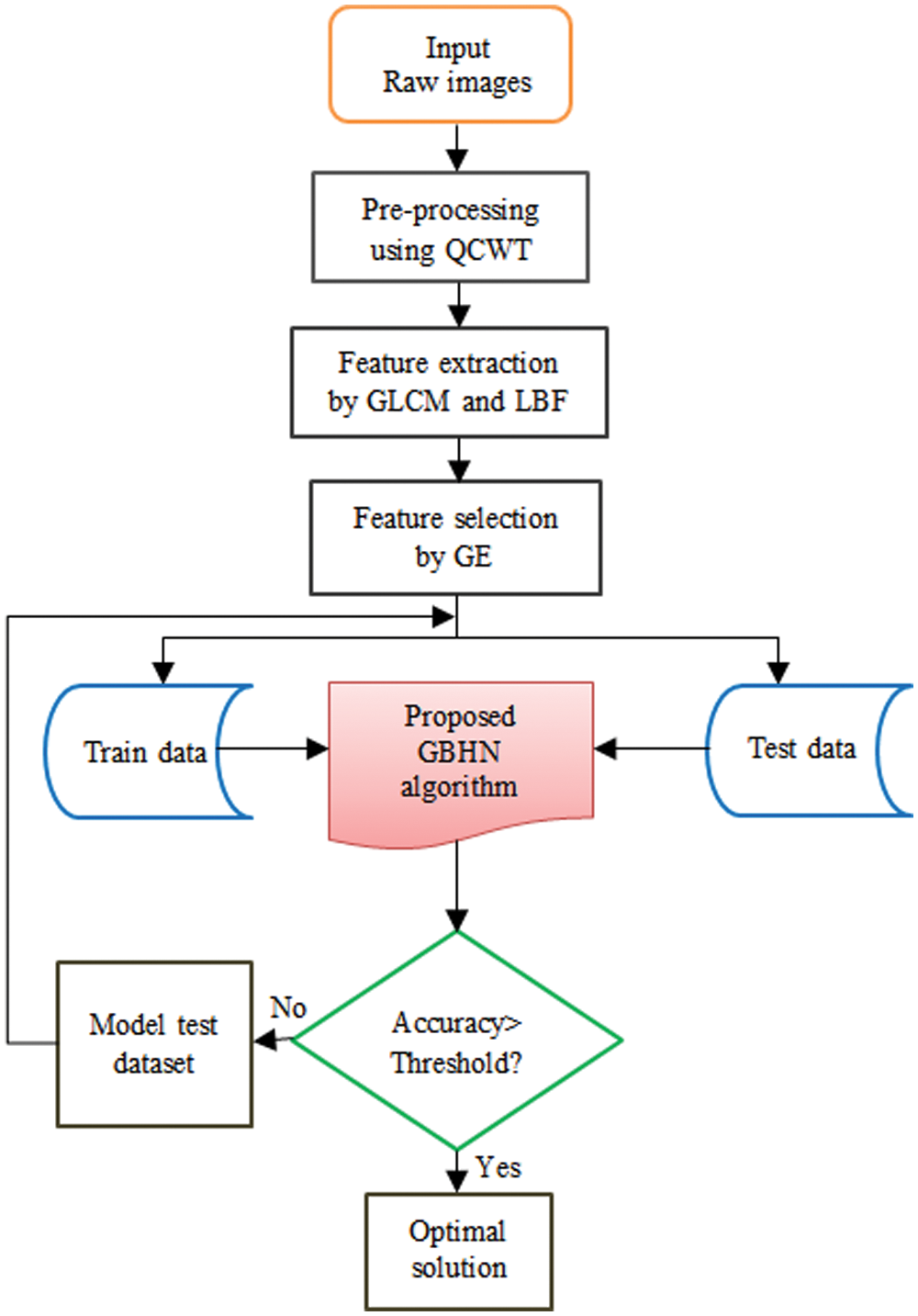
Figure 3: Flowchart of proposed medical image segmentation
This section is enclosed with the result and discussion of proposed approaches for medical image segmentation. The proposed method has been validated on the medical datasets of UCI machine learning 2017. The implementation of this work is carried out using MATLAB 2018b software in the windows platform. Moreover, the performance of the developed model is compared with the conventional methods for validating the efficiency of the proposed approaches in medical image segmentation.
The main aim of this research is to segment the tumors from the different phases using optimized deep learning methods. The performance of proposed system is estimated by various evaluation measures.
Also, the results from the developed method have been compared with the different conventional methods related to this research. The sample images collected from the data set for proper validations, which is illustrated in Fig. 4.
Figure 4: Sample medical images from the datasets (a) Brain tumor (b) Spinal cord tumor (c) Kidney tumor (d) Prostate and (e) Lung cancer
In the primary step of preprocessing stage takes place using the QCWT based noise reduction method. The noise from the images are removed by this approach. The accurate medical image segmentation is achieved by the proposed GHBN method. Then, the exact features are extracted using the GLCM and LBF approaches. Consequently, the optimal features are selected using the GEO method. At last, the GHBN method has segmented the medical images accurately. The achieved segmented results for the input medical images by the proposed GHBN method are illustrated in Fig. 5.
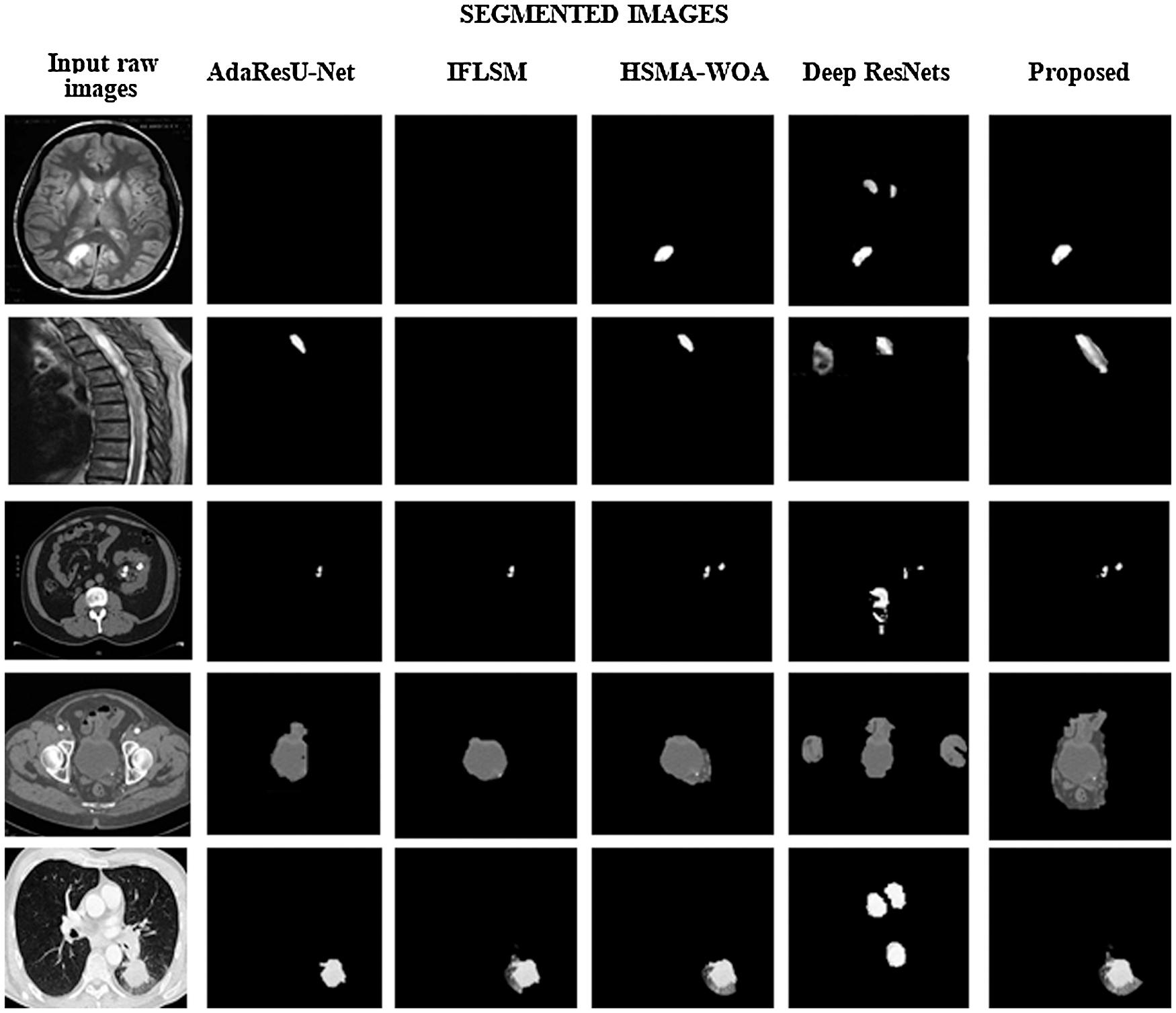
Figure 5: Comparitive analysis of segmented images
The comparative analysis is explained in this section. The proposed approach is compared with different conventional methods such as IFLSM [23], AdaResU-Net [24], WO-SMA [25] and Deep Residual Net- works (Deep ResNets) [26]. The comparision of different method and the segmented output is detailed in Fig. 5. The performance of the system is validated with the various parameters such as Accuracy, Sensitivity, Positive Predictive Value (PPV), Specificity, and Negative Predictive Value (NPV), False Negative value (FNV) and False Positive Value (FPV).
Specificity: The specificity value is estimated based on the relation of true negative amount to the overall of false positive and true negative values.
Sensitivity: The sensitivity value is defined as the relation of true positive numbers to the overall of false negatives as well as true positive values.
Accuracy: The value of accuracy for the medical image segmentation is estimated based on the metrics obtained from the specificity as well as sensitivity.
FPR: The FPR value is estimated based on the relation of false positive prediction to the overall negative values.
FNR: The FNR value is estimated based on the relation of false negative prediction to the overall negative values.
NPV: The value of NPV is defined as the relation of negative true value to the addition of overall negative false and true rate.
PPV: The value of PPV is estimated by the ratio of positive results obtained by the developed methodology.
Moreover, the Performance evaluations of segmentation results are detailed in Fig. 6. The conventional Deep ResNets has attained 0.92 accuracy, AdaResU-Net with 0.89 and HSMA-WOA with 0.90, which is less over the proposed method. However, the proposed method has attained superior performance than the traditional method; the suggested method has achieved 0.999 of accuracy. Similarly, the observation shows that the proposed method has attained more sensitivity and specificity than the earlier segmentation techniques.
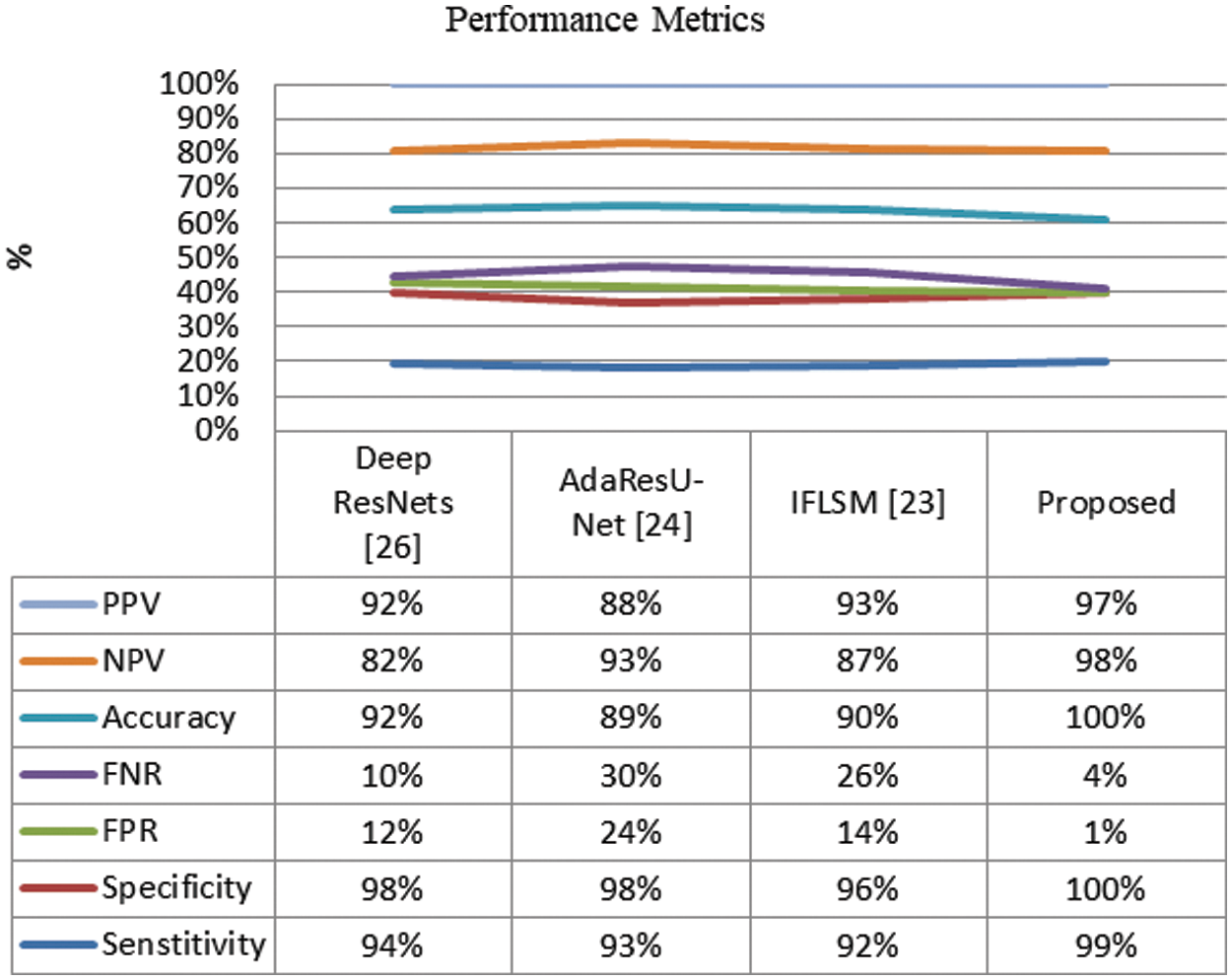
Figure 6: Performance evaluations of segmentation results
Furthermore, the FNR and FPR of the system is compared with the conventional methods like are illustrated in Fig. 6. It shows that the proposed system has attained more reliable than the earlier methods.
The observation shows that the conventional methods have certain limitation over the proposed method. The conventional Deep ResNets [26] method has consume more time over the other methods and also the AdaResU-Net [24] method has attained high skill but the computational problem of this method is more. Moreover, the IFLSM [23] has attained higher error rate over other methods. The reliability of the proposed system is very high compared with the earlier segmentation strategies. Thus, the overall comparative analysis demonstrates that the proposed method has achieved finest performance in image segmentation system while compared with the conventional methods. Also, it proves that the proposed method has consumed more accuracy, sensitivity, specificity and reduced false segmentation. Therefore, the results show the effective performance of proposed method in medical image segmentation.
The objective is to determine which part of the brain is afflicted from the input MRIs and CT pictures. This work proposes an optimized deep learning framework consisting of deep belief and heap optimization that achieves state-of-the-art segmentation performances while using the full training data and golden eagle optimization is used for feature selection to significantly reduce the burden of manual labeling in medical image segmentation tasks. It is the primary goal of this research to distinguish between the normal tissue around the damaged region and the abnormal tumor. Unique value measures are used to assess the display. As a result, this framework produces better results than current state-of-the-art segmentation performance.
Acknowledgement: The author would like to express her gratitude to King Khalid University, Saudi Arabia for providing administrative and technical support.
Funding Statement: This research is financially supported by the Deanship of Scientific Research at King Khalid University under research Grant Number (RGP.2/202/43).
Conflicts of Interest: The author declares that they have no conflicts of interest to report regarding the present study
1. Z. Gu, J. Cheng, H. Fu, K. Zhou, H. Hao et al., “CE-Net: Context encoder network for 2D medical image segmentation,” IEEE Transactions on Medical Imaging, vol. 38, no. 10, pp. 2281–2292, 2019. [Google Scholar]
2. Z. Zhou, M. M. Siddiquee, N. Tajbakhsh and J. Liang, “UNet++: A nested U-Net architecture for medical image segmentation,” Lecture Notes in Computer Science, vol. 11045, pp. 3–11, Springer, Cham. 2018. [Google Scholar]
3. G. Wang, W. Li, M. A. Zuluaga, R. Pratt, P. A. Patel et al., “Interactive medical image segmentation using deep learning with image-specific fine tuning,” IEEE Transactions on Medical Imaging, vol. 37, no. 7, pp. 1562–1573, 2018. [Google Scholar]
4. J. Cho, K. Lee, E. Shin, G. Choy and S. Do, “How much data is needed to train a medical image deep learning system to achieve necessary high accuracy? arXiv e-prints,” 2015. [Online]. Available: https://ui.adsabs.harvard.edu/#abs/2015arXiv151106348C. [Google Scholar]
5. T. Ren, H. Wang, H. Feng, C. Xu, G. Liu et al., “Study on the improved fuzzy clustering algorithm and its application in brain image segmentation,” Applied Soft Computing, vol. 81, pp. 105503, 2019. [Google Scholar]
6. E. A. Maksoud, M. lmogy and R. Awadi, “Brain tumor segmentation based on a hybrid clustering technique,” Egyptian Informatics Journal, vol. 16, no. 1, pp. 71–81, 2015. [Google Scholar]
7. M. H. Hesamian, W. Jia, X. He and P. Kennedy, “Deep learning techniques for medical image segmentation: Achievements and challenges,” Journal of Digital Imaging, vol. 32, no. 4, pp. 582–596, 2019. [Google Scholar]
8. J. Chang, L. Zhang, N. Gu, X. Zhang, M. Ye et al., “A mix-pooling CNN architecture with FCRF for brain tumor segmentation,” Journal of Visual Communication and Image Representation, vol. 58, no. 13, pp. 316–322, 2019. [Google Scholar]
9. N. Arunkumar, M. A. Mohammed, M. K. A. Ghani, D. A. Ibrahim, E. Abdulhay et al., “K-means clustering and neural network for object detecting and identifying abnormality of brain tumor,” Soft Computing, vol. 23, no. 19, pp. 9083–9096, 2019. [Google Scholar]
10. M. J. Sheller, G. A. Reina, B. Edwards, J. Martin and S. Bakas, “Multi-institutional deep learning modeling without sharing patient data: A feasibility study on brain tumor segmentation,” in Int. MICCAI Brain Lesion Workshop. Cham: Springer, 2018. [Google Scholar]
11. M. Mittal, L. M. Goyal, S. Kaur, I. Kaur, A. Verma et al., “Deep learning based enhanced tumor segmentation approach for MR brain images,” Applied Soft Computing, vol. 78, no. 10, pp. 346–354, 2019. [Google Scholar]
12. R. Thillaikkarasi and S. Saravanan, “An enhancement of deep learning algorithm for brain tumor segmentation using kernel based CNN with M-SVM,” Journal of Medical Systems, vol. 43, no. 4, pp. 1–7, 2019. [Google Scholar]
13. L. Li, X. Zhoa, W. Lu and S. Tan, “Deep learning for variational multimodality tumor segmentation in PET/CT,” Neurocomputing, vol. 392, pp. 277–295, 2020. [Google Scholar]
14. K. R. Laukamp, F. Thiele, G. Shakirin, D. Zopfs, A. Faymonville et al., “Fully automated detection and segmentation of meningiomas using deep learning on routine multiparametric MRI,” European Radiology, vol. 29, no. 1, pp. 124–132, 2019. [Google Scholar]
15. C. Chen, “Image segmentation for lung lesions using ant colony optimization classifier in chest CT,” in Advances in Intelligent Information Hiding and Multimedia Signal Processing. Cham: Springer, 2017. [Google Scholar]
16. H. Fang, H. Fan, S. Lin, Z. Qing and F. R. Sheykhahmad, “Automatic breast cancer detection based on optimized neural network using whale optimization algorithm,” International Journal of Imaging Systems and Technology, vol. 31, no. 1, pp. 425–438, 2021. [Google Scholar]
17. M. Sharif, J. Amin, M. Raza, M. Yasmin and S. C. Satapathy, “An integrated design of particle swarm optimization (PSO) with fusion of features for detection of brain tumor,” Pattern Recognition Letters, vol. 129, pp. 150–157, 2020. [Google Scholar]
18. M. A. Mohammed, M. K. A. Ghani, N. Arunkumar, R. I. Hamed, M. K. Abdullah et al., “A real time computer aided object detection of nasopharyngeal carcinoma using genetic algorithm and artificial neural network based on Haar feature fear,” Future Generation Computer Systems, vol. 89, pp. 539–547, 2018. [Google Scholar]
19. M. Balaji, S. Saravanan, M. Chandrasekar, G. Rajkumar and S. Kamalraj, “Analysis of basic neural network types for automated skin cancer classification using Firefly optimization method,” Journal of Ambient Intelligence and Humanized Computing, vol. 12, no. 7, pp. 7181–7194, 2021. [Google Scholar]
20. P. Sathish and N. M. Elango, “Exponential cuckoo search algorithm to radial basis neural network for automatic classification in MRI images,” Computer Methods in Biomechanics and Biomedical Engineering: Imaging & Visualization, vol. 7, no. 3, pp. 273–285, 2019. [Google Scholar]
21. T. Eelbode, J. Bertels, M. Berman, D. Vandermeulen, F. Maes et al., “Optimization for medical image segmentation: theory and practice when evaluating with Dice score or Jaccard index,” IEEE Transactions on Medical Imaging, vol. 39, no. 11, pp. 3679–3690, 2020. [Google Scholar]
22. M. B. Calisto and S. Yuen, “AdaEn-Net: An ensemble of adaptive 2D–3D fully convolutional networks for medical image segmentation,” Neural Networks, vol. 126, pp. 76–94, 2020. [Google Scholar]
23. R. Radha and R. Gopalakrishnan, “A medical analytical system using intelligent fuzzy level set brain image segmentation based on improved quantum particle swarm optimization,” Microprocessors and Microsystems, vol. 79, pp. 103283, 2020. [Google Scholar]
24. M. B. Calisto and S. Yuen, “AdaResU-Net: Multiobjective adaptive convolutional neural network for medical image segmentation,” Neurocomputing, vol. 392, pp. 325–340, 2020. [Google Scholar]
25. A. B. Mohamed, C. Victor and M. Reda, “HSMA_WOA: A hybrid novel Slime mould algorithm with whale optimization algorithm for tackling the image segmentation problem of chest X-ray images,” Applied Soft Computing, vol. 95, pp. 106642, 2020. [Google Scholar]
26. L. Shehab, O. M. Fahmy, S. M. Gasser and M. Mahallawy, “An efficient brain tumor image segmentation based on deep residual networks (ResNets),” Journal of King Saud University-Engineering Sciences, vol. 33, no. 6, pp. 404–412, 2020. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |