DOI:10.32604/iasc.2023.023474

| Intelligent Automation & Soft Computing DOI:10.32604/iasc.2023.023474 |  |
| Article |
Pre-Trained Deep Neural Network-Based Computer-Aided Breast Tumor Diagnosis Using ROI Structures
Department of Electronics and Communication Engineering, SRM Institute of Science and Technology, Kattankulathur, Tamilnadu, 603203, India
*Corresponding Author: Venkata Sunil Srikanth. Email: vm5484@srmist.edu.in
Received: 09 September 2021; Accepted: 09 February 2022
Abstract: Deep neural network (DNN) based computer-aided breast tumor diagnosis (CABTD) method plays a vital role in the early detection and diagnosis of breast tumors. However, a Brightness mode (B-mode) ultrasound image derives training feature samples that make closer isolation toward the infection part. Hence, it is expensive due to a meta-heuristic search of features occupying the global region of interest (ROI) structures of input images. Thus, it may lead to the high computational complexity of the pre-trained DNN-based CABTD method. This paper proposes a novel ensemble pre-trained DNN-based CABTD method using global- and local-ROI-structures of B-mode ultrasound images. It conveys the additional consideration of a local-ROI-structures for further enhancing the pre-trained DNN-based CABTD method’s breast tumor diagnostic performance without degrading its visual quality. The features are extracted at various depths (18, 50, and 101) from the global and local ROI structures and feed to support vector machine for better classification. From the experimental results, it has been observed that the combined local and global ROI structure of small depth residual network ResNet18 (0.8 in %) has produced significant improvement in pixel ratio as compared to ResNet50 (0.5 in %) and ResNet101 (0.3 in %), respectively. Subsequently, the pre-trained DNN-based CABTD methods have been tested by influencing local and global ROI structures to diagnose two specific breast tumors (Benign and Malignant) and improve the diagnostic accuracy (86%) compared to Dense Net, Alex Net, VGG Net, and Google Net. Moreover, it reduces the computational complexity due to the small depth residual network ResNet18, respectively.
Keywords: Computer-aided diagnosis; breast tumor; B-mode ultrasound images; deep neural network; local-ROI-structures; feature extraction; support vector machine
In the last 5 years, the diagnosis of breast tumor patient rate is going way high beyond the considerable number. Significantly, countries like America, Europe, Russia have launched many live screening centers to identify disabilities and also provide necessary treatment for patients carrying abnormalities. A breast tumor is a lump or mass in the breast which may happen due to irregular health issues includes late menopause, early menarche, obesity, etc. are common risk factors for breast cancer [1]. In 2020, the number of total cancer incidents was 32.1 million, of which 11.6% is due to breast cancer [2]. The estimated global burden of top-5 cancers has shown in Fig. 1. It covers significant women cancer types are breast, colorectum, lung, cervix uteri, and thyroid. It is inferred that breast cancer is the leading cancer type in women worldwide [3]. If one can detect nonmetastatic breast cancer early, then it is possible to cure breast cancer in almost 70 to 80% of the cases [4]. Hence, early diagnosis of breast cancer is needed to achieve potential benefits that are widely acceptable by combining physical examinations, imaging, and biopsy [5].
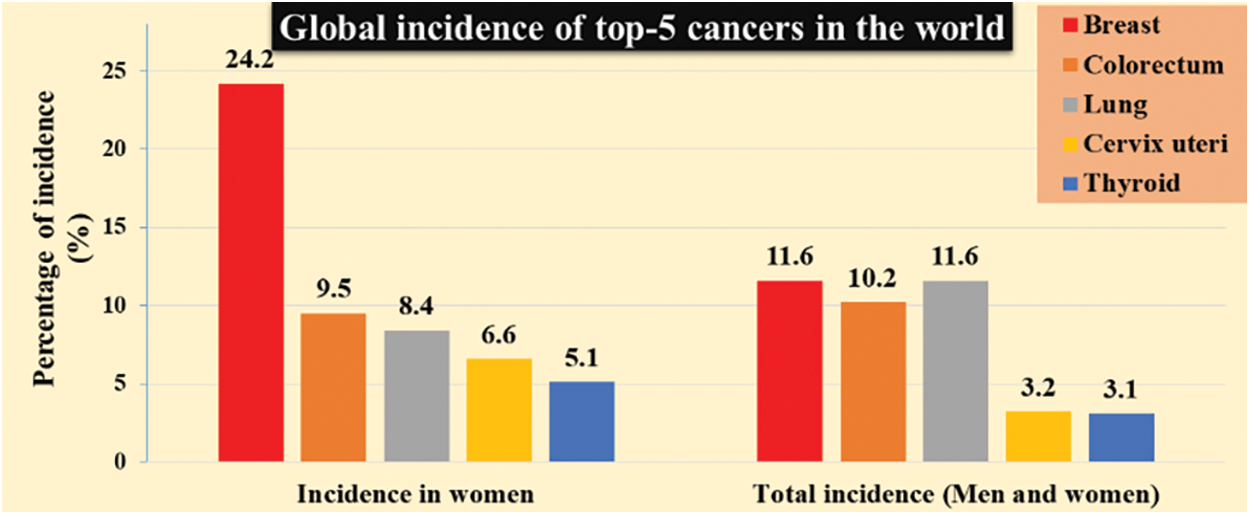
Figure 1: The global burden of cancer worldwide [2,3]
There are possible breast tumor screening methods are available such as magnetic resonance imaging (MRI), digital mammography, ultrasound, computerized tomography, breast-specific gamma imaging, positron emission tomography, and also thermography [6]. Initially, the standard screening method is adapted for breast cancer was mammography. However, it is ineffective for women with dense breasts [7]. Subsequently, MRI ultrasounds are often used with mammography to ensure better diagnostic performance [8]. Moreover, it is less expensive, convenient as well as less time-consuming in daily clinical practice. Still, it needs high human interactions to capture B-mode ultrasound images in a different position to acquire their desired structural content [9]. The acquisition process of such photos involves noise in the form of speckles as a consequence the quality of the acquired ultrasound image is mostly operator dependent. Recently, the computer-aided breast tumor diagnosis (CABTD) method has been used for the anomaly diagnostic process [10]. Later, Zonderland et al. [11] have concentrated only on specific growing regions with the help of the quasi-Monte Carlo method, which is a type of nonlinear mapping. It includes local adaptive algorithm operated under seed selection process collects the feature extraction by mapping fuzzy sets along with extended into accessing coordinate of the probability model involved in the expectation-maximization (EM) algorithm performs clustering operation [12]. However, its captures low-quality B-mode ultrasound images degrade the system performance that may be considered a limitation. To resolve this problem, machine learning-based CABTD methods are often preferred to achieve better diagnostic performance [13]. It is mainly target-oriented, classifying the given input ultrasound breast tumor image into either benign or malignant. Fig. 2 shows the sample of B-mode ultrasound images with benign and malignant lesions. It is noticed that the benign lesions have homogenous texture as well as border, but malignant lesions have heterogeneous textures also fuzzy borders [14]. However, it undergoes nonlinear adjustment of pixel intensity, which leads to appearing artifacts more over degrade the image’s visual quality. Currently, the use of deep neural networks (DNN) in CABTD methods has been increased. It has a transfer learning approach that is advantageous due to the minimum over-fitting issues in small datasets. However, more expensive computational resources for training (fine-tuning) have become a significant limitation to DNN-based CABTD methods [15]. It is observed that the depth of DNN in CABTD methods has been increasing to enhance the diagnostic performance [16–18]. However, increasing the depth of DNN further increases the computational complexity and the accuracy is strongly dependent on the depth of DNN. Therefore, developing a DNN-based CABTD method is required to improve accuracy without increasing the network depth. It is addressed by a possibility-based fuzzy approach that minimizes the noisy occupancy levels at the end of classification [19,20]. It improves the neighboring pixel intensity, which maintains the quality of the segmented portion. Further, enhanced the support vector machines (SVM) classification operation, fuzzy C-means (FCM) algorithm is presented along with updates its membership function by evaluating fuzzy factor and kernel weigh metric [21]. This paper presents a pre-trained DNN-based CABTD method using the state-of-art residual network with fewer layers (ResNet18). In contrast to existing CABTD methods [10,15,16], the proposed method combines local and global region of interest (ROI) structures to improve diagnostic accuracy. It consists of three stages, namely preprocessing, feature extraction, and classification. First, extract the local and global ROI structure from the preprocessing stage, followed by image resizing operation. The features are extracted from global- and local-ROI-structures using pre-trained ResNet18. The extracted features are then given to the support vector machine (SVM) for better classification. Effectively analyzing the proposed pre-trained DNN-based CABTD method is considering two publicly available ultrasound image class-balanced datasets such as BUSI [22] and UDIAT [23] datasets. The components are removed from the global and local ROI structures at various depths (18, 50, and 101) fed to support vector machine (SVM) for better classification.
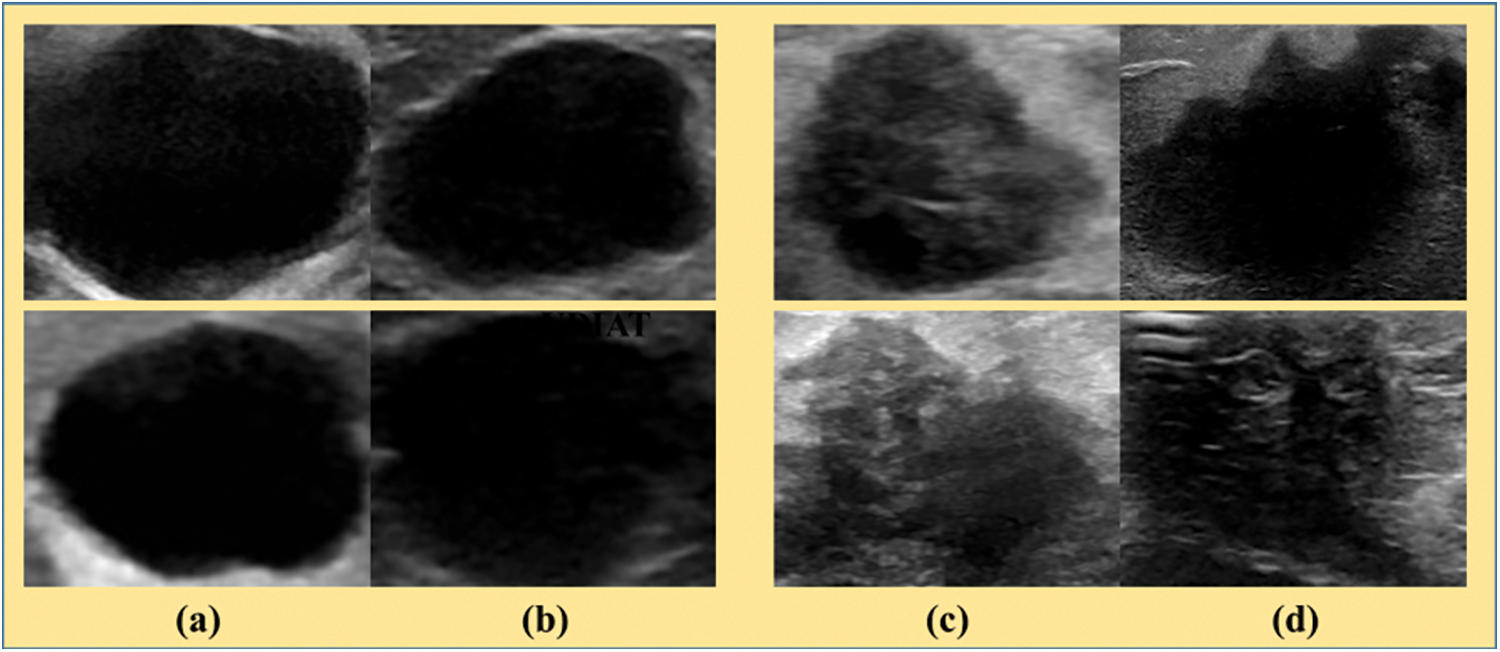
Figure 2: The B-mode ultrasound images with benign and malignant lesions. (a) Benign lesions from BUSI dataset [24], (b) Benign lesions from UDIAT dataset [25], (c) Malignant lesions from BUSI dataset [24] and (d) Malignant lesions from UDIAT dataset [25]
The main contribution of this paper is stated as follows: (i) It reduces the false-negative and false-positive problems while doing mammography screening tests. (ii) The pre-trained DNN-based CABTD method holds effective decomposition of the foreground as well as background region separation carried by DNN layers. Hence, it supports for early detection of breast cancer cell deformity levels. (iii) It is suitable for all the abnormal structural problems, especially, Benign and Malignant, associated with a mammography screening test. (iv) It provides an enhanced image of mammograms that accurately measures the abnormalities by extracting a feature vector from local and global ROI structures in B-mode ultrasound images. (v) It has a high capability of measuring infection spread. Moreover, it takes a quick recovery time to establish the newly infected parts compared with a previous track recorded. (vi) It contains hyperbolic regularization, which provides the suitable fuzzy sets for obtaining the best possible solution for pixel adjustment and image visual quality enhancement to standardize the predefined attribute levels. (vii) It induces membership grade modification by intuitionist fuzzy logic sets for accurate detection of infection spreads.
The rest of the paper is organized as: Section 2 describes the recent methodology adapted for breast tumor diagnostic analysis. In Section 3, the authors elaborate on the preprocessing, image resizing, feature extraction, and classification technique proposed in the research. Section 4 briefs the proposed method’s performance compared to two publicly available ultrasound image class-balanced datasets such as BUSI [22] and UDIAT [23] datasets availed in the literature. Finally, discussion, conclusion, and future scope are given in Sections 5 and 6, respectively.
The research on the computer-aided diagnosis of breast tumors using B-mode ultrasound images has been performed by several researchers. A detailed description of various CABTD methods from the last two decades is available [25,26]. The existing CABTD methods are broadly divided into two categories: traditional machine learning-based and pre-trained DNN-based strategies. The description of existing CABTD methods is as follows:
2.1 Traditional Machine Learning-Based CABTD Methods
The features are extracted manually from the input B-mode ultrasound images in traditional machine learning-based CABTD methods [10,19–23]. Then, the extracted features have been used to design classifiers such as SVM, backpropagation neural network (BPNN), etc. In Ref. [19,20], the authors used fractal analysis and k-means classification to diagnose breast tumors. The performance obtained in Ref. [19,20] is improved further by combining k-means classification with BPNN [10]. The independent morphological features have been extracted using principal component analysis (PCA) in Ref. [21]. It is observed that the extraction of morphological features is computationally expensive. To reduce computational complexity, the morphological solidity features associated with auto-covariance texture features have been extracted and then given to the SVM classifier for better classification [22]. In Ref. [23], the optimal set of textural, fractal, and histogram-based features are selected based on the stepwise regression method. The fuzzy SVM is then used to achieve better diagnostic performance. The genetic algorithm has been used to determine the significant auto-covariance texture and morphological features obtained from input ultrasound images [26]. The multiple instances learning-based CABTD method is proposed in [27].
The distribution of frequency components is analyzed using 1-D discrete wavelet transform to identify malignancy of breast tumor lesions in Ref. [28]. In Ref. [29], the authors have proposed a CABTD method using shear let-based texture features. The congruency-based binary patterns are extracted as features and then given to SVM in Ref. [30]. In Ref. [31], the authors have performed decision fusion using multiple-ROI nonoverlapping patches combined with morphological features obtained from input ultrasound images. The feature extraction methods such as textural-, fractal-, and morphological-based methods are combined to improve the diagnostic performance of CABTD methods in [32–34]. The multifractal dimension features are given to BPNN for better classification in Ref. [35]. In Ref. [36], the ROI regions segmented from the active contour-based and segmentation-based methods are then used to extract texture features.
2.2 Pre-Trained DNN-Based CABTD Methods
The use of manual feature extraction techniques in traditional machine learning-based CABTD methods [10,19–33] often requires domain expertise. In contrast to manual feature extraction techniques, the pre-trained DNN-based CABTD methods [16–18,37–43] extract the features automatically are effective and accurate when compared with traditional machine learning-based CABTD methods.
Several authors have proposed various CABTD methods using different DNN architectures [37]. The commonly used DNNs in B-mode ultrasound image-based CABTD methods are Alex Net, VGG, Google Net, ResNet, and Dense Net [37]. The transfer learning-based CABTD approach is proposed in Ref. [38]. In Ref. [39], histogram equalization has been used to enhance the input ultrasound images. Then the pre-trained Google Net is fine-tuned for better breast tumor diagnosis [39]. In Ref. [16], fine-tuned ResNet152 has been used to diagnose breast cancer using ultrasound images. The automatic feature extraction technique using pre-trained Google Net followed by the adaptive neuro-fuzzy classification is used for breast tumor diagnosis [40]. In Ref. [41], the effectiveness of the CABTD method using fine-tuned inception v2 has been verified subjectively in association with three human readers. The computer-aided diagnosis of breast cancer using an ensemble of DNNs such as VGG-like network, ResNet, and Dense Net is proposed [17]. In Refs. [42–49], the features extracted from the last three layers of pre-trained Alex Net are combined and then given an SVM classifier to improve the accuracy of the CABTD method. In Ref. [18], the features extracted from the global average pooling layer of pre-trained ResNet101 are then given to SVM for better diagnostic performance. Unlike existing DNN methods, a custom DNN-based CABTD method using a single convolutional layer has been proposed [42].
In this section, the proposed CABTD method is explained. The presented method consists of preprocessing, feature extraction, and classification stages, as shown in Fig. 3. The detailed description and illustration of each step involved in the proposed CABTD method are explained as follows:
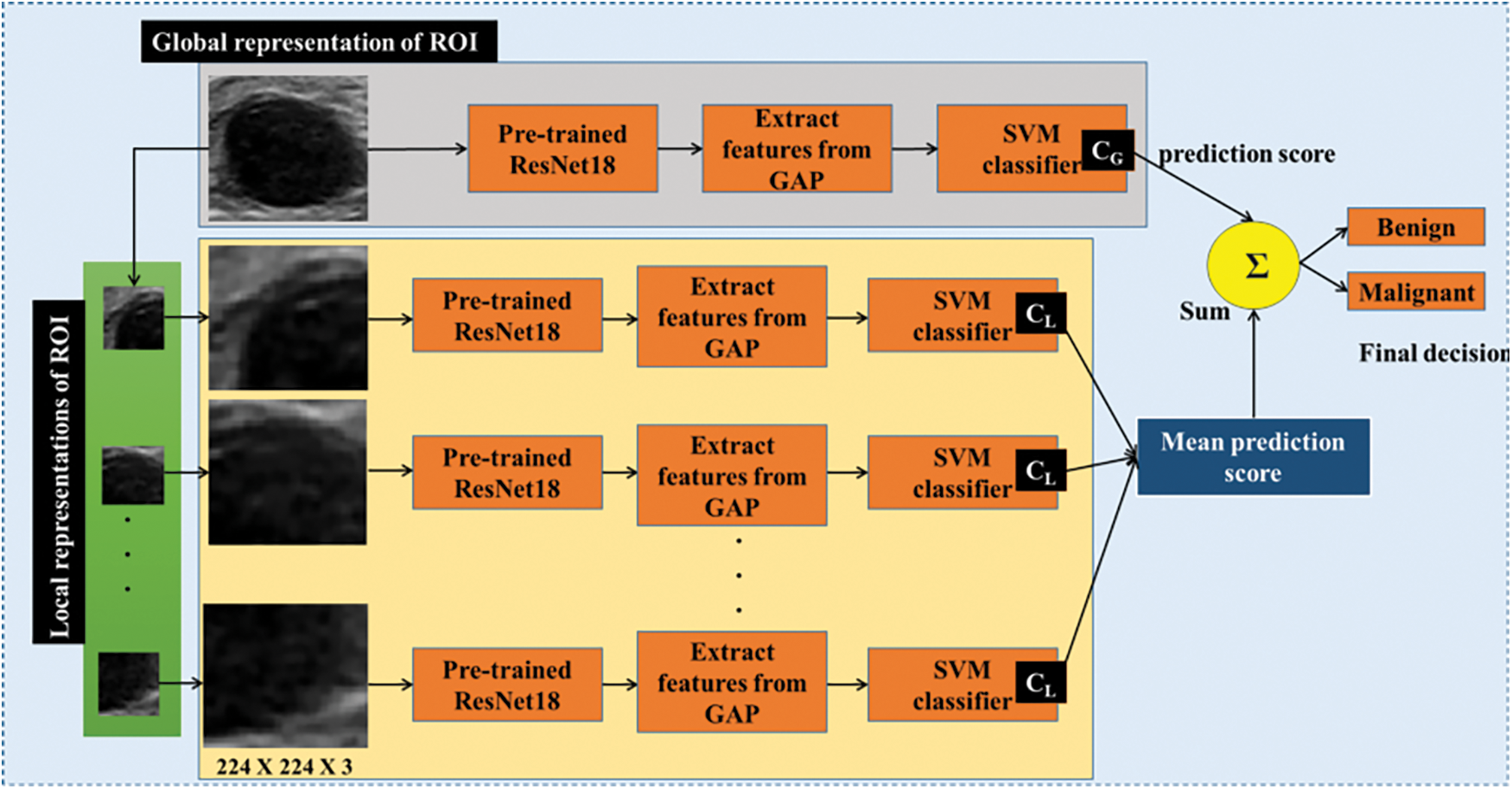
Figure 3: The framework of the proposed CABTD method
Let us consider two distinct and class-balanced datasets “and.” Where “and” represents the ROI B-mode ultrasound images of breast tumor in the spatial coordinates “u and.” Here, M and N denote the number of images in the datasets P and Q, respectively. In this work, datasets P and Q consist of ROI images (called global-ROI-structures) of size 224 × 224. Thereby, hidden information is continuously traced out and extracted feature vector sets from the hidden layers of the pre-trained DNN.
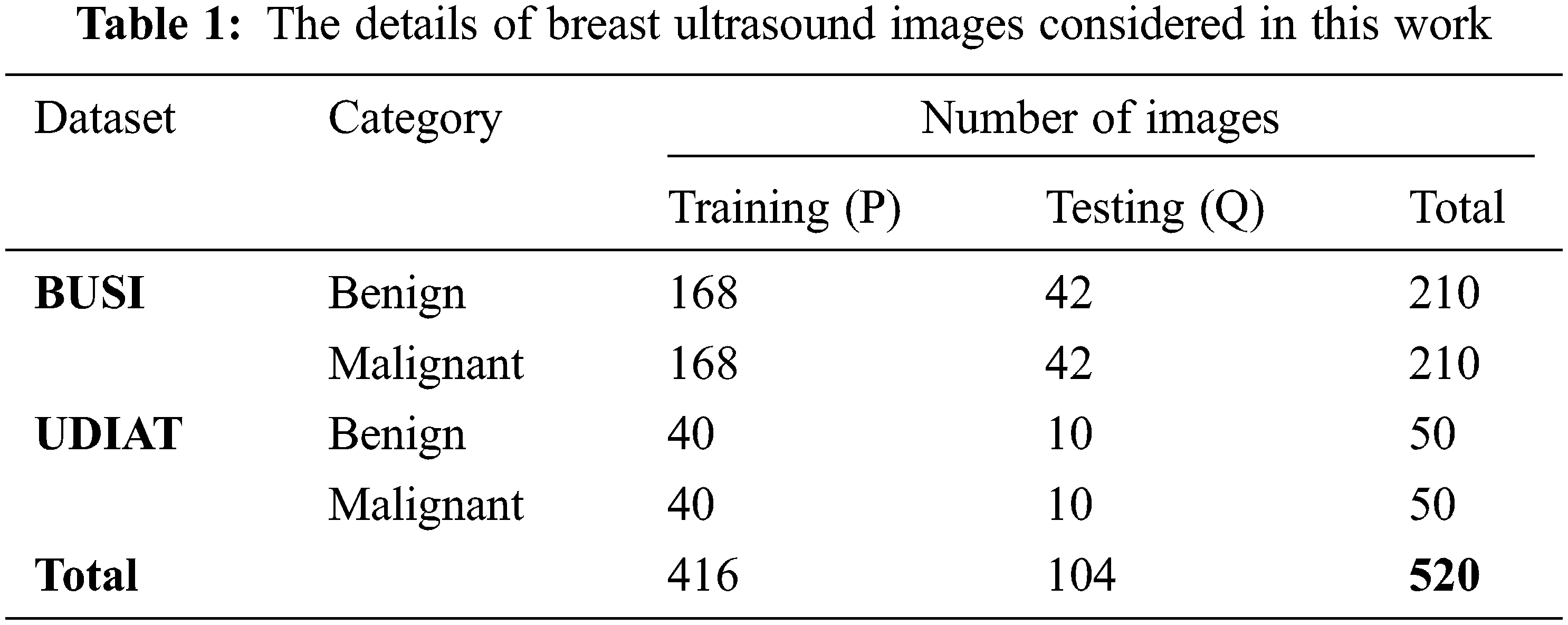
This work collected 520 B-mode ultrasound images from the publicly available datasets BUSI [22] and UDIAT [23]. In the preprocessing stage, extraction of ROI and image resizing are performed. The preprocessed ultrasound images are divided into 5-folds in which 4-folds are used in training (denoted with P) and the remaining 1-fold is used in the testing phase (indicated with Q). The details of datasets used in this work are presented in Tab. 1.
The preprocessing stage consists of ROI extraction (global-ROI-structure), image resizing, and the generation of local-ROI-structures from global-ROI-structures. The preprocessing steps involved in the proposed CABTD method are explained as follows:
3.2.1 Extraction of Global-ROI-Structure
The global-ROI structure is the ROI of a breast tumor cropped from the original ultrasound image, as shown in Fig. 4. The resolution of the original B-mode ultrasound images in the datasets BUSI [24] and UDIAT [25] are comparatively large compared to the exact ROI of breast tumors. As the detection of ROI from the original ultrasound image is crucial for better tumor diagnosis. However, in this paper, we focused mainly on breast tumor diagnosis and not on ROI detection. So, the ROIs from the original ultrasound images are extracted manually concerning available mask images using MATLAB crop tools. The process of removing the global-ROI structure is illustrated in Fig. 4.

Figure 4: The process of extracting global- and local-ROI-structures
3.2.2 Extraction of Local-ROI-Structures
The global-ROI-structure extracted from the original ultrasound image concerning mask image is further divided into multiple sub-ROIs called local-ROI-structures. Initially, the extracted global-ROI structure is resized to 128 × 128 for uniform analysis. Then, the 128 × 128 global-ROI structure is divided into multiple 64 × 64 ROIs overlapping as shown in Fig. 4. The extracted 64 × 64 sub-ROIs from the global-ROI-structures are termed as local-ROI-structures. In this work, each global-ROI structure consists of nine local-ROI structures, as illustrated in Fig. 4.
The extracted 128 × 128 global-ROI-structures and 64 × 64 local-ROI-structures are resized to 224 × 224 to meet the input image size requirement of the proposed CABTD approach. The extracted local-ROI structures from the corresponding global-ROI structure are then formed as datasets “and.” The dataset consists of local-structures corresponding to global-structures present in the dataset. Similarly, the dataset consists of local-structures corresponding to global-structures present in the dataset.
The proper feature extraction technique is highly required to solve problems like CABTD. In this work, the pre-trained residual network with 18 layers (ResNet18) [50] is considered for feature extraction. The pre-trained ResNet18 is initially trained with the ImageNet [51] natural image database against 1000 classes. The weights or activations of the pre-trained ResNet18 are used as features in the proposed CABTD method. The pre-trained ResNet18 consists of 18 layers, including convolutional layers and the global average pooling (GAP) layer. The architecture of the ResNet18 is shown in Fig. 5.

Figure 5: The architecture of ResNet18
The GAP layer in the ResNet18 contains 512 activations, which are used as features in this work. The features that are extracted from datasets
In this work, two SVM classifiers such as

Figure 6: The training process of SVM classifiers
In the classification stage, the binary SVM classifier [49] is used. The binary SVM classifier is designed by solving the optimization problem is:
Subject to
Here, the weight vector is denoted by
The proposed CABTD method is evaluated using the datasets “and.” In Fig. 3, the global-ROI structure is applied to the proposed CABTD method, along with its local-ROI structure. Then, the global-ROI-structure is classified using and the corresponding local-ROI-structures are classified using CABTD. The average of decision scores obtained from local-ROI structures is added with the decision score obtained from the corresponding global-ROI structure to get the final decision, as shown in Fig. 3. The evaluation metrics such as accuracy, precision, and recall are used in the evaluation. The expressions of the evaluation metrics are given as.
where,
In this section, we describe the experimental setup, data characteristics, and performance of the proposed method. The effectiveness of the proposed method is illustrated by comparing the proposed method with pre-trained DNN-based CABTD methods and existing methods in the literature.
All the experiments are performed using a personal computer with specifications; Intel Core-i7, 16 GB RAM, and 64-bit operating system. The software platform MATLAB R2019b is chosen to validate experiments. Using deep learning toolboxes and statistics and machine learning toolboxes, the proposed method is implemented and then executed. The existing CABTD methods are re-executed using the same dataset used in this work without changing the hyperparameter setting.
4.2 Characteristics of the Dataset
In this paper, the proposed method is evaluated using two open-access available datasets such as BUSI [24] and UDIAT [25]. The original images in the datasets are initially cropped exactly to match breast lesions, using mask images available with the collected dataset and the class balance is maintained by randomly considering the equal number of images per class from each dataset to evaluate the proposed method effectively. The details of the collected dataset are presented in Tab. 2.
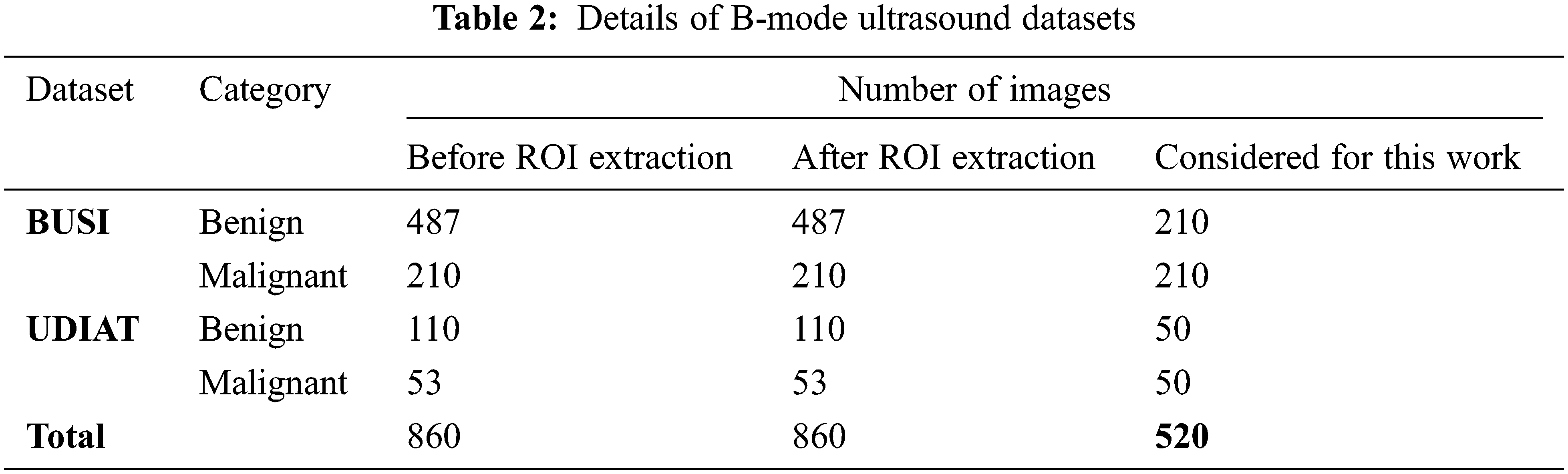
The dataset BUSI [22] contains 780 B-mode ultrasound images in total, in which 487 images with a benign tumor, 210 images with a malignant tumor, and 133 normal (no tumor) images. The ultrasound systems used for this dataset are LOGIQ E9 and LOGIQ E9 Agile with 1–5 MHz ML6-15-D Matrix linear probe transducer [52]. The original resolution of the images using the LOGIQ E9 and LOGIQ E9 Agile ultrasound systems is 1280 × 1024. However, dataset images are saved in portable network graphics format with an average resolution of 500 × 500.
Another dataset considered in this work is UDIAT [23], consisting of 53 malignant and 110 benign B-mode ultrasound breast tumor images. The ultrasound system used for this dataset is Siemens ACUSON. This dataset has belonged to the UDIAT Diagnostic Centre of the Parc Tauli Corporation, Sabadell (Spain). The principal authors, Dr. Robert Marti and Dr. Moi Hoon Yap used this dataset to develop a deep-learning-based method for breast tumor segmentation [53,54]. The request has been made to these authors to permit the use of this dataset for this work.
4.3 Performance of Proposed Method
The proposed CABTD method intends to improve the diagnostic accuracy using residual networks with small depth. For this purpose, the residual network ResNet18 is chosen in this work. To achieve enhanced performance, local-ROI structures are incorporated into the design. The performance of the proposed approach using residual networks at depths 18, 50, and 101 are analyzed using global-ROI-structure and local-ROI-structures exclusively, as shown in Tab. 3.
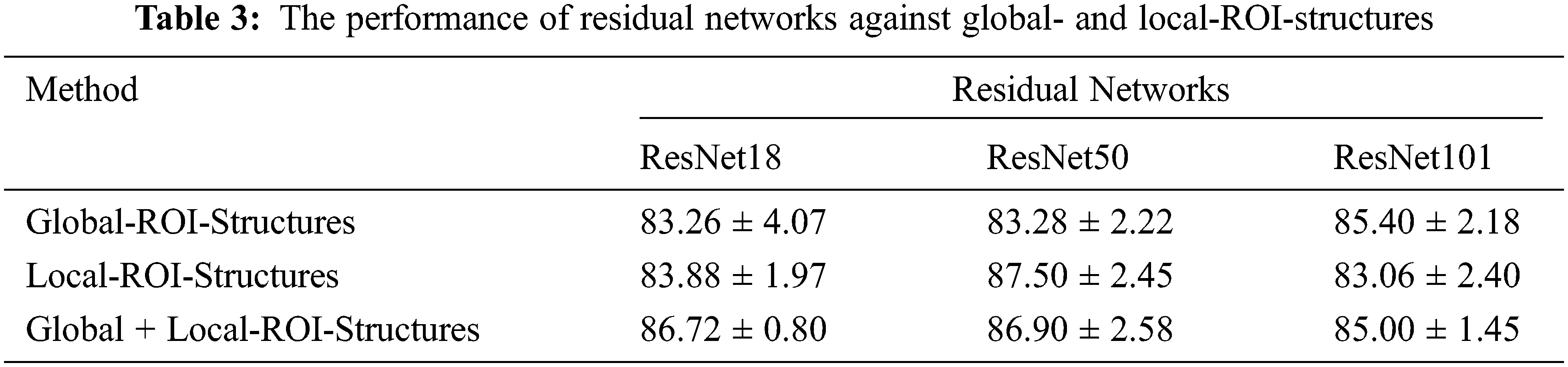
From Tab. 3, it is clear that the performance of global-ROI-based CABTD methods in terms of accuracy is increasing with respect to the depth of the residual network. The proposed CABTD approach is evaluated using 5-fold cross-validation and the confusion matrices for each fold are provided in Fig. 7 for reference.

Figure 7: The confusion matrices of the proposed method in (a) Fold 1, (b) Fold 2, (c) Fold 3, (d) Fold 4, and (e) Fold 5
Similarly, the performance of CABTD methods using local-ROI-structures is significantly good when compared with global-ROI-structure-based methods. However, the combination of global- and local-ROI-structures improved the performance considerably for small-depth networks compared to higher-depth networks. It is observed that there is no significant improvement in the performance of the proposed method using ResNet101. However, the proposed method using ResNet18 showed better performance than global-ROI-based methods and achieved effectiveness in terms of computational complexity and accuracy.
4.4 Comparison with Existing Methods
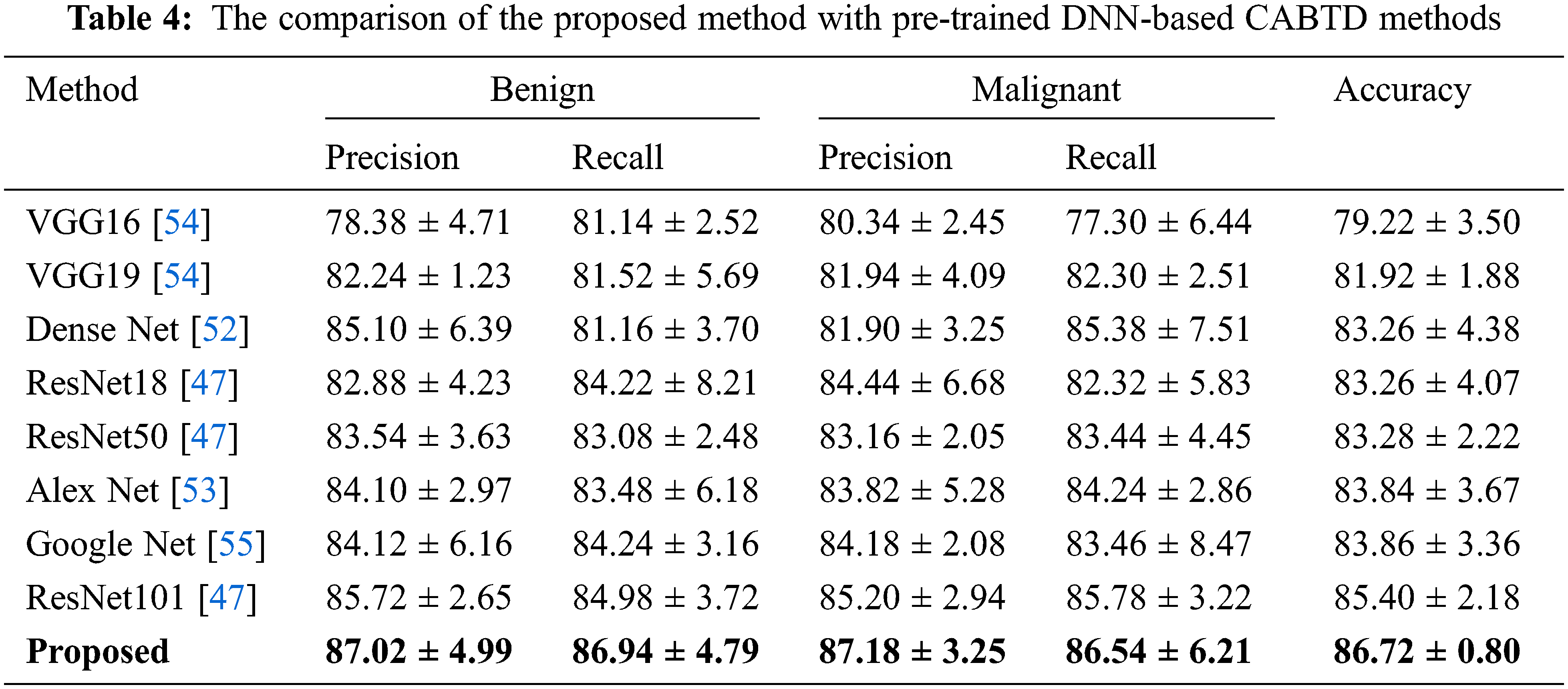
The proposed method is compared with pre-trained DNN-based CABTD methods such as ResNet [50], Dense Net [52], Alex Net [53], VGG Net [54], and Google Net [55]. The comparison of the proposed CABTD method with pre-trained DNN-based CABTD methods is shown in Tab. 4.
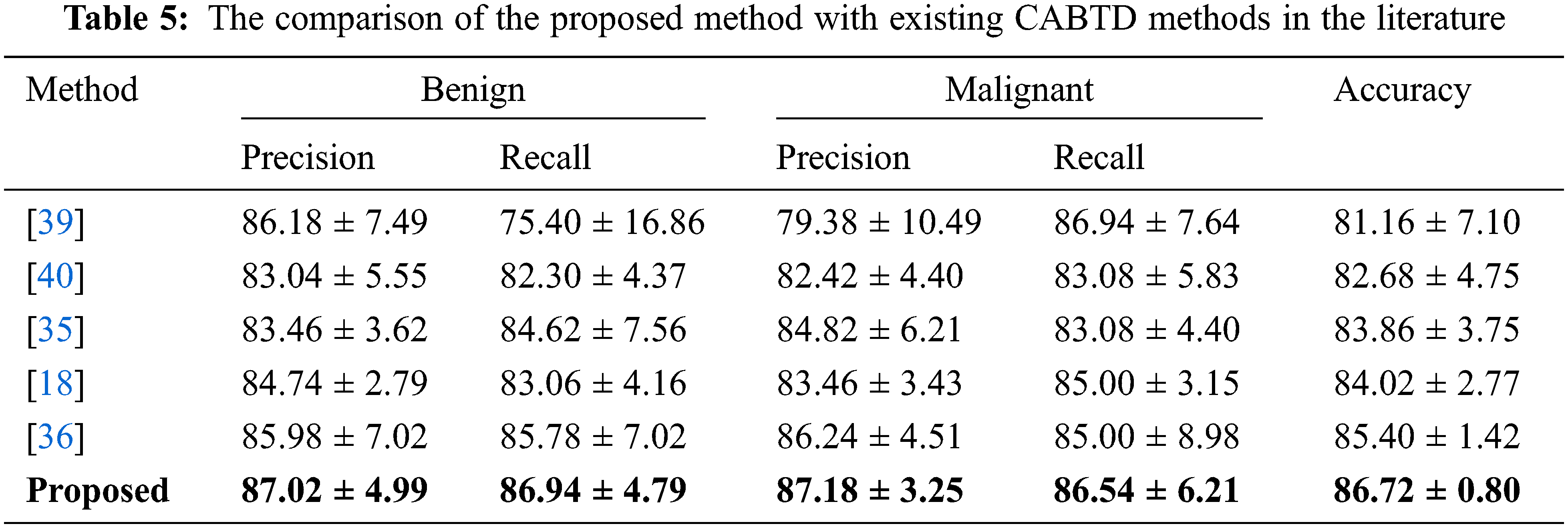
The proposed CABTD method is compared with existing methods in the literature. The performance comparison in terms of precision, recall, and accuracy with the existing CABTD methods is shown in Tab. 5.
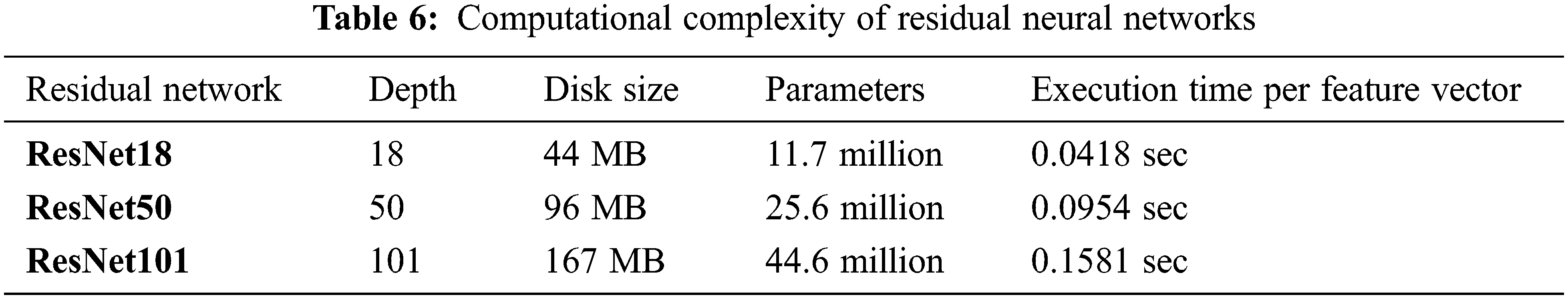
The computational complexity of the proposed CABTD technique is evaluated for residual networks ResNet18, ResNet50, and ResNet101 in terms of depth, disc size, parameters, and execution time per feature vector, as shown in Tab. 6. ResNet18’s computational complexity is minimal when compared to ResNet50 and ResNet101, as seen in Tab. 6. As a result, the suggested CABTD approach takes into account the residual network ResNet18.
The manual feature extraction techniques often extract texture-, fractal-, and also morphological-based features from medical ultrasound images [10,19–33,46]. However, the use of manual feature extraction techniques involves the knowledge of expertise. Hence, DNN-based CABTD methods [39] are preferred. In DNN-based CABTD methods, the features are extracted automatically compared to manual feature extraction, thereby achieving better diagnostic performance. The DNN-based CABTD methods are only effective with the following major requirements.
1. Large annotated image dataset to avoid over-fitting and convergence issues.
2. Huge computational resources for the training process.
In practice, the collection of a large annotated dataset is tedious and costly. The pre-trained DNN-based CABTD methods [16–18,34–38,40] use the pre-trained networks with a large dataset such as ImageNet. This strategy is known as transfer learning. The training of pre-trained DNN (transfer learning with fine-tuning) requires a sufficient amount of training data for proper convergence with less error rate. Using pre-trained DNN directly (transfer learning without fine-tuning) as a feature extraction technique followed by a traditional classifier such as SVM is beneficial and more applicable to the problem of breast tumor diagnosis where the availability of a dataset is scarce.
The use of local-ROI-structures in association with the networks of small depth such as ResNet18 is advantageous in terms of accuracy and computational complexity. Hence, the proposed CABTD method extracts feature from global- and local-ROI-structures using pre-trained ResNet18, thereby achieving better diagnostic performance with less computational resources.
Fig. 8 shows the ROC interpretation curve of the proposed pre-trained DNN-based CABTD method on B-mode ultrasound image ResNet18 datasets matched with BUSI [22] and UDIAT [23], respectively. It is identified that the ROC value of 0.7906 is obtained by comparing both the expected and predicted threshold value (true-positive rate [TPR] and false-positive rate [FPR]) of the PSO-RNN. It is observed in Fig. 8(c). Similarly, the ROC values of 0.7407 and 0.7889 are obtained for BUSI [22] and UDIAT [23]. It is conveyed that TPR and FPR show significant improvement. The trade-off ratio between two publicly available ultrasound image class-balanced datasets is observed in Figs. 8(a) and 8(b), respectively.

Figure 8: ROC interpretation curve of different class-balanced datasets (a) BUSI [41], (b) UDIAT [42], and (c) ResNet18 [proposed]
The computer-aided diagnosis method for breast tumors using B-mode ultrasound images is proposed in this paper. The presented method effectively utilized the global- and local-ROI structures achieved better accuracy and computational complexity. The analysis of residual neural networks at various depths 18, 50, and 101 are performed and tabulated exclusively. The performance of the proposed CABTD method is evaluated experimentally verified using a class-balanced dataset. From the experimental results, it is clear that the proposed CABTD method is effective in terms of accuracy and computational complexity compared with existing literature methods.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. T. J. Key, P. K. Verkasalo and E. Banks, “Epidemiology of breast cancer,” Lancet Oncology, vol. 2, no. 3, pp. 133–140, 2001. [Google Scholar]
2. J. Ferlay, “Estimating the global cancer incidence and mortality in 2018 GLOBOCAN sources and methods GLOBOCAN 2018 sources and methods,” International Journal of Cancer, vol. 144, no. 8, pp. 1941–1953, 2019. [Google Scholar]
3. WHO, “WHO report on cancer: Setting priorities, investing wisely and providing care for all,” World Health Organization, 2020. [Online]. Available: https://www.who.int/publications/i/item/who-report-on-cancer-setting-priorities-investing-wisely-and-providing-care-for-all. [Accessed: 03-Sep-2021]. [Google Scholar]
4. N. Harbeck, “Breast cancer,” Nature Reviews Disease Primers, vol. 5, no. 1, pp. 66, 2019. [Google Scholar]
5. L. J. Esserman, “New approaches to the imaging, diagnosis and biopsy of breast lesions,” Cancer Journal, vol. 8, pp. S1–14, 2002. [Google Scholar]
6. S. Iran Makani, “A review of various modalities in breast imaging: Technical aspects and clinical outcomes,” Egyptian Journal of Radiology and Nuclear Medicine, vol. 51, no. 1, pp. 1–22, 2020. [Google Scholar]
7. P. Autier and M. Boniol, “Mammography screening: A major issue in medicine,” European Journal of Cancer, vol. 90, no. 10, pp. 34–62, 2018. [Google Scholar]
8. M. L. Giger, “Computerized analysis of images in the detection and diagnosis of breast cancer,” Seminars in Ultrasound, CT and MRI, vol. 25, no. 5, pp. 411–418, 2004. [Google Scholar]
9. K. Zheng, T.-F. Wang, J.-L. Lin and D.-Y. Li, “Recognition of breast ultrasound images using a hybrid method,” in IEEE/ICME Int. Conf. on Complex Medical Engineering, Beijing, pp. 640–643, 2007. [Google Scholar]
10. K. Doi, “Computer-aided diagnosis in medical imaging: Historical review, current status and future potential,” Computerized Medical Imaging and Graphics, vol. 31, no. 4–5, pp. 198–211, 2007. [Google Scholar]
11. H. M. Zonderland, E. G. Coerkamp, J. Hermanus, M. J. van de Vijver and A. E. van Voorthuisen, “Diagnosis of breast cancer: Contribution of US as an adjunct to mammography,” Radiology, vol. 213, no. 2, pp. 413–422, 1999. [Google Scholar]
12. E. Y. Chae, H. H. Kim, J. H. Cha, H. J. Shin, H. Kim et al., “Evaluation of screening whole-breast sonography as a supplemental tool in conjunction with mammography in women with dense breasts,” Journal of Ultrasound in Medicine, vol. 32, no. 9, pp. 1573–1578, 2013. [Google Scholar]
13. L. J. Brattain, B. A. Telfer, M. Dhyana, J. R. Grajo and A. E. Samir, “Machine learning for medical ultrasound: Status methods and future opportunities,” Abdominal Radiology (New York), vol. 43, no. 4, pp. 786–799, 2018. [Google Scholar]
14. D. R. Chen, R. Chang and Y. L. Huang, “Computer-aided diagnosis applied to US of solid breast nodules by using neural networks,” Radiology, vol. 213, no. 2, pp. 407–412, 1999. [Google Scholar]
15. H. Greenspan, B. van Ginnken and R. M. Summers, “Guest editorial deep learning in medical imaging: Overview and future promise of an exciting new technique,” IEEE Transactions on Medical Imaging, vol. 35, no. 5, pp. 1153–1159, 2016. [Google Scholar]
16. X. Xie, F. Shi, J. Niu and X. Tang, “Breast ultrasound image classification and segmentation using convolutional neural networks,” in Advances in Multimedia Information Processing. Cham: Springer International Publishing, pp. 200–211, 2018. [Google Scholar]
17. W. K. Moon, Y.-W. Lee, H.-H. Kee, S. H. Lee, C.-S. Huang et al., “Computer-aided diagnosis of breast ultrasound images using ensemble learning from convolutional neural networks,” Computer Methods and Programs in Biomedicine, vol. 190, pp. 105361, 2020. [Google Scholar]
18. W.-C. Shia and D.-R. Chen, “Classification of malignant tumors in breast ultrasound using a pretrained deep residual network model and support vector machine,” Computerized Medical Imaging and Graphics, vol. 87, pp. 101829, 2021. [Google Scholar]
19. R.-F. Chang, C.-J. Chen, M.-F. Ho, D.-R. Chen, W. K. Moon et al., “Breast ultrasound image classification using fractal analysis,” in Fourth IEEE Sym. on Bioinformatics and Bioengineering, Taichung, pp. 100–107, 2004. [Google Scholar]
20. D. R. Chen, “Classification of breast ultrasound images using fractal feature,” Clinical Imaging, vol. 29, no. 4, pp. 235–245, 2005. [Google Scholar]
21. Y.-L. Huang, D.-R. Chen, Y.-R. Jiang, S.-J. Kuo, H.-K. Wu et al., “Computer-aided diagnosis using morphological features for classifying breast lesions on ultrasound,” Ultrasound in Obstetrics and Gynecology, vol. 32, no. 4, pp. 565–572, 2008. [Google Scholar]
22. W. Al-Dhabyani, M. Gomaa, H. Khaled and A. Fahmy, “Dataset of breast ultrasound images,” Data in Brief, vol. 28, pp. 104863, 2020. [Google Scholar]
23. M. H. Yap, “Automated breast ultrasound lesions detection using convolutional neural networks,” IEEE Journal of Biomedical Health Informatics, vol. 22, no. 4, pp. 1218–1226, 2018. [Google Scholar]
24. S. Han, “A deep learning framework for supporting the classification of breast lesions in ultrasound images,” Physics in Medicine and Biology, vol. 62, no. 19, pp. 7714–7728, 2017. [Google Scholar]
25. J. V. Kriti and R. Agarwal, “Deep feature extraction and classification of breast ultrasound images,” Multimedia Tools Applications, vol. 79, no. 37–38, pp. 27257–27292, 2020. [Google Scholar]
26. D.-R. Chen and Y.-H. Hsiao, “Computer-aided diagnosis in breast ultrasound,” Journal of Medical Ultrasound, vol. 16, no. 1, pp. 46–56, 2008. [Google Scholar]
27. K. He, X. Zhang, S. Ren and J. Sun, “Deep residual learning for image recognition,” in IEEE Conf. on Computer Vision and Pattern Recognition (CVPR), Las Vegas, pp. 2818–2826, 2016. [Google Scholar]
28. J. Deng, W. Dong, R. Socher, L.-J. Li, L. Fei-Fei et al., “ImageNet A large-scale hierarchical image database,” in IEEE Conf. on Computer Vision and Pattern Recognition, Miami, pp. 248–255, 2009. [Google Scholar]
29. W.-J. Wu and W. K. Moon, “Ultrasound breast tumor image computer-aided diagnosis with texture and morphological features,” Academic Radiology, vol. 15, no. 7, pp. 873–880, 2008. [Google Scholar]
30. X. Shi, H. D. Cheng, L. Hu, W. Ju, J. Tian et al., “Detection and classification of masses in breast ultrasound images,” Digital Signal Processing, vol. 20, no. 3, pp. 824–836, 2010. [Google Scholar]
31. H. D. Cheng, J. Shan, W. Ju, Y. Guo, L. Zhang et al., “Automated breast cancer detection and classification using ultrasound images: A survey,” Pattern Recognition, vol. 43, no. 1, pp. 299–317, 2010. [Google Scholar]
32. W.-J. Wu, S.-W. Lin and W. K. Moon, “Combining support vector machine with genetic algorithm to classify ultrasound breast tumor images,” Computerized Medical Imaging and Graph, vol. 36, no. 8, pp. 627–633, 2012. [Google Scholar]
33. J. Ding, H. D. Cheng, J. Huang, J. Liu, Y. Zhang et al., “Breast ultrasound image classification based on multiple-instance learning,” Journal of Digital Imaging, vol. 25, no. 5, pp. 620–627, 2012. [Google Scholar]
34. H.-W. Lee, B.-D. Liu, K.-C. Hung, S.-F. Lei and P.-C. Wang, “Breast tumor classification of ultrasound images using wavelet-based channel energy and ImageJ,” IEEE Journal of Selected Topics Signal Processing, vol. 3, no. 1, pp. 81–93, 2009. [Google Scholar]
35. S. Zhou, J. Shi, J. Zhu, Y. Cai and R. Wang, “Shear let-based texture feature extraction for classification of breast tumor in ultrasound image,” Biomedical Signal Processing and Control, vol. 8, no. 6, pp. 688–696, 2013. [Google Scholar]
36. L. Cai, X. Wang, Y. Wang, Y. Guo, J. Yu et al., “Robust phase-based texture descriptor for classification of breast ultrasound images,” Biomedical Engineering Online, vol. 14, no. 1, pp. 26, 2015. [Google Scholar]
37. M. Abdel-Nasser, J. Melendez, A. Moreno, O. A. Omer, D. Puig et al., “Breast tumor classification in ultrasound images using texture analysis and super-resolution methods,” Engineering Application of Artificial Intelligence, vol. 59, no. 7, pp. 84–92, 2017. [Google Scholar]
38. T. Prabhakar and S. Poonguzhali, “Automatic detection and classification of benign and malignant lesions in breast ultrasound images using texture morphological and fractal features,” in 10th Biomedical Engineering Int. Conf., Hokkaido, pp. 1–5, 2017. [Google Scholar]
39. M. Wei, “A benign and malignant breast tumor classification method via efficiently combining texture and morphological features on ultrasound images,” Computational and Mathematics Methods in Medicine, vol. 2020, pp. 5894010, 2020. [Google Scholar]
40. M. A. Mohammed, B. Al-Khateeb, A. N. Rashid, D. A. Ibrahim, M. K. Abd Ghani et al., “Neural network and multi-fractal dimension features for breast cancer classification from ultrasound images,” Computers & Electrical Engineering, vol. 70, no. 1, pp. 871–882, 2018. [Google Scholar]
41. S. Pavithra, R. Vanitha Mani and J. Justin, “Computer aided breast cancer detection using ultrasound images,” Materials Today: Proceedings, vol. 33, no. 4, pp. 4802–4807, 2020. [Google Scholar]
42. Z. Cao, L. Duan, G. Yang, T. Yue and Q. Chen, “An experimental study on breast lesion detection and classification from ultrasound images using deep learning architectures,” BMC Medical Imaging, vol. 19, no. 1, pp. 51, 2019. [Google Scholar]
43. B. Huynh, K. Drunker and M. Giger, “MO-DE-207B-06 computer-aided diagnosis of breast ultrasound images using transfer learning from deep convolutional neural networks,” Medical Physics, vol. 43, no. 6 Part30, pp. 3705, 2016. [Google Scholar]
44. T. Fujioka, “Distinction between benign and malignant breast masses at breast ultrasound using deep learning method with convolutional neural network,” Japanese Journal of Radiology, vol. 37, no. 6, pp. 466–472, 2019. [Google Scholar]
45. M. Masud, A. E. Eldin Rashed and M. S. Hossain, “Convolutional neural network-based models for diagnosis of breast cancer,” Neural Computing and Application, vol. 11, no. 1, pp. 1–12, 2020. [Google Scholar]
46. M. I. Daoud, S. Abdel-Rahman and R. Alazrai, “Breast ultrasound image classification using a pre-trained convolutional neural network,” in 15th Int. Conf. on Signal-Image Technology & Internet-Based Systems, Sorrento, pp. 167–171, 2019. [Google Scholar]
47. T. G. Debelee, F. Schwenker, A. I. Benthal and D. Yohannes, “Survey of deep learning in breast cancer image analysis,” Evolving Systems, vol. 11, no. 1, pp. 143–163, 2020. [Google Scholar]
48. M. I. Daoud, T. M. Bdair, M. Al-Najar and R. Alazra, “A fusion-based approach for breast ultrasound image classification using multiple-ROI texture and morphological analyses,” Computational and Mathematics Methods in Medicine, vol. 2016, pp. 6740956, 2016. [Google Scholar]
49. C. Cortes and V. Vapnik, “Support-vector networks,” Machine Learning, vol. 20, no. 3, pp. 273–297, 1995. [Google Scholar]
50. W. Al-Dhabyani, M. Gomaa, H. Khaled and A. Fahmy, “Deep learning approaches for data augmentation and classification of breast masses using ultrasound images,” International Journal of Advanced Computer Science and Applications, vol. 10, no. 5, pp. 1–11, 2019. [Google Scholar]
51. M. H. Yap, “Breast ultrasound lesions recognition: End-to-end deep learning approaches,” Journal of Medical Imaging, vol. 6, no. 1, pp. 011007, 2019. [Google Scholar]
52. G. Huang, Z. Liu, L. Van Der Maaten and K. Q. Weinberger, “Densely connected convolutional networks,” in 2017 IEEE Conf. on Computer Vision and Pattern Recognition, Honolulu, pp. 4700–4708, 2017. [Google Scholar]
53. A. Krizhevsky, I. Sutskever and G. E. Hinton, “ImageNet classification with deep convolutional neural networks,” Communications of the ACM, vol. 60, no. 6, pp. 84–90, 2017. [Google Scholar]
54. K. Simonyan and A. Zisserman, “Very deep convolutional networks for large-scale image recognition,” ArXiv preprint arXiv 1409.1556, 2014. [Google Scholar]
55. C. Szegedy, “Going deeper with convolutions,” in IEEE Conf. on Computer Vision and Pattern Recognition, Boston, pp. 1–9, 2015. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |