DOI:10.32604/iasc.2022.026601

| Intelligent Automation & Soft Computing DOI:10.32604/iasc.2022.026601 |  |
| Article |
A Hybrid Deep Features PSO-ReliefF Based Classification of Brain Tumor
Department of Radiology, College of Medicine, Qassim University, Buraidah, 52571, Saudi Arabia
*Corresponding Author: Alaa Khalid Alduraibi. Email: al.alderaibi@qu.edu.sa.com
Received: 30 December 2021; Accepted: 31 January 2022
Abstract: With technological advancements, deep machine learning can assist doctors in identifying the brain mass or tumor using magnetic resonance imaging (MRI). This work extracts the deep features from 18-pre-trained convolutional neural networks (CNNs) to train the classical classifiers to categorize the brain MRI images. As a result, DenseNet-201, EfficientNet-b0, and DarkNet-53 deep features trained support vector machine (SVM) model shows the best accuracy. Furthermore, the ReliefF method is applied to extract the best features. Then, the fitness function is defined to select the number of nearest neighbors of ReliefF algorithm and feature vector size. Finally, the particle swarm optimization algorithm minimizes the fitness function to determine the optimal feature vector for model training. The proposed approach is validated by using the available online dataset. The proposed approach enhances the classification accuracy to 97.1% using the optimal concatenated deep features of DenseNet-201and DarkNet-53. Therefore, owing to the high accuracy of the proposed approach, it can be helpful to use real-time applications in the future.
Keywords: Artificial intelligence; brain tumor classification; soft computing; deep learning model; tumor classification; machine learning
The human brain contains over billions of nerve cells; having trillions of interconnections (synapses) is one of the body’s most complex organs [1]. Moreover, the brain works as the central control/command center of the body to regulate the body’s organs. Therefore, any abnormality/mass existence due to brain cells’ abnormal proliferation has a severe impact on the brain and is known as a brain tumor. According to a report published by World Health Organization (WHO) [2], over ten million deaths are reported due to cancer in 2020, the second leading cause of death globally. Therefore, early classification and detection of a brain tumor enhance survival chances.
Brain tumors can be categorized in various ways. For instance, the most commonly known categorization type is to classify it as benign or malignant, which depends upon the location, progression stage, nature, and rate of growth [3,4]. The tumor developed inside the skull but outside the brain tissue is classified as a benign brain tumor. The benign brain tumor rarely affects its neighbor healthy cells and has distinct boundaries and a slow progression rate. On the other hand, the malignant brain tumor harms its neighbor healthy cells (spinal code and brain). Therefore, it has vast boundaries and a high progression rate compared to a benign brain tumor. The classification of brain tumors can also be done based upon their origin (primary and secondary) [5]. The brain tumor that originates in the brain is a primary brain tumor. If it emerges from the other body parts of the brain, it is known as a secondary brain tumor. Brain tumors can also be classified into four stages (stage 0, 1, 2, and 3) based upon severity, boundary, and rate of growth, according to WHO [6–8]. Therefore, early detection and classification of brain tumors into meningioma, pituitary, and glioma brain tumors is vital for patient treatment.
In current medical science advancements, brain tumor detection and classification still depend on histopathological analysis of biopsy specimens [9], the final diagnosis made after using the complete interpretation of medical imaging modalities (MIM) such as positron emission tomography (PET), computed tomography (CT), and magnetic resonance imaging (MRI). However, unlike other tumors (tumors in other parts of the body), brain tumor biopsy is mostly avoided before definitive brain surgery. Therefore, non-invasive approaches like MIM are preferred for brain tumor diagnosis.
In the last few years, artificial intelligence and deep learning networks greatly impacted the image processing field [10–14]. Many researchers used various traditional classifiers to deep learning networks to classify brain tumors using brain MRI images [15]. In traditional classification methods, the model’s classification accuracy depends on brain MRI images’ extracted features. For example, Kumari et al. [16], computed the gray-level co-occurrence matrix of brain MRI images to classify it using a support vector machine (SVM) model. As a result, they classify brain MRI images into two classes (normal and abnormal). The reported classification accuracy is reasonable, but the model consumes high time for training. To address this issue, Singh et al. [17], applied the dimension reduction method to reduce the training time of the model. Their proposed model only classifies the brain MRI images into two classes. However, the model’s accuracy in differentiating the various tumor types is low due to the same brain MRI images’ appearance (texture, size, intensity, etc.). The deep learning networks automatically computed the optimal features of brain MRI images and showed high accuracy [18]. The convolutional neural network (CNN) was designed to classify the brain MRI whole and mask images into two classes [19]. The proposed CNN classifies the whole-brain and brain masks images with the accuracy of 92.9% and 89.5%, respectively. Abiwinanda et al. [20], proposed a simple architecture of the CNN to classify MRI images into glioma, meningioma, and pituitary tumor classes. However, the model has an accuracy of only 84.19%. Recently, Badza et al. [9], proposed a 22-layer CNN to classify the brain tumor MRI images into three classes. The online available brain tumor dataset is utilized to validate their proposed model [21]. The data augmentation method is also applied to enhance the model’s accuracy. Their proposed model shows an accuracy of 96.56% using the 10-fold approach for the augmented dataset. The results of augmentation approaches are not considered very reliable for real-time application. In another recent study [22], the 25-layers CNN model was designed to classify the brain MRI images into five classes. The accuracy of their proposed network is 92.66% for five classes. The various researchers used pre-trained networks such as GoogleNet and ResNet-50 to classify the brain MRI images into subclasses [23,24]. The reported accuracy of these networks is very high (98% and 97.2%), but it consumes much time for the model’s training. To tackle this issue, in a study, researchers calculated the deep features using pre-trained networks and used them to train the traditional classifiers such as SVM, tree, Nave Bayes (NB), etc [25]. They found that the DenseNet-169, ShuffleNet V2, and MnasNet ensembled deep features trained SVM has the accuracy of 93.72% for four class classification of brain MRI images (no tumor, glioma tumor, meningioma tumor, and pituitary tumor). However, the authors also highlighted that the size of training feature vector is very large, resulted in high training time for the model. To address this issue, further research is needed to reduce the dimension of the training feature vector size.
In this paper, the deep features from various pre-trained CNNs (EfficientNet-b0, DarkNet-19, GoogLeNet365, AlexNet, ResNet-101, GoogLeNet, Inception-ResNet-v2, ResNet-18, MobileNet-v2, SqueezeNet, NASNet-Mobile, Inception-v3, DenseNet-201, NASNet-Large, ShuffleNet, ResNet-50, DarkNet-53, and Xception) are computed to train the classical classifiers. The ReliefF dimension reduction algorithm reduces the feature vector size and extracts the useful deep features. The particle swarm optimization (PSO) is applied to optimize the defined cost function to find the nearest neighbors parameter of ReliefF and training feature vector size. The available online dataset is used to check the performance of the proposed approach.
The paper’s organization is as follows: Section 2 explains the methods used to form the proposed framework. Then, the data description and results are presented in Section 3. In Section 4, the results are compared and discussed with the literature. Finally, the paper is concluded in the last section.
This section discusses all the methodologies used to form the proposed framework in detail. The proposed framework consists of several parts: MRI brain images pre-processing, extraction of pre-trained CNNs deep features, ReliefF algorithm, PSO, and machine learning classifiers.
2.1 MRI Brain Images Pre-Processing
All the brain MRI images contain undesired information (area and spaces), leading to poor classification. Therefore, it is compulsory to crop the images to remove those areas and noise to extract useful information for brain tumor classification. In this work, the cropping method is utilized to compute the extreme points, and the noise is removed using dilation and erosions operations; further detail about brain MRI images pre-processing can be found in [25,26]. All the brain MRI images are re-sized according to the requirement of the pre-trained networks.
2.2 Extraction of Brain MRI Image Features Using Pre-Trained CNN
The image features are the main variables used to classify the images into various classes. Therefore, extracting critical features (which have varying characteristics of the brain MRI images) is very important to enhance classification performance. The extraction of brain MRI images can be done manually or using CNN layers. The manual extraction of brain MRI images took much time. Its accuracy mainly depends upon high variation in brain MRI images. However, in CNNs, various convolutional, pooling, and fully connected layers are used to design the model. When the size of the dataset is not so large, then various pre-trained CNNs such as EfficientNet-b0, DarkNet-19, GoogLeNet365, AlexNet, ResNet-101, GoogLeNet, Inception-ResNet-v2, ResNet-18, MobileNet-v2, SqueezeNet, NASNet-Mobile, Inception-v3, DenseNet-201, NASNet-Large, ShuffleNet, ResNet-50, DarkNet-53, and Xception can extract deep features using the transfer learning concept [27,28]. The pre-trained network transfer learning concept is illustrated in Fig. 1.
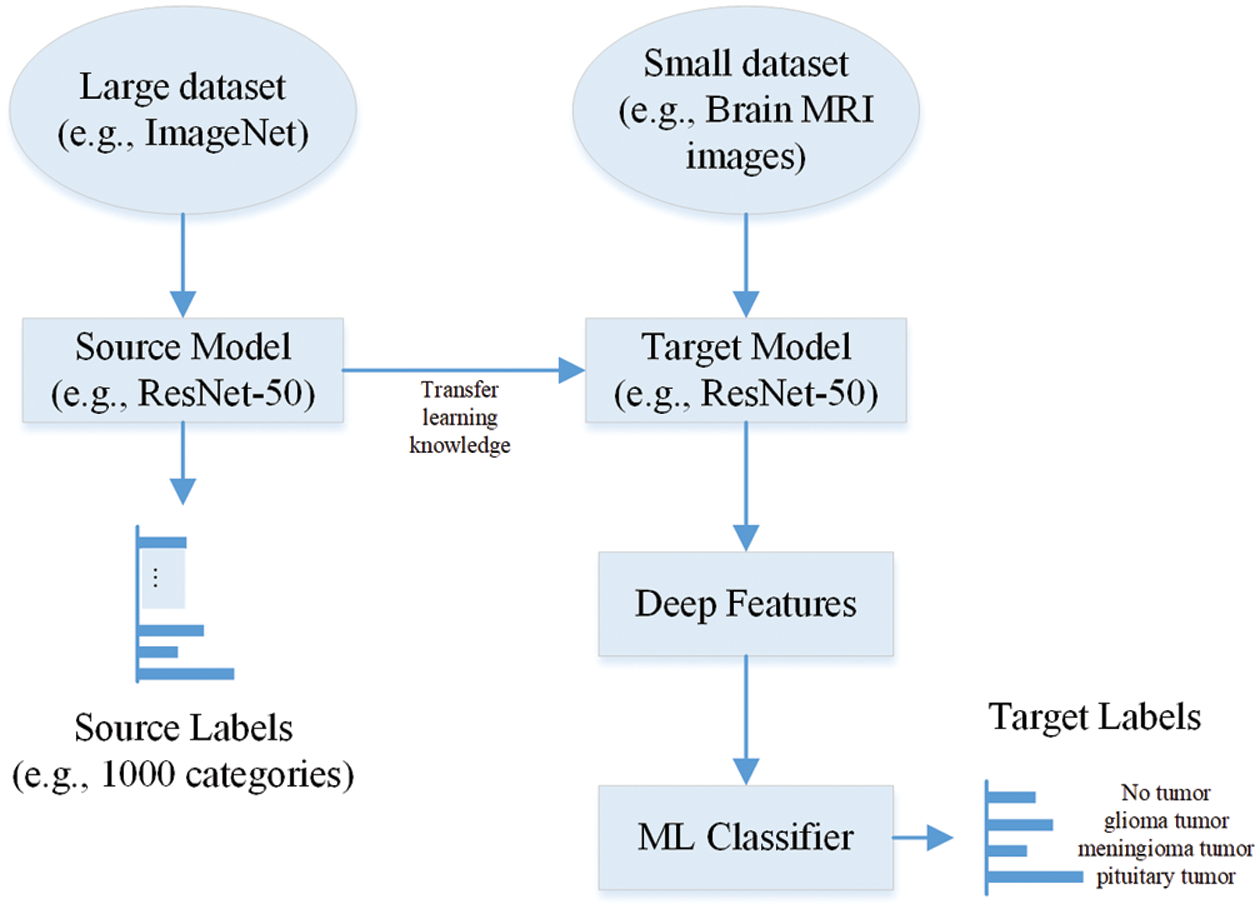
Figure 1: Extraction of deep features using pre-trained CNN
2.3 ReliefF Based Dimension Reduction Approach
The attribute/feature quality is the most critical part of all learning-based classification methods. Also, the number of attributes/features used to depict the objects is important in machine learning applications. It is tough to perform accurately for machine learning approaches using large number of attributes/features, because some of the irrelevant attributes/features only provide very little information for classification and lead to poor accuracy. Therefore, extracting the tiny subset of valuable features (most relevant) to describe the target classes is essential to enhance the model’s classification accuracy.
To deal with it, the authors proposed an approach to select the most relevant features of the object for binary class-based classification problems using instance-based learning [29]. In the consequent study [30], the author proposed an extension of the Relief algorithm for multiclass problems, known as ReliefF. The performance of the algorithm is satisfactory under a perturbed environment. The working of the ReleifF algorithm is shown in Fig. 2.
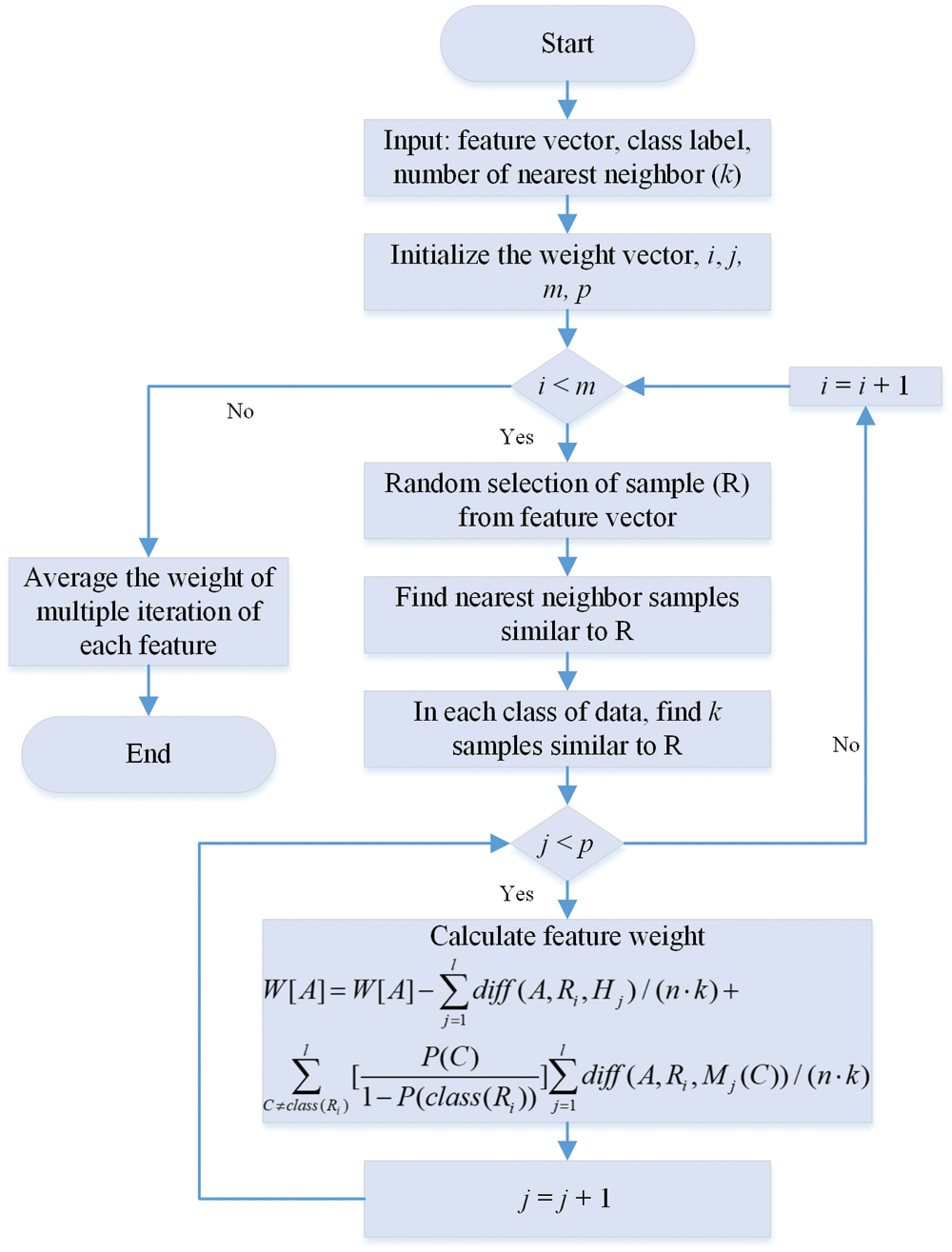
Figure 2: Flowchart of ReliefF method [31,32]
In the ReliefF method, the algorithm initializes the weight vector (W[A]), iteration count (i), iteration feature count (j), and nearest neighbor (k). Then, the algorithm randomly selects the sample R from the feature vector in each iteration. After that, it selects the k nearest sample for the same feature vector sample. After that, it computed the weight vector using the formula shown in Fig. 2. Further detail about the working of the ReliefF algorithm can be found in [31–33]. The nearest neighbor (k) parameter number selection significantly impacts the weight adjustment. After finding all the weights, the number of features for model training also substantially impacts training accuracy. In this work, the PSO is utilized to find the optimal value of k and the size of the feature vector.
2.4 Particle Swarm Optimization
It is a population-based optimization algorithm inspired by the social behavior of fish schooling or birds flocking [34,35]. The PSO estimates the optimal solution by maximizing or minimizing the problem. In this algorithm, the knowledge is shared through group communication while searching for the food in the area. Although each individual is unaware of the exact food site, they all reached the exact point based on information sharing. The complete flow chart of the PSO is shown in Fig. 3. Further detail can of PSO be found in [36,37].

Figure 3: Flowchart of PSO algorithm to compute the optimal value of k and feature vector size
For ReliefF based method, in the PSO, the population (value of k and size of feature vector) are randomly initialized in n-dimension according to the limits of the feature vector. After that, the fitness function is computed for the individual particle to check its fitness value for the function using Eq. (1).
Based upon the fitness value of each particle, compute the best position of each particle (Pbest), which is the individual best position of each particle, and updated iteratively. Then find out the global best (Gbest) position of each particle by comparing all the (Pbest) values, which is also updated iteratively. The following equation can be used to compute the velocity of each particle based upon (Pbest) and (Gbest).
where
The described process will continue till all the particles converge to a single point or the algorithm reaches the maximum iteration criteria. The all PSO parameters values are following:
In this section, the working of the proposed framework is discussed in detail. The architecture of the proposed model to classify the brain MRI images is illustrated in Fig. 4.

Figure 4: A proposed framework to classify Brain MRI images
First, the input images of brain MRI images are pre-processed to remove the noise and unwanted information, as discussed in Section 2.1. Then, after pre-processing, the brain MRI images feed to the pre-trained network to compute the deep features, as discussed in Section 2.2. Next, the PSO-based ReliefF algorithm is applied to compute the optimal k and features vector size to enhance the classification performance, as discussed in Sections 2.3 and 2.4. Finally, the optimal feature vector feed to the classifiers such as tree [38], NB [39], SVM [40], k-nearest neighbors (KNN) [41], ensemble, and neural network (NN) to classify the brain MRI images.
3 Experimental Dataset and Results
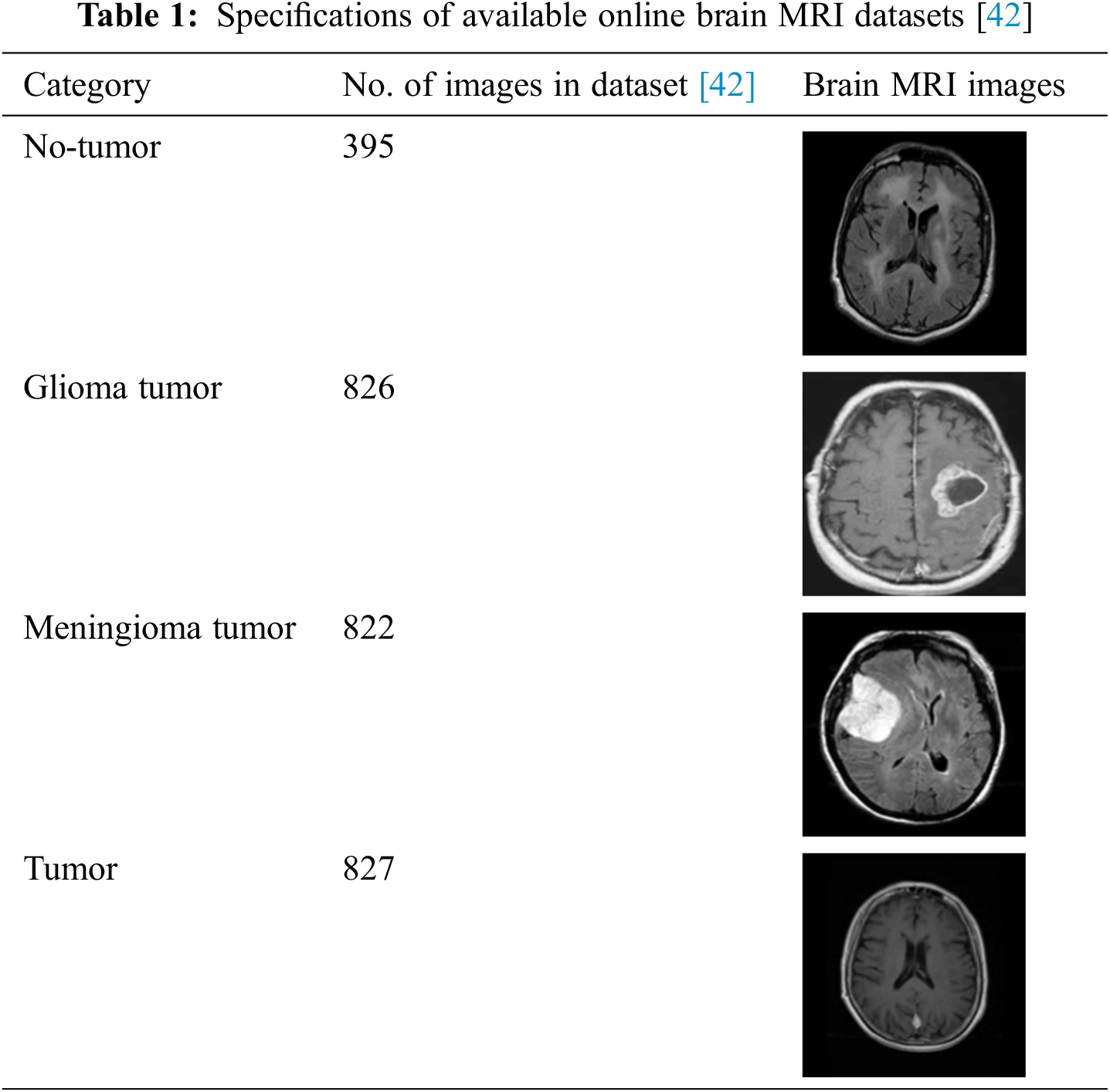
This work utilizes an online dataset of brain MRI images to validate the proposed framework [42]. The details of brain MRI datasets are provided in Tab. 1.
The MATLAB 2021® is used to perform all the computations. In addition, the personal computer has the following specifications: Intel(R) Core (TM) i7-10700 CPU @ 2.90 GHz processor with 32 GB RAM, 1 TB SSD, and a 64-bit Windows 10 Pro operating system (OS). The dataset is divided into 0.8 and 0.2 ratios to train and test all the models. All the deep features of EfficientNet-b0, DarkNet-19, GoogLeNet365, AlexNet, ResNet-101, GoogLeNet, Inception-ResNet-v2, ResNet-18, MobileNet-v2, SqueezeNet, NASNet-Mobile, Inception-v3, DenseNet-201, NASNet-Large, ShuffleNet, ResNet-50, DarkNet-53, and Xception pre-trained models are extracted before SoftMax layer for dataset. The number of extracted features of each pre-trained is shown in Fig. 5. All the computed deep features of the pre-trained model are used to train tree, NB, SVM, KNN, ensemble, and NN. The classification accuracy is the only metric used to check and compare the performances of each model. The results are illustrated in Fig. 6.

Figure 5: Total number of the features of various pre-trained CNNs for brain MRI images
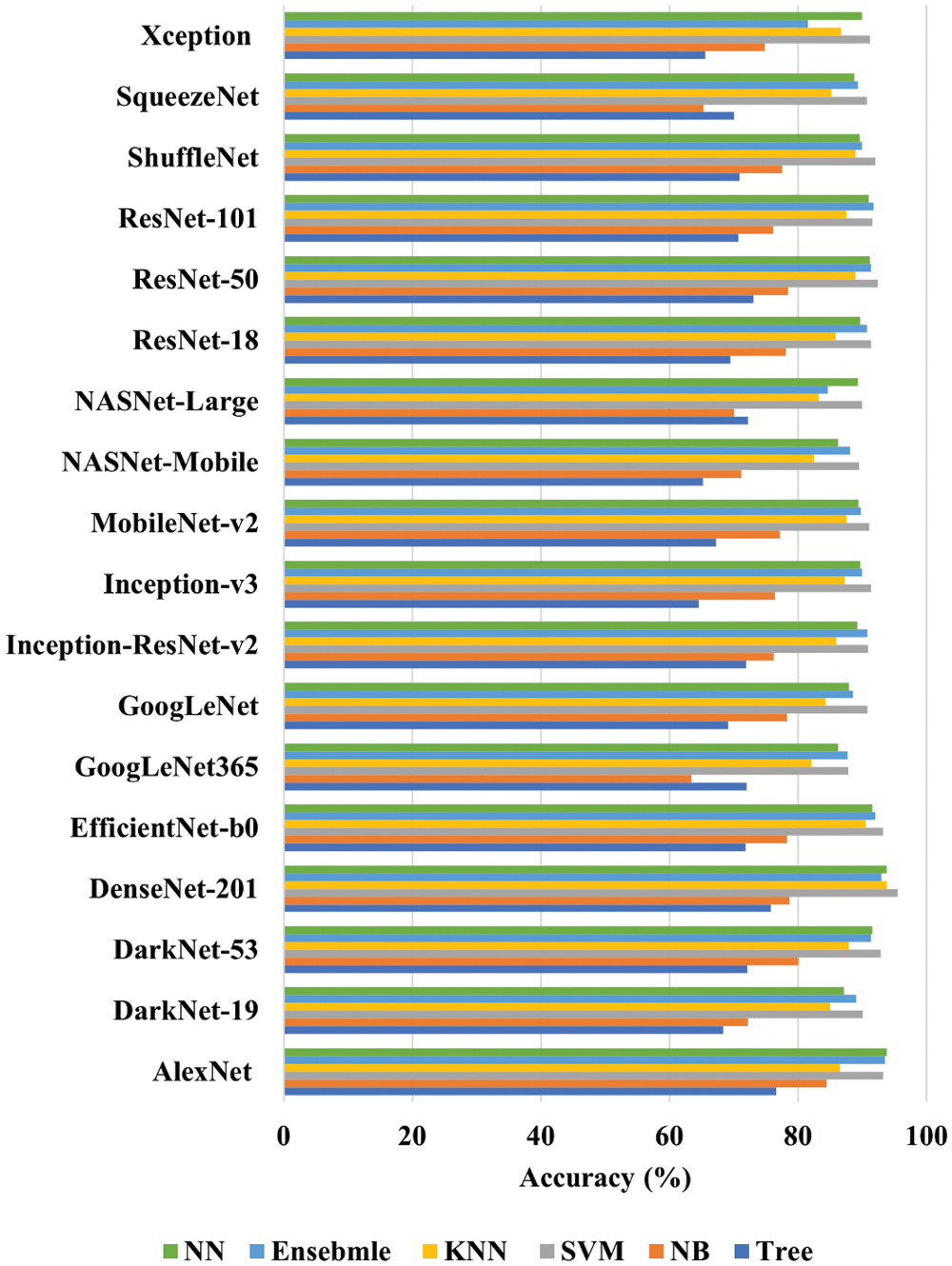
Figure 6: Performance of each machine learning algorithm for the deep features of each pre-trained CNN
After carefully analyzing the performances, it is found that DenseNet-201, EfficientNet-b0, and DarkNet-53 trained SVM have the highest accuracy of 95.5%, 93.3%, and 93.1%, as shown in Fig. 6. In addition, the AlexNet trained SVM and NN model also shows good performance and have accuracies of 92.8% and 92.9%, respectively. However, the AlexNet trained models took almost 125.88 and 63.21 s to train both models due to large feature vector size (9216), as shown in Fig. 5. Moreover, the true positive rate (TPR), false-negative rate (FNR), positive predictive value (PPV), and false discovery rate (FDR) are used for the detailed analysis of the three best deep features trained SVM (DenseNet-201, EfficientNet-b0, and DarkNet-53). The grid search algorithm is used to select the hyper-parameters of SVM. The detailed results of these mentioned classifiers are presented in Tab. 2.
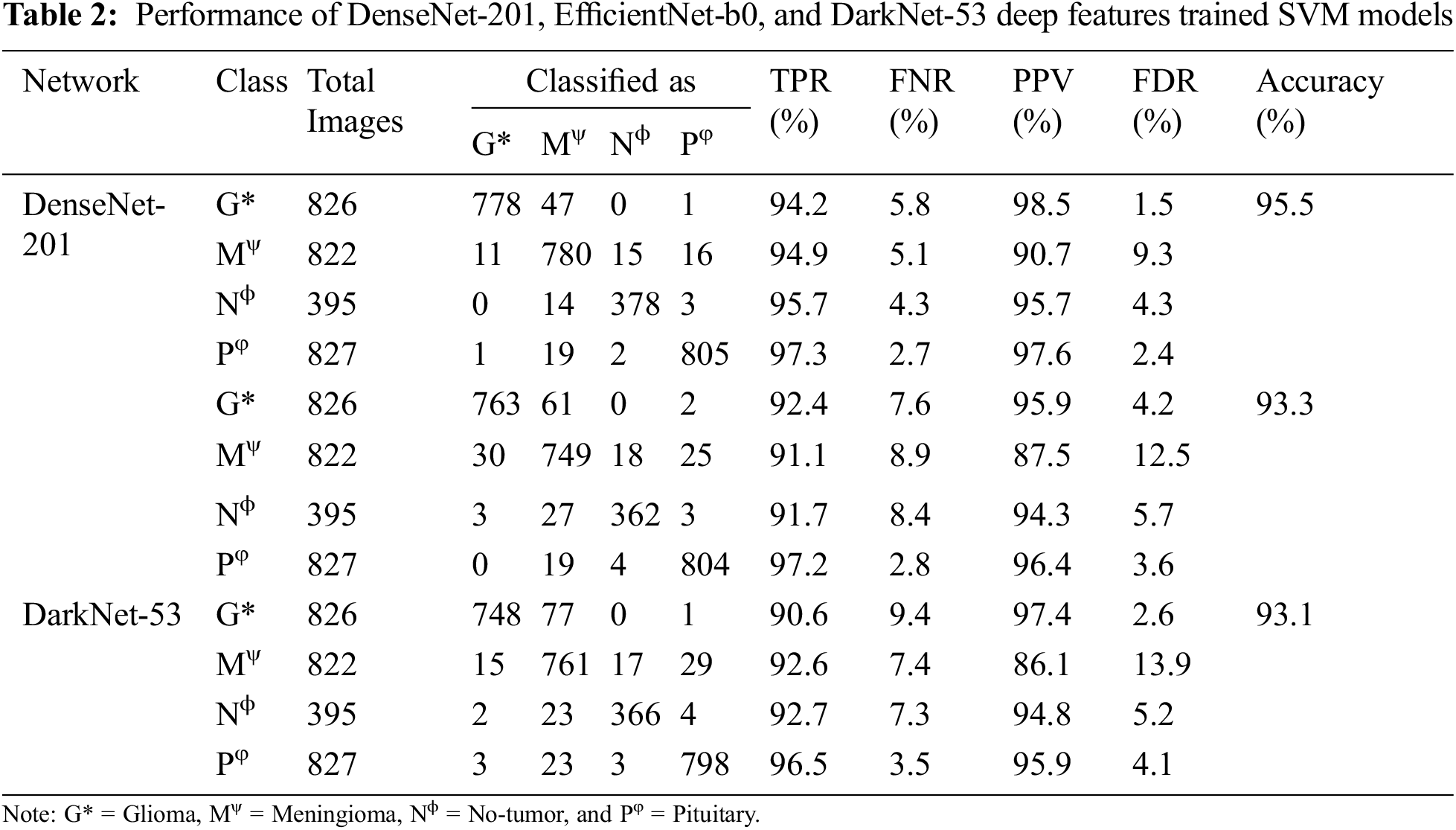
After analyzing the results, it is found that the training time of all the mentioned models is high due to a large set of training features. Therefore, the PSO-based ReliefF algorithm is applied to find the optimal features for each network to reduce the training time and enhance the classification accuracy. The convergence curves of each deep CNN feature are presented in Fig. 7. The performance of DenseNet-201, EfficientNet-b0, and DarkNet-53 deep features trained SVM is presented in Tab. 3.

Figure 7: Convergence Curves
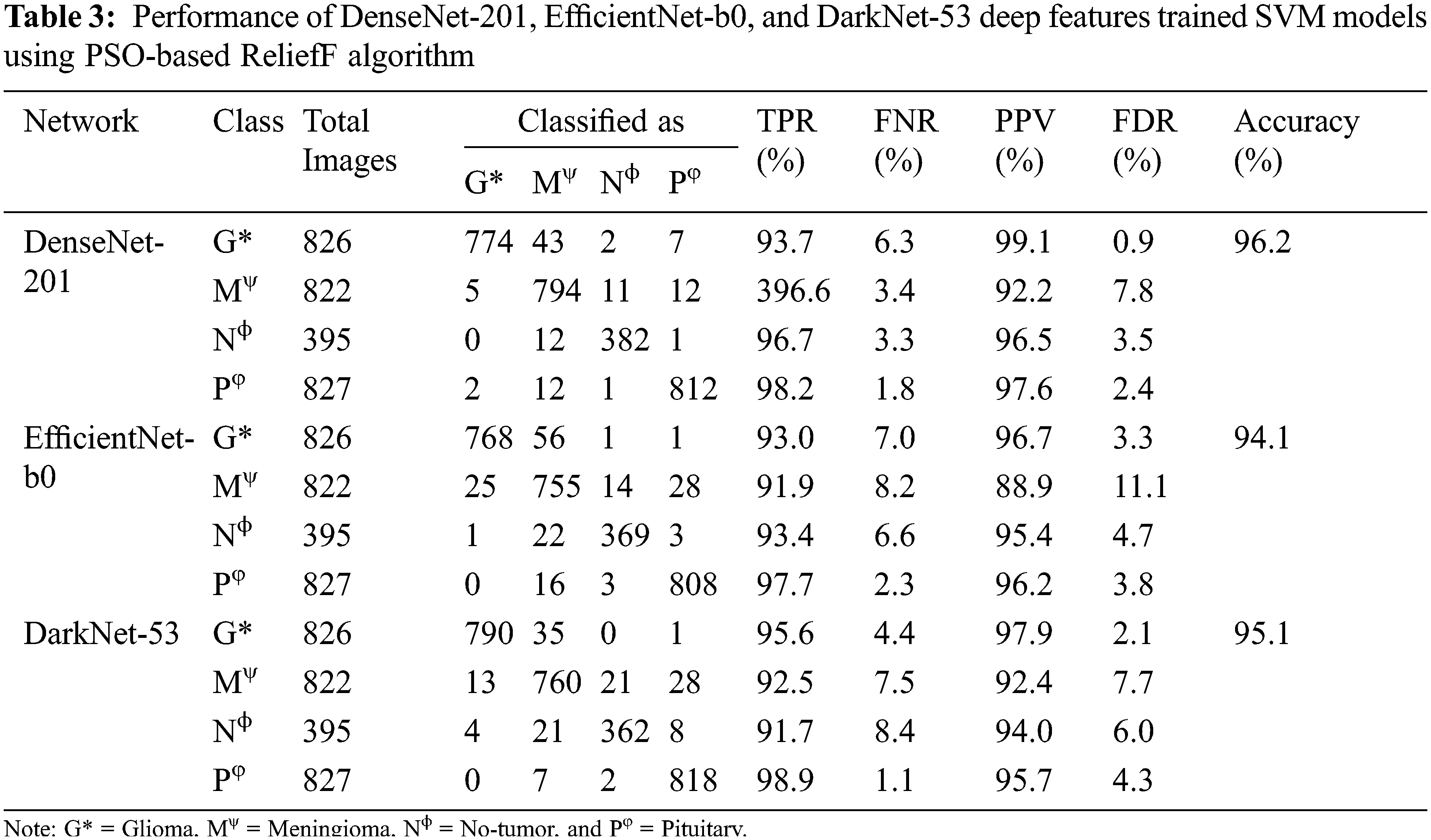
The PSO-based ReliefF dimension reduction algorithm enhances the performance of each algorithm by finding the optimal features, as shown in Tab. 3. For example, the PSO-based ReliefF DenseNet-201 deep feature trained SVM converges at fitness value of 0.038 and has 17 and 192 k nearest neighbors and feature vector size values, respectively. Similarly, EfficientNet-b0 and DarkNet-53 have the fitness value of 0.059 and 0.049 (see Fig. 7), whereas their corresponding k nearest neighbors values are 27 and 8, respectively. And the size of 226 and 44 deep optimal features is used for training the models, respectively. After that, the DenseNet-201 and DarkNet-53 hybrid optimal feature model is trained to analyze the classification performance of the brain MRI images. The results of the hybrid model are presented in Tab. 4. The classification accuracy of brain MRI images increases with an optimal hybrid features model. The TPR of the glioma tumor class is 95.9%, whereas the pituitary tumor class has the highest TPR of 98.8%. It is also found that the glioma tumor class has the best PPV among all the classes (99.1%), as shown in Tab. 4. The comparison of the proposed approach is presented in Tab. 5.


The use of machine learning in medical diagnostic using medical imaging has increased tremendously in recent years. As a result, many researchers have presented several training strategies for brain MRI image classification [9,22,25,43–45].
The use of deep learning networks to classify brain MRI images has very high accuracy [9,22]. Irmak [22], proposed three distinct CNN models, the first one for brain tumor detection, the second one for differentiation in various types of tumor, and the last one for tumor stage. They claimed classification accuracy of 92.66% for categorizing the tumor into subclasses. In another study [43], authors utilized the concept of transfer learning to classify the brain MRI images using pre-trained networks. Their reported accuracy for the various pre-trained model is shown in Tab. 5. Although the accuracy of the models is very high, it took more than 42 min to train each network. Kang et al. [25], computed the deep features from DenseNet-169, Shufflenet, and MnasNet CNNs, and ensembled them to train the SVM model. They reported a classification accuracy of 93.72% for brain MRI images. They also discussed the issue of large feature vector size for training. In this work, 18 pre-trained models of deep features are used to classify the brain MRI images to address this issue (see Fig. 6). After extensive training and testing of the models, the three best deep features (DenseNet-201, EfficientNet-b0, and DarkNet-53) are selected based on their classification accuracy (see Tab. 2).
Then, the PSO-based ReliefF algorithm is applied to find the optimal feature vector. The PSO-based ReliefF algorithm reduces the feature vector size by selecting the most appropriate features, increasing the classification performance (see Tab. 3). The PSO-based ReliefF algorithm is separately applied on each model, since the ReliefF feature score is based on identifying feature value differences across neighboring instance pairs. Each model calculated its features in distinct domains. If pre-trained models are concatenated before using the PSO-based ReliefF method, it only examines the feature of a single model. After finding the performance of each model, the best two models’ features merged to enhance the classification accuracy (97.1%), as shown in Tab. 5. As a result, the suggested technique has the potential to play a critical role in assisting doctors and radiologists in the early detection of brain cancers.
In this work, the deep features of 18 pre-trained CNNs are calculated to check their classification performance for brain MRI images. After extensive training and testing, DenseNet-201, EfficientNet-b0, and DarkNet-53 deep feature trained SVM have the best classification accuracy for brain MRI images. Then, the ReliefF algorithm is applied to extract the most relevant features. Finally, the cost function is defined to select the best parameter (nearest neighbors) and feature vector size for the optimal training feature vector. As a result, the PSO-based ReliefF algorithm increases the accuracy of DenseNet-201 from 95.5% to 96.2% by extracting the most relevant features. Similarly, the increase of 0.7% and 2% is noted in the classification accuracy of EfficientNet-b0 and DarkNet-53, respectively. Finally, DenseNet-201 and DarkNet-53 deep concatenated features trained SVM has the highest accuracy of 97.1% for brain MRI images. The proposed approach has the potential to help doctors and radiologists to detect brain tumors early. In the future, to enhance reliability and accuracy, researchers may utilize the big MRI brain images dataset or the data augmentation approach to train the proposed model.
Funding Statement: The author would like to thank the Deanship of Scientific Research, Qassim University for funding the publication for this project.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. D. N. Louis, A. Perry, G. Reifenberger, A. von Deimling, D. Figarella-Branger et al., “The 2016 world health organization classification of tumors of the central nervous system: A summary,” Acta Neuropathologica, vol. 131, no. 6, pp. 803–820, 2016. [Google Scholar]
2. W. H. Organization, Cancer, 2021. [Online]. Available: https://www.who.int/news-room/fact-sheets/detail/cancer. [Google Scholar]
3. A. C. Society, Cancer, 2021. [Online]. Available: https://www.cancer.org/cancer.html. [Google Scholar]
4. Cancer.Net, Brain Tumor: Diagnosis, 2021. [Online]. Available: https://www.cancer.net/cancer-types/brain-tumor/diagnosis. [Google Scholar]
5. G. S. Tandel, M. Biswas, O. G. Kakde, A. Tiwari, H. S. Suri et al., “A review on a deep learning perspective in brain cancer classification,” Cancers, vol. 11, no. 1, pp. 111, 2019. [Google Scholar]
6. S. Viral and K. Pratiksha, “Brain cancer: Implication to disease, therapeutic strategies and tumor targeted drug delivery approaches,” Recent Patents on Anti-Cancer Drug Discovery, vol. 13, no. 1, pp. 70–85, 2018. [Google Scholar]
7. S. Ahmed, K. M. Iftekharuddin and A. Vossough, “A. efficacy of texture, shape, and intensity feature fusion for posterior-fossa tumor segmentation in MRI,” IEEE Transactions on Information Technology in Biomedicine, vol. 15, no. 2, pp. 206–213, 2011. [Google Scholar]
8. S. Deorah, C. F. Lynch, Z. A. Sibenaller and T. C. Ryken, “Trends in brain cancer incidence and survival in the United States: Surveillance, epidemiology, and End results program, 1973 to 2001,” Neurosurgical Focus FOC, vol. 20, no. 4, pp. E1, 2006. [Google Scholar]
9. M. M. Badža and M. C. Barjaktarović, “Classification of brain tumors from MRI images using a convolutional neural network,” Applied Sciences, vol. 10, no. 6, pp. 1999, 2020. [Google Scholar]
10. W. Ahmed, A. Hanif, K. D. Kallu, A. Z. Kouzani, M. U. Ali et al., “A. photovoltaic panels classification using isolated and transfer learned deep neural models using infrared thermographic images,” Sensors, vol. 21, no. 16, pp. 5668, 2021. [Google Scholar]
11. M. U. Ali, S. Saleem, H. Masood, K. D. Kallu, M. Masud et al., “A. early hotspot detection in photovoltaic modules using color image descriptors: An infrared thermography study,” International Journal of Energy Research, vol. 46, no. 2, pp. 774–785, 2021. [Google Scholar]
12. K. Doi, “Computer-aided diagnosis in medical imaging: Historical review, current status and future potential,” Computerized Medical Imaging and Graphics, vol. 31, no. 4–5, pp. 198–211, 2007. [Google Scholar]
13. K. Munir, H. Elahi, A. Ayub, F. Frezza and A. Rizzi, “Cancer diagnosis using deep learning: A bibliographic review,” Cancers, vol. 11, no. 9, pp. 1235, 2019. [Google Scholar]
14. M. U. Ali, K. D. Kallu, H. Masood, K. A. K. Niazi, M. J. Alvi et al., “Kernel recursive least square tracker and long-short term memory ensemble based battery health prognostic model,” iScience, vol. 24, no. 11, pp. 103286, 2021. [Google Scholar]
15. A. Wadhwa, A. Bhardwaj and V. S. Verma, “A review on brain tumor segmentation of MRI images,” Magnetic Resonance Imaging, vol. 61, pp. 247–259, 2019. [Google Scholar]
16. R. Kumari, “SVM classification an approach on detecting abnormality in brain MRI images,” International Journal of Engineering Research and Applications, vol. 3, no. 4, pp. 1686–1690, 2013. [Google Scholar]
17. D. Singh and K. Kaur, “Classification of abnormalities in brain MRI images using GLCM, PCA and SVM,” International Journal of Engineering and Advanced Technology (IJEAT), vol. 1, no. 6, pp. 243–248, 2012. [Google Scholar]
18. M. Nazir, S. Shakil and K. Khurshid, “Role of deep learning in brain tumor detection and classification (2015 to 2020A review,” Computerized Medical Imaging and Graphics, vol. 91, pp. 101940, 2021. [Google Scholar]
19. S. Pereira, R. Meier, V. Alves, M. Reyes and C. A. Silva, “Automatic Brain Tumor Grading from MRI Data Using Convolutional Neural Networks and Quality Assessment,” in Understanding and Interpreting Machine Learning in Medical Image Computing Applications, Cham: Springer, pp. 106–114, 2018. [Google Scholar]
20. N. Abiwinanda, M. Hanif, S. T. Hesaputra, A. Handayani and T. R. Mengko, “Brain tumor classification using convolutional neural network,” in World Congress on Medical Physics and Biomedical Engineering 2018, Singapore, Springer, pp. 183–189, 2019. [Google Scholar]
21. C. Jun, Brain tumor dataset, 2017. [Online]. Available: https://figshare.com/articles/dataset/brain_tumor_dataset/1512427. [Google Scholar]
22. E. Irmak, “Multi-classification of brain tumor MRI images using deep convolutional neural network with fully optimized framework,” Iranian Journal of Science and Technology, Transactions of Electrical Engineering, vol. 45, pp. 1015–1036, 2021. [Google Scholar]
23. S. Deepak and P. M. Ameer, “Brain tumor classification using deep CNN features via transfer learning,” Computers in Biology and Medicine, vol. 111, pp. 103345, 2019. [Google Scholar]
24. A. Çinar and M. Yildirim, “Detection of tumors on brain MRI images using the hybrid convolutional neural network architecture,” Medical Hypotheses, vol. 139, pp. 109684, 2020. [Google Scholar]
25. J. Kang, Z. Ullah and J. Gwak, “MRI-Based brain tumor classification using ensemble of deep features and machine learning classifiers,” Sensors, vol. 21, no. 6, pp. 2222, 2021. [Google Scholar]
26. A. Rosebrock, Finding extreme points in contours with Open CV, 2016. [Online]. Available: https://www.pyimagesearch.com/2016/04/11/finding-extreme-points-in-contours-with-opencv/. [Google Scholar]
27. I. M. Baltruschat, H. Nickisch, M. Grass, T. Knopp and A. Saalbach, “A. comparison of deep learning approaches for multi-label chest X-ray classification,” Scientific Reports, vol. 9, no. 1, pp. 6381, 2019. [Google Scholar]
28. J. Kang and J. Gwak, “Ensemble of instance segmentation models for polyp segmentation in colonoscopy images,” IEEE Access, vol. 7, pp. 26440–26447, 2019. [Google Scholar]
29. K. Kira and L. A. Rendell, “A practical approach to feature selection,” in Machine Learning Proceedings 1992, USA: Elsevier, pp. 249–256, 1992. [Google Scholar]
30. I. Kononenko, “Estimating attributes: Analysis and extensions of RELIEF,” in Proc. ECML, Berlin, Germany, pp. 171–182, 1994. [Google Scholar]
31. M. Robnik-Šikonja and I. Kononenko, “Theoretical and empirical analysis of ReliefF and RReliefF,” Machine Learning, vol. 53, no. 1, pp. 23–69, 2003. [Google Scholar]
32. R. J. Urbanowicz, M. Meeker, W. La Cava, R. S. Olson and J. H. Moore, “Relief-based feature selection: Introduction and review,” Journal of Biomedical Informatics, vol. 85, pp. 189–203, 2018. [Google Scholar]
33. Z. Wu, X. Wang and B. Jiang, “Fault diagnosis for wind turbines based on ReliefF and eXtreme gradient boosting,” Applied Sciences, vol. 10, no. 9, pp. 3258, 2020. [Google Scholar]
34. S. Ekinci and B. Hekimoğlu, “Improved kidney-inspired algorithm approach for tuning of PID controller in AVR system,” IEEE Access, vol. 7, pp. 39935–39947, 2019. [Google Scholar]
35. J. Mannan, M. A. Kamran, M. U. Ali and M. M. N. Mannan, “Quintessential strategy to operate photovoltaic system coupled with dual battery storage and grid connection,” International Journal of Energy Research, vol. 45, no. 15, pp. 21140–21157, 2021. [Google Scholar]
36. N. Anwar, A. Hanif, M. U. Ali and A. Zafar, “Chaotic-based particle swarm optimization algorithm for optimal PID tuning in automatic voltage regulator systems,” Electrical Engineering & Electromechanics, vol. 1, pp. 50–59, 2021. [Google Scholar]
37. M. U. Ali, B. Habib and M. Iqbal, “Fixed head short term hydro thermal scheduling using improved particle swarm optimization,” The Nucleus, vol. 52, pp. 107–114, 2015. [Google Scholar]
38. S. R. Safavian and D. Landgrebe, “A survey of decision tree classifier methodology,” IEEE Transactions on Systems, Man, and Cybernetics, vol. 21, no. 3, pp. 660–674, 1991. [Google Scholar]
39. K. A. K. Niazi, W. Akhtar, H. A. Khan, Y. Yang and S., Athar, “Hotspot diagnosis for solar photovoltaic modules using a Naive Bayes classifier,” Solar Energy, vol. 190, pp. 34–43, 2019. [Google Scholar]
40. M. U. Ali, H. F. Khan, M. Masud, K. D. Kallu and A. Zafar, “A machine learning framework to identify the hotspot in photovoltaic module using infrared thermography,” Solar Energy, vol. 208, pp. 643–651, 2020. [Google Scholar]
41. N. Ali, D. Neagu and P. Trundle, “Evaluation of k-nearest neighbour classifier performance for heterogeneous data sets,” SN Applied Sciences, vol. 1, pp. 1–15, 2019. [Google Scholar]
42. S. Bhuvaji, A. Kadam, P. Bhumkar, S. Dedge and S. Kanchan, Brain Tumor Classification (MRI) Dataset, 2020. [Online]. Avalialabe: https://www.kaggle.com/sartajbhuvaji/brain-tumor-classification-mri. [Google Scholar]
43. A. Rehman, S. Naz, M. I. Razzak, F. Akram and M. Imran, “A deep learning-based framework for automatic brain tumors classification using transfer learning,” Circuits, Systems, and Signal Processing, vol. 39, vol. 2, pp. 757–775, 2020. [Google Scholar]
44. Z. N. K. Swati, Q. Zhao, M. Kabir, F. Ali, Z. Ali et al., “Brain tumor classification for MR images using transfer learning and fine-tuning,” Computerized Medical Imaging and Graphics, vol. 75, pp. 34–46, 2019. [Google Scholar]
45. J. Cheng, W. Yang, M. Huang, W. Huang, J. Jiang et al., “Retrieval of brain tumors by adaptive spatial pooling and fisher vector representation,” PLOS ONE, vol. 11, no. 6, pp. e0157112, 2016. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |