DOI:10.32604/iasc.2022.020386

| Intelligent Automation & Soft Computing DOI:10.32604/iasc.2022.020386 |  |
| Article |
Classification Framework for COVID-19 Diagnosis Based on Deep CNN Models
1Department of Electronics and Electrical Communications, Faculty of Electronic Engineering, Menoufia University, Menouf, 32952, Egypt
2Department of Information Technology, College of Computer and Information Sciences, Princess Nourah Bint Abdulrahman University, Riyadh, 84428, Saudi Arabia
3Department of Industrial Electronics and Control Engineering, Faculty of Electronic Engineering, Menoufia University, Menouf, 32952, Egypt
4Department of Electronics and Communications Engineering, Faculty of Engineering, Zagazig University, Zagazig, 44519, Egypt
*Corresponding Author: Abeer D. Algarni. Email: adalqarni@pnu.edu.sa
Received: 22 May 2021; Accepted: 11 July 2021
Abstract: Automated diagnosis based on medical images is a very promising trend in modern healthcare services. For the task of automated diagnosis, there should be flexibility to deal with an enormous amount of data represented in the form of medical images. In addition, efficient algorithms that could be adapted according to the nature of images should be used. The importance of automated medical diagnosis has been maximized with the evolution of COVID-19 pandemic. COVID-19 first appeared in China, Wuhan, and then it has exploded in the whole world with a very bad impact on our daily life. The third wave of COVID-19 in the third world is really a disaster in current days, especially with the emergence of the delta variant of COVID-19 that is widespread. Required inspections should be carried out to monitor the COVID-19 spread in daily life and allow primary diagnosis of suspected cases, and long-term clinical laboratory monitoring. Healthcare professionals or radiologists can exploit AI (Artificial Intelligence) tools to quickly and reliably identify the cases of COVID-19. This paper introduces a DCNN (Deep Convolutional Neural Network) framework for chest X-ray and CT image classification based on TL (Transfer Learning). The objective is to perform multi-class and binary classification of the images in order to determine pneumonia and COVID-19 case. The TL is feasible, when using a small dataset by transferring knowledge from natural image classification to medical image classification. Two types of TL are used. The first type is fine-tuning of the DenseNet121, Densenet169, DenseNet201, ResNet50, ResNet152, VGG16, and VGG19 models. The second type is deep tuning of the LeNet-5, AlexNet, Inception naïve v1, and VGG16 models. Extensive tests have been carried out on datasets of chest X-ray and CT images with different training/testing ratios of 80%:20%, 70%:30%, and 60%:40%. Experimental results on 9,270 chest X-ray ray and 2,762 chest CT images acquired from different institutions show that the TL is effective with an average accuracy of 98.49%.
Keywords: Medical image diagnosis; DL classification; CT and X-ray images; healthcare applications
Apparently, at the end of 2019, the world witnessed an outbreak of COVID-19 (Coronavirus). It can be easily transferred from person to person in the world leading to a very harmful effect on the whole world. Statistics revealed by the WHO (World Health Organization) ensure that quarantine action is badly needed in most countries. In addition, there is a need for efficient tools that can be used for the automated diagnosis of COVID-19 to take fast decisions about suspected cases. Identifying clinical symptoms of COVID-19 from CT and X-ray images is essential to detect the really-infected cases. Medical symptoms may include fever, coughing, and shortness of breath [1]. Good detection results should be obtained from the CT and X-ray images for the infected COVID-19 cases. The virus RNA sequence detection is also a diagnostic tool for COVID-19. Nevertheless, this approach is time-consuming, and may be inappropriate with the wide spread of COVID-19. In addition, its exact rate of diagnosis ranges from 30% to 50%. Different diagnosis tools can be used in order to obtain accurate decisions [2]. In most third-world countries, due to the lack of human resources and funding for healthcare services, the treatment of COVID-19 cases is inefficient. Hence, automated medical diagnosis based on AI tools is recommended to solve the healthcare system problems due to the high accuracy of automated diagnosis, ease of implementation, and low cost.
The need to interpret radiographic images has inspired a series of deep learning AI systems [3], which have shown promising accuracy results in detecting COVID-19 cases from radiographic imagery [4,5]. Most of the AI systems that have come to practical applications on chest radiographic images were designed to classify pneumonia and normal cases. Hence, there is a need for more generalized classification systems that can also identify COVID-19 cases. The reason is that the X-ray and CT patterns corresponding to COVID-19 cases differ from those of normal and pneumonia cases [2].
With the number of infected COVID-19 patients increasing, radiologists find it increasingly difficult to finish the diagnosis process, promptly [6]. They find it necessary to urge the research community to develop highly-efficient software tools that can work on chest images of both X-ray and CT types in order to come to fast diagnosis decisions that may be suitable to the explosive scenario of COVID-19 spread. Recently, much research has been introduced for medical image analysis based on DL (Deep Learning) tools. In this context, DL algorithms [7] have performed better in pneumonia detection compared to traditional machine learning algorithms that depend on feature extraction and neural classification. The CNNs (Convolutional Neural Networks) are good DL candidates to work on chest X-ray and CT images. In such CNNs, the feature extraction is an embedded step that is performed through the use of convolutional masks. In addition, feature reduction is performed through appropriate pooling scenarios. Finally, the classification task is performed through a fully-connected neural network.
The DL models that can be developed for COVID-19 detection and for the classification of COVID-19, pneumonia, and normal cases need to be embedded in commercial software as soon as possible. They need to be used by the medical community, nowadays. That is why several DL models and algorithms need to be introduced and compared for the selection of the best of them in order to build a CAD (Computer-Aided Diagnosis) system for different types of viral infections including COVID-19 infection. Building such models from scratch may be a tedious task now requiring several design strategies in addition to large datasets of different types and different optimization tools. A good alternative to the design from scratch is the utilization of TL.
Deep TL is an efficient machine learning strategy that is used to speed up training and get better performance. In TL, a pre-trained model for a certain application is developed to be appropriate for another application. Some researchers have adopted deep TL to deal with the COVID-19 diagnosis problem to save the consumed time in designing a model from scratch and training it due to the tiny available datasets of chest X-ray and CT images [8–10]. Any TL model comprises three stages. The appropriate pre-trained model with large data is selected in the first stage. In the second stage, the output layers are modified to fit the new task. Finally, the TL model is adapted and tuned during training until it accomplishes the highest accuracy for the task of interest.
This research introduces different scenarios of TL models based on fine-tuning and deep-tuning processes. The fine-tuning process aims to train some layers by changing the learning parameters to achieve a high classification accuracy with networks such as DenseNet121, Densenet169, DenseNet201, ResNet50, ResNet152, VGG16, and VGG19. The deep-tuning process is presented to retrain all layers in the employed TL models to improve the classification accuracy using networks such as LeNet-5, AlexNet, Inception naïve v1, and VGG16. Two different datasets are examined in this work for COVID-19 diagnosis with different training/testing ratios of 80%:20%, 70%:30%, and 60%:40%. The obtained results demonstrate that the suggested CNN models achieve an average accuracy of 98.49% for multi-class classification, and 97.64% for binary classification to detect COVID-19. We can summarize the contributions of this paper is the following points:
• Different full-trained and pre-trained CNN models are investigated for the detection of COVID-19.
• These models are tested on two different datasets of X-ray and CT images.
• Extensive comparisons are presented between the different models for COVID-19 detection with different training/testing ratios in order to select the best candidates to be involved in CAD diagnosis systems.
The rest of this research work is coordinated as follows. The recent related studies are discussed in Section 2. The suggested classification framework is presented in Section 3. Simulation results and a comparison are given in Section 4. The concluding remarks are summarized in Section 5.
In [8], the authors proposed a model for the classification of chest CT images in order to detect COVID-19 cases. In this model, TL is utilized based on a pre-trained DenseNet201 architecture for deep feature extraction. Three activation functions (ReLU, PReLU, and TanhReLU) are adopted in this structure. In addition, an ELM (Extreme Learning Machine) classifier is utilized for performance evaluation. Finally, the decision is taken with a the majority voting algorithm that is applied on the generated results from the three activation functions. This model achieved an accuracy of 98.36%.
In [9], a model for automated diagnosis of COVID-19 from chest X-ray images was introduced. In this model, deep TL is exploited to enhance the detection performance compared to that obtained from the state-of-the-art models. Unfortunately, this model has been investigated on a small dataset of images. In addition, the authors of [9] presented a modified ResNet18 model for the classification of normal, pneumonia, and COVID-19 cases. With this modification, an accuracy level of 96.73% has been achieved. This level is better than those of the traditional ResNet18 and ResNet50 models.
In [10], the authors investigated the utilization of eight pre-trained models to classify chest X-ray images into healthy, bacterial, viral, COVID-19, and SARS classes. These models have been tested on the ImageNet dataset. Simulation results prove that DenseNet121 achieves the best accuracy, F1-score, recall, and precision levels. In [11], the authors introduced a DL model from scratch called COVID-Net for the classification of pneumonia and COVID-19 cases. It achieved an accuracy of 92.43%.
For DL-based COVID-19 detection from X-ray images, the authors of [12] developed a COVIDX-Net platform comprising seven CNN frameworks. In [13], the authors used an SVM (Support Vector Machine) and various CNN models for X-ray image classification. The best accuracy has been achieved with the ResNet50 model using an SVM classifier. An efficient DL classification model for binary and ternary classification scenarios was introduced in [14]. This model achieved accuracy levels of 98.75% for binary classification and 93% for ternary classification. The authors of [15] achieved an accuracy level of 98% for the classification of chest X-ray images with the ResNet50 model.
In [16], the authors presented an automatic COVID-19 detection model to speed up the interpretation process of lung CT images in order to allow fast and accurate diagnosis of COVID-19 cases. This model allows fusion of the features of CT scans with time information about these scans to follow up the patient case. The decision-tree method has been used for binary classification of the fused data. The LinkNet network has been utilized with a lesion segmentation network in order to obtain a high classification accuracy. The obtained accuracy, precision, recall, and F1-score results of this model were 94.4%, 96.7%, 95.2%, and 96.0%, respectively.
The authors of [17] studied the classification of CT scans based on TL using five pre-trained networks: DenseNet, Inception, MobileNet, ResNet50, and VGG16. The TL was adopted in these networks to enhance the performance as the authors worked on small datasets in order to eliminate the network training from scratch and avoid the large complexity of training. The feature extraction and classification are performed using the embedded CNN in each model to determine the infected and non-infected cases. The simulation experiments achieved an accuracy of 94%, a sensitivity of 96%, and a specificity of 92%.
In [18], the authors introduced a framework to classify X-ray images based on the pre-trained GoogLeNet model. The traditional GoogLeNet is adapted by modifying the final network layers and adopting 20-fold cross-validation to reduce the over-fitting problem. The hyperparameters of this framework are optimized using a multi-objective genetic algorithm to obtain a good performance. The experiments proved outstanding results that are suitable for real-time COVID-19 diagnosis.
An efficient COVID-19 detection model was presented in [19] with an FKNN (Fuzzy K-Nearest Neighbor) classifier and HHO (Harris Hawks Optimization). This model determines the COVID-19 severity degree as it performs the feature extraction operation and finds the best parameters, simultaneously. In [20], the authors introduced the DeTraC network to diagnose COVID-19 from chest X-ray images. This model adapts class boundaries to accommodate for any irregularities in X-ray images. It achieved an accuracy of 93.1% with a sensitivity of 100%.
In [21], the authors presented a hybrid framework for COVID-19 diagnosis from X-ray images to allow treatment in real clinical centers. Firstly, the CLAHE (Contrast-Limited Adaptive Histogram Equalization) CLAHE and Butterworth bandpass filter are utilized to improve the image contrast and remove noise. Then, the classification is performed on fused features extracted with two pre-trained models: ResNet34 and HRNet. Such framework achieves an outstanding performance with an accuracy of 99.99%, a sensitivity of 99.98%, a specificity of 100%, and a precision of 100%. In [22], the authors utilized CLAHE and Butterworth bandpass filter for image enhancement, while merging DBN (Deep Belief Network) and CDBN (Convolutional Deep Belief Network) models in a unified framework for binary X-ray image classification. The experimental results revealed an accuracy of 99.93% and a sensitivity of 99.90%.
3 Proposed Automated COVID-19 and Pneumonia Diagnosis Framework
In this section, an automated diagnosis framework is suggested based on DL and TL algorithms to distinguish non-COVID-19 cases from COVID-19 cases. As demonstrated in Fig. 1, the framework comprises three stages: image acquisition, pre-processing, and classification. The image acquisition stage aims to collect images from various sources. The second pre-processing stage reads the dataset and resizes it using a selected interpolation technique, because the images have different sources and sizes. The input image size is modified to 224 × 224 × 3 to be suitable for the input size of the employed TL models. The third stage is employed for classification using TL models.
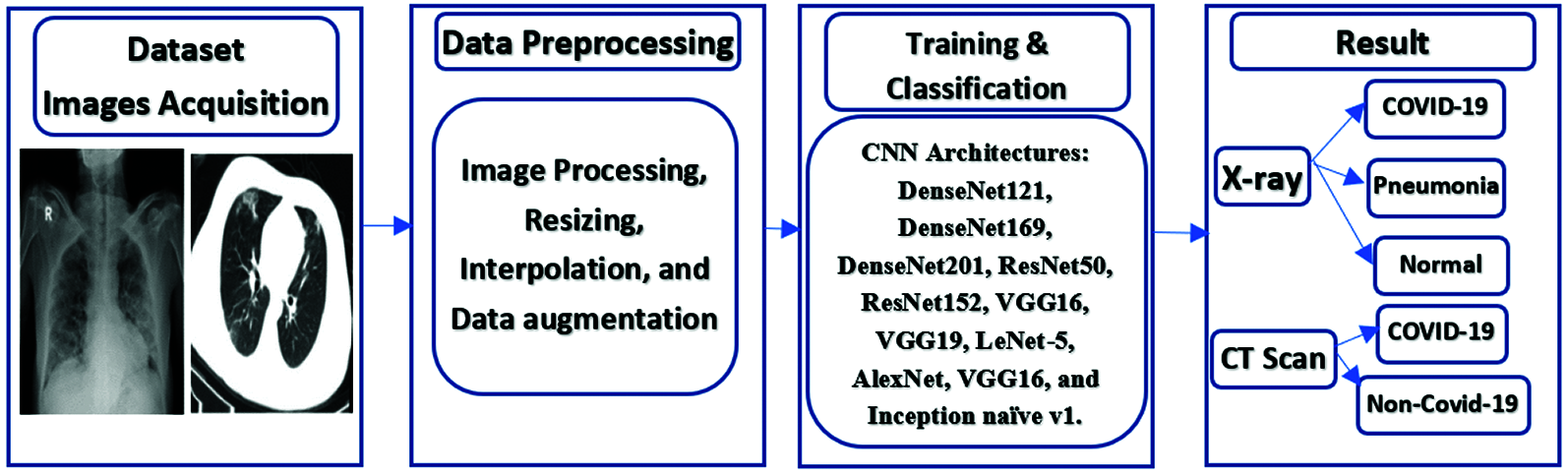
Figure 1: The proposed TL-based COVID-19 diagnosis framework
Recently, DL techniques have exhibited tremendous capabilities in computer vision and image processing applications. They have been used on various medical image modalities with superior efficiency of classification, identification, and segmentation. The CNN is motivated by the operation principles of the visual cortex of the human brain. The availability of large datasets leads to the success of CNN operations. Unfortunately, to guarantee high classification results with CNNs, large computational resources are required for the training process. In addition, deep CNNs with a large number of parameters need to work on massive datasets to prevent over-fitting during the training process. Due to the unavailability of large-enough datasets for COVID-19 cases, our objective is to use TL scenarios. The rationale behind the utilization of TL is to transfer knowledge from generic image classification to medical image classification. In addition, the computational cost is reduced and the over-fitting problem is eliminated, while achieving high accuracy and stability.
There are two adopted scenarios in this paper. The first one is fine-tuning of the upper layers of the DenseNet121, Densenet169, DenseNet201, ResNet50, ResNet152, VGG16, and VGG19 networks. DenseNet versions are distinguished with their high ability to deal with X-ray and CT scans and differentiate between various diseases that affect the lung. ResNet is a deep network that depends on skip connection, which makes it suitable to work on all image features in order to achieve the highest possible efficiency. The VGG is used, because it has several parameters, which can control its behavior. The second scenario is the deep tuning of all layers with the last layer changed to be suitable for the number of categories of LeNet-5, AlexNet, VGG16, and Inception naïve v1. Inception is used to have a small number of parameters, while achieving a relatively high efficiency.
4 Experimental Results and Comparison
Our contributions in this paper for automatic binary and multi-class classification are implemented on chest CT and X-ray datasets [23–26]. The subsequent sub-sections reveal in detail the main phases of the proposed work.
We worked on the subsequent image datasets:
1. COVID-19 CT dataset on GitHub [23]. This dataset includes 349 COVID-19 and 397 non-COVID-19 images.
2. COVID-19 collection on GitHub [24]. This is a growing array of globally identified CT and CXR (chest X-ray) scans for COVID-19 patients.
3. Chest X-ray images for pneumonia known as being a challenge detection dataset [25]. This dataset is present on Kaggle. It is divided into three different folders, and it includes subfolders for every normal/pneumonia group of images. It consists of 5,863 JPEG X-ray images and two (Normal/Pneumonia) groups.
4. Augmented and extensive COVID-19 chest CT and X-ray image dataset [26]. This data collection is available on Mendley. It consists of both X-ray and CT images for non-COVID-19 and COVID-19 instances.
All input images are pre-processed to have the same size. They are resized to the size of 224 × 224 × 3, to be appropriate for the TL models. The INTER_AREA method from the OpenCV library is used in oeder to resample the images using a pixel area relation. This library gives moiré-free results and eliminates the artifacts resulting from image zooming with the INTER_NEAREST method. Therefore, local information can be improved. To prevent over-fitting, because the number of CT scans is small, we have employed data processing strategies like arbitrary transformations. Such transformations accommodate for the range of rotation, the range of change in width, and the range of brightness. The transformation parameters are generated randomly for each training sample, and the augmentation is applied identically for every slice in the tested digital image. Additional aspects about the utilized medical images are given in Tab. 1.
For data splitting, we adopted different ratios for training and testing (80%:20%, 70%:30%, and 60%:40%), to ensure that the models work well in different scenarios. We guarantee that the medical images selected for cross-validation are not utilized throughout the training step to execute multi-class and binary classification, effectively. Furthermore, it is noticed that the utilized X-ray dataset is imbalanced. Indeed, 46.1% of the images represent the pneumonia class, 41.4% of the images represent the normal class, and 12.5% of the images represent the COVID-19 class. The dataset imbalance problem causes over-fitting. To solve this problem, we used a dropout strategy with all employed TL models. On the other hand, the utilized CT dataset is balanced. Samples for the utilized medical images are demonstrated in Fig. 2.
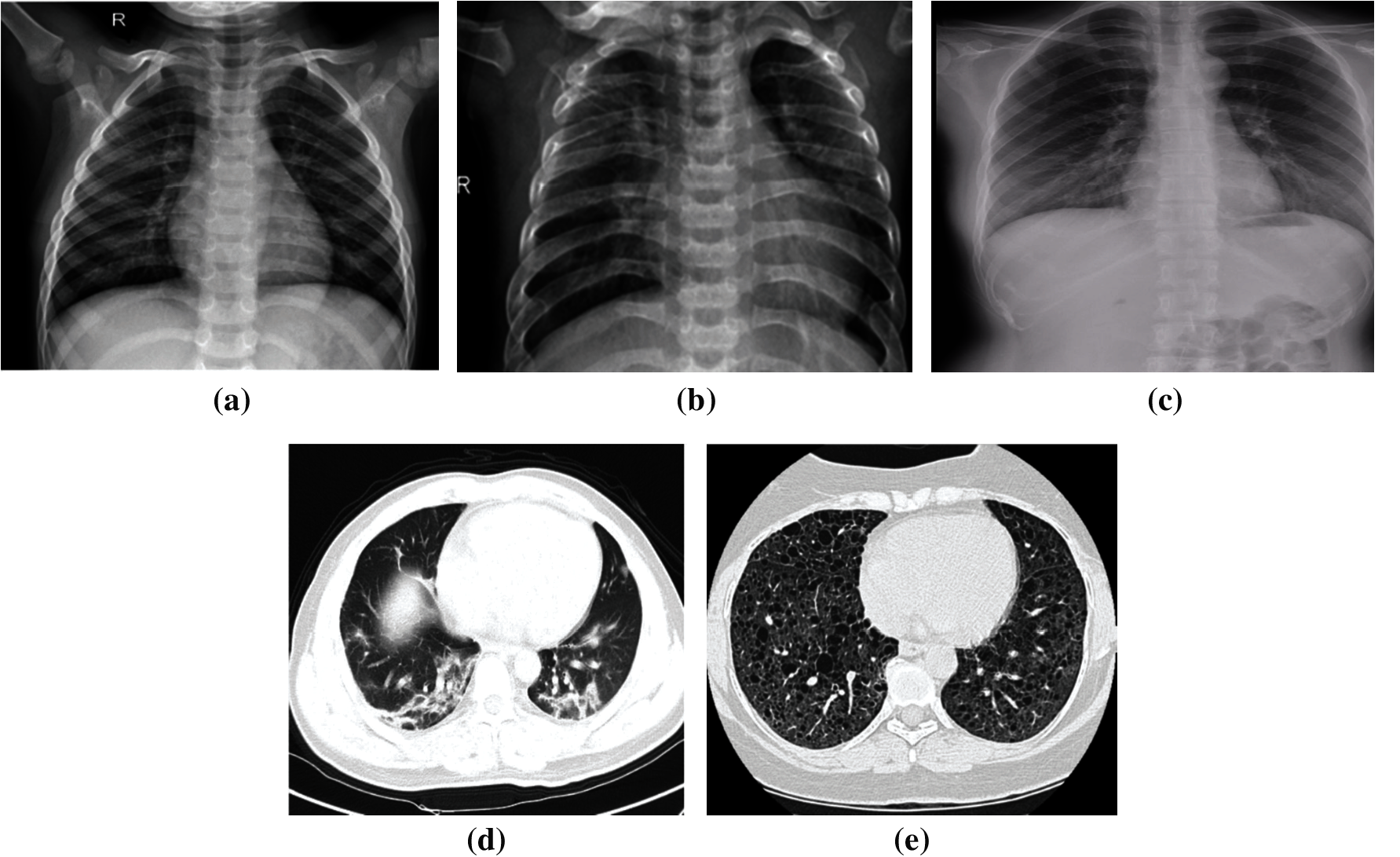
Figure 2: Samples of chest X-ray and CT scans (a) Normal X-ray. (b) Pneumonia X-ray. (c) COVID-19 X-ray. (d) COVID-19 CT (e) Non-COVID-19 CT
4.3 Training/Classification Dataset
After using data augmentation, splitting, and pre-processing procedures, the size of the training dataset is enlarged to accommodate for the operation of TL models to allow proper feature extraction. To construct the feature vector, the extracted attributes of each suggested TL model are floored, collectively. To categorize every medical image into a related group, the produced feature vector is forwarded to a multi-layer perceptron. Finally, the efficiency of the suggested model is assessed on test images. We replicate every experiment six times and estimate the average outcomes.
We have performed binary and multi-class classification on chest CT and X-ray datasets. The utilized medical images are resized to 224 × 224 to be appropriate for the classification models. Different batch sizes are considered with different numbers of epochs. The validation and training processes are performed with different ratios. The Adam's optimizer with β2 = 0.999, and β1 = 0.9 is utilized for the optimization purpose within the classification models. The learning rate is firstly set to 0.0001 and reduced to 0.000001.
Furthermore, we have used a dropout strategy to decrease the probability of over-fitting of the TL models. The realization of the suggested CNN models is accomplished using a Kaggle platform that provides notebook editors with free access to NVIDIA TESLA P100 GPUs and 13 GB RAM operating on professional Microsoft Windows 10 (64-bit). For simulation tests, Python 3.7 is utilized, and TensorFlow and Keras are employed as DL backend.
After obtaining the proper features, the final stage is to categorize the obtained data [27]. Hence, we measure the efficiency of the classification stage. Throughout our assessment, we use the accuracy, recall, F1-score, and precision shown in Eqs. (1)–(4) [27]. In addition, the AUC (Area Under Curve) of the ROC (Receiver Operating Characteristic) curve [28], cross-entropy loss, aka logistic loss, and log loss [29] given in Eq. (5), are used to evaluate the proposed CNN models.
where TN is True Negative, FN is False Negative, TP is True Positive and FP is False Positive. yT represents ground-truth (correct) labels, and yP represents predicted labels.
The binary and multi-class classification results are presented in this section for chest CT and X-ray images for the full-training of all layers of the LeNet-5, AlexNet, VGG16, and Inception naïve v1 and the fine-tuning of the top layers of the DenseNet121, Densenet169, DenseNet201, ResNet50, ResNet152, VGG16 and VGG19. Additionally, to verify the robustness and performance of every utilized CNN model, numerous tests have been performed on the chest CT and X-ray datasets.
The training curve is estimated on the training dataset. It indicates how properly the employed CNN model is learned, while the test curve or validation curve is estimated on a validation hold-out dataset, which shows how properly the employed framework is over-simplified. The loss of validation and training is characterized as the number of mis-classifications resulting from every instance of training or validation. To sum up, the best CNN model is the one that performs well with neither over-fitting nor under-fitting. Besides, the confusion matrix demonstrates a comprehensive interpretation of what happens after grading with images [11].
When it is needed to visualize or check the multi-class or binary classification performance, the AUC-ROC curve [28] is used. It is one of the essential assessment metrics for examining the performance of any classification model. The ROC curve is a likelihood curve, and the AUC signifies the measure or degree of separability. It reveals how much the CNN model can discriminate between different classes. The ROC curve is mapped with the FPR against the TPR, where FPR is on the x-axis and TPR is on the y-axis. The FPR is the False Positive Rate and the TPR is the True Positive Rate. Precision-Recall curve [28] is a useful predictive indicator of performance, while the groups are incredibly imbalanced. Precision is an assessment of the importance of the results in information retrieval, while recall is an assessment of how often genuinely valid data are retrieved. Therefore, the curve of precision-recall indicates the compromise between the accuracy of various thresholds and their recall. A large area under the curve means high precision and high recall, where high precision is related to a low FPR, and high recall is associated with a high FPR.
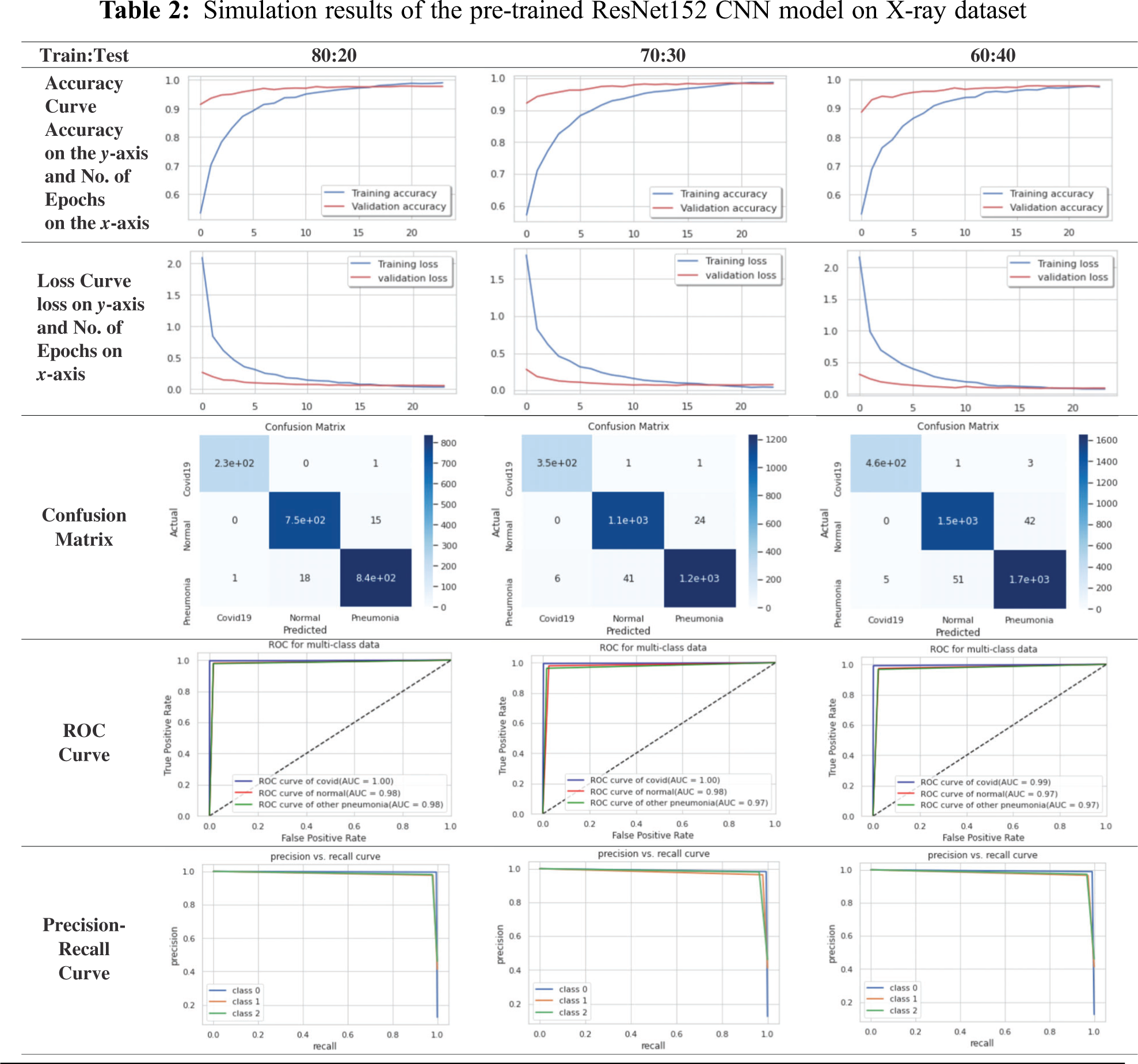
For simplicity, we will present the best obtained curves for the CNN models. For the X-ray dataset, the ResNet152 model achieves a superior performance compared to those of other examined models. According to Tab. 2, the ResNet152 for the X-ray dataset has high stability and high accuracy for different training and testing ratios. For the 80:20 ratio, the training accuracy equals 98.95%, the testing accuracy equals 98.11%, the training loss equals 0.0360, and the testing loss equals 0.0573 for 24 epochs. For the confusion matrix, the true positives equal 230 for COVID-19 case, 750 for normal case, and 840 for pneumonia case. The ROC curve indicates that the TPR and FPR nearly equal 98%. Precision-recall curve has a high AUC with high precision and high recall. The computational time of the training process is 38 sec for every epoch. Hence, the total time of all epochs is 15.5 min.
For simplicity, we will present the best obtained curves for the tested CNN models. For the CT dataset, the DenseNet201 model achieves superior results compared to those of other examined models. According to Tab. 3, the DenseNet201 on the CT dataset has high stability and high accuracy for different training and testing ratios. For the 80:20 ratio, the training accuracy equals 99.00%, the testing accuracy equals 96.61%, the training loss equals 0. 0272, and the testing loss equals 0.0719 for 49 epochs. For the confusion matrix, the true positives equal 260 for the COVID-19 case and 280 for the normal case. The ROC curve indicates that TPR and FPR nearly equal 97%. Precision-recall curve has high AUC with high precision and high recall. The computation time of the training process is 10 sec for every epoch. So, the total time of all epochs is 8.2 min.
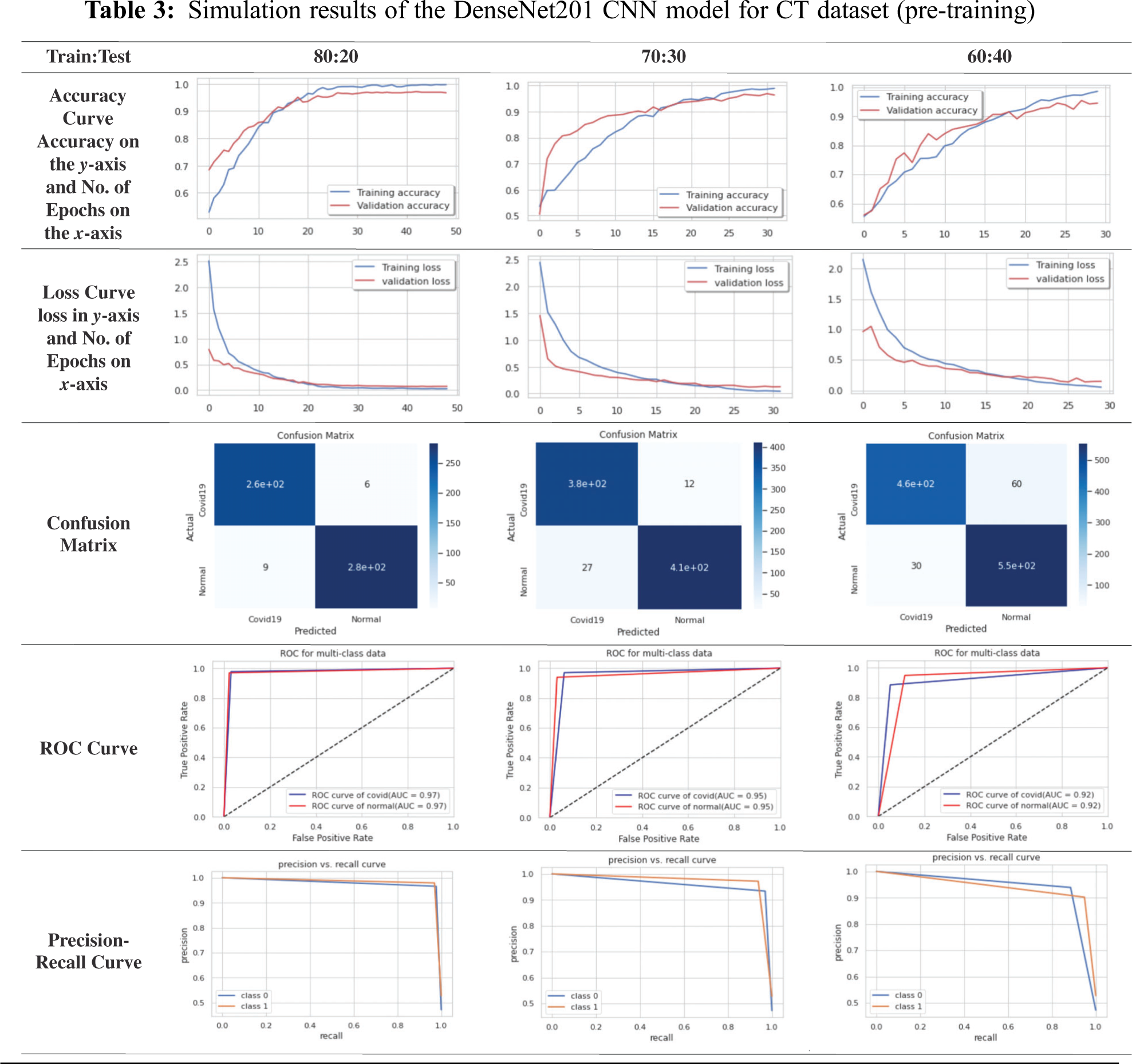
4.7 Comparison Between TL Models on X-ray and CT Datasets
The comparison outcomes in terms of F1-score, loss, log loss, recall, accuracy, and precision of various CNN models of multi-class and binary classification scenarios on the X-ray and CT datasets at different training/testing ratios (80:20, 70:30, 60:40) are presented in Tabs. 4 and 5. It is observed from Tabs. 4 and 5 that the fine-tuned DensNet121, DensNet169, DensNet201, ResNet50, ResNet152, and VGG19 TL models achieve better outcomes in terms of the validation and training accuracies on the CT and X-ray datasets. They outperform other deep-tuning LeNet-5, AlexNet, VGG16, Inception naïve v1, ResNet50, and VGG16 TL models.
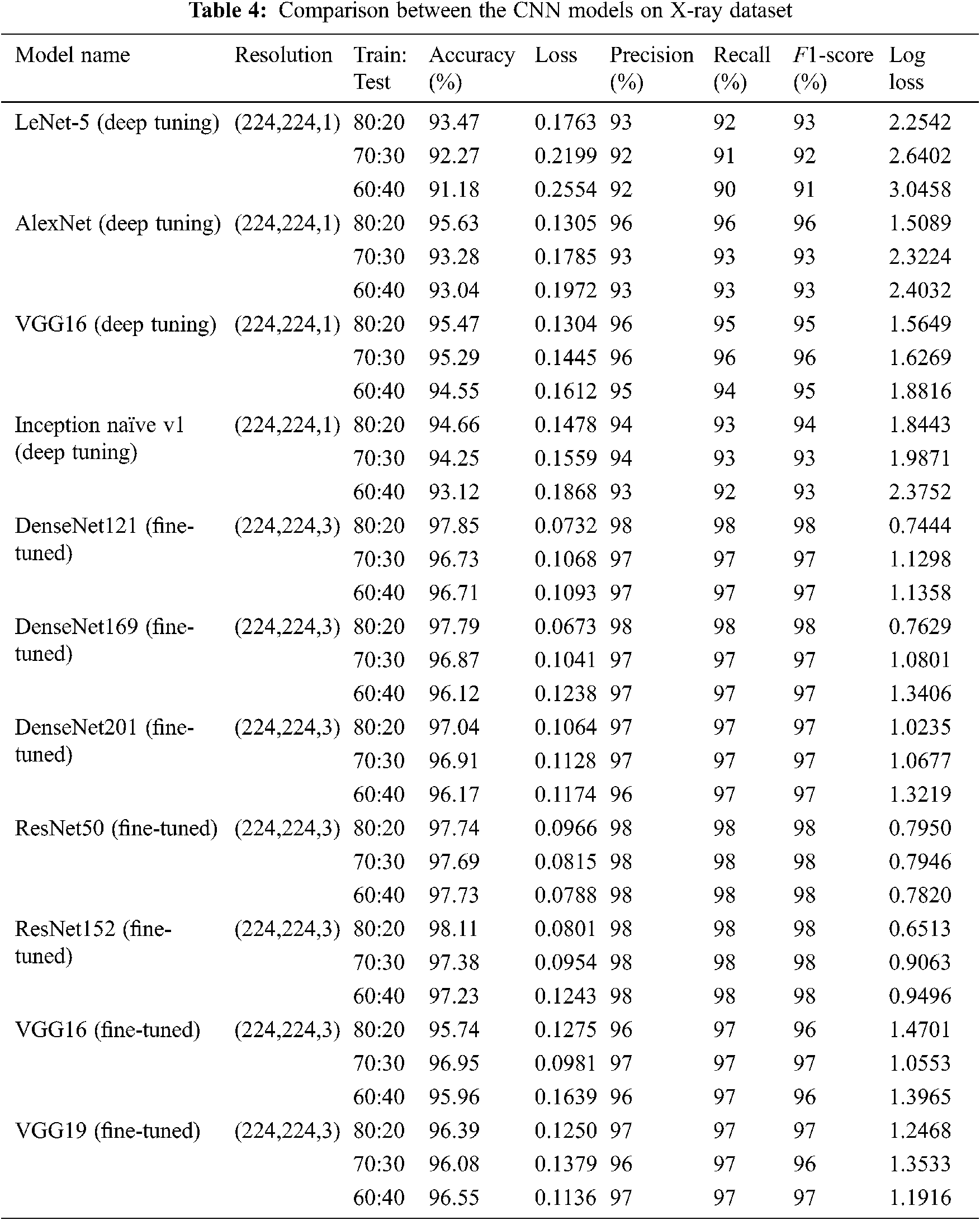
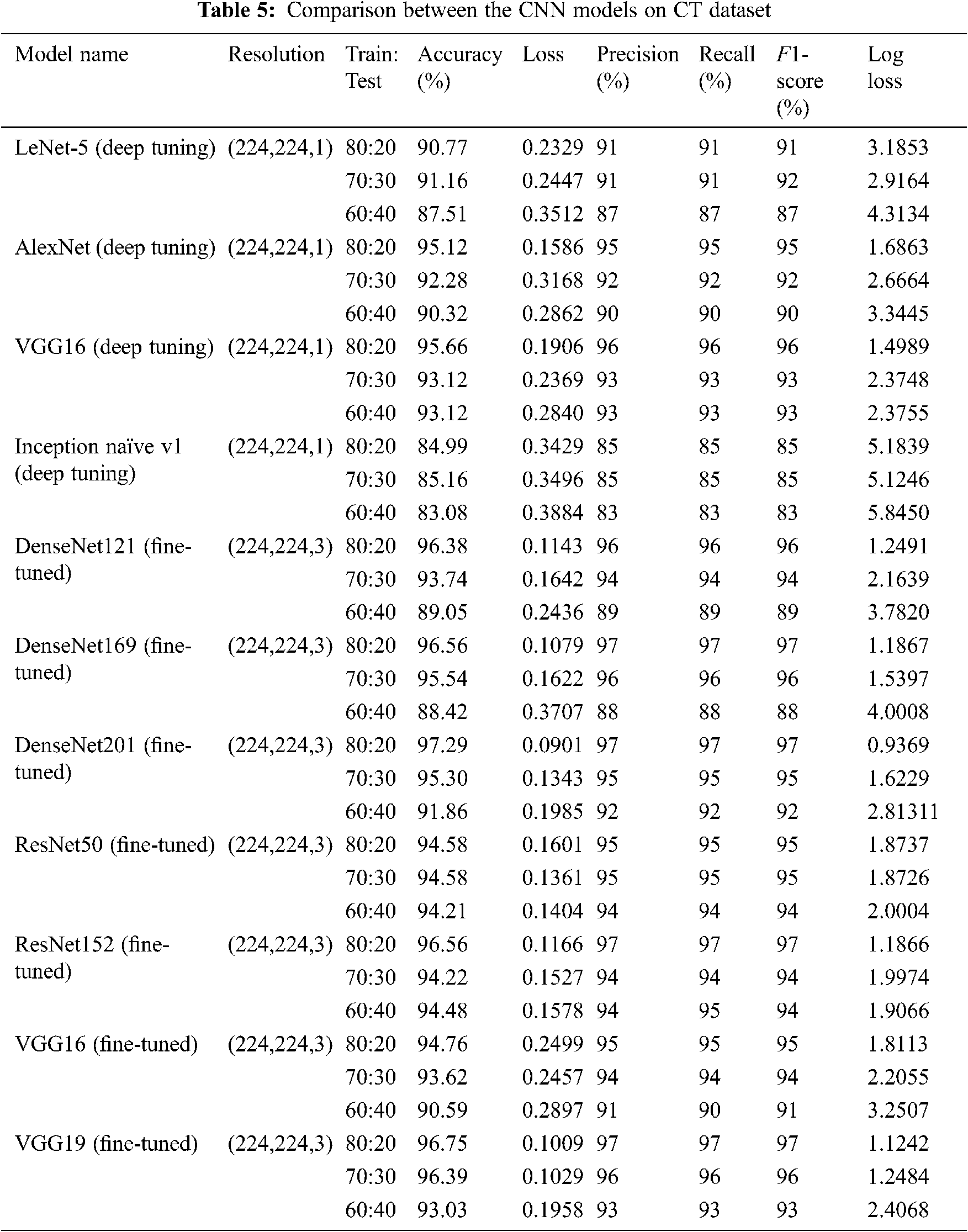
Fig. 3 presents a summary of the accuracy results of the TL models on the X-ray dataset compared to the results of the TL models on the CT dataset. It is noticed that the TL models in the case of X-ray dataset reveal superior classification accuracies compared to those obtained on CT images by obtaining accuracy levels greater than 98% on X-ray images and accuracy levels greater than 96% on CT images.
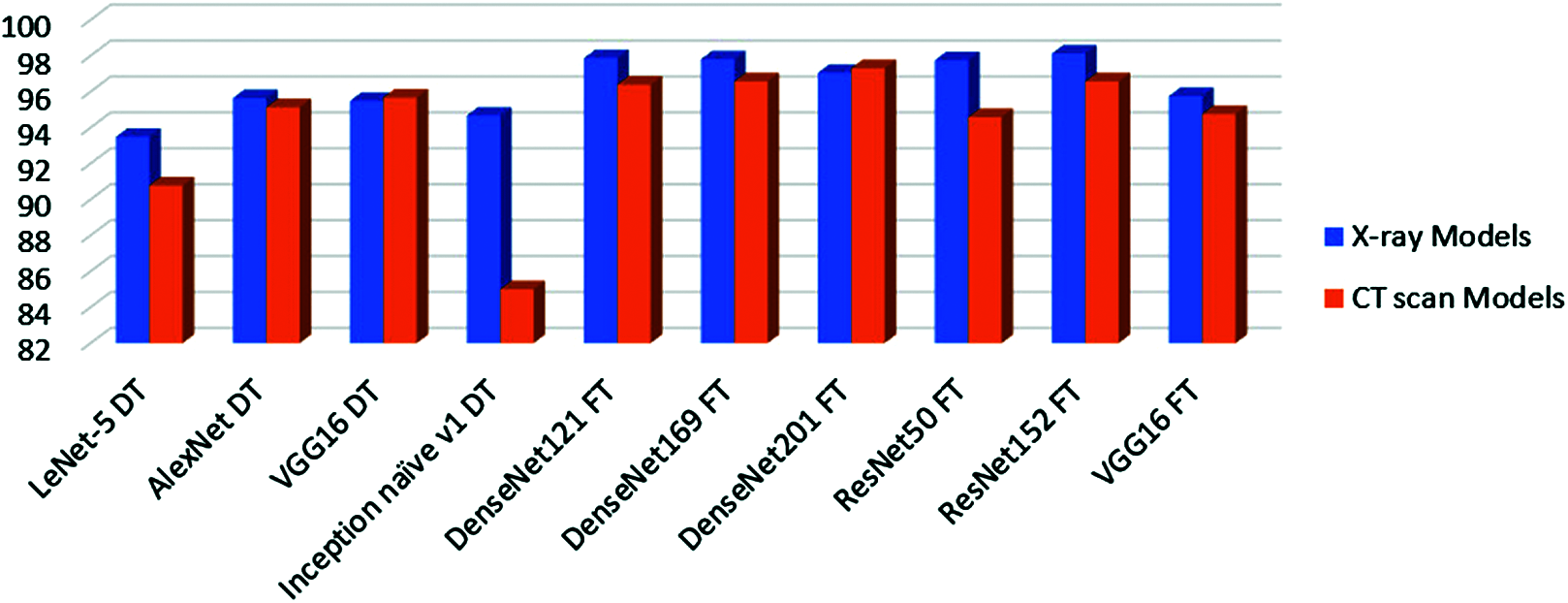
Figure 3: Accuracy of all tested models on X-ray and CT datasets. DT means deep-tuning and FT means fine-tuning
This paper presented different CNN models for the binary (COVID-19, Normal) and multi-class (COVID-19, Normal, and Pneumonia) classification of COVID-19 cases. These models have been investigated on different CT and X-ray datasets with TL concepts considering deep-tuning and fine-tuning scenarios. A comprehensive comparison has been presented between TL models including LeNet-5, AlexNet, VGG16, and Inception naïve v1 as deep-tuning models, and the DenseNet121, DenseNet169, DenseNet201, ResNet50, ResNet152, VGG16, and VGG19 as fine-tuning models. Simulation experiments have been performed on chest CT and X-ray datasets containing 12,032 images (2,466 COVID-19, 4,273 pneumonia, and 5,293 normal). All models have been assessed with numerous classification assessment metrics. The ResNet152 and DenseNet201 achieved superior results compared to other TL models for the examined X-ray and CT images. Future research work will be directed to a complete viral infection diagnosis system through deep-learning. Moreover, the classification performance can be improved using more datasets, more advanced techniques for extracting DL features, such as VDSR (Very-Deep Super-Resolution) and other optimized generative models for single-image super-resolution.
Acknowledgement: The authors extend their appreciation to the Deputyship for Research & Innovation, Ministry of Education in Saudi Arabia for funding this research work through the project number (PNU-DRI-Targeted-20-027).
Funding Statement: The authors extend their appreciation to the Deputyship for Research & Innovation, Ministry of Education in Saudi Arabia for funding this research work through the Project Number (PNU-DRI-Targeted-20-027).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. N. Zhang, L. Wang, X. Deng, R. Liang, M. Su et al., “Recent advances in the detection of respiratory virus infection in humans,” Journal of Medical Virology, vol. 92, no. 4, pp. 408–417, 2020. [Google Scholar]
2. E. Chorin, M. Dai, E. Shulman, L. Wadhwani, R. Bar-Cohen et al., “The QT interval in patients with COVID-19 treated with hydroxychloroquine and azithromycin,” Nature Medicine, vol. 26, no. 6, pp. 808–809, 2020. [Google Scholar]
3. Y. Oh, S. Park and J. C. Ye, “Deep learning covid-19 features on cxr using limited training data sets,” IEEE Transactions on Medical Imaging, vol. 39, no. 8, pp. 2688–2700, 2020. [Google Scholar]
4. S. Vaid, R. Kalantar and M. Bhandari, “Deep learning COVID-19 detection bias: Accuracy through artificial intelligence,” International Orthopaedics, vol. 44, pp. 1539–1542, 2020. [Google Scholar]
5. C. Jin, W. Chen, Y. Cao, Z. Xu, Z. Tan et al., “Development and evaluation of an artificial intelligence system for COVID-19 diagnosis,” Nature Communications, vol. 11, no. 1, pp. 1–14, 2020. [Google Scholar]
6. H. Li, R. Mukundan and S. Boyd, “Robust texture features for breast density classification in mammograms,” in Proc. 2020 16th IEEE Int. Conf. on Control, Automation, Robotics and Vision (ICARCVShenzhen, China, pp. 454–459, 2020. [Google Scholar]
7. A. Bhandary, G. A. Prabhu, V. Rajinikanth, K. P. Thanaraj, S. C. Satapathy et al., “Deep-learning framework to detect lung abnormality–A study with chest X-ray and lung CT scan images,” Pattern Recognition Letters, vol. 129, pp. 271–278, 2020. [Google Scholar]
8. M. Turkoglu, “COVID-19 detection system using chest CT images and multiple kernels-extreme learning machine based on deep neural network,” Innovation and Research in BioMedical Engineering, vol. 5, pp. 1–22, 2021. [Google Scholar]
9. R. A. Al-Falluji, Z. D. Katheeth and B. Alathari, “Automatic detection of COVID-19 using chest X-ray images and modified resnet18-based convolution neural networks,” Computers, Materials, & Continua, vol. 66, no. 2, pp. 1301–1313, 2021. [Google Scholar]
10. K. KC, Z. Yin, M. Wu and Z. Wu, “Evaluation of deep learning-based approaches for COVID-19 classification based on chest X-ray images,” Signal, Image and Video Processing, vol. 11, pp. 1–15, 2021. [Google Scholar]
11. L. Wang, Z. Lin and A. Wong, “Covid-net: A tailored deep convolutional neural network design for detection of covid-19 cases from chest x-ray images,” Scientific Reports, vol. 10, no. 19549, pp. 1–12, 2020. [Google Scholar]
12. M. Jamshidi, A. Lalbakhsh, J. Talla, Z. Peroutka, F. Hadjilooei et al., “Artificial intelligence and COVID-19: Deep learning approaches for diagnosis and treatment,” IEEE Access, vol. 8, pp. 109581–109595, 2020. [Google Scholar]
13. J. Liu, H. Yu and S. Zhang, “The indispensable role of chest CT in the detection of coronavirus disease 2019 (COVID-19),” European Journal of Nuclear Medicine and Molecular Imaging, vol. 47, no. 7, pp. 1638–1639, 2020. [Google Scholar]
14. I. D. Apostolopoulos and T. A. Mpesiana, “Covid-19: Automatic detection from x-ray images utilizing transfer learning with convolutional neural networks,” Physical and Engineering Sciences in Medicine, vol. 43, no. 2, pp. 635–640, 2020. [Google Scholar]
15. M. Pandit, S. Banday, R. Naaz and M. Chishti, “Automatic detection of COVID-19 from chest radiographs using deep learning,” Radiography, vol. 27, no. 2, pp. 483–489, 2021. [Google Scholar]
16. T. Li, W. Wei, L. Cheng, S. Zhao, C. Xu et al., “Computer-aided diagnosis of COVID-19 CT scans based on spatiotemporal information fusion,” Journal of Healthcare Engineering, vol. 3, pp. 1–13, 2021. [Google Scholar]
17. A. Oluwasanmi, M. Aftab, Z. Qin, S. Ngo, T. Doan et al., “Transfer learning and semisupervised adversarial detection and classification of COVID-19 in CT images,” Complexity, vol. 5, pp. 1–19, 2021. [Google Scholar]
18. P. Shukla, J. Sandhu, A. Ahirwar, D. Ghai, P. Maheshwary et al., “Multi-objective genetic algorithm and convolutional neural network based COVID-19 identification in chest X-ray images,” Mathematical Problems in Engineering, vol. 20, pp. 1–9, 2021. [Google Scholar]
19. H. Ye, P. Wu, T. Zhu, Z. Xiao, X. Zhang et al., “Diagnosing coronavirus disease 2019 (COVID-19Efficient harris hawks-inspired fuzzy k-nearest neighbor prediction methods,” IEEE Access, vol. 9, pp. 17787–17802, 2021. [Google Scholar]
20. A. Abbas, M. Abdelsamea and M. Gaber, “Classification of COVID-19 in chest X-ray images using deTraC deep convolutional neural network,” Applied Intelligence, vol. 51, no. 2, pp. 854–864, 2021. [Google Scholar]
21. A. Al-Waisy, S. Al-Fahdawi, M. Mohammed, K. Abdulkareem, S. A. Mostafa et al., “COVID-Chexnet: Hybrid deep learning framework for identifying COVID-19 virus in chest X-rays images,” Soft Computing, vol. 7, pp. 1–16, 2020. [Google Scholar]
22. A. Al-Waisy, M. Mohammed, S. Al-Fahdawi, S. Maashi, B. Garcia-Zapirain et al., “COVID-Deepnet: Hybrid multimodal deep learning system for improving COVID-19 pneumonia detection in chest X-ray images,” Computers, Materials and Continua, vol. 67, no. 2, pp. 2409–2429. 2021. [Google Scholar]
23. “COVID dataset,” https://github.com/UCSD-AI4H/COVID-CT, (last Access on 25-3-2021). [Google Scholar]
24. “COVID dataset,” https://github.com/ieee8023/covid-chestxray-dataset, (last Access on 25-3-2021). [Google Scholar]
25. “COVID dataset,” https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia, (last Access on 25-3-2021). [Google Scholar]
26. “COVID dataset,” https://data.mendeley.com/datasets/8h65ywd2jr/1?fbclid=IwZLb04fZMx4CX7fU1B6Ln1Do, (last Access on 25-3-2021). [Google Scholar]
27. T. RN and R. Gupta, “A survey on machine learning approaches and its techniques,” in Proc. IEEE Int. Students’ Conf. on Electrical, Electronics and Computer Science (SCEECSBhopal, India, pp. 1–6, 2020. [Google Scholar]
28. D. Hand and R. Till, “A simple generalisation of the area under the ROC curve for multiple class classification problems,” Machine Learning, vol. 45, no. 2, pp. 171–186, 2020. [Google Scholar]
29. S. Sharma and V. Kumar, “Low-level features based 2D face recognition using machine learning,” International Journal of Intelligent Engineering Informatics, vol. 8, no. 4, pp. 305–330, 2020. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |