DOI:10.32604/iasc.2022.025021

| Intelligent Automation & Soft Computing DOI:10.32604/iasc.2022.025021 |  |
| Article |
A Novel Convolutional Neural Networks-Fused Shallow Classifier for Breast Cancer Detection
Department of Radiology, College of Medicine, Qassim University, Buraidah, 52571, Saudi Arabia
*Corresponding Author: Sharifa Khalid Alduraibi. Email: sharifadurraibbi@gmail.com
Received: 08 November 2021; Accepted: 13 December 2021
Abstract: This paper proposes a fused methodology based upon convolutional neural networks and a shallow classifier to diagnose and differentiate breast cancer between malignant lesions and benign lesions. First, various pre-trained convolutional neural networks are used to calculate the features of breast ultrasonography (BU) images. Then, the computed features are used to train the different shallow classifiers like the tree, naïve Bayes, support vector machine (SVM), k-nearest neighbors, ensemble, and neural network. After extensive training and testing, the DenseNet-201, MobileNet-v2, and ResNet-101 trained SVM show high accuracy. Furthermore, the best BU features are merged to increase the classification accuracy at the cost of high computational time. Finally, the feature dimension reduction ReliefF algorithm is applied to address the computational complexity issue. An online publicly available dataset of 780 BU images is utilized to validate the proposed approach. The dataset was further divided into 80 and 20 percent ratios for training and testing the models. After extensive testing and comprehensive analysis, it is found that the DenseNet-201 and MobileNet-v2 trained SVM has an accuracy of 90.39% and 94.57% for the original and augmented BU images online dataset, respectively. This study concluded that the proposed framework is efficient and can easily be implemented to help and reduce the workload of radiologists/doctors to diagnose breast cancer in female patients.
Keywords: Artificial intelligence; machine learning; soft computing; breast cancer detection; classification
Cancer is a term related to a group of disordered dysregulated cells growth, leading to the tumor’s development [1]. Among women, one of the most frequently diagnosed cancers is breast cancer worldwide. According to World Health Organization (WHO), nearly 10 million deaths were reported in 2020 due to cancer and is the second leading cause of death [2]. Thus, women’s breast cancer is one of the leading causes of death globally (685,000 deaths in 2020). In 2020, 2.26 million new cases of breast cancer were reported, which were highest compared to other causes of cancer [2]. Therefore, early detection and differentiation between benign and malignant breast cancer lesions are important for patient treatment [3]. In case of delayed detection of breast cancer may lead lower chance of survival. Therefore, it is vital for both the treatment and the prognosis to detect breast cancer at early stages, enabling more effective treatments and significantly improving survival rates. The most established modality for early detection of breast cancer is mammography, but it has low sensitivity in detecting breast cancer in young women with dense breasts [4].
On the other hand, Breast Ultrasonography (BU) is most widely used as a primary imaging modality for early breast cancer diagnosis [5,6]. It is because BU offers the benefits like non-invasiveness, non-radioactive, and cost-effectiveness compared to others. However, despite its aforementioned advantages, it is still difficult to interpret BU images due to considerable intra-reader variability, leading to increased false-positive findings with low specificities and low positive predictive values, unnecessary biopsies, and significant discomfort patients [7]. Therefore, fast computer-aided methods are required for automatic detection of breast cancer with high accuracy [8,9].
The excellent performance of machine learning and deep learning methods in the various image recognition applications has been gaining massive attention in recent years [10–12]. Recently, several deep learning-based frameworks were developed and employed for mass breast differentiation in clinical practice [13]. In a recent study [14], the authors comprehensively discussed the developed deep learning-based automated breast cancer detection approaches. They also provided a deep insight into available online public datasets. Kwon et al. [15] evaluated the performance of two view scan techniques (2-VST) and three-view scan techniques (3-VST). They found that the 2-VST model works better as compared to others. In another study [16], CNN models were developed to classify the BU images into two classes (i.e., benign and malignant). The area under the curve (AUC) of 95% for their classifier was noted. In a subsequent study [17], an optimized deep learning model was constructed to classify the BU images. The dataset of 3739 images was used to validate their proposed model. The accuracy of their proposed model was 92.86%. Zhang et al. [18] proposed a hybrid model by combining the graph convolution network and CNN. The proposed model shows promising results and has a high accuracy of 96.10%, but the authors still think more research is needed to implement their proposed model for real-time application. In another study [19], the authors used a pre-trained network (InceptionV3, VGG16, ResNet50, and VGG19) to classify the BU images into two classes. The results show the high accuracy of pre-trained networks. However, the main shortcoming of the pre-trained model is the high computational time for the training of the predictors. Furthermore, the authors only classify the BU images into two classes (benign and malignant). However, the classifier must classify the BU images in real-time scenarios into three categories (normal, benign, and malignant). Moon et al. [20] proposed a 3-D convolution neural network model to classify the BU images. Their proposed model shows a very high accuracy of 94.62% at the cost of increased training time. To overcome the issue related to training time, shallow classifiers such as support vector machines (SVM) are also trained using morphological features of the BU images [21]. The authors only classify the BU images into benign and malignant; normal BU images were neglected for classification. Araújo et al. [22] calculated the deep features using CNN and utilized it for SVM training for four and two classes. The accuracy of 77.8% and 83.3% was noted for four and two classes, respectively. Recently Shia et al. [23] calculated the features for SVM training using the pre-trained ResNet-101 network. The AUC is utilized as an accuracy measurement metric. The reported AUC of 0.938 for the classification of BU images into two classes. A similar drawback of two-class categorization is also noted for this trained model. Therefore, more research is needed to reduce the training time with high accuracy for all three classes (benign, malignant, and normal) for real-time identifications.
In this work, BU images are utilized to diagnose cancer by fusing the advantages of convolution neural networks and shallow classifiers. BU images features are extracted by using the various pre-trained convolutional neural network layers such as AlexNet, DarkNet-19, DarkNet-53, DenseNet-201, EfficientNet-b0, GoogLeNet365, GoogLeNet, Inception-ResNet-v2, Inception-v3, MobileNet-v2, NASNet-Mobile, NASNet-Large, ResNet-18, ResNet-50, ResNet-101, ShuffleNet, SqueezeNet, and Xception. The different shallow classifiers like tree, naïve Bayes (NB), SVM, k-nearest neighbors (KNN), ensemble, and neural network (NN) are trained using extracted features to classify the BU images into three classes (benign, malignant, and normal). The relief-based filter feature selection method is used to reduce the feature dimensionality and to increase the accuracy. After comprehensive training, testing, and analysis best feature are fused to train the shallow classifier to predict the BU images.
The online available BU images dataset is used to train and validate the models in this work [24]. The dataset contained 780 ultrasound images of 500 × 500 pixels of 600 women aged 25–75 years and was collected at Behaye hospital, Cairo, Egypt. Details about the dataset are provided in Tab. 1. Further information about the dataset can be found in [24].

2.2 Pre-Trained Convolutional Neural Network Model’s Feature Extraction
Features are the variables that vary from one class image to second class image and so on. The selection of the most prominent, i.e., highly varying characteristics of images, increases the classification accuracy. Relevant features extraction from images is the most important, which can be done manually or using convolutional neural networks layers. The manual feature extraction is a time-consuming task, and accuracy depends on the variation in the images. On the other hand, the convolutional neural network is a class of deep neural networks. It uses various convolutional, pooling, and fully connected layers to build the architect of the model. The convolutional neural network shows high accuracy in the presence of a large dataset. When the training data size is small, then some of the pre-trained networks, such as AlexNet, DarkNet-19, DarkNet-53, DenseNet-201, EfficientNet-b0, GoogLeNet365, GoogLeNet, Inception-ResNet-v2, Inception-v3, MobileNet-v2, NASNet-Mobile, NASNet-Large, ResNet-18, ResNet-50, ResNet-101, ShuffleNet, SqueezeNet, and Xception can be used for the extraction of the features. In various medical imaging applications, pre-trained models are used to classify images [25,26]. The Initial letter of each notional word in all headings is capitalized.
2.3 Dimensionality Reduction: ReliefF Based Feature Selection Method
In all machine learning algorithms, the quality of attributes or features is one of the most crucial concerns. Likewise, in all learning applications, thousands of potential features are used to describe the object. Unfortunately, most learning methods cannot perform accurately in these circumstances because of irrelevant and noisy features, which provide very little information for the class. So, the feature selection is a critical task to choose a small subset of features, which contains a small set of most relevant features, used to describe the target class.
In 1992, Kira et al. [27] inspired by instance-based learning, presented a Relief algorithm as a feature selection method to train the machine learning algorithm for two-class problems. Then, Kononenko [28] introduced the extension/variant of the Relief algorithm known as ReliefF to address the issues related to the binary classification problem. It works robustly in the presence of perturbated and incomplete data. The ReliefF algorithm is illustrated in Fig. 1.

Figure 1: ReliefF algorithm [29,30]
To understand the concept of working of ReliefF algorithm, randomly initialize the instance (Ri). Then, it will search for k to its nearest neighbor for the same class (known as nearest hits (Hj)) and also for each of other classes (known as nearest misses Mj(C)). Finally, it will update the equations presented at points 7, 8, and 9 of Fig. 1; further details related to the ReliefF algorithm can be found in [29,30].
In this work, the ultrasonography machine is utilized to capture the images of a woman’s breasts. These captured images are fed to the pre-trained deep neural networks (AlexNet, DarkNet-19, DarkNet-53, DenseNet-201, EfficientNet-b0, GoogLeNet365, GoogLeNet, Inception-ResNet-v2, Inception-v3, MobileNet-v2, NASNet-Mobile, NASNet-Large, ResNet-18, ResNet-50, ResNet-101, ShuffleNet, SqueezeNet, and Xception) to compute the related features. These features are fed to shallow classifiers like the tree [31], NB [32], SVM [33], KNN [34], ensemble, and NN for the categorization. Before feeding these features to the classifiers, the ReliefF filter is used for dimensionality reduction. The effect of the feature dimension reduction ReliefF method on the prediction accuracy is also investigated. The ReliefF algorithm is separately applied for both models because each model calculated the deep features in different feature spaces. If the algorithm is applied after merging features, one model’s features are neglected due to changes in the dimensions of the feature. After the extensive testing and comprehensive analysis of all the trained models, the proposed framework for cancer diagnosis is illustrated in Fig. 2.

Figure 2: Proposed framework for the diagnostic of breast cancer
In this work, the available online dataset of breast cancer is used to validate the proposed method [24]. The details about the dataset are already presented in Section 2.1. The MATLAB 2021® environment is used to implement all the models. The personal computer has the following specifications: Intel(R) Core (TM) i7-10700 CPU @ 2.90 GHz processor with 32 GB RAM, 1 TB SSD, and a 64-bit Windows 10 Pro operating system (OS). The dataset for each class is equally divided into 80% and 20% for all the models’ training and testing, respectively.
All the shallow classifier models are trained using all the features of 18 pre-trained networks. The features are collected before the SoftMax layer of each pre-trained network. The total number of features of each network is shown in Fig. 3.

Figure 3: Total number of the feature of each pre-trained model for BU images
After these features computation, all the features are fed to the shallow classifiers. The validation accuracy is used as a comparison metric for all the models. The accuracy of all the trained models with each pre-trained model feature using 5-fold cross-validation are presented in Fig. 4.

Figure 4: Accuracy of each trained model against every pre-trained network feature
After carefully analyzing the training and testing results, it is found that the SVM trained networks show the highest accuracy compared to a tree, NB, KNN, Ensemble, and NN models. Furthermore, although the training time of each model was not taken into account for comparison, the SVM model is trained in the least time compared to others. It also noted that the SVM model trained with DenseNet-201, MobileNet-v2, and ResNet-101 shows the accuracy of 86.92%, 85.51%, and 85%, respectively. The performance of all these three SVM-trained models is presented in Tab. 2. The grid search algorithm is utilized to optimize the hyperparameters of the SVM. The true positive rate (TPR), false-negative rate (FNR), positive predictive value (PPV), and false discovery rate (FDR) are utilized to evaluate the models further. The details about calculating the TPR, FNR, PPV, and FDR are given in Eqs. (1)–(4).

The result presented in Tab. 2 reveals that the DenseNet-201 trained SVM model correctly predicted the 409 out of 437 benign BU images, whereas ResNet-101 trained SVM most correctly predicts the normal BU images. All the models have the same classification accuracy for the malignant BU images. The training time of the model is considered one of the essential factors for real-time applications. It is evident from the results that as the training features size increases, the training time of the model increases (see Fig. 3 and Tab. 2). MobileNet-v2 has the least training time as it has the smallest feature subset (1280 features per image) among these three (see Fig. 3).
After analyzing the accuracies of the entire feature subset of each network, the ReliefF algorithm is applied to each model feature subset to determine the best-related features using cross-validation. The performance of each model for the first 200 and 400 features subset is presented in Tabs. 3 and 4.


After carefully analyzing Tabs. 2–4, the accuracy of the model trained with the ReliefF feature reduction method is changed compared to the full feature trained model. It is important to note that the 200 DenseNet-201 features trained SVM model correctly predicted the 413 out of 437 benign images (see Tab. 3). However, its TPR decreases in the case of malignant class compared to the complete feature set. The accuracy of the 200 MobileNet-v2 trained SVM model is similar to full trained features, but the TPR of the malignant class is increased compared to the full features trained model. It means that the hybridization/fusion of the features of different models may result in better accuracy. The accuracy of all the models decreased when trained with the ReliefF feature reduction method using 400 features of each pre-trained model (see Tab. 4).
The results of the fused/hybrid full features set and ReliefF based feature set (200 features/model) trained model are presented in Tabs. 5 and 6.


It is evident from Tabs. 5 and 6 that the accuracy of the hybrid/fused features trained model increased as compared to the single feature trained model. The maximum efficiency of all hybrids features trained model (DenseNet-201 + MobileNet-v2 + ResNet-101) is 87.56% (see Tab. 5). In the case of the ReliefF feature reduction trained model, the DenseNet-201 + MobileNet-v2 trained model shows the highest accuracy of 90.39%. This model correctly predicts the 417 BU images out of 437. The TPR of the malignant and normal classes is also increased in this case.
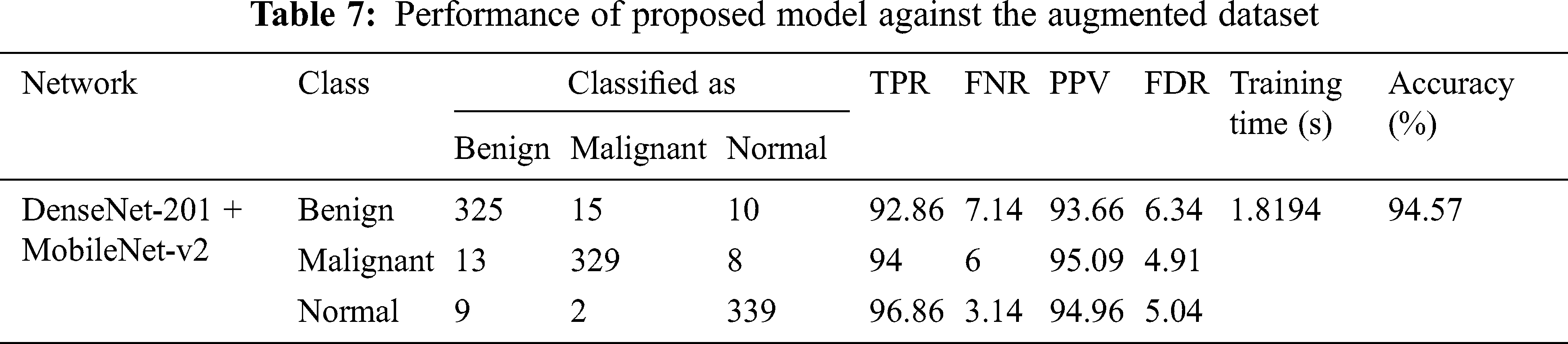
The classification accuracy of the benign class is high compared to others because the malignant and normal class has fewer images (210 and 133, respectively). In addition, augmentation techniques can be applied to increase the model accuracy [35]. This study applies the rotation operation to balance malignant and normal classes images. After augmentation, each class contains 350 BU images, augmented images are used to train the model, and the models are validated using original images. The results of augmented dataset images for DenseNet-201 + MobileNet-v2 ReliefF features trained SVM model are presented in Tab. 7.
Women BU imaging is an indispensable technique to evaluate breast lesions in the presence of a palpable mass or pain in the breast. The most established modality for early detection of breast cancer is mammography, but it has low sensitivity in detecting breast cancer in young women with dense breasts. Additionally, to prevent the risk of radiation in mammography, the BU is the most preferred diagnostic method for female patients [36]. According to a report [37], the BU imaging has a 100% accuracy for detecting palpable breast masses in 30–39 years old female patients, compared to mammography. In addition to this, BU imaging provides complete information related to solid lesions. Cysts are the primary benign lesion of the women’s breast. In BU imaging, the anechoic, thin-walled, and well-circumscribed lesions can easily be seen. Besides the non-invasiveness, non-radioactive, and cost-effectiveness of BU imaging, it can also be easily tolerated by women patients. With the advancement of deep learning methodologies, it has become a tool to assist the doctors for diagnosis of breast cancers [38–40].
In a study [41], the concept of transfer learning is utilized for training the pre-trained model for the classification of BU images. The ResNet-50 and MobileNet pre-trained models showed 97.03% and 94.42% accuracy and took almost 114.57 and 192.4 min for training, respectively. Zhang et al. [42] also proposed a deep learning model for breast cancer diagnosis using BU images. Their proposed models shows the accuracy of 92.86%. In this work, a hybrid feature reduction methodology is proposed to classify the BU images correctly. Various convolution neural network models’ features are used to train the shallow classifier model to check the classification accuracy. The SVM trained model shows the best results among all (see Tab. 2). Furthermore, the ReliefF dimension reduction method is applied to reduce the size of the feature set. It is noted that the features set with 200 sizes show better results compared to the feature set with 400 lengths (see Tabs. 3 and 4). It is because the first 200 features are more relevant for the categorization of each class. The next 200 features reduce the model’s accuracy because these features have some similarities, which reduce the classification accuracy. After that, full features are fused to increase the accuracy. It was noted that all three full feature hybrid model (DenseNet-201 + MobileNet-v2 + ResNet-101) has the accuracy of 87.56% with the training time of 15.768 s. It is noted that as the size of the feature set increases, the computational complexity of the system also increases (see Tab. 5). The ReleifF feature reduction trained model shows the best accuracy of 90.39% against the DenseNet-201 + MobileNet-v2 trained SVM model. The size of the training features dataset is only 400, which is thirteen times lesser than the full feature hybrid dataset (1920 + 1280 + 2048 = 5248), resulting in ten times lesser computational complexity. As the BU images of malignant and normal classes are lesser in quantity, the data augmentation technique is applied to balance the data resulted in high accuracy of 94.57% (see Tab. 7). As discussed in Section 1, most of the studies found in literature only worked for benign and malignant images classification. This work shows a high accuracy of 94.57% for all three classes. It concluded that the proposed methodology is robust and has high accuracy for detecting breast cancer in women as compared to [20,41]. It means that it can be implemented for real-time applications to reduce the workload and help the doctor diagnose breast cancer. However, further research is still needed to optimize the size of feature vector by applying optimization algorithm.
Due to advancements and high efficiency in medical imaging, machine learning methodologies get so much attention to manage doctors’ workload, save money and time in the hospital. In this work, the BU images-based framework is designed using deep neural networks feature trained shallow classifier (SVM) to diagnose breast cancer in women. The proposed model computed the features of BU images using the DenseNet-201 and MobileNet-v2 networks. Furthermore, the ReliefF dimension reduction algorithm is employed to reduce the computational complexity and enhances classification accuracy to the SVM model. As a result, the proposed model shows a high classification accuracy to classify the BU images in benign, malignant, and normal classes. With this high true positive rate of 94.57%, the proposed approach is suitable for diagnosing breast cancer in females.
Funding Statement: The author would like to express their gratitude to Deanship of Scientific Research, Qassim University Saudi Arabia for providing research funds for publication.
Conflicts of Interest: The author declares no conflicts of interest to report regarding the present study.
1. S. G. Kandlikar, I. Perez-Raya, P. A. Raghupathi, J. -L. Gonzalez-Hernandez, D. Dabydeen et al., “Infrared imaging technology for breast cancer detection–current status, protocols and new directions,” International Journal of Heat and Mass Transfer, vol. 108, pp. 2303–2320, 2017. [Google Scholar]
2. W. H. Organization, Cancer, 2021. [Online]. Available: https://www.who.int/news-room/fact-sheets/detail/cancer. [Google Scholar]
3. J. Anitha and J. D. Peter, “Mammogram segmentation using maximal cell strength updation in cellular automata,” Medical & Biological Engineering & Computing, vol. 53, no. 8, pp. 737–749, 2015. [Google Scholar]
4. S. Y. Kim, Y. Choi, E. K. Kim, B. -K. Han, J. H. Yoon et al., “Deep learning-based computer-aided diagnosis in screening breast ultrasound to reduce false-positive diagnoses,” Scientific Reports, vol. 11, no. 1, pp. 395, 2021. [Google Scholar]
5. M. H. Yap, G. Pons, J. Marti, S. Ganau, M. Sentis et al., “Automated breast ultrasound lesions detection using convolutional neural networks,” IEEE Journal of Biomedical and Health Informatics, vol. 22, no. 4, pp. 1218–1226, 2018. [Google Scholar]
6. W. A. Berg, L. Gutierrez, M. S. NessAiver, W. B. Carter, M. Bhargavan et al., “Diagnostic accuracy of mammography, clinical examination, US, and MR imaging in preoperative assessment of breast cancer,” Radiology, vol. 233, no. 3, pp. 830–849, 2004. [Google Scholar]
7. J. M. Lee, R. F. Arao, B. L. Sprague, K. Kerlikowske, C. D. Lehman et al., “Performance of screening ultrasonography as an adjunct to screening mammography in women across the spectrum of breast cancer risk,” JAMA Internal Medicine, vol. 179, no. 5, pp. 658–667, 2019. [Google Scholar]
8. T. Xiao, L. Liu, K. Li, W. Qin, S. Yu et al., “Comparison of transferred deep neural networks in ultrasonic breast masses discrimination,” BioMed Research International, vol. 2018, pp. 4605191, 2018. [Google Scholar]
9. W. -X. Liao, P. He, J. Hao, X. -Y. Wang, R. -L. Yang et al., “Automatic identification of breast ultrasound image based on supervised block-based region segmentation algorithm and features combination migration deep learning model,” IEEE Journal of Biomedical and Health Informatics, vol. 24, no. 4, pp. 984–993, 2019. [Google Scholar]
10. M. U. Ali, S. Saleem, H. Masood, K. D. Kallu, M. Masud et al., “Early hotspot detection in photovoltaic modules using color image descriptors: An infrared thermography study,” International Journal of Energy Research, 2021. [Google Scholar]
11. W. Ahmed, A. Hanif, K. D. Kallu, A. Z. Kouzani, M. U. Ali et al., “Photovoltaic panels classification using isolated and transfer learned deep neural models using infrared thermographic images,” Sensors, vol. 21, no. 16, pp. 5668, 2021. [Google Scholar]
12. Z. Rezaei, “A review on image-based approaches for breast cancer detection, segmentation, and classification,” Expert Systems with Applications, vol. 182, pp. 115204, 2021. [Google Scholar]
13. M. Di Segni, V. de Soccio, V. Cantisani, G. Bonito, A. Rubini et al., “Automated classification of focal breast lesions according to S-detect: Validation and role as a clinical and teaching tool,” Journal of Ultrasound, vol. 21, no. 2, pp. 105–118, 2018. [Google Scholar]
14. X. Yu, Q. Zhou, S. Wang and Y. -D. Zhang, “A systematic survey of deep learning in breast cancer,” International Journal of Intelligent Systems, vol. 37, no. 1, pp. 152–216, 2022. [Google Scholar]
15. B. R. Kwon, J. M. Chang, S. Y. Kim, S. H. Lee, S. -Y. Kim et al., “Automated breast ultrasound system for breast cancer evaluation: Diagnostic performance of the two-view scan technique in women with small breasts,” Korean Journal of Radiology, vol. 21, no. 1, pp. 25–32, 2020. [Google Scholar]
16. Q. Sun, X. Lin, Y. Zhao, L. Li, K. Yan et al., “Deep learning vs. radiomics for predicting axillary lymph node metastasis of breast cancer using ultrasound images: Don’t forget the peritumoral region,” Frontiers in Oncology, vol. 10, pp. 53, 2020. [Google Scholar]
17. X. Zhang, H. Li, C. Wang, W. Cheng, Y. Zhu et al., “Evaluating the accuracy of breast cancer and molecular subtype diagnosis by ultrasound image deep learning model,” Frontiers in Oncology, vol. 11, pp. 606, 2021. [Google Scholar]
18. Y. -D. Zhang, S. C. Satapathy, D. S. Guttery, J. M. Górriz and S. -H. Wang, “Improved breast cancer classification through combining graph convolutional network and convolutional neural network,” Information Processing & Management, vol. 58, no. 2, pp. 102439, 2021. [Google Scholar]
19. H. Zhang, L. Han, K. Chen, Y. Peng and J. Lin, “Diagnostic efficiency of the breast ultrasound computer-aided prediction model based on convolutional neural network in breast cancer,” Journal of Digital Imaging, vol. 33, no. 5, pp. 1218–1223, 2020. [Google Scholar]
20. W. K. Moon, Y. -S. Huang, C. -H. Hsu, T. -Y. Chang Chien, J. M. Chang et al., “Computer-aided tumor detection in automated breast ultrasound using a 3-D convolutional neural network,” Computer Methods and Programs in Biomedicine, vol. 190, pp. 105361, 2020. [Google Scholar]
21. C. D. L. Nascimento, S. D. D. S. Silva, T. A. d. Silva, W. C. d. A. Pereira, M. G. F. Costa et al., “Breast tumor classification in ultrasound images using support vector machines and neural networks,” Research on Biomedical Engineering, vol. 32, pp. 283–292, 2016. [Google Scholar]
22. T. Araújo, G. Aresta, E. Castro, J. Rouco, P. Aguiar et al., “Classification of breast cancer histology images using convolutional neural networks,” PLOS ONE, vol. 12, no. 6, pp. e0177544, 2017. [Google Scholar]
23. W. -C. Shia and D. -R. Chen, “Classification of malignant tumors in breast ultrasound using a pretrained deep residual network model and support vector machine,” Computerized Medical Imaging and Graphics, vol. 87, pp. 101829, 2021. [Google Scholar]
24. W. Al-Dhabyani, M. Gomaa, H. Khaled and A. Fahmy, “Dataset of breast ultrasound images,” Data in Brief, vol. 28, pp. 104863, 2020. [Google Scholar]
25. I. M. Baltruschat, H. Nickisch, M. Grass, T. Knopp and A. Saalbach, “Comparison of deep learning approaches for multi-label chest X-ray classification,” Scientific Reports, vol. 9, no. 1, pp. 6381, 2019. [Google Scholar]
26. J. Kang and J. Gwak, “Ensemble of instance segmentation models for polyp segmentation in colonoscopy images,” IEEE Access, vol. 7, pp. 26440–26447, 2019. [Google Scholar]
27. K. Kira and L. A. Rendell, “A practical approach to feature selection,” in Machine Learning Proceedings, 1992, Elsevier, pp. 249–256, 1992. [Google Scholar]
28. I. Kononenko, “Estimating attributes: Analysis and extensions of RELIEF,” in European Conference on Machine Learning, 1994, Springer, pp. 171–182, 1994. [Google Scholar]
29. M. Robnik-Šikonja and I. Kononenko, “Theoretical and empirical analysis of ReliefF and RReliefF,” Machine Learning, vol. 53, no. 1, pp. 23–69, 2003. [Google Scholar]
30. R. J. Urbanowicz, M. Meeker, W. La Cava, R. S. Olson and J. H. Moore, “Relief-based feature selection: Introduction and review,” Journal of Biomedical Informatics, vol. 85, pp. 189–203, 2018. [Google Scholar]
31. S. R. Safavian and D. Landgrebe, “A survey of decision tree classifier methodology,” IEEE Transactions on Systems, Man, and Cybernetics, vol. 21, no. 3, pp. 660–674, 1991. [Google Scholar]
32. K. A. K. Niazi, W. Akhtar, H. A. Khan, Y. Yang and S. Athar, “Hotspot diagnosis for solar photovoltaic modules using a Naive Bayes classifier,” Solar Energy, vol. 190, pp. 34–43, 2019. [Google Scholar]
33. M. U. Ali, H. F. Khan, M. Masud, K. D. Kallu and A. Zafar, “A machine learning framework to identify the hotspot in photovoltaic module using infrared thermography,” Solar Energy, vol. 208, pp. 643–651, 2020. [Google Scholar]
34. N. Ali, D. Neagu and P. Trundle, “Evaluation of k-nearest neighbour classifier performance for heterogeneous data sets,” SN Applied Sciences, vol. 1, no. 12, pp. 1–15, 2019. [Google Scholar]
35. A. Gudigar, J. Samanth, U. Raghavendra, C. Dharmik, A. Vasudeva et al., “Local preserving class separation framework to identify gestational diabetes mellitus mother using ultrasound fetal cardiac image,” IEEE Access, vol. 8, pp. 229043–229051, 2020. [Google Scholar]
36. E. S. Kim, N. Cho, S. -Y. Kim, B. R. Kwon, A. Yi et al., “Comparison of abbreviated MRI and full diagnostic MRI in distinguishing between benign and malignant lesions detected by breast MRI: A multireader study,” Korean Journal of Radiology, vol. 22, no. 3, pp. 297–307, 2021. [Google Scholar]
37. V. Sorin, R. Faermann, Y. Yagil, A. Shalmon, M. Gotlieb et al., “Contrast-enhanced spectral mammography (CESM) in women presenting with palpable breast findings,” Clinical Imaging, vol. 61, pp. 99–105, 2020. [Google Scholar]
38. D. Sheth and M. L. Giger, “Artificial intelligence in the interpretation of breast cancer on MRI,” Journal of Magnetic Resonance Imaging, vol. 51, no. 5, pp. 1310–1324, 2020. [Google Scholar]
39. K. J. Geras, R. M. Mann and L. Moy, “Artificial intelligence for mammography and digital breast tomosynthesis: Current concepts and future perspectives,” Radiology, vol. 293, no. 2, pp. 246–259, 2019. [Google Scholar]
40. E. P. V. Le, Y. Wang, Y. Huang, S. Hickman and F. J. Gilbert, “Artificial intelligence in breast imaging,” Clinical Radiology, vol. 74, no. 5, pp. 357–366, 2019. [Google Scholar]
41. A. Abdelli, R. Saouli, K. Djemal and I. Youkana, “Combined datasets for breast cancer grading based on multi-CNN architectures,” in 2020 Tenth International Conference on Image Processing Theory, Tools and Applications (IPTA), pp. 1–7, 2020. [Google Scholar]
42. X. Zhang, H. Li, C. Wang, W. Cheng, Y. Zhu et al., “Evaluating the accuracy of breast cancer and molecular subtype diagnosis by ultrasound image deep learning model,” Frontiers in Oncology, vol. 11, pp. 606, 2021. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |