DOI:10.32604/iasc.2022.019360

| Intelligent Automation & Soft Computing DOI:10.32604/iasc.2022.019360 |  |
| Article |
A Deep Learning to Distinguish COVID-19 from Others Pneumonia Cases
1Laboratory of Advanced Technology and Intelligent Systems, National Engineering School, University of Sousse, Sousse, 4054, Tunisia
2Laboratory of Computer engineering and systems, Faculty of Science and Technologies, Cadi Ayyad University, Marrakech, 40000, Morocco
*Corresponding Author: Sami Gazzah. Email: sami.gazzah@gmail.com
Received: 11 April 2021; Accepted: 18 May 2021
Abstract: A new virus called SARS-CoV-2 appeared at the end of space 2019 in Wuhan, China. This virus immediately spread throughout the world due to its highly contagious nature. Moreover, SARS-CoV-2 has changed the way of our life and has caused a huge economic and public health disaster. Therefore, it is urgent to identify positive cases as soon as possible and treat them as isolated. Automatic detection of viruses using computer vision and machine learning will be a valuable contribution to detecting and limiting the spread of this epidemic. The delay introduction of X-ray technology as diagnostic tool limited our ability to distinguish COVID-19 from other pneumonia cases. These images can be feed into a machine learning system that can be trained to detect lung infections, which can arise in acute pneumonia. However, some efforts attempt to detect SARS-CoV-2 using binary data and such combinations, which may leading to confusion. Indeed, in this case, the classifier’s training minimizes the intra-class similarity between pneumonia in COVID-19 and typical pneumonia. Due to the fact that in addition to SARS-CoV-2 pneumonia, there are two other common types: viral and bacterial pneumonia. They all appear in similar shapes, thus making them difficult to distinguish. This research proposed a deep multi-layered classification of pneumonia based on a neural network using chest X-ray images. The experiment was conducted using the combination of two images from an open-source dataset collected by Cohen GP and Kagel. The data consists of 4 categories: normal pneumonia, bacterial pneumonia, viral pneumonia, and COVID19, for 2,433 images. We considered two architectures: Xception and ResNet50. In addition, a comparison was made between the two models. The pre-trained Xception model in 20 epochs provided a classification accuracy of 86%.
Keywords: COVID-19; pneumonia; deep learning; diagnosis level
Pneumonia is an infection that attacks the respiratory organs in humans; the infection inflames the air sacs in one or both lungs. The alveoli in the lungs are filled with pus or fluid, causing difficulty breathing and limits the quantity of oxygen intake. Clinical signs and radiographic tests can confirm or disprove the diagnosis of pneumonia. However, it is difficult to distinguish between bacterial and viral pneumonia, especially with the lack of a specialist in rural areas. Accurate and rapid diagnostics will help to reduce the global mortality rates in children under five, the elderly, and people with present health problems or weakened immune systems. Pneumonia is the primary cause of mortality in underaged children and is a major cause of child mortality in all regions over the world [1].
Pneumonia can be caused by viruses, bacteria, and fungi. In this study, we will mainly focus on detecting of a novel virus, named SARS-CoV-2 or coronavirus, which may cause pneumonia. This virus can lead to severe or life-threatening illness, and even death. The COVID-19 is an infectious disease caused by the SARS-CoV-2 virus, firstly detected in December 2019 in China. The World Health Organization recognized it as a global pandemic on 11 March 2020, after that, it widely spread worldwide killing thousands of people. The five pandemics since 1918 share the main character that they are highly contagious. The most important measures to take are social distancing and quarantine. The European Centre for Disease Prevention and Control reported that the number of confirmed cases between 31 December 2019 and 07 May 2020 rose to 3 713 796 cases of COVID-19 reached 263 288 deaths [2]. The scientific community has an obligation to provide prompt responses to the urgent need to fight against this huge threat and mitigate its spread. Computer vision and Artificial Intelligence (AI) researchers got immediately involved in the fight against this pandemic. Much attention has been given to areas of research that tackle some of COVID 19 prevention, detection, and understanding the transmission of the pandemic [3,4]. However, the highly responsive and efficient diagnosis of COVID-19 pneumonia applications detect all pneumonia types similar in form, making it difficult to distinguish between them. We performed a set of research-based on coronavirus classification using Chest X-ray Images data, and most of the articles are limited to only 2 or 3 categories, which is due to the inability of the proposed model to classify more than that.
In this paper, we are mainly interested in the contribution of machine learning and computer vision on COVID-19 at the diagnosis level to detect coronavirus pneumonia and distinguish them from other types of pneumonia caused by other viruses and bacteria.
The rest of this paper is structured as follows: we first present a literature review followed by the methodology to adopt and categorize pneumonia subtypes in Section 2. Section 3 and 4 are dedicated to experimentation and results. Conclusion and future directions can be found in Section 5.
2 Emerging Trends and Literature Review
In this section, we present three emerging trends arising from the COVID-19 pandemics, then we describe the related works for COVID-19 diagnosis based on CT and X-ray images and in the end, we review the most popular CT and X-ray images datasets used for pandemic diagnosis.
2.1 Emerging Trends Arising from the COVID-19 Pandemic
The COVID-19 pandemics proposed new challenges to the research community, (1) the rapid spread of COVID-19 requires the immediate search for a rapid solution. Mainly in the case of novel pandemics in which we do not have enough data to train a Machine learning system, vary, but we dispose of a trained system in close syndrome: i.e., Pneumonia in COVID-19 and other typical viral pneumonia is different but shared some common characteristics [5]. At this level, it is judicious to investigate how to use “stored back knowledge” gained while dealing with typical viral pneumonia and try to transfer the knowledge learned from the source domain whose examples are generic toward learning a classifier in the target domain-containing few data as and almost different to target domain (Fig. 1) [6,7].
In addition, transfer learning (TL) in COVID-19 remains an interesting research trend because the SARS-CoV-2 virus is constantly mutating during the pandemic. New variants of the virus appear and emerge over time in different countries. Accordingly, our trained systems need to quickly adapt the learned knowledge to the multiple variants of the virus while maintaining high accuracy level of the results.

Figure 1: TL system proposed by [7] based on SMC specialization approach
(2) Because the virus is extremely contagious, it spreads and continuously evolves in the population. Urgent steps have been taken, including social distancing and keeping the population across the country in lockdown. In this context, governments worldwide have imposed barrier measures during the spread phase of the pandemic. Computer vision systems can be deployed in these situations to ensure safety and prevent contamination due to short distancing. A valuable contribution to the prevention level is to measure the distance between pedestrians in public places since many datasets are already available [7,8]. Moreover, a mask-wearing verification system can represent another interesting contribution since there is an available dataset. This dataset includes 1376 RGB facial images; among them, 690 are with the mask on this link https://github.com/prajnasb/observations/tree/master/experiements/data.
(3) The other interesting trend is person identification in a challenging situation when he has been wearing a mask: bottom occlusions made by a mask can significantly change the appearance of a face and a trained face detector will fail to detect such faces [9].
Among all emerging trends mentioned above in tackling COVID-19, we focus on pandemic diagnosis based on Chest X-ray images and how to distinguish COVID-19 from others pneumonia cases. This represents a first step towards the automatic reuse of trained typical viral pneumonia to develop a new model on COVID-19.
Alsaade et al. [10] have proposed a recognition system for classifying COVID-19 using chest X-ray/CT images. Three classification algorithms are considered for this research: SVM, KNN, and CNN. The first two classifiers are used to combine handcrafted features from 2D-DWT with a gray level co-occurrence matrix to feed a SVM and a KNN classifier. In the last classification system, a deep-learning convolutional Neural Networks (ResNet 50) is explored and compared to the handcraft based methods. The different experimentation shows that the CNN based methods outperform the two others approaches and achieve an accuracy of 97.14%.
Waisy et al. [11] have proposed an automated system for diagnosing COVID-19 using chest X-ray (CX-R) chest radiography images. A deep learning based approach, named COVID-DeepNet, is deployed to distinguish between healthy and COVID-19 infected subjects. A preprocessing step is performed to enhance the contrast and reduce the noise levels in images. The reported results exhibited a detection accuracy rate of 99.93%.
In the same vein, Ozturk et al. [12] have achieved a classification accuracy of 98.8% by using two classes but dropping to 87.02 % in a multiclass context. A deep learning based model called DarkNet is applied to detect and classify COVID-19 cases.
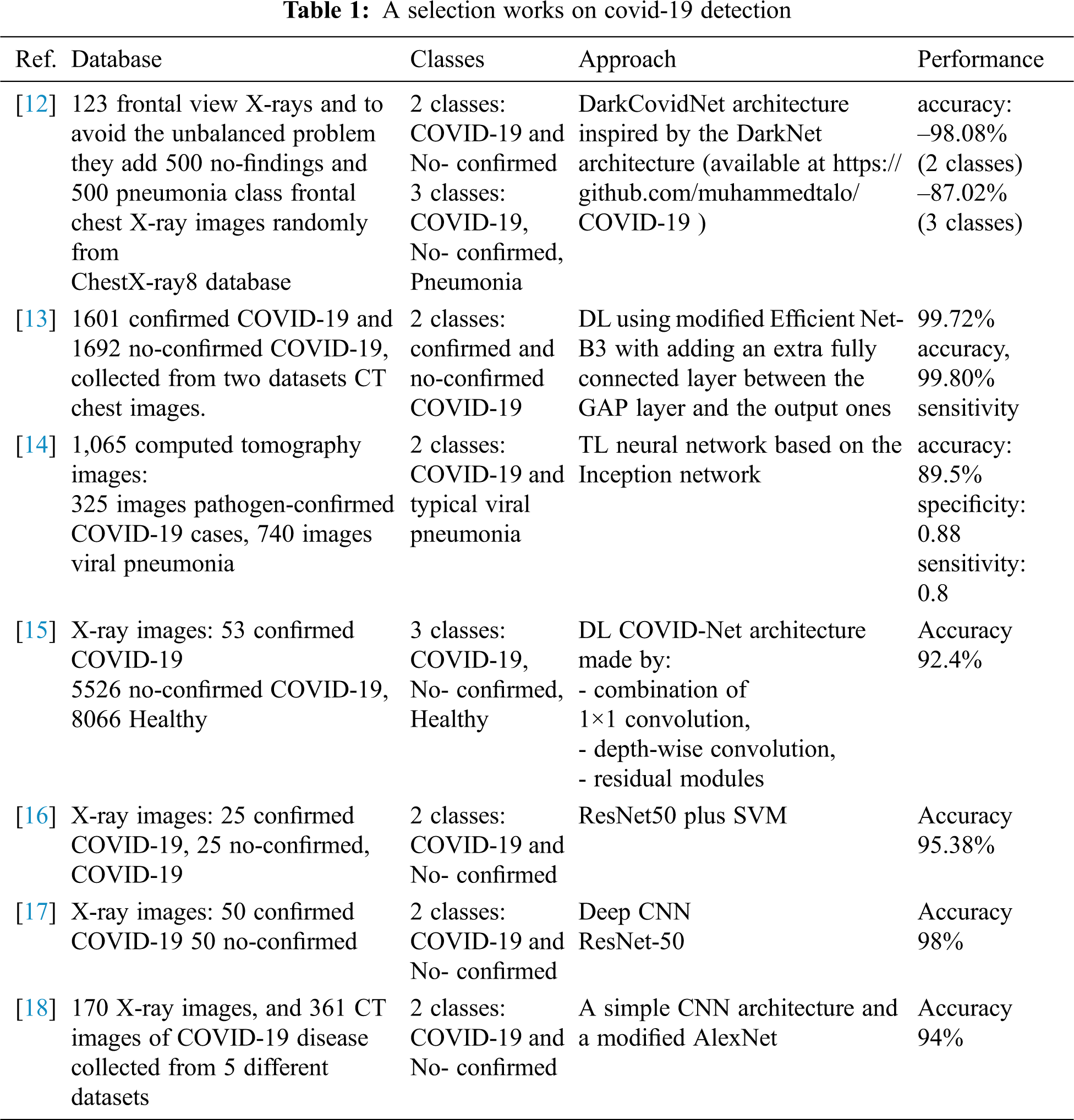
The following table (Tab. 1) synthesizes selected approaches studied in several works in the literature. Almost all the proposed approaches have considered two or three classes based on deep learning with promising results. The classification rate drops when considering three classes compared to binary classification.
Since the outbreak of the COVID-19 virus, researchers and societies of radiology are still working on collecting of CT and X-ray images from infected patients. Almost all available datasets are small and containing a limited number of X-rays or/and CT images.
The fact that the training phase requires a large number of examples to learn intra-class variability. Almost all authors have tried to collect images from different available small datasets to construct a larger one. This measure sounds very interesting because the coronavirus is mutating, and we have identified distinctive variants from different countries. Therefore, the collection of images from different countries and sources is important to take into account the intra-variability of its effects. The researches behind Nextstrain.org counts until March 15, 2021 more than hundreds of thousands of SARS-CoV-2 genomes and this number rises every day (Fig. 2) [19]. New variants arise and others disappear across different waves of coronavirus infections.
Alqudah et al. [20] collected augmented X-ray Images for COVID-19 datasets. These datasets result from the merger of two online available datasets : (https://github.com/ieee8023/covid-chestxray-dataset and https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia). The actual version 3 of the dataset contains two folders: the first one contains COVID-19 augmented images while Non-COVID-19 images are augmented; the other folder contains augmented images of both COVID-19 and Non-COVID-19.

Figure 2: SARS-CoV-2 Genome mutation explained by (a) a phylogenetic tree of SARS-CoV-2 illustrates the transformation of the virus over time that is represented by branching and mutation events associated with each node, and the (b) shows the periodical mutation [19]
Authors in [18] also collected images from five different datasets of 170 X-ray images and 361 CT images of COVID-19 disease. The Italian Society of Medical and Interventional Radiology COVID-19 makes available x-ray referenced images related to 68 patients (SIRM) COVID-19 Database, references for each image are provided in the metadata, the respective classes are annotated and case review [21].
Since the SARS-CoV-2 virus attacks the lungs of infected people [22], the French Society of Radiology (SFR) announced on April2, 2020, the creation of a French Imaging Database against Coronavirus («FIDAC»). It indicates having recorded “6235 scanners out of 16762 (37%) chest scanners for the exploration of a possible attack by the coronavirus in one week”, coming from 315 French centers already registered in the project, as well as Belgian, Swiss centers, Maghreb, Africa, and others. “This project aims to quickly collect several thousand scan files made for suspected lung damage related to SARS-CoV-2.
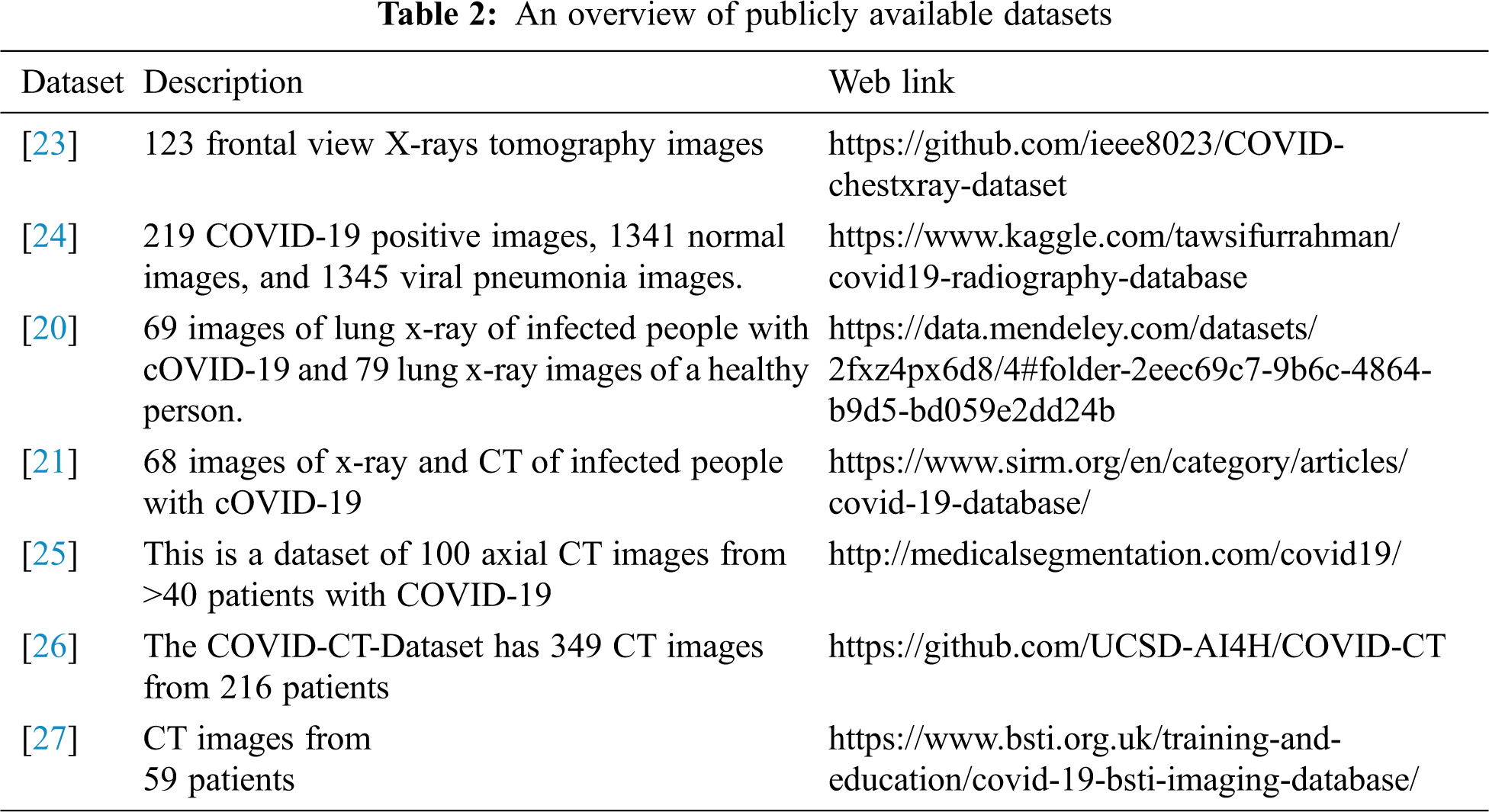
Tab. 2 gives an overview of publicly available datasets with appropriate references.
Two methods for distinguishing between COVID-19 and others pneumonia cases are described, based on the investigation of two modified versions of the original Xception and ResNet50 models.
Deep learning models are used with success in computer-aided detection systems that help clinicians interpret medical images [28]. Imaging techniques in X-ray diagnostics have achieved acceptable results and have opened up a field for scientific research in medicine. For this, the Convolutional Neural Network (CNN) has been efficiently compared to approaches based on handcraft-feature extraction, and it has been widely applied in medical image analysis, which contributes significant support in the development of health research. CNN or ConvNet is a multi-layered deep artificial neural network that is an expert at processing data with more precision and reducing costs.
To process data more accurately and reduce calculation costs, the strength of these networks lies in their robust semantics and the value they generate from the input data. Since medical imaging datasets are unavailable in some cases, we use transfer learning (TF) that contains the most efficient architectures of the most widely used deep convolutional networks. This permits us to use a pre-trained model from ImageNet, allowing us to transfer the learned model from generic data into a new specific model, including VGG, ResNet, DenseNet, Inception, and Xception. In this study, we proposed Xception and ResNet50 architectures. The main target of this work is to categorize pneumonia subtypes: SARS-Cov-2, bacterial and viral pneumonia, and differentiate it from normal lung, using x-ray images.
• Xception model [29] is a deep convolutional neural network architecture based on a deep separable wrapping layer developed by Francois Chollet of Google. This model is divided into three flows: an entry flow, a middle flow, and an exit flow. Used the default input image size of 299 × 299, so the images are not that size, we will need to resize them before feeding them as input. Contains Multiple convolutional layers, max pooling, separable convolution, ReLU, and global average pooling are among the features. The normalized green filter images were fed into the CNN model, and 2048 features were extracted from each image before being fed into the logistic regression model. The outcomes, on the other hand, were relatively minor. Apart from the extracted features from the green filter images, additional features such as correlation and mean square error between the green filter image and the red, yellow, and blue filter images were added to the feature vector list. The findings of logistic regression thus obtained were overfitting. As a result, CNN softmax was employed. There were now two wholly linked layers, a dropout layer and a softmax in the model. This yielded the best results, so the hybrid Xception model was applied, and the results were checked using K fold cross-validation. in our modified model, for the exit flow, we replace the linear regression Fig. 3a by the outputs layers that contained flatten layer, dense layer with ‘Relu’ activation and dense layers with softmax activation Fig. 3b.
• ResNet50 [30] is a classic neural network very similar to that of VGGnet with several 3 * 3 filters, and this model takes an input image of size 224 * 224. The ResNet50 containing 50 layers, consists of two blocks: identity block and convolutional block. It has a global average pooling followed by a classification layer. Before ResNet training, deep neural networks were complex due to the problem of fading gradients. The ResNet model is one of the improved versions of the Convolutional Neural Network (CNN). The significant advantage of ResNet is that it adds shortcuts between layers to overcome this problem. In addition, it prevents the distortion that occurs as the mesh becomes deeper and more complex. ResNet-50 is a Convolutional Neural Network with 48 Wrap Layers with 1 MaxPool and 1 Medium Pool Layer, and the pre-trained network can classify images into 1000 categories Fig. 4a. For the output layers in this model, we add the flatten layer, two dense layers with Relu activation, and the final layer with softmax activation Fig. 4b.

Figure 3: Illustrate the (a) original model, (b) modified model, and (c) and the final retained architecture for Xception

Figure 4: Illustrate the (a) original pre-trained model, (b) modified model and (c) and the final retained architecture for ResNet
Since the size of the available datasets is relatively small and the need for a rapid implementation of coronavirus diagnostic tools, pre-trained models are widely used for assessing COVID-19 patients. These models demonstrate a solid ability to generalize to new datasets outside the original one via transfer learning. Phan in Pham [31] considered that the fine-tuned pretrained models outperform several other base-line approaches for the classification of CIVID-19 in term of performance and computation efficiency.
In this work, we are evaluated and compared the transfer learning performance in two pretrained models, which are Xception and ResNet, used as a fixed feature extraction mechanism. Initially, layers of two models are pretrained on the ImageNet dataset, we put forward a modification in the last output layers according to the number of classes in the new dataset. Therefore, we assigned random weights to the included layers. Therefore, the model weights are updated during the training.
In this study, two open-source datasets compiled by Cohen JP (https://github.com/ieee8023/covid-chestxray-dataset) which currently includes around 142 x-ray images of COVID-19 positive patients are used. For normal healthy patients and other pneumonia, Kaggle’s chest x-ray images by Paul Money (https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia) consisting of 5863 images are considered. These x-ray images are divided into three different class values Normal, Bacterial & Viral pneumonia.
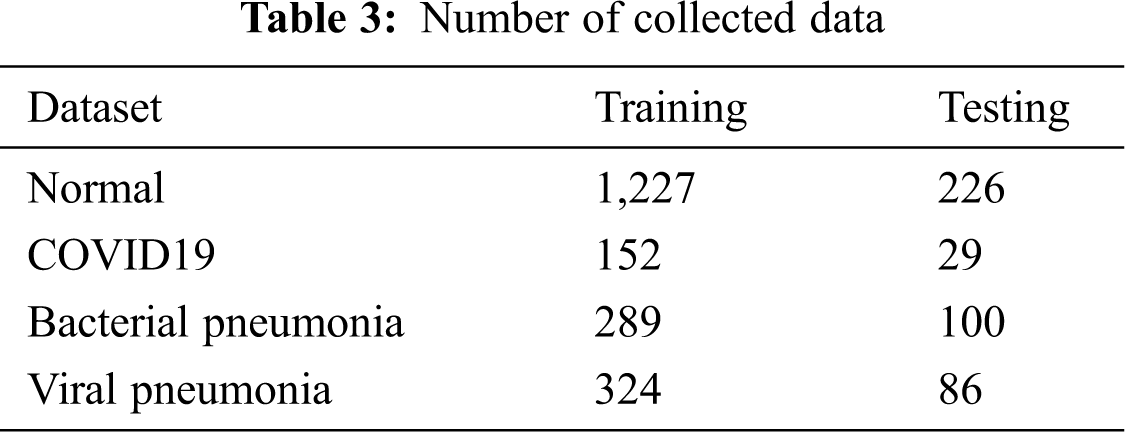
From this dataset, we selected approximately 2,498 x-ray images divided into four classes Fig. 5. This data set is then converted to training and testing with the distribution of 70% of the data for training the proposed deep learning model and 30% of the data for testing Tab. 3.

Figure 5: X-ray radiographic images for (a) normal, (b) COVID-12 positive, (c) Bacterial pneumonia and (d) viral pneumonia
For training, the model must be fed with a set of input images of the same size. Since we have collected images from different small datasets to construct larger ones, images were in different sizes. Indeed, we brought back all images to the same size of 224 x 224 pixels. We used data augmentation to train all models, including non-deep learning algorithms. Data augmentation is a common technique for improving the quality of trained models, mainly by increasing the effective amount of disaggregated data available. We applied the random transformation, zooming, rotation, and random flipping of images. We utilized hyperparameter optimization to find optimal parameters for the above operations: minimum and maximum number of shifted and enlarged pixels, and range of rotation angle, we have added some additions to the images Fig. 5. Central features and std_normalization, zoom “0.1”, rotation range “20”, constant fill mode, and 0.2 for validation split are applied.
The selected dataset consists of 1531 training images, 440 test images and about 381 images are reserved to evaluate the performance of models with different hyperparameter values of size 32 x 32.
In this section, the experimental setup and obtained results for both flat and hierarchical approaches are presented. The Xception & ResNet framework, including deep learning classifiers, has been carried using Python and the Keras package with TensorFlow2 on Intel(R) Core (TM) i7-3.60 GHz processor. In addition, the experiments have been executed using the graphical processing unit (GPU) NVIDIA GTX 1050 Ti and RAM with 8 GB.
Moreover, to evaluate the performances of the proposed deep learning model, various metrics are considered to measure truth and/or misclassification for diagnosing COVID-19 in X-ray images. The confusion matrix contains four predicted outcomes as follows [32]:
• True-positive (TP) measures the proportion of normal cases correctly predicted.
• True-Negative (TN) measures the proportion of abnormal cases correctly predicted.
• False-positive (FP) measures the proportion of abnormal cases classified as a normal categories.
• False-negative (FN) measures the proportion of normal cases considered as infected categories.
To verify the effectiveness of the established model, the known performances measures are used in terms of:
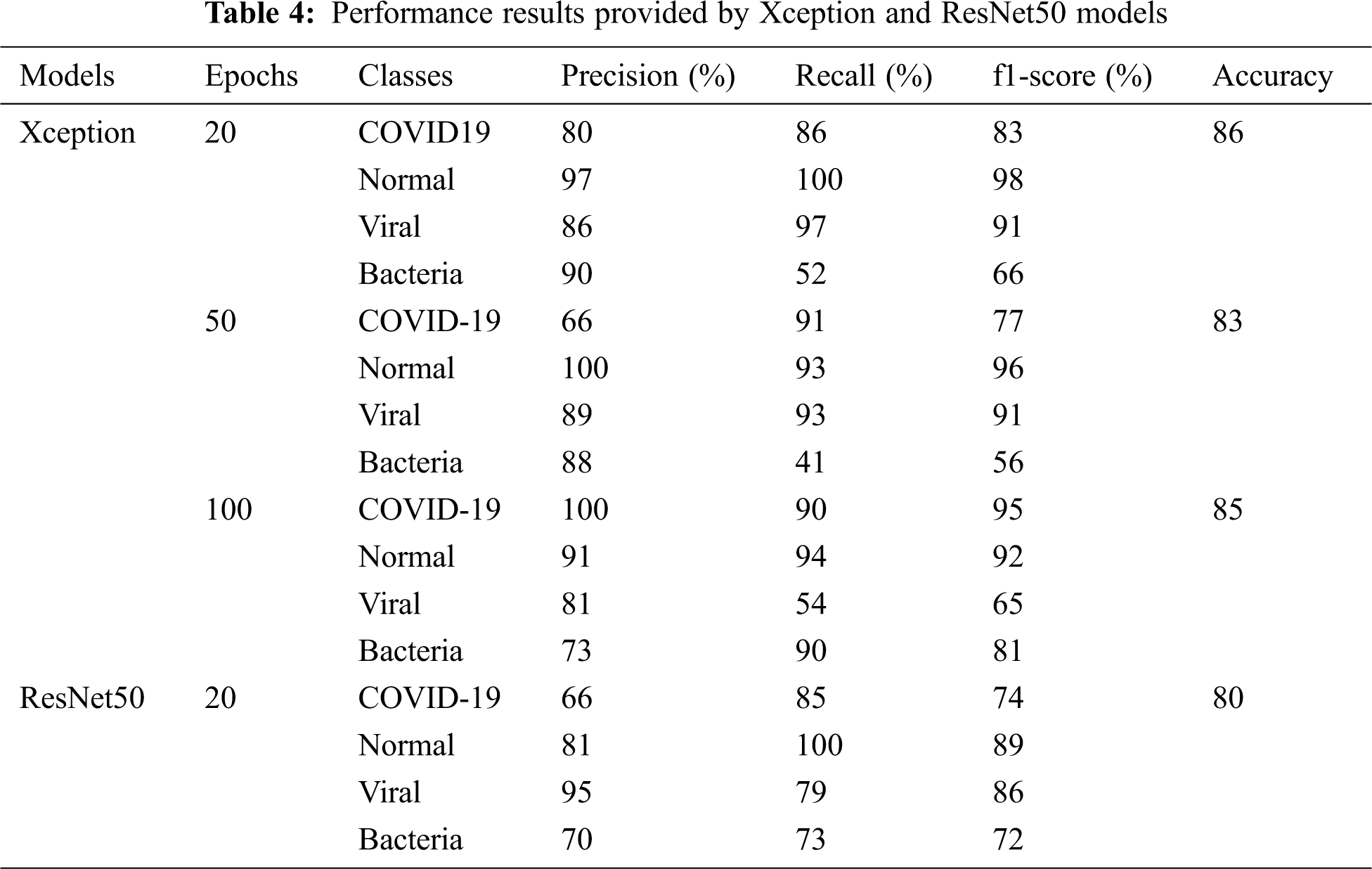
In this part, chest X-ray images to predict patients with Coronavirus (COVID-19) are considered. Pre-trained models such as Xception and ResNet50 are trained and tested on chest X-ray images. The training accuracy and loss values of the previously trained models are respectively reported in Fig. 6. During the training phase, we consider (1) between 20 and 100 epochs for Xception, wherein every epoch we increased data by changing the rotation range and zoom, and (2) 20 epochs for ResNet50 to avoid overfitting all pre-trained models. In this case, it can be observed that the highest training accuracy is obtained with the Xception model that is perform better than ResNet model. We remarked that ResNet50 exhibits a fast-training process compared to the other models. The confusion matrix in Fig. 6 illustrated that both models, There are similar results for the two classes: COVID-19=29 and Bacteria=86. However, in normal class the Xception model provides a best accuracy, which reached 80% (Tab. 4).
Next, for the Xception model, we added in the proposed novel model a dense layer with a dropout of 0.2. Then, we augmented the number of epochs to 50 (c) and 100 (d), and we adjusted the rotation range to 0. In the classification stage, the same result, are obtained with some changes in viral pneumonia classes.

Figure 6: Accuracy and loss curve and confusion matrix of Xception and ResNet50 proposed model (a) Accuracy and loss of Xception model (b) Accuracy and loss of ResNet50 model (c) Accuracy and loss of Xception model with 50 Epochs (d) Accuracy and loss of Xception model with 100 Epochs
Tab. 4 summarized the performance results provided by the two deep convolutional neural networks. The results showed that the pre-trained Xception in 20 epochs outperforms the ResNet50 model with a recognition rate of 86% accuracy vs. 80%. We noticed that the Xception hierarchical model ranks normal COVID-19 better. However, it decreases for viral and bacterial pneumonia category. One hypothesis is that both categories of pneumonia and COVID-19 are similar and share key features. Thus, the lack of normal images at the second classification level reduces the diversity of the training set, which interferes with the model’s training.
• Comparison between Xception and ResNet50 models: We have noticed from the confusion matrix that the Xception model performed better at detecting COVID-19, not detecting false cases of COVID-19, and producing better overall accuracy. Although there are a small number of cases of COVID19, the technology that we have suggested, can improve the detection of COVID-19 and the detection of other categories. It gave the accuracy of the category of COVID-19 at 99%, compared to some other research that worked to discover COVID-19 from X-ray images. As we explained previously, we have only 31 cases of COVID-19 and 419 cases from other categories, so the false positives for the COVID-19 category were shown as favorable compared to the other categories.
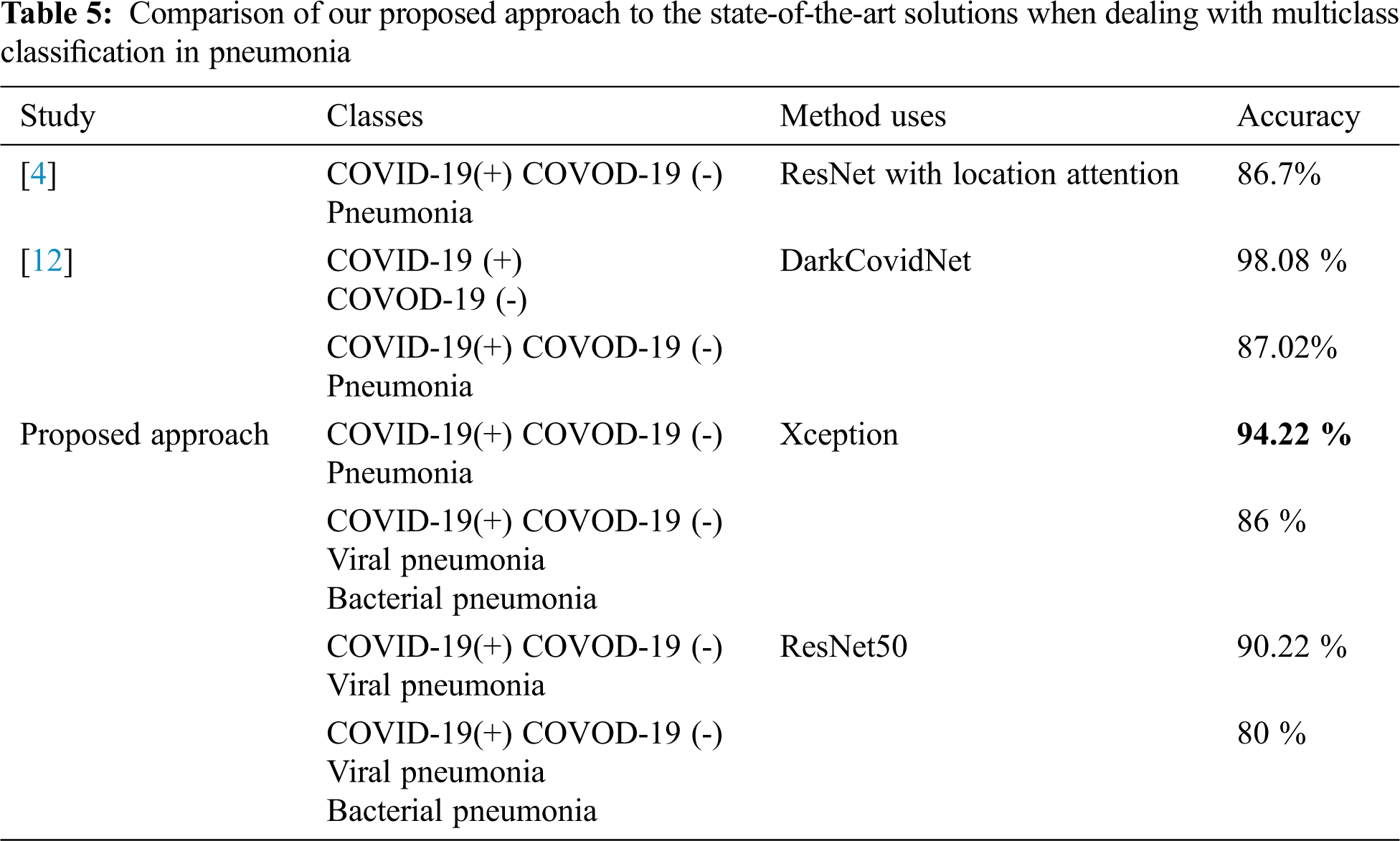
Comparison with state of the art: In other studies, we have seen results presented with 2 or 3 classes, the overall performances show acceptable scores when dealing with binary classification or even adding a healthy class. However, when dealing with a multi-class classification including viral pneumonia images the performance drops significantly. That means that the COVID-19 shows partially similar aspects with another pneumonia virus [32]. Tab. 5 gives the comparison of our approach with other state-of-the-art ones. As shown, our proposed method achieved an acceptable performance in terms of accuracy and illustrates the effect of the number of classes augmentation on the classifier performance: reducing the number of classes increases the classifier’s precision.
According to our bibliographic research, we have not found who considered four classes. In order to be able to make a significant comparison of the performance of our approaches, we start the second series of experiments considering three classes. We considered the same conditions, and same features size and composition, as performed in [12]. Results provided by Xception and reported in Tab. 5 shows that our method outperforms those reported in [5] and [12].
In this study, we attempted to work on a multi-layered classification and introduced both Xception and ResNet50, which are deep convolutional neural network designs for detecting SARS-CoV-2 cases from chest X-ray images. We modified two models by removing the latest layer and modified them by our top layers, then, we examined how both models made accurate predictions. The system uses an annotation method to gain deeper insights into the critical factors associated with COVID cases, which can help clinicians improve screening and speed medical analysis. It should be noted that working on four categories: normal, bacterial pneumonia, viral pneumonia, and COVID-19 allowed us to improve the classification rate.
Future directions for how computer vision responds to the urgent need to contribute to epidemics and to investigate many aspects of novel viral reproduction and disease pathogenesis. First, we need to create a suitable dataset for machine learning training. Second, when we do not have enough data to train a machine learning system but are eliminating a trained system with a close syndrome, it is wise to investigate how to use the “stored background knowledge” acquired while dealing with typical viral pneumonia and try to transfer the knowledge learned from the source domain.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. A. Ulhaq, A. Khan, D. Gomes and M. Paul, “Computer vision for COVID-19 control: A survey,” arXiv Prepr., arXiv:2004.09420, pp. 1–24, 2020. [Google Scholar]
2. A. B. Abdulkareem, N. S. Sani, S. Sahran, Z. Abdi, A. Adam et al., “Predicting covid-19 based on environmental factors with machine learning,” Intelligent Automation & Soft Computing, vol. 28, no. 2, pp. 305–320, 2021. [Google Scholar]
3. S. I. B. Al-Khateeb, M. Mahmood and B. Garcia-Zapirain, “An enhanced convolutional neural network for covid-19 detection,” Intelligent Automation & Soft Computing, vol. 28, no. 2, pp. 293–303, 2021. [Google Scholar]
4. X. Xu, X. Jiang, C. Ma, P. Du, X. Li et al., “Deep learning system to screen coronavirus disease 2019 pneumonia,” Engineering, vol. 6, no. 10, pp. 1122–1129, 2020. [Google Scholar]
5. H. Gunraj, L. Wang and A. Wong, “COVIDNet-CT: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest CT images,” arXiv Prepr. arXiv:2009.05383, pp. 1–12, 2020. [Google Scholar]
6. H. Maâmatou, T. Chateau, S. Gazzah, Y. Goyat and N. E. Ben Amara, “Transductive transfer learning to specialize a generic classifier towards a specific scene,” in Proc. VISAPP, Roma, Italy, 4, pp. 411–422, 2016. [Google Scholar]
7. A. Mhalla, T. Chateau, H. Maâmatou, S. Gazzah and N. E. Ben Amara, “SMC faster R-CNN: Toward a scene-specialized multi-object detector,” Computer Vision and Image Understanding, vol. 164, no. 2, pp. 3–15, 2017. [Google Scholar]
8. A. Mhalla, H. Maamatou, T. Chateau, S. Gazzah and N. E. Ben Amara, “Faster R-CNN scene specialization with a sequential Monte-Carlo framework,” in Proc. DICTA, Gold Coast, QLD, Australia, pp. 1–7, 2016. [Google Scholar]
9. P. Bouges, T. Chateau, C. Blanc and G. Loosli, “Handling missing weak classifiers in boosted cascade: Application to multiview and occluded face detection,” EURASIP Journal on Image and Video Processing, vol. 55, no. 1, pp. 671, 2013. [Google Scholar]
10. F. Waselallah Alsaade, T. H. H. Aldhyani and M. Hmoud Al-Adhaileh, “Developing a recognition system for classifying COVID-19 using a convolutional neural network algorithm,” Computers, Materials & Continua, vol. 68, no. 1, pp. 805–819, 2021. [Google Scholar]
11. A. S. Al-Waisy, M. A. Mohammed, S. Al-Fahdawi, M. S. Maashi, B. Garcia-Zapirain et al., “Covid-deepnet: Hybrid multimodal deep learning system for improving covid-19 pneumonia detection in chest x-ray images,” Computers, Materials & Continua, vol. 67, no. 2, pp. 2409–2429, 2021. [Google Scholar]
12. T. Ozturk, M. Talo, E. A. Yildirim, U. B. Baloglu, O. Yildirim et al., “Automated detection of COVID-19 cases using deep neural networks with X-ray images,” Computers in Biology and Medicine, vol. 121, no. 103792, pp. 1–11, 2020. [Google Scholar]
13. H. Alhichri, “CNN ensemble approach to detect COVID-19 from computed tomography chest images,” Computers, Materials & Continua, vol. 67, no. 3, pp. 3581–3599, 2021. [Google Scholar]
14. S. Wang, B. Kang, J. Ma, X. Zeng, M. Xiao et al., “A deep learning algorithm using CT images to screen for Corona virus disease (COVID-19),” European Radiology, vol. 25, no. 1, pp. 278, 2021. [Google Scholar]
15. L. Wang and A. Wong, “COVID-Net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray Images,” arXiv Prepr. arXiv: 2003.09871, pp. 1–12, 2020. [Google Scholar]
16. P. K. Sethy and S. K. Behera, “Detection of coronavirus disease (covid-19) based on deep features,” Preprints, vol. 2020030300, pp. 1–9, 2020. [Google Scholar]
17. A. Narin, C. Kaya and Z. Pamuk, “Automatic detection of coronavirus disease (COVID-19) using X-ray images and deep convolutional neural networks,” arXiv Prepr., arXiv:2003.10849, pp. 1–31, 2020. [Google Scholar]
18. H. S. Maghdid, A. T. Asaad, K. Z. Ghafoor, A. S. Sadiq and M. K. Khan, “Diagnosing COVID-19 pneumonia from X-cay and CT images using deep learning and transfer learning algorithms,” arXiv Prepr., arXiv: 2004.00038, pp. 1–8, 2020. [Google Scholar]
19. J. Hadfield, C. Megill, S. M. Bell, J. Huddleston, B. Potter et al., “NextStrain: Real-time tracking of pathogen evolution,” Bioinformatics, vol. 34, no. 23, pp. 4121–4123, 2018. [Google Scholar]
20. A. M. Alqudah and S. Qazan, “Augmented COVID-19 X-ray images dataset,”Mendeley Data, 2020. [Online]. Available: https://data.mendeley.com/datasets/2fxz4px6d8/4. [Google Scholar]
21. Italian Society of Medical and Interventional Radiology, “COVID-19 database SIRM,” [Online]. Available: https://www.sirm.org/en/category/articles/covid-19-database/. [Accessed Nov. 14, 2020]. [Google Scholar]
22. webinaire SIT : imagerie du COVID-19 - synthèse des connaissances | SFR e-bulletin, [Online]. Available: https://ebulletin.radiologie.fr/actualites-covid-19/webinaire-sit-imagerie-du-covid-19-synthese-connaissances. [Accessed Apr. 01, 2021]. [Google Scholar]
23. J. P. Cohen, P. Morrison, L. Dao, K. Roth, T. Q. Duong et al., “COVID-19 image data collection: Prospective predictions are the future,” Journal of Machine Learning for Biomedical Imaging, vol. 2, pp. 1–38, 2020. [Google Scholar]
24. COVID-19 radiography database | kaggle, [Online]. Available: https://www.kaggle.com/tawsifurrahman/covid19-radiography-database. [Accessed Nov. 14, 2020]. [Google Scholar]
25. COVID-19 CT segmentation dataset. [Online]. Available: http://medicalsegmentation.com/covid19/. [Accessed Nov. 14, 2020]. [Google Scholar]
26. X. Yang, X. He, J. Zhao, Y. Zhang, S. Zhang et al., “COVID-CT-dataset: A CT scan dataset about COVID-19,” [Online]. Available: http://arxiv.org/abs/2003.13865. [Accessed: Nov. 14, 2020]. [Google Scholar]
27. COVID-19 British Society of Thoracic Imaging Database, The British Society of Thoracic Imaging, [Online]. Available: https://www.bsti.org.uk/training-and-education/covid-19-bsti-imaging-database/. [Accessed Nov. 14, 2020]. [Google Scholar]
28. Y. J. Gaona, M. J. R. Álvarez and V. Lakshminarayanan, “Deep-learning-based computer-aided systems for breast cancer imaging: A critical review,” Applied Sciences, Special Issue Medical Digital Image: Technologies and Applications, vol. 10, no. 22, pp. 1–28, 2020. [Google Scholar]
29. F. Chollet, “Xception: Deep learning with depthwise separable convolutions,” in Proc. CVPR, Honolulu, HI, USA, pp. 1800–1807, 2017. [Google Scholar]
30. K. He, X. Zhang, S. Ren and J. Sun, “Deep residual learning for image recognition,” in Proc. CVPR, Las Vegas, NV, USA, pp. 770–778, 2016. [Google Scholar]
31. T. D. Pham, “Classification of COVID-19 chest X-rays with deep learning: New models or fine tuning?,” Health Information Science and Systems, vol. 9, no. 2, pp. 1–11, 2021. [Google Scholar]
32. M. C. Belavagi and B. Muniyal, “Performance evaluation of supervised machine learning algorithms for intrusion detection,” Procedia Computer Science, vol. 89, no. 9, pp. 117–123, 2016. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |