DOI:10.32604/iasc.2022.023994

| Intelligent Automation & Soft Computing DOI:10.32604/iasc.2022.023994 |  |
| Article |
Improved U-Net-Based Novel Segmentation Algorithm for Underwater Mineral Image
1School of Information and Engineering, Minzu University of China, Beijing, 100081, China
2Key Laboratory of Marine Environmental Survey Technology and Application, Ministry of Natural Resource, Guangzhou, 510300, China
3National Language Resource Monitoring & Research Center of Minority Languages, Beijing, 100081, China
4School of Ocean Science, China University of Geosciences, Beijing, 100191, China
5Department of Buoy Engineering, South China Sea Marine Survey and Technology Center of State Oceanic Administration, Guangzhou, 510300, China
*Corresponding Author: Wei Song. Email: songwei@muc.edu.cn
Received: 29 September 2021; Accepted: 08 November 2021
Abstract: Autonomous underwater vehicle (AUV) has many intelligent optical system, which can collect underwater signal information to make the system decision. One of them is the intelligent vision system, and it can capture the images to analyze. The performance of the particle image segmentation plays an important role in the monitoring of underwater mineral resources. In order to improve the underwater mineral image segmentation performance, some novel segmentation algorithm architectures are proposed. In this paper, an improved mineral image segmentation is proposed based on the modified U-Net. The pyramid upsampling module and residual module are bring into the U-Net model, which are called JPU-Net, JPMU-Net and ResU-Net. These models combined the power of the residual block and the pyramid upsampling in the encoder part and in the decoder part respectively. The proposed models are tested on the Electron Microscopy images (EM) dataset and the underwater mineral image dataset. The experimental results show that JPU-Net has superior performance on the EM dataset, and JPMU-Net has a better segmentation result than existing convolutional neural network on the underwater mineral image dataset.
Keywords: Autonomous underwater vehicle (AUV); image segmentation; deep learning; underwater mineral image
The ocean occupies 70% of the earth’s total area and is rich in mineral resources. The exploration and effective mining of solid ore resources such as polymetallic sulfides, polymetallic nodules and cobalt nodules in the deep-sea can effectively alleviate the current shortage of land resources. The development of deep-sea mineral resources has become a new strategic goal for all countries. The traditional method based on cableless grab sampling or multi-frequency detection cannot study the continuous distribution law of nodule mines in deep-sea mining areas due to insufficient sampling samples, and the mining area resource evaluation accuracy is relatively low; the deep tow system is equipped with an underwater vision optical device, which uses Non-contact photographic detection makes continuous visual sampling of minerals in the mining area possible without destroying the seabed environment, and obtains a large number of rich images of deep-sea minerals. Fig. 1 is the deep-sea mining system. They can be classified into two parts. The first one is the shallow sea area, and the other one is the deep sea area. The former light which the sensor captured is the sunshine, which can make the images generated by the underwater vision sensor clear. The captured images in the second condition are different from the first because the light is artificial light. However, so far, the research on the segmentation algorithm of these deep-sea mineral image information is not sufficient and in-depth.
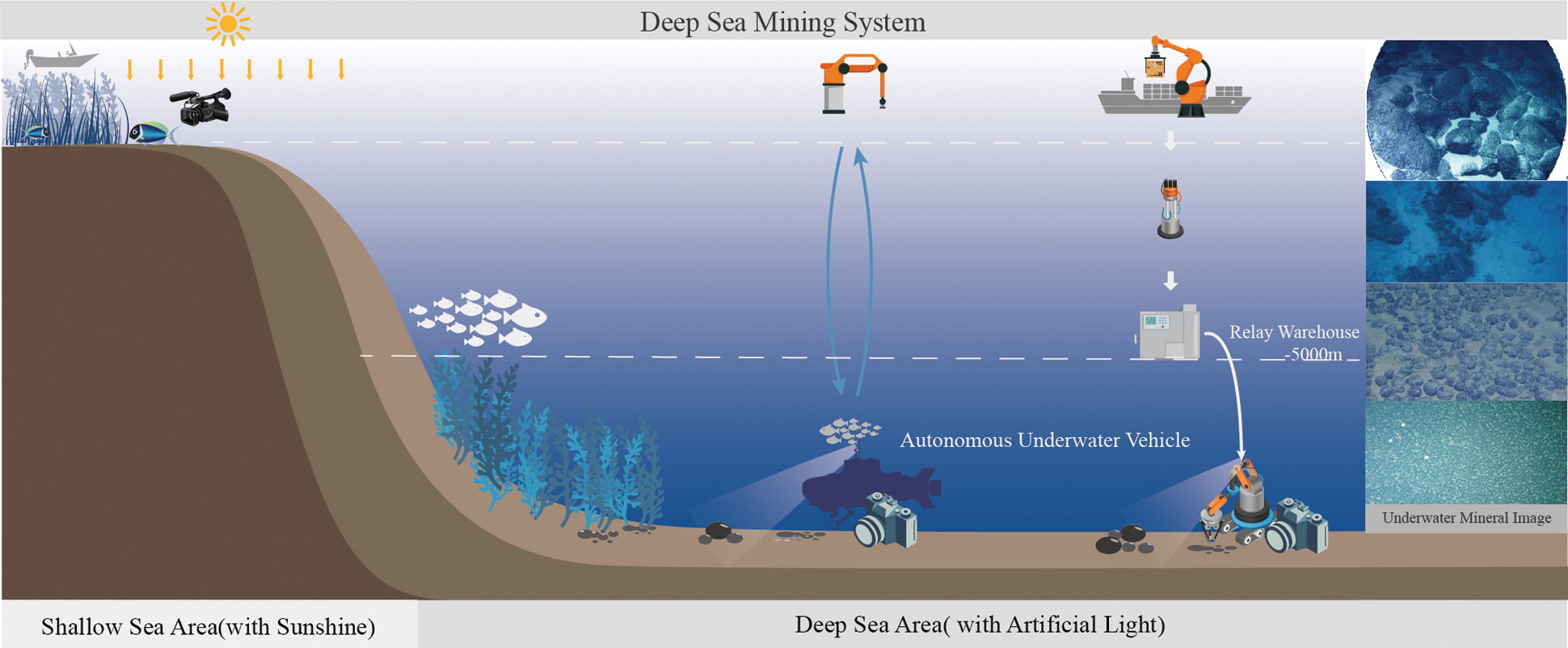
Figure 1: The deep-sea mining system. Which can be divided into two parts: shallow sea area with the sunshine and the deep sea area with the artificial light
Image semantic segmentation algorithm has been widely used in real life. For example, in the medical images processing area, the aneurysm tumor melanoma should be located for better excision, accurate segmentation of heart and brain tissue can improve the reliability of the diagnosis of related diseases and the treatment [1,2]. In the field of safety, the iris cannot be modified, so iris recognition can accurately confirm the identity [3], the segmentation of the cold trap device in the nuclear reaction can confirm the concentration of impurities and ensure the stable operation of the reactor [4]. In the field of surveillance, specific objects should be segmented for pedestrian detection, traffic detection, and so on [5]. So, there are many different challenges in the fields of computer vision and medical imaging [6–8].
With the development of smart collaborative technology [9], intelligent networks are becoming more developed, more and more intelligent means to get underwater pictures. The illumination environment of underwater minerals changes greatly, it has an adverse effect on segmentation. Meanwhile, the shapes of mineral particles are varied, and the diameter of the particles is also different, varied shapes and sizes are difficult for measuring accurately. And due to the uneven distribution of the underwater particles, the foreground color of the underwater image is similar to the background color. Under natural conditions, some particles were burial, the images reflect the phenomenon of particle adhesion. Reference [10] shows that the higher accuracy of the underwater mineral image segmentation can accelerate the rapid development of underwater mineral resources.
The development of deep learning algorithms like convolutional neural networks or deep learning not only affected typical tasks like object classification but are also efficient in other related tasks like object detection, localization, tracking, or as in this case image segmentation. Since 2012, several deep convolutional neural network models have been proposed such as AlexNet [11], VGG [12], GoogLeNet [13], DenseNet [14] and CapsuleNet [15]. In most cases, for very large-scale datasets like ImageNet [11], models are explored and evaluated using classification tasks, where the output of the classification tasks is a single label or probability values. Alternatively, small architecturally variant models are used for semantic image segmentation tasks. For example, variants of the encoder-decoder architecture like U-Net [16] and fully-connected convolutional neural network (FCN) [17] provide state-of-the-art results for image segmentation tasks in computer vision. Another variant of FCN was also proposed which is called SegNet [18]. U-Net first extracts the features of the image, then restores the image resolution by upsampling, and simultaneously connects to the same stage by skipping connection during the upsampling process. The features of the encoder part are merged with the features of the decoder part, the fusion makes the segmentation map more precise in detail, and the network structure has a good performance on the grayscale Electron Microscopy images dataset (EM dataset).
Although U-Net performed well, due to the limitation in different segmentation tasks, a series of improved networks based on U-Net were proposed. A V-Net network [19] was proposed and this method extends the two-dimensional structure to three-dimensional, the three-dimensional image segmentation was solved, end-to-end training on the prostate MRI volume, and learn to predict the entire volume. The segmentation performed well on the PROMISE 2012 dataset. H-DenseUNet [20] was proposed to fuse the on-chip representation and inter-slice features of liver tumor images through a mixed feature fusion layer, and this scheme achieved excellent results in the liver tumor segmentation challenge. MDU-Net [21] acquired better results than U-Net at MICCAI 2015 Gland segmentation task, the method proposed three different multi-scale densely connected U-shaped structure encoders, decoders and jump connections, the dense connection directly merged the high-level and low-level adjacent scale feature maps and enhanced the propagation of current layer features. Isensee et al. [22] proposed a robust adaptive network based on two-dimensional and three-dimensional ordinary U-shaped networks, namely nnU-Net, which removes redundant parts of the network and pays more attention to the performance and generalization of the constituent methods. In the case where the dataset is not manually adjusted, good results are obtained in a plurality of medical image segmentation tasks. A hierarchical probability U-Net segmentation network [23] was proposed, this method is based on conditional variational autoencoder (cVAE) to high fidelity sample and reconstructs segments. Meanwhile, this structure provides the flexibility of learning the distribution of cross-scale complex structures and has a good performance for fuzzy medical image segmentation. Hasan et al. [24] modify the U-Net architecture named UNetPlus, by introducing a pre-trained encoder and re-design the decoder part, by replacing the transposed convolution operation with an upsampling operation based on nearest-neighbor (NN) interpolation.
Based on the vigorous performance of the U-Net, residual structure and pyramid structure, an improved U-Net for the underwater mineral image segmentation task is proposed, and the main contributions can be summarized as follows: Three new models JPU-Net, ResU-Net and JPMU-Net are proposed for EM dataset segmentation. Experiments demonstrate the effectiveness of these three segmentation networks on EM datasets and compare the best-performing model. Successfully migrated the improved model to the underwater mineral image dataset, and the experimental comparison yielded the best performing model for the dataset.
In recent years, a lot of models have been proposed that have proved that deeper networks are better for recognition and segmentation tasks [12]. However, it is difficult to train very deep models because of the vanishing gradient problem. This problem can be solved by executing modern activation functions such as Rectified Linear Units (ReLU) or Exponential Linear Units (ELU) [13]. In addition, in order to solve the problem that deeper networks are difficult to train, 2015, He et al. [25] proposed a residual learning framework. This framework is a good solution to the problem of the inability to converge due to the gradient explosion caused by the deepening of the network depth. Residual learning framework explicitly reformulates the layers as learning residual functions with reference to the layer inputs, instead of learning unreferenced functions. Therefore, using the residual structure can simplify the design of the network, more layers can be designed to obtain more advanced semantic information. MultiResUNet [26] was presented and this method added MultiRes block to the U-Net, the MultiRes block can augment U-Net with the ability of multi-resolution analysis and formulate a compact analogous structure similar to Inception [13]. Alom et al. [27] added a recurrence convolution neural network and a recurrence residual structure based on U-Net to form RU-Net and R2U-Net respectively, R2U-Net represents the characteristics of the segmentation task by feature accumulation better, the residual structure in the R2U-Net can help design deeper network. This achievement has better segmentation performance on the three reference datasets of retinal image blood vessel segmentation, skin cancer segmentation and lung damage segmentation.
Before the spatial pyramid pooling, existing deep convolutional neural networks (CNNs) require a fixed-size input image. Fixed-size is “artificial” and may reduce the recognition accuracy for images of arbitrary size. To solve this problem, He et al. [28] proposed a new pooling strategy, “spatial pyramid pooling”, and this new network structure can generate a fixed-length representation regardless of image size. Pyramid pooling is also robust to object deformations. Therefore, it is widely used in the field of computer vision.
As for the underwater mineral image dataset, Reference [29] has proposed an improved U-Net, in the decoder part, the features are fused by different scale up-sampled operations to obtain the final segmentation map. For convenience, in this paper, the fusion structure is called the merge module and the network is named MU-Net.
The whole architecture is shown in Fig. 2. In general, the proposed semantic segmentation network could be seen as an encoder-decoder structure. As discussed above, residual structure and joint pyramid upsampling structure are added to the based model U-Net.
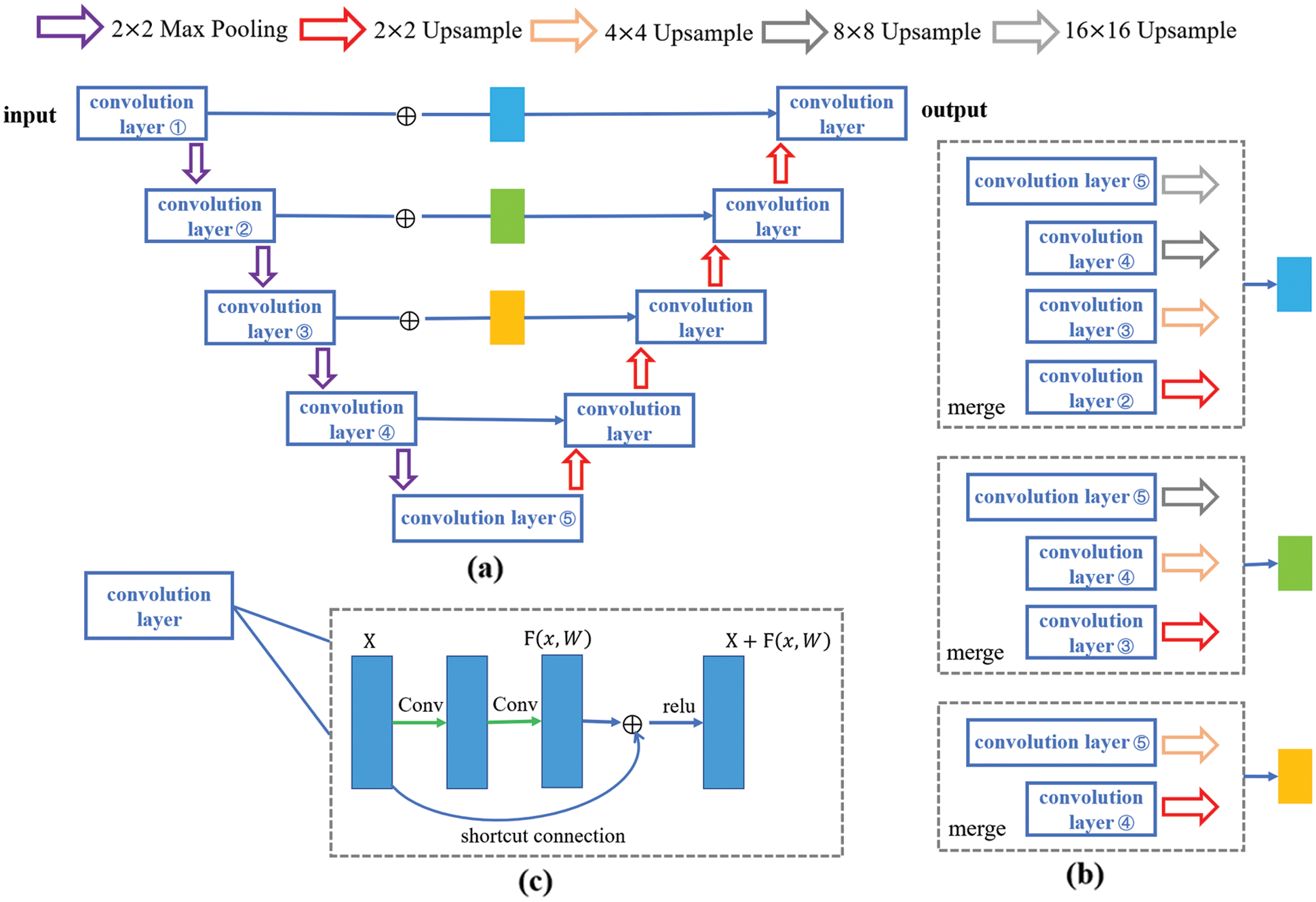
Figure 2: The whole network architecture. (a) is the whole encoder-decoder structure and expresses the mode of adding modules. (b) is the joint pyramid upsampling module. (c) is the residual module
The model employing joint pyramid upsampling structure (JPU-Net) is (a). In the encoder part of U-Net, the input image goes through five convolutional layers and four pooling layers. And in the decoder part of U-Net, features go through four convolutional layers and four upsampling layers. An improved network adds a joint pyramid upsampling structure to skip connection. And (b) shows the specific structure. Each block is a fusion of different scale feature maps by upsampling. So, first merge the feature maps which upsampled from different scales obtained from the last three convolution layers, and then use the merged features in the decoder part for subsequent upsampling operations. Given an input image noted as X ∈ ℝC×W×H, in the encoder part, the feature obtained from each convolution layer recorded as Xi ∈ ℝC×W×H, i = 1, 2, …, 5, and the upsampling is bilinear inter polation. Mathematically, bilinear interpolation is a linear interpolation extension of an interpolation function with two variables. The core idea is to perform linear interpolation in two directions. Suppose the goal is to get the value of the unknown function f at the point P = (x, y), and the values of the function f at point Q11 = (x1, y1), Q12 = (x1, y2), Q21 = (x2, y1) and Q22 = (x2, y2) is known. First linear interpolation in the x direction:
Then linear interpolation in the y direction:
So, we can get the wanted result f(x, y):
The picture zoom j times by bilinear interpolation noted as srcj, and the joint pyramid upsampling module is defined as follows:
This structure can magnify deep semantic information for better detail segmentation. The model employing joint pyramid upsampling structure and merge module simultaneously named JPMU-Net. The model employing residual structure is the ResU-Net. In the whole encoder-decoder structure, add residual structure to each layer. The specific structure of each convolution layer is shown in (c). Each residual module can be expressed as:
where xl+1 is the output, xl is the input, F(xl, Wl) represents the convolution operation. Due to the addition of the residual structure, the number of convolutions per layer is increased by one, that is, each layer undergoes three or more 3 × 3 convolution operations. By means of shortcut connections, pass the input directly to the output as the initial result.
4 Experimental Setup and Results
In order to evaluate the performance of the model, verify it on a challenging benchmark: the gray-scale Electron Microscopy images (EM dataset), and then apply it to the underwater mineral image dataset to find the relatively suitable segmentation model.
EM dataset is comprised of consecutive ventral nerve cord gray-scale images from different drosophila first instar larvae. The dataset contains 30 annotated images that can be used as the train dataset and 30 unannotated images as the test dataset, all images have a resolution of 512 × 512. Following the setting of the EM dataset, annotate 49 mineral images taken from the underwater under different lighting conditions, and randomly pick out 4 images with different visual effects to add to test dataset, the test dataset has 34 different images, which all have a resolution of 4000 × 3000. Fig. 3 shows the examples of the EM dataset and Fig. 4 shows the examples of the underwater mineral image dataset.
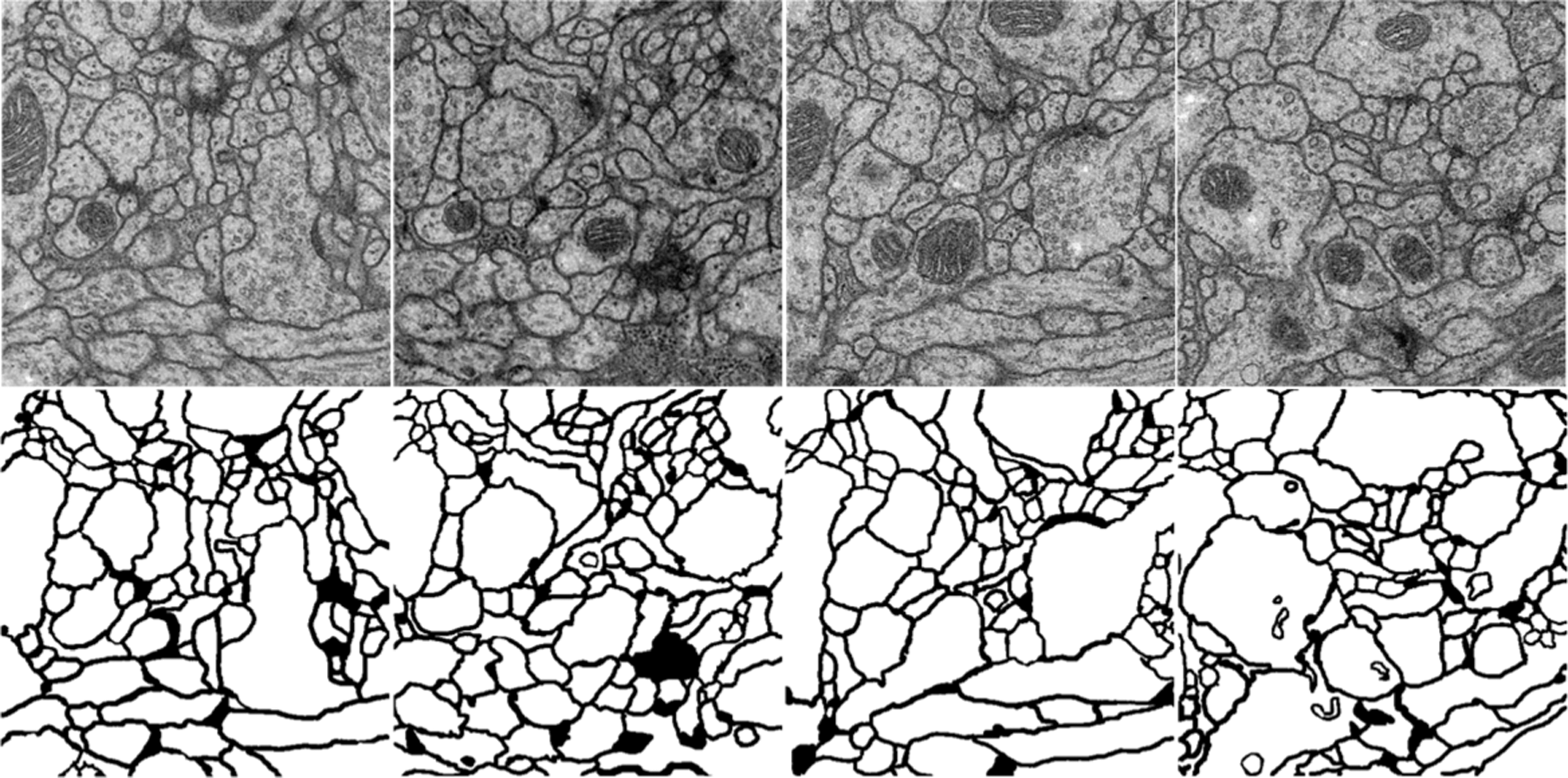
Figure 3: The examples of the EM dataset. The first line is the train image, and the second line is the corresponding ground truth. The goal of the challenge is to transform a grayscale EM image into an accurate boundary map
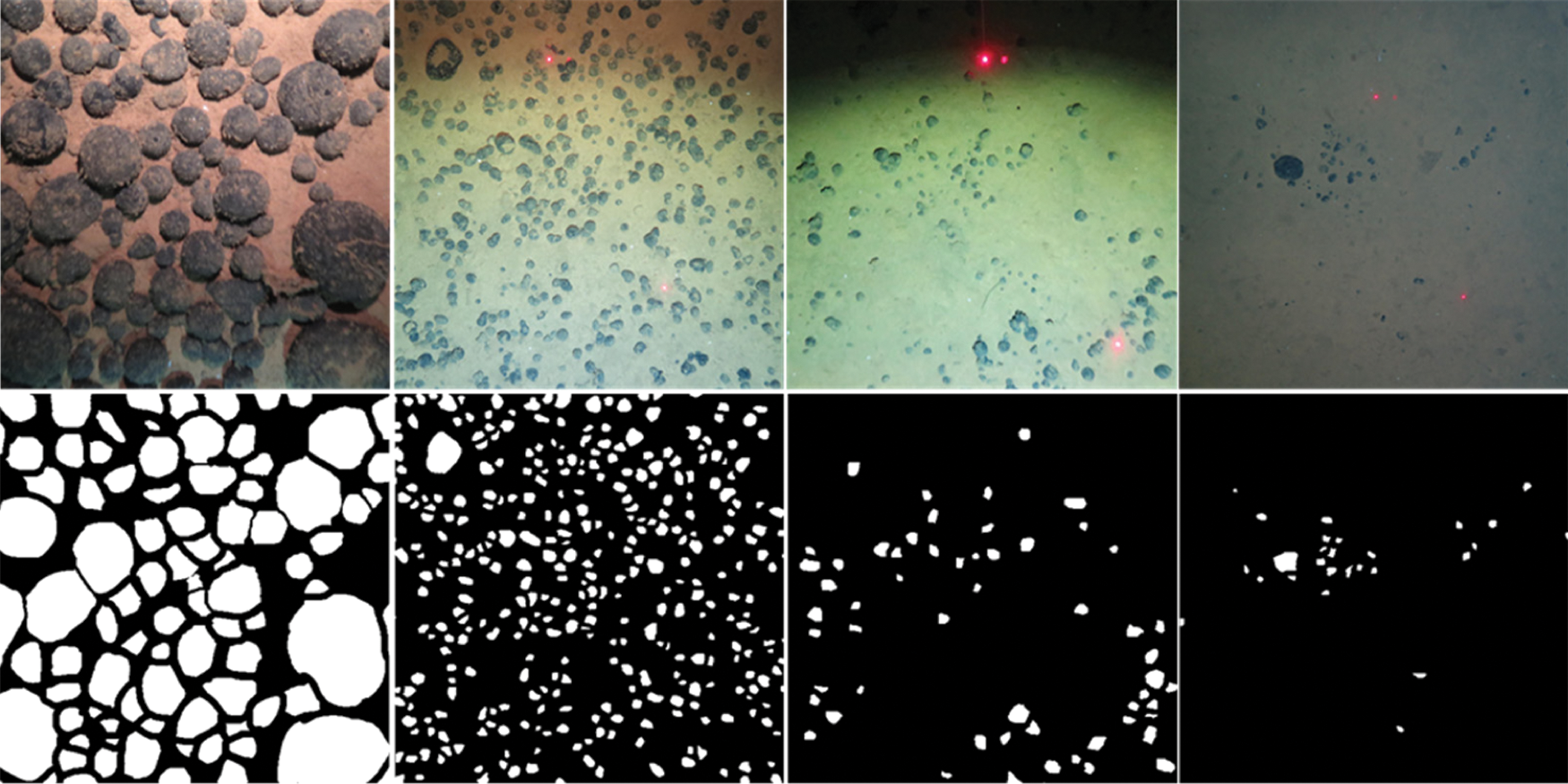
Figure 4: The examples of the EM dataset. The first line is the train image, and the second line is corresponding ground truth. The goal of the challenge is to transform a grayscale EM image into an accurate boundary map
The training strategies are as follows. Stochastic gradient descent (SGD) with batch size 1, optimizer Adam, and learning rate 1e-4. The cross-entropy error at each pixel over the categories is applied as our loss function. In addition, the annotated images are too few to train the networks, date augmentation contains random horizontal flip, random rotation with range 10, random horizontal and vertical pan with range 0.005, random shear and random zoom into a fixed size for training.
This paper employs the joint pyramid module, residual structure and merge module respectively at the bottom of the U-Net for better scene understanding. To verify which module combined with U-Net have better performance. We conduct experiments with different structure settings in Tab. 1.
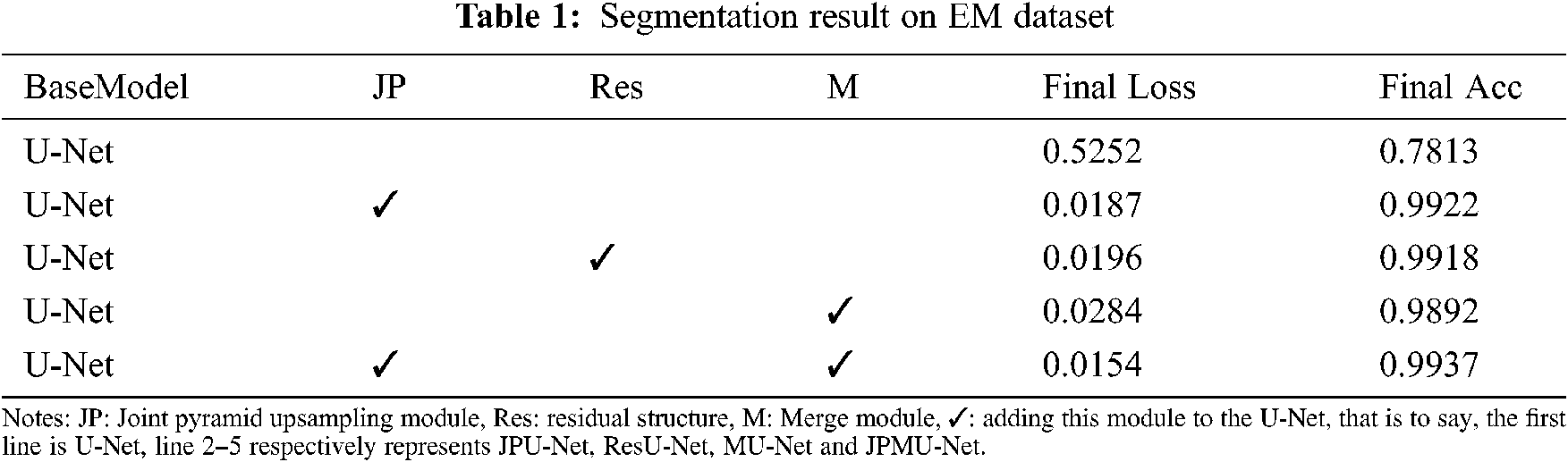
As shown in Tab. 1, all of these modules improve the performance remarkably. Compared with the base model (U-Net), employing the joint pyramid upsampling module yields an accuracy result of 0.9922 and a loss result of 0.0187, which brings 0.2109 improvements and 0.5065 loss reduction. Meanwhile, employing residual structure individually outperforms the baseline by 0.2105 on accuracy and 0.5056 on loss. When we integrate the joint pyramid upsampling module and merge module together, the performance further improves to accuracy 0.9937 and loss 0.0154. These results show the combination of joint pyramid upsampling module and merge module with U-Net bring great benefit to segmentation for EM dataset.
When adding merge module on U-Net, the performance has a certain improvement. Meanwhile, the model convergence faster. Figs. 5 and 6 show the accuracy and the loss comparison varies epoch. It can be concluded that JPMU-Net performs best, JPU-Net is next and ResU-Net followed, which all convergence faster than MU-Net.
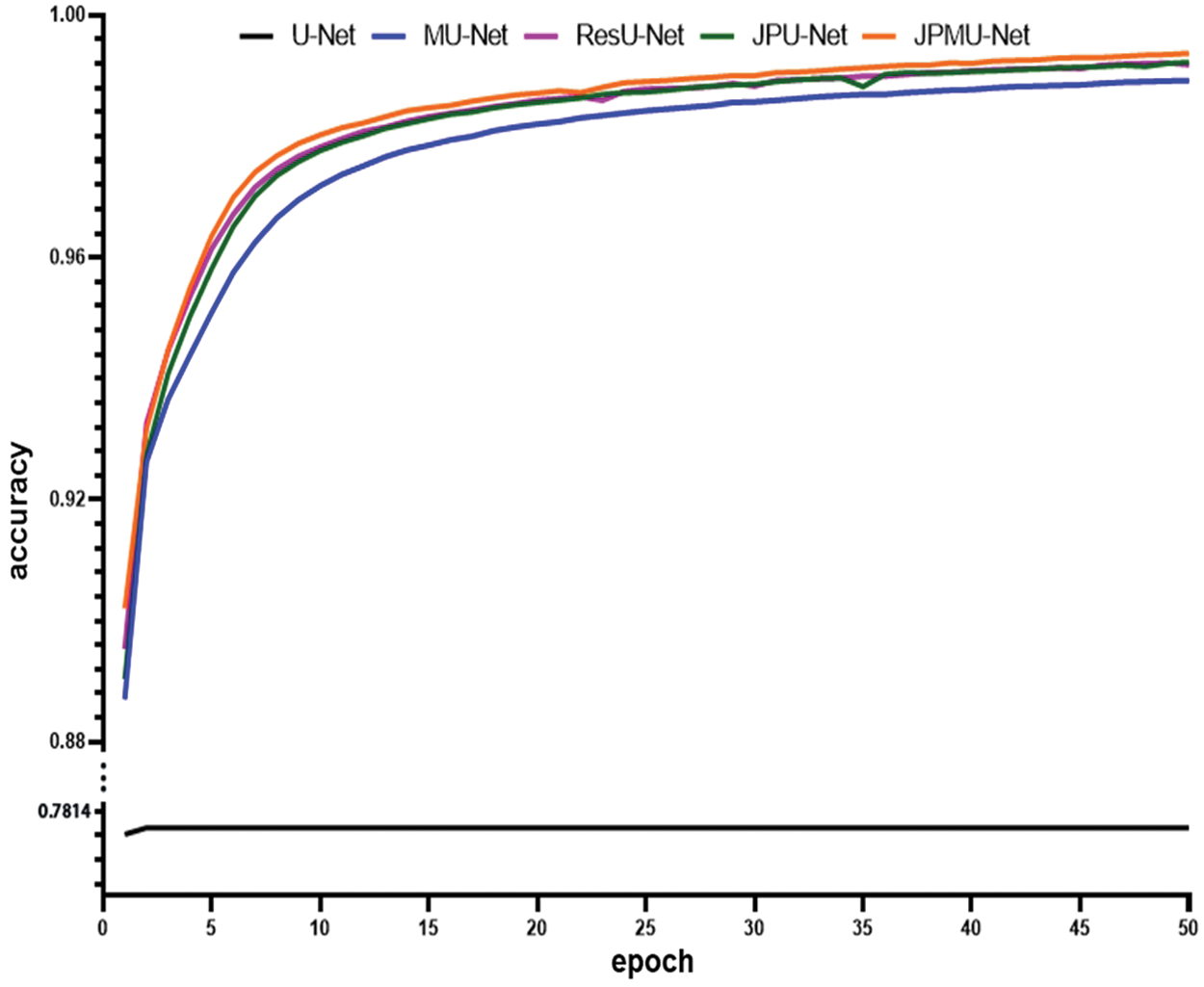
Figure 5: The accuracy comparison of these networks varies epoch
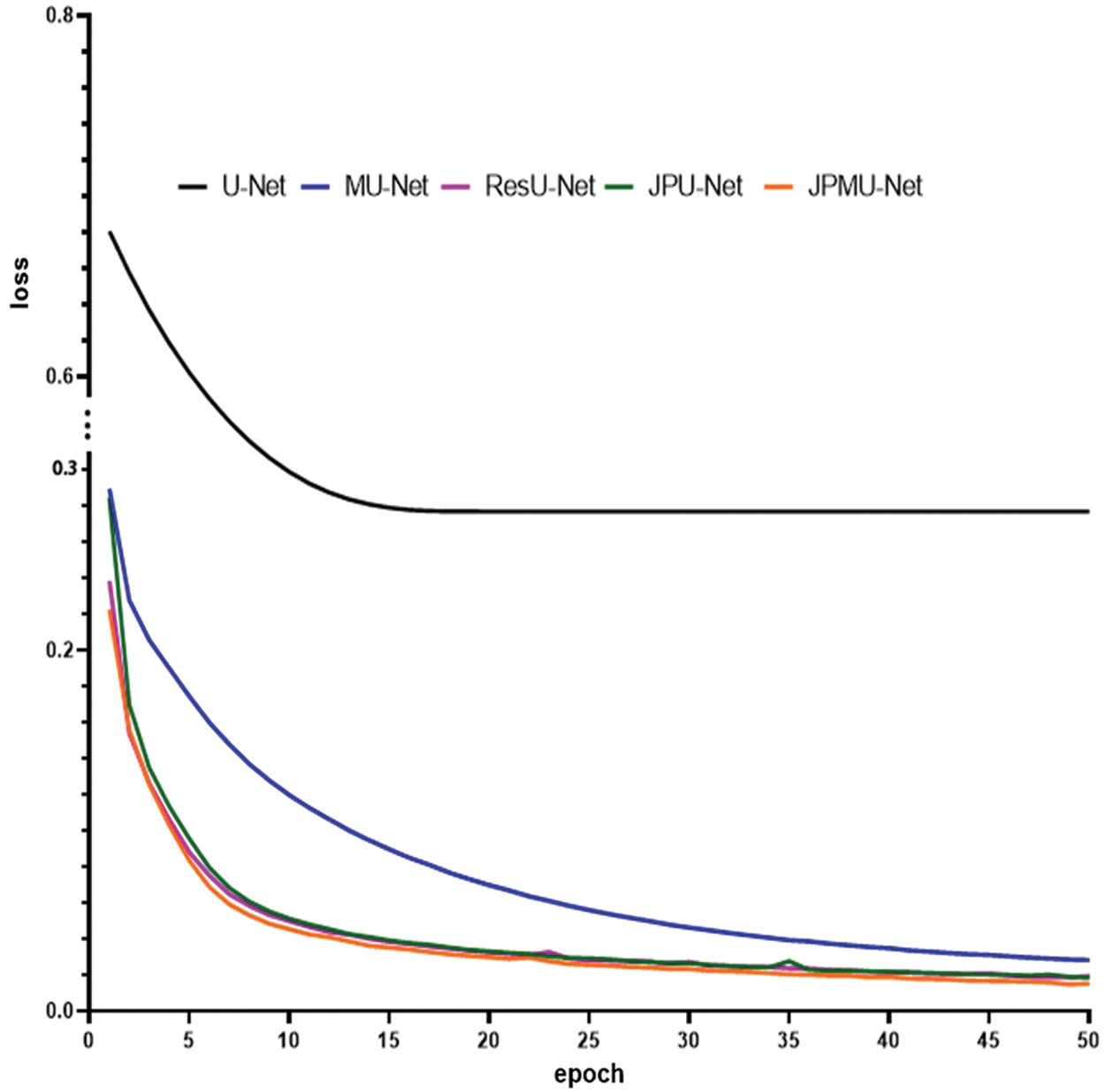
Figure 6: The loss comparison of these networks varies epoch
In the decoder part of the MU-Net network, the final segmentation result image is fused by different scale-up-sampled features. But the upsampled features of the decoder part are obtained by upsampling and convolution, in the part of the upsampling from low-level features, much semantic information has been lost. So, although some semantic information lost by U-Net is compensated by adding the fusion operation, it is not enough. JPU-Net network upsamples features of different scales in the encoder part and then merge them together. In the encoder part, different scale features are obtained after multiple convolution and pooling operations. First of all, we upsample the small-scale features to the size of this convolution layer feature, and merge the features from convolution and upsampling, and then merge these features to the same scale features of the decoder part. In this process, we repeatedly add low-level semantic information to the decoder part, so that it can notice more details in the image segmentation. Our ResU-Net network adds residual structure in each convolution layer, it can increase the number of convolution layers, so more features are extracted. But the results of JPMU-Net are not as good as the ideal, because when the network becomes more complicated, the training data does not increase, resulting in over-segmentation. The visible segmentation results validate the effectiveness of our network again.
Fig. 7 displays the portion random results of segmentation. We marked the contrasting area with red boxes. Boundary detection is always challenging because many boundaries look fuzzy and ambiguous. Furthermore, only boundaries between neurites should be detected, and those of intracellular organelles like mitochondria and synaptic vesicles should be ignored [23], for example, the area inside the red boxes is the mitochondria or synaptic vesicles inside the cell in the original picture, and it should not be segmented.
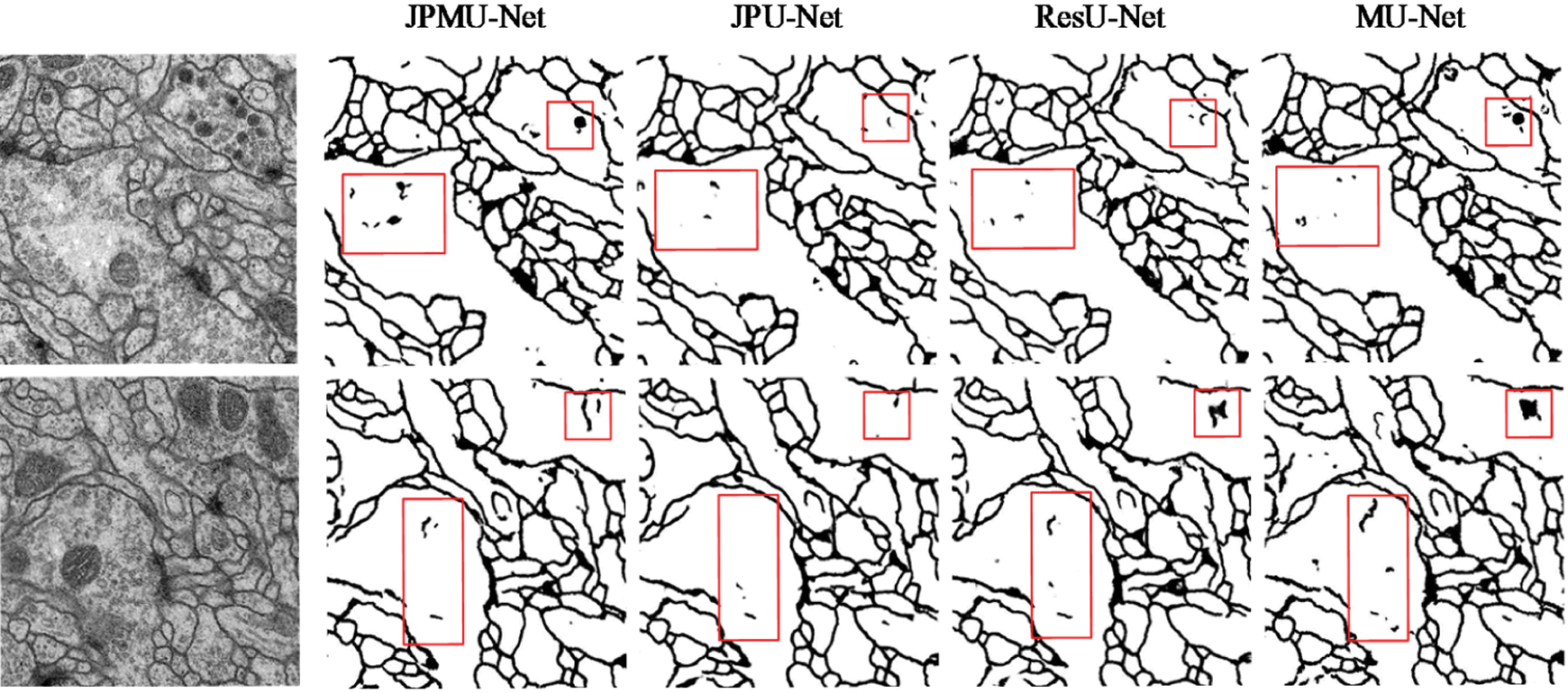
Figure 7: Visible results on EM dataset. The first line is input images, and lines 2∼5 display the output of each network
As can be seen from Fig. 7, although the MU-Net segmentation result is better than U-Net, it also has certain defects. In the segmentation details, the ResU-Net network and the JPU-Net network both have a less obvious situation of over-segmentation compared with the MU-Net network. However, the output of the JPMU-Net network is worse than the result of the MU-Net network. Therefore, from the visualization results of segmentation, we can summarize that for the EM dataset, JPU-Net has the best performance.
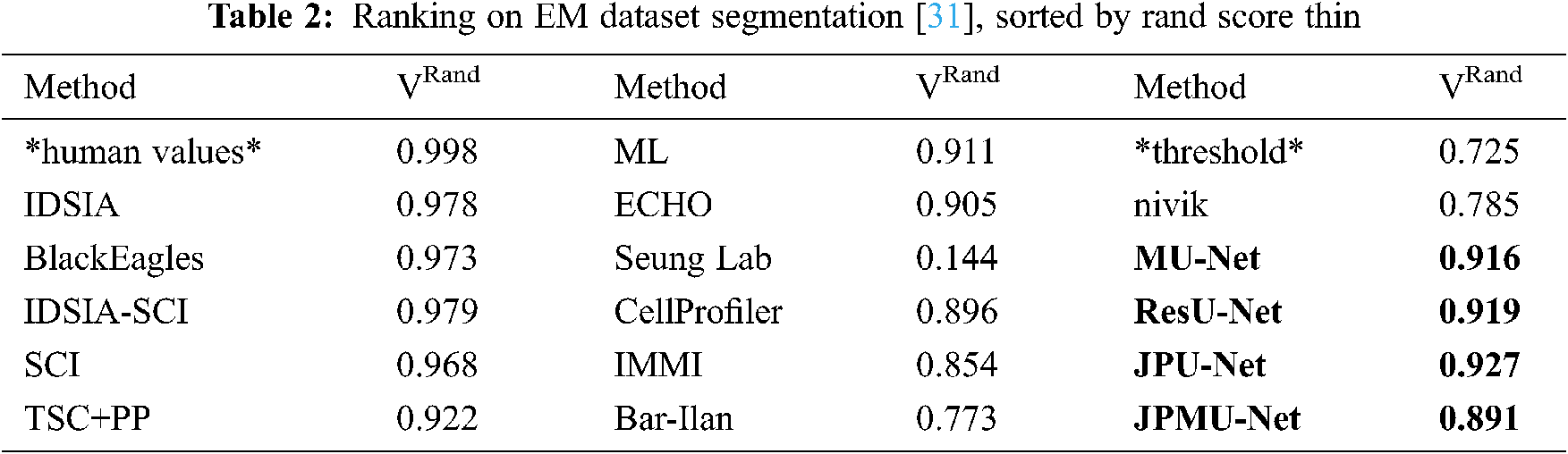
To further verify the effectiveness of the network, we compare the results using specially normalized versions of the Rand error and Variation of Information. Because of the robustness, we choose the VRand scores as a reference, and we get the VRand scores within Fiji using the open script [30].
Tab. 2 shows the average of all Rand scores and compare it with other teams. The result is the same as the visible results. JPU-Net has the best performance, and ResU-Net is little worse than JPU-Net, but they are both better than MU-Net. The JPMU-Net has the worst performance.
4.4 Results on Underwater Mineral Image Dataset
Similarly, we apply these network structures to the underwater mineral image dataset. Tab. 3 shows the quantification results of joint pyramid upsampling module, residual structure and merge module-based U-Net. Thus further verify the validity of the network structure.
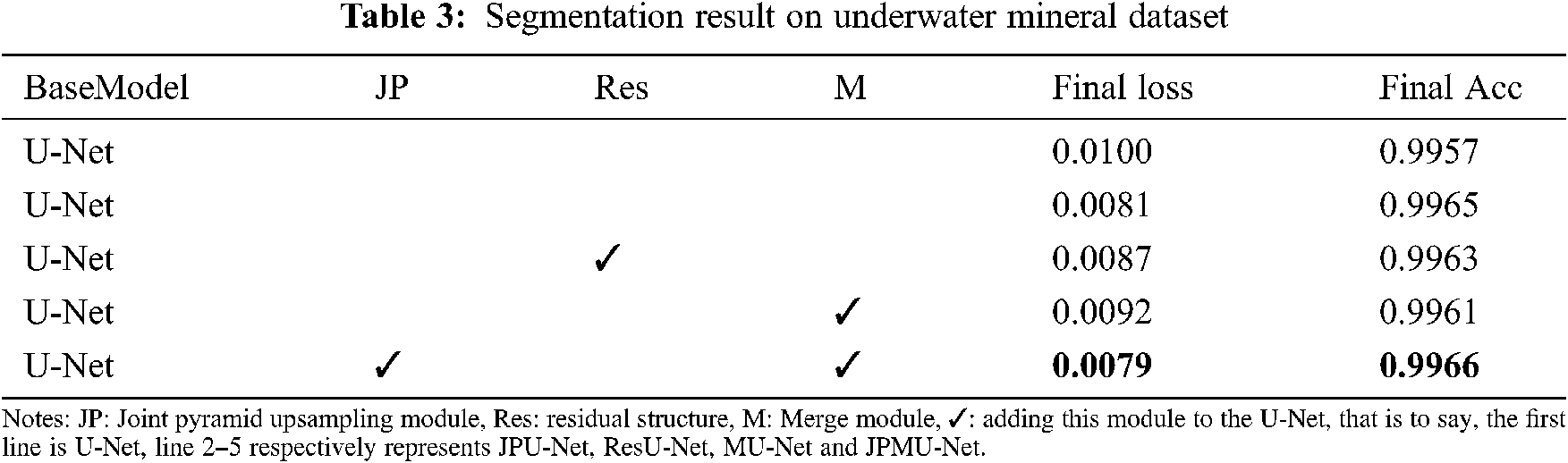
As shown in Tab. 3, all of these modules improve the performance. Compared with the base model U-Net, employing the joint pyramid upsampling module yields an accuracy result of 0.9965 and a loss result of 0.0081, which brings 0.0008 improvements and 0.0019 loss reduction. Meanwhile, employing residual structure individually outperforms the baseline by 0.0006 on accuracy and 0.0013 on loss. When we integrate the joint pyramid upsampling module and merge module together, the performance further improves to accuracy 0.9966 and loss 0.0079. These results show the combination of joint pyramid upsampling module and merge module with U-Net bring great benefit to segmentation for underwater mineral image dataset.
When adding merge module on U-Net, the performance has a certain improvement. Meanwhile, the model convergence faster. Figs. 8 and 9 show the accuracy and the loss comparison varies epoch. It can be concluded that the initial accuracy of MU-Net is higher than JPU-Net and ResU-Net, but it is lower than JPMU-Net. Meanwhile, the initial loss of JPMU-Net is the lowest. That is to say, for the underwater mineral image dataset, the JPMU-Net convergence is faster than others.
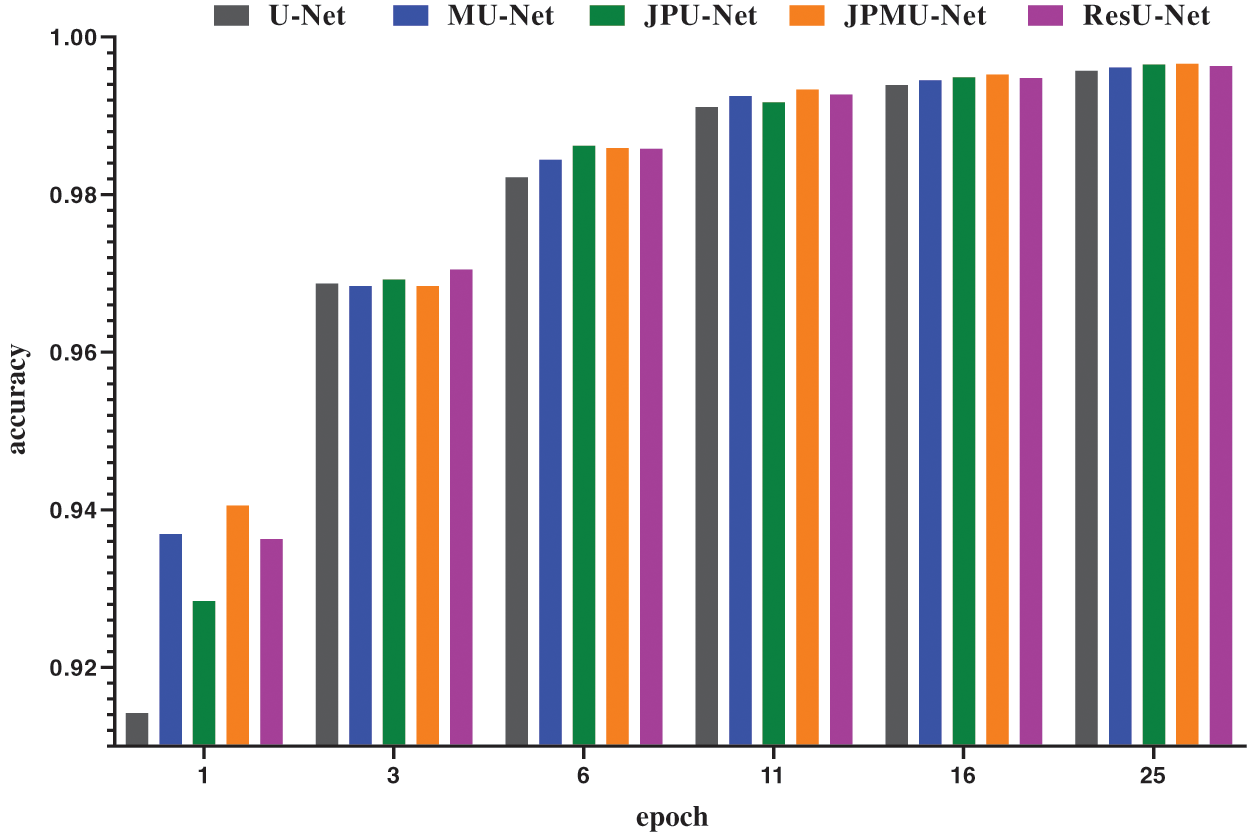
Figure 8: The accuracy and loss comparison of these networks varies epoch
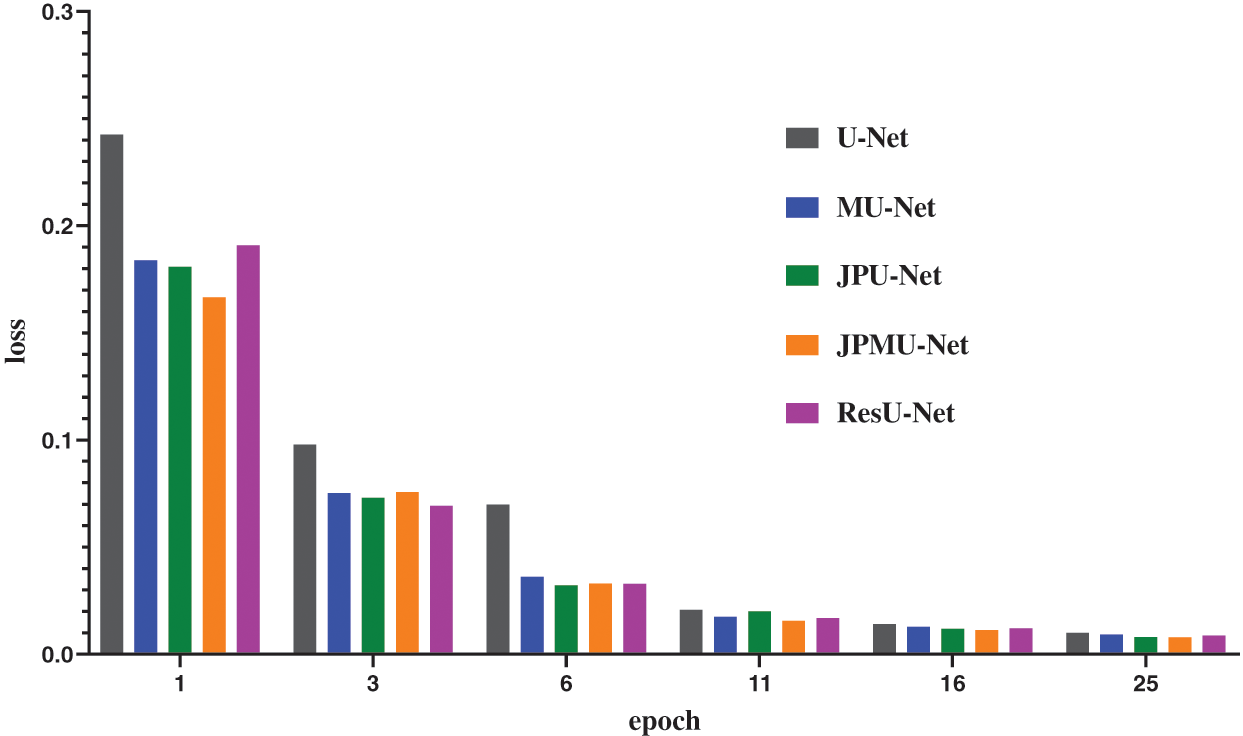
Figure 9: The accuracy and loss comparison of these networks varies epoch
Similarly, then analyze the visualization results of the segmentation results. Fig. 10 displays the portion of random segmentation maps. In order to analyze the segmentation results more clearly, we randomly crop part of the results and compare the results with the hand-marked labels. The red box in the first line should have segmented two underwater mineral particles, but only the segmentation map of JPMU-Net is suitable with the label. This shows that employing the joint pyramid upsampling structure and merge module simultaneously on the U-Net can extract more detailed features. Line 2 has more differences. In the two red boxes, there are both 3 underwater mineral particles. The JPMU-Net only missed one, the MU-Net and the JPU-Net are the same, this indicates the joining the joint pyramid upsampling module is effective. But the result of ResU-Net missed half of the particles. In summary, on the underwater mineral image dataset, the effect of MU-Net and JPU-Net is the same and both better than the ResU-Net, the JPMU-Net performs best. It further shows that the network we proposed is valid.
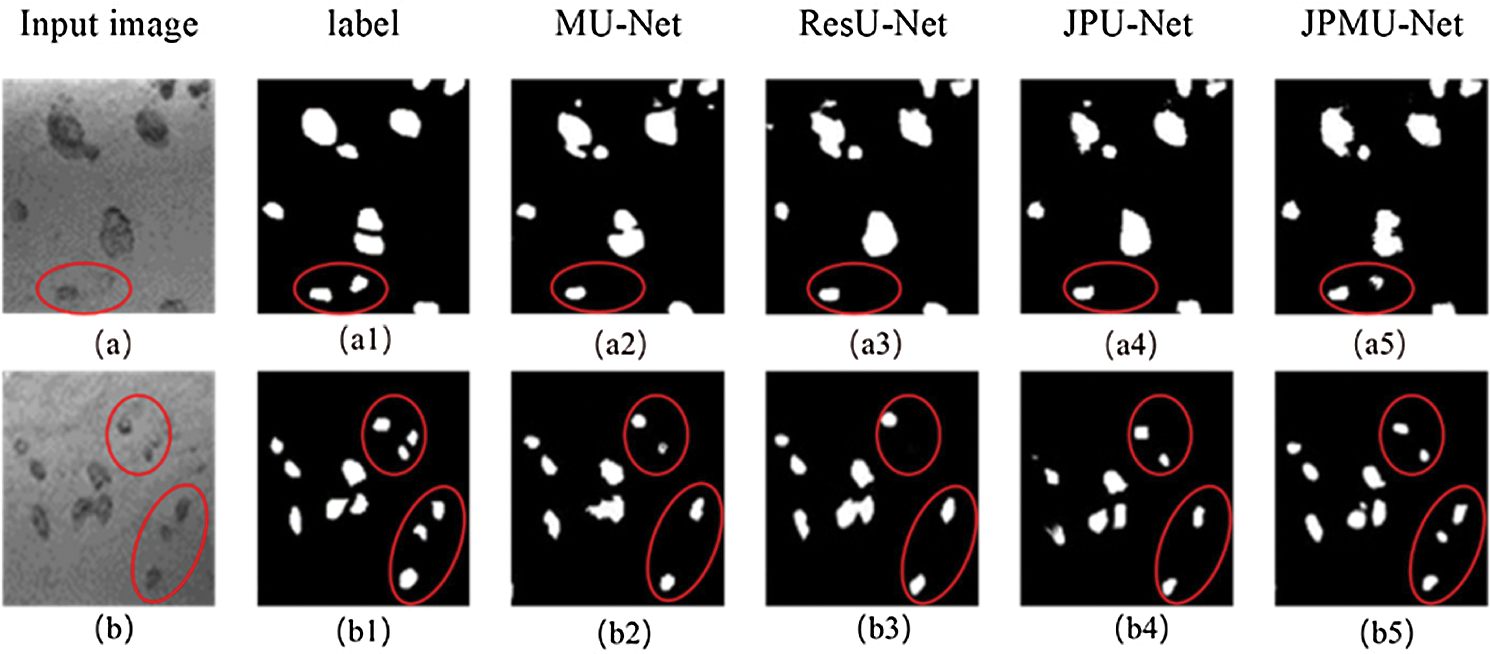
Figure 10: Visible results on the underwater mineral image dataset, where (a),(b) represent two different input images, the (a1)–(a5) and (b1)–(b5) represent the corresponding segmentation results using five different methods for the two images (a) and (b). Note that we have circled the areas that need to be highlighted in red.
In this paper, three networks for the mineral image captured by the underwater vision sensor segmentation are presented, JPU-Net, JPMU-Net and ResU-Net, which respectively employ joint pyramid upsampling module, merge module and joint pyramid upsampling module simultaneously, and residual module based on the U-Net. The joint pyramid upsampling module merges the feature maps of different scales by upsampling in the encoder part, then adds to the skip connection. The residual module is a residual block, and add the residual module to each convolution layer. The network achieves outstanding performance consistently on the EM dataset and underwater mineral image dataset. In addition, it is important to decrease the computational complexity and enhance the robustness of the model, which will be studied in future work.
Acknowledgement: Thanks to other students in the Media Computing Laboratory of Minzu University of China and anonymous reviewers for their valuable comments and contributions to this research.
Funding Statement: This work was supported in part by national science foundation project of P.R. China under Grant No. 52071349, U1906234, partially supported by the Open Project Program of Key Laboratory of Marine Environmental Survey Technology and Application, Ministry of Natural Resource MESTA-2020-B001, Young and Middle-aged Talents Project of the State Ethnic Affairs Commission, the crossdiscipline research project of Minzu University of China (2020MDJC08), the Graduate Research and Practice Projects of Minzu University of China.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. A. Renugambal and K. S. Bhuvaneswari, “Image segmentation of brain MR images using otsu’s based hybrid WCMFO algorithm,” Computers, Materials & Continua, vol. 64, no. 2, pp. 681–700, 2020. [Google Scholar]
2. C. Luo, C. H. Shi, X. J. Li, X. Wang, Y. C. Chen et al., “Multi-task learning using attention-based convolutional encoder-decoder for dilated cardiomyopathy CMR segmentation and classification,” Computers, Materials & Continua, vol. 63, no. 2, pp. 995–1012, 2020. [Google Scholar]
3. F. Mallouli, “Robust EM algorithm for iris segmentation based on mixture of Gaussian distribution,” Intelligent Automation & Soft Computing, vol. 25, no. 2, pp. 243–248, 2019. [Google Scholar]
4. B. Thamotharan, B. Venkatraman and S. Chandrasekaran, “Identification and segmentation of impurities accumulated in a cold-trap device by using radiographic images,” Intelligent Automation & Soft Computing, vol. 26, no. 2, pp. 335–340, 2020. [Google Scholar]
5. Y. H. Sun, Y. Mu, Q. Feng, T. L. Hu, H. Gong et al., “Deer body adaptive threshold segmentation algorithm based on color space,” Computers, Materials & Continua, vol. 64, no. 2, pp. 1317–1328, 2020. [Google Scholar]
6. G. J. Brostow, J. Fauqueur and R. Cipolla, “Semantic object classes in video: A high-definition ground truth database,” Pattern Recognition Letters, vol. 30, no. 2, pp. 99–97, 2009. [Google Scholar]
7. S. Song, S. P. Lichtenberg and J. X. Xiao, “SUN Rgb-d: A RGB-d scene understanding benchmark suite,” in 2015 IEEE Conf. on Computer Vision and Pattern Recognition (CVPRBoston, MA, USA, pp. 567–576, 2015. [Google Scholar]
8. M. Kistler, S. Bonaretti, M. Pfahrer, R. Niklaus and P. Büchler, “The virtual skeleton database: An open access repository for biomedical research and collaboration,” Journal of Medical Internet Research, vol. 15, no. 11, pp. 1–14, 2013. [Google Scholar]
9. Z. Y. Zhang, Y. T. Zhou and F. Song, “A smart collaborative routing protocol for reliable data diffusion in IoT scenarios,” Sensors, vol. 18, no. 6, pp. 1–26, 2018. [Google Scholar]
10. C. Cai, H. Qiu, B. Cao and J. X. Xia, “Experimental studies on passing characteristics of coarse particles in lifting pump of deep-sea mining system,” The Ocean Engineering, vol. 34, no. 2, pp. 64–70, 2016. [Google Scholar]
11. A. Krizhevsky, I. Sutskever and G. E. hinton, “ImageNet classification with deep convolutional neural networks,” in 2012 Advances in Neural Information Processing Systems (NIPS), Lake Tahoe, NV, USA: Curran Associates, Inc, pp. 1097–1105, 2012. [Google Scholar]
12. K. Simonyan and A. Zisserman, “Very deep convolutional networks for large-scale image recognition,” in 2015 Int. Conf. on Learning Representations (ICLRSan Diego, CA, USA, pp. 1–14, 2015. [Google Scholar]
13. C. Szegedy, W. Liu, Y. Q. Jia, P. Sermanet, S. Reed et al., “Going deeper with convolutions,” in 2015 IEEE Conf. on Computer Vision and Pattern Recognition (CVPRBoston, MA, USA, pp. 1–9, 2015. [Google Scholar]
14. G. Huang, Z. Liu, L. V. D. Maaten and K. Q. Weinberger, “Densely connected convolutional networks,” in 2017 IEEE Conf. on Computer Vision and Pattern Recognition (CVPRHonolulu, Hawaii, USA, pp. 4700–4708, 2017. [Google Scholar]
15. S. Sabour and N. Frosst, “Dynamic routing between capsules,” in 2017 31st Conf. on Neutal Information Processing Systems (NIPSLong Beach, California, USA, pp. 3859–3869, 2017. [Google Scholar]
16. O. Ronneberger, P. Fischer and T. Brox, “U-Net: Convolutional networks for biomedical image segmentation,” in 2015 Int. Conf. on Medical Image Computing and Computer Assisted Intervention (MICCAIMunich, Germany, pp. 1–8, 2015. [Google Scholar]
17. J. Long, E. S. U. Berkeley and T. Darrell, “Fully convolutional networks for semantic segmentation,” in 2015 IEEE Conf. on Computer Vision and Pattern Recognition (CVPRBoston, MA, USA, pp. 3431–3440, 2015. [Google Scholar]
18. V. Badrinarayanan, A. Kendall and R. Cipolla, “Segnet: A deep convolutional encoder-decoder architecture for image segmentation,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 39, no. 12, pp. 2481–2495, 2017. [Google Scholar]
19. F. Milletari, N. Navab and S. Ahmadi, “V-Net: Fully convolutional neural networks for volumetric medical image segmentation,” in 2016 Fourth Int. Conf. on 3D Vision (3DVStanford, California, USA, pp. 565–571, 2016. [Google Scholar]
20. X. M. Li, H. Chen, X. J. Qi, Q. Dou, C. W. Fu et al., “H-DenseUNet: Hybrid densely connected UNet for liver and tumor segmentation from CT volumes,” in IEEE Transactions on Medical Imaging, vol. 37, no. 12, pp. 2663–2674, 2018. [Google Scholar]
21. J. W. Zhang, Y. Z. Jin, J. L. Xu, X. W. Xu and Y. C. Zhang, “MDU-Net: Multi-scale densely connected U-net for biomedical image segmentation,” in 2018 IEEE Conf. on Computer Vision and Pattern Recognition (CVPRSalt Lake City, Utah, USA, pp. 1–10, 2018. [Google Scholar]
22. F. Isensee, J. Petersen, A. Klein, D. Zimmerer, P. F. Jaeger et al., “nnU-Net: Self-adapting framework for U-Net-based medical image segmentation,” in Bildverarbeitung für die Medizin, Berlin, Germany: Springer Vieweg, Wiesbaden, pp. 22, 2019. [Google Scholar]
23. S. A. A. Kohl, B. Romera-Paredes, K. H. Maier-Hein, D. J. Rezende, S. M. A. Eslami et al., “A hierarchical probabilistic U-net for modeling multi-scale ambiguities,” in 2019 IEEE Conf. on Computer Vision and Pattern Recognition (CVPRLong Beach, CA, USA, pp. 1–25, 2019. [Google Scholar]
24. S. M. K. Hasan and C. A. Linte, “U-NetPlus: A modified encoder-decoder U-net architecture for semantic and instance segmentation of surgical instrument,” in 2019 41st Int. Conf. of the IEEE Engineering in Medicine and Biology Society (EMBCBerlin, Germany, pp. 7205–7211, 2019. [Google Scholar]
25. K. M. He, X. Y. Zhang, S. Q. Ren and J. Sun, “Deep residual learning for image recognition,” in 2016 IEEE Conf. on Computer Vision and Pattern Recognition (CVPRLas Vegas, NV, USA, pp. 770–778, 2016. [Google Scholar]
26. N. Ibtehaz and M. S. Rahman, “MultiResUNet: Rethinking the U-net architecture for multimodal biomedical image segmentation,” Neural Networks, vol. 121, pp. 74–87, 2020. [Google Scholar]
27. M. Z. Alom, M. Hasan, C. Yakopcic, T. M. Taha and V. K. Asari, “Recurrent residual convolutional neural network based on U-Net (R2U-Net) for medical image segmentation,” Journal of Medical Imaging, vol. 6, no. 1, pp. 1–16, 2019. [Google Scholar]
28. K. M. He, X. Y. Zhang, S. Q. Ren and J. Sun, “Spatial pyramid pooling in deep convolutional networks for visual recognition,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 37, no. 9, pp. 1904–1916, 2015. [Google Scholar]
29. W. Song, N. Zheng, X. C. Liu, L. R. Qiu and R. Zheng, “An improved U-net convolutional networks for seabed mineral image segmentation,” IEEE Access, vol. 7, pp. 82744–82752, 2019. [Google Scholar]
30. I. Arganda-Carreras, S. C. Turage, D. R. Berger, D. Ciresan, A. Giusti et al., “Crowdsourcing the creation of image segmentation algorithms for connectomics,” Frontiers in Neuroanatomy, vol. 9, pp. 1–13, 2015. [Google Scholar]
31. I. Arganda-Carrerasm, S. Seung, A. Cardona and J. Schindelin, “ISBI Challenge: Segmentation of neuronal structures in EM stacks,” ISBI Data, 2012. [Online]. Available: http://brainiac2.mit.edu/isbi_challenge. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |