DOI:10.32604/iasc.2022.024495

| Intelligent Automation & Soft Computing DOI:10.32604/iasc.2022.024495 |  |
| Article |
Detection of Microbial Activity in Silver Nanoparticles Using Modified Convolution Network
1Department of Biotechnology, Udaya School of Engineering, Kanyakumari, Tamil Nadu, 629204, India
2Department of ECE, Ponjesly College of Engineering, Nagercoil, Tamil Nadu, 629003, India
*Corresponding Author: D. Devina Merin. Email: merindevina@gmail.com
Received: 19 October 2021; Accepted: 08 December 2021
Abstract: The Deep learning (DL) network is an effective technique that has extended application in medicine, robotics, biotechnology, biometrics and communication. The unique architecture of DL networks can be trained according to classify any complex tasks in a limited duration. In the proposed work a deep convolution neural network of DL is trained to classify the antimicrobial activity of silver nanoparticles (AgNP). The process involves two processing steps; synthesis of silver nanoparticles and classification (SEM) of AgNP based on the antimicrobial activity. AgNP images from scanning electron microscope are pre-processed using Adaptive Histogram Equalization in the networking system and the DL classification model Deep convolution neural network (DCNN) with modified activation function Leaky ReLy is designed particularly to classify the antibacterial activity of AgNP in various concentrations. DCNN analyses the absorption maxima AgNP and categorizes the activity of microbes. The experimental analysis of the proposed method shows that the AgNP shows the maximum absorption value of 0.56 at 450 nm. The overall technique yields an accuracy of 94% through the DCNN technique. The methodology used here to develop silver particles at the nanoscale is easy and economic.
Keywords: Networking; green extract; silver nanoparticles; adaptive histogram equalization; deep convolution neural network; anti-bacterial activity
Green nanotechnology has recently developed novel and sustainable methods for the production of nanoparticles. Nanoparticles (NP) are current and distinct in terms of size, distribution and shape. Nanoparticles provide significance in diagnostics, antagonistic studies, nano electronics and the detection of extremely sensitive biomolecules due to their unique properties. Modern bio nanotechnology may create unique distinctive nanoparticles. Nanoparticles are used in huge quantities in biomolecule labels, cell labels and cancer treatment. The nanoparticles display unique optical, magnetic and catalytic properties [1]. To develop and synthesize the nanoparticles with the required size, form and operations, one requires a detailed investigation of the chemical, physical and biological properties of a particular component [2]. The usage of natural plant and micro-organisms contains plenty of hydrogen atomic particles which is helpful in the synthesis of nanoparticles [3]. Moreover, the process is biocompatible, natural reductive and low cost in comparison to the conventional process. The synthesis of nanoparticles is allowed to interact with the natural microorganism to produce the anti-bacterial agents [4]. The majority of antibacterial drugs are chemically modified natural substances. These genetically modified antibacterial agents resist the recurrence of infection caused by particular micro-organisms.
Microorganisms are generally single-celled or multicellular (dense group) living organisms known as biofilms that are invisible to the naked eyes [5]. The biofilm gives motorized integrity and adherence to biotic and abiotic surfaces, including human tissue, surgical equipment, industrial equipment and implants [6]. Considering the significance of biofilm in antibiotic resistance, eliminating bacteria requires multiple medicines which may have possible adverse effects on people and surroundings, the increased toxicity, expense and length of therapeutic drug therapies are due to the requirement for high levels of common disinfectants and antibiotics [7].
NP is extensively utilized in medical drugs because of its peculiar physical, size and chemical (p-chem) features [8]. The improvement of antibacterial properties makes them a viable antibiotic substitute [9]. NPs have been investigated on their ability to suppress microbial infections and prevent bacterial colonization by eliminating biofilms in different surface devices including catheters and prostheses [10].
Fig. 1 shows the SEM image of agglomerated silver nanoparticles. Silver and silver oxides (AgO) are auspicious antimicrobials against a wide range of bacteria, including drug-resistant types. Surface modification, intrinsic features, content and bacterial species all influence the toxify mechanism of NP against microbes [11]. Some of the major influence mechanism includes: (1) electrostatic contact causes mechanical damage to the cell wall; (2) production of reactive oxygen species induces oxidative; and (3) metal cation release causes disruption of cell and protein structures. Among the silver nanoparticles (Ag-NP), silver nitrate (AgNO3) is more suitable for the study of green nanoparticles [12]. It has an oligodynamic effect which can accumulate the bacterial cell walls and bind with biomolecule membranes [13]. It produces free radicals and reactive, oxygen species and acts as a signal modulator for microorganisms. Bacterial type and its P-chem property decide the antibacterial property of nanoparticles [14].

Figure 1: SEM image of agglomerated silver nanoparticles
For a better analysis of nanoparticles, computational assist deep learning (DL) tools can be implemented to decrease the error rate and experimental trials [15]. Deep learning, the branch of artificial intelligence that learns multiple algorithms and data to solve a complex problem without humans indeed [16]. The main capability of DL includes decision-making, problem-solving and object recognition [17]. Furthermore, these technologies are efficient and low-cost and they rely on data input. They can also estimate the impact of materials that haven't been synthesized yet, leading to safe-by-design approaches [18]. The use of deep learning for the prediction of NP toxicity profiles and the development of novel antibiotics has been successful.
Based on our study on deep learning, there is no detailed study for the prediction of the Antagonistic nature of silver nanoparticles. In the proposed work, we develop Convolution neural network model to analyze the antibacterial activity of pathogenic organisms such as Escherichia coli, Proteus vulgarizes, Klebsiella pneumonia, Pseudomonas aeruginosa. The objective of the proposed work is to design a deep learning model to evaluate the antimicrobial activity of silver nanoparticles. The extract is collected from four different species such as Cocos nucifera, Emblica officianalis, Carica papaya and Azadirachta Indica and mixed with AgNO3 at different concentrations for the synthesis of nanoparticles. The SEM (scanning electron microscope) output of nanoparticles is processed and classified using a deep learning algorithm to test the Antagonistic nature of silver nanoparticles. The main need and objective for this study are to develop a deep learning technology to synthesize nanoparticles eco-friendly. The physical and chemical modes of synthesizing silver nanoparticles need very high temperature and pressure as operating conditions. To agglomerate the synthesized nanoparticles, a stabilizer needs to be added. This biological method needs no toxic chemicals and energy-intensive methods.
The proposed study focuses on the detection of anti-bacterial, anti-fungal, anti-malarial and anti-oxidant activity of the micro-organism in different concentrations of AgNO3. The process of the proposed method involves two main steps (i) Data collection and synthesis of silver Nanoparticle, (ii) classification of Scanning Electron Microscope (SEM) Nanoparticle images using Deep convolution Neural network (DCNN). The classification of SEM images includes several steps such as pre-processing, model development, validation and classification. The overall block diagram of the proposed work is shown in Fig. 2.
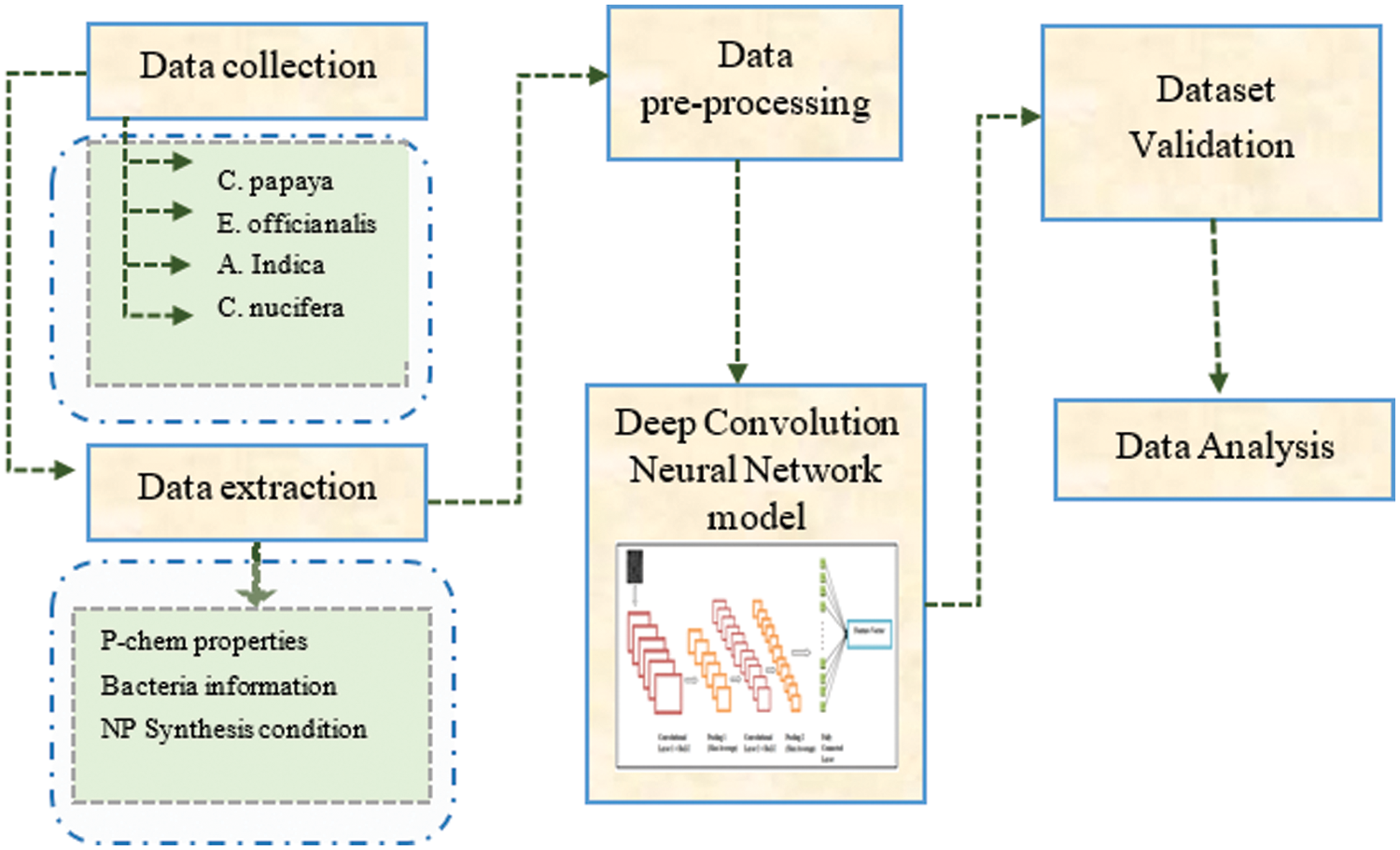
Figure 2: Overall block diagram of proposed work based on DCNN network
The SEM images of silver nanoparticles in the proposed method are produced at a specialized microbiology laboratory of the biotechnology department. The description of materials is given in the following steps. The chemical silver nitrate prepared by High Purity Laboratory Chemicals, Mumbai with 99.8% purity is used in the experiment. Leaves of Azadirachta indica, fresh fruits of Carica papaya, Emblica officianalis and the seed of Cocos nucifera are collected and confirmed by an official from the Department of Horticulture for the scientific nomenclature.
2.2 Extraction and Synthesis of Nanoparticle
The plants extracts were collected and filtered from A. indica and C. nucifera for the synthesis of the nanoparticle. All the plant extracts are mixed with silver nitrate solution individually in the ratio 9:1, 8:2, 7:3, 6:4 and 5:5 to analyze and optimize the concentrations. Then the solution is kept separately at the optimized condition for 5 h. Then the solution is tested in different medium such as SEM, TEM (Transmission electron microscope), Ultra Violet and XRD (X-ray powder diffraction). The detailed description of nanoparticles extraction is shown in Fig. 3.

Figure 3: Extraction of silver nanoparticles (AgNO3) from the plant extract
The output image of the SEM image is processed in the DCNN network to detect the anti-bacterial activity of AgNP’s. In this step, the SEM image is processed to remove the artifacts and to enhance the quality of the image. The size of the SEM image is reduced to 256 × 256 pixels and enhanced using Contrast Limited Adaptive Histogram Equalization. It is the contrast augmentation technique, broadly used in microscopic image analysis because of its ability to progress the contrast and brightness of the input image. Microscopic image analysis is not similar to the normal image analysis as it has a limited number of datasets. It causes over-fitting and irregular classification. To decrease the mentioned impacts, information extension has been performed through grouping; zooming multiple times. The mathematical expression of histogram equalization (HE) is given in Eq. (1).
Here nk, denotes the number of adjacent pixels and w is denoted as 1 in the correlation position of the relationship histogram; or else, w is denoted as 0 and gathered constantly. The Processed image is directly fed into the DCNN network to detect bacterial activity. Fig. 4 Shows the Contrast-enhanced SEM image of silver nanoparticles.

Figure 4: Contrast-enhanced SEM image of silver nanoparticles
3 Proposed Deep Convolution Neural Network (DCNN)
The deep learning convolution architecture is widely recommended in medical imaging because of its outstanding classification accuracy. The proposed DCNN consists of 10 layers: 1 input, 8 hidden (3 convolution layers, 2 pooling layers, 2 fully connected layers and 1 softmax layer) and 1 output. The SEM image of AgNO3 fed into the convolution layer. The dataset has different images of AgNP at different concentrations and to detect the antibacterial properties. Here the target class of AgNP is classified into four categories such as anti-bacterial, anti-fungal, anti-malarial and anti-oxidant activity. In this work, we developed a novel DCNN architecture with a modified additional feature map and activation layer to classify agglomerated silver nanoparticles. The detailed description of the proposed work along with its architecture is shown in Fig. 5.

Figure 5: DCNN architecture
3.1 Architecture Design of DCNN
Fig. 5 illustrates the basic function steps of a deep convolution neural network. The DCNN architecture used in the proposed study is explained in the following locution. The input layer first transforms the 28 × 28 pixel into a vector, resulting in a 784-pixel-wide feature space. This DCNN’s first layer contains 256 neurons linked to 512 weights, resulting in 512 * 256 + 256 = 1,31,328 weights to compute, including the bias. In total, there are 1,31,328 coefficients. The leaky ReLU function was employed for activation. There are now two convolutional layers based on 3 × 3 filters with average pooling. As a result, the feature space is decreased from 32 × 32 to 6 × 6. The probabilities are computed using the softmax layer, which is followed by two hidden and deep layers of 60 and 42 neurons. Through a thick convolution route, the convolution process separates background information in many scales. The softmax layer that distributes the class label numbers has received the output of the final linked layer. To perform the layer transformation, the transition block is provided. All nanoparticles were standardized and statistics were generated on the training set.
3.2 Classification Procedure of DCNN
The target class identification of the SEM images is labelled in each image. Each variable is labelled with a predetermined length (22 ms) as specified in the design. The input target is a vector probability of the target classes and the output target is an audio signal frame (x) from the sample of length (l). In the DCNN, there are 25 filters of size 3 × 3 and stride = 2. The Leaky RELU (Rectified Linear Unit) is used to convert characteristics from a linear to a non-linear space. The size of the product is unaffected by the feed-in size. When the input is negative, it may be thought of as a threshold function with a zero product.
Classification of Sub segments: The feature map
Here
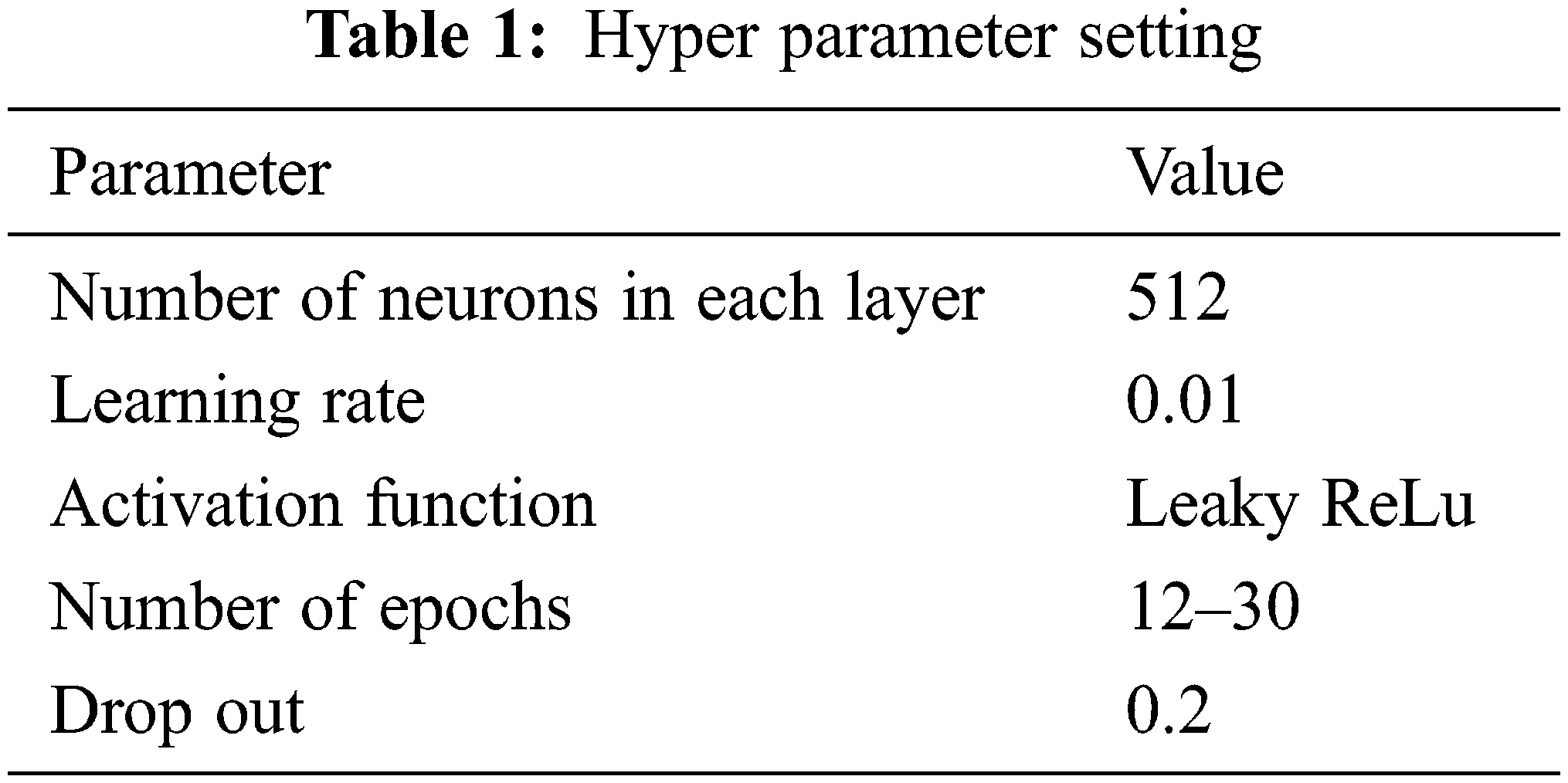
The hyper parameter setting of the proposed work is given in Tab. 1.
According to some estimation, this study necessitates a spatially-alternating activation function that alternates between space coordinates and serves as a space activation function for the content in the first half and as Wavelet domain activation in the second half. Using conventional 2D max-pooling lowers the volume sizes of the convolutional feature maps. This network classifies the four target class of silver nanoparticles based on the level of microbial activity. The experimental result of the SEM image is given in Section 3.
The AgNP activity of micro-organism is trained and tested using Matlab 2019b. The process includes two major steps such as synthesis of silver nanoparticles from Different plants extract and classification of nanoparticles based on the activity of microbes using the DCNN network. This section elaborates the experimental analysis of the proposed technology and provides a comparative analysis for the proposed DL technique.
4.1 Data Collection and Extraction of Nanoparticle
Antagonistic nature of silver particles at nano scale is studied using E. coli, P. vulgaricus, K. pneumoniae and P. aeruginosa. The Study is conducted using spread plate, pour plate, disc diffusion method. The inhibition zone is measured only after it is kept for 24 h in incubation The respiration function and permeability of microbes are drastically affected by the silver nanoparticles of size 1–10 m. Cell wall of microbe is penetrated by silver nanoparticles and the growth of microorganism is inhibited by silver nanoparticles. Further these attached silver nanoparticles, penetrate inside the cell and interact with compounds. Finally, the silver nanoparticles destroy microbe by releasing silver ions which are fatal. The signal transduction of bacteria gets affected by modulating phosphor tyrosine of peptides which lead to prevention in the growth of microorganisms. The Ultra Violet-Visible spectra reading indicates that the plant extract added at different concentrations shows the maximum absorption values at 450–550 nm. Fig. 6 shows the maximum absorbance of silver nanoparticles synthesized from different plant extract at various concentrations.

Figure 6: Maximum absorbance of silver nanoparticles synthesized from (a) A. indica, (b) C. nucifera milk, (c) C. papaya, (d) E. officianalis and (e) C. nucifera water
In the 60 + 40 concentrations, the absorbance maximum of silver nanoparticles from C. papaya is produced at 450 nm. When compared to other concentrations, silver nanoparticles produced from E. officianalis had the greatest absorbance of 0.59 at 550 nm. Absorption shows that the maximum peak for 50 + 50 concentrations with silver nitrate solution is 0.56 at 450 nm is found to be the maximum absorption found in silver nanoparticles synthesized by A. indica. The highest absorption at 450 nm is determined to be 0.42 at 50 + 50 concentration.
The absorption spectra of silver nanoparticles from plant extract at 450 nm reveal that the C. papaya fruit extract has the highest peak at 60 + 40 concentration. At 50 + 50 concentration, E. officianalis fruit extract outperforms C. papaya in terms of absorption. Fig. 7 reveals that C. Papaya and E. officianalis had high absorbance of about 0.612 and 0.61 at 450 nm respectively. When the silver nitrate solution and extract are in equal amounts, it is evident that silver nanoparticles can form quickly.

Figure 7: Absorbance maxima of silver nanoparticles synthesized from various plant sources at 450 nm
The development of microbial colonies on plates of Muller Hinton agar gets decreased and the efficiency of silver nanoparticles antagonistic nature against microbes is analyzed by dispersion of particles using a disc. The silver particles of nanoscale attach on the surface of bacteria is based on the availability of surface area. When the silver molecules get reduced to the nanoscale, the surface area of silver nanoparticles gets increased and hence this leads to the attachment of more silver nanoparticles on the cell membrane of bacteria and disturbs its function. The bactericidal quality of silver nanoparticles increases with an increase in the surface area of nanoparticles. Fig. 8 shows Antagonistic action by silver particles at nano-scale against pathogenic organisms.

Figure 8: Antagonistic action by silver particles at nano-scale against pathogenic organisms
In this section, we evaluate the performance of the DCNN network for detecting the micro-bacterial activity of silver nanoparticles. The SEM images of the silver nanoparticles are evaluated individually to measure the accuracy of the proposed DCNN technique to detect bacterial activity. Each dataset's output is compared to the ground truth images. For performance evaluation, the most commonly utilized evaluation criteria such as accuracy, selectivity and sensitivity were used. These figures are influenced by the TP, FN, TN and FP figures.
Here, TP implies the True Positive, TN-True Negative, FP-False Positive and FN-False Negative respectively. Tab. 2 describes the performance efficiency of the proposed method.
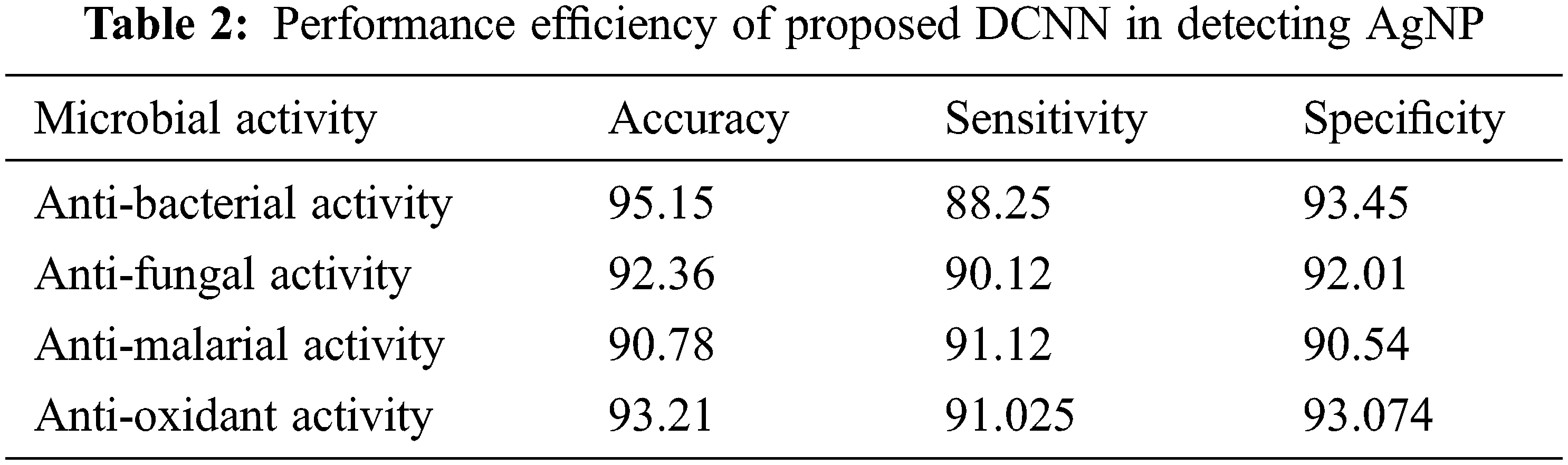
Tab. 2 elaborates the performance efficiency of the proposed Deep convolution neural network in detecting the microbial activity of silver nanoparticles. The proposed model calculate the four different microbial activity from the selected microorganism in the silver nanoparticle interaction. The microbial activity such as anti-bacterial, anti-fungal, anti-malarial and anti-oxidant. Herein, the performance measures such as accuracy, sensitivity and specificity values are calculated for the different activity medium. From the table analysis, it is observed that the proposed network had shown an overall accuracy of 92.875, 90.128 sensitivity, 92.268 specificity in detecting the microbial activity using the silver Nanoparticles.
Figs. 9–11 show the comparative analysis of the proposed technique with the existing machine learning techniques. The comparison graph of the accuracy, specificity and sensitivity projects that the proposed method has better efficiency in detecting the microbial activity of silver nanoparticles.

Figure 9: Accuracy of proposed DCNN compared to existing techniques

Figure 10: Specificity of proposed DCNN compared to existing techniques

Figure 11: Sensitivity of proposed DCNN compared to existing techniques
In Tab. 3, the average DCNN values obtained has been compared with the existing technique to evaluate the improvement percentage.
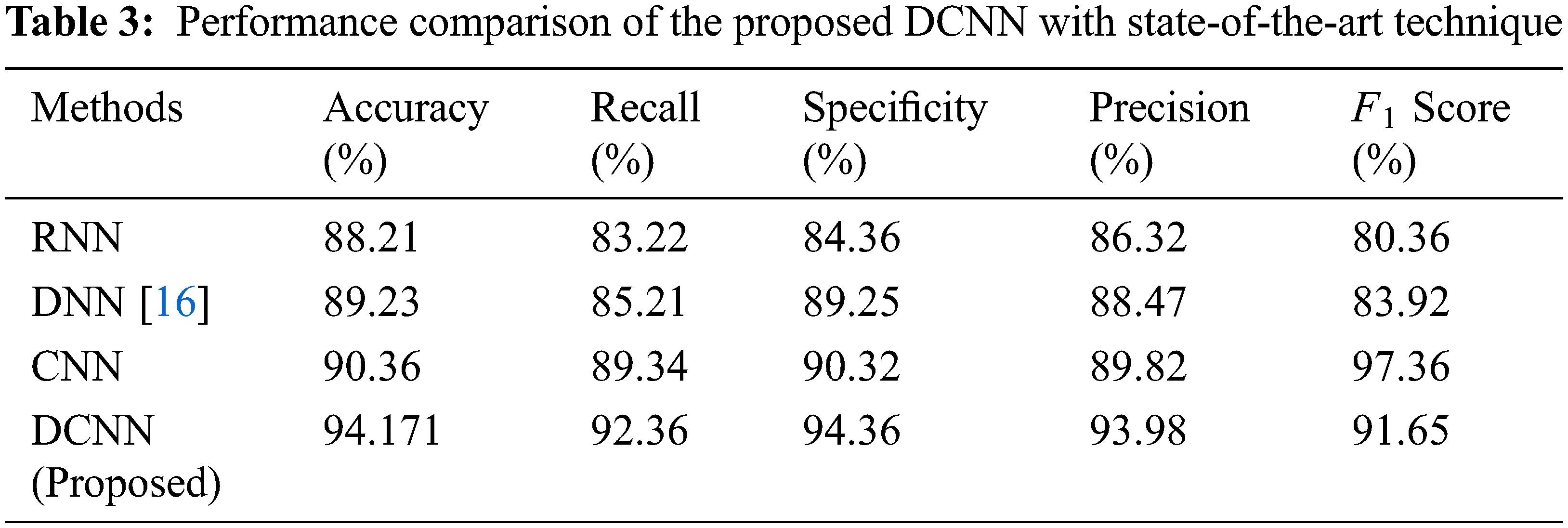
Tab. 3, denotes the comparative analysis of the proposed technique with the existing State-of-the-art Technique such as CNN, RNN and DNN. The methodology used here to develop silver particles at the nanoscale is easy and economic. The nanoparticles thus produced are constantly stable, reproducible and also eco-friendly. The overall technique yields an accuracy of 94.171%, 92.36% sensitivity and 94.36% specificity through the DCNN technique.
The proposed work focuses on the synthesis of silver nanoparticles from the green extract and determines its antibacterial activity. Silver nanoparticles show the absorbance maxima at 450 nm and when the concentration of silver particles in the nanoscale gets increased, the growth of bacterial colonies on agar plates gradually declined. The quantity of silver nanoparticles increases with an increase in the amount of silver nitrate solution added. In addition, the microbial activity of nanoparticles is determined using a deep convolution neural network by classifying the absorption activity. The deep learning network DCNN is designed with leaky ReLU activation function is designed to reduce the dying pixels. The experimental result of the DCNN network indicates that the extract of A. indica. C. papaya and E. officianalis showed the maximum absorption spectra 0.56 at 450 nm in 60 + 40 concentration and 50 + 50 concentrations respectively. 0.32 at 420 nm is the maximum absorbance of C. nucifera. The DCNN technique yields accuracy of 94.171%, 92.36% sensitivity and 94.36% specificity in detecting microbial activity. From the research, we detected that the growth of Escherichia coli and Klebsiella pneumoniae are very effectively controlled and the application of these silver nanoparticles synthesized is tremendous.
Acknowledgement: The authors with a deep sense of gratitude would thank the supervisor for his guidance and constant support rendered during this research.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. N. Suganthy, V. S. Ramkumar, A. Pugazhendhi, G. Benelli and G. Archunan, “Biogenic synthesis of gold nanoparticles from Terminalia arjuna bark extract: Assessment of safety aspects and neuroprotective potential via antioxidant, anticholinesterase and anti-amyloidogenic effects,” Environmental Science and Pollution Research, vol. 25, no. 11, pp. 10418–10433, 2018. [Google Scholar]
2. P. Boomi, R. M. Ganesan, G. Poorani, H. G. Prabu, S. Ravikumar et al., “Biological synergy of greener gold nanoparticles by using Coleus aromaticus leaf extract,” Materials Science and Engineering: C, vol. 99, pp. 202–210, 2019. [Google Scholar]
3. G. V. Khutale and A. Casey, “Synthesis and characterization of a multifunctional gold-doxorubicin nanoparticle system for pH triggered intracellular anticancer drug release,” European Journal of Pharmaceutics and Biopharmaceutics, vol. 119, pp. 372–380, 2017. [Google Scholar]
4. Z. Khan, J. I. Hussain, A. A. Hashmi and S. A. A. Thabaiti, “Preparation and characterization of silver nanoparticles using aniline,” Arabian Journal of Chemistry, vol. 10, pp. S1506–S1511, 2017. [Google Scholar]
5. P. Laokul, V. Amornkitbamrung, S. Seraphin and S. Maensiri, “Characterization and magnetic properties of nanocrystalline CuFe2O4, NiFe2O4, ZnFe2O4 powders prepared by the aloe vera extract solution,” Current Applied Physics, vol. 11, no. 1, pp. 101–108, 2011. [Google Scholar]
6. N. Saifuddin, C. W. Wong and A. A. Yasumira, “Rapid biosynthesis of silver nanoparticles using culture supernatant of bacteria with microwave irradiation,” E-journal of Chemistry, vol. 6, no. 1, pp. 61–70, 2009. [Google Scholar]
7. M. Kowshik, W. Vogel, J. Urban, S. K. Kulkarni and K. M. Paknikar, “Microbial synthesis of semiconductor PbS nano crystallites,” Advanced Materials, vol. 14, pp. 815–818, 2002. [Google Scholar]
8. M. Kowshik, S. Ashtaputre, S. Kharrazi, W. Vogel, J. Urban et al., “Extracellular synthesis of silver nanoparticles by a silver-tolerant yeast strain MKY3,” Nanotechnology, vol. 14, no. 1, pp. 95, 2002. [Google Scholar]
9. N. Ahmad, S. Sharma, V. N. Singh, S. F. Shamsi, A. Fatma et al., “Biosynthesis of silver nanoparticles from desmodium triflorum: A novel approach towards weed utilization,” Biotechnology Research International, vol. 2011, pp. 1–13, 2011. [Google Scholar]
10. S. Naqvi and S. J. S. Flora, “Nanomaterial’s toxicity and its regulation strategies,” Journal of Environmental Biology, vol. 41, no. 4, pp. 659–671, 2020. [Google Scholar]
11. E. K. Elumalai, T. N. V. K. V. Prasad, J. Hemachandran, S. V. Therasa, T. Thirumalai et al., “Extracellular synthesis of silver nanoparticles using leaves of Euphorbia hirta and their antibacterial activities,” Journal of Pharmaceutical Sciences and Research, vol. 2, no. 9, pp. 549–554, 2010. [Google Scholar]
12. M. Safaepour, A. R. Shahverdi, H. R. Shahverdi, M. R. Khorramizadeh and A. R. Gohari, “Green synthesis of small silver nanoparticles using geraniol and its cytotoxicity against fibrosarcoma-wehi 164,” Avicenna Journal of Medical Biotechnology, vol. 1, no. 2, pp. 111, 2009. [Google Scholar]
13. R. A. Jose and P. Jagatheeswari, “Neutroscopic data synthesis of biodegradable polymer for industrial bio robots,” Cognitive Systems Research, vol. 56, no. 10, pp. 72–81, 2019. [Google Scholar]
14. D. D. Merin, S. Prakash and B. V. Bhimba, “Antibacterial screening of silver nanoparticles synthesized by marine micro algae,” Asian Pacific Journal of Tropical Medicine, vol. 3, no. 10, pp. 797–799, 2010. [Google Scholar]
15. A. C. Mejías, R. A. Calvo, H. Gavilan, J. Crespo and V. Maojo, “A deep learning approach using synthetic images for segmenting and estimating 3D orientation of nanoparticles in EM images,” Computer Methods and Programs in Biomedicine, vol. 202, no. 2, pp. 105958, 2021. [Google Scholar]
16. F. M. Berrada, Z. Ren, T. Huang, W. K. Wong, F. Zheng et al., “Two-step machine learning enables optimized nanoparticle synthesis,” NPJ Computational Materials, vol. 7, no. 1, pp. 1–10, 2021. [Google Scholar]
17. M. Mirzaei, I. Furxhi, F. Murphy and M. Mullins, “A machine learning tool to predict the antibacterial capacity of nanoparticles,” Nanomaterials, vol. 11, no. 7, pp. 1774, 2021. [Google Scholar]
18. A. Appathurai, R. Sundarasekar, C. Raja, E. J. Alex, C. A. Palagan et al., “An efficient optimal neural network-based moving vehicle detection in traffic video surveillance system,” Circuits Systems and Signal Processing, vol. 39, no. 2, pp. 734–756, 2020. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |