DOI:10.32604/iasc.2022.025046

| Intelligent Automation & Soft Computing DOI:10.32604/iasc.2022.025046 |  |
| Article |
A Deep Learning Framework for COVID-19 Diagnosis from Computed Tomography
1Department of Computer Science, Applied College, University of Ha’il, KSA
2ReDCAD Laboratory, Sfax University, Sfax, Tunisia
3Department of Engineering, College of Engineering and Applied Sciences, American University of Kuwait, Kuwait
4Department of Information and Computer Science, College of Computer Science and Engineering, University of Ha’il, KSA
*Corresponding Author: Nabila Mansouri. Email: n.mansouri@uoh.edu.sa
Received: 09 November 2021; Accepted: 29 January 2022
Abstract: The outbreak of novel Coronavirus COVID-19, an infectious disease caused by the SARS-CoV-2 virus, has caused an unprecedented medical, economic, and social emergency that requires data-driven intelligence and decision support systems to counter the subsequent pandemic. Data-driven models and intelligent systems can assist medical researchers and practitioners to identify symptoms of COVID-19 infection. Several solutions based on medical image processing have been proposed for this purpose. However, the most shortcoming of hand craft image processing systems is the lower provided performances. Hence, for the first time, the proposed solution uses a deep learning model that is applied to Computed Tomography (CT) images for the efficient extraction of COVID-19 features. Since there are few patients in the COVID-CT-Dataset, the Convolutional Neural Network (CNN) model cannot undergo further learned to enhance performances. Therefore, the proposed solution works as a pipeline framework involving two steps: (A) baseline classification is provided by a CNN model; (B) baseline results are re-ranked using distances to features vectors of CT image parts. A re-ranking framework is used as additional means of COVID-19 symptom identification. These steps exploit the diversity of different parts of CT images to enhance classification performance. Evaluations of the proposed solution are driven by real world data based on clinical findings in the form of COVID-CT-Dataset images. The results of the evaluation illustrate the streamlined efficiency and accuracy of the proposed solution to the image-based diagnosis of COVID-19 patients. Our findings support smart healthcare solutions–specifically addressing COVID-19 challenges–and provide guidelines to engineer and develop intelligent and autonomous systems.
Keywords: COVID-19; deep to counter the current and futuristic waves of the pandemic; decision support system; health informatics; convolutional neural network
Throughout human history, pandemics (i.e., epidemics of infectious diseases that spread across geographical continents) have ravaged global progress, affecting economic growth, disrupting social interactions, and affecting human lives [1]. COVID-19, as the most recent pandemic, has imposed a widespread lockdown on human activities, with approximately 225 million recorded infections and 4.6 million deaths to date [2], resulting in a medical and economic emergency. Consequently, states, communities, medics, researchers, and health practitioners around the world face a war-like situation with an invisible enemy that has infected the human population in an unprecedented manner [3]. In this context, researchers, and practitioners across various scientific domains, including the medical sciences, engineering, and social sciences, are in continuous pursuit of models and solutions that can minimize the impacts of the pandemic on the local and global population [4,5]. From a technical perspective, the computing community has proposed several solutions, including but not limited to contact tracing [6], infection modelling [7], and medical recommendation systems [8], to tackle the pandemic. Despite the existing solutions, and the emergence of new ones, one of the critical challenges is to answer how computing technologies and information systems can be exploited to develop innovative solutions that can eradicate or minimize the impacts of the pandemic. Information systems or decision support models as a class of computing systems can empower medics and stakeholders to exploit Information and Communication Technologies (ICTs) across a multitude of scenarios to monitor, analyze, and curtail the spread of COVID-19 infections [5,9].
Artificial Intelligence (AI) systems are a specific genre of intelligent and autonomous computing technologies, represented as expert or decision support systems for data-driven decision making, imitating human intelligence for problem solving [10]. AI methods and techniques have been used to propose several COVID-19 solutions, such as infection modeling [11] or the prediction of infectious transmission in social interactions [7]. AI-based decision support systems can empower medical practitioners to make informed decisions based on automated identification of potential infections, recommendation of prescriptions and streamlining of causes, all of by machine intelligence that learns from historical data [12]. More specifically, deep learning solutions can be developed and trained using COVID-19 specific historical data to predict emerging and future patterns and trends in COVID-19 infection [13,14]. Deep learning solutions, as a sub-class of AI systems, are being continuously explored to assist medical researchers and practitioners to identify symptoms of COVID-19 infections from health critical data (e.g., medical images and health records) in an automated and efficient manner [15–19]. Although medical imaging and health datasets have been used to train AI or deep learning systems to identify the potential symptoms of COVID-19 infections, however, such methods face some problems. These problems include but are not limited to lack of accuracy (e.g., identifying false positive) and efficiency (i.e., system performance) reflecting critical challenges to develop and operationalize such class of systems [20].
Challenges and Solution overview: The proposed research aims to develop a deep learning solution–an artificially intelligent function–that can be applied to Computed Tomography (CT) images to discover recurring patterns from COVID-19 infection data. The infection data is specifically referred to as a COVID-CT-Dataset and is based on clinical findings from CT images from COVID-19 infected patients. We focus on the research challenge about “how to fully exploit the information involved in a single image to improve the retrieval accuracy, while no other CT images are available”. Hence, the re-ranking framework for COVID-19 symptom identification is proposed in this paper. It exploits the diversity from different parts of a CT image. More specifically, given an image, we first divide it into sub-parts. Then the distance between each sub-feature of the query and gallery images is iteratively encoded into a new distances vector. Finally, a majority vote between the classes of the Top-5 distances is adopted to identify the new class of query image. The solution works as a pipeline framework and consists of two steps: (A) baseline classification is provided by a CNN model, (B) baseline results are re-ranked using an efficient re-ranking algorithm that focuses on details in the parts of the CT image. The proposed solution is evaluated based on real-world data from clinical findings concerning infected patients via CT images. The solution evaluation accumulated a total of 349 CT images that were obtained from clinical findings concerning 216 COVID-19 patients. The results of the evaluation highlight that the proposed solution, based on re-ranking, considerably enhances the baseline results. The primary contributions of this research are:
• The exploitation of image processing techniques with the application of deep learning to discover recurring patterns of COVID-19 infection using Computed Tomography images. Convolutional Neural Networks are used to recognize and classify the potential symptoms of infections effectively and efficiently.
• Enhanced results, despite the lack of a large dataset. Therefore, we propose an extra classification step based on a re-ranking algorithm. Since transfer learning can fine-tune CNN models and provide extra samples for deeper learning steps, this method can cause overfitting. However, the proposed re-ranking algorithm is an intelligent way to enhance results without overfitting the baseline model. Also, a re-ranking approach based on divide and fusion proposed algorithm introduces new local features from CT images, which can improve performance.
• Application of method, that can use local features from CT images (potential COVID-19 infections), to support performance.
• Leveraging of the visual information available in a single image by dividing and classifying parts of a single image to detect the symptoms of a potential infection. This enables the solution to achieve the efficiency of infection detection, while preserving its accuracy in infection detection based on analyzing the granularity and diversity of the image.
Our proposed solution applies deep learning techniques to medical imaging datasets and provides a decision support system that can assist medics to identify potential COVID-19 infections. The evaluations achieved complement existing research and development on engineering intelligence and autonomous systems that aim to counter the COVID-19 pandemic. The findings of this research may be beneficial to:
• Medical practitioners and stakeholders, who can rely on AI-driven smart systems that will provide them with data-driven intelligence to manage the impacts of the pandemic. The proposed solution will empower medics and public health workers to identify potential infections in an automated and efficient manner.
• Researchers and developers in computing and medical sciences who are interested in exploring and applying Information and Communication Technologies (ICT) in health informatics to counter the current wave of the pandemic. The role of AI systems, specifically deep learning solutions, advances the state-of-the-art in autonomic computing technologies in the context of smart healthcare.
Organization of the paper: The remainder of this paper is organized as follows; Section 2 contextualizes the proposed solution and explains the technical background and research method. Section 3 presents related research to justify the scope and contributions of the proposed solution. Section 4 provides an overview of the implementation of the solution. Section 5 presents the results of the evaluation. Section 6 concludes the paper by revisiting the key contributions, identifying the potential limitations, and suggesting dimensions for future research into the proposed solution.
2 Research Context and Methodology
First, we contextualize the proposed research (Section 2.1) by providing background information and explaining the core concepts that enable us to elaborate technical details of the solution later in the paper. Second, we outline the proposed research methodology (Section 2.2) that has been adopted to conduct this research. The concepts and terminologies introduced in this section are used throughout the paper.
Here we, (a) detail the research context in terms of intelligent systems and image analysis, and (b) streamline the role of deep learning in the image-based analysis of potential infections. The discussion in this subsection is guided by the illustrations in Fig. 1.
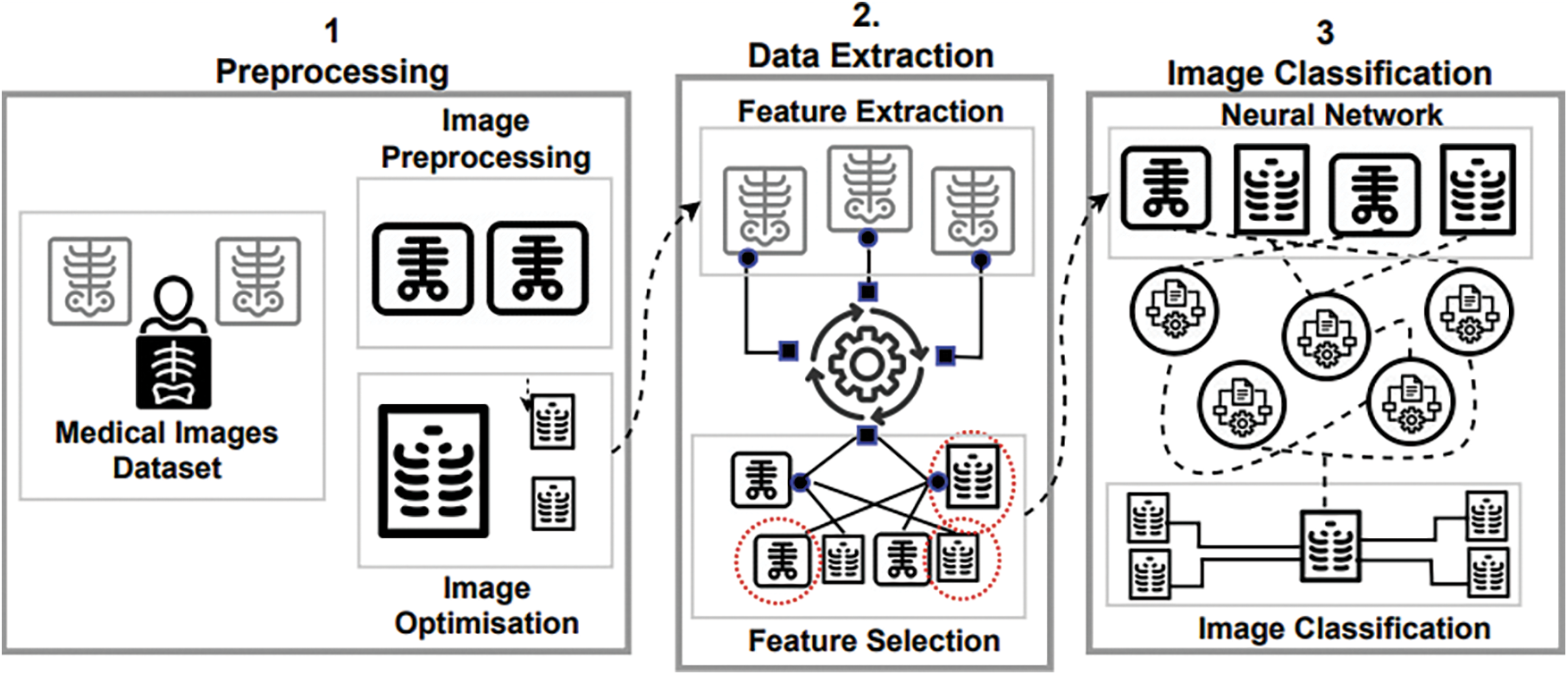
Figure 1: A process-centred view of medical image classification. ([1] Preprocessing, [2] Data Extraction, and [3] Image Classification)
2.1.1 AI and Deep Learning for Image Analysis
An increased adoption of smart systems and technologies in urban planning, transportation, and healthcare can be attributed to intelligent systems that learn from historical data (data and pattern mining) and operational context (context-aware computing) to adapt a system’s behaviour in an autonomic way [12]. One of the central features of computational intelligence is the need to understand the principles that make intelligent behavior possible, in natural or artificial systems [21]. In fact, Artificial Intelligence (AI) techniques aim to enable the operationalization of intelligent agents, where an intelligent agent is a system that surveys and learns from its environment to accumulate expertise to make intelligent decisions with minimal to no human supervision. Intelligent and autonomic computing systems have been proven to be successful in analyzing images without human intervention, to process, identify, and extract visually relevant information accurately and efficiently [22].
AI methods and techniques such as deep learning exploit (deep) artificial neural networks to address computationally complex problems across a multitude of real-world scenarios including, but not limited to, image analytics and natural language processing [23]. Specifically, deep learning, as a computer vision model, refers to the design of deep CNN, providing machines with vision in an attempt to imitate human behavior in problem solving [23]. Hence, in several high-profile image analysis benchmarks, deep learning has outperformed other methods. For example, in 2012, in one of the famous ImageNet Large-Scale Visual Recognition Challenges (ILSVRC) [24], a deep learning model achieved the second-best error rate for the image classification task. Deep learning techniques for image classification consist of roughly three complementary steps [25], namely: (i) pre-processing of image data (i.e., analysis); (ii) extraction of features from the data (i.e., identification); and (iii) classification (i.e., computational decision making), as illustrated in Fig. 1.
2.1.2 The Role of Deep Learning in Analyzing COVID-19 Symptoms
The success of deep learning techniques in many pattern recognition problems, like natural language processing, visual data processing, and social network analysis [26], has made deep learning tools and algorithms one of the most appropriate means of addressing the challenges associated with the image-based identification of disease symptoms [20]. Deep learning techniques exploit medical imaging that includes but is not limited to X-ray, ultrasound, Computerized Tomography (CT), Magnetic Resonance Imaging (MRI), and single photon emission computed tomography (SPECT) to detect various diseases (e.g., tuberculosis, cancer, etc.) and perform medical check-ups. For example, deep learning algorithms such as multilayer perceptron have been used successfully in the detection of tumours [27]. In the context of COVID-19 symptom detection, deep learning techniques that rely on deep architectural design can effectively manage and analyze COVID-19 infection datasets through the segmentation of CT images and categorization of various infection types [13].
In addition to supporting the required functionality (i.e., automated detection of COVID-19 infections), deep learning solutions must also ensure the desired quality (i.e., accuracy and efficiency) of infection detection. Instead of proposing completely new (often less stable, less tested) solutions, existing tools and methods can be leveraged for image analytics in the context of detecting COVID-19 infections. As an example, to analyze CT images for COVID-19 image interpretation, existing techniques such as [28,29], that utilize CNN, can be tailored to address the COVID-19 detection problem. Performing a deep and recurrent process with a deep learning model for pre-processing, feature extraction, and selection and image classification (see Fig. 1) is the key to success in deep learning with medical image processing in general, and COVID-19 symptom detection specifically.
We now detail the three-step methodology (see Fig. 2) that was used as a systematic approach to planning, conducting, and validating the proposed research.
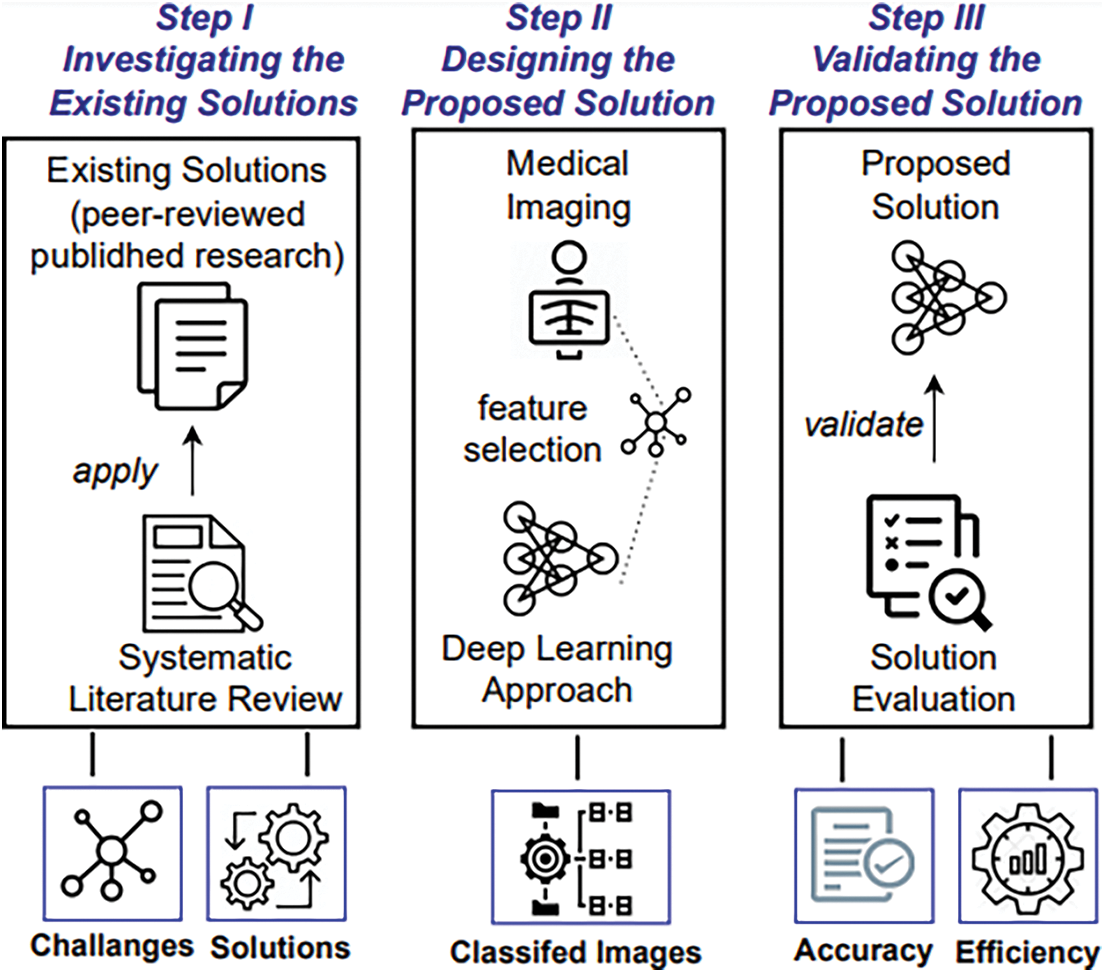
Figure 2: A step-wise view of the research methodology guideline
2.2.1 Investigating the Existing Solutions
As the first step of the methodology, we needed to investigate the state-of-the-art of the automated and machine-driven detection of COVID-19 symptoms. Specifically, we identified and analyzed the existing solutions (peer-reviewed published research) by following the guidelines for Systematic Literature Reviews (SLRs) [30]. Based on conducting the SLR, we analyzed the current solutions, their limitations and suggested directions for future research, as detailed in Section 3. Our findings from Step 1 helped us to design the proposed solution, which is detailed in the next section (Section 3) of this paper.
2.2.2 Designing the Proposed Solution
The next step in the methodology was to design a solution that would process a dataset of clinical images to discover potential symptoms of COVID-19 infection. We collected CT and X-ray clinical images that could be used in datasets to train the proposed solution. More specifically, we extracted features from COVID-19 related images and processed them to classify the symptoms of infection from those images. Further details about the proposed solution are given in Section 4.
2.2.3 Validating the Proposed Solution
Finally, we needed to validate the proposed solution to evaluate its usefulness in terms of functionality and quality of the system. To achieve this we used the ISO/IEC/9126 model for software quality [31]. The quality model helped with the criteria-based evaluation of the proposed solution in terms of its efficiency and accuracy. The results of the evaluation are presented in Section 5.
In this section, we review the most relevant existing work as per Step I of the research method (i.e., investigating the existing solutions, as shown in Fig. 2). Specifically, we review the existing CNNs (Section 3.1) and re-ranking models (Section 3.2) for image classification and COVID-19 symptom detection. As per the outlined methodology, investigating the existing solutions helped us to develop foundations for the proposed solution and to justify its scope and contribution.
3.1 Deep Learning Techniques for COVID-19 Detection
3.1.1 CNN Approaches based on Transfer Learning for COVID-19 Image Analysis
Convolutional Neural Networks (CNNs), as a deep learning technique, have been recently used to detect potential symptoms of COVID-19 infections [13]. For example, the research in [32] used a procedure called Transfer Learning to assess the performance of some existing CNN models when classifying COVID-19 related scans. The authors collected two different sets of data to investigate whether deep learning with Xray scans could help with the extraction of significant bio-information related to COVID-19. The authors in [28] employed deep learning algorithms to increase the accuracy of identification of confirmed COVID-19 cases, as well as to predict those who were infected from their X-ray images. Their solution relied on the use of CNN models that had been used as the basis for uncovering hidden patterns to identify abnormal structures. Makris et al. [29] employed 9 existing CNNs to classify X-ray images for individuals infected with COVID-19, individuals with pneumonia, and individuals with normal health. The results showed that the proposed CNN method was good at detecting COVID-19 symptoms.
Aslan et al. [33] have proposed two deep learning architectures to automatically detect COVID-19 infected cases in an X-ray dataset. Both include the AlexNet architecture which uses a transfer learning procedure. They also account for the temporal properties via a dedicated layer in their model.
In [34], the authors introduced a two steps CNN-based approach. First, they pre-processed X-ray images to eliminate some portions of the diaphragm. Second, they processed the original X-ray images again using various techniques, including histogram equalization and a low-pass filter. The resulting images were then classified using a Transfer Learning CNN architecture into either COVID-19 infected or non-COVID-19 infected.
3.1.2 Analyzing X-Ray Images for Symptom Detection
An ACGAN-based model was developed in [35]. Known as CovidGAN, it was implemented on a repository of chest X-ray scans from COVID infected patients and healthy individuals. The authors studied the performance of the proposed approach with a synthetic data augmentation model, where synthetic data augmentation was used to enlarge the dataset. CovidGAN has been used to provide synthetic X-ray scans. The authors observed a better classification performance when the model was trained using both actual data and synthetic augmentation.
In [36], a method of examining chest X-ray images to detect COVID-19 symptoms was presented. The solution consists of three predicting layers: the prophet algorithm, the autoregressive integrated moving average model, and the long short-term memory neural network. The proposed method can help to predict the number of confirmed COVID-19 cases, the number of recovered cases, and the number of deaths over the next few days. Empirical findings showed that the technique was very useful and an effective method of detecting COVID-19.
The authors of [37] proposed a deep learning framework that combined a CNN model and long short-term memory to identify COVID-19 infected cases from chest X-ray scans in an automated fashion. The CNN model and long short-term memory were used to perform a deep feature extraction and detection using the extracted features respectively.
Abraham et al. [38] have also investigated the possibility of combining various pre-trained CNN models for COVID-19 detection from chest X-ray scans. Their proposed approach involved combining features extracted from a multi-CNN model with a correlation-based feature selection technique and Bayesnet classifier. The developed system was evaluated using two publicly available repositories.
Like the work in [37,38], Ismael et al. [39] adopted several popular deep learning approaches to classify chest X-ray images into COVID-19 infected and non-COVID-19 infected. They used deep feature extraction techniques, pre-trained CNN model, and end-to-end training of a developed CNN model. To elaborate, they used pre-trained CNN models such as ResNet18, ResNet50, ResNet101, VGG16, and VGG19 for deep feature extraction. They also used the Support Vector Machine for the classification of the deep features. Moreover, a CNN model with end-to-end training was proposed.
The authors of [40] devised a five convolutional-layer network to help train the CNN model from scratch. Then, the developed CNN model was exploited to perform a deep feature extraction. The extracted features were used as input to the k-nearest neighbour, support vector machine, and decision tree algorithms.
3.2 Methods for Re-ranking COVID-19 Queries
Research re-ranking methods are used as an additional processing layer in many information retrieval systems. Upon running a query, the initial ranking list is sorted based on the similarities between the query and occurrences in the data collection [41]. The initial ranking list is then refined based on the neighbourhood relations among all identified instances.
The authors of [42] presented the zero-shot ranking algorithm (SLEDGE-Z) which is an adaptation of a ranking framework for COVID-19 scientific literature searches. The proposed method was based on filtering the training data down to medical-related queries. It used a neural re-ranking model that was pre-trained using scientific text and filtered the target document collection using a contextualized language model.
Su et al. [43] developped a real-time question answering (QA) and multi-document summarization system called CAiRE-COVID. Their approach aimed to tackle some of the challenges related to mining the huge number of scientific documents focused on COVID-19 and to provide answers to the most important and urgent queries from the community. Their solution combined information extraction with the most advanced query focused multi-document summarization techniques.
The work in [44] proposed an end-to-end document re-ranking model called PARADE and demonstrated its effectiveness on some ad-hoc benchmark datasets (i.e., TREC-COVID). The authors also investigated how model size affects performance and analyzed dataset characteristics to identify when representation aggregation strategies are more effective.
MacAvaney et al. [45] developed a search engine called Neural Covidex that incorporated the latest neural ranking models to grant access to the COVID-19 Open Research Repository maintained by the Allen Institute for AI. The proposed work suffered from a lack of evaluation.
Conclusive Summary: As discussed in Section 3.1, many CNN-based proposals in the literature have used Transfer Learning methods for COVID-19 detection [28–29,32–34]. All surveyed proposals, such as the ones reported in [35,42] suffer from a lack of appropriate COVID-19 datasets, which could lead to a problem with the quality of the results. In contrast to state-of-the-art [28,32,34], our proposed approach develops a deep learning solution to discover recurring patterns in COVID-19 infection data. The solution works as a pipeline framework that involves two stages: a baseline classification using a CNN model, and a re-ranking framework for COVID-19 symptom identification. The latter exploits the diversity of different parts of CT images. Baseline results are re-ranked using distances to CT image parts’ features vectors. The re-ranking method is added to improve the overall discriminatory power. It provides an extra verification phase in the identification process by utilizing the semantics of existing information. Therefore, our proposed solution outperforms existing approaches [32–38] in the sense that it fully exploits the information involved in a single image to improve the retrieval accuracy, when no other CT images are available.
Having reviewed the state-of-the-art, we now detail the proposed solution–based on computed tomography in patients–for detecting symptoms that are suggestive of COVID-19 infection. First, we present some preliminaries (summaries of the necessary baselines) in Section 4.1, followed by details of the proposed image re-ranking method in Section 4.2.
The solution works by exploiting a deep learning method that predicts whether an individual is affected with COVID-19 by analyzing his/her CTs based on 275 CT scans. One of the central features of the proposed solution is to totally use the information encapsulated in a single image to improve the recognition accuracy, while no other CT images are available. To ensure the accuracy of infection detection from a single image, a re-ranking framework for COVID-19 symptom identification is used that takes advantage of the diversity of different parts of a CT image. Specifically, given an image, we first divide it into sub-parts. Then, the distance between each sub-feature of the query and the gallery images is iteratively encoded into a new distance vector. Finally, a majority vote between the classes of the Top-5 distances is taken to identify the new class of query image.
As shown in Fig. 3, based on the above considerations, the proposed approach is a pipeline of two steps.
1. First, a baseline CNN that provides the first classification list of COVID-19 images.
2. The second step consists of a re-ranking process (features division and fusion) based on the sub-features parts (see Fig. 3). It aims to generate more positive samples to verify the first diagnosis and provide the best classification.
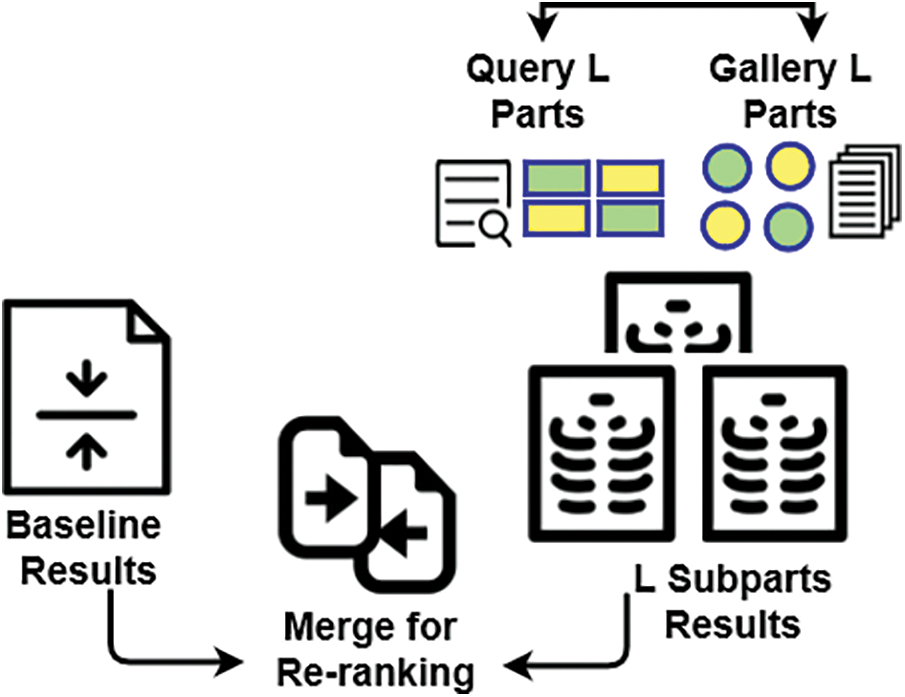
Figure 3: Illustrative overview of the proposed approach
The baseline results are enhanced based on a re-ranking algorithm relying on the similarity between probe L parts and most L neighbors’ parts of gallery images, as will be explained in Section 4.2.
4.1 Backbones and Study Preparations
Let S = (x1, y1),…,(xn, yn) be the person’s ground truth diagnosis [46] (e.g., no-pneumonia, influenza A/B, COVID-19) based on a chest Computed Tomography (CT) test, where xi and yi denote the ith image and its CT test label, respectively. It is important to note that, to evaluate the effectiveness of our re-ranking method, we use baseline Neural Networks architectures that is DenseNet169 with and without a transfer learning step.
• Baseline 1, DenseNet169 [47]. It has been shown in the literature that convolutional networks can be not only substantially deeper but also more accurate, and efficient to train, if they include shorter connections between layers close to the input and those close to the output [48]. Due to this observation the Dense Convolutional Network (DenseNet) [47] was introduced. DenseNet-169 was chosen because despite this CNN has very depth layers, about of 169 layers. Also, compared to other models DenseNet is relatively low in parameters, and its architecture well addresses the vanish gradient problem [47]. DenseNet-169 connects each layer to every other layer in a feed-forward way. However, traditional architectures with L layers have L connections–one between each layer and its next layer–whereas the DenseNet169 network has L(L + 1)/2 direct connections. For each layer, the feature-maps of all preceding layers are used as inputs, and its own feature-maps are used as inputs into all subsequent layers. Generally, DenseNets have important compelling advantages such as they address the critical gradient problem, strengthen feature propagation, reuse features, and greatly reduce the number of parameters.
• Baseline 2, Self-Trans [47]. Due to the lack of COVID-19 images, hence, to tune the network weights pre-trained on original data a self-supervised transfer learning approach can be applied to transfer learning process. In Self-Supervised Learning (SSL), an auxiliary task on CT images is performed. Hence, the authors introduce supervised labels in these tasks from the images themselves without any human contribution. Then, network weights are fine-tuned by applying these auxiliary tasks. Self-Trans model is pre-trained on ImageNet that performs SSL on Lung Nodule Analysis (LUNA) without using labels. Then, perform SSL on the COVID-CT-Dataset without using labels of COVID-19 CT, and finally adjust the COVID-19 CT using labels.
4.2 Proposed Re-ranking Method
The ability of re-ranking methods, such as features division, to perform an additional verification phase in order to enhance the initial classification list ranks, is the major motivation of our proposition in this work. This advantage allows us to perform an extra verification step using semantic local information. In this phase of the solution, we simulate the medics’ (e.g., doctors, nurses, paramedics) intervention as additional information to assist in a COVID-19 first diagnosis. Hence, the proposed approach aims to divide the initial probe CT image features into small sets. The positive and negative gallery CT images are also divided into the same small feature sets. The final classification decision is generated based on the classes of the Top-5 distances between probe image parts and positive and negative gallery CT image samples. The assumption of this proposition is that local features (features from image parts) may be more informative than the global features in the case of CT image analysis.
Feature is divided into L parts, namely sub-features (more details are presented in Section 4.2.2) as long as L is relatively small and informative. Consider a probe image p and the gp and gn positive and negative gallery CT images respectively. We divide the features of each image (p, gp and gn) into sub-features. Let xpi be the i sub-features of the probe image and xgpj, xgnj be the the j sub-features of the positive and negative gallery CT images respectively. The initial positive distance between each features part Lpi of the probe image and y, the corresponding features parts Lgpj and Lgnj of positive and negative gallery CT images respectively. These distances are generated as per the mathematical notations in the following Eqs. (1) and (2).
where dp and dn are distances between positive (pi) and negative (nj) sub-features of probe image (x) and gallery image (g) for the full dimension of the sub-features vector.
Then, the probe image represents similarities of gallery images and sub-features are created by sorting already generated distances (see Fig. 4). To match the probe image in the positive or negative CT image category, a majority vote between the top-5 distances’ category is used. Using this re-ranking method, the new class of the probe image is predicted. A binary classification rate is then re-ranked, and the class of the instance x (probe image) is re-predicted based on the class of the Top-5 ranked images.
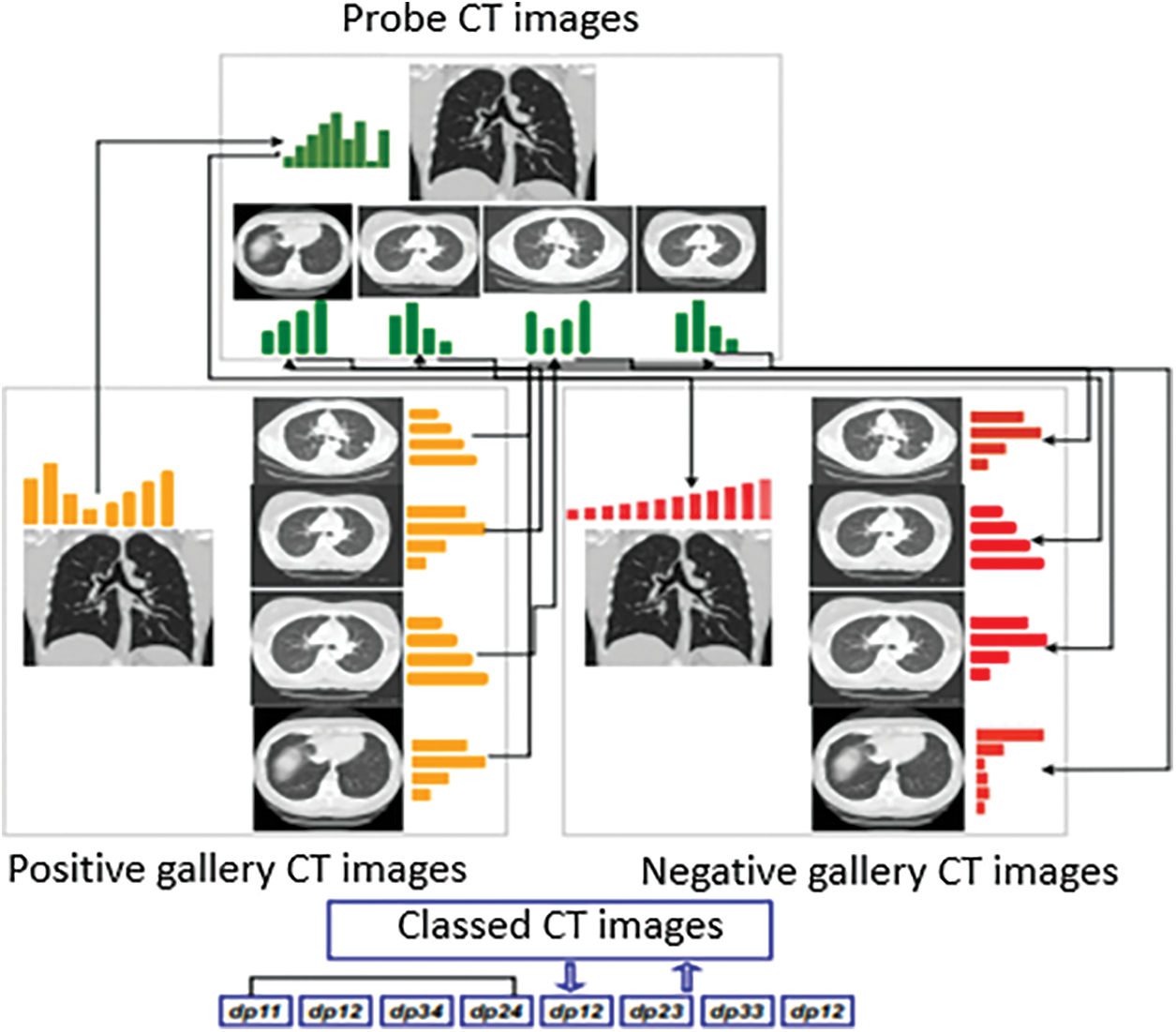
Figure 4: Overview of feature division for re-ranking
In the above process of creating the instantaneous set, re-ranking and prediction are performed in the test set and involve only two categories. This post-processing of the initial classification list adds only small computational cost as compared to other re-ranking methods. The proposed re-ranking process is explained in Fig. 4.
Accurate CT image segmentation is very important and is the initial step in our proposition. The segmented areas are easily affected by their surroundings because of the high degree of similarity between the gray values in the CT image. Limitations such as this lead to the loss of semantic information [49]. Hence, despite the divide-and-fuse re-ranking approaches, we do not adopt a feature division scheme. To split an image, elements can be selected manually, randomly, or also based on image segmentation techniques. In this work, we use the manual approach for clinical reasons, as explained above. The element selection from CT images is achieved by dividing the initial image into four parts which guarantees no loss of any important information. Generally, by dividing the CT image into L parts and computing L sub-features vectors, we will have in result sub features with more different, that permits to highlight COVID-19 location and features.
For the complexity analysis of our re-ranking approach several mean parameters will be studied: i) sub features parts (L) similarity; ii) the image size; and iii) parts number. Considering that image size is N and in binary classification process we complete a pairwise distance comparison for all probe and positive and negative gallery CT image pairs, the computation complexity for the initial distances calculation and for the sub features parts similarity calculation are both Θ(N × L × 2). The ranking process computation complexity is Sp × Θ(N × L × 2), where Sp is the size of the probe set. To reduce this complexity, in practical applications, sub-features maps of gallery positive and negative CT images (xgpj and xgnj) can be calculated offline. Therefore, for a new probe image p, we first extract its features and compose its sub-features parts list, and then we compute the distances between xpi and xgpj, xgnj. However, the computation complexity of the ranking process for final distance is then is not considered.
5 Experimental Evaluations and Results
Below we present the results of the experimental evaluations carried out to validate the proposed solution. Section 5.1 provides information on the dataset used for evaluation, Section 5.2 deals with the implementation details, and the evaluation of the proposed method for COVID-19 symptom detection is covered in Section 5.3.
The COVID-CT-Dataset consisted of 349 CT images, all of which were COVID-19 related clinical findings relevant to 216 patients. These CT images were of different sizes, resolutions, and dimensions. The minimum, average, and maximum heights of the images were 153, 491, and 1853 pixels respectively. Image dimensions were 124, 383, and 1485 pixels for minimum, average, and maximum width respectively [47]. Fig. 5 shows several samples of the COVID-19 CT images. For those patients annotated as positive CT subjects, 169 had age information and 137 had gender information. Fig. 6 (left) presents the age distribution of those who had COVID-19. Fig. 6 (right) illustrates the gender distribution of patients annotated as COVID-19 positive subjects. It is clear that there were more male than female patients (86 and 51 respectively).
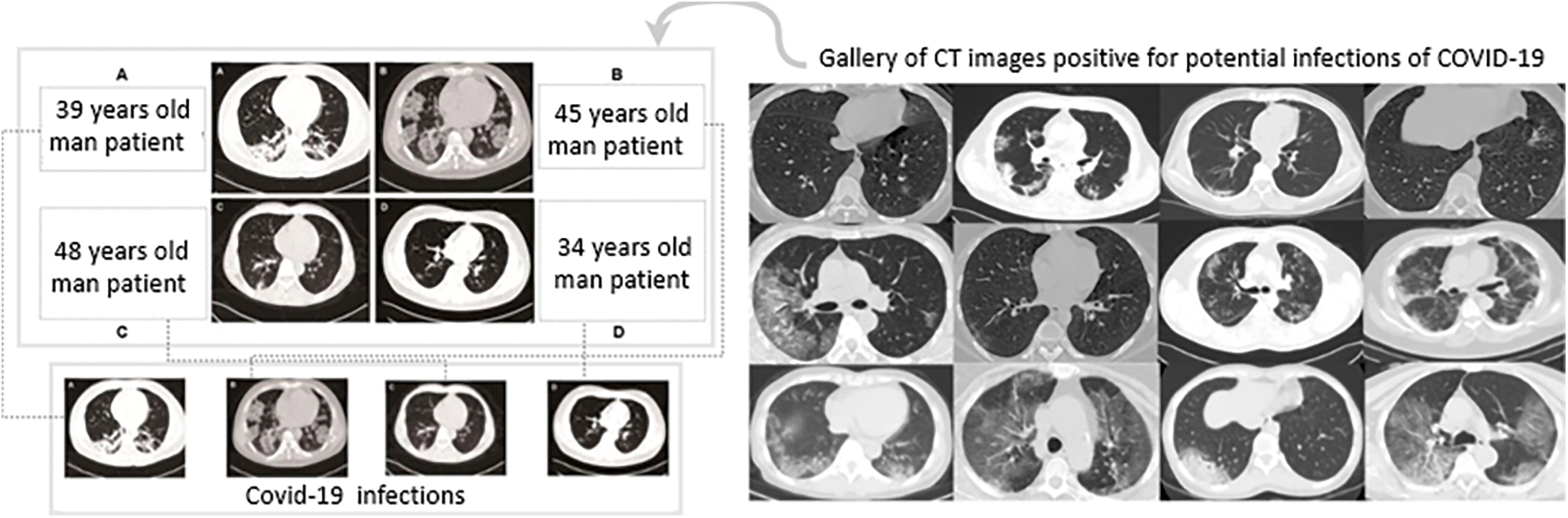
Figure 5: (Left) Any figure that contained multiple CT images as sub-figures was manually split into separate CTs: (A, B) Patients with bilateral ground glass opacities, (C, D) Patients with patchy shadows. (Right) Examples of CT images that were positive for COVID-19 [47]
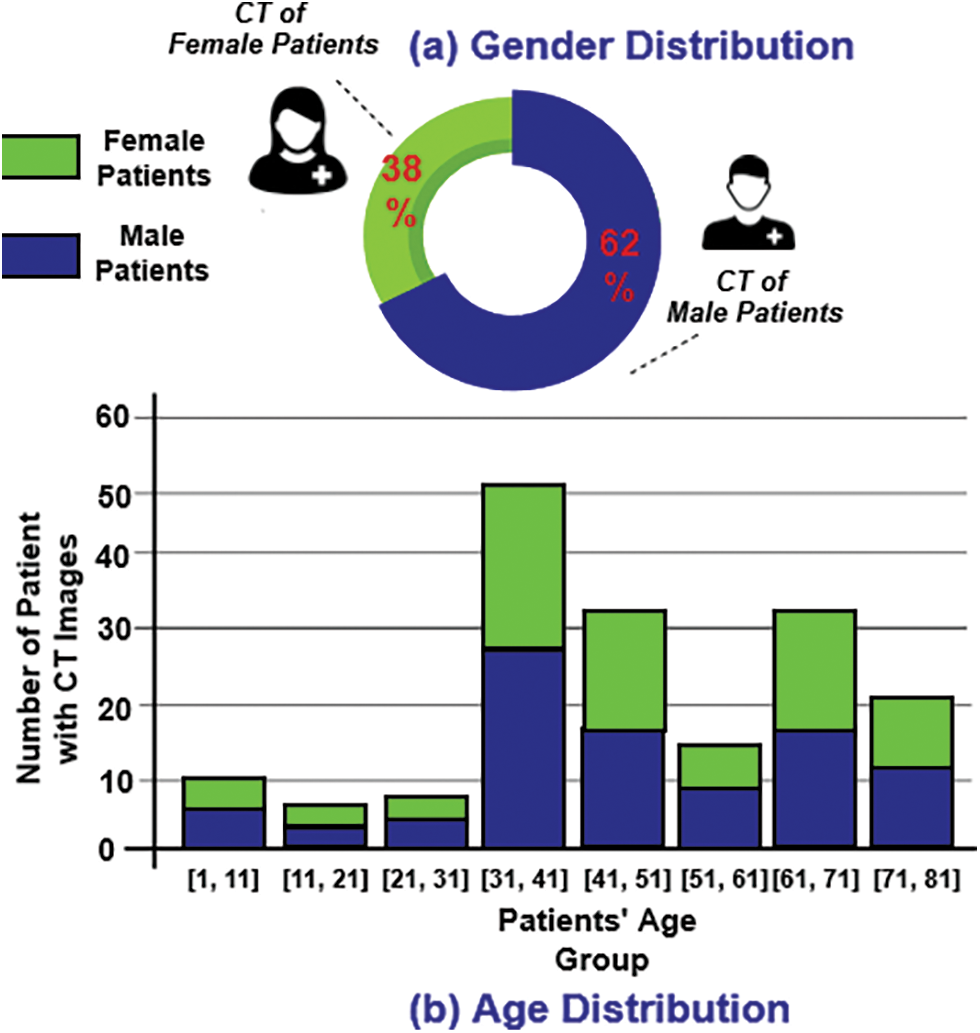
Figure 6: (a) Gender distribution of COVID-19 patients. (b) Age distribution of COVID-19 patients
First of all, we used PyTorch to implement CNNs models. Second, to make the CNNs faster and more stable, we normalized the layer inputs by re-centring and re-scaling. Since we were addressing a binary classification problem (COVID and non-COVID CT images), binary cross-entropy was used as the loss function. The networks were trained with four Giga Texel Shader eXtreme (GTX) 1080Ti Graphics Processing Units (GPUs) using data parallelism. Hyper-parameters were tuned on the validation set. To improve training performances with additional features we adopted a data augmentation step. To generate, and improve the database with other samples, various random transformations, such as random cropping with a scale of 0.5 and horizontal flip, were applied to each image in the COVID-CT-Dataset. Color jittering was also applied with random contrast and random brightness with a factor of 0.2. Furthermore, the Adam optimizer [50] was used with an initial learning rate of 0.0001 and a mini-batch size of 16 to train the classifier. The learning rate was adjusted, by applying the cosine annealing scheduler on the optimizer, to a period of 10 during the training process.
5.3 Evaluation of the Proposed Re-ranking Method
5.3.1 Test Protocol and Evaluation Metrics
To conduct the experiments and illustrate the performance of the proposed re-ranking approach, CT images were first resized to 224 × 224. Tab. 1 shows the statistics for the three sets in terms of images number. We also split the dataset into a training set, a validation set, and a test set by patient. Hence, training, validation and test portions were about 57%, 16%, and 27% respectively, as presented in Tab. 2.


Furthermore, we adopted 3 evaluation metrics, namely: (1) Accuracy (ACC), which is a measure of the percentage of positive predictions; (2) F1-score, present how much precision goes with recall; and (3) Area Under the roc Curve (AUC), which is the area under the curve showing the inversely proportional relation between false positive rate and true positive rate.
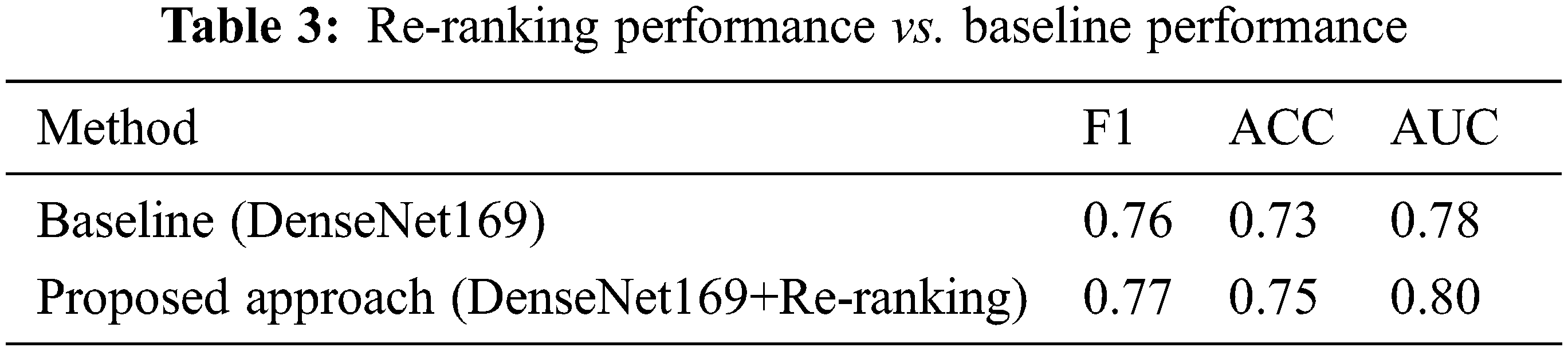
In this section, we evaluate the performance of the proposed re-ranking approach and compare it to the baseline systems. Tab. 3 summarizes all the results. It is clear from Tab. 3 that the re-ranking process enhances the performance of the baseline model by 0.01%, 0.02% and 0.02% for the F1 score, ACC, and AUC respectively.
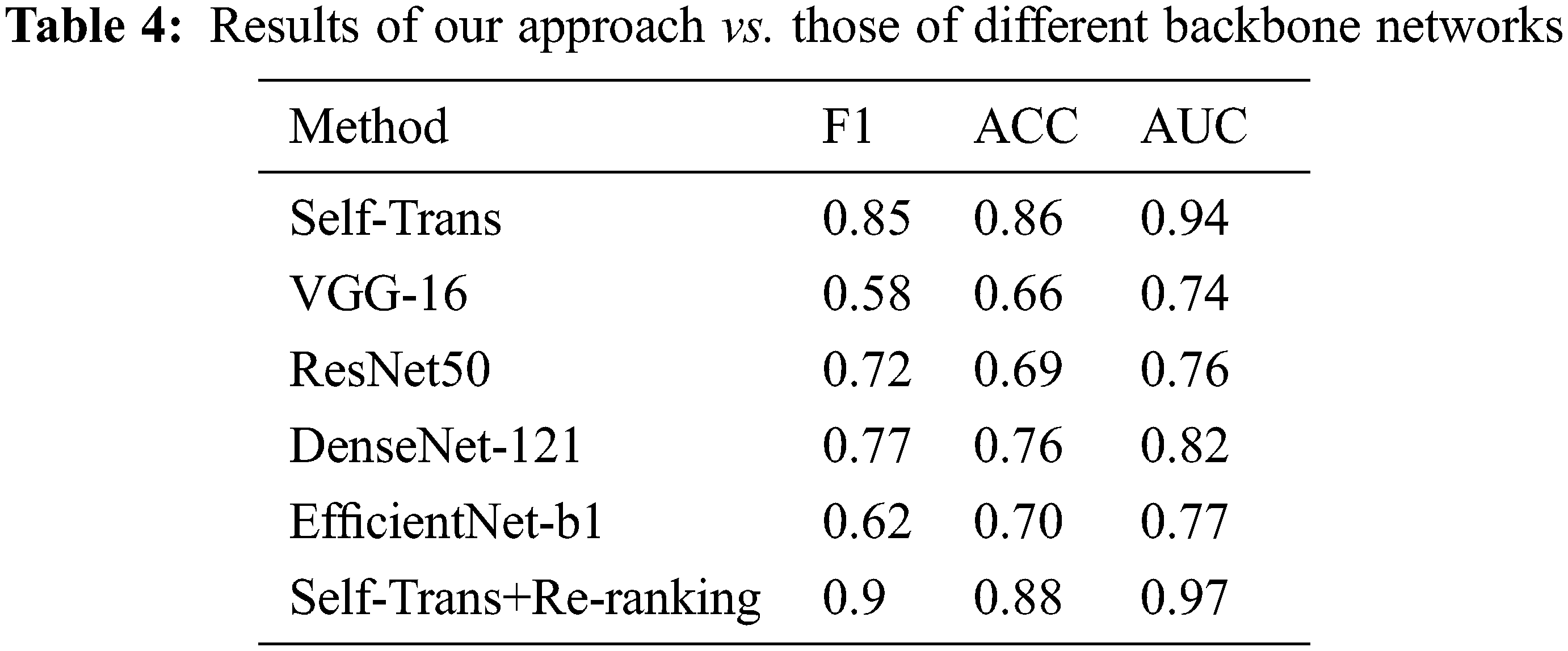
In order to highlight the performance of our proposed process, we compared it to several state-of-the-art approaches, like [47,51–54]. Concretely, to study the effect of re-ranking factor in neural architectures, we experimented with different backbone networks, including Self-Trans [47], VGG16 [51], ResNet50 [52], DenseNet-121 [53], and EfficientNet-b1 [54]. Tab. 4 illustrates the performance of these approaches in terms of F1 score, ACC, and AUC. This comparative study clearly shows that our proposed method outperforms the existing approaches in terms of these three measures. The proposed approach is also better than the Self-Trans model. Hence, the effectiveness of the re-ranking process is better than that of the hard learning transformation process which is used to build the Self-Trans model. The comparative study demonstrates the ability of the proposed method to predict whether a person is infected with COVID-19 by analyzing his/her CTs images. Also, the adopted re-ranking method based on dividing query image into sub-features allows us to overcome the lack of CT images and enhance the learning accuracy of the CNN models. Thus, this study has shown that it is possible to fully exploit the features encapsulated in one CT image to enhance classification accuracy when there are no other CT images available.
AI techniques, data-driven models, and image processing can provide decision support as well as increase the efficiency and effectiveness of diagnosing potential symptoms of COVID-19 infections. Accordingly, in this paper, we explored the methods used to assist the diagnoses of COVID-19. Most previous work has adopted CNN techniques for classification. Since Deep learning models perform well in medical image classification tasks, CNNs need huge datasets to learn effectively. Available CT-COVID-19 datasets are not yet large enough, so the research question addressed by this work is “how to fully exploit the information involved in a single image to improve the retrieval accuracy, while no other CT images are available”.
Primary contributions and implications: The core contribution of the proposed solution includes using a pipeline process composed of 2 steps: i) a CNN model is used to provide a baseline classification of CT images into COVID-19 affected and non-COVID-19 affected images; ii), and a re-ranking to improve baseline results is carried out without using an extra fine-tuning phase.
The proposed solution and its evaluation can have implications for research and practices. Specifically, medical practitioners and stakeholders who can rely on AI-driven smart systems that can provide them data-driven intelligence to manage the impacts of the pandemic. The proposed solution can empower medics and public health workers to identify the potential infections in an automated and efficient manner. Moreover, researchers and developers in computing and medical sciences who are interested in exploring and applying the role of Information and Communication Technologies (ICT) in health informatics to encounter the current wave of pandemic. The role of AI systems, specifically deep learning solutions, advances the state-of-the-art in autonomic computing technologies in the context of smart healthcare.
Experimental results show that this model provides competitive performance in comparison with state-of-the-art methods. Binary cross-entropy was used as the loss function and the Adam optimizer was used with an initial learning rate of 0.0001 and a mini-batch size of 16 to train the classifier. The findings of this research could be beneficial to medical practitioners, who can rely on data-driven autonomous systems to identify potential infections in a non-invasive manner, and to research and development engineers in information systems and autonomic computing who rely on artificially intelligent models and systems to support smart healthcare in the context of the COVID-19 pandemic.
Potential limitations and vision for future research: Future work is to explore other re-ranking algorithms that may enhance results. Also, more deep baselines should be tested.
• Exploiting image processing techniques with application of deep learning to discover recurring patterns of COVID-19 infections via Computed Tomography images. Application of re-ranking algorithm, based on divide and fusion method, that can utilize local features from CT images (potential COVID-19 infections), to support solution performance.
• Leveraging the visual information available in a single image, dividing, and classifying parts of a single image to detect the symptoms of a potential infection.
Funding Statement: The authors would like to acknowledge the support and funding for this project received from the Deanship of Scientific Research at the University of Ha’il, Kingdom of Saudi Arabia, through Project Number RG-20155.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. D. L. Heymann, T. Prentice and L. T. Reinders, “The world health report 2007: A safer future: Global public health security in the 21st century,” World Health Organization, 2007. [Google Scholar]
2. World Health Organization. WHO Coronavirus (COVID-19) Dashboard. 2021, URL https://covid19.who.int/. [Google Scholar]
3. Z. A. A. Alyasseri, M. A. Al-Betar, I. A. Doush, M. A. Awadallah, A. K. Abasi et al., “Review on COVID-19 diagnosis models based on machine learning and deep learning approaches,” Expert Systems, vol. 39, pp. e12759, 2021. [Google Scholar]
4. S. L. Pan and S. Zhang, “From fighting COVID-19 pandemic to tackling sustainable development goals: An opportunity for responsible information systems research,” International Journal of Information Management, vol. 55, pp. 102196, 2020. [Google Scholar]
5. C. Boldrini, A. Ahmad, M. Fahmideh, R. Ramadan and M. Younis, “Special issue on IoT for fighting COVID-19,” Pervasive and Mobile Computing, vol. 77, pp. e101492, 2021. [Google Scholar]
6. L. Garg, E. Chukwu, N. Nasser, C. Chakraborty and G. Garg, “Anonymity preserving IoT-based COVID-19 and other infectious disease contact tracing model,” Ieee Access, vol. 8, pp. 159402–159414, 2020. [Google Scholar]
7. S. Kumar, R. D. Raut and B. E. Narkhede, “A proposed collaborative framework by using artificial intelligence-internet of things (AI-IoT) in COVID-19 pandemic situation for healthcare workers,” International Journal of Healthcare Management, vol. 13, no. 4, pp. 337–345, 2020. [Google Scholar]
8. S. S. Vedaei, A. Fotovvat, M. R. Mohebbian, G. M. Rahman, K. A. Wahid et al., “COVIDSAFE: An IoT-based system for automated health monitoring and surveillance in post-pandemic life,” Ieee Access, vol. 8, pp. 188538–188551, 2020. [Google Scholar]
9. W. He, Z. J. Zhang and W. Li, “Information technology solutions, challenges, and suggestions for tackling the COVID-19 pandemic,” International Journal of Information Management, vol. 57, pp. 102287, 2021. [Google Scholar]
10. F. Provost and T. Fawcett, “Data science and its relationship to big data and data-driven decision making,” Big Data, vol. 1, no. 1, pp. 51–59, 2021. [Google Scholar]
11. T. T. Nguyen, Q. V. H. Nguyen, D. T. Nguyen, E. B. Hsu, S. Yang et al., “Artificial intelligence in the battle against coronavirus (COVID-19A survey and future research directions,” arXiv preprint arXiv:2008.07343, 2020. [Google Scholar]
12. F. Shaikh, J. Dehmeshki, S. Bisdas, D. R. Dupont, O. Kubassova et al., “Artificial intelligence-based clinical decision support systems using advanced medical imaging and radiomics,” Current Problems in Diagnostic Radiology, vol. 50, no. 2, pp. 262–267, 2021. [Google Scholar]
13. J. Nayak, B. Naik, P. Dinesh, K. Vakula, P. B. Dash et al., “Significance of deep learning for covid-19: State-of-the-art review,” Research on Biomedical Engineering, vol. 2021, pp. 1–24, 2021. [Google Scholar]
14. C. Shorten, T. M. Khoshgoftaar and B. Furht, “Deep learning applications for COVID-19,” Journal of Big Data, vol. 8, no. 1, pp. 1–54, 2021. [Google Scholar]
15. S. Bhattacharya, P. K. R. Maddikunta, Q. V. Pham, T. R. Gadekallu, C. L. Chowdhary et al., “Deep learning and medical image processing for coronavirus (COVID-19) pandemic: A survey,” Sustainable Cities and Society, vol. 65, pp. 102589, 2021. [Google Scholar]
16. A. S. Al-Waisy, M. A. Mohammed, S. Al-Fahdawi, M. S. Maashi, B. Garcia-Zapirain et al., “COVID-DeepNet: Hybrid multimodal deep learning system for improving COVID-19 pneumonia detection in chest X-ray images,” Computers, Materials and Continua, vol. 67, no. 2, pp. 2409–2429, 2021. [Google Scholar]
17. M. A. Mohammed, K. H. Abdulkareem, B. Garcia-Zapirain, S. A. Mostafa, M. S. Maashi et al., “A comprehensive investigation of machine learning feature extraction and classification methods for automated diagnosis of COVID-19 based on X-ray images,” Computers, Materials, and Continua, vol. 66, no. 3, pp. 3289–3310, 2021. [Google Scholar]
18. A. S. Al-Waisy, S. Al-Fahdawi, M. A. Mohammed, K. H. Abdulkareem, S. A. Mostafa et al., “COVID-CheXNet: Hybrid deep learning framework for identifying COVID-19 virus in chest X-rays images,” Soft Computing, vol. 24, pp. 1–16, 2020. [Google Scholar]
19. N. M. Kumar, M. A. Mohammed, K. H. Abdulkareem, R. Damasevicius, S. A. Mostafa et al., “Artificial intelligence-based solution for sorting COVID related medical waste streams and supporting data-driven decisions for smart circular economy practice,” Process Safety and Environmental Protection, vol. 152, pp. 482–494, 2021. [Google Scholar]
20. T. Alafif, A. M. Tehame, S. Bajaba, A. Barnawi and S. Zia, “Machine and deep learning towards COVID-19 diagnosis and treatment: Survey, challenges, and future directions,” International Journal of Environmental Research and Public Health, vol. 18, no. 3, pp. 1117, 2020. [Google Scholar]
21. S. N. Deepa and B. A. Devi, “A survey on artificial intelligence approaches for medical image classification,” Indian Journal of Science and Technology, vol. 4, no. 11, pp. 1583–1595, 2011. [Google Scholar]
22. Y. Guo, Y. Liu, A. Oerlemans, A. Lao, S. Wu et al., “Deep learning for visual understanding: A review,” Neurocomputing, vol. 187, pp. 27–48, 2016. [Google Scholar]
23. T. Young, D. Hazarika, S. Poria and E. Cambria, “Recent trends in deep learning based natural language processing,” IEEE Computational Intelligence Magazine, vol. 13, no. 3, pp. 55–75, 2018. [Google Scholar]
24. A. Krizhevsky, I. Sutskever and G. E. Hinton, “ImageNet classification with deep convolutional neural networks,” Advances in Neural Information Processing Systems, vol. 55, pp. 1097–1105, 2012. [Google Scholar]
25. I. Goodfellow, Y. Bengio and A. Courville, Deep Learning. Cambridge, Massachusetts: MIT press, 2016. [Google Scholar]
26. S. Pouyanfar, S. Sadiq, Y. Yan, H. Tian, Y. Tao et al., “A survey on deep learning: Algorithms, techniques and applications,” ACM Computing Surveys (CSUR), vol. 51, no. 5, pp. 1–36, 2018. [Google Scholar]
27. K. Sharma, A. Kaur and S. Gujral, “Brain tumor detection based on machine learning algorithms,” International Journal of Computer Applications, vol. 103, no. 1, pp. 7–11, 2014. [Google Scholar]
28. S. Vaid, R. Kalantar and M. Bhandari, “Deep learning COVID-19 detection bias: Accuracy through artificial intelligence,” International Orthopaedics, vol. 44, pp. 1539–1542, 2020. [Google Scholar]
29. A. Makris, I. Kontopoulos and K. Tserpes, “COVID-19 detection from chest X-ray images using deep learning and convolutional neural networks,” in The 11th Hellenic Conf. on Artificial Intelligence, Athens, Greece, pp. 60–66, 2020. [Google Scholar]
30. D. Budgen and P. Brereton, “Performing systematic literature reviews in software engineering,” in The Proc. of the 28th Int. Conf. on Software Engineering, Shanghai, China, pp. 1051–1052, 2006. [Google Scholar]
31. P. Botella, X. Burgués, J. Carvallo, X. Franch, G. Grau et al., “Iso/iec 9126 in practice: What do we need to know,” Software Measurement European Forum, vol. 2004, pp. 307–312, 2004. [Google Scholar]
32. I. D. Apostolopoulos and T. A. Mpesiana, “Covid-19: Automatic detection from x-ray images utilizing transfer learning with convolutional neural networks,” Physical and Engineering Sciences in Medicine, vol. 43, no. 2, pp. 635–640, 2020. [Google Scholar]
33. M. F. Aslan, M. F. Unlersen, K. Sabanci and A. Durdu, “CNN-Based transfer learning–BiLSTM network: A novel approach for COVID-19 infection detection,” Applied Soft Computing, vol. 98, pp. 106912, 2020. [Google Scholar]
34. M. Heidari, S. Mirniaharikandehei, A. Z. Khuzani, G. Danala, Y. Qiu et al., “Improving the performance of cnn to predict the likelihood of COVID-19 using chest X-ray images with preprocessing algorithms,” International Journal of Medical Informatics, vol. 144, pp. 104284, 2020. [Google Scholar]
35. A. Waheed, M. Goyal, D. Gupta, A. Khanna, F. Al-Turjman et al., “Covidgan: Data augmentation using auxiliary classifier gan for improved covid-19 detection,” Ieee Access, vol. 8, pp. 91916–91923, 2020. [Google Scholar]
36. M. Alazab, A. Awajan, A. Mesleh, A. Abraham, V. Jatana et al., “COVID-19 prediction and detection using deep learning,” International Journal of Computer Information Systems and Industrial Management Applications, vol. 12, pp. 168–181, 2020. [Google Scholar]
37. M. Z. Islam, M. M. Islam and A. Asraf, “A combined deep CNN-LSTM network for the detection of novel coronavirus (COVID-19) using X-ray images,” Informatics in Medicine Unlocked, vol. 20, pp. 100412, 2020. [Google Scholar]
38. B. Abraham and M. S. Nair, “Computer-aided detection of COVID-19 from X-ray images using multi-CNN and bayesnet classifier,” Biocybernetics and Biomedical Engineering, vol. 40, no. 4, pp. 1436–1445, 2020. [Google Scholar]
39. A. M. Ismael and A. Sengür, “Deep learning approaches for COVID-19 detection based on chest X-ray images,” Expert Systems with Applications, vol. 164, pp. 114054, 2020. [Google Scholar]
40. M. Nour, Z. Cömert and K. Polat, “A novel medical diagnosis model for COVID-19 infection detection based on deep features and Bayesian optimization,” Applied Soft Computing, vol. 97, pp. 106580, 2020. [Google Scholar]
41. V. Jain and M. Varma, “Learning to re-rank: Query-dependent image re-ranking using click data,” in The Proc. of the 20th Int. Conf. on World Wide Web, Hyderabad, India, pp. 277–286, 2021. [Google Scholar]
42. S. MacAvaney, A. Cohan and N. Goharian, “SLEDGE-Z: A zero-shot baseline for COVID-19 literature search,” Empirical Methods in Natural Language Processing, pp. 4171–4179, 2020. [Google Scholar]
43. D. Su, Y. Xu, T. Yu, F. B. Siddique, E. J. Barezi et al., “CAiRE-COVID: A question answering and query-focused multi-document summarization system for covid-19 scholarly information management,” arXiv preprint arXiv:2005.03975, 2020. [Google Scholar]
44. C. Li, A. Yates, S. MacAvaney, B. He and Y. Sun, “PARADE: Passage representation aggregation for document reranking,” arXiv preprint arXiv:2008.09093, 2020. [Google Scholar]
45. E. Zhang, N. Gupta, R. Nogueira, K. Cho and J. Lin, “Rapidly deploying a neural search engine for the covid-19 open research dataset,” Preliminary Thoughts and Lessons Learned, arXiv preprint arXiv:2004.05125, 2020. [Google Scholar]
46. L. Peel, D. B. Larremore and A. Clauset, “The ground truth about metadata and community detection in networks,” Science Advances, vol. 3, no. 5, pp. e1602548, 2017. [Google Scholar]
47. X. He, X. Yang, S. Zhang, J. Zhao, Y. Zhang et al., “Sample-efficient deep learning for COVID-19 diagnosis based on CT scans,” Medrxiv, 2020. [Google Scholar]
48. G. Huang, Z. Liu, G. Pleiss, L. V. D. Maaten and K. Weinberger, “Convolutional networks with dense connectivity,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 2019, 2019. [Google Scholar]
49. H. Xia, W. Sun, S. Song and X. Mou, “Md-net: Multi-scale dilated convolution network for CT images segmentation,” Neural Processing Letters, vol. 51, no. 3, pp. 2915–2927, 2020. [Google Scholar]
50. D. Kingma and J. Ba, “Adam: A method for stochastic optimization,” in Int. Conf. on Learning Representations, San Diego, California, 2014. [Google Scholar]
51. K. Simonyan and A. Zisserman, “Very deep convolutional networks for large-scale image recognition,” arXiv preprint arXiv, pp. 1409–1556, 2020. [Google Scholar]
52. K. He, X. Zhang, S. Ren and J. Sun, “Deep residual learning for image recognition,” in The Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, Honolulu, HI, USA, pp. 770–778, 2017. [Google Scholar]
53. G. Huang, Z. Liu, L. V. D. Maaten and K. Q. Weinberger, “Densely connected convolutional networks,” in The Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, Long Beach, CA, USA, pp. 4700–4708, 2019. [Google Scholar]
54. M. Tan and Q. V. Le, “Efficientnet: Rethinking model scaling for convolutional neural networks,” in Int. Conf. on Machine Learning, California, USA, pp. 6105–6114, 2019. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |