DOI:10.32604/iasc.2022.019970

| Intelligent Automation & Soft Computing DOI:10.32604/iasc.2022.019970 |  |
| Article |
Deep Learning Model to Detect Diabetes Mellitus Based on DNA Sequence
1Information Systems Department, Faculty of Computers and Artificial Intelligence, Benha University, Benha, 13518, Egypt
2Mathematics and Computer Science Department, Faculty of Science, Port Said University, 42525, Egypt
3Computer Science Department, Faculty of Computers and Information, Damietta University, 34517, Egypt
*Corresponding Author: Noha E. El-Attar. Email: noha.ezzat@fci.bu.edu.eg
Received: 03 May 2021; Accepted: 08 June 2021
Abstract: DNA sequence classification is considered a significant challenge for biological researchers to scientifically analyze the enormous volumes of biological data and discover different biological features. In genomic research, classifying DNA sequences may help learn and discover the new functions of a protein. Insulin is an example of a protein that the human body produces to regulate glucose levels. Any mutations in the insulin gene sequence would result in diabetes mellitus. Diabetes is one of the widely spread chronic diseases, leading to severe effects in the longer term if diagnosis and treatment are not appropriately taken. In this research, the authors propose a hybrid deep learning model based on Long Short-Term Memory (LSTM) and Convolutional Neural Network (CNN) to classify the DNA sequence for the insulin gene to predict type 2-diabetes based on gene sequence mutation. To evaluate the proposed models, we used several performance indexes such as accuracy, precision, sensitivity, recall, and F1 score. The experiments shown in the paper reveal that the proposed model accomplished the best results. The overall accuracy of the learning process is recorded as 99% for the proposed hybrid LSTM-CNN model while it is recorded as 97.5%, and 95% for CNN, and LSTM, respectively.
Keywords: Deep learning; DNA sequence; diabetes mellitus
Artificial intelligence, particularly deep learning, is ushering in a new era in clinical medicine. The main idea of the AI system is to train the health data that has been previously labeled and interpreted by human experts. This training process assists the AI system in learning and performing the interpretation process on new incoming data of the same kind. This interpretation may be to detect or predict a disease state. There are several AI interpretation tasks, to name a few, time series analysis for health data provided by the electrocardiogram, computer vision applications for interpreting radiological images, and natural language processing to extract meaningful information from health records or DNA sequences [1].
Machine learning (ML) and deep learning (DL) are part of artificial intelligence, extensively utilized in several genomics studies. DL is an evolution of ML, recently considered an attractive solution for genome classification and prediction problems. For instance, the Deep Neural Network (DNN) structure comprises multiple layers of non-linear transformations, making it more scalable and flexible in dealing with massive amounts of data and identifying the complex patterns in the feature-rich datasets [2]. Convolutional Neural Networks (CNN), Recurrent Neural Networks (RNN), Restricted Boltzmann Machine (RBM), and Long Short-Term Memory (LSTM) are considered the most common architectures of DNN [3].
In recent decades, the role of Deoxyribonucleic Acid (DNA) sequencing has been manifested in disease prediction. Practically, it is possible to screen the expression of all genes in the genome simultaneously. However, the issue now is interpreting such massive data to obtain a deep understanding of the biological processes and human disease mechanisms [4]. The DNA genes are responsible for encoding the protein molecules considered the “workhorses” of the cell that carry out all the functions necessary for life [5]. In a simplified sense, expressing a gene means constructing its corresponding protein. One of the produced proteins in the human body is insulin. The mutation in the insulin gene sequence may lead to an imbalance in the hormone insulin production, resulting in diabetes mellitus (DM). Diabetes is among the most widespread chronic diseases globally, leading to severe effects in the longer term if diagnosis and treatment are not appropriately taken [1]. According to the World Health Organization (WHO), DM is classified into four types, as shown in Fig. 1 [6].
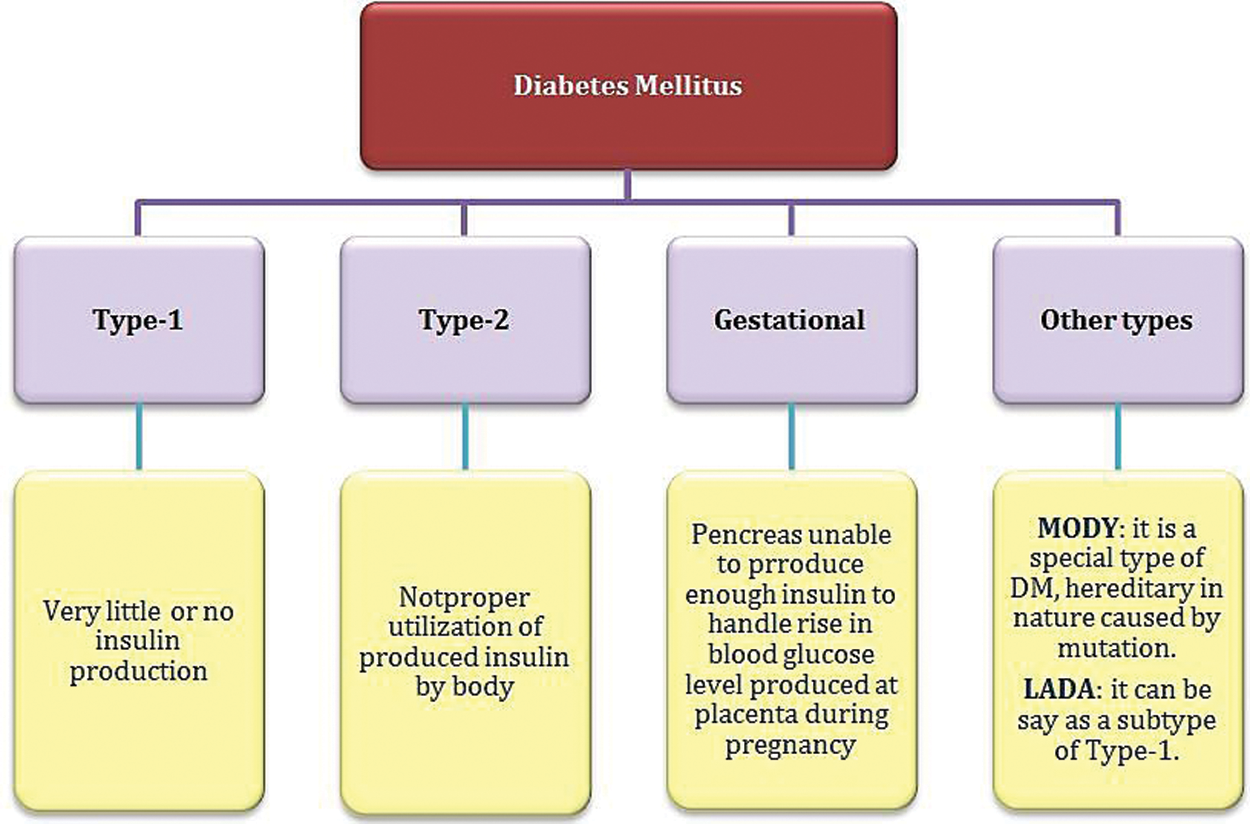
Figure 1: Diabetes mellitus classification according to WHO [6]
The main contribution of this study is to produce a diabetes mellitus prediction model based on a deep hybrid network of long short-term memory (LSTM) and convolutional neural network (CNN). The target of the proposed LSTM-CNN model is to classify the potential Insulin sequences across various human specimens into Diabetic and Non-diabetic. After that, the trained model would identify any new insulin sequences as normal or mutated quickly. The rest of this paper is organized as follows; Section 2 represents the materials and methods used in this experiment. Section 3 displays a literature review on a different machine and deep learning algorithms used to predict diabetes mellitus. The experiment details and dataset description are presented. Section 4 concludes the results and evaluation. Finally, the conclusion is reviewed in Section 5.
AI is a broad term for computer systems that mimic human intelligence. In the context of medical diagnostics, AI is defined as a computer system capable of correctly interpreting health data, especially in its native shape observed by physicians. These AI frameworks can speed up the analysis of massive, complex health datasets [7].
Machine learning is a branch of AI that involves algorithms that can interpret data, learn from it, and then implement what they have learned to produce better decisions. ML is beneficial in identifying hard-to-discern patterns from large, noisy, or complex data sets. It employs various statistical, probabilistic, and optimization techniques to enable a computer system to “learn” from past models [8]. In general, ML can be classified into two categories: supervised and unsupervised. In supervised learning (i.e., classification or regression), the relations between a set of inputs are constructed based on the variables or labels extracted from the training instances; these relations can be used to predict the outputs of new instances. Neural network, Support vector machine, Linear and logistics regression, Classification trees, and random forest are examples of supervised learning algorithms [9].
On the other hand, unsupervised learning methods (i.e., clustering and principal component analysis) are used in inferring the patterns from data sets without defining the labeled responses. Cluster algorithms, K-means algorithms, dimensionally reduction algorithms, and anomaly detection algorithms are examples of unsupervised algorithms. The intrinsic goal of several ML algorithms is to improve the performance of the learning model on new independent datasets (i.e., generalization performance) rather than available data (i.e., training performance). Thus, ML must balance model flexibility and training data volume, which is practically very hard due to underfitting or overfitting, which may occur [10]. The flexible structure of DNNs has radically transformed the outlook of many research fields by promising high-accuracy outcomes, especially in dealing with medical and health data [11].
2.1 Convolutional Neural Network (CNN)
CNN is a powerful model for large Neural Networks inspired by the visual mechanism of living organisms. The CNN function is manifested in eliciting the higher-level abstraction features from the features extracted from the previous layers. In CNN, the artificial neurons are analyzed across the input matrix to identify translation-invariant patterns at each input position. After that, CNN computes the locally weighted sum and produces the expected output values [5,10]. The raw dataset's intrinsic features are extracted through the multiple layers structure, representing the different abstraction levels of features [12]. As shown in Fig. 2, the CNN architecture commonly consists of an input layer, convolution Layer, ReLU or Rectified Linear Unit, pooling layer, and a fully connected neural network layer. The Convolution layer consists of a set of filters whose parameters need to be learned. Each filter slides across the input matrix, producing a smaller dimension than the input matrix. The pooling layer works separately on each feature map produced from the convolution layer to create a new set of pooled feature maps. The final output layer is a fully connected neural network layer that produces the output based on the activation function [2].
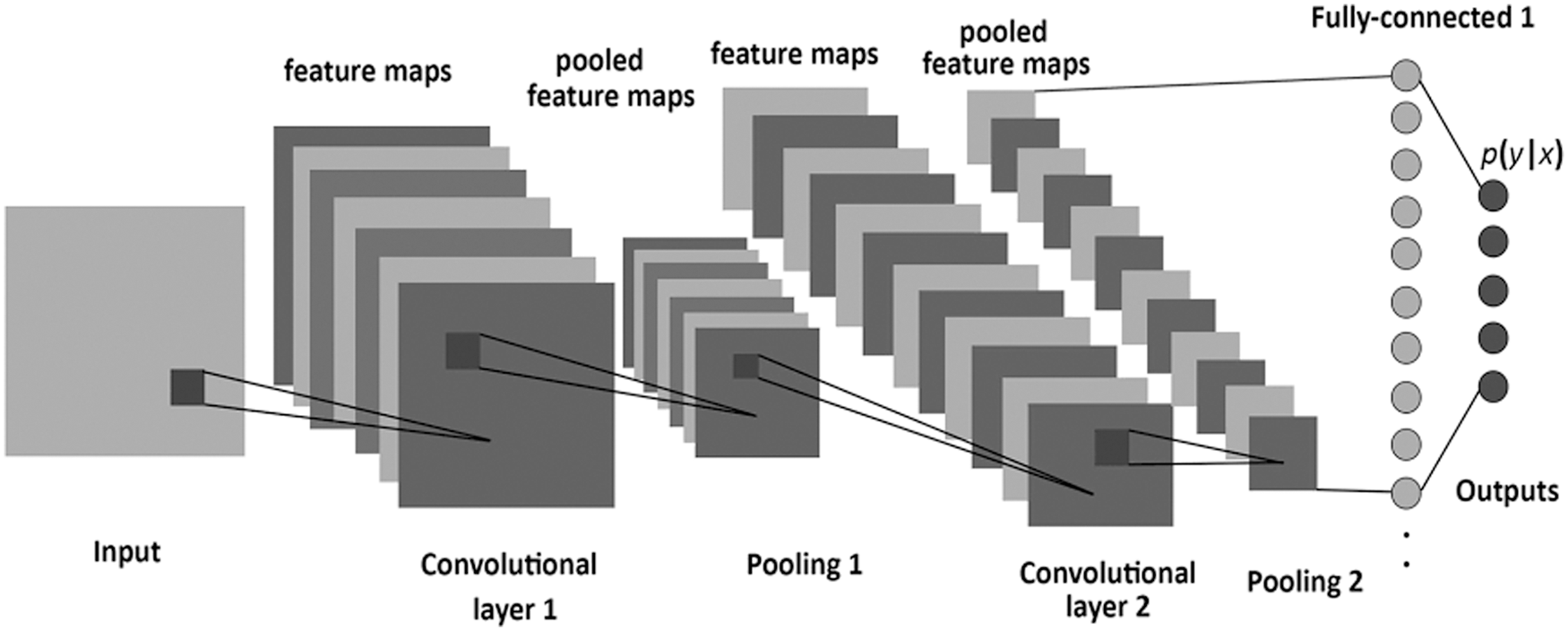
Figure 2: Architecture of convolutional neural network
CNNs have been widely used in handling several image processing problems. The presence of several layers of neurons helps in identifying image signatures that enhance the classification process. CNNs have also been considered as an attractive technique to classify text based on characters. Thus, CNNs have been used in analyzing DNA sequences to figure out many features such as promoters and binding sites [13].
2.2 Long Short-Term Memory (LSTM)
Long Short-Term Memory is an evolutionary type of Recurrent Neural Networks (RNN). LSTM is well-versed in dealing with the RNN's well-known drawbacks by adding additional interactions to each module. As a default, it is designed to learn long-term dependencies and recall data for future periods [14]. The LSTM modules, which comprise a set of gates, are commonly referred to as cells rather than neurons [15]. The default structure of the LSTM is shown in Fig. 3 [14]. The development of the LSTM contains unique units called memory blocks in the repetitive hidden layer. These memory blocks contain self-association memory cells that store the earlier states. There are also multiplicative units known as gates that control data progression. A forget gate considers the new input and the hidden state to determine which cell state information can be safely ignored. Therefore, the input gate specifies which information from the new input stream should indeed be applied to the remembered cell state. Eventually, the output gate produces the output for the current time phase using information from the cell state, input, and hidden state. The LSTM network is considered an ideal technique for detecting longer-term data patterns by remembering information over several iterations. On the other hand, the LSTM cell uses a hidden state to have a short-term memory. LSTM networks are an efficient tool for time series data forecasting [15].
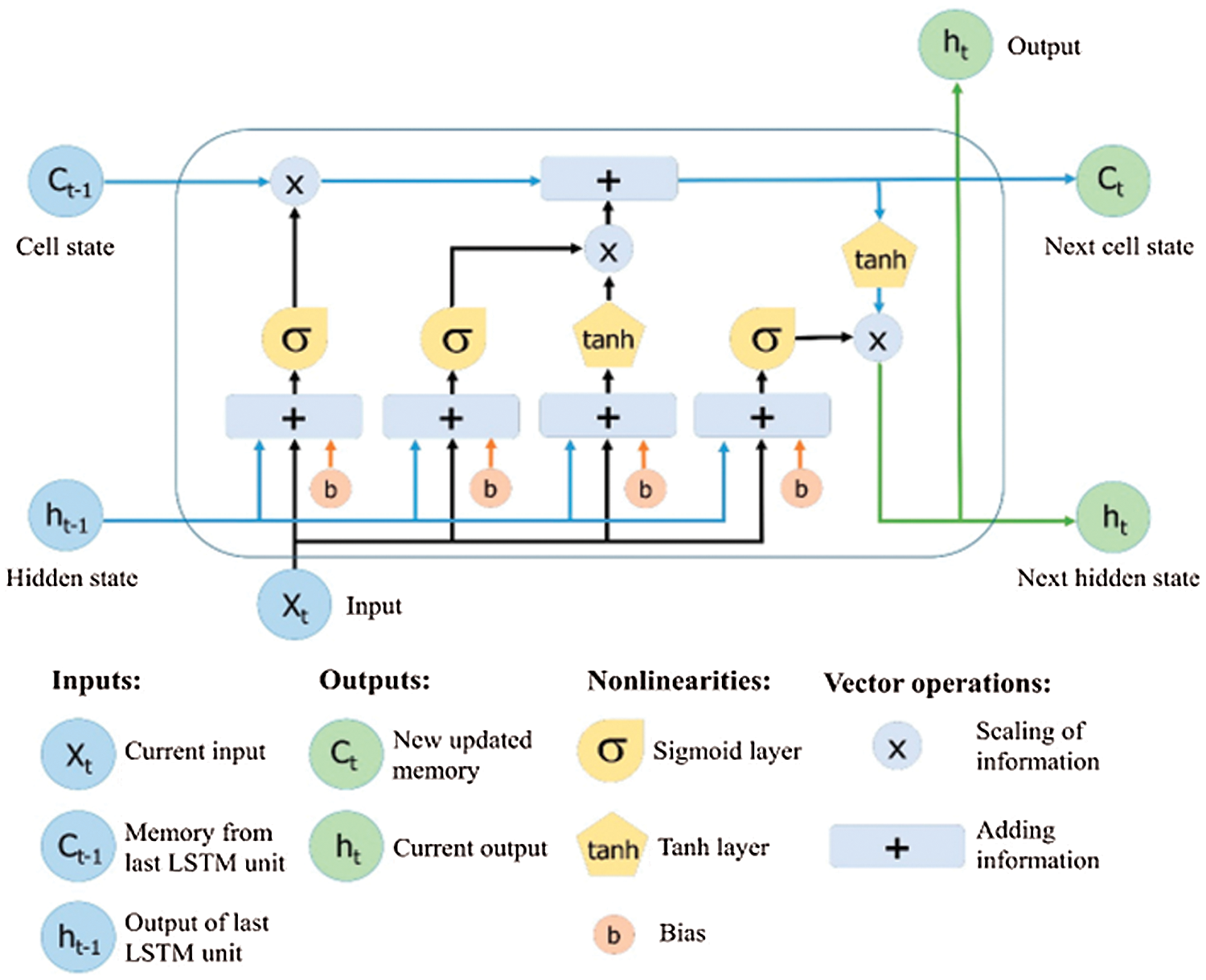
Figure 3: Architecture of long short-term memory [14]
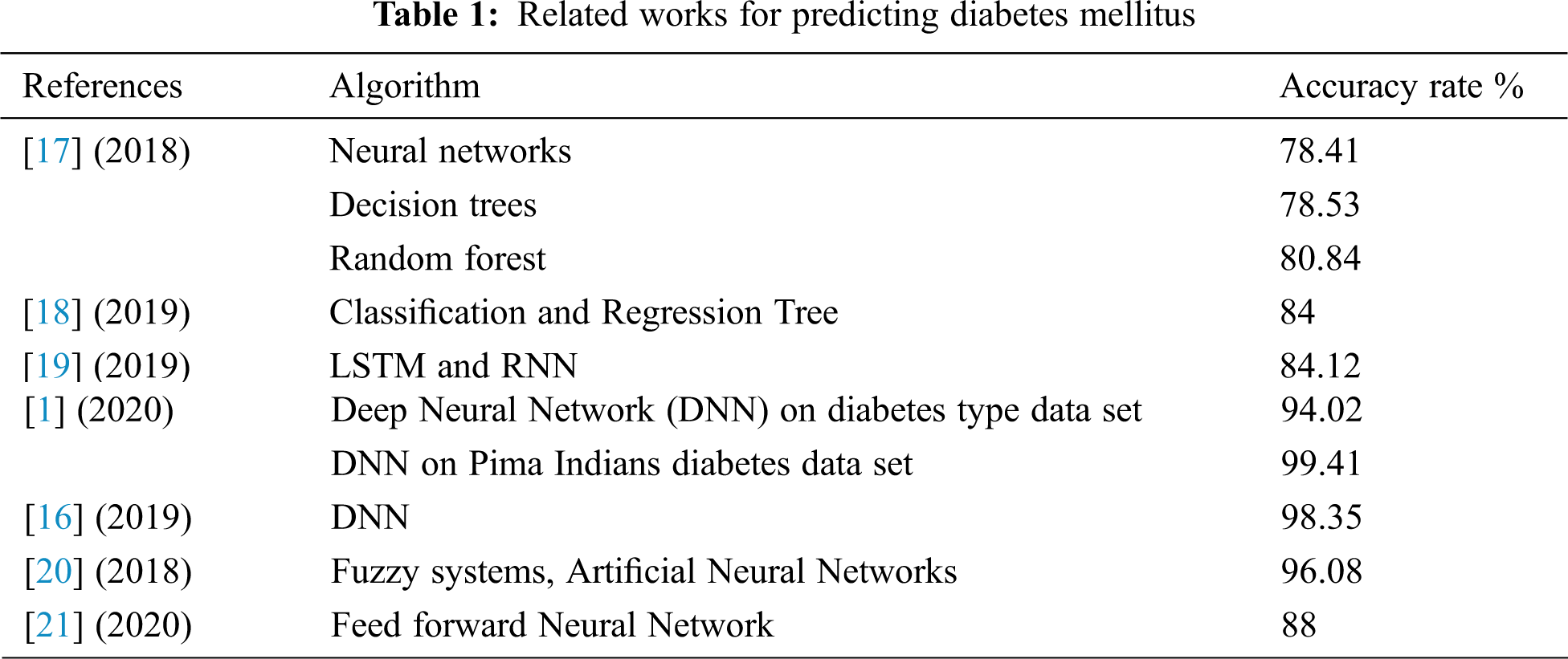
Diabetes is one of the world's most dangerous and common diseases. It affects blood glucose levels and triggers various diseases such as renal failure, heart disease, blindness, and others [16]. In recent decades, deep learning has gained popularity due to its supremacy in terms of accuracy for predicting various types of disease. In this context, some researchers have adopted AI techniques such as data mining, ML, DL, and fuzzy systems in diagnosing and predicting the types of diabetes mellitus (DM). Zou et al. [17] have adopted ML techniques, neural networks, decision trees, and random forest to predict diabetes mellitus. The prediction indicator used in this study was fasting glucose, but they found that this indicator is not enough to give accurate results. The best accuracy was 80.48%, which is recorded by the random forest algorithm. Hathaway et al. [18] also applied some popular machine learning algorithms to sequencing, biochemical, and physiological data for 50 patients (30 non-diabetic and 20 type2-diabetic). They have used Linear Discriminant Analysis, Gaussian Naïve Bayes, Logistic Regression, Support Vector Machine (SVM), and Classification and Regression Tree. They used these ML algorithms to recognize various biomarkers that are known to be chaotic in disease-related systems and affect diabetic status. The high recorded accuracy in this study was 84%. Another direction to predict the glucose level has been adopted by Hasib et al. [19]. They have used deep learning to forecast levels of glucose dynamically every day for type 2-diabetic patients. The forecasting model was designed based on their day before recorded mobile health data such as physical activities, weight, diet, and glucose level. In this study, they have adapted LSTM-based recurrent neural network to predict the glucose levels for the next day for ten individual patients. According to this experiment, the accuracy of predicted values was 84.12% [19]. Zhou et al. [1] have used an enhanced deep neural network called diabetes type prediction model to determine the type of the disease that the patient is suffering from. The experiment was done on two datasets: the diabetes type dataset and the Pima Indians diabetes data set. The recorded results were 94.02% for the first dataset and 99.4% for the second one. Likewise, Ayon et al. [16] have developed a deep neural network (DNN) model to identify diabetes types based on several medical factors. They trained the attributes of the proposed DNN in a five-fold and ten-fold cross-validation manner. The experiment showed that by using five-fold cross-validation, their proposed model produced promising results where it recorded an accuracy rate of 98.35%. One more prediction model for type 2-diabetes has been presented by Alby et al. [20]. They have developed an adaptive neuro-fuzzy interface system integrated with genetic algorithms and recorded a 96.08% accuracy rate.
As we have mentioned in this literature, most researchers have used health care indicators in predicting or classifying, but few have depended on DNA analysis in predicting the DM, although this type of data may give more accurate results. By using polymerase chain reaction and DNA sequencing, Ma'mon et al. [21] developed an artificial neural networks model to predict type 2-diabetes. The recorded accuracy of this proposed model was 88%. Tab. 1 summarizes the recent prediction models used for DM.
The significant impact of deep learning algorithms in handling biological problems encouraged us to propose a deep learning model to predict type 2-diabetes by classifying the insulin gene sequence into normal or mutated. In this research, two deep learning algorithms were adopted to build the classifier model based on hybrid DL algorithms: LSTM and CNN.
The construction of DNA is based on nucleotide units. Each nucleotide unit consists of three sub-units: a nitrogenous base, a five-carbon sugar, and at least one phosphate group. There are four types of nitrogenous bases in DNA; Adenine (A), Guanine (G), Cytosine (C), and Thymine (T). The sequence of these nitrogenous bases encodes biological information, and variations in these nucleotide sequences lead to biological diversity among human beings and every living organism [22].
The mechanism by which proteins are created from their related genes is a two-step process; transcription and translation, as displayed in Fig. 4 [23]. In the transcription step, a particular segment of DNA is copied into a temporary mRNA molecule. Both DNA and RNA are nucleic acids, which utilize base pairs of nucleotides as a corresponding language. In the translation step, the resulting mRNA, a single-stranded copy of the gene, is translated into a protein molecule. These two steps are called gene expression [5,24]. Insulin is a protein produced by pancreatic β-cells to regulate glucose levels in the blood [25]. In the body's normal state, whenever the blood glucose levels begin to rise (e.g., during the digestion process), β-cells rapidly react by emitting some of their stored insulin and increasing the creation of the hormone simultaneously. But ruefully, sometimes the β-cells become unable to produce a sufficient amount of insulin needed to control the blood glucose levels, as in diabetes type-2 [26].
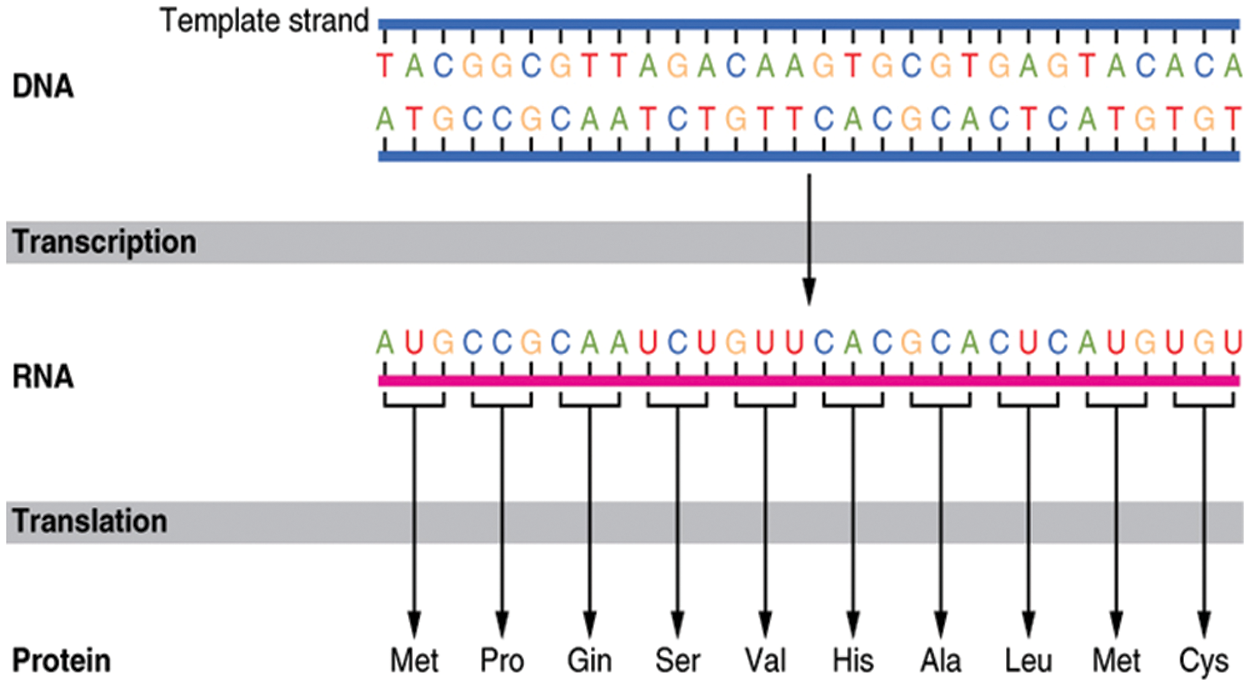
Figure 4: DNA to protein transaction process [23]
Like all other proteins, mutations in the insulin gene sequence will lead to an imbalance in the hormone insulin production, resulting in diabetes. Fig. 5 shows a segment of DNA sequence for a normal human insulin gene versus a mutated sequence [27]. The proposed experiment study is done on a human insulin dataset collected from GenBank [28]. Three hundred sequences of human insulin are randomly selected from Genbank to perform the learning process. The length of a single human insulin gene sequence is about 5112 long.
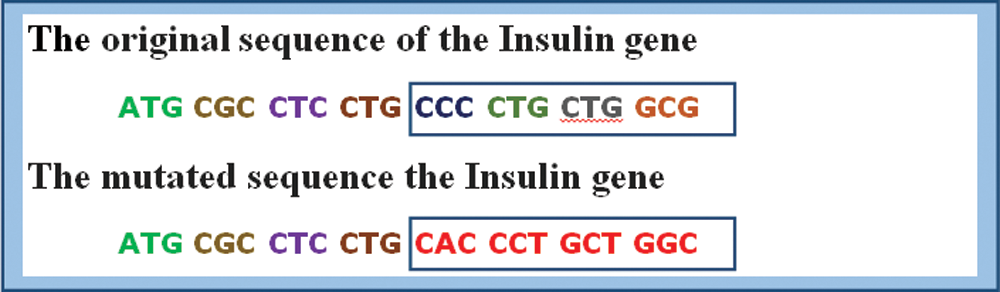
Figure 5: Normal human insulin gene sequence vs. mutated sequence
Initially, the dataset is divided into 70% and 30% for the training and testing processes. Before beginning the learning process, the input data must be prepared to be processed. As shown in Fig. 6, the DNA sequences are groupings of successive letters without space, and this form is hard to understand by the deep networks. Thus, the insulin DNA sequence is encoded to an understandable form through a data preprocessing (i.e., data encoding) phase. This phase aims to apply a similar representation procedure on the DNA text data without losing position data of every nucleotide in sequences. DNA contains four main amino acid letters (G, A, T, and C). The proposed encoding process is based on translating each amino acid letter in the Insulin DNA sequence into its corresponding binary digits (i.e., zeros and ones) based on the one-hot vector encoding method.
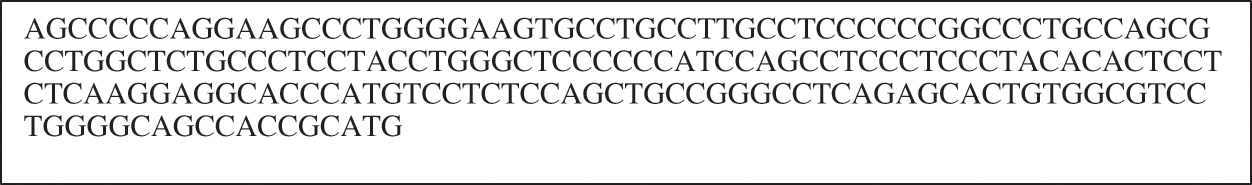
Figure 6: Example of DNA sequence
The four binary representations for each amino acids G, A, T, C are [1,0,0,0], [0,1,0,0], [0,0,1,0], and [0,0,0,1] respectively [10]. Fig. 7 presents a sample of translating a part of an insulin DNA sequence into its corresponding binary vector. This extracted binary vector is then applied to the learning process as the input for the proposed CNN, LSTM, and ANN models.
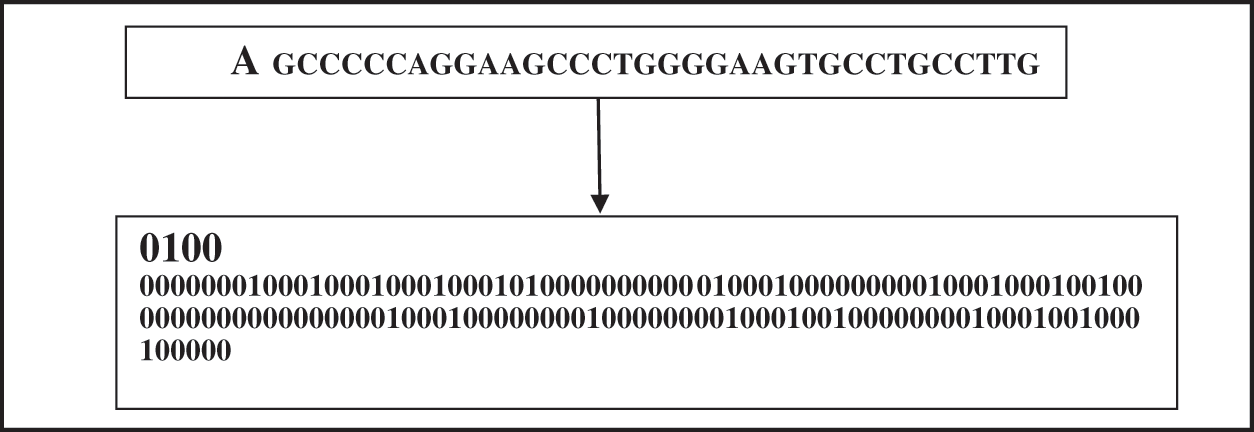
Figure 7: Data encoding phase–translating a DNA sequence into its binary vector
The proposed classification model for type 2 diabetes prediction is based on two deep learning algorithms, CNN and LSTM. In the following, we explain the proposed architecture for each algorithm individually. After that, the proposed hybrid LSTM-CNN architecture is explained according to our predefined architecture of LSTM and CNN. Fig. 8 displays a comprehensive flowchart for the stages of the proposed algorithms.
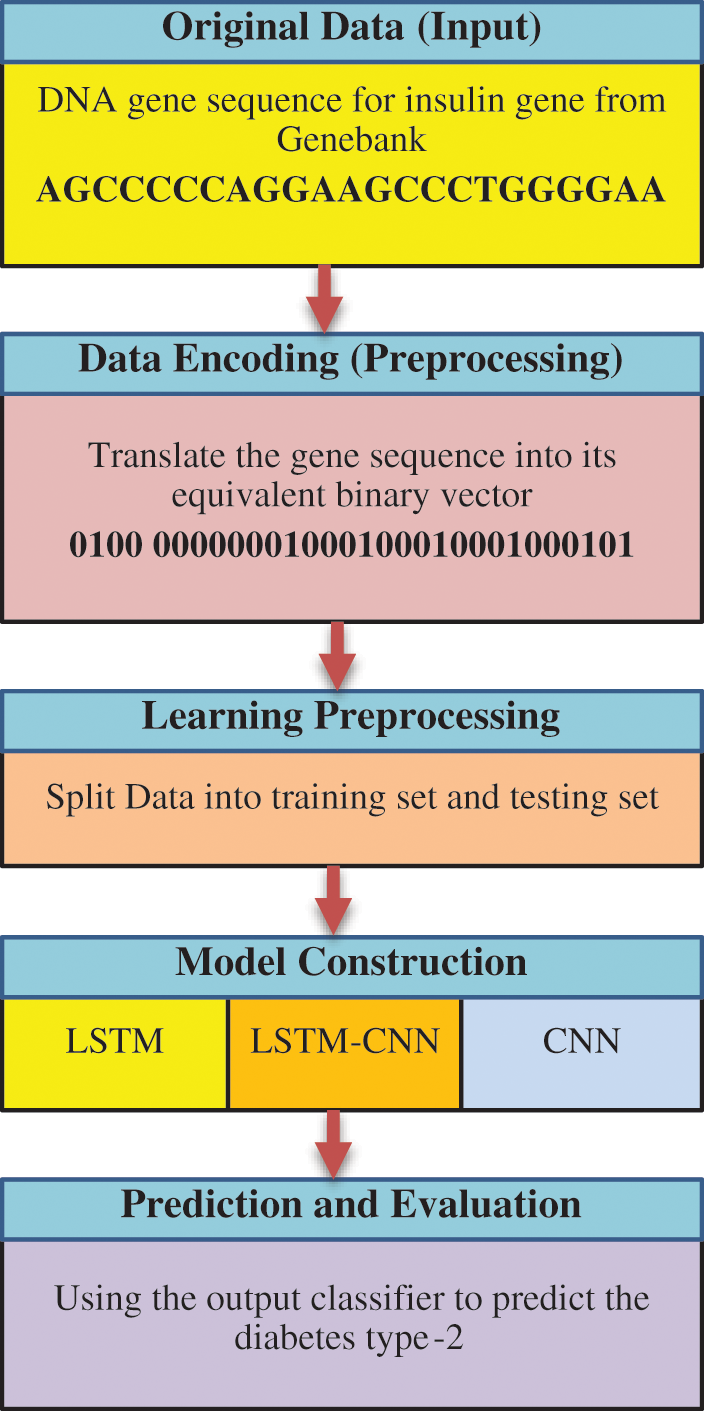
Figure 8: A comprehensive flowchart for the stages of the proposed algorithms
4.3.1 CNN Classification Model
The CNN classification model utilized in this study is a modified version of the LeNet-5 developed by LeCun et al. [12]. This LeNet-5 is a network that contains four processing layers divided into one convolution layer, one max-pooling layer, and two fully connected Multi-Layer Perceptron (MLP) processing layers. As shown in Fig. 9, the input of the convolution layer is a one-dimensional binary vector of size (20448 × 1) that represents the encoded DNA sequence. After feeding the CNN with the input, the convolutional process begins by applying 12 kernels of size (400 × 1) over four strides on the original input. To reduce the overfitting of the training data, a (4 × 1) max-pooling layer is applied to each filter over a stride 2. In the max-pooling layer, the input matrix is partitioned into a set of non-overlapping regions. For each sub-region, the maximum value is considered the output value of the max-pooling layer. The final output of the convolution and pooling processes is 48 kernels size (200 × 1).
These kernels are grouped into a one-dimensional vector called flatten layer, which will be the input for the fully connected MLP layers. The MLP is composed of two hidden layers. The first hidden layer uses the Relu function and the second layer produces the output based on the sigmoid activation function. The Relu or Rectified Linear Unit, which can be calculated by Eq. (1), is an activation function characterized by its simplicity and efficiency in avoiding and rectifying vanishing gradient problems. Its primary function is to neglect any negative values by converting them to zero and passing only the positive values to the next layer. The second layer, the final hidden layer, uses the sigmoid function presented in Eq. (2) to perform a non-linear transformation, producing the network classifiers' probability [29].
4.3.2 LSTM Classification Model
Another type of deep learning algorithm used to classify the insulin gene sequence is LSTM [14]. LSTM uses its gates to control the memorization process. Initially, the forget gate uses the sigmoid function presented in Eq. (3) to determine the unrequired information and erases it from the memory cell. In this step, the inputs of the sigmoid function are
where,
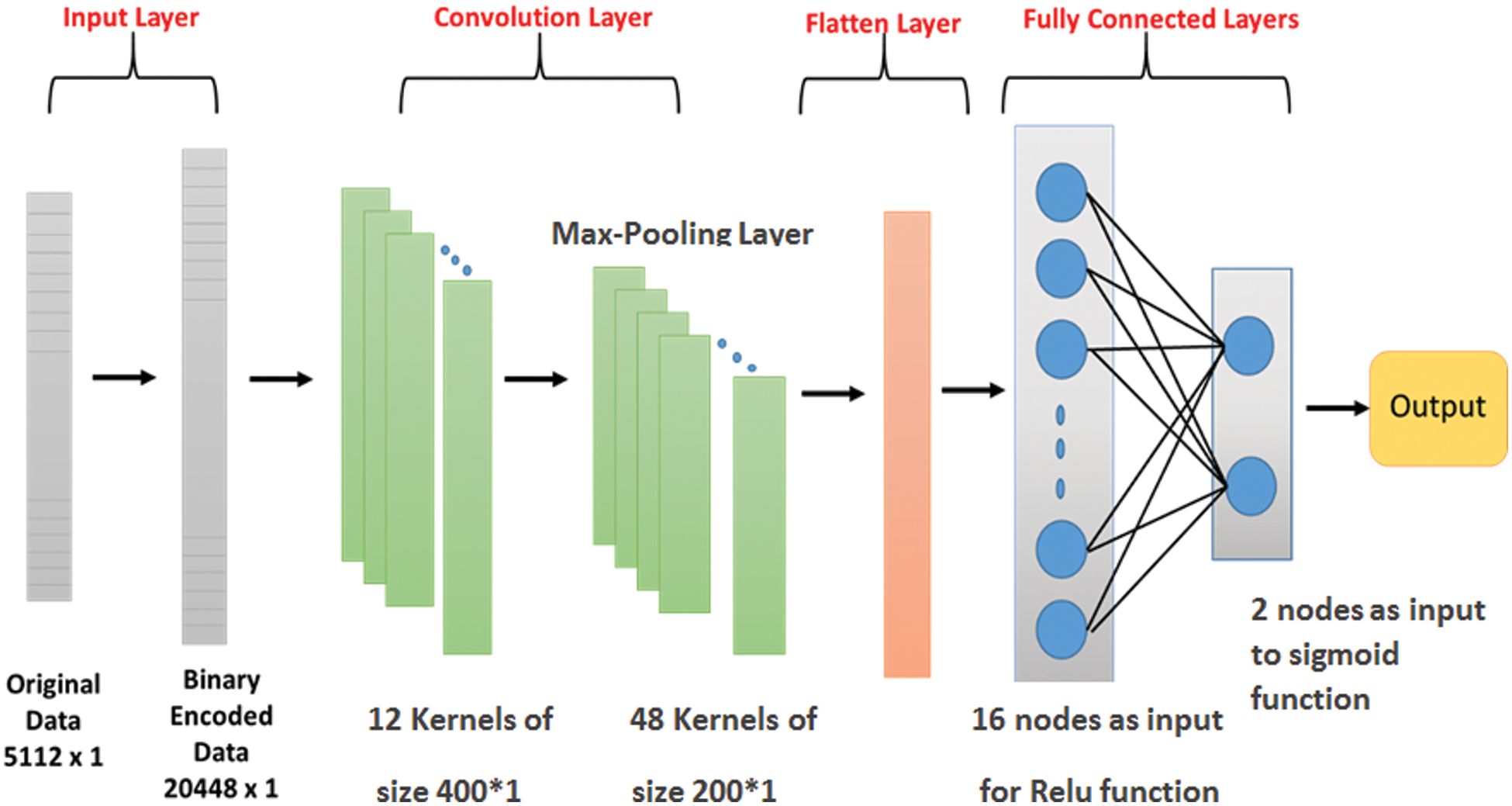
Figure 9: The proposed architecture for CNN
The second stage in the LSTM architecture is selecting and storing important information from the new input Xt to upgrade the state of the memory cell. This stage is based on two functions composed of two layers, the sigmoid layer and the tanh layer. The function of the sigmoid layer is to determine if the new information should be adjusted or ignored according to its value (i.e., 0 or 1). Following that, the tanh function assigns weights between [–1, 1] to all of the passed values to define their significance. The values resulting from sigmoid and tanh layers are calculated by Eqs. (4) and (5). The following cell state Ct is calculated by Eq. (6) [14].
Eventually, the last layer is a modified version of the sigmoid and tanh layers to produce the outcome (ht) as calculated in Eq. (7).
4.3.3 The LSTM-CNN Hybrid Model
According to the benefits of CNNS in extracting the essential features, it is a good option for processing large amounts of new data. On the other hand, LSTM is characterized by remembering the long-term dependencies and shape of the input sequence and producing a particular pattern. In this proposed methodology, the LSTM network and a CNN network are combined to build a hybrid LSTM-CNN classification model. This model uses the LSTM to characterize the potentially complex order in insulin gene sequences conveniently. It uses the CNN and max-pooling layers to develop filters that interpret the gene sequence patterns. By extracting the information from every moderate hidden value of BLSTM and CNN, this hybrid network can capture both long and short dependency information of gene sequences.
The LSTM-CNN classification model architecture is displayed in Fig. 10. The network layers begin with the binary encoding layer. The second layer is a BLSTM layer which will apply the input to every cell block to calculate the value of the hidden cell state based on the previous state. This layer continues working till the last nucleic acid is reached, then the last LSTM block decides the final output. The output from the LSTM layer is directly entered into the CNN layers (i.e., convolutional layer, max-pooling layer, and fully connected layer).
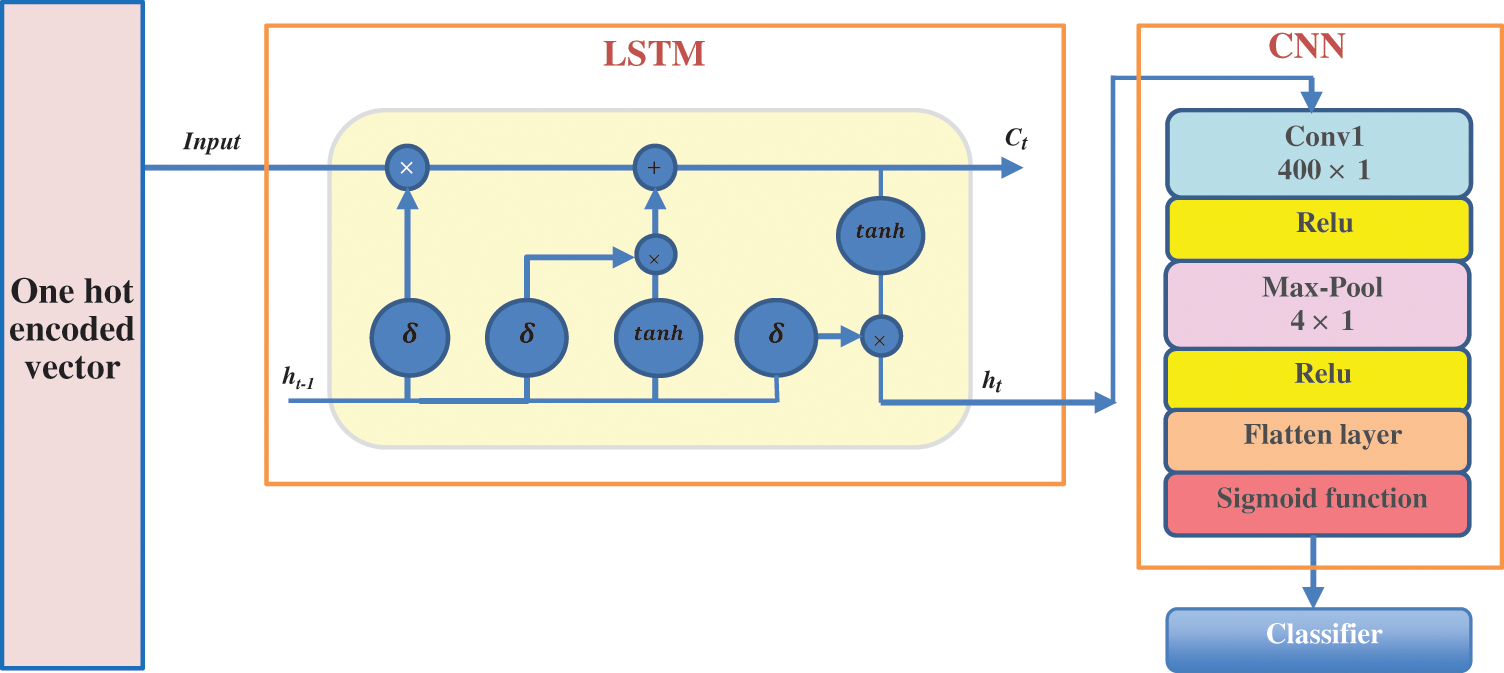
Figure 10: The architecture of the proposed hybrid LSTM-CNN classification model
The experiment study on the LSTM-CNN model trains the datasets as batches, with each batch having a size of 64. Each batch contains several sequences, and only one patch is trained at a time. All the proposed models CNN, LSTM, and LSTM-CNN are implemented and executed using the Keras 0.2.0 library, integrated into the TensorFlow open-source library [30]. The initial parameters in this experiment are optimized as follows: 100 iterations, learning rate between [0.001,0.01], and random weights initialized by Keras default values.
To assess the performance of the proposed LSTM-CNN model, experiments based on the same insulin gene against LSTM, CNN are performed. The evaluation metrics used in this study are sensitivity, accuracy, sensitivity, specificity, precision, F1 Score, Mathew's correlation coefficient (MCC), and mean square error (MSE). These metrics are calculated by Eqs. (8)–(14) [31].
where y is the original output and
Tab. 2 shows a performance comparison among the three models, LSTM, CNN, and LSTM-CNN, based on the above performance metrics. The three experiments' accuracy and loss models are shown in Fig. 11.
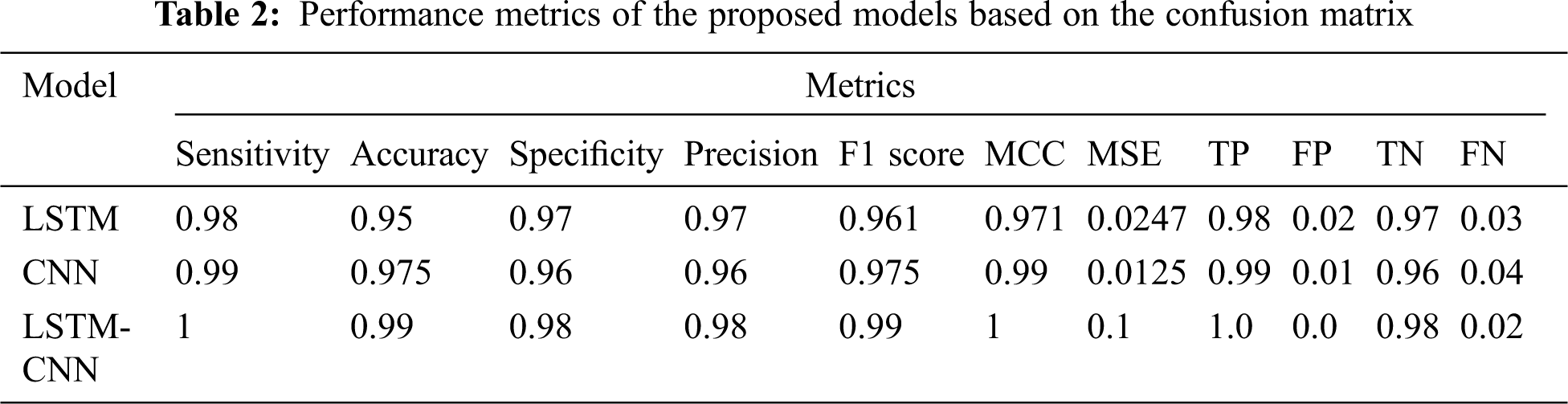
According to the results in Tab. 2, we can conclude that the proposed hybrid LSTM-CNN in the training process is superior to CNN and LSTM algorithms. The recorded overall accuracy of the validation process for LSTM, CNN, and LSTM-CNN is 73%, 94%, and 98%, respectively.
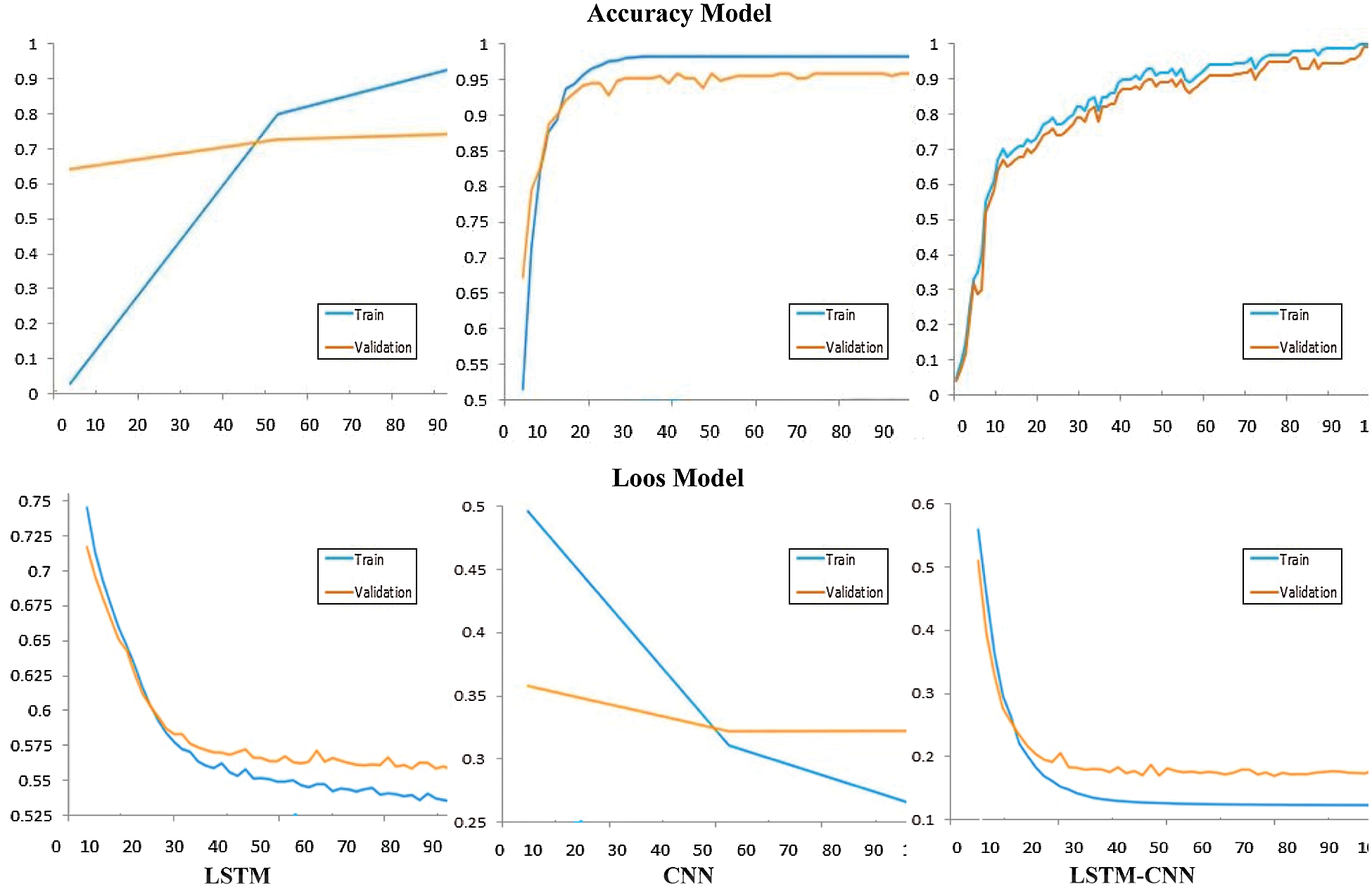
Figure 11: Comparison between performance model of CNN, LSTM, and LSTM-CNN
The significance of AI, especially deep learning algorithms, has been increasing day after day in medical and biological research. Several deep learning algorithms have demonstrated the ability to interpret feature-rich biological data, such as DNA sequences correctly. This study aims to detect diabetes mellitus type 2 based on DNA sequence analysis of insulin gene. According to the proposed study, three classification models based on deep learning algorithms are proposed, a hybrid LSTM-CNN, CNN, and LSTM. According to the experiment results, the hybrid model LSTM-CNN outperforms CNN and LSTM in the accuracy rate for the trial process, which is recorded as 99%, 97.5%, and 95, respectively. In addition, the validation process also clarifies the superiority of LSTM-CNN. The resulting accuracy of the validation process is 98%, 94%, and 73% for LSTM-CNN, CNN, and LSTM, respectively.
Funding Statement: This project was supported financially by the Academy of Scientific Research and Technology (ASRT), Egypt, Grant No. 6415.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. H. Zhou, R. Myrzashova and R. Zheng, “Diabetes prediction model based on an enhanced deep neural network,” EURASIP Journal on Wireless Communication Networks, vol. 148, pp. 1–13, 2020. [Google Scholar]
2. N. E. El Attar, M. K. Hassan, O. A. Alghamdi and W. A. Awad, “Deep learning model for classification and bioactivity prediction of essential oil producing plants from Egypt,” Scientific Reports, vol. 10, no. 1, pp. 1–10, 2020. [Google Scholar]
3. A. Shrestha, A. Mahmood and S. Member, “Review of deep learning algorithms and architectures,” IEEE Access, vol. 7, pp. 53040–53065, 2019. [Google Scholar]
4. W. Kinsner, “Towards cognitive analysis of DNA,” in Proc. 9th IEEE Int. Conf. on Cognitive Informatics, Beijing, China, 2010. [Google Scholar]
5. N. G. Nguyen, V. A. Tran, D. L. Ngo and D. Phan, “DNA sequence classification by convolutional neural network,” Journal of Biomedical Science Engineering, vol. 9, no. 5, pp. 280–286, 2016. [Google Scholar]
6. R. Patgiri, A. Biswas and P. Roy, “Health informatics: A computational perspective in healthcare,” in Studies in Computational Intelligence. Singapore: Springer Nature, 2021. [Google Scholar]
7. R. Dias and A. Torkamani, “Artificial intelligence in clinical and genomic diagnostics,” Genome Medicine, vol. 11, no. 70, pp. 1–12, 2019. [Google Scholar]
8. J. A. Cruz and D. S. Wishart, “Applications of machine learning in cancer prediction and prognosis,” Cancer Informatics, vol. 2, no. 3, pp. 59–77, 2006. [Google Scholar]
9. C. Xu and S. A. Jackson, “Machine learning and complex biological data,” Genome Biology, vol. 20, no. 1, pp. 1–4, 2019. [Google Scholar]
10. J. Zou, M. Huss, A. Abid, P. Mohammadi, A. Torkamani et al., “A primer on deep learning in genomics,” Nature Genetics, vol. 51, no. 1, pp. 12–18, 2019. [Google Scholar]
11. M. Mohammadpoor and M. Sheikhi, “A deep learning algorithm to detect coronavirus (COVID-19) disease using CT images,” PeerJ Computer Science, vol. 3, pp. 1–12, 2021. [Google Scholar]
12. Y. LeCun, Y. Bengio, and G. Hinton “Deep Learning Nature,” Intelligent Control and Automation, vol. 521, no. 7553, pp. 436–444, 2015. [Google Scholar]
13. Y. Sun, S. Zhu, K. Ma, W. Liu, Y. Yue et al., “Identification of 12 cancer types through genome deep learning,” Scientific Reports, vol. 9, pp. 1–9, 2019. [Google Scholar]
14. X. Le, H. V. Ho, G. Lee and S. Jung, “Application of long short-term memory (LSTM) neural network for flood forecasting,” Water, vol. 11, no. 7, pp. 1–19, 2019. [Google Scholar]
15. N. Halpern-wight, M. Konstantinou and A. G. Charalambides, “Training and testing of a single-layer LSTM network for near-future solar forecasting,” Applied Science, vol. 10, no. 17, pp. 1–9, 2020. [Google Scholar]
16. S. I. Ayon and M. Islam, “Diabetes prediction: A deep learning approach,” International Journal of Information Engineering and Electronic Business, vol. 11, no. 2, pp. 21–27, 2019. [Google Scholar]
17. Q. Zou, K. Qu, Y. Luo, D. Yin, Y. Ju et al., “Predicting diabetes mellitus with machine learning techniques,” Frontiers in Genetics, vol. 9, pp. 1–10, 2018. [Google Scholar]
18. Q. A. Hathaway, S. M. Roth, M. V. Pinti, D. C. Sprando, A. Kunovac et al., “Machine learning to stratify diabetic patients using novel cardiac biomarkers and integrative genomics,” Cardiovascular Diabetology, vol. 18, no. 1, pp. 1–16, 2019. [Google Scholar]
19. S. Hasib, A. Faruqui, Y. Du, R. Meka and A. Alaeddini, “Development of a deep learning model for dynamic forecasting of blood glucose level for type 2 diabetes mellitus: secondary analysis of a randomized controlled trial,” JMIR Mhealth and Uhealth, vol. 7, no. 11, pp. 1–14, 2019. [Google Scholar]
20. S. Alby and B. Shivakumar, “A prediction model for type 2 diabetes using adaptive neuro-fuzzy interface system,” Biomedical Research, Special Issue: S69–S74, pp. 1–6, 2018. [Google Scholar]
21. M. H. Ma’mon, S. M. Abderrahman, W.Nimer, Z.Al-Eisawi, H. J. Al-Ameer et al., “Artificial neural networks model for predicting type 2 diabetes Mellitus based on VDR gene foki polymorphism, lipid profile and demographic data,” Biology (Basel), vol. 9, no. 8, pp. 1–17, 2020. [Google Scholar]
22. M. Hashiyada, “DNA biometrics,” in Biometrics, IntechOpen, pp. 139–154, 2011. [Google Scholar]
23. Protein synthesis.[Online]. Available:https://oerpub.github.io/epubjs-demo-book/content/m46032.xhtml#fig-ch03_04_01. [Google Scholar]
24. S. Clancy and W. Brown, “Translation: DNA to mRNA to protein,” Nature Education, vol. 1, no. 1, pp. 2–6, 2008. [Google Scholar]
25. A. De Gaetano, C. Gaz, P. Palumbo and S. Panunzi, “A unifying organ model of pancreatic insulin secretion,” PLoS One, vol. 10, no. 11, pp. 1–34, 2015. [Google Scholar]
26. P. Rorsman and F. M. Ashcroft, “Pancreatic β-Cell electrical activity and insulin secretion: Of mice and men,” Physcological Review, vol. 98, no. 1, pp. 117–214, 2018. [Google Scholar]
27. M. Nishi and K. Nanjo, “Insulin gene mutations and diabetes,” Journal of Diabetes Investigation, vol. 2, no. 2, pp. 92–100, 2011. [Google Scholar]
28. E. W. Sayers, M. Cavanaugh, K. Clark, J. Ostell, K. D. Pruitt et al., “Genbank,” Nucleic Acids Research, vol. 48, pp. 84–86, 2020. [Google Scholar]
29. S. Sharma, S. Sharma and A. Athaiya, “Activation functions in neural networks,” International Journal of Engineering Applied Sciences and Technology, vol. 4, no. 12, pp. 310–316, 2020. [Google Scholar]
30. M. Abadi, A. Agarwal, B. Barham, E. Brevdo, Z. Chen et al., “TensorFlow: Large-scale machine learning on heterogeneous distributed systems,” in Proc. 12th USENIX Conf. on Operating Systems Design and Implementation, CA, United States, pp. 265–283, 2015. [Google Scholar]
31. D. Chicco and G. Jurman, “The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation,” BMC Genomics, vol. 21, no. 1, pp. 1–13, 2020. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |