DOI:10.32604/iasc.2021.018350

| Intelligent Automation & Soft Computing DOI:10.32604/iasc.2021.018350 |  |
| Article |
Detection of COVID-19 Using Deep Learning on X-Ray Images
1Department of Computer Science, Shaqra University, Shaqra, Saudi Arabia
2Department of Information Technology, and Sensor Networks and Cellular Systems Research Center (SNCS), University of Tabuk, Tabuk, Saudi Arabia
*Corresponding Author: Munif Alotaibi. Email: munif@su.edu.sa
Received: 05 March 2021; Accepted: 22 April 2021
Abstract: The novel coronavirus 2019 (COVID-19) is one of a large family of viruses that cause illness, the symptoms of which range from a common cold, fever, coughing, and shortness of breath to more severe symptoms. The virus rapidly and easily spreads from infected people to others through close contact in the absence of protection. Early detection of COVID-19 assists governmental authorities and healthcare specialists in reducing the chain of transmission and flattening the curve of the pandemic. The widespread form of the COVID-19 diagnostic test lacks a high true positive rate and a low false negative rate and needs a particular piece of hardware. Multifariously, computed tomography (CT) and X-ray images indicate evidence of COVID-19 disease. However, it is difficult to diagnose COVID-19 owing to the overlap of its symptoms with those of other lung infections. Thus, there is an urgent need for precise diagnostic solutions based on deep learning algorithms, such as convolutional neural networks, to quickly detect positive COVID-19 cases. Therefore, we propose a deep learning method based on an advanced convolutional neural network architecture known as EffencientNet pre-trained on ImageNet dataset to detect COVID-19 from chest X-ray images. The proposed method improves the accuracy of existing state-of-the-art techniques and can be used by medical staff to screen patients. The framework is trained and validated using 1,125 chest raw X-ray images (i.e., these images are not in regular shape and require image preprocessing) and compared with existing techniques. Three sets of experiments were carried out to detect COVID-19 early utilizing raw X-ray images. For the three-class (i.e., COVID-19, pneumonia, and no-findings) classification set of experiments, our method achieved an average accuracy of 89.60%. For the binary classification two set of experiments, our method yielded an average accuracy of 99.04% and 99.11%. The proposed method has achieved superior results compared to state-of-art methods. The method is a candidate solution for accurately detecting positive COVID-19 cases as soon as they occur using X-ray images. Additionally, the method does not require extensive preprocessing or feature extraction. The proposed technique can be utilized in remote areas where radiologists are not available and can detect other chest-related diseases, such as pneumonia.
Keywords: COVID-19; deep learning; convolutional neural networks; X-ray images; machine learning; computer vision
The novel coronavirus 2019 (COVID-19) disease was first discovered at the end of 2019 in Wuhan, China [1,2]. COVID-19 is one of a large family of viruses that can rapidly and easily spread from infected people to others through close contact in the absence of protection [3–5]. COVID-19 has rapidly spread across the world, affecting more than 200 countries, which makes early diagnosis vital. In the United States, the first COVID-19 case was announced on January 20, 2020 [6], and by June 2020, the number of cases amounted to approximately 2,000,000 cases, including approximately 100,000 deaths. COVID-19 causes symptoms that range from the common cold, fever, coughing, and shortness of breath to more severe indications [7–9]. This disease is correlated with high admissions to the intensive care unit and an increasing number of human fatalities [10]. Early diagnosis of positive cases assists governmental authorities and healthcare specialists in decreasing the chain of transmission by isolating patients and flattening the curve of the pandemic [11].
The most common diagnostic test for COVID-19, reverse transcription polymerase chain reaction (RT-PCR), is used to evaluate the viral ribonucleic acid from a nasopharyngeal swab or sputum [12]. Regrettably, this diagnostic test is associated with a low true positive rate and a high false negative rate [13]. Specific equipment is also required to perform the tests and may be difficult to obtain [14,15]. Since positive COVID-19 cases should be diagnosed and isolated as quickly as possible, the use of RT-PCR for diagnostic purposes is not practical, as the results take a long time to obtain. Moreover, there are concerns about the accuracy and availability of this approach, as well as the lengthy time taken to obtain the results. In certain cases, patients who have undergone RT-PCR testing for COVID-19 have initially been diagnosed as negative, but when retested later, they tested positive, or a clear alternative diagnosis was made [16]. Computed tomography and X-ray images can indicate signs of COVID-19. However, the overlap between COVID-19 symptoms and other lung infections makes it difficult to diagnose the former. Thus, new X-ray-based diagnostic solutions using deep learning algorithms have recently been proposed as a method for rapidly detecting positive COVID-19 cases.
Deep learning algorithms have been used successfully in many medical applications, particularly for the diagnosis and prediction of diseases [17]. Thus, they have the potential to provide a solution to controlling the further spread of the COVID-19 pandemic. Discovering and detecting COVID-19 in its early stages, with high accuracy, is essential to reducing and preventing the widespread dissemination of the virus. The aim of this research is to design a framework to facilitate the identification of COVID-19 with high accuracy from the chest X-rays of infected individuals. The proposed model can detect COVID-19 in its early stages and thus prevent the extensive spread of the virus. As indicated in the literature, deep learning algorithms have the ability to diagnose COVID-19 from the radiography images of infected patients. Thus, the plan in the current study is to develop an improved advanced deep learning model that has not previously been investigated in the literature by building upon existing approaches. The proposed method is an end-to-end framework that uses the prominent self-feature extraction capability of deep learning models to extract and work on meaningful features of X-ray images. The framework is trained and validated using 1,125 chest raw X-ray images (i.e., these images are not in regular shape and require image pre-processing) and compared with existing techniques. The proposed framework can identify COVID-19 accurately in its early stages and thus prevent spread of the virus.
This research paper is organized as follows. The related work is surveyed in Section 2. Section 3 introduces the proposed deep learning framework and presents the experimental settings and the information of the dataset used to validate the proposed method. Section 4 shows the proposed method results. Section 5 discusses the results of the proposed method and the comparison with closely related methods. Section 6 concludes the research paper.
Recently, deep learning algorithms have been demonstrated to be invaluable in many medical applications, and they have been proposed as a method of predicting and identifying COVID-19 infection based on its symptoms. Severe respiratory symptoms and pneumonia, characteristic symptoms of COVID-19, present as unique graphical manifestations and characteristics on chest computerized tomography (CT) scans and X-rays. Thus, Wang et al. [18] proposed a screening model with the aim of detecting and controlling the spread of COVID-19 using chest CT images and a deep learning algorithm. The suggested prediction framework consisted of phases, the selection of an region of interest (ROI) in specific samples, the extraction of meaningful features using a convolutional neural network (CNN) architecture, and classification using a fully connected layer. The authors developed and validated their technique utilizing a dataset of 453 CT images of 99 subjects.
Unlike CT image-based approaches, some methods have been reported that utilize X-ray images since the results can be rapidly obtained; in addition, the methodology is easy, more cost-effective, and less harmful than CT. Automatic detection models, using X-ray images and deep CNN, have also been utilized for COVID-19 detection [19–24]. Narin et al. [19] investigated the early detection of COVID-19 using three types of CNN architectures: residual networks 50 (ResNet50), InceptionV3, and Inception-ResNetV2, which were used to classify X-ray images of COVID-19 patients and normal samples. The authors found that the ResNet outperformed the two other methods in terms of accuracy. However, the lack of sufficient samples (i.e., there were only 20 samples) was identified as a limitation to training the deep learning model. This issue was overcome with the application of a transfer learning technique, which led to a promising result. Farooq et al. [20] proposed the use of ResNet to distinguish between COVID-19 cases and those with similar diseases, such as pneumonia. The authors applied three techniques, discriminative learning, progressive resizing, and cyclical learning, to improve ResNet accuracy and make it less complex.
Xu et al. [25] proposed a three-dimensional CNN model to detect COVID-19 by subdividing the X-ray images into image patches. The proposed method was able to differentiate COVID-19 from influenza A viral pneumonia and healthy cases. It utilized four steps: (1) preprocessing to extract valuable features from specific regions, (2) the use of three-dimensional CNN architecture to subdivide the samples into possible meaningful image patches (i.e., by segmenting the center of the image and two neighboring pixels for each patch), (3) the classification of each subdivided patch, and (4) analysis that utilized a noisy-OR Bayesian function. Similarly, Gozes et al. [26] suggested the utilization of a CNN model by medical staff to differentiate between COVID-19 patients and uninfected individuals.
Luz et al. [27] suggested the use of a customized deep learning model that utilized certain components that were initially introduced to the ResNet architecture to enhance the performance of the technique; there were also fewer parameters to monitor using this approach compared to other techniques. To overcome the limited number of samples, the authors applied transfer learning using a pretrained model of ImageNet instances. Oh et al. [28] evaluated the use of a CNN approach in the detection of COVID-19 in a patchwise rather than pixelwise manner to overcome the lack of sufficient available samples required to train deep learning-based techniques. In other research, a deep anomaly detection technique was investigated for its efficacy in detecting COVID-19-positive samples [16]. The proposed method utilized stochastic gradient descent for training, and it showed promising results and had a reasonable detection time.
Apostolopoulos et al. [29] evaluated the performance of state-of-the-art CNN architectures such as visual geometry group 19 (VGG19), MobileNet, Inception, extreme version of Inception (Xception), and Inception-ResNet that have been applied for medical image classification and have adopted transfer learning to detect COVID-19. To evaluate their methods, the authors utilized a collection of two datasets that consisted of three classes, namely, COVID-19 class, normal class, and bacterial pneumonia class. The proposed method achieved promising results of 96.78% accuracy, 98.66% sensitivity, and 96.46% specificity, suggesting that applying deep learning to detect COVID-19 using X-ray images might extract meaningful COVID-19 biomarkers. The authors found that MobileNet outperformed the other CNN architectures in terms of specificity.
Toğaçar et al. [30] presented a deep learning technique consisting of two convolutional neural network architectures, namely, SqueezeNet and MobileNetV2, to train stacked images and a support vector machine (SVM) to detect COVID-19 in early stages. The authors validated their idea using a dataset of three classes: normal class, pneumonia class, and COVID-19 class. The fuzzy color technique was applied as a preprocessing step to restructure the classes. Therefore, the restructured images along with the original images were bundled to be fed into the deep learning architectures. Thereafter, the bundled images were passed to the SqueezeNet and MobileNetV2 models for training purposes. Afterward, the features generated by the models were processed using optimization methods known as social mimic. Finally, the features processed by the optimization method were combined and passed to the SVM to classify the three classes.
Khan et al. [31] introduced a deep CNN model based on Xception deep CNN architecture to diagnose COVID-19 through X-ray images. The proposed method was pre-trained using ImageNet dataset to address the limited number of samples limitation. The authors performed various set of experiments utilizing raw X-ray images collected from two different public repositories to evaluate their method using five evaluation metrics. The proposed method achieved promising results on four classes, three classes, and two classes set of experiments employing 4-fold cross validation technique.
Ozturk et al. [32] proposed an automated method based on the Darknet model to detect COVID-19 early using X-ray images. The model was validated using a dataset of 1,125 chest X-ray images. The dataset that the authors utilized to validate their method consisted of three classes, namely, no-findings class, COVID-19 class, and pneumonia class. Two data splits were performed for evaluation measures, a data split for binary classification and a data split for multiclass classification. The proposed technique achieved promising results in both data splits, in which 98.08% was achieved in the binary classification data split and 87.02% was achieved in the three-class classification.
This section introduces the information of the X-ray image dataset utilized to evaluate the proposed method and the proposed deep EfficientNet-based method.
To evaluate the proposed method, we utilized the Python programming language. Two Python libraries designed for developing machine learning models, namely, Keras [33] and TensorFlow [34], were used to train the model and validate the results. The proposed model was trained by employing a collaborative environment that provides access to graphics processing units (GPUs) and tensor processing units (TPUs) (i.e., Google colaboratory (Colab)). A dataset provided by Ozturk et al. [32] was used to validate the proposed method. This dataset, which contains chest X-ray images, is designated for the early diagnosis of COVID-19. The X-ray images were collected from two different sources. The first set of X-ray images was collected by Cohen et al. [35] using images from various open access sources. The number of female COVID-19-positive cases is 43, while the number of male COVID-19-positive cases is 82. The average age of only 26 positive cases is 55 years. Some of the X-ray images samples present in this dataset are shown in Fig. 1. The other dataset was published by Wang et al. [36] for classifying common thorax diseases. The total number of samples collected from the two sources was 1,125 X-ray images. These samples belong to three classes: the first class, i.e., COVID-19, consists of 125 X-ray images, the second class, i.e., pneumonia, consists of 500 X-ray images, and the third class, i.e., the no-findings class, consists of 500 X-ray images.
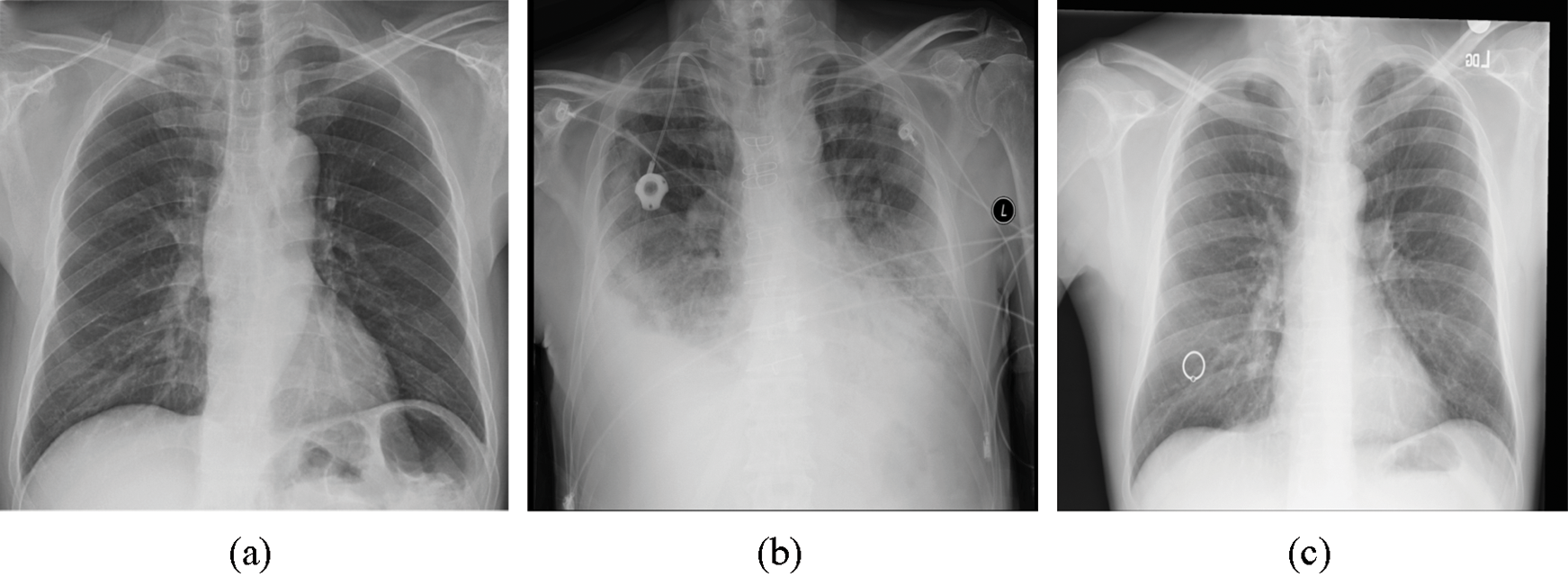
Figure 1: Three images present in the dataset representing three classes: (a) COVID-19, (b) pneumonia, and (c) no-findings
To compare our proposed method with a recent related work, i.e., Ozturk et al. [32], we used the same dataset of 500 chest X-ray images with no-findings cases, 500 X-ray images with pneumonia cases, and 125 chest X-ray images with COVID-19 cases.
Artificial intelligence has been revolutionized since the emergence of deep learning techniques [37,38]. The term deep is introduced because the size of the neural network increases in which more layers are added to extend the learning process. One of the most successful deep learning algorithms that has attracted the attention of the research community due to the considerable success in precisely classifying or recognizing objects is CNN. A typical CNN architecture comprises three main types of layers: a convolution layer that receives the input and applies a number of filters to extract meaningful features, a pooling layer that reduces the complexity of the CNN architecture by downsampling the features, and a fully connected layer that deduces the decision of final classification.
Most effective deep learning architectures have been constructed using already proven models. Therefore, it is more reasonable to build a model based on an already proven model instead of developing a new deep learning model from scratch. As a starting point, a recent CNN architecture known as EfficientNet [39] is investigated. EfficientNet uses a very effective scaling method, named the compound coefficient, that solves the issue of scaling up all dimensions of width, depth, and resolution to the available resources in a more wise and principled manner while maintaining model efficiency. Fig. 2 explains the idea of the compound scaling method. It uniformly scales up all dimensions with a constant ratio.
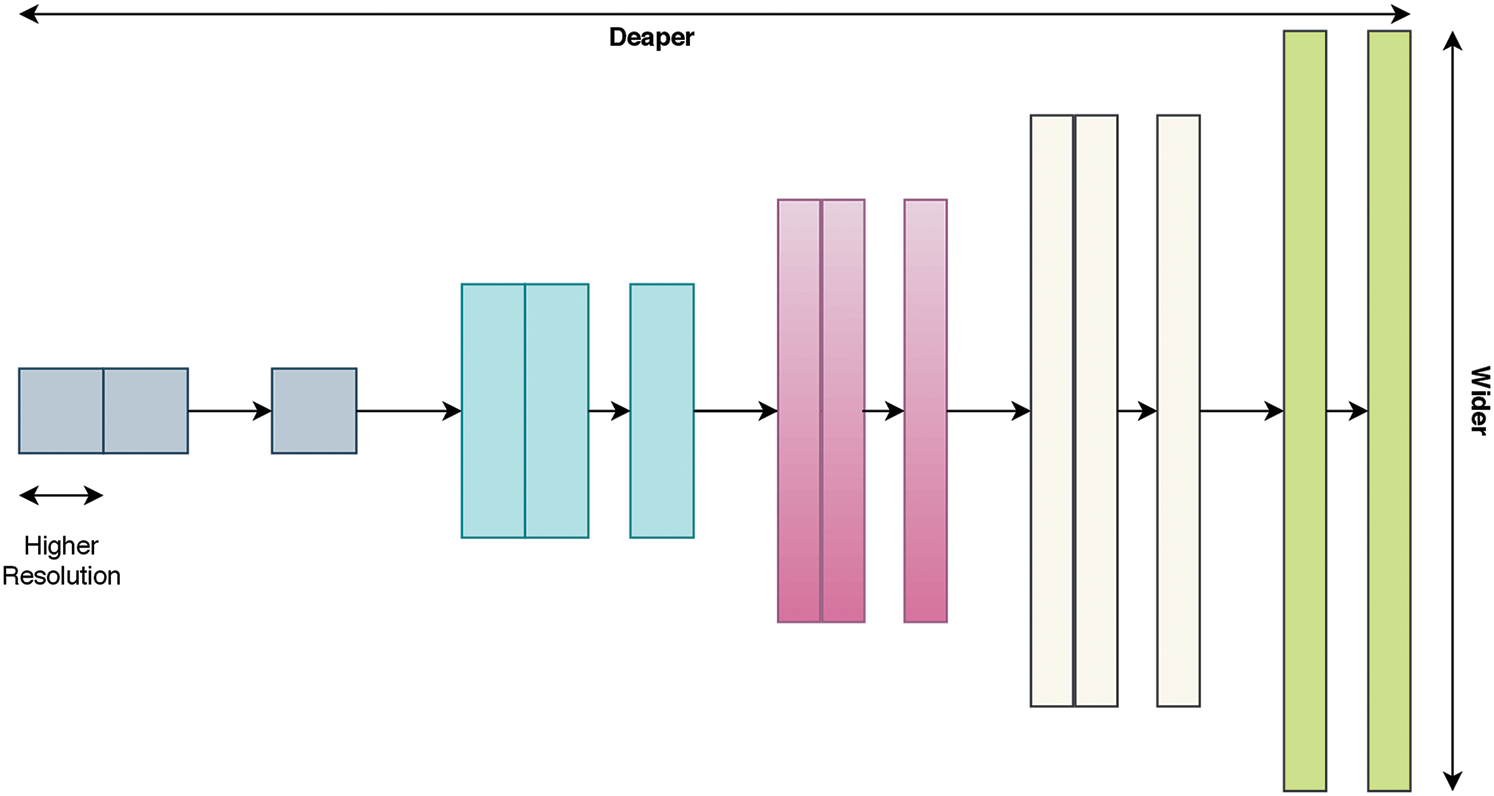
Figure 2: Compound scaling method as in Tan et al. [39]; uniformly scales up the width, depth, and resolution with a constant ratio
The compound scaling method highly depends on the baseline architecture. Thus, Tan et al. [39] used neural architecture search as the baseline architecture. EfficientNet models are based on automated machine learning (AutoML) [40] and the compound scaling technique. The authors utilized the AutoML mobile neural architecture search (MNAS) model, suitable for mobile devices, to develop a new mobile-size architecture, named EfficientNet-B0. Moreover, they used the compound scaling technique to scale up this baseline to obtain a family of models (EfficientNet-B1 to EfficientNet-B7).
EfficientNet models were able to achieve much better accuracy and efficiency than previous CNN architectures. EfficientNet-B7 achieved the state-of-the-art on the ImageNet dataset. It has the ability to capture richer and more complex features.
In this paper, we resized all of the X-ray images to 250 × 250 and then utilized the EfficientNet-B2 model to detect COVID-19 from raw X-ray images as shown in Fig. 3. We normalize the data of X-ray images so that they are of approximately the same scale. We added a global max pooling layer, followed by the dropout technique with a rate of 0.2 at the end of the model to prevent any overfitting. We used the RmsProp optimizer for the optimization of the model. We also used a softmax classifier at the end of the model for the classification. Our model trained with 40 epochs, using a batch size of 20, a learning rate of 0.001 and momentum of 0.
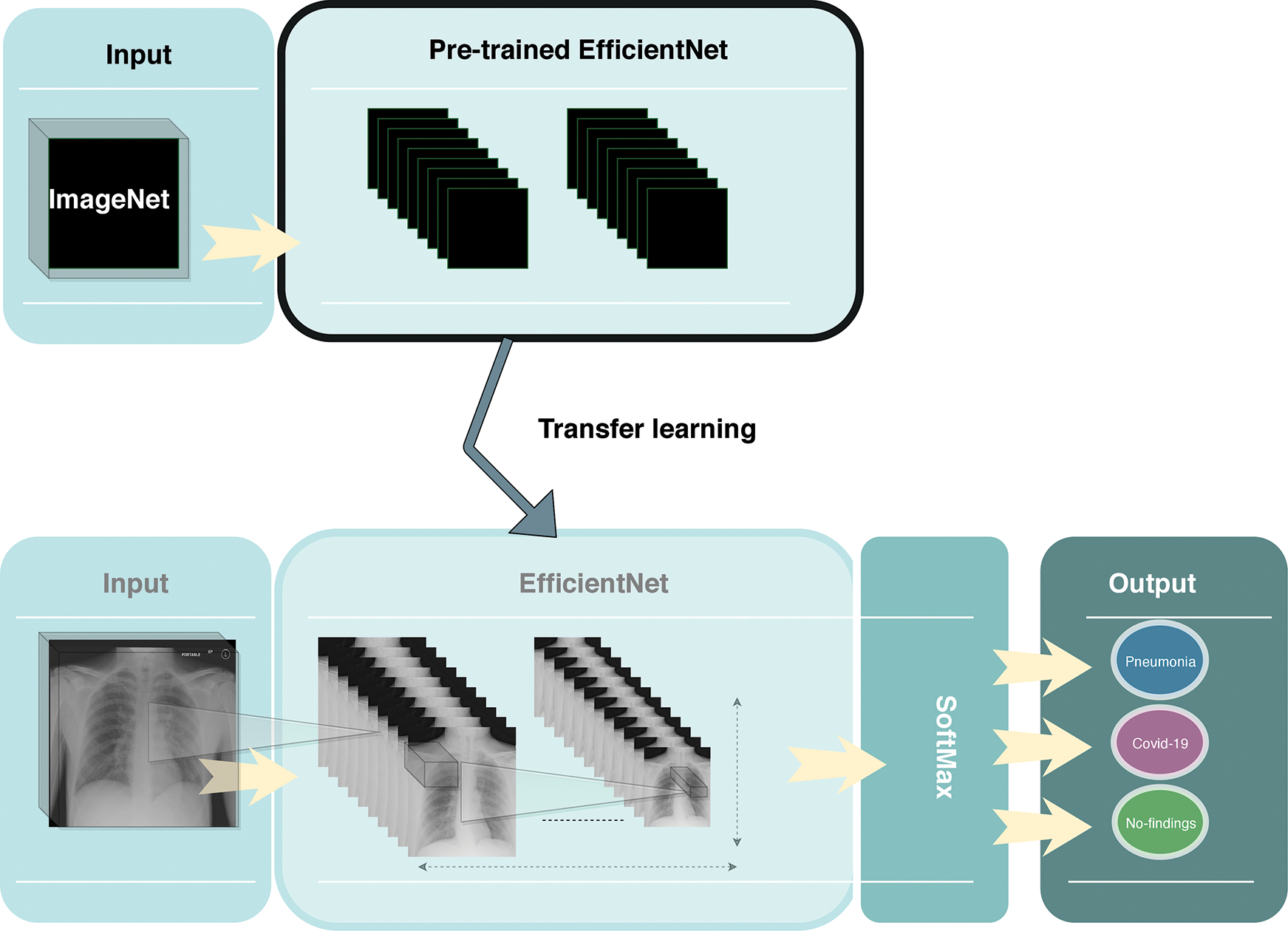
Figure 3: Overview of the proposed framework
Moreover, we used the transfer learning technique to improve the performance of our model. Due to the limited number of COVID-19 X-ray images, transfer learning was utilized to enhance the learning process of our method by transmitting knowledge from a related domain. Thus, we used the weights of the EfficientNet-B2 model that was pretrained on the ImageNet database and finetuned the new dataset, which helped to achieve better performance and decrease the training time.
Three sets of experiments were carried out to detect COVID-19 early utilizing raw X-ray images. In the first set of experiments, the deep EfficientNet-based approach was trained in multiclass classification settings using the three classes: no finding, COVID-19, and pneumonia. In the second set of experiments, the proposed method was trained in binary classification settings using two classes: non-COVID (pneumonia and no-findings) and COVID-19. In the third set of experiments, the proposed method was trained in binary classification settings using two classes: no-findings and COVID-19. To validate the generalization of the proposed model and compare it with the DarkNet-based approach [32], the 5-fold cross-validation technique was adopted for the three sets of experiments (i.e., multiclass and both binary classification settings). For both the multiclass and binary classification, the training of 80% of the data and validation of 20% of the data were iterated 5 times for each fold. The 5-fold prediction was averaged to induce the final results. The number of epochs used to train the model for each fold was 40.
The confusion matrices for both the three-class and binary classifications are shown in Figs. 4–6, respectively. The confusion matrix of each fold for three-class classification presents the percentage of X-ray images that were correctly classified and misclassified by our proposed method. The confusion matrix was evaluated using four performance measures: true positive (TP), false positive (FP), true negative (TN), and false negative (FN). The TP represents the number of COVID-19 positive instances that were correctly classified as positive. The FP represents the number of COVID-19-positive samples that were mistakenly misclassified. The TN represents the number of negative instances (i.e., no-findings and pneumonia instances) that were correctly classified as negative. The FN represents the number of negative instances that were classified mistakenly as positive COVID-19 instances.
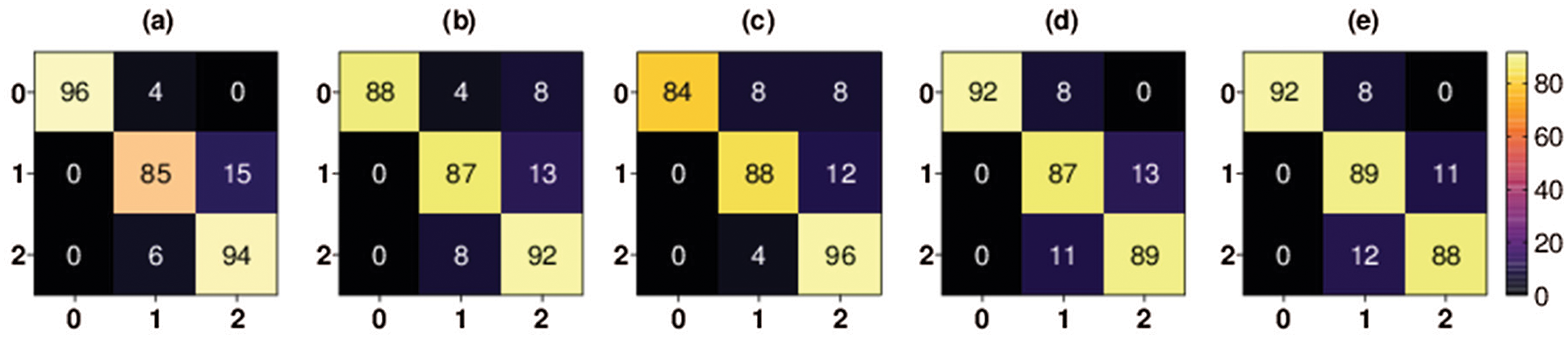
Figure 4: Multiclass classification 5-fold confusion matrices. Class 0 denotes COVID-19 class, class 1 denotes pneumonia class, and class 2 denotes no-findings class, (a) Fold-1, (b) Fold-2, (c) Fold-3, (d) Fold-4, (e) Fold-5
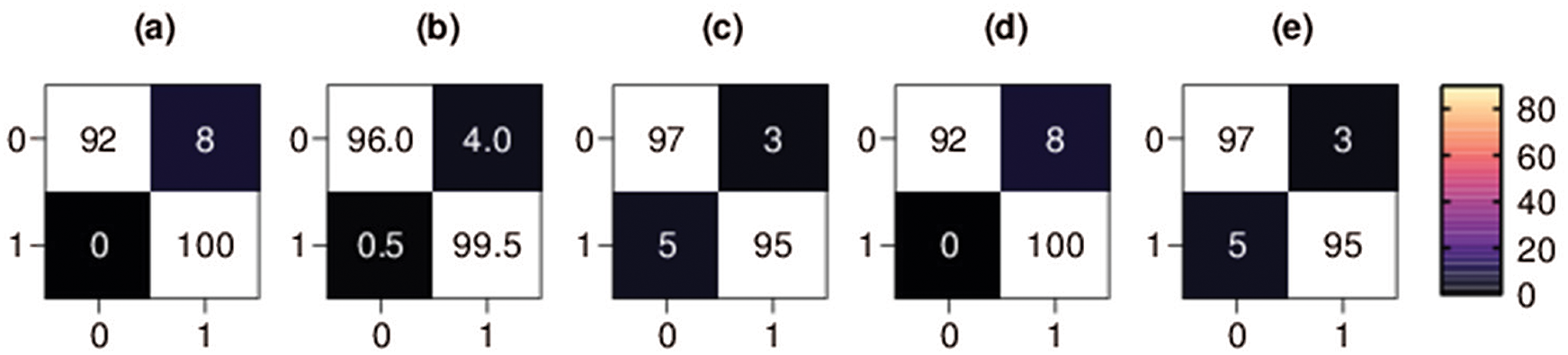
Figure 5: Binary classification 5-fold confusion matrices. Class 0 denotes the COVID-19 class, and class 1 denotes the non-COVID class, (a) Fold-1, (b) Fold-2, (c) Fold-3, (d) Fold-4, (e) Fold-5
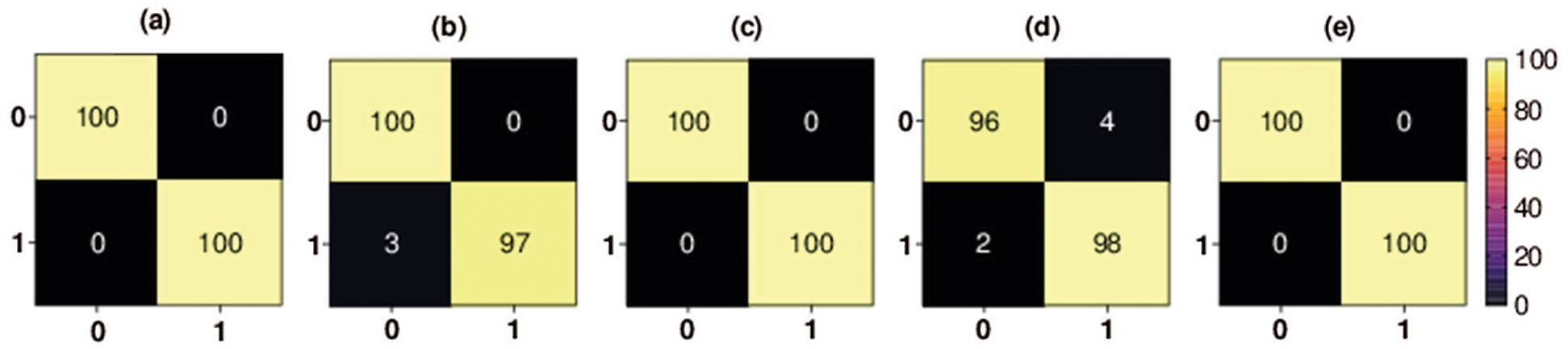
Figure 6: Binary classification 5-fold confusion matrices. Class 0 denotes the COVID-19 class, and class 1 denotes the no-findings class, (a) Fold-1, (b) Fold-2, (c) Fold-3, (d) Fold-4, (e) Fold-5
As shown in Fig. 4, the percentage of COVID-19 X-ray images TP rate in the first fold is 84% and the percentage of COVID-19 FP rate is 16%. The percentage of COVID-19 X-ray images TP rate in the second fold is 84% and the percentage of COVID-19 FP rate is 16%. The percentage of COVID-19 X-ray images TP rate in the third fold is 92% and the percentage of COVID-19 FP rate is 8%. The percentage of COVID-19 X-ray images TP rate in the fourth fold is 96% and the percentage of COVID-19 FP rate is 4%. Finally, the percentage of COVID-19 X-ray images TP rate in the fifth fold is 96% and the percentage of COVID-19 FP rate is 4%.
The confusion matrix of each fold for first binary classification presents the percentage of X-ray images that were correctly classified and misclassified by our proposed method, as shown in Fig. 5. The percentage of COVID-19 X-ray images TP rate in the third fold and fifth fold (i.e., the best scoring folds of the second classification set of experiments in terms of TP rate) was 97%, the percentage of COVID-19 FP rate was only 3%, the percentage of COVID-19 X-ray images FN rate was 5% and the percentage of COVID-19 TN rate was 95%. The percentage of COVID-19 X-ray images TN rate in the first and fourth folds (i.e., the best scoring folds of the binary classification set of experiments in terms of TN rate) was 100%, the percentage of COVID-19 FN rate was zero, the percentage of COVID-19 TP rate was 92%, and the percentage of COVID-19 FP rate was 8%.
The confusion matrix of each fold for second binary classification set of experiments is shown in Fig. 6. The percentage of COVID-19 X-ray images TP rate in the first fold, third fold, and fifth fold (i.e., the best scoring folds of the third classification set of experiments in terms of TP rate) was 100%, the percentage of COVID-19 FP rate was 0%, the percentage of COVID-19 X-ray images FN rate was 0% and the percentage of COVID-19 TN rate was 100%. The percentage of COVID-19 X-ray images TN rate in the second fold was 97%, the percentage of COVID-19 FN rate was 3%, the percentage of COVID-19 TP rate was 100%, and the percentage of COVID-19 FP rate was 0%. The percentage of COVID-19 X-ray images TN rate in the fourth fold was 98%, the percentage of COVID-19 FN rate was 2%, the percentage of COVID-19 TP rate was 96%, and the percentage of COVID-19 FP rate was 4%.

In the first set of experiments, as shown in Tab. 1(a), where the target was to classify three classes, the accuracy of the first fold was 90.22%, the accuracy of the second fold was 89.33%, the accuracy of the third fold was 91.11%, the accuracy of the fourth fold was 91.11%, and the accuracy of the last fold was 88.89%. The average accuracy of the proposed method was 89.60%.
In the second set of experiments, as shown in Tab. 1(b), the accuracy of the first fold was 99.11%, the accuracy of the second fold as 99.11%, the accuracy of the third fold was 98.22%, the accuracy of the fourth fold was 99.56%, and the accuracy of the last fold was 99.56%. The average accuracy of the proposed method was 99.11%. In the third set of experiments, as shown in Tab. 1(c), the accuracy of the first fold was 100%, the accuracy of the second fold as 97.60%, the accuracy of the third fold was 100%, the accuracy of the fourth fold was 97.60%, and the accuracy of the last fold was 100%. The average accuracy of the proposed method was 99.04%.
We used the same dataset utilized by a recent related work to compare our proposed method with this closely related method. To compare our proposed method with a recent related work [32] that achieved promising results, we used the same number of samples (i.e., 1,125 X-ray images in which 125 images belonged to the COVID-19 class, 500 images belonged to the pneumonia class, and 500 images belonged to the no-findings class). We also used a set of experiments identical to Ozturk et al. [32] for binary and three-class classifications utilizing a 5-fold cross-validation technique. Our method yielded outstanding results compared to the closely related method in terms of the four evaluation measures. As shown in Fig. 7, for the three-class classification set of experiments, our method achieved an average accuracy of 89.60%, which is better than the method that was proposed by Ozturk et al. [32] by 2.58% (i.e., their method yielded an average accuracy of 87.02%).
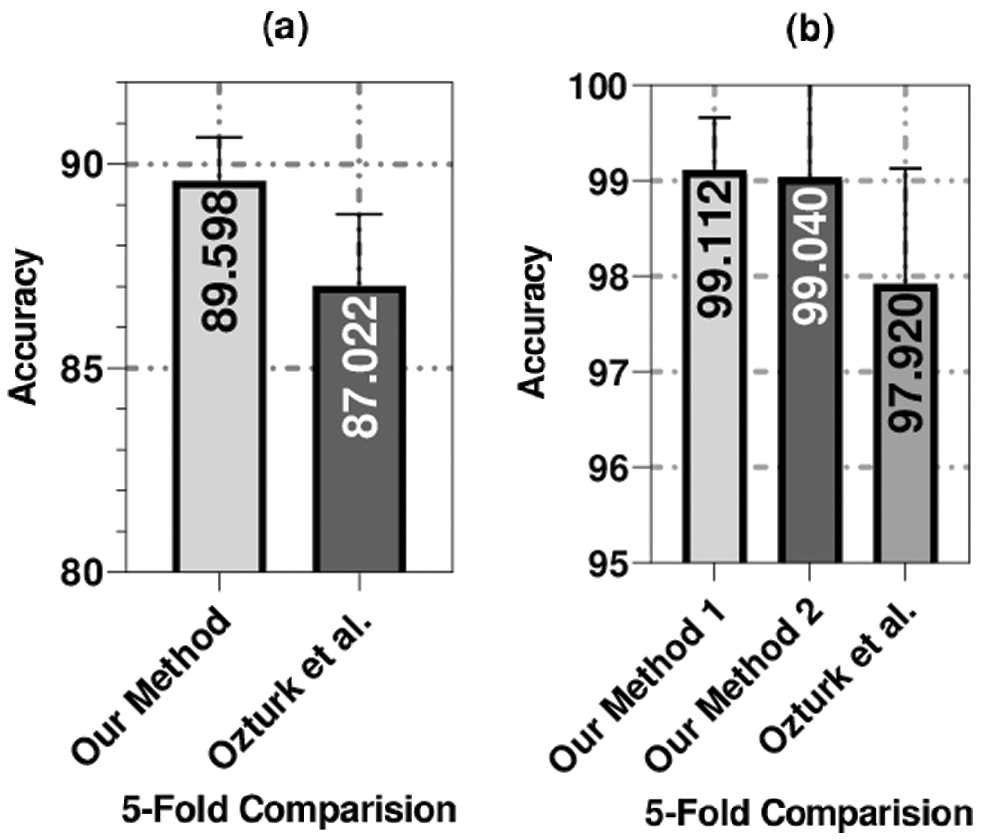
Figure 7: The average accuracy of the 5-fold validation technique used to compare our methods (our method 1 was applied on COVID-19 and non-COVID data split and method 2 was applied on COVID-19 and no-Findings data split) with the method of Ozturk et al. [32]
For the binary classification set of experiments, our method achieved an average accuracy of 99.04%, which is better than the method that was proposed by Ozturk et al. [32] by almost 1% (i.e., their method achieved an average accuracy of 98.08%). The proposed method outperformed the other method by five-fold in the three-class set of experiments, as shown in Fig. 7. Additionally, the proposed method outperformed the other method in three out of five folds and had identical performance in the other two folds in the binary classification set of experiments.
The dataset that was used in this research has a limited number of samples. As shown in Tab. 2 and similar to the previously proposed methods, the limited number of samples is a common issue. Several related works used fewer samples than we used to validate their methods. For instance, Hemdan et al. [41] used 50 images, Wang et al. [42] used 300 images, Narin et al. [19] used a total of 100 images, and Sethy et al. [43] used a total of 50 images to validate their approaches. To overcome this limitation, we used transfer learning to improve the performance of the proposed method in terms of the four evaluation metrics. As more COVID-19 X-ray images are publicly released, we contemplate incorporating more X-ray images to further generalize the proposed method.
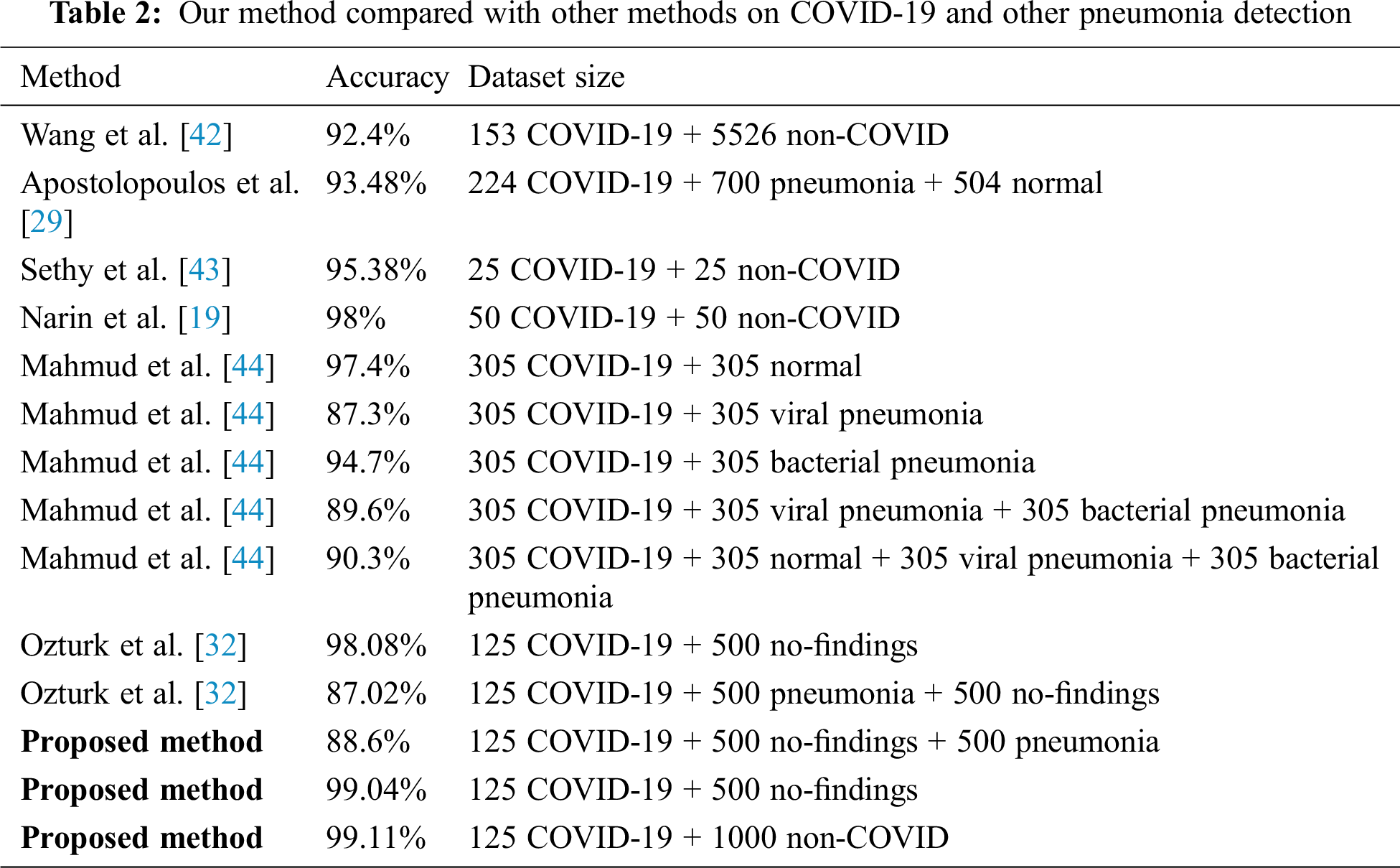
As shown in Tab. 2, our method is compared with other methods under different splits and size of the dataset. Similar to our porposed methods, these methods used deep learning architectures to detect COVID-19 using X-ray images. As indicated in this table, our method has achieved promising results compared to state-of-art methods. The proposed method is a candidate solution for accurately detecting positive COVID-19 cases as soon as they occur using X-ray images. Additionally, the method does not require extensive preprocessing or feature extraction. Moreover, the proposed method can detect COVID-19-positive cases within milliseconds.
COVID-19 disease has affected millions of people and remains a threat to the lives of many of people. In this research, a deep EffecientNet-based approach is proposed to automatically detect COVID-19-positive cases from X-ray images. This approach is an end-to-end framework that involves a transfer learning technique to overcome the limited number of samples. The proposed method avoids extensive preprocessing steps and handcrafted feature extractions and utilization. The framework was evaluated using two sets of experiments: three-class classification and binary classification set of experiments using four evaluation metrics, namely, accuracy, recall, precision, and F1-score. Our technique achieved outstanding results using 1,125 X-ray images compared to recent related work. The average accuracies of our technique using three-class classification and binary classification are 89.60% and 99.11%, respectively. The proposed technique can be utilized in remote areas where radiologists are not available and can detect other chest-related diseases, such as pneumonia.
Acknowledgement: The authors wish to express their appreciation to the reviewers for their helpful suggestions which greatly improved the presentation of this paper.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. X. W. Xu, X. X. Wu, X. G. Jiang, K. J. Xu, L. J. Ying et al., “Clinical findings in a group of patients infected with the 2019 novel coronavirus (SARS-Cov-2) outside of Wuhan, China: Retrospective case series,” BMJ, vol. 368, pp. m606, 2020. [Google Scholar]
2. L. Brunese, F. Mercaldo, A. Reginelli and A. Santone, “Explainable deep learning for pulmonary disease and coronavirus COVID-19 detection from X-rays,” Computer Methods and Programs in Biomedicine, vol. 196, no. 20, pp. 105608, 2020. [Google Scholar]
3. S. Amrane, H. Tissot-Dupont, B. Doudier, C. Eldin, M. Hocquart et al., “Rapid viral diagnosis and ambulatory management of suspected COVID-19 cases presenting at the infectious diseases referral hospital in Marseille, France, – January 31st to March 1st, 2020: A respiratory virus snapshot,” Travel Medicine and Infectious Disease, vol. 36, pp. 101632, 2020. [Google Scholar]
4. L. T. Phan, T. V. Nguyen, Q. C. Luong, T. V. Nguyen, H. T. Nguyen et al., “Importation and human-to-human transmission of a novel coronavirus in Vietnam,” New England Journal of Medicine, vol. 382, no. 9, pp. 872–874, 2020. [Google Scholar]
5. E. Cuevas, “An agent-based model to evaluate the COVID-19 transmission risks in facilities,” Computers in Biology and Medicine, vol. 121, no. 223–241, pp. 103827, 2020. [Google Scholar]
6. M. L. Holshue, C. DeBolt, S. Lindquist, K. H. Lofy, J. Wiesman et al., “First case of 2019 novel coronavirus in the United States,” New England Journal of Medicine, vol. 382, no. 10, pp. 929–936, 2020. [Google Scholar]
7. W. J. Guan, Z. Y. Ni, Y. Hu, W. H. Liang and C. Q. Ou, “Clinical characteristics of coronavirus disease 2019 in China,” New England Journal of Medicine, vol. 382, no. 18, pp. 1708–1720, 2020. [Google Scholar]
8. B. R. Beck, B. Shin, Y. Choi, S. Park and K. Kang, “Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug-target interaction deep learning model,” Computational and Structural Biotechnology Journal, vol. 18, no. 3, pp. 784–790, 2020. [Google Scholar]
9. R. M. Pereira, D. Bertolini, L. O. Teixeira, C. N. Silla Jr and Y. M. G. Costa, “COVID-19 identification in chest X-ray images on flat and hierarchical classification scenarios,” Computer Methods and Programs in Biomedicine, vol. 194, no. 17, pp. 105532, 2020. [Google Scholar]
10. Y. M. Arabi, S. Murthy and S. Webb, “COVID-19: A novel coronavirus and a novel challenge for critical care,” Intensive Care Medicine, vol. 46, no. 5, pp. 833–836, 2020. [Google Scholar]
11. L. O. Gostin, E. A. Friedman and S. A. Wetter, “Responding to COVID-19: How to navigate a public health emergency legally and ethically,” Hastings Center Report, vol. 50, no. 2, pp. 8–12, 2020. [Google Scholar]
12. A. A. Ardakani, A. R. Kanafi, U. R. Acharya, N. Khadem and A. Mohammadi, “Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: Results of 10 convolutional neural networks,” Computers in Biology and Medicine, vol. 121, no. 10229, pp. 103795, 2020. [Google Scholar]
13. F. Shi, J. Wang, J. Shi, Z. Wu, Q. Wang et al., “Review of artificial intelligence techniques in imaging data acquisition, segmentation, and diagnosis for COVID-19,” IEEE Reviews in Biomedical Engineering, vol. 14, pp. 4–15, 2021. [Google Scholar]
14. T. Ai, Z. Yang, H. Hou, C. Zhan, C. Chen et al., “Chen etal, Correlation of chest CT and RT-PCR testing for coronavirus disease 2019 (COVID-19) in China: A report of 1014 cases,” Radiology, vol. 296, no. 2, pp. E32–E40, 2020. [Google Scholar]
15. P. Afshar, S. Heidarian, F. Naderkhani, A. Oikonomou, N. Plataniotis et al., “COVID-CAPS: A capsule network-based framework for identification of COVID-19 cases from X-ray images,” Pattern Recognition Letters, vol. 138, no. 4, pp. 638–643, 2020. [Google Scholar]
16. Y. Fang, H. Zhang, J. Xie, M. Lin, L. Ying et al., “Sensitivity of chest CT for COVID-19: Comparison to RT-PCR,” Radiology, vol. 296, no. 2, pp. E115–E117, 2020. [Google Scholar]
17. J. A. M. Sidey-Gibbons and C. J. Sidey-Gibbons, “Machine learning in medicine: A practical introduction,” BMC Medical Research Methodology, vol. 19, no. 1, pp. 64, 2019. [Google Scholar]
18. S. Wang, B. Kang, J. Ma, X. Zeng, M. Xiao et al., “A deep learning algorithm using CT images to screen for Corona virus disease (COVID-19),” European Radiology, pp. 1–9, 2021. [Google Scholar]
19. A. Narin, C. Kaya and Z. Pamuk, “Automatic detection of coronavirus disease (COVID-19) using X-ray images and deep convolutional neural networks,” arXiv preprint, arXiv:2003.10849, 2020. [Online]. Available: https://arxiv.org/abs/2003.10849. [Google Scholar]
20. M. Farooq and A. Hafeez, “COVID-ResNet: A deep learning framework for screening of COVID19 from radiographs,” arXiv preprint, arXiv:2003.14395, 2020. [Online]. Available: https://arxiv.org/abs/2003.14395. [Google Scholar]
21. M. Mahmoud Al Rahhal, Y. Bazi, R. M. Jomaa, M. Zuair and N. Al Ajlan, “Deep learning approach for COVID-19 detection in computed tomography images,” Computers Materials & Continua, vol. 67, no. 2, pp. 2093–2110, 2021. [Google Scholar]
22. S. M. Elghamrawy, A. Ella Hassnien and V. Snasel, “Optimized deep learning-inspired model for the diagnosis and prediction of COVID-19,” Computers Materials & Continua, vol. 67, no. 2, pp. 2353–2371, 2021. [Google Scholar]
23. A. S. Al-Waisy, M. Abed Mohammed, S. Al-Fahdawi, M. S. Maashi, B. Garcia-Zapirain et al., “COVID-DeepNet: Hybrid multimodal deep learning system for improving COVID-19 pneumonia detection in Chest X-ray images,” Computers Materials & Continua, vol. 67, no. 2, pp. 2409–2429, 2021. [Google Scholar]
24. M. Attique Khan, N. Hussain, A. Majid, M. Alhaisoni, S. Ahmad Chan Bukhari et al., “Classification of positive COVID-19 CT scans using deep learning,” Computers Materials & Continua, vol. 66, no. 3, pp. 2923–2938, 2021. [Google Scholar]
25. X. Xu, X. Jiang, C. Ma, P. Du, X. Li et al., “A deep learning system to screen novel coronavirus disease 2019 pneumonia,” Engineering (Beijing, China), vol. 6, no. 10, pp. 1122–1129, 2020. [Google Scholar]
26. O. Gozes, M. Frid-Adar, H. Greenspan, P. D. Browning, H. Zhang et al., “Rapid AI development cycle for the coronavirus (COVID-19) pandemic: Initial results for automated detection & patient monitoring using deep learning CT image analysis,” arXiv preprint, arXiv:2003.05037, 2020. [Online]. Available: https://arxiv.org/abs/2003.05037. [Google Scholar]
27. E. Luz, P. L. Silva, R. Silva, L. Silva, G. Moreira et al., “Towards an effective and efficient deep learning model for COVID-19 patterns detection in X-ray images,” arXiv preprint, arXiv: 2004.05717, 2020. [Online]. Available: https://arxiv.org/abs/2004.05717. [Google Scholar]
28. Y. Oh, S. Park and J. C. Ye, “Deep learning COVID-19 features on CXR using limited training data sets,” IEEE Transactions on Medical Imaging, vol. 39, no. 8, pp. 2688–2700, 2020. [Google Scholar]
29. I. D. Apostolopoulos and T. A. Mpesiana, “COVID-19: Automatic detection from X-ray images utilizing transfer learning with convolutional neural networks,” Physical and Engineering Sciences in Medicine, vol. 43, no. 2, pp. 635–640, 2020. [Google Scholar]
30. M. Toğaçar, B. Ergen and Z. Cömert, “COVID-19 detection using deep learning models to exploit social mimic optimization and structured chest X-ray images using fuzzy color and stacking approaches,” Computers in Biology and Medicine, vol. 121, pp. 103805, 2020. [Google Scholar]
31. A. I. Khan, J. L. Shah and M. M. Bhat, “CoroNet: A deep neural network for detection and diagnosis of COVID-19 from chest X-ray images,” Computer Methods and Programs in Biomedicine, vol. 196, no. 18, pp. 105581, 2020. [Google Scholar]
32. T. Ozturk, M. Talo, E. A. Yildirim, U. B. Baloglu, O. Yildirim et al., “Automated detection of COVID-19 cases using deep neural networks with X-ray images,” Computers in Biology and Medicine, vol. 121, no. 7798, pp. 103792, 2020. [Google Scholar]
33. F. Chollet, Keras: The Python deep learning library. Astrophysics Source Code Library, California, USA, 2018. [Google Scholar]
34. M. Abadi, A. Agarwal, P. Barham, E. Brevdo, Z. Chen et al., “TensorFlow: Large-scale machine learning on heterogeneous distributed systems,” TensorFlow, 2015. [Online]. Available: https://www.tensorflow.org/. [Google Scholar]
35. J. P. Cohen, P. Morrison and L. Dao, “COVID-19 image data collection,” arXiv preprint, arXiv: 2003.11597, 2020. [Online]. Available: https://arxiv.org/abs/2003.11597. [Google Scholar]
36. X. Wang, Y. Peng, L. Lu, Z. Lu, M. Bagheri et al., “ChestX-Ray8: Hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases,” in 2017 IEEE Conf. on Computer Vision and Pattern Recognition (CVPRHonolulu, HI, USA, pp. 3462–3471, 2017. [Google Scholar]
37. Y. LeCun, Y. Bengio and G. Hinton, “Deep learning,” Nature, vol. 521, no. 7553, pp. 436–444, 2015. [Google Scholar]
38. A. Krizhevsky, I. Sutskever and G. E. Hinton, “Imagenet classification with deep convolutional neural networks,” in Advances in Neural Information Processing Systems 25 (NIPS 2012). Lake Tahoe, NV, USA, pp. 1097–1105, 2012. [Google Scholar]
39. M. Tan and Q. V. Le, “EfficientNet: Rethinking model scaling for convolutional neural networks,” arXiv preprint, arXiv:1905.11946, 2019. [Online]. Available: https://arxiv.org/abs/1905.11946. [Google Scholar]
40. M. Tan, B. Chen, R. Pang, V. Vasudevan, M. Sandler et al., “MnasNet: Platform-aware neural architecture search for mobile,” in 2019 IEEE/CVF Conf. on Computer Vision and Pattern Recognition (CVPRLong Beach, CA, USA, pp. 2815–2823, 2019. [Google Scholar]
41. E. E. D. Hemdan, M. A. Shouman and M. E. Karar, “COVIDX-Net: A framework of deep learning classifiers to diagnose COVID-19 in X-ray images,” arXiv preprint, arXiv:2003.11055, 2020. [Online]. Available: https://arxiv.org/abs/2003.11055. [Google Scholar]
42. L. Wang, Z. Q. Lin and A. Wong, “COVID-Net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images,” Scientific Reports, vol. 10, no. 1, pp. 19549, 2020. [Google Scholar]
43. P. K. Sethy and S. K. Behera, “Detection of coronavirus disease (COVID-19) based on deep features,” Preprints, 2020030300, 2020 [Online]. Available: 10.20944/preprints202003.0300.v1. [Google Scholar]
44. T. Mahmud, M. A. Rahman and S. A. Fattah, “CovXNet: A multi-dilation convolutional neural network for automatic COVID-19 and other pneumonia detection from chest X-ray images with transferable multi-receptive feature optimization,” Computers in Biology and Medicine, vol. 122, no. 1, pp. 103869, 2020. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |